Abstract
Purpose of review
The purpose of this study is to summarize recent studies of the lower respiratory microbiome in asthma, the role of innate immunity in asthma and strategies to understand complex microbiome–immune interactions in asthma.
Recent findings
Recent evidence indicates that the composition of lower respiratory microbiota in asthmatic individuals, across a spectrum of disease severity, is altered compared with healthy individuals. Attributes of this altered airway microbiome have been linked to clinical and inflammatory features of asthma. The importance of innate immune cells and mucosal defense systems in asthma is increasingly appreciated and may be dysregulated in the disease.
Summary
Interactions between the respiratory microbiome and innate mucosal immunity in asthma are complex and a challenge to dissect. Multiple avenues of investigation, leveraging a variety of methodologies, will need to be pursued to understand functional relationships to clinical and inflammatory phenotypes seen in asthma.
Keywords: airway, asthma, lung, microbiota
INTRODUCTION
The increased prevalence of asthma in industrialized countries has suggested that environmentally derived cofactors are important in its development and persistence. Modern interpretations of the originally proposed ‘hygiene hypothesis’ [1] centre around the concept that low exposure to microbes in cleanly environments, particularly during early infancy, decreases the stimulation necessary for proper development of the immune system and immune tolerance. Associations have been reported between asthma (and related conditions such as allergic sensitization or childhood wheeze), and particular microbes detected from the environment, gut or respiratory tract [2–5]. To understand possible mechanisms contributory to asthma, microbial–host interactions have been intensely studied in model systems. However, the majority of such investigations continue to be designed around the single species-host response paradigm [6,7,8▪▪,9–11]. Typically, these studies have involved introducing infection by individual organisms and evaluating subsequent inflammatory responses, rather than infection by a collection of organisms simultaneously, which could conceivably yield different findings.
A conceptual frameshift is occurring in pulmonary medicine due to the recognition that rather than being completely sterile, the lower airways are populated by microbial populations (microbiota), whose abundance and types differ greatly in the setting of chronic inflammatory airway disease, including asthma [12,13,14▪,15]. This substantially increases the complexity of understanding polymicrobial– host interactions in the airway microbiome and mechanistic relationships to asthma. The innate immune system is at the frontline of initial host responses to external stimuli, and its potential role in asthma has garnered increasing attention. This article will summarize recent advances in our understanding of the airway microbiome in asthma, aspects of innate immunity relevant to asthma and discuss strategies to understand complex microbiome–immune interactions that likely contribute, along with other factors such as exposures and comorbidities, to the clinical heterogeneity of asthma.
THE AIRWAY MICROBIOME IN ASTHMA
The role of microbial exposures and respiratory infections in the pathogenesis of asthma have long been of interest. In the current era of microbiomeoriented investigations, it is now known that alterations in the airway microbiome are a feature of asthma in some patients [13,14▪,15,16]. This can coexist with allergic sensitization. In murine models, it has been observed that the presence of certain airway microbiota can modulate expression of allergic airway inflammation, which could also be relevant in human asthma [6,17▪▪].
To date, there have been very few clinical studies of the airway microbiome in asthma; the results of these investigations are summarized here. In a study that included 11 adults and 20 children with asthma controlled by inhaled with or without oral corticosteroid therapy, Hilty et al. [12] noted that compared with healthy controls, asthmatic individuals possessed more potentially pathogenic bacteria belonging to the Proteobacteria phylum in lower airway samples obtained by bronchoscopy. Healthy controls by contrast demonstrated greater prevalence of members of the Bacteroidetes phylum. Marri et al. [14▪] also observed in their analysis of cell-free sputum that Proteobacteria members were more prevalent in patients with mild asthma than in healthy controls. This small but important study is notable for the asthmatic individuals examined who were not on maintenance corticosteroid therapy. It thus provides evidence that altered airway microbiota composition can be a feature associated with asthma itself and not solely a reflection of concurrent inhaled steroid treatment. Whether there exist differences in the lower airway microbiome associated with atopy alone in the absence of asthma is unknown.
Studies involving larger numbers of well characterized asthmatic individuals have observed important relationships between attributes of the airway microbiome and disease features. The largest investigation to date examined the bronchial microbiome in 65 adults with mild but suboptimally controlled asthma, in whom the prevalence of particular microbiota members, again many Proteobacteria members, correlated with greater degrees of bronchial hyper-responsiveness. Airway bacterial diversity was also associated with improved bronchial hyper-responsiveness after treatment with clarithromycin for 16 weeks [13]. More recently, in patients with severe asthma, relationships have been reported between the prevalence of particular airway microbiota and measures of asthma control, BMI [15], as well as sputum neutrophils and interleukin (IL)-8 concentration [18]. The impact of exposures such as cigarette smoke or medications on the lower airway microbiome in asthmatic individuals is not known. However, studies of smokers without lung disease and nonsmokers suggest that lung microbiota, as sampled by bronchoalveolar lavage, do not differ by smoking status, whereas oropharyngeal-associated microbiota do [19–21]. Collectively, investigations to date across a spectrum of asthma cohorts with differences in disease severity, control status, comorbidities and medication usage suggest that a number of interactions between the airway microbiome, host immunity and potential exposures are likely to be mechanistically relevant in asthma. These relationships likely influence the type and degree of airway inflammation seen and could explain some of the phenotypic heterogeneity seen in asthma.
INNATE IMMUNITY AND ASTHMA
The innate immune system comprises a range of host defense systems that generate nonspecific responses to environmental triggers. It encompasses cellular and noncellular components (Table 1). The airway epithelium and mucosal layer also provide innate immune functions beyond serving as mechanical barriers. Innate immune cells include dendritic cells, innate lymphoid cells (ILCs) of which there are several types [22▪▪], and leukocytes such as macrophages, neutrophils and eosinophils. Patternrecognition receptors such as Toll-like receptors (TLRs), retinoic acid inducible gene I (RIG-I)-like receptors and nucleotide-binding oligomerization domain (NOD)-like receptors recognize different ligand motifs and are important in mediating early immune responses that subsequently shape adaptive immunity [20]. Noncellular components of innate immunity are also important in host defense and include a variety of secreted factors such as lactoferrin, interferons, defensins, secretory leukocyte protease inhibitor (SLPI) and LL-37 (cathelicidin), all of which have antimicrobial effects.
Table 1.
Components of innate immunity likely to mediate airway microbial–immune interactions in asthma
| Anatomic | Cellular | Soluble |
|---|---|---|
| Epithelium | Granulocytes | Antimicrobial peptides |
| Airway mucus | Macrophages | Lactoferrin |
| Eosinophils | Interferons | |
| Neutrophils | Defensins | |
| Basophils | LL-37 (cathelicidin) | |
| Secretory leukocyte protease inhibitor | ||
| Other cell types | ||
| Dendritic cells | ||
| Innate lymphoid cells | ||
| Group 1 (natural killer cells) | ||
| Group 2 (natural helper cells) | ||
| Cellular receptors | ||
| Toll-like receptors | ||
| NOD-like receptors | ||
| RIG-I like receptors |
Multiple aspects of the innate immune system have been studied in asthma. A comprehensive review of this is beyond the scope of this review, which will highlight certain elements that have drawn more recent attention [23▪▪, 24]. The respiratory tract is continually exposed to airborne matter, which translates into frequent opportunity for interaction with mucosa-associated immunity. In addition to air, aspirated material from the upper respiratory or gastrointestinal tracts also serve as potential sources of immune stimuli, including microbes and bile salts. The mucus-lined epithelium, dendritic cells with their interdigitating processes that contact the airway lumen, and secreted antimicrobial peptides, all are involved in the initial contact and responses to external triggers.
In addition to being an anatomic barrier, the airway epithelium is an important coordinator of immune function [25]. Secretory epithelial cells produce antimicrobial peptides, including lysozyme, defensins and IgA [26]. Epithelial cells also shape type 2 mucosal immune responses, through their production of thymic stromal lymphopoeitin (TSLP), IL-25 and IL-33. These epithelial cell derived cytokines stimulate other cell types to promote downstream Th2 inflammation. The potential clinical relevance of epithelial-driven Th2 responses was demonstrated in a recent proof-of-concept study of TSLP inhibition in asthma [27▪]. Treatment with an anti-TSLP antibody attenuated allergen-induced bronchoconstriction and decreased markers of Th2-related airway inflammation such as sputum eosinophils and fraction of exhaled nitric oxide. Other cytokines produced by epithelial cells include IL-10 and transforming growth factor (TGF)-beta, which regulate the activity of other immune cells. The airwayepitheliumtherefore is adynamic orchestrator of immune defense, straddling innate and adaptive immunity.
Airway mucus is a complex entity with important properties beyond its contribution to normal mucociliary clearance [26]. Mucus gel is composed of mostly water and nonaqueous components such as mucins and nonmucin proteins. Mucins are very large glycoproteins, the predominant types found in human airways being MUC5AC and MUC5B. Recent evidence in mice suggests an interesting and critical role for MUC5B, but not for MUC5AC, in airway defense [28▪▪]. In this study, it was found that the absence of MUC5B leads to lung inflammation, impaired immune homeostasis and chronic infection with multiple bacterial species [28▪▪]. In addition, antimicrobial proteins are present in airway mucus, wherein microorganisms can become trapped and either are removed by normal mucociliary clearance or propagate. The latter occurs prominently in cystic fibrosis (CF) wherein chronic infection by Pseudomonas aeruginosa is common. In contrast to the phenotype of P. aeruginosa that can cause acute infections such as pneumonia, chronic P. aeruginosa infection in CF is characterized by a noninvasive phenotype that is highly resistant to eradication. Recent studies suggest that in CF, the density of mucus along with high levels of neutrophil elastase may interact to promote the growth of P. aeruginosa aggregates that are resistant to antimicrobial therapy [29]. Another mucus–microbial interaction of potential interest involves bacteriophages, viruses that infect specific bacterial species. Bacteriophages appear to be capable of binding to exposed mucin glycans that result in reduced bacterial adherence to mucus-producing airway epithelial cells [30▪▪]. The investigators speculate that phage adherence to airway mucus may present a form of nonhost-derived protection against bacterial infection.
Dendritic cells are also an important bridge between the innate and adaptive immune systems, including the development of allergic inflammation and responses to viruses [31,32]. As antigen-presenting and processing cells, they present fragments of microbes that can initiate a broad range of regulatory and adaptive T-cell responses, including Th1, Th2, Th17 pathways. The role of lung dendritic cells in asthma has been intensely studied and reviewed recently [31]. The programmatic breadth of dendritic cells, in addition to the epithelium, enables them to be critical in activating a range of immune cell types, including T cells and ILCs.
ILCs comprise a family of immune cells present at barrier surfaces and have the capacity to respond quickly to environmental signals, including commensal bacteria [22▪▪, 23▪▪]. Although they do not express classic lineage markers, ILCs can manifest a diverse repertoire of responses. In particular, Group 1 ILCs, also known as conventional natural killer cells, play a role in antiviral responses, while Group 2 ILCs, or natural helper cells, produce IL-4, IL-5 and IL-13. As with dendritic cells, microbiota can engage ILCs leading to multiple pathways of immune activation that are not necessarily mutually exclusive.
Finally, the influx of leukocytes into bronchial tissue is a histologic hallmark of airway inflammation seen in asthma. Phagocytic macrophages have an important role in host defense against pathogens, but granulocytic eosinophils and neutrophils are often increased in sputum samples from asthmatic individuals. Importantly, the relative prevalence of sputum eosinophils varies by patient, by asthma phenotype and even over time [33]. Although eosinophilic inflammation is often observed, many other asthmatic individuals do not consistently demonstrate, if at all, airway eosinophils in sputum [33]. Thus, ‘noneosinophilic asthma’ characterizes a significant and important proportion of patients who tend to be less responsive to current therapies, the majority of which target aspects of Th2 inflammation. A variety of asthma phenotypes have been described [34], including several not associated with eosinophilic or Th2-driven inflammation. Underlying mechanisms for these ‘non-Th2’ phenotypes in general remain poorly understood.
DISSECTING AIRWAY MICROBIOME– IMMUNE INTERACTIONS IN ASTHMA
There is an increasing interest in understanding ‘non-Th2’ immune pathways and their etiologic role in asthma or specific asthma phenotypes. Irrespective of whether the apparent dominant immune phenotype in a given patient is ‘Th2’ or ‘non-Th2’, it remains unclear what the key stimuli are that drive aberrant lung immune responses that lead to asthma, or perpetuate airway inflammation in chronic asthma. The recent recognition that airway dysbiosis (i.e. an imbalance in the microbial community of a given niche) exists in asthma invites speculation that airway microbiota could play a role. Microbiota could shape lung-specific immune responses that result in either homeostatic or detrimental inflammatory effects. This relationship could also be bi-directional, whereby the composition of airway microbiota is shaped by the dominant immune axis present in an individual. Although there is currently limited insight into relationships between the airway microbiome and airway inflammation in asthma, recent reports linking particular communities of airway microbiota to different clinical characteristics among asthmatic individuals [13,15,18] suggest that different inflammatory pathways may mediate these observations.
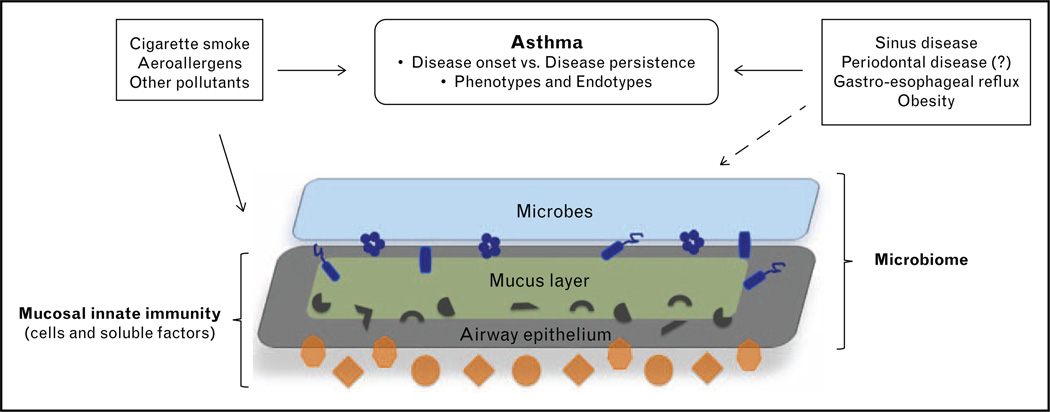
Elucidating mechanistic relationships between the airway microbiome and lung immunity in asthma is a challenge. At the crux of this challenge is the inherent complexity and heterogeneity in each aspect of this system: asthma itself, the nature of the microbiome, environmental exposures and host immunity, genetics and comorbidities that interact with asthma (Fig. 1). In particular, the effects of factors such as smoking, prescribed therapies, obesity and gastro-esophageal reflux disease need to be considered concurrently in this system. The lack of firm consensus or validated definitions of asthma phenotypes, much less putative endotypes, speaks to the clinical heterogeneity of the disease [34,35] and the likelihood of different underlying pathophysiologic mechanisms. For the microbiome, beyond the challenges of microbiota characterization and determining relevant clinical relationships, there is the even greater hurdle of understanding functionality of the lung microbiome, such as collective gene expression profiles, their metabolic products and mechanisms by which microbiota cause or shape manifestations of lung disease. In this framework, one must recognize that the natural state of microbial community systems is responsiveness to their surrounding milieu, leading to a multitude of interspecies and environmental interactions. Thus, it is possible that the collective effects of a polymicrobial community, rather than that of a single species, are most functionally relevant to the pathogenesis of disease or particular disease phenotypes.
Figure 1.
Schematic of airway microbiome and innate immune interactions at the mucosal barrier. Each component shown in this system: asthma itself, exposures, comorbidities, microbiota (bacterial, viruses, fungi) and innate immunity (e.g. dendritic cells, innate lymphoid cells, granulocytes, antimicrobial peptides) are complex entities. Understanding microbiome–host interactions in asthma will require multiple and integrated cross-disciplinary approaches.
This ecological perspective of the respiratory microbiome is essential to informing our future understanding of its role in asthma and other airway diseases [36▪▪]. Appreciation for this concept was demonstrated in a recent study that assessed lung microbiota composition in relation to the development of allergic airway inflammation in neonatal mice [17▪▪]. Despite the presence of high numbers of a regulatory T-cell population detected soon after birth, this did not afford protection against airway hyper-responsiveness after exposure to house dust mite allergen. Rather, changes in bacterial load and lung microbiota composition following birth (from a predominance of Gamma-Proteobacteria and Firmicutes to Bacteroidetes) were associated with decreased aeroallergen responsiveness. This tolerance was linked to a different T-reg subset, whose emergence required microbial colonization, and developed only during a very early window soon after birth. This study provides first-line evidence that the composition of airway microbiota can shape immune responses and modulate development of allergic airway inflammation, adding to existing evidence that gut microbiota are important in shaping immune development in early life [37]. Additional evidence that airway microbiota can shape lung-specific immune responses has been demonstrated in the context of viral infection [38].
Current evidence indicates that interactions between respiratory microbiota and host immunity are bi-directional and both seem likely relevant in asthma [39]. Studies are also needed to understand the role of nonbacterial microbiota in this context, along with studies that incorporate the evaluation of other likely contributory factors such as fumes and cigarette smoke exposure, therapies and comorbidities associated with asthma. To understand mechanisms, future investigative efforts in this field will need to integrate a variety of different approaches. This necessarily spans human and animal studies, so that clinically driven research questions lead to the leveraging of ‘omic’ tools, systems biology approaches and other bench and computational methods to understand microbiome–host interactions that shape asthma.
CONCLUSION
The presence of airway dysbiosis in asthma suggests that characteristics of the respiratory microbiome may shape immune function that lead to the development of asthma or of particular asthma phenotypes. As microbiome–immune interactions are likely bi-directional, thoughtfully designed studies driven by clinically relevant questions, and that leverage a variety of investigative methods, will be necessary to dissect the complexity of these relationships to asthma.
KEY POINTS.
The lower respiratory microbiome in asthma patients across a spectrum of disease severity is characterized by an altered microbial (bacterial) composition, compared with healthy persons.
The innate immune system plays a critical role in initial responses to microbial and other stimuli and shapes subsequent inflammatory and adaptive immune responses seen in asthma.
This airway dysbiosis along with altered lung mucosal immunity can lead to augmented specific inflammatory responses that contribute to asthma development or determine particular asthma phenotypes.
Acknowledgements
Financial support and sponsorship
Support for this work was provided by NIH/NHLBI HL105572.
Footnotes
Conflicts of interest
The author declares no conflicts of interest related to the content of this article.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
▪ of special interest
▪▪ of outstanding interest
- 1.Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989;299:1259–1260. doi: 10.1136/bmj.299.6710.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lynch SV, Wood RA, Boushey H, et al. Effects of early-life exposure to allergens and bacteria on recurrent wheeze and atopy in urban children. J Allergy Clin Immunol. 2014;134:593.e12–601.e12. doi: 10.1016/j.jaci.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heederik D, von Mutius E. Does diversity of environmental microbial exposure matter for the occurrence of allergy and asthma? J Allergy Clin Immunol. 2012;130:44–50. doi: 10.1016/j.jaci.2012.01.067. [DOI] [PubMed] [Google Scholar]
- 4.Brar T, Nagaraj S, Mohapatra S. Microbes and asthma in the elderly: the missing cellular and molecular links. Curr Opin Pulm Med. 2012;18:14–22. doi: 10.1097/MCP.0b013e32834dccc0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang YJ, Boushey HA. The microbiome and asthma. Ann Am Thorac Soc. 2014;11(Suppl 1):S48–S51. doi: 10.1513/AnnalsATS.201306-187MG. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Essilfie AT, Simpson JL, Dunkley ML, et al. Combined Haemophilus influenzae respiratory infection and allergic airways disease drives chronic infection and features of neutrophilic asthma. Thorax. 2012;67:588–599. doi: 10.1136/thoraxjnl-2011-200160. [DOI] [PubMed] [Google Scholar]
- 7.Larsen JM, Steen-Jensen DB, Laursen JM, et al. Divergent proinflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS One. 2012;7:e31976. doi: 10.1371/journal.pone.0031976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fujimura KE, Demoor T, Rauch M, et al. House dust exposure mediates gut microbiome Lactobacillus enrichment and airway immune defense against allergens and virus infection. Proc Natl Acad Sci U S A. 2014;111:805–810. doi: 10.1073/pnas.1310750111. A bacterium identified from house dust as associated with decreased allergy was cultured from dust material and inoculated into a mouse model to demonstrate in-vivo protection against allergic airway inflammation.
- 9.Dulek DE, Newcomb DC, Goleniewska K, et al. Allergic airway inflammation decreases lung bacterial burden following acute Klebsiella pneumoniae infection in a neutrophil and CCL8-dependent manner. Infect Immun. 2014;82:3723–3739. doi: 10.1128/IAI.00035-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liang Z, Zhang Q, Thomas CM, et al. Impaired macrophage phagocytosis of bacteria in severe asthma. Respir Res. 2014;15:72. doi: 10.1186/1465-9921-15-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sagar S, Morgan ME, Chen S, et al. Bifidobacterium breve and Lactobacillus rhamnosus treatment is as effective as budesonide at reducing inflammation in a murine model for chronic asthma. Respir Res. 2014;15:46. doi: 10.1186/1465-9921-15-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hilty M, Burke C, Pedro H, et al. Disordered microbial communities in asthmatic airways. PLoS One. 2010;5:e8578. doi: 10.1371/journal.pone.0008578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang YJ, Nelson CE, Brodie EL, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. 2011;127:372–381. doi: 10.1016/j.jaci.2010.10.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marri PR, Stern DA, Wright AL, et al. Asthma-associated differences in microbial composition of induced sputum. J Allergy Clin Immunol. 2013;131:346–352. doi: 10.1016/j.jaci.2012.11.013. Important evidence which indicated that altered airway microbiota composition is present even among asthmatic individuals not taking inhaled corticosteroid therapy.
- 15.Huang YJ, Nariya S, Lynch SV, et al. The airway microbiome in severe asthma. Ann Am Thorac Soc. 2014;11(Suppl):S78. [Google Scholar]
- 16.Huang YJ. Asthma microbiome studies and the potential for new therapeutic strategies. Curr Allergy Asthma Rep. 2013;13:453–461. doi: 10.1007/s11882-013-0355-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gollwitzer ES, Saglani S, Trompette A, et al. Lung microbiota promotes tolerance to allergens in neonates via PD-L1. Nat Med. 2014;20:642–647. doi: 10.1038/nm.3568. This study examined the nature of and change in lung microbiota composition from birth onward in neonatal mice and its significance in allergic airway inflammation.
- 18.Green BJ, Wiriyachaiporn S, Grainge C, et al. Potentially pathogenic airway bacteria and neutrophilic inflammation in treatment resistant severe asthma. PLoS One. 2014;9:e100645. doi: 10.1371/journal.pone.0100645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Segal LN, Alekseyenko AV, Clemente JC, et al. Enrichment of lung microbiome with supraglottic taxa is associated with increased pulmonary inflammation. Microbiome. 2013;1:19. doi: 10.1186/2049-2618-1-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morris A, Beck JM, Schloss PD, et al. Lung HIV Microbiome Project. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013;187:1067–1075. doi: 10.1164/rccm.201210-1913OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jaspers I. Cigarette smoke effects on innate immune mechanisms in the nasal mucosa. Potential effects on the microbiome. Ann Am Thorac Soc. 2014;11(Suppl 1):S38–S42. doi: 10.1513/AnnalsATS.201306-154MG. [DOI] [PubMed] [Google Scholar]
- 22. Philip NH, Artis D. New friendships and old feuds: relationships between innate lymphoid cells and microbial communities. Immunol Cell Biol. 2013;91:225–231. doi: 10.1038/icb.2013.2. A thorough review of ILC types, their development and role in microbial interactions.
- 23. Hansel TT, Johnston SL, Openshaw PJ. Microbes and mucosal immune responses in asthma. Lancet. 2013;381:861–873. doi: 10.1016/S0140-6736(12)62202-8. An excellent review and integrated perspective of the role of respiratory microbes and mucosal immunity in atopy and asthma.
- 24.Lambrecht BN, Hammad H. Asthma: the importance of dysregulated barrier immunity. Eur J Immunol. 2013;43:3125–3137. doi: 10.1002/eji.201343730. [DOI] [PubMed] [Google Scholar]
- 25.Hallstrand TS, Hackett TL, Altemeier WA, et al. Airway epithelial regulation of pulmonary immune homeostasis and inflammation. Clin Immunol. 2014;151:1–15. doi: 10.1016/j.clim.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 26.Fahy JV, Dickey BF. Airway mucus function and dysfunction. N Engl J Med. 2010;363:2233–2247. doi: 10.1056/NEJMra0910061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gauvreau GM, O’Byrne PM, Boulet LP, et al. Effects of an anti-TSLP antibody on allergen-induced asthmatic responses. N Engl J Med. 2014;370:2102–2110. doi: 10.1056/NEJMoa1402895. A proof-of-concept study of the potentially clinically relevant role of the epithelial-derived cytokine TSLP in allergen-related airway responses.
- 28. Roy MG, Livraghi-Butrico A, Fletcher AA, et al. Muc5b is required for airway defence. Nature. 2014;505:412–416. doi: 10.1038/nature12807. Initial evidence that the airway mucin, specifically muc5B, in mice is important for host defense against bacterial infection.
- 29.Staudinger BJ, Muller JF, Halldórsson S, et al. Conditions associated with the cystic fibrosis defect promote chronic Pseudomonas aeruginosa infection. Am J Respir Crit Care Med. 2014;189:812–824. doi: 10.1164/rccm.201312-2142OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Barr JJ, Auro R, Furlan M, et al. Bacteriophage adhering to mucus provide a nonhost-derived immunity. Proc Natl Acad Sci U S A. 2013;110:10771–10776. doi: 10.1073/pnas.1305923110. A study that incorporated viral metagenomic analysis to identify a large bacteriophage population in CF sputum, demonstrating that such bacterial viruses bind to airway mucin and potentially contribute to airway defense against bacteria.
- 31.van Helden MJ, Lambrecht BN. Dendritic cells in asthma. Curr Opin Immunol. 2013;25:745–754. doi: 10.1016/j.coi.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 32.Kim TH, Lee HK. Differential roles of lung dendritic cell subsets against respiratory virus infection. Immune Netw. 2014;14:128–137. doi: 10.4110/in.2014.14.3.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McGrath KW, Icitovic N, Boushey HA, et al. Asthma Clinical Research Network of the National Heart, Lung, and Blood Institute. A large subgroup of mild-to-moderate asthma is persistently noneosinophilic. Am J Respir Crit Care Med. 2012;185:612–619. doi: 10.1164/rccm.201109-1640OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wenzel SE. Asthma phenotypes: the evolution from clinical to molecular approaches. Nat Med. 2012;18:716–725. doi: 10.1038/nm.2678. [DOI] [PubMed] [Google Scholar]
- 35.Chung KF, Adcock IM. How variability in clinical phenotypes should guide research into disease mechanisms in asthma. Ann Am Thorac Soc. 2013;10(Suppl):S109–S117. doi: 10.1513/AnnalsATS.201304-087AW. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dickson RP, Erb-Downward JR, Huffnagle GB. Towards an ecology of the lung: new conceptual models of pulmonary microbiology and pneumonia pathogenesis. Lancet Respir Med. 2014;2:238–246. doi: 10.1016/S2213-2600(14)70028-1. An important discussion of the new conceptual perspectives necessary to understand the nature and role of the respiratory microbiome in lung health and disease.
- 37.West CE, Jenmalm MC, Prescott SL. The gut microbiota and its role in the development of allergic disease: a wider perspective. Clin Exp Allergy. 2014 doi: 10.1111/cea.12332. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 38.Wang J, Li F, Sun R, et al. Bacterial colonization dampens influenza-mediated acute lung injury via induction of M2 alveolar macrophages. Nat Commun. 2013;4:2106. doi: 10.1038/ncomms3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Holt PG, Strickland DH, Hales BJ, Sly PD. Defective respiratory tract immune surveillance in asthma: a primary causal factor in disease onset and progression. Chest. 2014;145:370–378. doi: 10.1378/chest.13-1341. [DOI] [PubMed] [Google Scholar]