Abstract
The genetic underpinnings that contribute to variation in olfactory perception are not fully understood. To explore the genetic basis of variation in olfactory perception, we measured behavioral responses to 14 chemically diverse naturally occurring odorants in 260400 flies from 186 lines of the Drosophila melanogaster Genetic Reference Panel, a population of inbred wild-derived lines with sequenced genomes. We observed variation in olfactory behavior for all odorants. Low to moderate broad-sense heritabilities and the large number of tests for genotype–olfactory phenotype association performed precluded any individual variant from reaching formal significance. However, the top variants (nominal P < 5×10−5) were highly enriched for genes involved in nervous system development and function, as expected for a behavioral trait. Further, pathway enrichment analyses showed that genes tagged by the top variants included components of networks centered on cyclic guanosine monophosphate and inositol triphosphate signaling, growth factor signaling, Rho signaling, axon guidance, and regulation of neural connectivity. Functional validation with RNAi and mutations showed that 15 out of 17 genes tested indeed affect olfactory behavior. Our results show that in addition to chemoreceptors, variation in olfactory perception depends on polymorphisms that can result in subtle variations in synaptic connectivity within the nervous system.
Key words: behavioral genetics, Drosophila melanogaster Genetic Reference Panel, genetic networks, genome-wide association analysis, olfactory behavior
Introduction
Most organisms depend on olfaction to evaluate their habitat and to detect food, toxins, predators, and mating partners. Thus, variation in the ability to perceive and respond to chemosensory information for survival and reproduction provides a target for natural selection and adaptive evolution. Whereas neural mechanisms of olfaction have been well studied in vertebrates (Su et al. 2009; Touhara and Vosshall 2009) and insects (Su et al. 2009; Hansson and Stensmyr 2011), the genetic basis of phenotypic variation in odor perception and odor-guided behavior is less well understood. Olfactory impairments have been well studied in humans (Doty 2005) along with individual variation in the organization of odorant receptor (Or) genes (Waszak et al. 2010), and, in rare cases, specific anosmias have been characterized at the gene level in humans (Keller et al. 2007) and in mice (Griff and Reed 1995). However, the genome-wide factors that contribute to subtle individual variations in perception and evaluation of odors in humans or model organisms remain to be elucidated.
Drosophila melanogaster represents an excellent model for investigating the genetic basis of phenotypic variation for olfactory behavior (Anholt 2010). The functional organization of its olfactory system is well characterized. The recent establishment of a population of wild-derived inbred fly lines with fully sequenced genomes, the Drosophila melanogaster Genetic Reference Panel (DGRP), enables genome-wide association (GWA) analyses (Mackay et al. 2012; Huang et al. 2014).
Olfaction in Drosophila is mediated by multigene families of odorant-binding proteins (Obps) (Galindo and Smith 2001; Hekmat-Scafe et al. 2002), Ors (Clyne et al. 1999; Gao and Chess 1999; Vosshall et al. 1999), ionotropic receptors (Irs) (Benton et al. 2009), and gustatory receptors (Grs) (Clyne et al. 2000; Scott et al. 2001; Weiss et al. 2011), of which Gr21a and Gr63a have been identified as carbon dioxide sensors (Jones et al. 2007; Kwon et al. 2007). Combinatorial interactions between odorants and Obps (Swarup et al. 2011) and Ors (de Bruyne et al. 2001; Fishilevich et al. 2005; Hallem and Carlson 2006) generate patterns of neural activity in chemosensory neurons that are relayed to the antennal lobes and translated into spatial and temporal patterns of glomerular activation that encode odor quality and concentration (Marin et al. 2002; Ng et al. 2002; Wilson et al. 2004). This information is relayed to the mushroom bodies and lateral horn of the protocerebrum, where olfactory perceptions are shaped (Marin et al. 2002; Wong et al. 2002; Wang et al. 2003; Jefferis et al. 2007). The mushroom bodies are associated with storage and retrieval of olfactory information and determining appropriate behavioral responses (Blum et al. 2009; Masse et al. 2009; Davis 2011). The distribution patterns of chemosensory neurons expressing individual Ors in the antenna and their projections to the antennal lobes have been delineated (Vosshall et al. 2000) and elegant electrophysiological studies have characterized the molecular response profiles of a large number of Ors (de Bruyne et al. 2001; Dobritsa et al. 2003; Hallem and Carlson 2006).
Previous studies have identified single nucleotide polymorphisms (SNPs) in the Obp99a-d group of Obp genes that were associated with phenotypic variation in responses to benzaldehyde (Wang et al. 2007) and the structurally closely related odorant, acetophenone (Wang et al. 2010). These studies and others (Arya et al. 2010; Swarup et al. 2011) showed that Obps recognize odorants in a combinatorial manner but that different SNPs in Obp genes generate odorant-specific individual variation in chemosensory behavior. Similarly, a study that examined the relationship between olfactory behavior and SNPs in 3 Or genes, known to mediate electrophysiological responses to the same odorants (Hallem and Carlson 2006), showed that different SNPs in Or genes also generate odorant-specific individual variation in chemosensory behavior (Rollmann et al. 2010). Studies in which expression of specific Obp genes was reduced by targeted RNAi showed that the relationships between Obps and Ors depend on complex functional mosaics of combinatorial recognition patterns (Swarup et al. 2011).
A previous GWA analysis using the DGRP Freeze 1 (Mackay et al. 2012) combined with extreme quantitative trait locus mapping of a DGRP-derived advanced intercross population identified SNPs associated with variation in olfactory behavior to a single odorant, benzaldehyde (Swarup et al. 2013). This study revealed a network of genes involved in cellular signaling and neural development associated with phenotypic variation and showed that epistatic interactions dominate the genetic architecture that underlies variation in olfactory behavior to this odorant (Swarup et al. 2013).
Here, we explore the genetic underpinnings for variation in olfactory behavior to a panel of 14 chemically diverse odorants. We analyzed genotype–phenotype relationships using 186 lines of the DGRP Freeze 2, which includes documentation of insertion–deletion polymorphisms and chromosomal inversions in addition to SNPs (Huang et al. 2014). Olfactory responses had generally low to moderate broad-sense heritabilities, which combined with the multiple-testing penalty incurred by testing millions of variants for 14 odorants limited our ability to identify individual variants associated with olfactory behavior. Therefore, we employed gene set enrichment analyses, identifying gene ontology (GO) categories and biological pathways significantly enriched for genes harboring the top associations. These analyses gave rise to a genetic network centered on nervous system development and function, similar to previous observations obtained for responses to a single odorant (Brown et al. 2013; Swarup et al. 2013). We functionally validated several of the genes implicated in the network. Our data indicate that subtle variations in neural connectivity associated with naturally occurring genetic perturbations may affect phenotypic variation in olfactory behavior to multiple chemically diverse odorants.
Materials and methods
Drosophila melanogaster stocks
DGRP flies were generated and are maintained in our laboratories (Mackay et al. 2012; Huang et al. 2014). We obtained the OK107-GAL4 line from Dr Tanouye (University of California, Berkeley) and the c739-GAL4, elav-GAL4, repo-GAL4 from the Bloomington Drosophila Stock Center. We also obtained Mi{ET1} lines (CG34113 MB04218, CG34113 MB04817, CG42313 MB08581, Cip4 MB03744, cv-c MB03489, cv-c MB03717, Dgk MB10383, Gefmeso MB10683, Pkc53E MB02781, Pkcδ MB00303, RhoU MB00991, side MB07679, trio MB09917) and their co-isogenic control from the Bloomington Drosophila Stock Center. We obtained UAS-RNAi lines (CdGAPr KK100409, CG30440 KK101642, CG6424 KK107381, Cip4 KK101912, cv-c K107255, RhoGAP68F KK102638, RhoU KK112816, robo KK108817, rut KK109441) and the progenitor control y w 1118 ;P{attP,y + w 3 } line from the Vienna Drosophila Resource Center (Dietzl et al. 2007). These lines were crossed to elav, repo, or mushroom body–specific GAL4 driver lines to induce targeted gene silencing. All flies were reared on cornmeal–molasses–agar–yeast medium at 25 °C, 70% humidity, and a 12-h light/dark cycle.
Behavioral assays
We measured olfactory behavior of 4- to 8-day-old mated flies in single-sex groups of 5 flies/replicate and 10 replicates/sex. Olfactory behavior was quantified against a panel of 14 odorants using the well-established “dipstick” assay (Anholt et al. 1996). Briefly, 5 flies of the same sex were food-deprived for 1h prior to the assay and placed in an empty culture vial. An odorant solution was introduced on the tip of a cotton wool swab wedged between the plug and the wall of the vial and, following a 15-s acclimatization period, the number of flies that moved to a marked compartment 3cm from the bottom of the tube was recorded at 5-s intervals. The average of 10 measurements was calculated as the response score of each individual trial and the averages of 10 trials on the same genotype and sex were recorded as the line means. Response scores greater than 2.5 indicate avoidance of the odorant, whereas scores lower than 2.5 indicate attraction. All assays were conducted between 1:00 and 4:00 pm. Pilot experiments on 5 DGRP lines established that a concentration of 0.3% (v/v) for hexanal and 3.5% (v/v) for all other odorants provided optimal resolution for evaluating variation in olfactory behavior. Response scores to acetophenone on the DGRP lines are those reported previously (Wang et al. 2010). To minimize environmental variation during the long period required to complete all the measurements, the lines were grouped into 6 blocks for measurements of responses to each odorant. All odorants used were of the highest purity available. Hexanal was purchased from MP Biomedicals, citral from Acros Organics, and all other odorants from Sigma-Aldrich.
Quantitative genetic analysis
We partitioned variance of responses to odorants among the lines by analysis of variance (ANOVA) according to the mixed model ANOVA Y = μ + B + S + B × S + L(B) + S × L(B) + E, where Y is the observed value, μ is the overall mean, B is the random effect of block, S is the fixed effect of sex, L(B) is the random effect of line within block, S × L(B) is the random effect of sex by line within block, and E represents environmental error. To correct for any significant block effects, we subtracted the average response of lines within the block from the mean responses of each individual line and added the average response of line across all blocks. We then ran the reduced models Y = μ + S + L + S × L + E for all odorants. We estimated broad-sense heritability as from the variance components. The cross-sex genetic correlations (r MF) for the traits for which the sex × line interaction term was significant were estimated as (Falconer and Mackay 1996). Correlations between odorants and between sexes within odorants were calculated using multivariate analyses in JMP10. Phenotypic correlations (r P) of r P = ±0.25 were significantly different from zero at P < 0.05. Statistically significant differences in olfactory responses between Mi{ET1} mutants and controls or between GAL4/UAS-RNAi F1s and GAL4/progenitor F1s were determined by Dunnett’s tests.
GWA analyses
Associations were computed for each odorant separately using line means for phenotypic scores, using 1890367 polymorphic markers with a minor allele frequency (MAF) > 0.05 (Huang et al. 2014). Line means were adjusted for the effects of Wolbachia infection and 5 major chromosomal inversions (In(2L)t, In(2R)NS, In(3R)P, In(3R)K, In(3R)Mo). The adjusted phenotypes were fitted using a linear mixed model Y = μ + M + g + E, where μ was the population mean, M was the fixed effect for marker effect, and g was a polygenic term with its covariance among lines determined by the genomic relationship matrix (Huang et al. 2014). The same analysis was performed for each sex separately and for sex average and sex difference of the adjusted phenotypes.
GO and bioinformatics analyses
GO analyses were performed using the DAVID algorithm (Huang et al. 2009), with the Benjamini correction for multiple tests. To identify ensembles of interacting gene products, we used the R-spider program in the BioProfiling.de web portal (Antonov et al. 2010). The R-spider algorithm incorporates data for approximately 2000 genes from the D. melanogaster genome and combines signaling and metabolic pathways from Reactome and Kyoto Encyclopedia of Genes and genomes (KEGG) databases to determine if interactions between the input genes are greater than expected by chance using a permutation test.
Functional validation
We performed functional validation using RNAi knockdown and analysis of mutations in selected genes in the network detected by the GWA study, for which reagents were available. We crossed UAS-RNAi constructs for CdGAPr, CG30440, CG6424, Cip4, cv-c, RhoGAP68F, and RhoU to elav-GAL4 and repro-GAL4 drivers, knocking down expression in neurons and glia, respectively. We assessed olfactory behavior in response to 1-hexanol, 2-heptanone, acetophenone, citral, ethyl acetate, and hexanal for males and females as described above. We crossed RNAi constructs for robo and rut to each of 2 mushroom body–specific drivers, OK107-GAL4 and c739-GAL4, and measured olfactory responses to 2-phenyl ethyl alcohol, acetophenone, benzaldehyde, citral, eugenol, l-carvone, and methyl salicylate. Finally, we assessed olfactory responses of Mi{ET1} insertional alleles of CG34113, CG42313, Cip4, cv-c, Dgk, Gefmeso, Pkc53E, Pkcδ, RhoU, side, and trio to 1-hexanol, 2-heptanone, acetophenone, citral, d-carvone, ethyl acetate, helional, and hexanal. All genotypes and their respective co-isogenic controls were evaluated with 10 replicate measurements per sex and odorant.
We performed factorial fixed-effect ANOVAs for crosses of RNAi to each GAL4 driver and for the Mi{ET1} mutants. The full models were Y = μ + S + G + O + S × G + S × O + G × O + S × G × O + ε, where S is the effect of S, G is genotype, O is odorant, and ε is the residual. We also performed reduced ANOVAs using the same model, but for which only the control and a single RNAi or mutant genotype was evaluated, pooled across sexes and odorants.
Results
Quantitative genetics of olfactory behavior in the DGRP
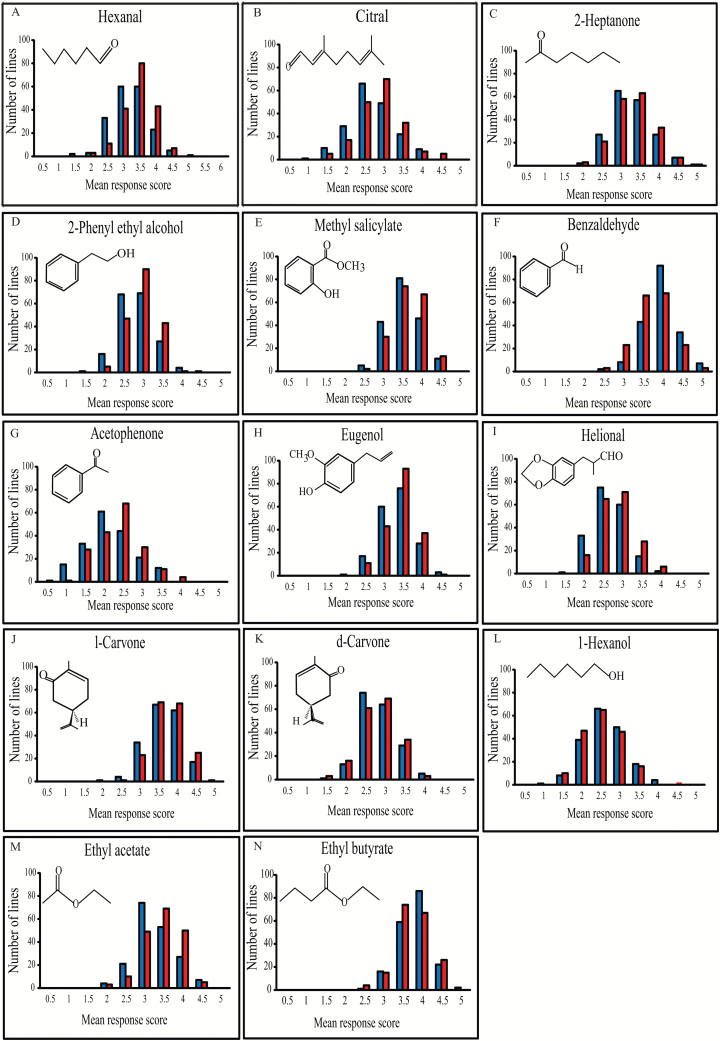
We measured variation in olfactory responses of 260400 flies from 186 DGRP lines to 14 odorants that naturally occur in fruits and plants and that belong to various chemical classes, including aldehydes, ketones, aromatics, alcohols, and esters. We observed extensive variation in olfactory behavior to all odorants; however, the means of the distributions of response scores to different odorants varied, with some odorants acting as general repellants, whereas others span the range from attractant to repellant (Figure 1; Supplementary Table 1). ANOVAs showed highly significant genetic variation in olfactory behavior for all odorants, but estimates of broad-sense heritabilities (H 2) (Falconer and Mackay 1996) were low to moderate, ranging from 0.14 to 0.33 (Supplementary Table 2). This is similar to previous estimates of olfactory behavior (Anholt et al. 1996; Mackay et al. 1996). Responses to 12 odorants showed significant mean differences between males and females, and there was significant variation in sex dimorphism for 8 of the odorants (Supplementary Table 2). Cross-sex genetic correlations were high and close to unity, indicating that largely the same variants affect olfactory behavior in males and females; however, the lack of perfect correlation between the sexes for many odorants means that sex-specific variation is also expected (Supplementary Table 2).
Figure 1.
Variation in behavioral responses to 14 odorants in 186 DGRP lines. Variation in olfactory responses are depicted for (A) hexanal, (B) citral, (C) 2-heptanone, (D) 2-phenyl ethyl alcohol, (E) methyl salicylate, (F) benzaldehyde, (G) acetophenone, (H) eugenol, (I) helional, (J) l-carvone, (K) d-carvone, (L) 1-hexanol, (M) ethyl acetate, and (N) ethyl butyrate. Responses of males are shown by blue bars and of females by red bars.
We next asked to what extent olfactory responses to different odorants were correlated, performing these analyses for sexes separately, because the responses of males and females are different genetically for many odorants (Supplementary Figure 1). Although many odorants showed significant positive correlations, these correlations were weak, indicating effective discrimination between these odorants. Only a single significant weak negative correlation was observed between 1-hexanol and ethyl butyrate (Supplementary Figure 1). Phenotypic correlations between sexes were greater than any correlations among odorants, ranging from 0.54 to 0.79 (Supplementary Figure 1).
Polymorphisms in chemoreceptor genes and natural variation in olfactory behavior
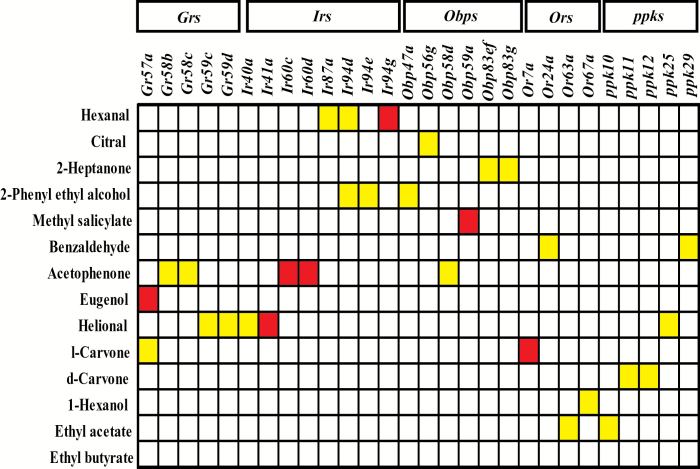
The chemosensory subgenome of Obp, Or, Ir, and Gr genes represents a priori candidate genes to test for associations with olfactory behavior. We performed association analyses for each of the 14 odorants for the 15637 polymorphisms with MAF > 0.05 in genes in these families. We corrected for effects of Wolbachia infection status, major polymorphic inversions, and polygenic relatedness in these analyses using a linear mixed model (Huang et al. 2014). We performed 4 association analyses for each of these polymorphisms: males, females, the sex-averaged response score, and the sex difference. Thus, even with this restricted set of polymorphic variants, we performed 875672 tests for association. Applying a strict Bonferroni correction for multiple tests and a 5% experiment-wide significance threshold would require individual variants to have a P value < 5.71×10−8. None of the variants tested achieved this level of individual significance. At an arbitrary reporting P value threshold < 5×10−5, we found 33 unique variants in 23 chemosensory genes, including 6 Obp genes (Obp47a, Obp45g, Obp58d, Obp59a, Obp83ef, and Obp83g), 4 Or genes (Or7a, Or24a, Or63a, and Or67a), 8 Ir genes (Ir40a, Ir41a, Ir60c, Ir60d, Ir87a, Ir94e, Ir94d, and Ir94g), and 5 Gr genes (Gr57a, Gr58b, Gr58c, Gr59c, and Gr59d) (Supplementary Table 3, Figure 2). Unfortunately, the complex correlation structure and large size of the data set precludes computing permutation-derived significance thresholds and false discovery rates (FDRs). If we assume that all tests are independent, 33 associations is less than would be expected by chance (43.8); however, if we assume that only the variants are independent, 33 associations correspond to an FDR of 0.024.
Figure 2.
Polymorphisms in chemosensory receptor genes associated with variation in olfactory behavior. The diagram represents polymorphic markers in 23 chemosensory genes associated with variation in olfactory behavior toward 14 odorants identified by GWA analysis. We performed 4 different GWA analyses for each variant; the P value represents the lowest of the 4 P values in cases where more than one analysis per variant or multiple variants in the same gene reached the reporting threshold. P < 10−5 (red); P < 5×10−5 (yellow).
GWA analyses for olfactory responses to a panel of odorants
We next performed the same linear mixed model association analyses genome wide to identify candidate genes associated with natural variation in olfactory behavior. We evaluated the effects of 1890367 polymorphic markers with MAF > 0.05 for males and females separately, and sex-averaged response score and sex difference for each odorant, that is, 105860552 (partially correlated) association analyses. Not surprisingly, no single variant was significant at a Bonferroni-corrected, 5% experiment-wide significance level (4.72×10−10) (Supplementary Table 4). At a P value threshold < 5×10−5, we found 3540 unique polymorphisms in or near 2154 genes. The vast majority of the polymorphisms were associated with only a single odorant; only 41 variants were associated with more than one odorant. Again, if we (too conservatively) assume that all tests are independent, 3540 polymorphisms is less than expected by chance (5293); whereas if we (too leniently) assume only the variants are independent, 3540 associations correspond to an FDR < 0.027 (assuming a uniform distribution of P values when there were no associations). Therefore, we infer that the top associations are significantly enriched for true positives and that we need to utilize a different method than single SNP association analyses to resolve this signal from the noise.
We note that, in addition to the chemoreceptor genes described above, we identified 5 members of the pickpocket (ppk) family of sodium channels, of which ppk11 has been implicated in taste perception (Liu et al. 2003) and a ppk11 mutant affects olfactory response to benzaldehyde (Swarup et al. 2011); ppk25 has been implicated in pheromone recognition and courtship behavior (Vijayan et al. 2014) and ppk29 in courtship behavior (Thistle et al. 2012).
GO and gene set enrichment analyses reveal networks of genes that harbor polymorphisms associated with variation in olfactory behavior
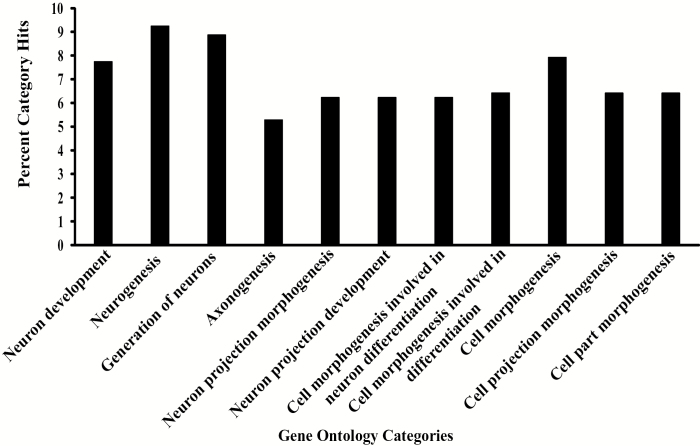
We performed GO enrichment analysis to assess to what extent the candidate genes identified as top associations in the GWA analyses were enriched for biological processes, molecular functions, and cellular components, using the DAVID algorithm (Huang et al. 2009). We found 170 GO terms that were significantly enriched for these genes using a Benjamini-corrected FDR < 0.05; 87 terms were significantly enriched using a Benjamini-corrected FDR < 0.001 (Supplementary Table 5). Among the top associated GO terms are biological process terms for development in general and more specifically the development and function of the nervous system. A total of 530 genes clustered in 11 GO categories with enrichment scores greater than 3 (Benjamini-corrected P < 0.05; Figure 3). These GO categories were highly enriched for genes involved in the development of the nervous system.
Figure 3.
GO analyses. Significantly enriched GO (Benjamini correction: P < 0.05) for genes with sequence variants associated with variation in behavioral responses to one or more odorants. The analysis is based on biological processes level 5 using the DAVID algorithm and software (Huang et al. 2009).
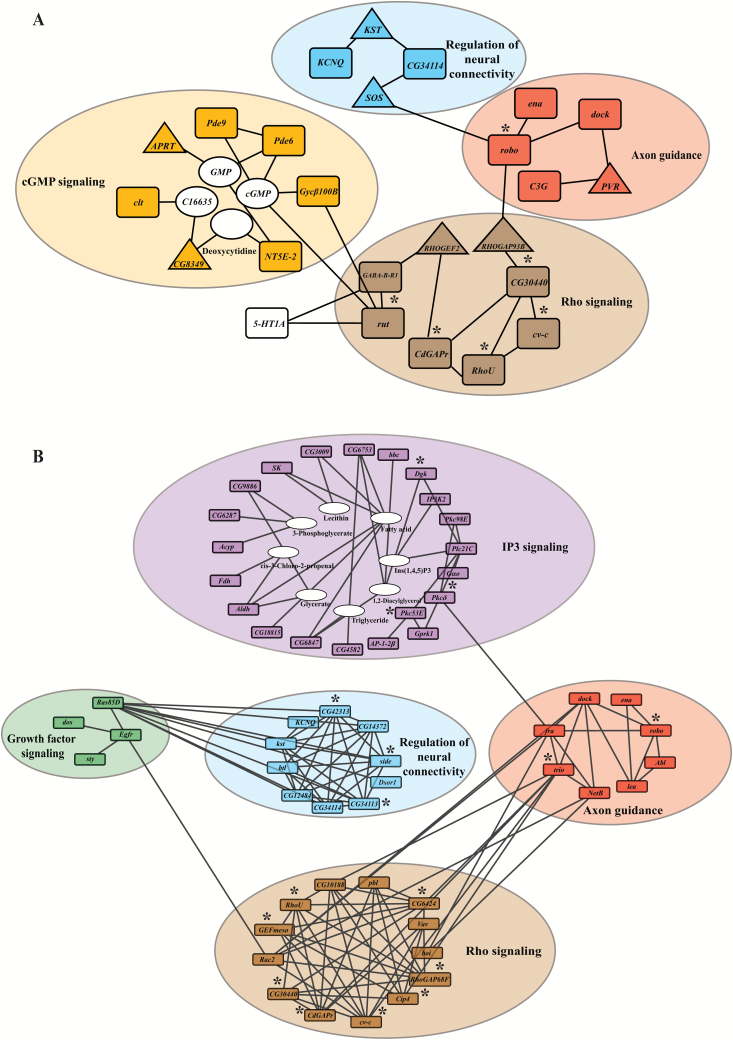
Next, we performed pathway enrichment analyses for the candidate genes identified as top associations in the GWA analyses using the R-spider algorithm (Antonov et al. 2010), which combines information from the Reactome and KEGG databases to build interactive networks, while determining whether interactions between gene products are greater than expected by chance. We first considered the 645 genes that contained at least one DNA variant showing associations at P < 10−5. Using a model that allows for one missing gene (i.e., a gene that interconnects with genes with associated SNPs but itself does not harbor polymorphisms associated with phenotypic variation) reveals a network of 21 genes related to cyclic guanosine monophosphate (GMP) metabolism and Rho signaling (permutation P < 0.04; Figure 4A). It is of interest to note the 5-HT1A gene, which encodes a serotonin receptor and is connected with rut encoding adenylyl cyclase and GABA-B-R1. Serotonin and GABA have been implicated in a mushroom body circuit that mediates olfactory learning and memory (Johnson et al. 2011; Lee et al. 2011) and in processing of olfactory information in the antennal lobes (Dacks et al. 2009).
Figure 4.
Genetic networks associated with variation in olfactory behavior. (A) A genetic network allowing one missing gene (permutation P < 0.04) derived from candidate genes detected at a nominal significance threshold in the GWA analysis of P < 10−5. The network was obtained with the R-spider algorithm (Antonov et al. 2010). Missing genes are indicated with triangles. (B) A genetic network with no missing genes (permutation P < 0.005) derived from candidate genes detected at a nominal significance threshold in the GWA analysis of P < 5×10−5. Candidate genes selected for subsequent mutational analysis or RNAi targeting are marked with an asterisk.
We then performed the same analysis on genes with variant associations at P < 5×10−5 using a model with no missing genes. This revealed a network of 56 genes (permutation P < 0.005; Figure 4B), including 7 genes that overlapped with the previous analysis (Figure 4A). This network consists of modules associated with inositol triphosphate signaling, growth factor signaling, regulation of neural connectivity, axon guidance, and Rho signaling, which plays a key role in neural development and formation of synaptic connectivity (Tolias et al. 2011).
We infer from these analyses that, although individual associations for variants affecting olfactory behavior do not achieve formal significance, cumulatively the associations considering all GWA analyses and all 14 odorants are significantly enriched for GO terms and pathways. Furthermore, these variants are likely to exert their effects by causing subtle differences in neuronal connectivity.
Effects of disruption of genetic network components on olfactory behavior
The advantage of the D. melanogaster model system is that we can use the publicly available toolkit of RNAi knockdown constructs to infer functional associations of candidate genes nominated by the GWA analysis and drive the knockdown of gene expression in different tissues using GAL4 tissue-specific drivers. In addition, mutations in many candidate genes have been induced in a common isogenic background and can hence be tested for subtle effects on olfactory behavior. We adopted this strategy here, using a combination of co-isogenic Mi{ET1} mutants and targeted RNAi to assess to what extent disruptions of candidate genes in the networks associated with olfactory perception affect olfactory behavior against our odorant panel, separately for males and females. For RNAi targeting, we used the panneuronal elav-GAL4 driver, the panglial repo-GAL4 driver, or mushroom body-specific drivers, OK107-GAL4, which drives expression throughout the mushroom bodies, and c739-GAL4, which targets the α and β lobes but spares the α′ and β′ lobes (Aso et al. 2009) (Supplementary Table 6).
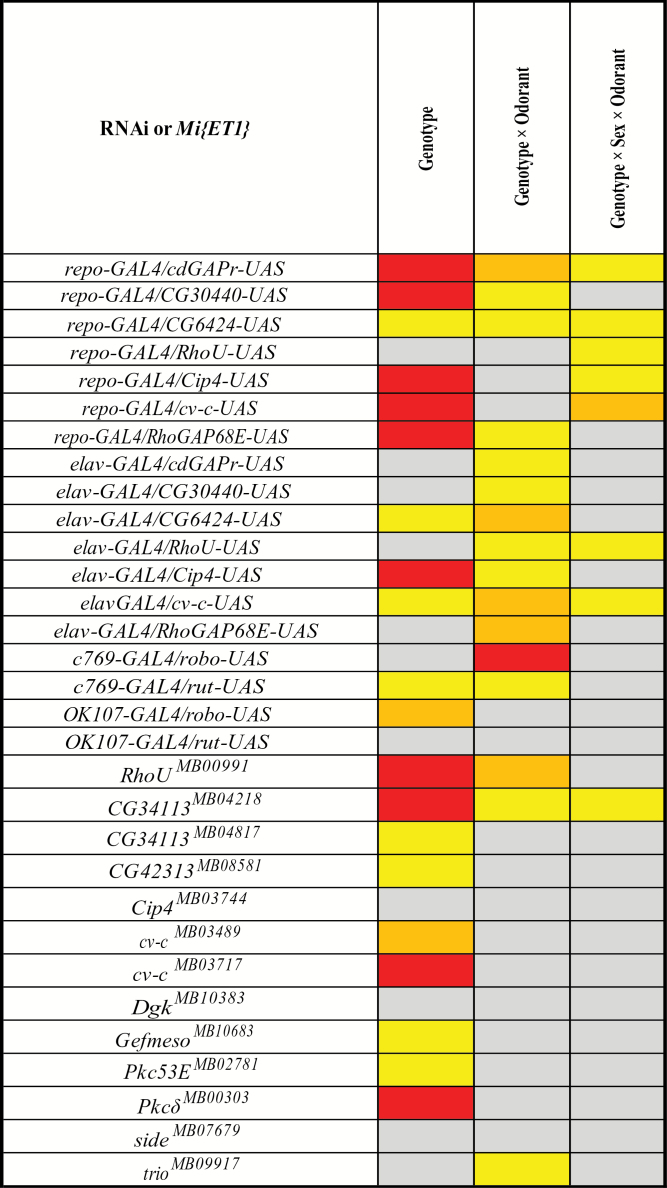
We first assessed whether there was significant genetic variation among all RNAi knockdown genotypes and their control, crossed to the same driver, and among all Mi{ET1} mutations and the control line (Supplementary Table 7). In all cases, the genotype term and/or genotype by odorant term was significant, indicating differential responses of at least one genotype in each group to at least one odorant. Next, we used the same ANOVA model to test whether each line differed from the control, pooled across sexes and odorants. In these analyses, significance of the genotype, genotype by odorant, and/or genotype by sex by odorant terms indicates the RNAi line or mutation is different from the control for at least one odorant in at least one of the 2 sexes. All but one of the RNAi constructs (OK107-GAL4/rut-UAS) were significant for one or more of these terms, and rut was significant when driven by c769-GAL4 (Figure 5, Supplementary Table 8). A total of 10 of the 13 Mi{ET1} mutations tested also exhibited differences from the control line (Figure 5, Supplementary Table 8). Of the 17 genes targeted for functional validation using RNAi, mutations, or both, 15 (88.2%) showed altered behavioral responses against distinct odorants. This confirms that the bioinformatically derived networks are indeed enriched for novel genes affecting olfactory behavior.
Figure 5.
Mutational analysis and RNAi targeting of candidate genes. The boxes indicate P values of the main effect of genotype and the genotype × odorant and genotype × sex × odorant terms from ANOVAs comparing the olfactory behavior of each mutation or RNAi construct and its control line across a battery of different odorants. P ≤ 0.001 (red); 0.001 < P ≤ 0.01 (orange); 0.01 < P ≤ 0.05 (yellow); P > 0.05 (gray).
Discussion
Understanding the mechanisms that give rise to individual variation in olfactory perception is a long-standing challenge in the chemical senses. There is wide variation in perception of odor quantity and quality among individuals in human populations. A classic example of this phenotypic variation is the perception of 5α-androst-16-en-3-one, to which a large percentage of the population is anosmic, whereas others perceive it either as an offensive or pleasant odor (Araneda and Firestein 2004). Individual variation in perception of this odorant has been attributed to polymorphisms in OR7D4 (Keller et al. 2007) and segregates with a near Mendelian inheritance pattern in the population. In contrast to 5α-androst-16-en-3-one, most odorants are recognized combinatorially by multiple Ors both in mammals (Malnic et al. 1999) and insects (Hallem et al. 2004) and variation in perception is subtle rather than all or none. The lack of control over genetic background and environmental conditions, including previous exposures and olfactory learning, make it challenging to study the complex genetic architecture that governs variation in olfactory perception in human populations. We have used the DGRP as a powerful model system to study the genetic underpinnings that give rise to variation in olfactory behavior and identified evolutionarily conserved neurogenetic networks that can be extrapolated across phyla.
Using the DGRP lines, we assessed the genetic architecture of olfactory behavior to 14 chemically diverse, naturally occurring odorants and simultaneously queried the effects of 1890367 common natural polymorphisms on olfactory responses. We found significant genetic variation in response to all odorants, but the correlations in responses to different odorants were low. We identified 3540 polymorphisms in or near 2154 genes associated with olfactory behavior at a nominal P < 5×10−5. Most of these variants were associated with variation in olfactory response to only one of the 14 test odorants; only 41 polymorphisms were associated with variation in responses to more than one odorant.
Functional redundancy in odorant recognition among peripheral chemoreceptors may obscure the effects of individual polymorphisms on phenotypic variation, requiring large effect sizes and large sample sizes with sufficient statistical power to resolve associations. Previously, we identified polymorphisms associated with variation in response to benzaldehyde and acetophenone in several Obp genes (Wang et al. 2007, 2010; Arya et al. 2010) and Or genes known to respond to these odorants (Rollmann et al. 2010). These polymorphisms were not recapitulated in the present study. The likely reason for this discrepancy is that the original association analyses on these Obp and Or genes were done prior to the availability of whole-genome sequences of the DGRP and were performed only on these particular Obp and Or genes and with more wild-derived inbred lines than those that ultimately comprised the DGRP, thereby significantly increasing power. When we performed association analyses for each previously identified SNP separately, we found that 3R_25540327_SNP (previously designated G67A) and 3R_25540338_SNP (previously designated T78G) in Obp99d (Wang et al. 2007) showed evidence for association with variation in olfactory behavior to benzaldehyde at P < 0.07 and P < 0.002, respectively. Similarly, 3R_25542126_SNP (previously designated C384T) in Obp99b (Wang et al. 2010) was associated with variation in olfactory behavior to acetophenone at P < 0.01; 3R_25540289_SNP, 3R_25540452_SNP, and 3R_25540623_SNP (previously designated G29A, T192G, and G363A, respectively) in Obp99d showed P values of P < 0.06, P < 0.06, and P < 0.05, respectively, for variation in olfactory response to acetophenone. Furthermore, alleles with large effects and present at low frequencies in the larger collection of initially surveyed lines may be present at too low a frequency in the DGRP to be detected by GWA.
Given the large number of tests for association performed and the subtle effects of naturally occurring variants on olfactory behavior, none of the polymorphisms were significant using a strict Bonferroni-corrected significance threshold. Further, given the size of the data set, permutation-derived FDRs were not possible. Nevertheless, the notion that the top associations were enriched for true positives is supported in that they included 67 genes previously implicated in variation in responses to benzaldehyde (Swarup et al. 2013), 244 genes previously implicated in variation in responses to 2,3-butanedione (Brown et al. 2013), and 9 genes in which insertion of a P[GT1] transposon disrupted olfactory behavior to benzaldehyde (Sambandan et al. 2006). We therefore used gene set enrichment analyses to assess which GO categories and known pathways were significantly overrepresented among the top GWA hits. GO categories associated with nervous system development and function were highly enriched. Furthermore, we could place the top candidate genes in interconnected networks affecting axon guidance, regulation of neural connectivity, and Rho, inositol triphosphate, cyclic GMP, and growth factor signaling pathways that are statistically significant when compared with the likelihood of such a network arising from the same number of randomly selected genes. The nature of the genes within the network suggests that polymorphisms that contribute to natural variation in olfactory behavior may do so by causing subtle variations in neural connectivity in the olfactory projection, similar to results from previous GWA studies in the DGRP that focused on single odorants, benzaldehyde (Swarup et al. 2013) and 2,3-butanedione (Brown et al. 2013).
It is of interest to note that the chemoreceptor genes identified as candidate genes in our GWA analyses include not only Ors but also members of the Gr, Ir, and Obp families. This suggests that members from all 4 chemosensory receptor families may contribute to shaping the behavioral responses toward odorants. Additionally, it is interesting to note that olfactory receptors that have been demonstrated to respond to the tested odorants using electrophysiological methods (Hallem et al. 2004; Hallem and Carlson 2006; e.g., Or47a, Or59b, and Or43b respond to ethyl acetate) were not among the top candidate genes in our GWA analyses. There are several possible and nonmutally exclusive explanations for this observation. 1) Naturally occurring polymorphisms in these receptors may not result in individual variation in olfactory responses. This could occur if these genes are under strong natural selection and hence functionally invariant in this sample of alleles. 2) Effect sizes of causal polymorphisms at these loci are too small to be detected given the resolution of the behavioral assay and the sample size. 3) Rare alleles at these loci, not interrogated in our analyses, affect natural variation in olfactory responses. 4) Functional redundancy in odorant recognition among peripheral chemoreceptors may obscure the effects of individual polymorphisms on phenotypic variation.
The advantage of Drosophila is that we can perform secondary screens using RNAi knockdown and mutations to confirm whether the genes that harbor polymorphisms associated with phenotypic variation indeed affect olfactory behavior toward a subset of odorants. Although the consequences of transposon insertions or RNAi on gene expression may not precisely quantitatively replicate the effects of naturally occurring mutations, we would expect them to mimic the effects of polymorphisms in the target genes on olfactory behavior, which would be reflected by differential changes in behavior against different odorants, although not necessarily the same odorants as those associated with phenotypic variation due to the naturally occurring polymorphism. This is indeed what we observed. We tested 17 of the genes in the network and found that 15 indeed affect olfactory behavior toward different odorants: cdGAPr, CG30440, CG34113, CG42313, CG6424, Cip4, cv-c, GEFmeso, Pkcδ, Pkc53E, RhoGAP68F, RhoU, robo, rut, and trio. Among these genes rut has been previously associated with olfaction (Dacks et al. 2009; Johnson et al. 2011; Lee et al. 2011) and cv-c, Pkc53E, and trio have been implicated in a previous GWA study for variation in olfactory behavior to benzaldehyde (Swarup et al. 2013). The high validation rate—nearly 90%—engenders confidence that functional tests of other candidate genes involved in development and function of the nervous system will identify new components of genetic networks affecting olfactory perception and also implies that natural variation in olfactory perception is highly polygenic.
Finally, we note that we have tested only a sample of the large number of candidate genes implicated by our GWA study, primarily focused on those genes that comprise an interconnected biologically informative network and were validated at a high rate. However, the GWA studies presented here are a hypothesis generating paradigm that lays the foundation for further focused studies. Thus, our results provide a framework for more extensive functional studies and a detailed dissection of allelic effects of candidate genes in future endeavors.
In summary, we conclude that, in addition to polymorphisms associated with peripheral chemoreceptor genes, polymorphisms embedded in genetic networks that are associated with neural connectivity contribute to phenotypic variation in olfactory perception.
Supplementary material
Supplementary material can be found at http://www.chemse.oxfordjournals.org/
Funding
National Institute of Health (GM059469 and GM45146 to R.R.H.A. and T.F.C.M.).
Conflict of interest
Ms Serrano-Negron and Dr Magwire both went to their current places of employment (National Institute of Health and Syngenta, respectively) following completion of their roles in this project. Syngenta Biotechnology has not been a sponsor and has not contributed funding for this project. There are no conflicting interests with regards to patent or stock ownership, membership of a company board of directors, membership of an advisory board or committee for a company, and consultancy for or receipt of speaker’s fees from a company.
Supplementary Material
References
- Anholt RRH. 2010. Making scents of behavioral genetics: lessons from Drosophila . Genet Res. 92:349–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anholt RR, Lyman RF, Mackay TF. 1996. Effects of single P-element insertions on olfactory behavior in Drosophila melanogaster . Genetics. 143:293–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonov AV, Schmidt EE, Dietmann S, Krestyaninova M, Hermjakob H. 2010. R spider: a network-based analysis of gene lists by combining signaling and metabolic pathways from Reactome and KEGG databases. Nucleic Acids Res. 38:W78–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araneda RC, Firestein S. 2004. The scents of androstenone in humans. J Physiol. 554:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arya GH, Weber AL, Wang P, Magwire MM, Serrano-Negron YL, Anholt RRH. 2010. Natural variation, functional pleiotropy and transcriptional contexts of Odorant binding protein genes in Drosophila melanogaster . Genetics. 186:1475–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Grübel K, Busch S, Friedrich AB, Siwanowicz I, Tanimoto H. 2009. The mushroom body of adult Drosophila characterized by GAL4 drivers. J Neurogenet. 23:156–172. [DOI] [PubMed] [Google Scholar]
- Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. 2009. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila . Cell. 136:149–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum AL, Li W, Cressy M, Dubnau J. 2009. Short- and long-term memory in Drosophila require cAMP signaling in distinct neuron types. Curr Biol. 19:1341–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown EB, Layne JE, Zhu C, Jegga AG, Rollmann SM. 2013. Genome-wide association mapping of natural variation in odour-guided behaviour in Drosophila . Genes Brain Behav. 12:503–515. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Carlson JR. 2000. Candidate taste receptors in Drosophila . Science. 287:1830–1834. [DOI] [PubMed] [Google Scholar]
- Clyne PJ, Warr CG, Freeman MR, Lessing D, Kim J, Carlson JR. 1999. A novel family of divergent seven-transmembrane proteins: candidate odorant receptors in Drosophila . Neuron. 22:327–338. [DOI] [PubMed] [Google Scholar]
- Dacks AM, Green DS, Root CM, Nighorn AJ, Wang JW. 2009. Serotonin modulates olfactory processing in the antennal lobe of Drosophila . J Neurogenet. 23:366–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL. 2011. Traces of Drosophila memory. Neuron. 70:8–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruyne M, Foster K, Carlson JR. 2001. Odor coding in the Drosophila antenna. Neuron. 30:537–552. [DOI] [PubMed] [Google Scholar]
- Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, Gasser B, Kinsey K, Oppel S, Scheiblauer S, et al. 2007. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila . Nature. 448:151–156. [DOI] [PubMed] [Google Scholar]
- Dobritsa AA, van der Goes van Naters W, Warr CG, Steinbrecht RA, Carlson JR. 2003. Integrating the molecular and cellular basis of odor coding in the Drosophila antenna. Neuron. 37:827–841. [DOI] [PubMed] [Google Scholar]
- Doty RL. 2005. Clinical studies of olfaction. Chem Senses. 30(Suppl 1):i207–i209. [DOI] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC. 1996. Introduction to quantitative genetics. 4th ed. Essex (UK): Longmans Green, Harlow. [Google Scholar]
- Fishilevich E, Domingos AI, Asahina K, Naef F, Vosshall LB, Louis M. 2005. Chemotaxis behavior mediated by single larval olfactory neurons in Drosophila . Curr Biol. 15:2086–2096. [DOI] [PubMed] [Google Scholar]
- Galindo K, Smith DP. 2001. A large family of divergent Drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 159:1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q, Chess A. 1999. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 60:31–39. [DOI] [PubMed] [Google Scholar]
- Griff IC, Reed RR. 1995. The genetic basis for specific anosmia to isovaleric acid in the mouse. Cell. 83:407–414. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Carlson JR. 2006. Coding of odors by a receptor repertoire. Cell. 125:143–160. [DOI] [PubMed] [Google Scholar]
- Hallem EA, Ho MG, Carlson JR. 2004. The molecular basis of odor coding in the Drosophila antenna. Cell. 117:965–979. [DOI] [PubMed] [Google Scholar]
- Hansson BS, Stensmyr MC. 2011. Evolution of insect olfaction. Neuron. 72:698–711. [DOI] [PubMed] [Google Scholar]
- Hekmat-Scafe DS, Scafe CR, McKinney AJ, Tanouye MA. 2002. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster . Genome Res. 12:1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, Peiffer J, Ràmia M, Tarone AM, Turlapati L, Zichner T, Zhu D, Lyman RF, et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24:1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 4:44–57. [DOI] [PubMed] [Google Scholar]
- Jefferis GS, Potter CJ, Chan AM, Marin EC, Rohlfing T, Maurer CR, Jr, Luo L. 2007. Comprehensive maps of Drosophila higher olfactory centers: spatially segregated fruit and pheromone representation. Cell. 128:1187–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson O, Becnel J, Nichols CD. 2011. Serotonin receptor activity is necessary for olfactory learning and memory in Drosophila melanogaster . Neuroscience. 192:372–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones WD, Cayirlioglu P, Kadow IG, Vosshall LB. 2007. Two chemosensory receptors together mediate carbon dioxide detection in Drosophila . Nature. 445:86–90. [DOI] [PubMed] [Google Scholar]
- Keller A, Zhuang H, Chi Q, Vosshall LB, Matsunami H. 2007. Genetic variation in a human odorant receptor alters odour perception. Nature. 449:468–472. [DOI] [PubMed] [Google Scholar]
- Kwon JY, Dahanukar A, Weiss LA, Carlson JR. 2007. The molecular basis of CO2 reception in Drosophila . Proc Natl Acad Sci U S A. 104:3574–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PT, Lin HW, Chang YH, Fu TF, Dubnau J, Hirsh J, Lee T, Chiang AS. 2011. Serotonin-mushroom body circuit modulating the formation of anesthesia-resistant memory in Drosophila . Proc Natl Acad Sci U S A. 108:13794–13799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Leonard AS, Motto DG, Feller MA, Price MP, Johnson WA, Welsh MJ. 2003. Contribution of Drosophila DEG/ENaC genes to salt taste. Neuron. 39:133–146. [DOI] [PubMed] [Google Scholar]
- Mackay TF, Hackett JB, Lyman RF, Wayne ML, Anholt RR. 1996. Quantitative genetic variation of odor-guided behavior in a natural population of Drosophila melanogaster . Genetics. 144:727–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster Genetic Reference Panel. Nature. 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malnic B, Hirono J, Sato T, Buck LB. 1999. Combinatorial receptor codes for odors. Cell. 96:713–723. [DOI] [PubMed] [Google Scholar]
- Marin EC, Jefferis GS, Komiyama T, Zhu H, Luo L. 2002. Representation of the glomerular olfactory map in the Drosophila brain. Cell. 109:243–255. [DOI] [PubMed] [Google Scholar]
- Masse NY, Turner GC, Jefferis GS. 2009. Olfactory information processing in Drosophila . Curr Biol. 19:R700–R713. [DOI] [PubMed] [Google Scholar]
- Ng M, Roorda RD, Lima SQ, Zemelman BV, Morcillo P, Miesenböck G. 2002. Transmission of olfactory information between three populations of neurons in the antennal lobe of the fly. Neuron. 36:463–474. [DOI] [PubMed] [Google Scholar]
- Rollmann SM, Wang P, Date P, West SA, Mackay TF, Anholt RR. 2010. Odorant receptor polymorphisms and natural variation in olfactory behavior in Drosophila melanogaster . Genetics. 186:687–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambandan D, Yamamoto A, Fanara JJ, Mackay TF, Anholt RR. 2006. Dynamic genetic interactions determine odor-guided behavior in Drosophila melanogaster . Genetics. 174:1349–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott K, Brady R, Jr, Cravchik A, Morozov P, Rzhetsky A. 2001. A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila . Cell. 104:661–673. [DOI] [PubMed] [Google Scholar]
- Su CY, Menuz K, Carlson JR. 2009. Olfactory perception: receptors, cells, and circuits. Cell. 139:45–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S, Huang W, Mackay TF, Anholt RR. 2013. Analysis of natural variation reveals neurogenetic networks for Drosophila olfactory behavior. Proc Natl Acad Sci U S A. 110:1017–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup S, Williams TI, Anholt RR. 2011. Functional dissection of Odorant binding protein genes in Drosophila melanogaster . Genes Brain Behav. 10:648–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thistle R, Cameron P, Ghorayshi A, Dennison L, Scott K. 2012. Contact chemoreceptors mediate male-male repulsion and male-female attraction during Drosophila courtship. Cell. 149:1140–1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolias KF, Duman JG, Um K. 2011. Control of synapse development and plasticity by Rho GTPase regulatory proteins. Prog Neurobiol. 94:133–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touhara K, Vosshall LB. 2009. Sensing odorants and pheromones with chemosensory receptors. Annu Rev Physiol. 71:307–332. [DOI] [PubMed] [Google Scholar]
- Vijayan V, Thistle R, Liu T, Starostina E, Pikielny CW. 2014. Drosophila pheromone-sensing neurons expressing the ppk25 ion channel subunit stimulate male courtship and female receptivity. PLoS Genet. 10:e1004238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosshall LB, Amrein H, Morozov PS, Rzhetsky A, Axel R. 1999. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 96:725–736. [DOI] [PubMed] [Google Scholar]
- Vosshall LB, Wong AM, Axel R. 2000. An olfactory sensory map in the fly brain. Cell. 102:147–159. [DOI] [PubMed] [Google Scholar]
- Wang P, Lyman RF, Mackay TF, Anholt RR. 2010. Natural variation in odorant recognition among odorant-binding proteins in Drosophila melanogaster . Genetics. 184:759–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lyman RF, Shabalina SA, Mackay TF, Anholt RR. 2007. Association of polymorphisms in odorant-binding protein genes with variation in olfactory response to benzaldehyde in Drosophila . Genetics. 177:1655–1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wong AM, Flores J, Vosshall LB, Axel R. 2003. Two-photon calcium imaging reveals an odor-evoked map of activity in the fly brain. Cell. 112:271–282. [DOI] [PubMed] [Google Scholar]
- Waszak SM, Hasin Y, Zichner T, Olender T, Keydar I, Khen M, Stütz AM, Schlattl A, Lancet D, Korbel JO. 2010. Systematic inference of copy-number genotypes from personal genome sequencing data reveals extensive olfactory receptor gene content diversity. PLoS Comput Biol. 6:e1000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Dahanukar A, Kwon JY, Banerjee D, Carlson JR. 2011. The molecular and cellular basis of bitter taste in Drosophila . Neuron. 69:258–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RI, Turner GC, Laurent G. 2004. Transformation of olfactory representations in the Drosophila antennal lobe. Science. 303:366–370. [DOI] [PubMed] [Google Scholar]
- Wong AM, Wang JW, Axel R. 2002. Spatial representation of the glomerular map in the Drosophila protocerebrum. Cell. 109:229–241. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.