Abstract
Water plays an essential role in the structure and function of proteins, lipid membranes and other biological macromolecules. Solid-state NMR heteronuclear-detected 1H polarization transfer from water to biomolecules is a versatile approach for studying water-protein, water-membrane, and water-carbohydrate interactions in biology. We review radiofrequency pulse sequences for measuring water polarization transfer to biomolecules, the mechanisms of polarization transfer, and the application of this method to various biological systems. Three polarization transfer mechanisms, chemical exchange, spin diffusion and NOE, manifest themselves at different temperatures, magic-angle-spinning frequencies, and pulse irradiations. Chemical exchange is ubiquitous in all systems examined so far, and spin diffusion plays the key role in polarization transfer within the macromolecule. Tightly bound water molecules with long residence times are rare in proteins at ambient temperature. The water polarization-transfer technique has been used to study the hydration of microcrystalline proteins, lipid membranes, and plant cell wall polysaccharides, and to derive atomic-resolution details of the kinetics and mechanism of ion conduction in channels and pumps. Using this approach, we have measured the water polarization transfer to the transmembrane peptide of the influenza M2 protein to obtain information on the structure of this tetrameric proton channel. At short mixing times, the polarization transfer rates are site-specific and depend on the pH, labile protons, sidechain conformation, as well as the radial position of the residues in this four-helix bundle. Despite the multiple dependences, the initial transfer rates reflect the periodic nature of the residue positions from the water-filled pore, thus this technique provides a way of gleaning secondary structure information, helix tilt angle, and the oligomeric structure of membrane proteins.
Keywords: chemical exchange, spin diffusion, ion channels, influenza M2 protein, heteronuclear correlation
1. Introduction
Water is one of the most essential molecules for the proper structure and function of proteins and other biological macromolecules. Protein hydration, protein folding, ion channel function, and lipid self-assembly, all rely on water interactions with these molecules [1-6]. Solid-state NMR (SSNMR) spectroscopy is an excellent method to study water interactions with non-crystalline and insoluble biological macromolecules. We are particularly interested in understanding how water interacts with proteins in phospholipid membranes, and to what extent water can serve as a probe of membrane protein structure. In this perspective article, we review SSNMR pulse sequences for measuring water-biomolecule polarization transfer and recent findings of the mechanism of water interactions with proteins, lipids and other biological solids. We focus on studies that detect protein signals to obtain site-specific information about water dynamics and binding, and do not attempt to cover the large literature of direct observation of water dynamics using relaxation NMR. We then present our new study of the water interaction with the four-helix bundle formed by the influenza M2 transmembrane peptide (M2TM). The goal is to explore the extent to which secondary structure and oligomeric structure of membrane proteins can be extracted from site-specific water-protein polarization transfer rates.
2. Solid-State NMR Techniques for Studying Water-Protein Interactions
The main SSNMR approach for probing water-biomolecule interactions is to transfer the water 1H polarization to biomolecules and detect the result of the transfer via 13C, 15N or other heteronuclear signals of the biomolecule. This polarization transfer technique has many pulse sequence variations. When implemented in a 1D fashion, a 1H polarization gradient needs to be established first, which is usually accomplished by a 1H T2 filter. The Hahn echo retains the polarization of the dynamic water, which has long T2, while destroying the polarization of the rigid molecules, which have much shorter T2's. A subsequent longitudinal mixing period (tmix) allows the water polarization to transfer to the biomolecule, followed by 1H-X (X=13C, 15N, etc) cross-polarization (CP) for detection (Fig. 1a). The CP step can be implemented in a spin-diffusion-free fashion by locking the 1H magnetization along the magic-angle direction (LG-CP), or can include spin diffusion using the conventional Hartman-Hahn (HH) CP. In the latter case, the longitudinal mixing period may be skipped to shorten the total polarization transfer time, if one wishes to selectively examine only the nearest water protons [7, 8].
Figure 1.
Representative pulse sequences used to measure water polarization transfer to biomolecules. (a) 1D T2-filtered experiment. (b) 2D 1H-undecoupled HETCOR experiment with spin diffusion mixing. (c) 2D dipolar-dephased MELODI-HETCOR experiment. (d) 1H-detected 1D XHH experiment for sensitivity-enhanced detection of biomolecule-water polarization transfer.
This polarization transfer framework is modified from the Goldman-Shen experiment [9, 10]. The heteronuclear signals reveal which sites are close to water, and the distance information can be made quantitative by measuring the heteronuclear signals as a function of tmix.
When water is not the only species that survives the 1H T2 filter and other dynamic species also exist, then the 1D experiment needs to be modified either by selective excitation of the water resonance [11] or by extending the experiment to 2D and encoding the 1H chemical shift before the mixing period [12, 13] (Fig. 1b). In the resulting 2D spin-diffusion heteronuclear correlation (HETCOR) experiment, 1H homonuclear decoupling is optional during the evolution period. Without 1H-1H decoupling, only highly mobile molecules such as water and liquid-crystalline lipids will be detected in the indirect dimension. When homonuclear decoupling is applied, the 1H signals of the rigid solid also become detectable, thus providing higher information content to the spectra. For the undecoupled 2D experiment, the 1H T2 filter is necessary for suppressing the rigid proton signals in the first t1 slice, while for the homonuclear-decoupled experiment, the T2 filter can be removed.
In practice, even with 1H chemical shift encoding, some solid protons such as protein Hα can resonate near the bulk-water chemical shift of 4.8 ppm to cause 1H resonance overlap. To address this problem, and to simplify the 2D HETCOR spectra, we developed a dipolar-filtered HETCOR experiment called MELODI (medium- and long-distance heteronuclear correlation) HETCOR, which detects only cross peaks resulting from non-bonded spin pairs [14, 15] (Fig. 1c). Before the 1H evolution period, a 1H-X REDOR dephasing period is inserted in which an 180° X-pulse is applied every half a rotor period to recouple the 1H-X dipolar interaction. A single 1H 180° pulse in the center of the dephasing period refocuses the 1H chemical shift. This REDOR filter destroys the magnetization of protons that are directly bonded to an X spin. The protons that survive this dipolar filter include water, protein protons without directly bonded 13C or 15N nuclei, and highly dynamic protons with weaker 1H-X dipolar couplings. Demonstration on natural-abundance amino acids confirmed that only two-bond cross peaks such as Hα-Cβ and Hβ-Cα remained in the 13C-dephased 13C-1H MELODI-HETCOR spectra [14]. For a selectively 13C-labeled model protein, ubiquitin, the 13C-dephased 13C-1H MELODI-HETCOR spectra contain cross peaks between labeled 13C and protons without directly bonded 13C, and cross peaks between labeled 13C and 15N-bonded protons such as backbone amides, Lys NH3 and Arg guanidinium Hη and Hε. For a uniformly 13C, 15N-labeled membrane protein, colicin Ia channel domain, 13C-dephased 13C-1H MELODI-HETCOR spectra exhibit 13C-HN cross peaks.
The MELODI-HETCOR technique is well suited to the study of water-protein interactions, since water is unaffected by all dephasing pulses (unless 17O pulses are applied). Simultaneous irradiation of 13C and 15N in the dipolar dephasing period will destroy all rigid protein proton signals and leave only water 1H cross peaks. Using this approach, we have investigated water interactions with arginine residues of a cationic antimicrobial peptide [16]. The data allowed the unambiguous assignment of cross peaks at a 1H chemical shift of 4.9 ppm to water-Arg correlations, rather than Arg Hα-sidechain correlations. This means that water solvates membrane-inserted guanidinium ions, which provides direct evidence of the long-suspected but rarely proven phenomenon that water penetrates into the lipid membrane to lower the free energy of insertion of these cationic membrane peptides.
To enhance the sensitivity of water-protein polarization transfer experiments, we developed a 1H-detected experiment called XHH, in which the 1H mixing period is preceded by dual CP steps: forward 1H-X CP and reverse X-1H CP. In this way, only protons directly attached to the X spins are selected to undergo polarization transfer [17] (Fig. 1d). Under moderate MAS frequencies and without 1H dilution by deuteration, such a CHH experiment detects only the 1H signals of mobile water and lipids as transferred from the 13C-bonded protein protons. Between the two CP steps, the X-spin magnetization is stored along the z-axis while the initial 1H magnetization is destroyed in the transverse plane. 90° 1H “purge” pulses during the dephasing period can be applied to ensure clean suppression of all initial 1H magnetization. The sensitivity enhancement factor of the CHH experiment depends on the 13C labeling level and the number of protein protons relative to the number of mobile water and lipid protons. For 13C-labeled colicin Ia channel domain, we showed that the 1D CHH experiment is faster than the 2D 13C-detected 1H spin diffusion experiment by a time factor of 180-350 fold and yields the same buildup curves as the 2D experiment.
A different way of transferring water polarization is via the nuclear Overhauser effect (NOE). By irradiating on the water 1H resonance for several seconds before applying a 90° pulse on the X channel, one can observe NOE enhancement of the X-spin signal, which depends on the water-protein proximity. Solid-state NOEs has been observed in model compounds by Takegoshi and Terao [18], and application to water-protein transfer shows that enhancement factors of 50-100% can be achieved [8].
3. Mechanisms of Water-Protein Polarization Transfer
As alluded to above, during the longitudinal and transverse mixing periods, three mechanisms can exist to transfer water polarization to biomolecules: chemical exchange between labile protons, spin diffusion, and the NOE (Fig. 2a). At neutral pH, chemical exchange rates range from ~100 s−1 for amide protons in protein backbones to ~1000 s−1 for amino protons of Lys, His and Arg, to ~10,000 s−1 for hydroxy protons of Tyr [19, 20]. Exchange rates decrease with temperature and also depend on pH. Spin diffusion is dipolar-mediated relayed magnetization transfer that is described by a diffusion equation [21, 22], and is the most efficient mechanism in rigid solids. The NOE is a dipolar cross relaxation process that requires motional modulation of two spins in close proximity [23, 24].
Figure 2.
Main mechanisms of water-biomolecule polarization transfer (a) and applications of solid-state NMR to investigations of water-protein and water-polysaccharide interactions (b-e). (a) Water protons can transfer polarization to biomolecules via chemical exchange, dipolar spin diffusion, and NOE. (b) Distinguishing liquid water and ice by solid-state NMR provide structural information on ice-binding proteins. (c) Water polarization transfer to ion channels has been used to investigate ion conduction kinetics, oligomeric structure of the channel, and to identify lipid-facing versus pore-facing residues. (d) Water-edited solid-state NMR experiments have been conducted to determine the topology of membrane proteins in terms of surface-exposed loops (left) and helices (right) versus membrane-embedded residues. (e) Distinct water interactions with different polysaccharides of the plant cell wall have been studied to provide information on the three-dimensional structure of the cell wall.
At ambient temperature, chemical exchange has been found to be important and even necessary for water polarization transfer to biomembranes. Griffin and coworkers [7] detected the 15N signals of bacteriorhodopsin using a 1D 1H T2-filtered 15N CP experiment. With a T2 filter of 1 ms, the backbone amide 15N peak was completely suppressed while the Schiff-base 15N peak and the free Lys NH3 peaks survived. The 1H T2 associated with the remaining 15N signals is similar to the bulk-water T2 of ~50 ms, while the 1H-15N CP was equilibrated by 0.5 ms, indicating solid-like character for the protons that cross-polarize to 15N. These results indicate that the Schiff base, although deeply buried in the active site of the protein, is accessible to bulk water and exchanges with it rapidly. Hong and coworkers showed that chemical exchange is necessary for water-lipid polarization transfer [12]. In 2D 31P-1H spin-diffusion HETCOR spectra, many lipid membranes, including POPE, POPG, POPC/cholesterol, and POPE/POPG containing membrane peptides, showed strong water 1H-31P cross peaks, while POPC exhibited no water cross peak even after a long mixing time of 225 ms. POPC is the only lipid without any labile protons among all the lipids examined, thus the result unambiguously shows that chemical exchange is necessary for water-lipid polarization transfer. The implication is that if labile protons are absent in the system, then water cross peaks will not be detectable by polarization transfer experiments at ambient temperature even if the membrane surface is hydrated.
Following chemical exchange, 1H spin diffusion is usually the dominant mechanism for polarization transfer within the rigid biomolecules. At moderate MAS frequencies, spin diffusion is readily observed, and its effect increases with decreasing temperature, which is opposite the temperature dependence of chemical exchange. Direct dipolar transfer from water to proteins is rare due to the rapid translational diffusion of water, except when water is tightly bound or confined. To investigate if there are solid-like water molecules with long residence times in proteins, Böckmann and Lesage applied a dipolar double-quantum filtered (DQF) HETCOR experiment to the microcrystalline protein, Crh [8]. Only water with residence times longer than a few hundred microseconds can survive this DQF. They did not observe any water-protein double-quantum peaks, indicating that water molecules associated with Crh undergo translational diffusion on a much shorter timescale than microseconds. Reif and coworkers used the spin-diffusion buildup of the 15N signals of perdeuterated SH3 protein to investigate the water dynamics in this microcrystalline protein [25]. Some residues were found to have buildup rates that are proportional to the water-HN dipolar couplings calculated from the crystal structure, indicating that these water molecules are sufficiently bound to the protein to give distance-dependent spin diffusion. On the other hand, surface residues, although in contact with a large number of water molecules, show slow transfer rates, indicating that the dynamic bulk water on the protein surface does not contribute significantly to spin diffusion. This finding is consistent with a Crh study in which two water pools, bulk and bound, were resolved in the 1H spectra [26]. Removal of the bulk water did not change the water-protein polarization transfer. Water peaks with fine structures that reflect different environments are also commonly seen in membrane proteins and other hydrated biomolecules [12, 27, 28].
Using Crh, Böckmann, Lesage and coworkers systematically investigated the relative importance of the three polarization transfer mechanisms and the conditions under which each mechanism dominates [8, 29]. They found that chemical exchange is the primary mechanism at room temperature but slows down at low temperature, even when water is prevented from freezing by cryoprotectants. Spin diffusion is the dominant mechanism within protein protons under moderate MAS, but ultrafast MAS (> 45 kHz) suppresses this contribution, allowing the NOE mechanism to be detected. At low temperature or ultrafast MAS, these authors observed negative water – protein cross peaks in Crh [30], which can only be assigned to rotating-frame nuclear Overhauser effects. Zilm and coworkers attributed HN-ubiquitin cross peaks in 1H-detected 15N-1H correlation spectra to NOE, based on the fact that the rate of the cross peak buildup with the longitudinal mixing time differs from the rate of buildup with a transverse spin-lock time [31]. The assignment of NOE cross peaks in these microcrystalline proteins suggests that a small number of water molecules are sufficiently bound to the protein sidechains to undergo correlated motion with the protein.
4. Water Interactions with Proteins, Lipid Membranes, and Carbohydrates
In this section we review recent applications of water polarization transfer and water-edited SSNMR experiments to different biomolecular systems (Fig. 2b-e). McDermott and coworkers investigated the water interaction of an antifreeze protein (AFP) to understand how proteins retain hydration shells and interact with ice [32]. 1H spin-lattice relaxation of AFP in frozen water at -35°C showed biexponential behavior, while the non-ice-binding protein, ubiquitin, showed single-exponential relaxation. This difference indicates that two proton baths exist near AFP and undergo intermediate exchange with each other. When D2O solution was used, the AFP relaxation became single-exponential, indicating that liquid water is responsible for the second proton bath. Cross-saturation NMR experiments provided insight into the protein-ice interaction. With a 1H presaturation pulse, the 13C intensities of ubiquitin decreased due to cross saturation, and the intensity reduction does not depend on the presaturation pulse length. In contrast, the 13C intensities of AFP decreased over a wider 1H frequency range as the presaturation pulse lengthened, indicating that there is spin diffusion between ice and AFP. Thus, without detecting the ice 1H signals, which is difficult, the authors were able to show that AFP is in contact with ice. AFP also shows cross peaks with the liquid-water 1H signal at 5 ppm in 2D 13C-1H HETCOR spectra, indicating that the protein retains a partial hydration shell. Taken together, these data indicate that a part of the AFP, presumably the ice-binding surface, is in contact with the ice lattice (Fig. 2b), in contrast to the non-ice-binding ubiquitin, which retains a complete hydration shell and is fully shielded from ice in the frozen solution.
The water polarization-transfer technique has been applied fruitfully to obtain structural information on membrane proteins. Baldus and coworkers showed that the intensity buildup of protein signals as a function of tmix depends on the water-exposed surface area of the protein [11]. Adapting the spin diffusion theory from polymer NMR [22], they analyzed the water-protein spin diffusion rates to extract information about structural changes of a potassium channel between high and low pH. The low-pH channel, which shows faster polarization transfer, corresponds to a 65% larger water-protein surface area. Similarly, Hong and coworkers measured the water-exposed surface area of the influenza M2 proton channel [33]. Comparison of the water-protein spin diffusion rates among three states, the low pH open state, the high pH closed state, and the high-pH drug-bound state, revealed that the open state has the largest water-exposed surface area, while the drug-bound state is the least hydrated, with a 2-fold smaller water-exposed surface area (Fig. 2c). Moreover, site-specific buildup rates identified the drug-binding site to be below V27 and above G34.
The residence time of water in ion channels is a question of significant interest. Recent MD simulations suggested that long-lived water in the selectivity filter of potassium channels might explain the slow inactivation of these channels after opening. Using ultrafast MAS and 1H detection experiments, Baldus and coworkers measured water cross peaks to the selectivity filter residues in KcsA [34]. They found that the conductive state of the selectivity filter has fewer water cross peaks than the inactivated state. The difference was interpreted in light of the MD prediction that buried and ordered water molecules lock the selectivity filter in the inactivated conformation, thus slowing its relaxation to the conductive state. However, the observed water-protein cross peaks appear at a 1H chemical shift of 4.8 ppm, indicating that the protein-bound water is in fast exchange with bulk water, reminiscent of water contact with the Schiff base in bacteriorhodopsin. Thus, these protein-bound water molecules are unlikely to have long (microsecond) residence times. In a related context, Hong and coworkers measured the chemical shifts of protons that are dipolar-coupled to His37 imidazole nitrogens in the influenza M2 proton channel [35], in order to distinguish between a histidine-histidine strong hydrogen-bond model [36] and a water-histidine exchange model [37] for proton conduction. At low temperature, they found HN chemical shifts of <15 ppm for both Nε2 and Nδ1, consistent with regular N-H hydrogen bonds, while at high temperature, the Nε2 and Nδ1-correlated 1H chemical shift collapsed to 4.8 ppm, indicating that His37 undergoes rapid exchange with a large number of water molecules to adopt the bulk water 1H chemical shift. Thus, these data support the water-histidine exchange model for proton conduction.
Water polarization transfer can also be used in a spectral-editing fashion to selectively detect the water-exposed protein residues. This approach has been used to detect loop residues in a seven-helix transmembrane receptor [38], cytoplasmic residues in the full-length M2 protein [39], and to determine the topology of colicin and gramicidin channels [10, 13, 40] (Fig. 2d). In a complementary approach, hydrogen/deuterium (H/D) exchange has been used extensively to determine water-exposed and water-exchanged residues in membrane proteins. The effect of H/D exchange can be manifested as attenuation of the amide 15N signals after 1H-15N cross polarization, as amide 2H spectra, and even as 15N-2H dipolar couplings. The exchange-induced 15N signal suppression can be readily incorporated into 2D and 3D 15N-13C MAS correlation experiments for protein resonance assignment, and this approach has been applied to several multi-span membrane proteins to obtain topology information [41-43]. H/D exchange-edited 2H NMR and 15N-2H dipolar coupling experiments have been employed by Cross and coworkers to study the structures of gramicidin [44, 45] and influenza M2 channels [46]. The exchange rates were found to be sufficiently residue-specific to give information about the structures of these membrane peptides.
Water interaction with the lipid bilayer itself has also been studied. The self-diffusion coefficient of water is known precisely to be 2.299 × 10−9 m2/s at 25°C [47]. Deviation from this value gives information about how biomolecules interact with water to retard or speed up its translational diffusion. Han and coworkers used the Overhauser dynamic nuclear polarization technique to study water translational diffusion within 1.5 nm of the surface of a DPPC bilayer [48, 49]. The self- and cross relaxations of water near a nitroxide spin label that is covalently attached to the lipid headgroup depends on the water residence time near the spin label. By observing the water signal enhancement under microwave irradiation of the spin label, these authors measured the relaxation rates to deduce the water correlation time. They found that the correlation time of the surface hydration water is 6-fold longer than that of bulk water, consistent with the expected membrane retardation of water diffusion. However, when the bulk-solution viscosity was increased 10-fold by using high osmolyte concentrations, the surface-water correlation times increased by only 2-fold. This small magnitude of change suggests that the surface topology and chemistry of the membrane, rather than bulk solvent characteristics, dominates the dynamics of the surface-hydration water. In other words, water diffusion on the membrane surface is largely decoupled from bulk-water diffusion.
In addition to proteins and lipids, the water polarization transfer technique has been applied to polysaccharides in plant cell walls by Hong and coworkers to understand the hydration of cellulose, pectins, and hemicellulose (Fig. 2e) [27]. Water-pectin polarization transfer was found to be much faster than water-cellulose transfer, but chemical extraction of the cell wall exerted a major impact on the spin diffusion rates. Removal of calcium ions and the consequent extraction of homogalacturonan dramatically slowed down water polarization transfer to all polysaccharides, while further extraction of matrix polysaccharides restored the 1H spin diffusion rates. This unexpected trend indicates that calcium ions and homogalacturonan gelation in the intact cell wall increase the amount of bound water, thus speeding up polarization transfer, while calcium removal disrupts the gel and increases water dynamics, thus slowing down polarization transfer. The recovery of spin diffusion rates after more extensive extraction is attributed to increased water-exposed surface areas of the polysaccharides. The correlation between water-pectin and water-cellulose spin diffusion rates supports a single-network model of plant primary cell walls, as it indicates that a significant fraction of the cellulose microfibril surface is covered with pectins [50].
5. Water Interactions with the Influenza M2 Transmembrane Channel
We now present new results on the influenza M2 protein that illustrate one specific type of structural information that can be obtained from SSNMR measurements of water interactions with integral membrane proteins. The M2 protein of influenza viruses forms an acid-activated tetrameric proton channel in the virus envelope to conduct protons from the acidic endosome into the virion. Recent SSNMR data and other biophysical and mutagenesis results [51] showed that the transmembrane domain conducts protons by a shuttle mechanism [52], in which a conserved histidine, H37, undergoes successive protonation and deprotonation events, accompanied by microsecond reorientations of the imidazole rings [37, 53, 54]. Water is important in this process because it delivers protons to H37 as well as removing protons from H37 to transfer into the virus, and water-H37 forms a mixed hydrogen-bonded chain. Thus, understanding water dynamics is integral to elucidating the structure and function of this prototypical proton channel.
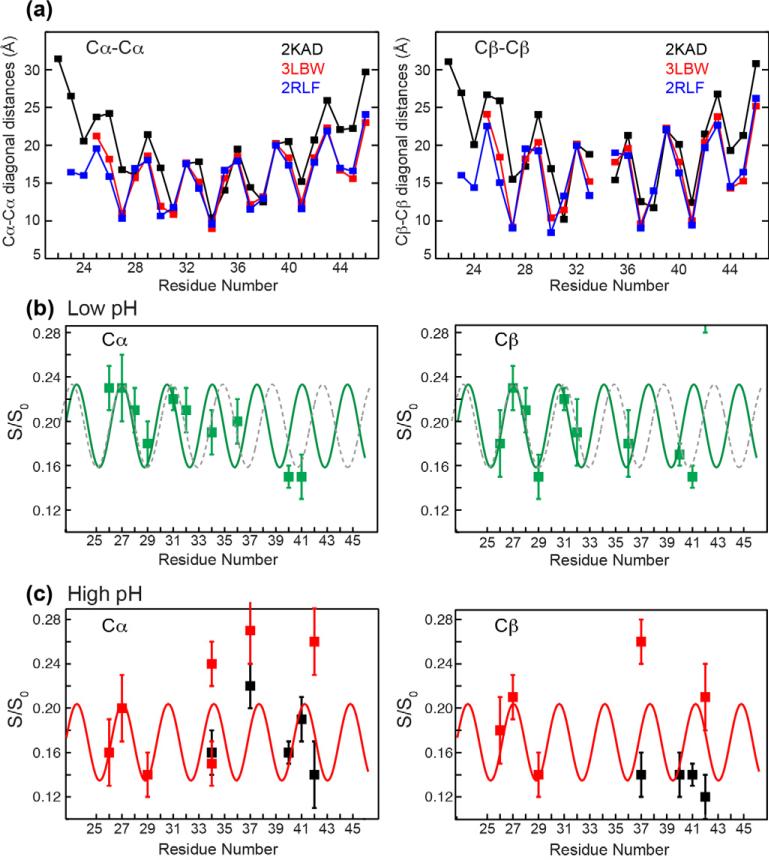
In addition to its functional role, water may also provide a way to determine the structure of the four-helix bundle. The distances of successi ve residues from the center of the pore should exhibit a periodicity reflecting the α-helical pitch of 3.6 residues per turn. These distances should also depend on the helix tilt angle. Fig. 3a shows the residue-specific amide 15N-15N distances between the two opposite helices of the tetramer, which are twice the 15N distances to the channel axis. The distances are extracted from three PDB structures of M2TM. The SSNMR structure (2KAD), measured in thin DLPC bilayers, has a 35° tilt angle and is believed to resemble the low-pH open-channel structure [55]. A 1.65 Å resolution crystal structure (3LBW), measured at pH 6.5, has an intermediate tilt angle [56]. A high-pH solution NMR structure (2RLF) has the smallest tilt angle of ~15° [57]. Fig. 3a shows that all three structures exhibit the same periodicity and “phase” in the 15N-water distance oscillation as a function of residue position, while the vertical displacement and amplitude of the oscillation differ. Residues near the two termini have longer distances to the water-filled pore than residues near the crossing point of the four-helix bundle, thus the distance profile droops in the middle. This periodic water-distance pattern partly resembles the dipolar waves of oriented membrane proteins [58]. Periodic structural features are also commonly exploited in EPR spectroscopy: paramagnetic broadening by lipid- or water-soluble spin labels has long been used to measure the structure of α-helical membrane proteins [59]. We thus ask the question whether water-protein polarization transfer rates can reveal the secondary structure and quaternary structure of ion channels. We measured the water-protein 1H spin diffusion of 12 residues in the 25-residue M2TM at low and high pH, and show that indeed periodic water-protein transfer rates can be observed, even though it is mixed with other structural factors.
Figure 3.
(a) Backbone N-N distances between two opposite helices in the tetrameric M2TM. The distances are extracted from three PDB structures of M2 with distinct helix tilt angles. The SSNMR structure 2KAD has a large tilt angle (~35°) that is expected for the low-pH state. The pH 6.5 crystal structure 3LBW has an intermediate helix tilt angle. The solution NMR structure 2RLF has the smallest tilt angle (~15°) and was measured at high pH. Periodic oscillation of the nitrogen distances to water, which is half the N-N distance plotted here, is seen for all structures. (b) Top view of the four-helix bundle formed by M2TM, showing the positions of the 12 labeled residues on the heptad repeat. Positions a and d (red) face the water-filled pore, e and g (blue) are interfacial, and c, f, and b (black) face lipids.
6. Experimental Methods
Membrane Samples
Four M2TM peptides were used in this study, each bound to cholesterol-containing virus-mimetic membranes at low and high pH, thus giving a total of eight samples that cover 12 residues of the transmembrane domain. The amino acid sequence corresponds to residues 22-46 of the Udorn strain of M2 (SSDPLVVAASIIGILHLILWILDRL). The labeled residues are: L26V27A29G34 (LVAG) [33], G34H37I42 (GHI) [37], V28S31I32L36 (VSIL) [60], and L40W41 (LW) [61]. LW-M2TM was bound to an unsaturated-lipid VM+ membrane consisting of POPC, POPE, sphingomyelin (SM) and cholesterol, while the other three peptides were bound to a saturated-chain virus-mimetic membrane (VM) consisting of DPPC, DPPE, SM and cholesterol [62, 63].
Solid-State NMR
SSNMR experiments were conducted on a Bruker DSX-400 MHz (9.4 T) and an AVANCE II 600 MHz (14.1 T) spectrometer using 4 mm MAS probes. Typical radiofrequency field strengths were 71-83 kHz for 1H and 50 kHz for 13C. 13C chemical shifts were referenced to the CH2 signal of adamantane at 38.48 ppm, while 1H chemical shifts were referenced to the Hγ peak of phosphocholine headgroup at 3.26 ppm on the TMS scale.
2D 1H-13C spin-diffusion HETCOR experiments (Fig. 1b) were used to measure water-M2 cross peaks. The 1H T2 filter time was 1.9 ms or 1.2 ms. For most samples, two mixing times, 4 ms and 100 ms, were used, and the samples were spun at either 7 kHz or 11 kHz. For the LVAG samples, additional spectra were measured with 1 ms and 2 ms mixing times under 10 kHz MAS to achieve more selective polarization transfer. All data were acquired at 293 K. The water 1H T1 relaxation time is about 500 ms in these samples. The water cross-peak intensity was extracted from the 2D spectra by summing over all cross sections of the water 1H linewidth (~0.5 ppm). The intensity ratios between the 4 ms and 100 ms spectra, corrected for 1H T1 relaxation, are denoted as S/S0 and compared among different residues. Error bars for S/S0 were propagated from the signal-to-noise ratios of the peak intensities.
7. Results and Discussion
The 12 labeled residues measured in this work represent half of the M2TM, and populate pore-facing, lipid-facing, and interfacial positions of the heptad repeat (Fig. 3b). The pore-facing a and d positions include all functionally important residues of the channel such as V27, G34, H37 and W41. The interfacial e and g positions include the drug-interacting S31 as well as L26 and L40, whereas the lipid-facing b, c, f positions include bulky hydrophobic residues V28, A29, I32, L36 and I42. Our hypothesis is that the water – protein polarization transfer rates of these residues may be periodic to reflect the structure of the helix as well as the structure of the tetramer. Fig. 4a, b show two representative 2D 1H-13C HETCOR spectra. With a 1.9 ms 1H T2 filter, signals of the most dynamic lipid moieties such as headgroup Hβ and chain CH2 are not completely suppressed, thus 2D spectra are necessary to resolve the water-peptide cross peaks from the lipid-peptide cross peaks. At 100 ms, protein-lipid cross peaks at 1H chemical shifts of ~1.3 ppm are observed, consistent with the membrane-spanning topology of this proton channel (Fig. 4b). Instead of measuring a full buildup curve, we chose two mixing times, 4 ms and 100 ms, and report their intensity ratio as an indicator of the initial buildup rates. Fig. 4c-f show the water 1H cross sections of three of the four peptides at 4 ms (red) and 100 ms (black) at both low and high pH. It can be seen that the 4 ms intensities are much higher at low pH than at high pH for all samples, and the low-pH GHI sample gives the highest initial intensities among all samples.
Figure 4.
Representative water-M2TM 1H spin diffusion spectra. (a) 2D spectrum of the pH 4.5 GHI sample with 4 ms mixing. (b) 2D spectrum of the pH 4.5 LW sample with 100 ms mixing. In addition to water-protein cross peaks, lipid-protein cross peaks are also observed at the 1H chemical shift of 1.3 ppm, consistent with the transmembrane orientation of the channel. (c) Water 1H cross sections of the low-pH GHI spectra with 4 ms (red) and 100 ms (black) mixing. (d-f) Low pH (left) and high pH (right) water 1H cross sections at 4 ms and 100 ms mixing for (d) GHI, (e) LVAG, and (f) LW samples. Only the aliphatic region is shown for clarity.
Fig. 5 plots the 4 ms versus 100 ms intensity ratios of all residues at Cα, Cβ, and Cγ. Two observations can be made. First, all residues show higher intensities at low pH than at high pH, consistent with the fact that the low-pH channel is open and better hydrated. This pH-induced S/S0 difference is the largest for GHI, and the imidazole Cε1 has the highest S/S0 value, about 1, among all sites (Fig. 6a). Second, the radial positions of the residues affect the initial spin diffusion rates and the pH dependence of the polarization transfer. Pore-facing residues such as V27, G34, and H37 have the largest S/S0 values as well as the largest intensity difference between low pH (average S/S0 = 0.28) and high pH (0.20). Interfacial residues at heptad repeat positions of e and g, such as L26 and S31, have comparable S/S0 values of 0.18 and 0.16 between low and high pH, whereas lipid-facing residues have S/S0 values of 0.23 and 0.18 at low and high pH. These S/S0 values have uncertainties of about ±0.02; thus, the pore-facing residues’ higher spin diffusion intensities compared to other radial positions are statistically significant. On the other hand, the pH-induced intensity difference for the lipid-facing residues is insignificant.
Figure 5.
Spin diffusion intensity ratios (S/S0) between 4 ms and 100 ms for all measured residues. (a) Cα, (b) Cβ, and (c) Cγ. Horizontal axis indicates the residue number as well as the heptad-repeat positions. Grey bars indicate residues that lie at the a and d positions. Data obtained at pH 4.5 (green), pH 7 or 7.5 (red) and pH 8.5 (black) are superimposed.
Figure 6.
Backbone to sidechain trends in the water polarization-transfer intensities, S/S0. (a) GHI sample. (b) LVAG sample. Open symbols in the V27 panel were data measured with shorter mixing times of 1 ms and 2 ms. (c) LW sample. Data measured at pH 4.5 are shown in green, at pH 7.0 and 7.5 in red, and at pH 8.5 in black. The residue's heptad positions are indicated.
The high polarization transfer rates of the GHI sample can be partly attributed to H37, since this sidechain is known to be exposed to water and hydrogen-bond to it [35, 37], and imidazole nitrogens have one of the largest chemical exchange rates among labile protons [19]. The high G34 spin diffusion rate can also be partly attributed to the pore-facing position of this residue. However, the high S/S0 values of the lipid-facing I42 is not expected, and suggests that sample preparation uncertainties such as lower actual pH than reported here may make these GHI-M2TM samples more efficient in receiving water magnetization. The G34 S/S0 value in the GHI sample is higher than the G34 intensity in the LVAG sample by ~0.2 at low pH and 0.1 at high pH, suggesting that these are the approximate correction factors to account for sample preparation variations.
To investigate how the atomic position in the sidechain affects spin-diffusion intensities, we plotted the S/S0 intensity ratios of different carbons within each residue (Fig. 6). Several trends emerge from this comparison. First, H37 and W41 are the only two residues where the backbone Cα intensities are lower than many sidechain intensities. This is consistent with the pore-facing positions of these two residues in the heptad repeat, and confirms that both residues directly contact water to carry out the essential functions of proton conduction and gating. The pH-induced intensity differences in the Trp41 indole are smaller than the intensity differences of the His37 imidazole. This is consistent with the experimentally determined rotameric structures of these two residues, which indicate that the Trp41 sidechain is less exposed to the channel pore than the H37 sidechain. 19F-19F and 13C-19F distance measurements constrained the Trp41 rotamer to be trans for χ1 and +90° for χ2, which points the six-membered benzene ring towards the helix-helix interface while leaving the five-membered pyrrole ring in the pore [61, 64]. In comparison, the His37 rotamer (trans-trans) points the imidazole ring straight into the pore [37]. Second, hydrophobic residues that face the lipids or the helix interface, such as I42, L26, A29 and L40, have either a flat or a decreasing intensity profile from the backbone to the sidechain tail, consistent with the fact that these sidechains point away from the water-filled pore. For the lowpH LVAG sample, both the interfacial L26 and the pore-facing V27 have higher S/S0 values than the pore-facing G34 (Fig. 6b). This trend does not correlate with the radial positions of these two residues but reflects the fact that L26 and V27 lie close to the membrane surface and thus interact with inter-bilayer water.
These analyses indicate that in addition to the radial position of the residues, the chemical exchange rates of labile protons, the sidechain conformation, and the vertical displacement from the membrane surface, all affect the polarization-transfer rates from water. To assess the extent to which the 13C-detected 1H polarization transfer rates can reveal the radial position of the residues and hence the helical secondary structure, we carried out a constrained fit of the Cα and Cβ S/S0 values (Fig. 7). We assume an ideal helical pitch of 3.6 residues per turn, and constrain the phase of the sinusoidal oscillation to be identical between Cα and Cβ and between low and high pH. The latter amounts to assuming that the rotation angle of the helix is unchanged by pH. The oscillation amplitude is allowed to vary since this parameter depends on the helix tilt angle and the intrinsic difference in the atomic positions of Cα and Cβ from the water-filled pore (Fig. 7a). Since the GHI samples show systematically higher S/S0 values due to sample-preparation variations, we do not consider their intensities in this global fit. Fig. 7b shows that a periodic pattern does exist, and reproduces the spin diffusion intensities of the N-terminal half of the helix relatively well. However, towards the C-terminal end, increasing discrepancy is seen between the experimental data and the sinusoidal wave. This is particularly pronounced for W41: the simulated wave predicts a high S/S0 value for W41, but the opposite was measured at low pH. In comparison, the high-pH data of W41 is better fit to the sine wave (Fig. 7c). Since the LW sample was the only one incorporated into the unsaturated VM+ membrane while all other peptides were reconstituted into the saturated VM membrane, these intensity deviations may reflect sample preparation differences. On the other hand, we cannot exclude the possibility that the C-terminal deviation from the simulated wave may reflect deviations of the peptide from an ideal helical structure. For example, a kink could occur between the N- and C-terminal regions to change the phase of the sinusoidal oscillation. Such helix kinks have often been observed in 15N- 1H dipolar waves and are diagnostic of the boundaries between two helices. The low-pH data could also be well fit by a significantly slower oscillation of 4 residues per turn (grey dashed lines in Fig. 7b). However, this periodicity departs significantly from an ideal α-helix structure, and predicts H37 intensities near the minimum of the wave, thus we exclude this alternative fit as unphysical.
Figure 7.
Residue-specific periodicity in the water polarization transfer to Cα and Cβ sites. (a) Extracted Cα-Cα and Cβ-Cβ diagonal distances in the M2TM tetramer, which correspond to twice the distances of these atoms to the center of the water-filled pore. Distances are extracted from three PDB structures with distinct helix tilt angles. (b) Low-pH S/S0 values of Cα and Cβ. Solid lines are empirical fits constrained by the ideal helical structure of 3.6 residues per turn, while grey dashed lines give an alternative fit with 4 residues per turn. (c) High-pH S/S0 values. Solid lines are empirical fits following the ideal helical structure. The GHI data are not included in these fits.
8. Conclusions
The initial polarization transfer rates from water to M2TM show site-specificity that depends on pH, chemical exchange rates, sidechain conformation, as well as the secondary structure of the helix backbone. Even with this confluence of factors, oscillatory patterns in the water-protein polarization transfer can be seen and reveal the underlying helical structure of the peptide. Pore-facing residues are identified both by their high initial buildup rates and their increasing transfer rates from the backbone to the sidechain carbons. These features are distinguishable from the characteristics of lipid-facing and interfacial residues. For the same samples and residues, intensity differences between low and high pH are not susceptible to sample-preparation differences and reliably report the pH-induced changes in the protein structure.
These water 1H polarization transfer experiments can be further optimized to increase the resolution of the structural information about membrane proteins. Using a single protein sample at each pH will yield more consistent residue-specific differences in the water contact. Conducting the experiments at moderate low temperatures where water remains liquid but chemical-exchange rates slow down will better emphasize distance-dependent spin diffusion as the polarization transfer mechanism. Use of shorter mixing times, as long as permitted by sensitivity, will also increase the contrast between pore-facing and lipid-facing residues. Experiments on the LVAG sample using 2 ms and 1 ms mixing times showed that the pore-facing V27 is the only residue with detectable intensities, whereas all other residues’ signals are suppressed (Fig. 6b). Thus, short polarization transfer times can provide better discriminate between the water proximities of different residues. Finally, 15N detection should provide independent and complementary information of the membrane protein structure.
Highlights.
Water polarization transfer reveals biomolecular structure and dynamics.
Heteronuclear detected water polarization transfer SSNMR is highly versatile.
Exchange, spin diffusion, and NOE are the mechanisms of polarization transfer.
Water-based SSNMR reveals membrane protein topology and ion channel kinetics.
Water SSNMR reports the four-helix-bundle structure of the flu M2 proton channel.
Acknowledgement
This work is supported by NIH grant GM088204 to M. H. The authors would like to thank Tuo Wang and Dr. Paul White for insightful discussions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Halle B. Protein hydration dynamics in solution: a critical survey. Phil. Trans. R. Soc. Lond. B. 2004;359:1207–1223. doi: 10.1098/rstb.2004.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer E. Internal water molecules and H-bonding in biological macromolecules: a review of structural features with functional implications. Protein Sci. 1992;1:1543–1562. doi: 10.1002/pro.5560011203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rand RP. Probing the role of water in protein conformation and function. Phil. Trans. R. Soc. Lond. B. 2004;359:1277–1284. doi: 10.1098/rstb.2004.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swanson JMJ, Maupin CM, Chen H, Petersen MK, Xu J, Wu Y, Voth GA. Proton Solvation and Transport in Aqueous and Biomolecular Systems: Insights from Computer Simulations. J. Phys. Chem. B. 2007;111:4300–4314. doi: 10.1021/jp070104x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Otting G, Liepinsh E, Wüthrich K. Protein hydration in aqueous solution. Science. 1991;254:974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- 6.Ernst JA, Clubb RT, Zhou HX, Gronenborn AM, Clore GM. Demonstration of positionally disordered water within a protein hydrophobic cavity by NMR. Science. 1995;267:1813–1817. doi: 10.1126/science.7892604. [DOI] [PubMed] [Google Scholar]
- 7.Harbison GS, Roberts JE, Herzfeld J, Griffin RG. Solid-state NMR detection of proton exchange between the bacteriorhodopsin Schiff base and bulk water. J. Am. Chem. Soc. 1988;110:7221–7223. [Google Scholar]
- 8.Lesage A, Emsley L, Penin F, Böckmann A. Investigation of dipolar-mediated water-protein interactions in microcrystalline Crh by solid-state NMR spectroscopy. J. Am. Chem. Soc. 2006;128:8246–8255. doi: 10.1021/ja060866q. [DOI] [PubMed] [Google Scholar]
- 9.Goldman M, Shen L. Spin-spin relaxation in LaF3. Phys. Rev. 1966;144:321–331. [Google Scholar]
- 10.Kumashiro KK, Schmidt-Rohr K, Murphy OJ, Ouellette KL, Cramer WA, Thompson LK. A novel tool for probing membrane protein structure: solid-state NMR with proton spin diffusion and X-nucleus detection. J. Am. Chem. Soc. 1998;120:5043–5051. [Google Scholar]
- 11.Ader C, Schneider R, Seidel K, Etzkorn M, Becker S, Baldus M. Structural rearrangements of membrane proteins probed by water-edited solid-state NMR spectroscopy. J. Am. Chem. Soc. 2009;131:170–176. doi: 10.1021/ja806306e. [DOI] [PubMed] [Google Scholar]
- 12.Doherty T, Hong M. 2D 1H–31P solid-state NMR studies of the dependence of inter-bilayer water dynamics on lipid headgroup structure and membrane peptides. J. Magn. Reson. 2009;196:39–47. doi: 10.1016/j.jmr.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huster D, Yao X, Hong M. Membrane protein topology probed by 1H spin diffusion from lipids Using solid-state NMR spectroscopy. J. Am. Chem. Soc. 2002;124:874–883. doi: 10.1021/ja017001r. [DOI] [PubMed] [Google Scholar]
- 14.Yao XL, Hong M. Dipolar filtered 1H-13C heteronuclear correlation spectroscopy for resonance assignment of proteins. J. Biomol. NMR. 2001;20:263–274. doi: 10.1023/a:1011251924874. [DOI] [PubMed] [Google Scholar]
- 15.Yao XL, Schmidt-Rohr K, Hong M. Medium- and long-distance 1H–13C heteronuclear correlation NMR in solids. J. Magn. Reson. 2001;149:139–143. [Google Scholar]
- 16.Li S, Su Y, Luo W, Hong M. Water-protein interactions of an arginine-rich membrane peptide in lipid bilayers investigated by solid-state nuclear magnetic resonance spectroscopy. J. Phys. Chem. B. 2010;114:4063–4069. doi: 10.1021/jp912283r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo W, Hong M. A 1D sensitivity-enhanced 1H spin diffusion experiment for determining membrane protein topology. Solid State Nucl. Magn. Reson. 2006;29:163–169. doi: 10.1016/j.ssnmr.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Takegoshi K, Terao T. C-13 nuclear Overhauser polarization nuclear magnetic resonance in rotating solids: Replacement of cross polarization in uniformly C-13 labeled molecules with methyl groups. J. Chem. Phys. 2002;117:1700–1707. [Google Scholar]
- 19.Liepinsh E, Otting G. Proton exchange rates from amino acid side chains--implications for image contrast. Magn. Reson. Med. 1996;35:30–42. doi: 10.1002/mrm.1910350106. [DOI] [PubMed] [Google Scholar]
- 20.Luz Z, Meiboom S. Kinetics of proton exchange in aqueous solutions of acetate buffer. J. Am. Chem. Soc. 1963;85:3923–3925. [Google Scholar]
- 21.Bloembergen N. On the interaction of nuclear spins in a crystalline lattice. Physica. 1949;15:386–426. [Google Scholar]
- 22.Schmidt-Rohr K, Spiess HW. Multidimensional Solid-State NMR and Polymers. 1st ed. Academic Press; San Diego: 1994. [Google Scholar]
- 23.Noggle JH, Shirmer RE. The Nuclear Overhauser Effect: Chemical Applications. Academic Press; New York: 1971. [Google Scholar]
- 24.Solomon I. Relaxation processes in a system of two spins. Phys. Rev. 1955;99:559. [Google Scholar]
- 25.Chevelkov V, Faelber K, Diehl A, Heinemann U, Oschkinat H, Reif B. Detection of dynamic water molecules in a microcrystalline sample of the SH3 domain of alpha-spectrin by MAS solid-state NMR. J. Biomol. NMR. 2005;31:295–310. doi: 10.1007/s10858-005-1718-z. [DOI] [PubMed] [Google Scholar]
- 26.Böckmann A, Gardiennet C, Verel R, Hunkeler A, Loquet A, Pintacuda G, Emsley L, Meier BH, Lesage A. Characterization of different water pools in solid-state NMR protein samples. J. Biomol. NMR. 2009;45:319–327. doi: 10.1007/s10858-009-9374-3. [DOI] [PubMed] [Google Scholar]
- 27.White PB, Wang T, Park YB, Cosgrove DJ, Hong M. Water – Polysaccharide Interactions in the Primary Cell Wall of Arabidopsis thaliana from Polarization Transfer Solid-State NMR. J. Am. Chem. Soc. 2014;136:10399–10409. doi: 10.1021/ja504108h. [DOI] [PubMed] [Google Scholar]
- 28.Yao H, Hong M. Conformation and lipid interaction of the fusion peptide of the paramyxovirus PIV5 in anionic and negative-curvature membranes from solid-state NMR. J. Am. Chem. Soc. 2014;136:2611–2624. doi: 10.1021/ja4121956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lesage A, Bockmann A. Water-protein interactions in microcrystalline Crh measured by H-1-C-13 solid-state NMR spectroscopy. J. Am. Chem. Soc. 2003;125:13336–13337. doi: 10.1021/ja036720y. [DOI] [PubMed] [Google Scholar]
- 30.Lesage A, Gardiennet C, Loquet A, Verel R, Pintacuda G, Emsley L, Meier BH, Böckmann A. Polarization transfer over the water-protein interface in solids. Angew. Chem. Int. Ed. Engl. 2008;47:5851–5854. doi: 10.1002/anie.200801110. [DOI] [PubMed] [Google Scholar]
- 31.Paulson EK, Morcombe CR, Gaponenko V, Dancheck B, Byrd RA, Zilm KW. High-sensitivity observation of dipolar exchange and NOEs between exchangeable protons in proteins by 3D solid-state NMR spectroscopy. J. Am. Chem. Soc. 2003;125:14222–14223. doi: 10.1021/ja037559u. [DOI] [PubMed] [Google Scholar]
- 32.Siemer AB, Huang K-Y, McDermott AE. Protein–ice interaction of an antifreeze protein observed with solid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 2010;107:17580–17585. doi: 10.1073/pnas.1009369107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo W, Hong M. Conformational changes of an ion channel detected through water-protein interactions using solid-state NMR spectroscopy. J. Am. Chem. Soc. 2010;132:2378–2384. doi: 10.1021/ja9096219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weingarth M, van der Cruijsen EAW, Ostmeyer J, Lievestro S, Roux B, Baldus M. Quantitative analysis of the water occupancy around the selectivity filter of a K+ channel in different gating modes. J. Am. Chem. Soc. 2014;136:2000–2007. doi: 10.1021/ja411450y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hong M, Fritzsching KJ, Williams JK. Hydrogen-bonding partner of the proton-conducting histidine in the influenza M2 proton channel revealed from 1H chemical shifts. J. Am. Chem. Soc. 2012;134:14753–14755. doi: 10.1021/ja307453v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath D, Zhou HX, Cross TA. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hu F, Luo W, Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Etzkorn M, Martell S, Andronesi OC, Seidel K, Engelhard M, Baldus M. Secondary structure, dynamics, and topology of a seven-helix receptor in native membranes, studied by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. Engl. 2007;46:459–462. doi: 10.1002/anie.200602139. [DOI] [PubMed] [Google Scholar]
- 39.Liao SY, Fritzsching KJ, Hong M. Conformational analysis of the full-length M2 protein of the influenza A virus using solid-state NMR. Protein Sci. 2013;22:1623–1638. doi: 10.1002/pro.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gallagher GJ, Hong M, Thompson LK. Solid-state NMR spin diffusion for measurement of membrane-bound peptide structure: gramicidin A. Biochemistry. 2004;43:7899–7906. doi: 10.1021/bi0356101. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y, Zhang Z, Tang X, Li J, Glaubitz C, Yang J. Conformation and Topology of Diacylglycerol Kinase in E.coliMembranes Revealed by Solid-state NMR Spectroscopy. Angew. Chem. Int. Ed. 2014:5624–5628. doi: 10.1002/anie.201311203. [DOI] [PubMed] [Google Scholar]
- 42.Shi L, Kawamura I, Jung K-H, Brown LS, Ladizhansky V. Conformation of a seven-helical transmembrane photosensor in the lipid environment. Angew. Chem. Int. Ed. 2011;50:1302–1305. doi: 10.1002/anie.201004422. [DOI] [PubMed] [Google Scholar]
- 43.Wang S, Shi L, Kawamura I, Brown LS, Ladizhansky V. Site-Specific Solid-State NMR Detection of Hydrogen-Deuterium Exchange Reveals Conformational Changes in a 7-Helical Transmembrane Protein. Biophys. J. 2011;101:L23–L25. doi: 10.1016/j.bpj.2011.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huo S, Arumugam S, Cross TA. Hydrogen exchange in the lipid bilayer-bound gramicidin channel. Solid State Nucl. Magn. Reson. 1996;7:177–183. doi: 10.1016/s0926-2040(96)01260-x. [DOI] [PubMed] [Google Scholar]
- 45.Cotten M, Fu R, Cross TA. Solid-state NMR and hydrogen-deuterium exchange in a bilayer-solubilized peptide: Structural and mechanistic implications. Biophys. J. 1999;76:1179–1189. doi: 10.1016/S0006-3495(99)77282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tian C, Gao PF, Pinto LH, Lamb RA, Cross TA. Initial structural and dynamic characterization of the M2 protein transmembrane and amphipathic helices in lipid bilayers. Prot. Sci. 2003;12:2597–2605. doi: 10.1110/ps.03168503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holz M, Heil SR, Sacco A. Temperature-dependent self-diffusion coefficients of water and six selected molecular liquids for calibration in accurate 1H NMR PFG measurements. Phys. Chem. Chem. Phys. 2000;2:4740–4742. [Google Scholar]
- 48.Franck JM, Scott JA, Han S. Nonlinear scaling of surface water diffusion with bulk water viscosity of crowded solutions. J. Am. Chem. Soc. 2013;135:4175–4178. doi: 10.1021/ja3112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Armstrong BD, Han S. Overhauser dynamic nuclear polarization to study local water dynamics. J. Am. Chem. Soc. 2009;131:4641–4647. doi: 10.1021/ja809259q. [DOI] [PubMed] [Google Scholar]
- 50.Dick-Pérez M, Zhang Y, Hayes J, Salazar A, Zabotina OA, Hong M. Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry. 2011;50:989–1000. doi: 10.1021/bi101795q. [DOI] [PubMed] [Google Scholar]
- 51.Hong M, DeGrado WF. Structural basis for proton conduction and inhibition by the influenza M2 protein. Protein Sci. 2012;21:1620–1633. doi: 10.1002/pro.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pinto LH, Dieckmann GR, Gandhi CS, Papworth CG, Braman J, Shaughnessy MA, Lear JD, Lamb RA, DeGrado WF. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc. Natl. Acad. Sci. U.S.A. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hu F, Schmidt-Rohr K, Hong M. NMR detection of pH-dependent histidine-water proton exchange reveals the conduction mechanism of a transmembrane proton channel. J. Am. Chem. Soc. 2012;134:3703–3713. doi: 10.1021/ja2081185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Williams JK, Tietze D, Wang J, Wu Y, DeGrado WF, Hong M. Drug-induced conformational and dynamical changes of the S31N mutant of the influenza M2 proton channel investigated by solid-state NMR. J. Am. Chem. Soc. 2013;135:9885–9897. doi: 10.1021/ja4041412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cady SD, Mishanina TV, Hong M. Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding. J. Mol. Biol. 2009;385:1127–1141. doi: 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Acharya A, Carnevale V, Fiorin G, Levine BG, Polishchuk A, Balannick V, Samish I, Lamb RA, Pinto LH, DeGrado WF, Klein ML. Structural mechanism of proton transport through the influenza A M2 protein. Proc. Natl. Acad. Sci. U. S. A. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, Fu R, Cross TA. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys. J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cuello LG, Cortes DM, Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306:491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 60.Su Y, Hu F, Hong M. Paramagnetic Cu(II) for Probing Membrane Protein Structure and Function: Inhibition Mechanism of the Influenza M2 Proton Channel. J. Am. Chem. Soc. 2012;134:8693–8702. doi: 10.1021/ja3026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Williams JK, Zhang Y, Schmidt-Rohr K, Hong M. pH-dependent conformation, dynamics, and aromatic interaction of the gating tryptophan residue of the influenza M2 proton channel from solid-state NMR. Biophys. J. 2013;104:1698–1708. doi: 10.1016/j.bpj.2013.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luo W, Cady SD, Hong M. Immobilization of the influenza A M2 transmembrane peptide in virus envelope-mimetic lipid membranes: a solid-state NMR investigation. Biochemistry. 2009;48:6361–6368. doi: 10.1021/bi900716s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cady S, Wang T, Hong M. Membrane-Dependent Effects of a Cytoplasmic Helix on the Structure and Drug Binding of the Influenza Virus M2 Protein. J. Am. Chem. Soc. 2011;133:11572–11579. doi: 10.1021/ja202051n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo W, Mani R, Hong M. Sidechain conformation and gating of the M2 transmembrane peptide proton channel of influenza A virus from solid-state NMR. J. Phys. Chem. 2007;111:10825–10832. doi: 10.1021/jp073823k. [DOI] [PubMed] [Google Scholar]