Abstract
Recurrent chromosomal translocations are found in numerous of tumor types, often leading to the formation and expression of fusion genes with oncogenic potential. Creating chromosomal translocations at the relevant endogenous loci, rather than just ectopically expressing the fusion genes, opens new possibilities for better characterizing molecular mechanisms driving tumor formation. In this chapter, we describe methods to create cancer translocations in human cells. DSBs or paired nicks generated by either wild-type Cas9 or the Cas9 nickase, respectively, are used to induce translocations at the relevant loci. Using different PCR-based methods, we also explain how to quantify translocation frequency and to analyze breakpoint junctions in the cells of interest. In addition, PCR detection of translocations is used as a very sensitive method to detect off-target effects, which has general utility.
Keywords: Chromosomal translocations, DNA double-strand breaks (DSBs), Cas9, paired nicks, nCas9, Cas9D10A, NPM-ALK, EWS-FLI1
1. INTRODUCTION
The discovery that a chromosomal translocation was associated with oncogenesis was a watershed event in tumor biology research (Chandra, Heisterkamp, Hungerford, Morrissette, Nowell, Rowley et al., 2011; Rowley, 1973). Hundreds of recurrent reciprocal translocations have now been found in a variety of human cancers, including hematological malignancies, sarcomas, and epithelial tumors (Mani & Chinnaiyan, 2010; Mitelman, Johansson & Mertens, 2007). These translocations are considered primary causes of many cancers and have been important for the development of targeted therapies. A typical consequence of a chromosomal translocation is the formation of a gene fusion that leads to expression of a novel protein with oncogenic potential; alternatively, translocations can lead to a gene coming under the control of a strong promoter, such that overexpression confers an oncogenic property. The cellular consequences of fusion gene expression have been widely investigated using ectopic expression in cells without the translocation or gene silencing in cells carrying the translocation. However, these studies are not optimal for several reasons, such that ectopic expression does not recapitulate the genetics of the disease and may lead to non-physiological levels of fusion gene expression and that silencing the fusion gene in tumor cells does not take into account the numerous other mutations that the tumor cells have acquired. Thus, having methods to induce translocations at will in a variety of cell types would provide a significant advantage to cancer researchers.
Translocations involve the breakage and aberrant joining of DNA ends. Two concurrent DNA double-strand breaks (DSBs), one on each chromosome, have been shown to induce translocations (Richardson & Jasin, 2000). Joining of the DNA ends typically involves some type of nonhomologous end-joining (NHEJ) repair (Weinstock, Elliott & Jasin, 2006). The simplest system for inducing concurrent DSBs is the expression of a nuclease which cleaves specific sites. Initial studies performed in model systems used the I-SceI endonuclease, a homing endonuclease from yeast, in which I-SceI sites were introduced at specific loci. However, the development of designer nucleases (ZFNs and TALENs) has allowed DSBs to be introduced into genomes without prior modification (Gaj, Sirk & Barbas, 2014; Urnov, Rebar, Holmes, Zhang & Gregory, 2010). As a result, chromosomal translocations could be readily induced in human and mouse cells at endogenous loci (Brunet, Simsek, Tomishima, DeKelver, Choi, Gregory et al., 2009; Piganeau, Ghezraoui, De Cian, Guittat, Tomishima, Perrouault et al., 2013; Simsek, Brunet, Wong, Katyal, Gao, McKinnon et al., 2011). These translocations included those associated with human tumors. EWS-FLI1 translocations associated with Ewing sarcoma were induced by ZFNs directed to the EWS and FLI1 loci in mesenchymal cells (Piganeau et al., 2013). NPM-ALK translocations associated with anaplastic large cell lymphoma (ALCL) were induced by TALENS directed to the NPM and ALK loci in Jurkat cells (Piganeau et al., 2013). Of note, the ALCL translocation could also be reversed in patient cell lines carrying the translocation, by TALENs directed to the NPM-ALK and ALK-NPM loci. The versatility of TALENs suggests that designer nucleases could be used to induce translocations involving any loci.
The most recently developed designer nucleases are RNA-guided, which as such are easiest to design. Currently, the most commonly used nuclease is Cas9 from S. pyogenes (Cong, Ran, Cox, Lin, Barretto, Habib et al., 2013; Hsu, Lander & Zhang, 2014; Mali, Yang, Esvelt, Aach, Guell, DiCarlo et al., 2013b). The guide RNA (gRNA) has ~20 nucleotides of sequence complementary to a target site, followed by a Protospacer Adaptator Motif (PAM) sequence (NGG) which is critical for binding to Cas9. When both Cas9 and the gRNA are expressed in cells, the target site is cleaved on both strands a few nucleotides away from the PAM, creating a DSB (Jinek, Chylinski, Fonfara, Hauer, Doudna & Charpentier, 2012). Because Cas9 has two active sites, each cleaving a defined strand, Cas9 can be converted to a nickase by mutation of one active site (nickase Cas9 or nCas9). For example, Cas9 D10A only cleaves the DNA strand complementary to the gRNA. When two gRNAs are provided which bind opposite DNA strands in close proximity, however, paired nicks can be introduced, also creating a DSB but with a long overhang (Mali et al., 2013b; Ran, Hsu, Lin, Gootenberg, Konermann, Trevino et al., 2013). Paired nicks are considered to have fewer potential off-target sites, since two distinct gRNAs are required for double-strand cleavage. Following the principles established with TALENs and ZFNs (Piganeau et al., 2013), Cas9 has also been used to induce EWS-FLI1 and other tumor-relevant translocations (Choi & Meyerson, 2014; Ghezraoui, Piganeau, Renouf, Renaud, Aallmyr, Ruis et al.; Torres, Martin, Garcia, Cigudosa, Ramirez & Rodriguez-Perales, 2014). More recently Cas9-induced paired nicks have also been used to induce translocations (Ghezraoui et al.). In this chapter, we detail methods for the induction of translocations by Cas9 and nCas9, using a PCR screen for translocation junctions (Brunet et al., 2009).
2. MATERIALS
2.1 Cas9, nCas9, and gRNA expression plasmid preparation
Expression plasmids can be obtained from Addgene (https://www.addgene.org/CRISPR/). We use pCas9_GFP (Addgene plasmid 44719) to induce DSBs and pCas9D10A_GFP to induce paired nicks (nCAS9) (Addgene plasmid 44720), together with gRNA expression plasmids derived from MLM3636 (Addgene plasmid 43860).
Competent bacteria, e.g., DH5α
LB agar plates with antibiotic (ampicillin for the plasmids above)
LB medium
PureLink® HiPure Plasmid Filter Maxiprep Kit (Invitrogen)
NanoDrop 2000c (Thermo Scientific)
2.2 Cell culture and transfection
RPE1 (hTert-RPE1) cells or Mesenchymal Stem Cells (MSC) are used in this chapter, although the approach is applicable to any other cell type that can be transfected.
Dulbecco’s Modified Eagle Medium: Nutrient Mixture F-12 (DMEM/F-12, Life Technologies) with 10% Fetal Bovine Serum (FBS) for RPE1 cells; alpha-Minimum Essential Eagle Medium (αMEM, Life Technologies), supplemented with 10% FBS and 2 ng/mL bFGF (Recombinant Human FGF basic (146 aa) 233-FB-025 R&D systems) for MSC
T150 flasks, 150 cm2
Cas9, nCas9, and gRNA expression plasmids, at a concentrations ≥2 μg/μL
6-well plates
48-well plates
96-well plates
0.05% Trypsin-EDTA (1X)
Dulbecco’s Phosphate-Buffered Saline (DPBS, Life Technologies)
Nucleofector II device (Lonza)
Cell Line Nucleofector Kit V with cuvettes (Lonza)
2.3 T7 endonuclease I assay
QIAamp DNA Mini Kit (Qiagen)
Primers. Primers may be designed using a variety of programs such as Primer3Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/). Settings are chosen to yield 22 bp primers with melting temperatures ~62°C.
Phusion High-Fidelity DNA Polymerase (Thermo Scientific)
T7 endonuclease I (New England Biolabs)
NEBuffer 2.1 (New England Biolabs)
2X T7 loading buffer: 50% sucrose, bromophenol blue, 260 μg/mL proteinase K
2.4% agarose gel with Ethidium Bromide (EtBr)
0.5X TBE running buffer (Life Technologies)
UV station
2.3 PCR detection of translocations
-
1
Primers for nested PCR (2 sets of primers). Settings are chosen to yield 20 bp primers with melting temperatures ~60°C.
-
1
FastStart Taq DNA polymerase (Roche)
-
2
1% agarose gel with EtBr
-
3
0.5X TBE running buffer
-
4
UV station
2.4 PCR quantification of translocations
10X Lysis buffer: 100 mM Tris-HCL pH 8, 4.5% NP40, 4.5% Tween20
Proteinase K (New England Biolabs)
2X Master Mix 1: 1X GC-RICH solution (Roche FastStart Taq), 2X PCR Buffer with 20 mM MgCl2 (Roche FastStart Taq), 400 μM dNTP mix (Roche FastStart Taq), 4% DMSO, 0.01% Tween20, 0.01% NP40. Store at 4°C for up to 2 weeks.
2X Master Mix 2: 1X GC-RICH solution (Roche FastStart Taq), 2X PCR Buffer with 20 mM MgCl2 (Roche FastStart Taq), 400 μM dNTP mix (Roche FastStart Taq), 4% DMSO, 0.01% Tween20, 0.01% NP40, 0.25 μl of SYBR® Green I (10,000x in DMSO, Sigma Aldrich) diluted in 2 mL 20% DMSO and Rox (concentrations depend on the real time PCR machine; 30 nM of Rox is used with MX3005P from Agilent). Store at 4°C for up to 2 weeks.
Primers for nested PCR
FastStart Taq DNA polymerase (Roche)
Real-time quantitative PCR machine (MX3005P, Agilent)
3. METHODS TO INDUCE AND DETECT CANCER TRANSLOCATIONS IN HUMAN CELLS
Both Cas9 and nCas9 can be used to generate translocations. We have found that Cas9 is somewhat more efficient (Ghezraoui et al.), but nCas9 has fewer off-target concerns. Two translocations are used to illustrate the methods described in this chapter: NPM-ALK found in ALCL (Elmberger, Lozano, Weisenburger, Sanger & Chan, 1995; Kuefer, Look, Pulford, Behm, Pattengale, Mason et al., 1997; Morris, Kirstein, Valentine, Dittmer, Shapiro, Saltman et al., 1994) (Fig. 1) and EWS-FLI1 found in Ewing sarcoma (May, Gishizky, Lessnick, Lunsford, Lewis, Delattre et al., 1993) (Fig. 2).
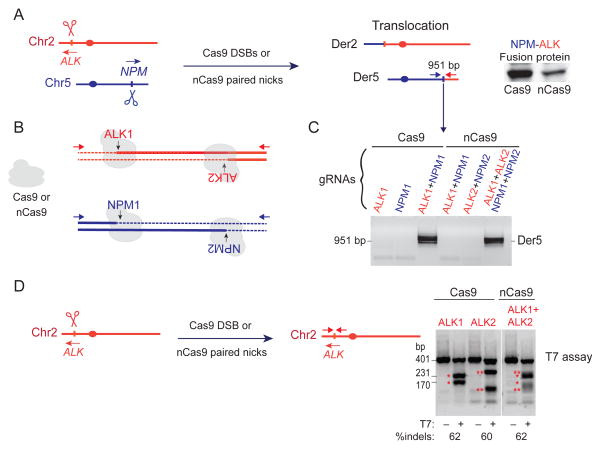
Figure 1. Induction of the NPM-ALK translocation by Cas9 DSBs or nCas9 paired nicks.
A. Induction of DSBs or paired nicks at the NPM and ALK loci can lead to chromosomal translocations and expression of the NPM-ALK fusion protein from Der5.
B. Position of the gRNAs. With wild-type Cas9, DSBs are formed using gRNAs NPM1 and ALK1. With nCas9, paired nicks are formed using NPM1+NPM2 (37 bp apart) and ALK1+ALK2 (41 bp apart), resulting in 5′ overhangs if the strands are separated.
C. Translocations are only detected when two DSBs or two paired nicks are introduced. PCR is performed with primers indicated in A.
D. Estimate of cleavage efficiency at the ALK locus using wild-type Cas9 and either the ALK1 or ALK2 gRNA and nCas9 with both the ALK1 and ALK2 gRNAs. The T7 assay detects 60% indel formation in all three instances.
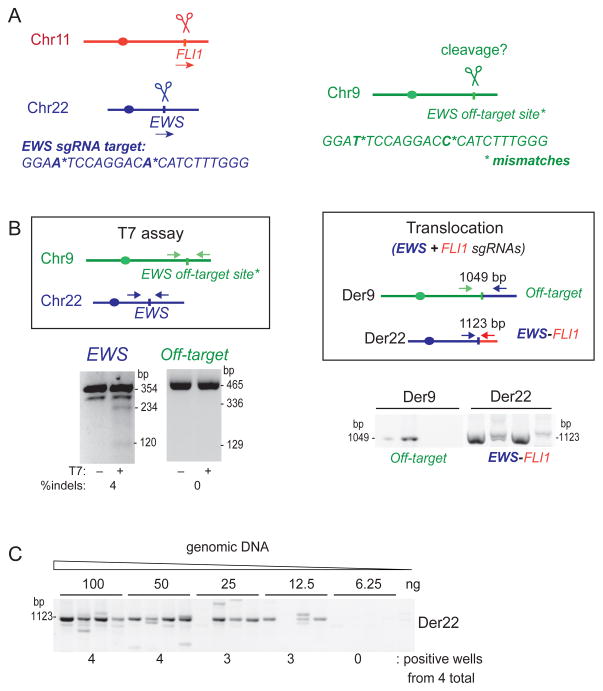
Figure 2. gRNA off-target analysis and estimation of translocation frequency by serial dilution.
A. The translocation of interest is EWS-FLI1. The EWS gRNA has a potential off-target site that contains two mismatches on another chromosome.
B. Using the T7 assay, the EWS gRNA with Cas9 gives detectable indel formation at the EWS locus, while no cleavage is observed at the off-target site. Using the translocation assay, the EWS and FLI1 gRNAs with Cas9 give a detectable signal in all 4 samples (150 ng DNA each); however, a signal is also observed for translocation between the off-target site and the EWS locus.
C. Estimate of translocation frequency using serial dilution of genomic DNA from RPE1 cells using Cas9 and gRNAs EWS and FLI1. In this example, the frequency of Der22 formation is ≥0.375 × 10−3, determined for 3 positive wells from ((4 total wells × 12.5 ng DNA)/6.25 ng) × 103 cells).
3.1. gRNA design and expression plasmid construction
-
For the translocation of interest, a good starting point for designing gRNA target sequences is close to reported breakpoint junctions in patients, if available. If the translocation generates a fusion gene, target sites within introns involved in the translocation are requisite. When using wild-type Cas9, design two gRNAs, one to each chromosome; when using nCas9 to induce paired nicks, design two pairs of gRNAs to each chromosome. The ~20 bp target sequence is located upstream of the requisite NGG PAM sequence. For a 20 bp target sequence, follow the simple rule N20NGG for Cas9 and CCNN20-spacer-N20NGG for nCas9. The spacer can be a few bp or longer (Mali, Aach, Stranges, Esvelt, Moosburner, Kosuri et al., 2013a; Ran et al., 2013). Specific websites can be used to minimize the number of off-target sites of the gRNAs (e.g., http://crispr.mit.edu/, http://zifit.partners.org/ZiFiT/Disclaimer.aspx, https://chopchop.rc.fas.harvard.edu/; see also (Montague, Cruz, Gagnon, Church & Valen, 2014; Xie, Shen, Zhang, Huang & Zhang, 2014)). The U6 promoter, which prefers G as the transcription start site, drives gRNA expression in MLM3636; thus, either the target sequence should begin with G or a G should be inserted before the target sequence.
NPM-ALK Cas9-generated DSBs with gRNAs NPM1 and ALK1; nCas9-generated paired nicks with gRNAs NPM1+NPM2 and ALK1+ALK2 (Fig. 1B). NPM genomic sequence followed by gRNA target sequences: - 5′-CCTCGAACTGCTACTGGGTTCACCTCAGCCTCTGGAATAGCTAGAACTACAGG-3′
- gRNA NPM1: 5′-GTGAACCCAGTAGCAGTTCG-3′
- gRNA NPM2: 5′-GCCTCTGGAATAGCTAGAACTAC-3′
ALK genomic sequence followed by gRNA sequences: - 5′-CCTCAGGTAACCCTAATCTGATCACGGTCGGTCCATTGCATAGAGGAGG-3′
- gRNA ALK1: 5′-GATCAGATTAGGGTTACCTG-3′
- gRNA ALK2: 5′-GTCGGTCCATTGCATAGAGG-3′
-
For each gRNA, order the synthesis of two DNA oligonucleotides (sense and antisense) that would anneal to BsmBI–linearized plasmid MLM3636. (http://zifit.partners.org/ZiFiT/CSquare9GetOligos.aspx).
Resuspend at 100 μM in annealing buffer (10 mM Tris pH 7.5, 1 mM EDTA, 50 mM NaCl).
gRNA NPM1 oligonucleotides: - 5′-ACACCGTGAACCCAGTAGCAGTTCGG-3′
- 5′-AAAACCGAACTGCTACTGGGTTCACG-3′
gRNA ALK1 oligonucleotides: - 5′-ACACCGATCAGATTAGGGTTACCTGG-3′
- 5′-AAAACCAGGTAACCCTAATCTGATCG-3′
Anneal the two oligonucleotides to generate a duplex. Add 10 μL each of the sense and antisense oligonucleotides to 80 μL of annealing buffer. Place tube in a standard heating block at 95°C for 5 min. Remove the heating block from the apparatus and allow to cool to room temperature on the workbench. Slow cooling to room temperature should take 45–60 min. Store on ice or at 4°C until ready to use.
Ligate the duplex into BsmBI-linearized gRNA plasmid MLM3636 vector.
Transform into competent bacteria. Spread on a LB agar plate and incubate overnight at 37°C.
Proceed to plasmid isolation with PureLink® HiPure Plasmid Filter Maxiprep Kit (Invitrogen). Because the BsmBI overhangs are distinct (GTGT and TTTT), a single duplex oligonucleotide should ligate in the correct orientation. Sequencing is recommended to confirm.
Measure the DNA concentration (e.g., using a NanoDrop 2000c spectrophotometer). The final DNA plasmid concentration should be >2 μg/μL.
Note: If the translocation rate is very low, gRNAs can be cloned directly in the same plasmid as Cas9 (pX330-U6-Chimeric_BB-CBh-hSpCas9, Addgene plasmid 42230) or nCas9 (pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A), Addgene plasmid 42335). Other gRNA plasmids contain markers such as GFP (Addgene plasmid PX458) for FACS sorting or the puromycin resistance gene (Addgene plasmid PX459) for selection to increase the recovery of transfected cells.
3.2 Cell transfections with gRNA and Cas9 or nCas9 expression plasmids
The protocol has been optimized for RPE1 and MSC cells, but any other cell line of interest can be used. Transfection can induce high cell lethality, which can be explained by (i) inappropriate cell culture conditions, (ii) inappropriate transfection program, (iii) lack of gRNA specificity, and (iv) induction of a lethal translocation. Check these parameters to reduce mortality.
RPE1 and MSC cells are cultured in a T150 flask in 20 mL of DMEM/F-12 medium and supplemented αMEM medium, respectively, at 37°C and 5% CO2. Passage every 2–3 days at a split ratio of 1:5 to 1:10, keeping them at ≤80% confluency.
The day before transfection, pass the cells without antibiotics (to decrease lethality during electroporation) to reach ~70–80% confluency on the day of transfection.
On the day of transfection, prepare Eppendorf tubes containing 3.5 μg of each gRNA plasmid and 3.5 μg of the Cas9 or nCas9 expression plasmid. As controls, transfect one of the two gRNA expression plasmids for Cas9 or two of the four gRNA expression plasmids for nCas9 and the same quantity of the gRNA expression plasmid in which no gRNA was cloned. For nCas9, you can transfect one gRNA each targeting the two chromosomes to exclude nick-induced translocations (Fig. 1C). The total volume of transfected DNA should not exceed 10 μL.
Pre-fill 2 to 3 wells of a 6-well plate with 1 mL medium and pre-warm to 37°C.
Pre-fill three 96-well plates with 50 μL medium per well and pre-warm to 37°C.
Trypsinize the RPE1 or MSC cells and resuspend in pre-warm supplemented medium DMEM/F-12 or αMEM, respectively (typically 10 mL). Do not use 4°C media at any time.
Count the cells and for each transfection put 7.5 × 105 cells in a 15-mL Falcon tube. The program and the number of cells must be adjusted for each cell line (refer to Lonza Nucleofector Protocols). Conditions described here are optimized for MSC and RPE1 cells.
Centrifuge 10 min at 90 g at room temperature.
Carefully remove the medium without aspirating the cells. FBS from the medium may inhibit transfection. A supplemental wash with DPBS may increase the transfection efficiency.
Carefully resuspend cells in 100 μL of Cell Line Nucleofector Kit V (Lonza) solution. To prevent cell lethality, avoid all unnecessary cell agitation.
Transfer the cells into the tube containing DNA and then transfer the cell/DNA mix into the Amaxa DNA cuvettes.
Electroporate using the Nucleofector II system (Lonza) on program B-016 for MSCs and X-001 for RPE1 cells.
Transfer transfected cells to 5 mL of pre-warmed medium.
Dilute the cells as follows to a final volume of 6 mL: 1.2 mL (1/5), 600 μL (1/10), and 300 μL (1/20). Plate 50 μL of each cell dilution into each well of a pre-warmed 96-well plate. In addition, transfer 1 mL of the 5mL of cells to a well of a 48-well plate for cell counts (see 3.6.1).
-
Transfer the rest of the cells into wells of a 6-well plate (3.2.4) for further DNA, protein, or chromosome analysis. If the translocation frequency is high enough, translocations can be directly detected by western blotting using antibodies directed against the fusion protein (frequency ≥ 10−3) or by microscopy using FISH probes (frequency ≥ 10−2) (Piganeau et al., 2013). Prepare supplementary wells during the transfection if you plan these additional analyses.
Induction of the NPM-ALK translocation is frequent enough that expression of the NPM-ALK fusion protein can be detected in the bulk population of transfected cells (Fig. 1A). Incubate cells at 37°C, 5% CO2.
Note: To estimate transfection efficiency, our experiments utilize pCas9_GFP and pCasD10A9_GFP in which GFP is expressed as a 2A fusion. The percent GFP positive cells is determined by flow cytometry 48 h after transfection. Poor transfection can result in low translocation efficiency. Test several programs to optimize the transfection efficiency for each cell line.
3.3 T7 endonuclease I assay to estimate cleavage efficiency
This assay estimates the efficiency of gRNAs to direct cleavage by quantifying insertions and deletions (indels) resulting from DSB repair via NHEJ (Guschin, Waite, Katibah, Miller, Holmes & Rebar, 2010). For nCas9 paired nicks, the estimate is done using each of the gRNAs separately with wild-type Cas9 as well as with the two gRNAs together with nCas9 (Fig. 1D).
Several gRNAs can be designed to target the same locus, such that gRNAs that are found to efficiently induce indels can be used to generate translocations.
5 days after transfection, trypsinize cells in the 6-well plate (from 3.2.15).
Centrifuge at 200 g at 4°C for 5 min.
Remove supernatant and wash the pellet with 1 mL of DPBS.
Repeat centrifugation.
Remove supernatant. Cell pellets can be stored at −80°C.
Extract genomic DNA with the QIAamp DNA Mini Kit (Qiagen). Genomic DNA can be stored at −20°C.
-
Design a set of primers amplifying a 300–500 bp region encompassing the target sites on non-translocated chromosomes. Cleavage sites should not be located in the middle of the amplicon, in order to obtain two distinct bands after T7 endonuclease I cleavage.
ALK locus primers generate a 401-bp fragment from the wild-type locus and smaller fragments from the modified locus (Fig. 1D): - ALK-F 5′-AGATGGGCAGAGGCTTGAAAAG-3′
- ALK-R 5′-TGAGGATGTTCTGGAAGGCAAA-3′
Perform a 35 cycle PCR on 50 ng of genomic DNA in a total of 25 μL to amplify regions encompassing the target sites. We typically use Phusion High-Fidelity DNA Polymerase (Thermo Scientific).
Verify the quality of the amplification by running 5 μL of the PCR reaction on a 1% agarose gel with EtBr in 0.5X TBE. Only one band should be amplified.
Mix 10 μL PCR reaction with 10 μL 2X NEB Buffer 2.1 in two different PCR tubes.
Melt and re-anneal amplicons by incubating as follows: 95°C for 5 min, 95°C to 25°C at −0.5°C/sec, and 15 min at 4°C. This step converts fragments with and without indels into mismatched heteroduplex DNA. Add 1.5 U of T7 endonuclease I in one of the two tubes. Add the same volume of buffer in the second one as a control.
Incubate at 37°C for 20 min. This step allows T7 endonuclease I to cleave mismatched DNA duplexes.
Add 10 μL 2X T7 loading buffer containing Proteinase K to 10 μL DNA. Incubate at room temperature for 5 min. In this step T7 endonuclease I is degraded by Proteinase K.
Load the PCR product on a 2.4% agarose gel with EtBr and run at 100 V for 20 min in 0.5X TBE buffer.
Capture the gel image with a UV imaging station (Fig. 1D). The amount of cleaved DNA estimates the Cas9-induced mutation rate. Each gRNA should induce detectable levels of indels when tested with Cas9. If the T7 endonuclease I assay reveals a low indel efficiency for a chosen gRNA, increase the amount of the gRNA expression vector or redesign the gRNA. If using the nCas9 approach, the four gRNAs must induce indels separately.
3.4 PCR-based translocation detection
-
With a low translocation frequency, a nested PCR approach is typically used. Design two sets of primers on each side of the translocation junction, the second set located within the first product (see 2.3). For a frequency >10−4, however, translocations can be detected with a single set of PCR primers. The amplified product should be 600–1000 bp to recover potentially long deletions from the DNA ends during translocation formation.
Der5, corresponding to the NPM-ALK fusion: External primers: - Der5-F 5′-CAGTTGCTTGGTTCCCAGTT-3′
- Der5-R 5′-AGGAATTGGCCTGCCTTAGT-3′
Internal primers: - Der5-NF 5′-GGGGAGAGGAAATCTTGCTG-3′
- Der5-NR 5′-GCAGCTTCAGTGCAATCACA-3′
Perform a 23 cycle PCR on 100–150 ng genomic DNA (extracted in 3.3.6) with the external set of primers. We classically use FastStart Taq DNA polymerase (Roche).
Perform a 40 cycle PCR on 0.5–1 μL of the first PCR reaction with the internal set of primers. Nested PCR is a highly sensitive method, presenting contamination risks. Work carefully in a dedicated place, wear gloves, take care not to contaminate a tube with already amplified PCR products, and clean up your bench and your material after each PCR.
Load the PCR product on a 1% agarose gel with EtBr in 0.5X TBE buffer.
Capture the gel image with a UV imaging station (Fig. 1C).
Note: Translocations require DSBs or paired nicks on both chromosomes (Fig. 1C). PCR amplification of translocation junctions may lead to larger or smaller products than expected due to indels generated during translocations formation (see below Fig. 2C), which can be confirmed by sequencing.
3.5. Quantification of potential off-target cleavage
When using Cas9, potential off-target sites should be determined during gRNA design, because mismatches to the ~20 nt target sequence does not necessary abrogate cleavage. Sequences containing up to 5 mismatches compared to the target sequence, especially in the PAM-distal region (Hsu, Scott, Weinstein, Ran, Konermann, Agarwala et al., 2013), should be considered as potential sites for cleavage by Cas9. Potential off-target effects from nCas9 and single gRNAs are significantly reduced since nicks are generated. However, off-target paired nicks are still possible with nCas9 if two gRNAs bind by chance relatively close to each other. Thus, all combinations of gRNAs should be considered for potential off-target binding.
| EWS-FLI1 |
EWS genomic sequence followed by gRNA sequence:
|
Potential off-target site for the gRNA EWS on Chr9 (2 mismatches) (Fig. 2A):
|
FLI1 genomic sequence followed by gRNA sequence:
|
-
After determining potential gRNA off-target sites, design a set of primers amplifying a 300–500 bp region encompassing the potential off target site. Then perform the T7 endonuclease I assay (section 3.3).
EWS locus primers generate a 354-bp fragment from the wild-type locus and smaller fragments from the modified locus (Fig. 2B): - EWS-F 5′-CCTCAGCCACCCAGAGTGTT-3′
- EWS-R 5′-TAGCTGCCTCCCCACTTTACAT-3′
EWS off-target locus primers generate a 474-bp fragment from the wild-type locus and smaller fragments from the modified locus - EWS-OFF-F 5′-ACTCACCTGGTTGGGTTGTCTT-3′
- EWS-OFF-R 5′-GTCCGTACTATGAAGGGGTCGT-3′
-
Design primers for detection of translocations between one of the target chromosomes and the potential off-target site. Then proceed to nested PCR to detect translocations (section 3.4)
Der22, corresponding to the EWS-FLI1 fusion (Fig. 2B): External primers: - Der22-F 5′-ATCCTACAGCCAAGCTCCAA-3′
- Der22-R 5′-GGCCTCATTGTTTCTGGCTA-3′
Internal primers: - Der22-NF 5′-CTACGGGCAGCAGAGTGAGT-3′
- Der22-NR 5′-TTCCTCAAGGCTCTGGAAAA-3′
Der9, corresponding to the Off-target-FLI1 fusion (Fig. 2B): External primers: - Der9-F 5′-TAGTGGGGGAAGGAGAGACA-3′
- Der9-B 5′-GCCAGGTTTCTTAGGGCTTT-3′
Internal primers: - Der9-NF 5′-GAAAAGGCTCCATTTCATGC-3′
- Der9-NB 5′-GGGCTGAGCTCCATAAATCA-3′
A positive signal in one of these two assays (presence of indels after T7 assay or translocation detection by PCR) will reveal off-target cleavage of the tested gRNA. However, the T7 assay is less sensitive than the translocation assay. The T7 assay sensitivity is ~1–2%, while translocations can be easily detected at a frequency ≥10−5. For example, in Fig. 2B a translocation between the target site and the off-target site can be amplified when no signal can be detect in the T7 assay.
Thus, PCR detection of translocation formation is a highly sensitive assay to detect off-target cleavage and can be used as a general method in other applications to evaluate off-target sites.
3.6 Quantification of translocation frequency using a 96-well plate screen
Translocation frequencies can be accurately determined using a 96-well plate format (Brunet et al., 2009; Piganeau et al., 2013). The basic strategy is that small pools of cells are placed in each well, such that most wells on a plate contain <1 translocation. The translocation frequency can be determined from the number of negative and positive wells, with correction for the number of wells with two or more translocations. Having a single event in most positive wells also means that unique translocation junctions can be scored.
48-well plate (3.2.14): Trypsinize cells in the well ≤24 h after transfection, before cells have had a chance to divide. Use a small volume of trypsin (typically 100 μL) and count the cells contained in this well. This number represents 1/5th the transfected cells surviving transfection. Do not exceed this time frame to ensure an accurate cell number to calculate translocation frequency.
96-well plates (3.2.14): Remove all of the culture medium 48 h to 5 days after transfection, depending on the growth of the cells. Plates can be stored wrapped in Parafilm at −80°C and thawed for later use.
Add Proteinase K to 2.5 mL 1X lysis solution at final concentration of 100 μg/mL. Place 25 μL Proteinase K solution into each well of the thawed 96-well plate.
Incubate at 55°C for 120 min in a humid chamber, such as a closed plastic container with moist towels at the bottom.
Transfer the lysates into 96 tubes on PCR tube strips.
Incubate at 95°C for 10 min to inactivate Proteinase K.
Add 200 nM of the external primers (same primers used in 3.4) to 5 mL of 1X PCR Master Mix 1, and add 25 μL of FastStart Taq polymerase. Perform the first round of PCR on 4–7 μL cell lysate in a total volume of 50 μL.
Add 200 nM of the internal primers (same primers used in 3.4) to 2 mL of 1X PCR Master Mix 2 (containing the SYBR Green), and add 10 μL of FastStart Taq polymerase. Perform the second round of PCR on 0.5 to 1 μL of the first PCR in a total volume of 20 μL. The PCR program has to contain a denaturation curve cycle.
Count the number of wells with a positive denaturation curve. Since nested PCR fragments corresponding to translocation junctions are designed to amplify typically 600–900 bp, all wells with a Tm >85 °C are considered to be positive, corresponding to fragments >100–150 bp.
≤12–14 positive wells per 96-well plate: Each well is estimated to contain no more than 1 translocation and the translocation frequency is (p), the number positive wells divided by the number of plated cells (determined in 3.6.1).
>12–14 positive wells per 96-well plate: The translocation frequency is determined using a correction following a beta cumulative distribution function k(x,a,b), where k = number of translocations per well, p = total number of positive wells per number of plated cells, n = number of cells per well, and x = 1 − p, a = n − k, b = k + 1. If the number of wells with two or more translocations is high to interfere with precise frequency measurements, calculate from a plate with fewer cells per well (3.2.14).
PCR products from positive wells can be sent for sequencing to analyze breakpoint junctions (e.g., Der5, Fig. 3 for Cas9 and Fig. 4 for nCas9). At the local sequence level, very different junctions are obtained for Cas9 and nCas9 due to the different DNA end structures. The small pool PCR means that with a unique translocation in a well, the reciprocal translocation junction can also be determined, and cells with a reciprocal translocation can be enriched by sib selection (Brunet et al., 2009).
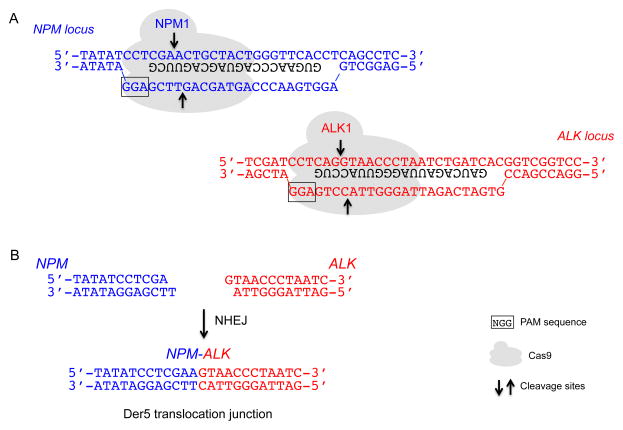
Figure 3. Cas9 cleavage at the NPM and ALK loci and example of a translocation junction sequence.
A. Cas9 cleavage at the NPM and ALK loci using the NPM1 and ALK1 gRNAs, respectively, leads to DSBs. Only the part of the gRNA that binds DNA is shown (black). PAM sequence, boxed; arrows, cleavage sites.
B. DNA ends from Cas9 cleavage that are relevant to Der5 formation are shown. NHEJ leads to a variety of Der5 translocation junctions, but a common junction (shown) is direct ligation of the DNA ends, presumably involving fill-in of the 1-base 5′ overhang.
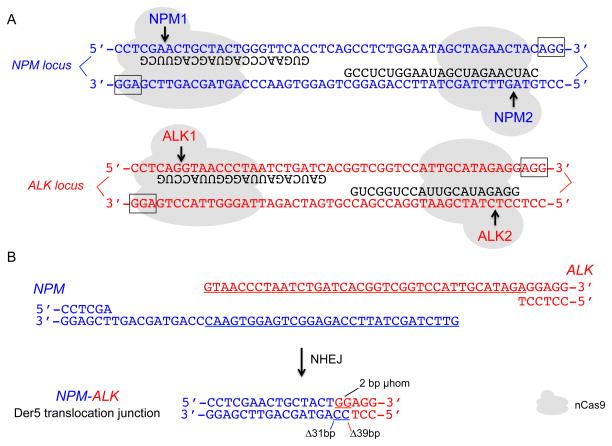
Figure 4. nCas9 cleavage at the NPM and ALK loci and example of a translocation junction sequence.
A. nCas9 cleavage at the NPM and ALK loci using the NPM1+NPM2 and ALK1+ALK2 gRNAs, respectively, leads to paired nicks. The relative position of the two gRNAs at each locus has been termed “PAMs out”. Only the part of the gRNA that binds DNA is shown (black). PAM sequence, boxed; arrows, cleavage sites.
B. The two DNA ends from nCas9 cleavage that are relevant to Der5 formation are shown. Long overhangs are predicted. NHEJ leads to a variety of Der5 translocation junctions, an example of which is shown. In this typical junction, deletions occur primarily in the overhangs at both DNA ends, although in other junctions deletions extend well into the double-stranded region on one or both sides.
3.7 Translocation frequency determination by serial dilution
The protocol described in 3.6 requires removal of medium from the 96-well plate. Thus, with non-adherent cells, cell loss is a potential problem. Here we suggest an alternative method for translocation frequency assessment when frequencies are >10−4, which is not atypical in our experience.
Perform serial dilutions on genomic DNA in quadruplicates starting from 100 ng to 1.56 ng DNA (extraction in 3.3.6).
Perform PCR on each dilution as in 3.4.3.
Load on a 1% agarose gel with EtBr.
Capture the gel image with a UV imaging station. Considering that one human diploid cell contains 6 pg of DNA, 6.25 ng represent about 103 cells. Consider all the quadruplicates to determine the translocation frequency (Fig. 2D for Der22).
4. CONCLUSIONS
In this chapter, we described a method to induce cancer translocations using Cas9 DSBs or nCas9 paired nicks in human cells. The ease at which potential target sites can be found at any locus of interest makes this approach readily adaptable for expanding the repertoire of possible cancer translocations that can be induced in any cell type of interest. Concerns about off-target effects are diminished by the use of nCas9. In addition, nCas9 will better recapitulate the types of breakpoints junctions found in patient cells (Ghezraoui et al.). Cells carrying the chosen translocations provide a more accurate model to understand mechanisms of tumor initiation than standard ectopic expression of a fusion protein. Results from such studies will allow the design of targeted therapies, for example, to avoid therapy-induced secondary tumors with specific translocations (Cowell, Sondka, Smith, Lee, Manville, Sidorczuk-Lesthuruge et al., 2012). Our approach can also be used to decipher the repair mechanisms that are involved in the translocation formation, because PCR screening of translocation junctions allows accurate frequency determination in parallel with the precise determination of breakpoint junctions (Ghezraoui et al.). Finally, translocation assays provide a highly sensitive method to detect off-target cleavage for any application, and so has general utility.
Acknowledgments
We thank Carine Giovannangeli and Jean-Paul Concordet (MNHN) and Matt Krawczyk (MSKCC) for helpful discussions. This work was supported by an ANR grant (BR), Le Canceropole IDF (MP), La Ligue Nationale contre le Cancer (HG), and NIH grant GM054668 (MJ).
References
- Brunet E, Simsek D, Tomishima M, DeKelver R, Choi VM, Gregory P, Urnov F, Weinstock DM, Jasin M. Chromosomal translocations induced at specified loci in human stem cells. Proc Natl Acad Sci USA. 2009;106:10620–10625. doi: 10.1073/pnas.0902076106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra HS, Heisterkamp NC, Hungerford A, Morrissette JJ, Nowell PC, Rowley JD, Testa JR. Philadelphia Chromosome Symposium: commemoration of the 50th anniversary of the discovery of the Ph chromosome. Cancer Genet. 2011;204:171–179. doi: 10.1016/j.cancergen.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi PS, Meyerson M. Targeted genomic rearrangements using CRISPR/Cas technology. Nat Commun. 2014;5:3728. doi: 10.1038/ncomms4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell IG, Sondka Z, Smith K, Lee KC, Manville CM, Sidorczuk-Lesthuruge M, Rance HA, Padget K, Jackson GH, Adachi N, et al. Model for MLL translocations in therapy-related leukemia involving topoisomerase IIbeta-mediated DNA strand breaks and gene proximity. Proc Natl Acad Sci USA. 2012;109:8989–8994. doi: 10.1073/pnas.1204406109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmberger PG, Lozano MD, Weisenburger DD, Sanger W, Chan WC. Transcripts of the npm-alk fusion gene in anaplastic large cell lymphoma, Hodgkin’s disease, and reactive lymphoid lesions. Blood. 1995;86:3517–3521. [PubMed] [Google Scholar]
- Gaj T, Sirk SJ, Barbas CF., 3rd Expanding the scope of site-specific recombinases for genetic and metabolic engineering. Biotechnol Bioeng. 2014;111:1–15. doi: 10.1002/bit.25096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghezraoui H, Piganeau M, Renouf B, Renaud J-B, Aallmyr A, Ruis B, Oh S, Tomkinson EA, Giovannangeli C, Jasin M, Brunet E. Chromosomal translocations in human cells are generated by canonical nonhomologous end-joining. under revision. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guschin DY, Waite AJ, Katibah GE, Miller JC, Holmes MC, Rebar EJ. A rapid and general assay for monitoring endogenous gene modification. Methods Mol Biol. 2010;649:247–256. doi: 10.1007/978-1-60761-753-2_15. [DOI] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuefer MU, Look AT, Pulford K, Behm FG, Pattengale PK, Mason DY, Morris SW. Retrovirus-mediated gene transfer of NPM-ALK causes lymphoid malignancy in mice. Blood. 1997;90:2901–2910. [PubMed] [Google Scholar]
- Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013a;31:833–838. doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013b;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mani RS, Chinnaiyan AM. Triggers for genomic rearrangements: insights into genomic, cellular and environmental influences. Nat Rev Genet. 2010;11:819–829. doi: 10.1038/nrg2883. [DOI] [PubMed] [Google Scholar]
- May WA, Gishizky ML, Lessnick SL, Lunsford LB, Lewis BC, Delattre O, Zucman J, Thomas G, Denny CT. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc Natl Acad Sci U S A. 1993;90:5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science. 1994;263:1281–1284. doi: 10.1126/science.8122112. [DOI] [PubMed] [Google Scholar]
- Piganeau M, Ghezraoui H, De Cian A, Guittat L, Tomishima M, Perrouault L, Rene O, Katibah G, Zhang L, Holmes M, et al. Cancer translocations in human cells induced by zinc finger and TALE nucleases. Genome Res. 2013;23:1182–1193. doi: 10.1101/gr.147314.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, et al. Double Nicking by RNA-Guided CRISPR Cas9 for Enhanced Genome Editing Specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson C, Jasin M. Frequent chromosomal translocations induced by DNA double-strand breaks. Nature. 2000;405:697–700. doi: 10.1038/35015097. [DOI] [PubMed] [Google Scholar]
- Rowley JD. Letter: A new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature. 1973;243:290–293. doi: 10.1038/243290a0. [DOI] [PubMed] [Google Scholar]
- Simsek D, Brunet E, Wong SY, Katyal S, Gao Y, McKinnon PJ, Lou J, Zhang L, Li J, Rebar EJ, et al. DNA Ligase III Promotes Alternative Nonhomologous End-Joining during Chromosomal Translocation Formation. PLoS Genet. 2011;7:e1002080. doi: 10.1371/journal.pgen.1002080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres R, Martin MC, Garcia A, Cigudosa JC, Ramirez JC, Rodriguez-Perales S. Engineering human tumour-associated chromosomal translocations with the RNA-guided CRISPR-Cas9 system. Nat Commun. 2014;5:3964. doi: 10.1038/ncomms4964. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Elliott B, Jasin M. A model of oncogenic rearrangements: differences between chromosomal translocation mechanisms and simple double-strand break repair. Blood. 2006;107:777–780. doi: 10.1182/blood-2005-06-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: A Software Package for Designing CRISPR sgRNA and Evaluating Potential Off-Target Cleavage Sites. PLoS One. 2014;9:e100448. doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]