Abstract
New insights in the study of virus and host biology in the context of viral infection are made possible by the development of model systems that faithfully recapitulate the in vivo viral life cycle. Standard tissue culture models lack critical emergent properties driven by cellular organization and in vivo–like function, whereas animal models suffer from limited susceptibility to relevant human viruses and make it difficult to perform detailed molecular manipulation and analysis. Tissue engineering techniques may enable virologists to create infection models that combine the facile manipulation and readouts of tissue culture with the virus-relevant complexity of animal models. Here, we review the state of the art in tissue engineering and describe how tissue engineering techniques may alleviate some common shortcomings of existing models of viral infection, with a particular emphasis on hepatotropic viruses. We then discuss possible future applications of tissue engineering to virology, including current challenges and potential solutions.
Keywords: cell-cell interactions, complexity, host-pathogen interactions, hepatotropic viruses, patterning, primary cells
MODELING VIRAL INFECTION
“It is not difficult to find the reason why the virologists have failed where the bacteriologists were so successful. They have been severely handicapped by the difficulties connected with the cultivation of viruses…. By giving the virologists a practicable method for the isolation and study of viruses you relieved them of a handicap, burdening them from the birth of their science and placed them for the first time on an even footing with other microbe hunters.”
—Sven Gard, in the 1954 Nobel Prize presentation speech
The Advent of Cell Culture and Modern Virology
Viruses are obligate intracellular pathogens; thus, it is not surprising that the field of virology continues to advance dramatically on the heels of cell culture technology. Accurate recapitulation of viral infection in a laboratory setting is critical for the dissection of virus-host interactions and ultimately for identification of potential therapeutic targets and subsequent drug development. Definition of the conditions under which cells can be maintained in culture was pioneered in the late nineteenth and early twentieth centuries (1,2) and enabled initial in vitro observation and attenuation of viruses in animal tissue cultures (3, 4). In 1949, Enders et al. (5) demonstrated the ability to grow poliomyelitis virus in nonneuronal human embryonic tissues, for which they were awarded the 1954 Nobel Prize in Physiology or Medicine. The subsequent isolation and serial propagation of the first human cell line—HeLa cells—which were shown to be susceptible to poliomyelitis virus and other viruses, provided a robust and convenient platform for large-scale virus amplification (6).
Where Do 3D Tissue-Engineered Models Fit on the Spectrum?
Since the 1950s, 2D cell culture models have produced a wealth of knowledge regarding the mechanisms of infection, and they still constitute the primary platform for viral propagation and pathogenesis studies. Despite these advances, standard monolayer or suspension cultures, though they are accessible, well characterized, and amenable to large-scale experimentation, do not adequately recapitulate the complexity of living organisms. In vivo, cells are integrated into higher-order structures, where they receive biochemical and mechanical cues via interaction with extracellular matrix (ECM), soluble signals, and other cells. The contributions of these components to cell state are pleiotropic and may often impact both the cells’ susceptibility and their response to viral infection.
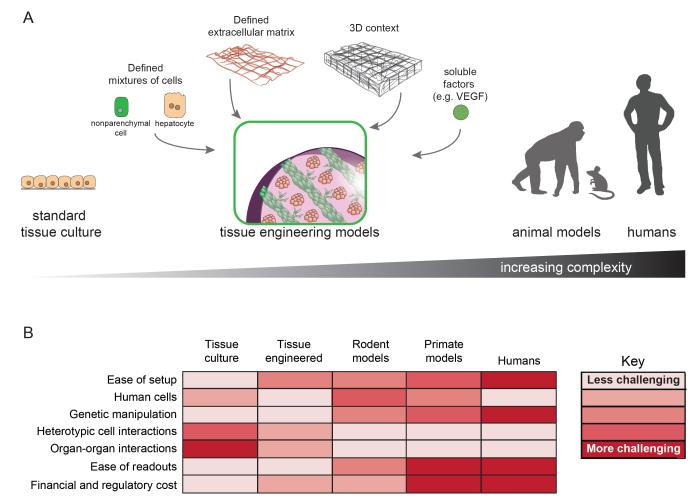
As researchers aim to culture newly discovered viruses and pinpoint cellular factors that impact the course of disease, additional systems that bridge the gap between traditional monolayer cultures and animal models are required. To this end, tissue engineering (TE) may have a significant role in moving virology into the next era of discovery. Owing to greater control over various orthogonal parameters, cells in these 2D or 3D systems can more accurately capture emergent tissue properties (e.g., multicellular architecture) and exhibit altered viability, morphology, differentiation status, proliferation capacity, and gene expression profiles, often yielding phenotypes that differ significantly from their counterparts in conventional culture. Maintenance of tissue-specific host factors, innate immune function, cell cycle status, and polarity that more closely resemble analogous cells in vivo may better support the natural course of viral infection than do cells in standard tissue culture, generating viral titers and persistence characteristics more closely resembling the in vivo scenario (see Figure 1).
Figure 1.
Tissue engineering (TE) approaches to recapitulate the complexity of the in vivo environment. (a) Schematic overview of tissue engineering approaches. Standard tissue culture consists of monocultures in monolayers. Tissues in vivo are made up of various cell types organized in specific 3D structures. TE approaches aim to model the emergent properties and architecture of the in vivo environment using combinations of relevant cell types, extracellular matrix proteins or mimics, and soluble signals (e.g., cytokines), organized into specific patterns using engineering tools, to promote cellular health in vitro or function upon implantation in vivo (e.g., vascularization). Because TE models replicate aspects of the in vivo environment, they may enhance virus permissiveness and result in more authentic host-virus interactions. TE models assembled with induced pluripotent stem cell (iPSC)-derived cells could also give rise to personalized models with defined and manipulable host genetics. (b) Chart comparing TE approaches with other in vitro and in vivo platforms. TE models bridge the simplicity of tissue culture with the completeness of animal models, while maintaining the capacity for easy manipulation and interrogation of cellular and viral inputs.
Through the definition of ECM niches from a physical, chemical, and biological perspective; the synthesis of biomaterials with controllable properties across many dimensions; and the establishment of microtechnological tools to enable precise cellular organization, TE has expanded the number and type of models that exhibit emergent properties (7, 8). It is possible to independently select the components of the system [cell type(s), matrix, etc.] and to define and scale both the geometry and quantity of individual models (even allowing for high-throughput applications), which offers advantages over laboratory animal models that are not always available, are costly, are more difficult to assay, and are even subject to ethical debate. TE models provide a fertile middle ground for experimentation. They are discussed in further detail below, along with their application in modern virology, specifically with respect to hepatotropic viruses.
TISSUE ENGINEERING: COMPONENTS AND CURRENT STATUS
Whereas isolated primary cells or lines lack access to the native environment that typically modulates their phenotype in vivo, TE offers the ability to control interactions between cell types of interest and other cells (8, 9), matrix components (10), and soluble growth factors/cytokines (11), as well as the cells’ exposure to physical forces and defined 3D contexts. Here, we briefly review each of these components.
Cell Sources for Tissue Engineering
Because of their robustness in culture and the relative ease of maintaining the viability of immortalized cells, cell lines such as HepG2 hepatoma cells have been used to engineer some epithelial tissues (13), and human umbilical vein endothelial cells (HUVECs) are often used in the TE of vascular constructs (14). Stromal cell lines can also be used to support primary or progenitor-derived cells in TE constructs (15). However, because primary cells are fully differentiated and exhibit higher levels of tissue-specific functionality than cell lines, they are a preferred cell source. For example, engineered tissues have been created with primary hepatocytes (16), neurons (17), and cardiomyocytes (18). Additionally, the flexibility in the choice of materials and soluble factors means that TE can be used to enable phenotypic maintenance of some cell types, such as hepatocytes (19), hepatic stellate cells (20), and neurons (17), whereas standard cell culture platforms cannot.
Finally, stem- and progenitor-derived cells are increasingly being used in TE. Though these cell types can be challenging to maintain in a differentiated state, and current differentiation protocols often yield immature cell types more closely related to fetal than to adult cells, they offer the promise of genetically tractable, expandable, and fully functional cells. TE strategies to recapitulate developmental niches (21) have resulted in successful early examples of several stem cell–derived tissues including liver (23), brain (24), intestine (25), heart (26), and vasculature, and numerous efforts to expand and combine these tissue types are underway.
Scaffolding Materials for Tissue Engineering
Biomaterials provide mechanical support and chemical cues to cells in TE scaffolds. Many biomaterials have been studied, and the choice of biomaterial depends upon the specifics of the fabricated tissue. Natural materials often contain endogenous domains that encourage cell attachment and proliferation, and they do not stimulate strong immune responses upon implantation, making them promising for transplant applications. Natural TE materials include fibrin, which promotes angiogenesis and vascular integration (27); collagen, a ubiquitous component of the ECM with numerous integrin-binding motifs (28); hyaluronic acid (29); alginate (30); and the basement membrane–mimicking Matrigel (31). However, natural biomaterials are less well characterized and are less amenable than synthetic materials to engineering and modification.
To alleviate these shortcomings, a wide variety of synthetic polymeric materials have been developed for TE applications. These include US Food and Drug Administration (FDA)-approved materials such as poly(lactic-co-glycolic acid) (PLGA)-based (33) and poly(ethylene glycol) (PEG)-based (34, 35) materials, as well as newer classes of polymers that exhibit better antifouling properties (36), modularity (37), or stimulus responsiveness (38). These materials have mechanical and gel-forming properties that can be tuned over wide ranges and have typically been designed to provide a biologically inert background upon which bioactive motifs such as RGD can be added (39).
As a way to combine the bioactive properties of natural materials with the modularity and control of synthetic materials, tissue engineers have generated many examples of hybrid materials, which contain natural and synthetic components that often provide orthogonal control or functionality. Gelatin methacrylate, synthesized from collagen, is a photo-cross-linkable biomaterial used in 3D bioprinting and stereolithography that maintains its base material’s cytocompatible properties (40), and numerous hyaluronic acid derivatives have been engineered to exhibit photoactivatable or constitutive cross-linking and allow for the display of tethered peptides or proteins (29). In a different strategy, Martino et al. (41) recently developed a PEG-fibrin hybrid in which minimal fibrinogen fragments are grafted onto tunable PEG backbones to impart fibrin’s proangiogenic and growth factor–binding properties.
Soluble and Tethered Signaling Factors in Tissue Engineering
In addition to cells of interest and scaffolding materials, TE models commonly incorporate growth factors to modulate cell function (11). VEGF (vascular endothelial growth factor) and PDGF (platelet-derived growth factor) have been commonly utilized to encourage angiogenesis and maturation of engineered vessels and skeletal muscle (42), BMP-2 (bone morphogenic protein 2) has been used to encourage bone tissue growth (43), and NGF (nerve growth factor) has been used to encourage neuronal growth (44). Unlike in standard 2D monolayer culture, transport and utilization of growth factors (45), passive sequestration by protein binding partners (41), and chemically induced tethering of growth factors (11) can all lead to gradient formation and distinct zones in engineered tissues (46) that better recapitulate heterogeneous organ microenvironments.
Other stimuli can also produce spatially distinct signaling effects in engineered tissues. Oxygen availability in densely packed tissues varies with distance to a perfused channel, and oxygen tension significantly affects numerous cellular pathways (47). Similarly, shear stress induced by fluid flow affects endothelial phenotype (48) and affects many epithelial cells (49) that are shear protected in vivo. These gradients can be modulated by varying vessel density and perfusion rates in constructs under flow, to provide both a physiological microenvironment and a platform for continuous sampling of engineered constructs.
Architecture of Tissue-Engineered Constructs
In vivo, tissue microarchitecture is intimately linked to its function; therefore, control over tissue architecture has been explored in engineered constructs. The ability to pattern cells using engineering techniques has been exploited in 2D engineered cultures. For example, cell shape has been examined, and spatially segregated cocultures of different cell types have been generated by microcontact printing with adhesive proteins (50).
Cellular patterning has also been extended to TE in 3D in order to control cell spreading and movement (17), spatially organize groups of cells (53), and control the production of vascular networks (54). Many techniques have been used to accomplish these goals: Photolithographic (40) or porosity-generating (55) techniques can modify the overall scaffold structure, and orthogonal chemical approaches have been used to pattern proteins and mechanical variations into these constructs (56). DNA (57) and shape-templated (58) approaches can also be used to hierarchically assemble small cell-laden building blocks into larger, multicomponent structures. Recent improvements in 3D printing of biological materials have also enabled the direct deposition of cells and matrix in defined configurations (59).
TISSUE ENGINEERING ADVANTAGES AND APPLICATIONS FOR MODELS OF VIRAL INFECTION
TE models have the potential to bridge the gap between viral infection in standard cell culture, which may lack higher-order tissue functions, and infection of animal models, which are less experimentally and genetically tractable and may make it difficult to isolate responses of interest. Here, we review the benefits of TE specific to the study of viral infections.
Advantages of Tissue-Engineered Constructs for Studying Viral Infection
Whereas standard cell culture models of viral infection are limited to single cell types, most viral infection in vivo involves multiple cell types. TE approaches have the potential to capture this cellular diversity and have been used in the liver, for example, to demonstrate the dependence of hepatocyte state on supporting cell types including endothelium (60, 61), fibroblasts (61a), and Kupffer cells (62). These interactions are important in hepatitis C virus (HCV) infection, as virion binding to liver sinusoidal endothelial cell (LSEC) receptors (63) can result in transcriptional changes that directly (through transinfection) and indirectly (through LSEC cytokine production) affect HCV infection of hepatocytes (64). Kupffer cell activation in HCV also modulates viral progression (65), and the incorporation of these two cell types into a liver-mimicking microenvironment could constitute a more relevant cellular milieu for understanding HCV infection in vitro. Hepatitis B virus (HBV) similarly modulates innate immune pathways in liver nonparenchymal cells (66) (which affect hepatocytes) in a way that engineered coculture systems might capture.
Beyond capturing relevant cell-cell interactions, TE offers avenues to study dynamic processes in host-pathogen interactions. Infection-relevant phenotypes such as endothelial barrier permeabilization (67) and immune cell trafficking (68) can be incorporated into engineered systems. For example, dengue virus (DENV) infection of endothelial cells leads to inflammation, increased permeability, and progression to septic shock upon immune cell recruitment to the infected endothelial cells (69). Modeling this phenomenon could help to predict and treat vascular complications.
The dynamic measurements enabled by TE extend to multicomponent tissues as well, as multiple tissue compartments can be fabricated in a spatially controlled manner at the microscale or mesoscale by microstructuring materials for cell encapsulation or assembly of microtissues containing distinct cell populations (19, 57). Increased control over materials chemistry and processing also allows the engineering of interfaces to smoothly connect different engineered tissue types (70), which may be important for studying viruses that affect multiple tissue compartments. For example, the multistage infection cycle of herpes simplex virus (HSV) is difficult to model in standard cell culture (71). By creating spatially defined mucosal and neural compartments in a manner amenable to time-lapse imaging and analysis, viral latency and reactivation could potentially be studied more directly (72). Even within tissues, spatial variation in cellular phenotype can affect viral infection. The cell type compartmentalization and positioning afforded by TE techniques may also be of use in understanding influenza infection. For example, engineered respiratory tracts could potentially be used to conclusively determine sialic acid–linked receptor specificity for avian and human viruses (73), and the effect of other important airway components such as the respiratory mucus layer (74) could be studied in an experimentally facile system.
Some of the effects detailed above can be adequately captured in animal models of infection. However, whereas intravital imaging techniques are improving in capability and accessibility, real-time monitoring of cellular and molecular parameters in some deeper organs can be difficult (75), and genetic manipulation in these systems requires time-consuming mouse engineering and breeding. TE couples the ability to generate relevant tissue complexity with the ability to easily manipulate input cells and capture biological and virological readouts. Cells in TE constructs are easily accessed by growth factors and cytokines (11), but they can also be manipulated genetically by lentiviral transduction (76, 77) or RNA interference (78), either prior to encapsulation or in situ in engineered tissues (76, 77). By tethering or trapping the viruses or nucleic acids in scaffold materials, this manipulation can also be spatially controlled (78) to target select subsets of cells. Combined with reporters easily introduced into encapsulated cells (79), numerous standard imaging modalities are available for analyzing TE constructs, including fluorescence tomography or multiphoton fluorescence compatible with thick engineered tissues, magnetic resonance, and electron microscopy (80). Many of these approaches can be used in real time to provide dynamic pictures of viral infection in engineered tissues.
Some Existing Examples of Tissue Engineering for Studying Viral Infection
Although the application of TE to viral models is still in its infancy, there are several existing examples of pathogen infection in engineered tissues. Those pertaining to the liver are covered in detail below. Outside of the liver, TE techniques have also been used to create an in vitro model of HSV-1 infection that captures latent neuronal infection and reactivation (72). In this model, a latently infected combined neuronal and dermal layer resides next to an epidermal layer at an air-water interface. Latent HSV DNA can be reactivated upon UV exposure, providing an in vitro reactivation model to complement animal studies. HSV has also been studied in the context of oncolytic virus therapy in TE tumor models: Viral infection and spread depend upon ECM degradation in brain tumors (81) and melanoma models (81a). Dynamic culture systems (82) can support microorganism growth as well, with a commensal bacterium modulating epithelial barrier integrity in a TE microfluidic device (83). Although this initial effort did not incorporate viruses, it could be adapted to studying enterovirus infection in this relevant intestinal model. Recent efforts to engineer in vitro bone marrow analogs may also aid the study of viral infection, as appropriate TE systems mimicking bone marrow microenvironments (84) have been shown to promote hematopoietic stem or progenitor cell differentiation, even resulting in B lymphocyte differentiation and influenza-specific antibody formation upon inoculation with the virus (84). These efforts highlight the potential of TE to provide more relevant viral model systems in the future.
HEPATOTROPIC VIRUS INFECTION IN MODELS WITH EMERGENT PROPERTIES
Hepatotropic viruses are globally distributed and cause both acute [e.g., hepatitis A virus (HAV) and hepatitis E virus (HEV)] and chronic (e.g., HBV and HCV) infection, leading to liver fibrosis, cirrhosis, hepatocellular carcinoma, and, ultimately, end-stage liver disease requiring liver transplantation (85–88). Although functional HAV, HBV, and, recently, HEV vaccines are available along with direct-acting antivirals targeting HBV or HCV, these pathogens remain prevalent in the human population. Furthermore, they continue to be a challenge to study in the laboratory setting due to a paucity of robust model systems that mimic clinical aspects of disease.
Current Limitations in the Study of Hepatotropic Viruses
Attempts to culture these viruses in vitro and in vivo have been limited by their restricted host range and hepatotropism. Although HEV has been found in various species in addition to humans (reviewed in 89), HAV infects only humans and nonhuman primate species, and only humans, chimpanzees, and tree shrews (Tupaia belangeri) are naturally susceptible to HCV and HBV (reviewed in 90). The requirement for liver-enriched factors such as the microRNA miR-122 for HCV (91) and transcription factors (e.g., HNF4α, PPARα, RXRα), some of which are liver enriched, for HBV (92) further restricts the cell type that will permit viral replication. In the absence of cell culture–adaptive mutations, slow viral replication and protein synthesis kinetics also remain an issue—in the case of wild-type HAV, antigen accumulation and particle production take weeks (93).
Further, although an efficient cell culture system was established for HCV with the isolation of a genotype (gt) 2a virus from a Japanese patient with fulminant hepatitis C (JFH-1) (94), this virus is genetically diverse, including six major genotypes and numerous subtypes that differ in their geographic distribution, response to antiviral therapy, and disease association (reviewed in 95). Currently, the study of genotype-specific phenotypes is restricted to replicons (96, 97), HCV pseudoparticles (98), and intergenotypic JFH-1-based recombinant viruses (99).
For HEV, cell culture systems exist for gt3 (100) and gt4 (101)—the two zoonotic genotypes of the four identified to date that infect humans (87). Still, the efficiency of HEV growth in cell culture is poor. Further, whereas cDNA clones of other HEV strains have been generated and shown to be infectious in either nonhuman primates (102) or pigs (103), in vitro studies are limited due to a lack of virus spread in cultured cells. Similarly, for HBV, hepatoma cells that replicate and assemble the virus (e.g., HepG2) have been identified (104), but these cells do not permit HBV entry without additional manipulation (105). Researchers therefore resort to systems that feature integrated HBV genomes or delivery of HBV DNA to cells via transfection.
Existing models of hepatotropic viral infection rely predominantly upon human hepatoma– or hepatocellular carcinoma–derived cell lines. The aberrant proliferative potential and polarity of these lines, combined with their dysfunctional drug metabolism and innate immune axis, can obscure important biology and make it critical to study viral infection in the authentic host cell (primary human hepatocytes). Additionally, both viral and host genetics have profound impacts on the outcome of infection, such as the influence of polymorphisms in the IL28B locus on spontaneous clearance of HCV and response to treatment (106, 107) or the highly varied success in achieving infection in primary hepatocytes of different human donors. Thus, there is great interest in establishing both in vitro and in vivo platforms for all of these viruses with pan-genotypic permissiveness, particularly those that feature the natural target cell of the virus and reflect the genetic diversity of the infected population (e.g., primary human hepatocytes, pluripotent stem cell– derived hepatocyte-like cells).
Polarization and Differentiation of Immortalized Cells
Manipulation of immortalized cells toward a more polarized or differentiated state has resulted in more permissive systems for hepatotropic viral infection and has yielded unique insights into viral entry mechanisms. Early evidence of productive HCV infection in culture came from the use of a human hepatocellular carcinoma–derived line (FLC4) cultured in 3D radial-flow bioreactors (108). More recently, Aly and coworkers demonstrated that HCV replication was increased in immortalized primary hepatocytes cultured in a 3D thermoreversible gelatin polymer (TGP) system (109) and that viral particle production was achieved upon challenge with HCV gt1b and gt2a (110) when these cells were cultured in a 3D hollow fiber system (111). Like the TGP system, the hollow fiber reactor is smaller scale than the radial-flow bioreactor, allowing easy access to both medium and cells for virological assessment. In a standard cell culture model, HepaRG cells were also shown to be permissive for gt3a serum-derived HCV during the proliferation stage, and once the cells were fully differentiated, they were able to replicate the virus and produce infectious particles, indicating that properties of both immature and mature hepatocytes may be beneficial for culture of HCV in vitro (112).
Additional polarized models, including HepG2 cells ectopically expressing miR-122 and CD81 (a receptor for the virus) and Huh-7/Huh-7.5 cells exposed to dimethyl sulfoxide (DMSO), in Matrigel, or in rotating wall vessels, have been shown to be permissive for HCV (113–115). These systems have demonstrated unique viral phenotypes including infectious particle production from a dicistronic gt1b HCV genome (116) and a shift in viral particle density compared with 2D-produced virus, suggesting assembly or association with host proteins and/or lipids may be altered in 3D (117). The HCV result has also been extended to 3D engineered tissues with HCV-permissive cell lines (117a). Notably, the addition of human serum (1–2%) to the medium in several systems had a beneficial impact—it promoted an increase in extracellular HCV RNA production in human adult hepatocytes (118) and more rapid viral penetration followed by more consistent detection of HCV RNA after inoculation of HepaRG cells with human serum-derived HCV (112). Steenbergen and colleagues (119) also reported growth arrest and increased expression of albumin, lipid metabolism–related genes, and cell-cell contact proteins, as well as HCV receptors, in Huh-7.5 cells exposed to human serum. These cells produced higher-titer, lower-density HCV, suggesting that serum factors impact cellular and viral phenotype.
Cell context has also recently been considered with the goal of increasing viral yields in HEV infection systems. Berto and colleagues (120) demonstrated detectable HEV RNA in the supernatants of PLC/PRF/5 cells cultivated in a rotating wall vessel but not in 2D cultures inoculated in parallel, and Rogée et al. (121) also demonstrated HEV RNA in supernatants of Matrigel-embedded HepaRG and PICM-19 cells (bipotent human and porcine lines, respectively, that differentiate into biliary or hepatocyte-like cells) cultured with murine embryonic fibroblasts. Direct comparison of RNA levels produced in these systems with those produced in primary hepatocytes or cell lines in standard culture is challenging given the differences in the source, titer, and quantity of inoculums used. However, these 3D systems may abrogate the requirement for high-titer inoculation to launch infection and may enable unique insight into infection pathways—particularly entry and egress.
For example, vectorial entry and release of HAV was shown to differ between enterocyte-derived, polarized Caco-2 cells on porous membrane supports (apical) (122) and a subclone of HepG2 cells (basolateral) (123), consistent with the expected route of exposure to HAV during transmission. For HCV, the entry process involves two tight junction proteins [claudin-1 (124) and occludin (125)], highlighting the importance of polarized cell models in defining precise entry mechanisms (126). Expression, organization, and dynamics of these receptors are altered upon polarization (127, 128), and several studies have noted reduced susceptibility to infection following polarization (127), suggesting cell junctions also act as physical barriers. Disruption of tight junctions may promote viral transmission (129), and the use of cellular proteins localized to this region may facilitate cell-cell spread (130).
HBV infection also depends on cell differentiation status. HepG2 cells normally support HBV replication and virus release following transfection with HBV DNA (104). Treating these cells with DMSO renders them permissive for HBV entry (105). Similarly, the differentiation state of HepaRG cells—mediated by DMSO and corticoid treatment— correlates with HBV susceptibility (131). The expression of the bile acid pump NTCP (sodium taurocholate cotransporting polypeptide)—a newly identified receptor for HBV— increases upon differentiation of HepaRG cells and decreases in primary tupaia hepatocytes after plating (132). Interestingly, DMSO treatment of either HepG2 or Huh-7 cells overexpressing NTCP robustly increases infection efficiency in those HBV-permissive cells (133). Collectively, these observations link cellular differentiation, entry factor expression, and viral susceptibility. HBV gene expression and replication are also linked to cellular differentiation status induced by culturing cells on ECM and treating with DMSO or dexamethasone (131). This is likely due to the increased expression of required host transcription factors, such as HNF4α and its regulator HNF1α, which control the HBV transcription machinery (134). TE techniques can modulate cell polarization and differentiation status through their control over cell-cell and cell-matrix interactions and soluble signals, and this modulation can strongly affect viral infectivity in these models.
Primary Cell Culture Models
Although differentiated, polarized cells yield more accurate insight into entry events, the host response to infection still differs in immortalized cells (compared with authentic host cells) due to their abnormal proliferation, dysregulated gene expression, and aberrant signaling. Thus, primary cell models may be useful for certain aspects of host-pathogen biology. The application of TE concepts to both primary human hepatocytes and induced pluripotent stem cell (iPSC)-derived cells has made these model systems more tractable, enabling in vitro interrogation of viral infection across cells with different genetic backgrounds.
Although primary hepatocytes are the principal target cells of hepatotropic viruses, and thus the most physiologically relevant model system, infection of these cells in vitro has been a challenge. Once dissociated from their native hepatic microenvironment, primary hepatocytes lose their polyhedral morphology, exhibit a decline in liver-specific function as determined by albumin secretion and cytochrome P-450 enzymatic activity, and become less susceptible to viral infection (136). Multiple groups have challenged primary hepatocytes [human fetal (137, 138) and adult (139, 140), chimpanzee (141), nonhuman primate (142), and tupaia (143, 144)] with tissue culture–derived virus or sera from infected HCV, HEV, or HBV patients. Results suggest that inoculation with high viral load is necessary but not sufficient and that amplification is typically low level and fluctuating.
Similar to induction of a differentiated state in immortalized cells, maintenance of this state in primary hepatocytes in vitro is critical for initial susceptibility to infection and efficient viral replication. Early reports noted increased yield and duration of HBV infection in the presence of DMSO or PEG (145); more recently, groups have employed nonparenchymal cells to promote hepatocyte function and subsequently viral infection in vitro (146). LDLR (low-density lipoprotein receptor) and EGFR (epidermal growth factor receptor), two host cofactors for HCV entry (147, 148), were more highly expressed when hepatocytes were cultured with LSECs on a single layer of collagen or in spheroid form on Matrigel (149). Further, spherical hepatocyte masses, initially cultured in the presence of preattached stellate cells, efficiently replicated HCV clones of gt1a, gt1b, and gt2a (150). Similarly, long-term cultures of primary human fetal hepatocytes were developed by plating a hepatic cell mixture at low density, allowing outgrowth of nonparenchymal cells and formation of hepatic islands that were subsequently cultured under DMSO-containing conditions to maintain characteristic hepatocytic features; these cultures were susceptible to HBV infection for up to 10 weeks in culture (151). In 3D engineered tissues, Bcl-2-transduced HUVECs have been used to modulate fetal hepatocyte phenotype and permit HCV replication (151a).
Adding microscale architecture to the concept of coculture systems, Ploss et al. (152) generated miniaturized hepatocyte islands of optimized dimensions, surrounded by supportive 3T3-J2 murine embryonic fibroblast cells, termed micropatterned cocultures (MPCCs). Unlike collagen gel sandwiches, Matrigel substratum, or random cocultures, MPCCs were able to sustain infection of a luciferase-expressing HCVcc. Infection frequency in this system was low with both HCVcc and plasma or sera from infected patients, but the authors demonstrated the potential application of this model as a physiological system for preclinical screening of antiviral therapeutics. Beyond HCV (152,153), this model also supports infection with malaria (154) and HBV (unpublished data). Toward recapitulation of complete tissue architecture, human adult ex vivo liver slice methods were recently adapted to virology. Common challenges of this system include poor viability and inherent variability (154a); however, Lagaye et al. (155) demonstrated productive infection of JFH-1 or JFH-1-based chimeric viruses as well as a gt1b primary isolate in cultures that remained viable for 10 days.
In theory, primary human hepatocytes provide the opportunity to correlate host genotype and viral phenotype under controlled experimental conditions. In practice, this has been difficult to achieve with statistical significance given the limited number of donors available and the inability to prescreen or select cells from patients of a particular genotype of interest. Alternatively, pluripotent stem cells have the capacity for infinite self-renewal and can be genetically altered at the pluripotent stage before differentiation into hepatocyte-like cells, enabling the introduction of a desired host phenotype and the parallel production of isogeneic controls. Importantly, human embryonic stem cell (hESC)- and iPSC-derived hepatocyte-like cells supported the complete HCV life cycle, evidenced by entry and replication in HCV-pseudotyped virus and subgenomic replicon assays, respectively (156), and by challenge with HCVcc (157–159) and gt1b- and gt1a-containing patient sera (HCVser) (whereas Huh-7.5 cells were not permissive for HCVser). In addition, short hairpin RNA–mediated knockdown of cyclophilin A or PI4KIIIa rendered these cells resistant to HCV, providing proof of concept for the use of genetic manipulation in these cells to interrogate host-pathogen interactions (157). Still, human hepatocytes derived from pluripotent stem cells or directly from fibroblasts via transdifferentiation are still functionally immature and exhibit a very different transcriptional profile from that of primary adult human hepatocytes (159a). Recent approaches to mature these cells using chemical (159b), genetic (159c), or combination approaches (159d) have yielded more adult-like phenotypes, which will be important for ensuring that the host-pathogen interactions explored in these systems are indicative of mature responses to infection.
Expansion of In Vivo Platforms
The restricted species tropism of hepatotropic viruses has led to the development of transgenic and humanized mouse models (e.g., uPA+/+-SCID, FAH−/−Fag2−/−Il2rg−/−, and AFC8) to launch infection of HBV and HCV in vivo (reviewed in 162, 163). Although chimeric human liver mouse models—in which genetic injury of mouse hepatocytes permits repopulation with and expansion of transplanted human hepatocytes—allow completion of the entire viral life cycle and have been instrumental in establishing rodent models of HBV and HCV, they are restricted by low breeding efficiency, high mortality, and the need for consistent, robust engraftment of human hepatocytes to achieve HCV viremia. This, along with variable chimerism, has led to difficulty in cost-effectively generating large cohorts of mice for viral challenge.
In attempt to alleviate these issues, efforts to generate additional mouse models have focused on supplementation of critical nonhomologous host factors (164), suppression of innate immunity (165), or viral adaptation (166). Alternatively, implantation of TE models featuring human cells into immunodeficient mice can stabilize cell phenotype, minimize engraftment variability, and serve as a platform to assess drug metabolism and toxicity in vivo (15). This work can be extended to 3D engineered tissue models of HCV, malaria, and HBV infection in primary hepatocytes, as a way to create ectopic human livers (15) permissive to infection in a small-animal model. These ectopic tissues may also provide an alternative to liver injury–based mouse models, allowing researchers to circumnavigate host barriers to infection in vivo.
FUTURE APPLICATIONS OF TISSUE ENGINEERING TO THE STUDY OF VIRAL INFECTION
Improved Control over Model System Inputs
To date, identification of TE model systems that promote desired cellular and virological phenotypes has largely been empirical. Rapid optimization of these models will benefit from platforms that allow systematic perturbations of the microenvironment that may result in prolonged cellular viability or expression of host proteins that are critical for viral propagation. Toward this goal, ECM arrays have been employed to determine optimal substrates for maintenance of liver-specific function in rat hepatocytes and for murine embryonic stem cell differentiation (167) and to test the combinatorial effects of ECM and soluble factors on cell fate (168). High-throughput platforms have also more recently been described for 3D constructs (169, 170). Such systems hold potential for drug toxicity screens, cell-ECM interaction studies, and investigation of cellular phenotypes that depend on cellular localization within 3D tissues, similar to the in vivo case. These systems provide methods for the identification of microenvironmental cues that yield desired cellular properties, and can be used in concert with genome-editing tools, including CRISPR-Cas (clustered, regularly interspaced short palindromic repeat–CRISPR-associated) (171) and TALEN (transcription activator–like effector nuclease) (172) approaches, which enable more precise control over the cell-autonomous aspects as well. By combining TE model systems with iPSC technology, researchers can now, in principle, generate personalized models of viral infection and follow up on clinical data or studies on host factors with significant clinical correlations identified through human genetic studies.
Capturing Cellular and Organ-Level Complexity
Tissue engineers exercise a great level of control over the types and positions of cells and scaffold materials. The use of TE to study viral infection can further be expanded by integrating dynamic reporters of cellular function and infection and establishing platforms for selective analysis of cellular subsets within engineered tissues. Fluorescent reporters for infection monitoring can be engineered by modifying either virus or host proteins (153), and these can be used in parallel with reporters of host pathways such as IRF3 (interferon regulatory factor 3) (173) or NFκB (nuclear factor κB) (79) to track infection spreading and responses in real time at unprecedented resolution. Cellular responses to infection can also modify local microenvironments through ECM remodeling and through local cytokine release, which can be tracked in real time by tethering reporters of protease activity (174) or biomolecule accumulation (175) into engineered tissues.
Integrating viral and cell state reporters into TE constructs may also aid in selecting subpopulations of interest for further analysis at the transcriptomic and proteomic level. Laser capture microdissection (176) can be used to selectively isolate infected cells from 2D engineered tissues (177), and a newly developed photo-uncaging approach can be used to isolate RNA from specific cells in 3D tissues (178). Additionally, the ability to selectively degrade certain scaffolding hydrogels with light (179) offers an orthogonal approach for accessing specific cells within engineered tissues.
Extending TE to the development of organ-on-a-chip systems will enable greater understanding of multiorgan interactions during viral infections. Advances in microfluidic on-chip cell culture (82) and assembly of microscale engineered tissue segments (57, 180) could enable multiorgan circuits consisting of cells in connected, functional organ modules that more accurately recapitulate in vivo biochemical, metabolic, and genetic activity. Pertinent to the development of viral models, these multiorgan systems could include engineered lymphoid components that serve as a dynamic reservoir for circulating immune cells (181) and constitute part of the host response to all viruses. The interconnections between organ modules can also be efficiently endothelialized to enable the study of endothelial responses either upon primary infection (63, 182–184) or secondary to systemic cytokine release, and the existence of multiple organ modules also offers the potential to predict off-target effects of new proposed antiviral therapies.
Dynamic Systems
Matrix remodeling, cell migration, chemical gradients, and mechanical forces (such as the laminar shear stress imposed by blood flow or phasic stress produced by breathing) represent dynamic aspects of living systems. Chemical gradients and mechanical forces modulate cell function (185, 186) and can impact the course of infection (187, 188); microfluidic devices represent versatile, miniaturized, and automatable platforms to generate these gradients and forces in vitro. The use of microfluidics in virology is in its infancy, but examples illustrate its potential for viral infection dynamics studies (189), viral quantification or diagnostics (190), imaging (191), and single-particle assays (192) as well as screen-based approaches to identify novel host-pathogen interactions (193).
CURRENT CHALLENGES AND NECESSARY BREAKTHROUGHS
The adoption of TE approaches to model viral infection will depend on making tissue engineering platforms robust and accessible and capturing as much relevant biology as possible. Below we review some major remaining challenges along this path.
Standardization of Tissue-Engineered Platforms
Although standard tissue culture approaches often lack key host functions, they are still widely used because of their experimental simplicity and robustness. Tissue engineers can similarly focus on standardization to make TE more accessible for studying viral infection. On the materials side, numerous approaches can reduce TE scaffold variability. First, large-batch synthesis or modification of scaffold materials can reduce variability in chemical modification percentages between synthesis runs. This problem can also be mitigated by using newly developed, high-efficiency orthogonal chemical reactions (194) for scaffold modification and cross-linking and by using controlled polymerization reactions for synthetic materials (195). Standardized sterilization or aseptic techniques for TE scaffold manipulation can also prevent the contamination that sometimes occurs in these more complex culture formats.
Toward this end, lessons can be learned from existing standardized TE platforms. MPCCs are used at an industrial scale for screening drugs for human liver toxicity (61a,196), and AggreWell plates can reliably produce defined-size cellular aggregates for stem cell culture and TE (197). Recently, a scalable platform for 3D pluripotent stem cell culture was also reported that uses a commercially available thermoresponsive material (38). As evidenced by these examples, focusing on simplicity in structure, material choice, and processing can aid in standardization of TE formats.
Transport and Data Collection in Tissue-Engineered Constructs
Unlike in standard monolayer cell culture, cells in TE constructs are subject to nutrient transport constraints due to the 3D nature of the culture format and impeded transport through materials with small pore sizes. In general, cells in TE constructs must be within 100–200 μm of a perfused channel to receive adequate levels of nutrients; strategies for vascularization in TE are extensively covered in recent reviews (e.g., 198). When using TE models to study viral infection, the vascularization requirement may be even more acute, as virions must be able to access host cells within the construct. Virions may traverse small-pore PEG hydrogels to host cells (117a), but this process is inefficient across long distances. Additionally, for viruses that utilize entry receptors of specific polarity, appropriate cell positioning and polarization must be maintained in TE constructs to facilitate viral entry. Transport is relevant to viral infection even outside of virion access, because the oxygen tension gradient generated by cell-dense TE constructs can affect viral replication through HIF-1α (hypoxia-inducible factor 1α)-dependent mechanisms (199).
Several methods exist to provide adequate transport within TE platforms, either through prefabrication of vascular channels using sacrificial materials (54), through directed cell seeding via microengineering (60) and bioprinting (59), or through stimulation of self-organizing endothelialized vascular networks (200, 201). For in vitro models of viral infection, new parallelizable bioprinting approaches (202) are favorable because they combine reproducible channel formation with the scalability to adequately power studies. Printing with multiple materials and cell types in defined positions (59) should enable patterned vascular network formation next to virally permissive parenchymal tissue components. In conjunction with adequate vascular access to tissue compartments, the continued development of orthogonal fluorescent (79, 153) and secreted (203, 204) reporters of viral and cell state will enable multiplexed, real-time analysis of viral infection in TE constructs.
Sustaining Cell Phenotype in Tissue-Engineered Constructs
Although cell lines are generally amenable to manipulation, the development of more physiologically relevant viral models requires the use of more fragile primary and stem- or progenitor-derived cells. The cell-cell signal density inherent in 3D culture systems can promote the phenotypic maintenance and function of some primary (15, 61) and progenitor (205) cells, but previously developed protocols for cell maintenance or directed differentiation of progenitor cells in 2D (206) may not directly translate to TE systems. These protocols can be performed prior to cell isolation and TE scaffold incorporation, but isolation from the native microenvironment often diminishes tissue-specific functions of differentiated cells. Further, in multicompartment TE models, the optimal culture medium formulation and other signals may vary from compartment to compartment.
Several approaches may be used to circumvent these problems. By optimizing protocols for long-term 3D culture in formats that allow atraumatic cell harvest, 2D culture steps can be eliminated altogether for TE with differentiated stem-derived cells (38). Development of 3D protocols for in situ directed differentiation of progenitors will be useful, aided by the inherent advantages of 3D differentiation for some lineages (207), the vast choice of cell fate–instructive scaffold materials (21, 37, 208), increased control over biomolecule presentation and valency in TE constructs (7), and new approaches for high-throughput arrayed 3D culture formats (209). Additionally, the precise control over mixing in microfluidic systems may allow spatial gradients in nutrient composition to control differentiation status and phenotypic maintenance (45, 46), enabling cross talk between organ modules while preventing off-target effects of culture medium additives.
CONCLUSIONS
The study of virology depends on adequate model systems to accurately capture the various stages of viral infection, from entry to replication to egress. Compared with standard cell culture techniques on one end, and animal models on the other, TE has the potential to recapitulate relevant higher-order interactions while preserving the ability to readily manipulate distinct components of the system. Continued advancements in TE will more readily allow the precise positioning of cells, matrix, and soluble factors, and interactions between these components, in a way that captures emergent properties important in viral infection. For example, TE approaches should enable the study of viral infection and spread in multiorgan models, the elucidation of host-pathogen interactions in authentic host cells, and the development of medium- and high-throughput analyses of viral replication in more physiological contexts.
SUMMARY POINTS.
Cell-cell and cell–extracellular matrix interactions and soluble signals impact cellular phenotype and thus susceptibility and response to viral infection.
Tissue-engineered models capture relevant biological complexity (e.g., cellular diversity, cell-cell interactions, chemical gradients) and are easier to miniaturize, multiplex, manipulate, and assay than in vivo models.
The application of tissue-engineered models in modern virology is in its infancy.
The initial implementation of tissue engineering concepts in the study of hepatotropic viruses has yielded systems that are more permissive for viral infection and has allowed unique insight into certain aspects of the viral life cycle.
Tissue engineering has enabled maintenance of primary cell function in vitro and thus may offer the opportunity to correlate host genotype and viral phenotype.
Generation and transplantation of 3D tissue-engineered models into small animals may expand current in vivo platforms and allow researchers to circumnavigate existing technical limitations and host barriers to infection.
FUTURE ISSUES.
High-throughput screening platforms to identify specific extracellular matrix and soluble factor combinations that promote cellular and virological phenotypes will enable optimization of current and novel tissue-engineered models.
Implementation of genome-editing tools and dynamic reporters of cellular function and infection will permit the development of personalized models of viral infection with real-time system monitoring capabilities.
Integration of multiple engineered tissues (e.g., organ-on-a-chip systems) and dynamic systems that allow cross talk between cellular compartments and recapitulate biological forces (e.g., microfluidic platforms) will allow interrogation of viral infection in models that capture additional aspects of relevant in vivo complexity.
Optimization of tissue engineering protocols to allow incorporation of primary cells and stem- or progenitor-derived cells into 3D models will expand the types of culture systems that feature physiologically relevant cell types with diverse host genetic backgrounds.
Modulation of biomaterials or formation of vascular channels within tissue-engineered constructs will improve nutrient transport and virion access to cells within 3D models.
The standardization of methodology and the use of widely accessible reagents in the construction of tissue-engineered platforms will make these systems more accessible to virologists.
ACKNOWLEDGMENTS
We are grateful to Amir Shlomai, Heather Fleming, and Kelly Stevens for critical reading of the manuscript. V.R. is supported by a Fannie and John Hertz Foundation Fellowship and a National Science Foundation Graduate Research Fellowship. T.P.S and M.A.S. were supported by National Research Service Awards F32 AI084448 and AI091207, respectively, from the National Institute of Allergy and Infectious Diseases. Our research is supported by National Institute for Diabetes and Digestive and Kidney Diseases grant DK085713 (to C.M.R. and S.N.B.), National Institute of Allergy and Infectious Diseases grants AI075099 and AI072613, National Cancer Institute grant CA057973 (to C.M.R.), and Skolkovo Institute of Science and Technology grant 022423-003 (to S.N.B.). Additional funding was provided by the Greenberg Medical Research Institute and the Starr Foundation (to C.M.R.). S.N.B. is a Howard Hughes Medical Investigator. We apologize to those whose work could not be cited due to space restrictions.
KEY TERMS AND DEFINITIONS
- DMSO
dimethyl sulfoxide
- ECM
extracellular matrix
- gt
genotype (of a given virus)
- HAV
hepatitis A virus
- HBV
hepatitis B virus
- HCV
hepatitis C virus
- HEV
hepatitis E virus
- LSEC
liver sinusoidal endothelial cell
- Microfluidics
systems (sometimes containing cells) in which small volumes of fluids are manipulated and guided through architectures created using engineering lithography tools
- MPCC
micropatterned coculture
- Organ-on-a-chip system
multiple engineered tissues integrated into a microfluidic system that enables fluid and soluble factor exchange between compartments
- PEG
poly(ethylene glycol)
- Scaffolding materials
natural materials (e.g., ECM components) or synthetic polymers used to provide mechanical support and biochemical cues to cells in engineered tissues Tissue engineering (TE): combining cells, scaffolding materials, and soluble factors in a defined architecture to create a living tissue mimic
Footnotes
DISCLOSURE STATEMENT The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Carrel A. On the permanent life of tissues outside of the organism. J. Exp. Med. 1912;15:516–28. doi: 10.1084/jem.15.5.516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carrel A. A method for the physiological study of tissues in vitro. J. Exp. Med. 1923;38:407–18. doi: 10.1084/jem.38.4.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steinhardt L, Israeli C, Lambert RA. Studies on the cultivation of the virus of vaccinia. J. Infect. Dis. 1913;13:294–300. [Google Scholar]
- 4.Haagen E, Theiler M. Studies of yellow fever virus in tissue culture. Proc. Soc. Exp. Biol. Med. 1932;29:435–36. [Google Scholar]
- 5.Enders JF, Weller TH, Robbins FC. Cultivation of the Lansing strain of poliomyelitis virus in cultures of various human embryonic tissues. Science. 1949;109:85–87. doi: 10.1126/science.109.2822.85. [DOI] [PubMed] [Google Scholar]
- 6.Gey GO, Coffman WD, Kubicek MT. Tissue culture studies of the proliferative capacity of cervical carcinoma and normal epithelium. Cancer Res. 1952;12:264–65. [Google Scholar]
- 7.Dvir T, Timko BP, Kohane DS, Langer R. Nanotechnological strategies for engineering complex tissues. Nat. Nanotechnol. 2011;6:13–22. doi: 10.1038/nnano.2010.246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Griffith LG, Naughton G. Tissue engineering—current challenges and expanding opportunities. Science. 2002;295:1009–14. doi: 10.1126/science.1069210. [DOI] [PubMed] [Google Scholar]
- 9.Bianco P, Robey PG. Stem cells in tissue engineering. Nature. 2001;414:118–21. doi: 10.1038/35102181. [DOI] [PubMed] [Google Scholar]
- 10.Carletti E, Motta A, Migliaresi C. Scaffolds for tissue engineering and 3D cell culture. Methods Mol. Biol. 2011;695:17–39. doi: 10.1007/978-1-60761-984-0_2. [DOI] [PubMed] [Google Scholar]
- 11.Lee K, Silva EA, Mooney DJ. Growth factor delivery–based tissue engineering: general approaches and a review of recent developments. J. R. Soc. Interface. 2011;8:153–70. doi: 10.1098/rsif.2010.0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deleted in proof
- 13.Griffith LG, Swartz MA. Capturing complex 3D tissue physiology in vitro. Nat. Rev. Mol. Cell Biol. 2006;7:211–24. doi: 10.1038/nrm1858. [DOI] [PubMed] [Google Scholar]
- 14.Khan OF, Sefton MV. Endothelialized biomaterials for tissue engineering applications in vivo. Trends Biotechnol. 2011;29:379–87. doi: 10.1016/j.tibtech.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen AA, Thomas DK, Ong LL, Schwartz RE, Golub TR, Bhatia SN. Humanized mice with ectopic artificial liver tissues. Proc. Natl. Acad. Sci. USA. 2011;108:11842–47. doi: 10.1073/pnas.1101791108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan J, Stevens KR, Trehan K, Underhill GH, Chen AA, Bhatia SN. Hepatic tissue engineering. In: Monga SPS, editor. Molecular Pathology of Liver Diseases. Springer; New York: 2011. pp. 321–42. [Google Scholar]
- 17.Schmidt CE, Leach JB. Neural tissue engineering: strategies for repair and regeneration. Annu. Rev. Biomed. Eng. 2003;5:293–347. doi: 10.1146/annurev.bioeng.5.011303.120731. [DOI] [PubMed] [Google Scholar]
- 18.Laflamme MA, Murry CE. Heart regeneration. Nature. 2011;473:326–35. doi: 10.1038/nature10147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stevens KR, Ungrin MD, Schwartz RE, Ng S, Carvalho B, et al. InVERT molding for scalable control of tissue microarchitecture. Nat. Commun. 2013;4:1847. doi: 10.1038/ncomms2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guvendiren M, Perepelyuk M, Wells RG, Burdick JA. Hydrogels with differential and patterned mechanics to study stiffness-mediated myofibroblastic differentiation of hepatic stellate cells. J. Mech. Behav. Biomed. Mater. doi: 10.1016/j.jmbbm.2013.11.008. In press. doi: 10.1016/j.jmbbm.2013.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dickinson LE, Kusuma S, Gerecht S. Reconstructing the differentiation niche of embryonic stem cells using biomaterials. Macromol. Biosci. 2011;11:36–49. doi: 10.1002/mabi.201000245. [DOI] [PubMed] [Google Scholar]
- 22.Deleted in proof
- 23.Takebe T, Sekine K, Enomura M, Koike H, Kimura M, et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature. 2013;499:481–84. doi: 10.1038/nature12271. [DOI] [PubMed] [Google Scholar]
- 24.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, et al. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–79. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–65. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 26.Stevens KR, Kreutziger KL, Dupras SK, Korte FS, Regnier M, et al. Physiological function and transplantation of scaffold-free and vascularized human cardiac muscle tissue. Proc. Natl. Acad. Sci. USA. 2009;106:16568–73. doi: 10.1073/pnas.0908381106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed TAE, Dare EV, Hincke M. Fibrin: a versatile scaffold for tissue engineering applications. Tissue Eng. B. 2008;14:199–215. doi: 10.1089/ten.teb.2007.0435. [DOI] [PubMed] [Google Scholar]
- 28.Parenteau-Bareil R, Gauvin R, Berthod F. Collagen-based biomaterials for tissue engineering applications. Materials. 2010;3:1863–87. [Google Scholar]
- 29.Burdick JA, Prestwich GD. Hyaluronic acid hydrogels for biomedical applications. Adv. Mater. 2011;23:H41–56. doi: 10.1002/adma.201003963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee KY, Mooney DJ. Alginate: properties and biomedical applications. Prog. Polymer Sci. 2012;37:106–26. doi: 10.1016/j.progpolymsci.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fischbach C, Chen R, Matsumoto T, Schmelzle T, Brugge JS, et al. Engineering tumors with 3D scaffolds. Nat. Methods. 2007;4:855–60. doi: 10.1038/nmeth1085. [DOI] [PubMed] [Google Scholar]
- 32.Deleted in proof
- 33.Pan Z, Ding JD. Poly(lactide-co-glycolide) porous scaffolds for tissue engineering and regenerative medicine. Interface Focus. 2012;2:366–77. doi: 10.1098/rsfs.2011.0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kloxin AM, Kasko AM, Salinas CN, Anseth KS. Photodegradable hydrogels for dynamic tuning of physical and chemical properties. Science. 2009;324:59–63. doi: 10.1126/science.1169494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lutolf MP, Hubbell JA. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 36.Zhang L, Cao Z, Bai T, Carr L, Ella-Menye JR, et al. Zwitterionic hydrogels implanted in mice resist the foreign-body reaction. Nat. Biotechnol. 2013;31:553–56. doi: 10.1038/nbt.2580. [DOI] [PubMed] [Google Scholar]
- 37.Anderson DG, Levenberg S, Langer R. Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells. Nat. Biotechnol. 2004;22:863–66. doi: 10.1038/nbt981. [DOI] [PubMed] [Google Scholar]
- 38.Lei Y, Schaffer DV. A fully defined and scalable 3D culture system for human pluripotent stem cell expansion and differentiation. Proc. Natl. Acad. Sci. USA. 2013;110:E5039–48. doi: 10.1073/pnas.1309408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown AC, Rowe JA, Barker TH. Guiding epithelial cell phenotypes with engineered integrin-specific recombinant fibronectin fragments. Tissue Eng. A. 2011;17:139–50. doi: 10.1089/ten.tea.2010.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gauvin R, Chen YC, Lee JW, Soman P, Zorlutuna P, et al. Microfabrication of complex porous tissue engineering scaffolds using 3D projection stereolithography. Biomaterials. 2012;33:3824–34. doi: 10.1016/j.biomaterials.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Martino MM, Briquez PS, Ranga A, Lutolf MP, Hubbell JA. Heparin-binding domain of fibrin(ogen) binds growth factors and promotes tissue repair when incorporated within a synthetic matrix. Proc. Natl. Acad. Sci. USA. 2013;110:4563–68. doi: 10.1073/pnas.1221602110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borselli C, Storrie H, Benesch-Lee F, Shvartsman D, Cezar C, et al. Functional muscle regeneration with combined delivery of angiogenesis and myogenesis factors. Proc. Natl. Acad. Sci. USA. 2010;107:3287–92. doi: 10.1073/pnas.0903875106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martino MM, Tortelli F, Mochizuki M, Traub S, Ben-David D, et al. Engineering the growth factor microenvironment with fibronectin domains to promote wound and bone tissue healing. Sci. Transl. Med. 2011;3:100ra89. doi: 10.1126/scitranslmed.3002614. [DOI] [PubMed] [Google Scholar]
- 44.Yang K, Lee JS, Kim J, Lee YB, Shin H, et al. Polydopamine-mediated surface modification of scaffold materials for human neural stem cell engineering. Biomaterials. 2012;33:6952–64. doi: 10.1016/j.biomaterials.2012.06.067. [DOI] [PubMed] [Google Scholar]
- 45.Yuen WW, Du NR, Chan CH, Silva EA, Mooney DJ. Mimicking nature by codelivery of stimulant and inhibitor to create temporally stable and spatially restricted angiogenic zones. Proc. Natl. Acad. Sci. USA. 2010;107:17933–38. doi: 10.1073/pnas.1001192107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chung BG, Flanagan LA, Rhee SW, Schwartz PH, Lee AP, et al. Human neural stem cell growth and differentiation in a gradient-generating microfluidic device. Lab Chip. 2005;5:401–6. doi: 10.1039/b417651k. [DOI] [PubMed] [Google Scholar]
- 47.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–61. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 48.Inoguchi H, Tanaka T, Maehara Y, Matsuda T. The effect of gradually graded shear stress on the morphological integrity of a HUVEC-seeded compliant small-diameter vascular graft. Biomaterials. 2007;28:486–95. doi: 10.1016/j.biomaterials.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 49.Powers MJ, Domansky K, Kaazempur-Mofrad MR, Kalezi A, Capitano A, et al. A microfabricated array bioreactor for perfused 3D liver culture. Biotechnol. Bioeng. 2002;78:257–69. doi: 10.1002/bit.10143. [DOI] [PubMed] [Google Scholar]
- 50.Ruiz SA, Chen CS. Microcontact printing: a tool to pattern. Soft Matter. 2007;3:168–77. doi: 10.1039/b613349e. [DOI] [PubMed] [Google Scholar]
- 51.Deleted in proof
- 52.Deleted in proof
- 53.Albrecht DR, Underhill GH, Wassermann TB, Sah RL, Bhatia SN. Probing the role of multicellular organization in three-dimensional microenvironments. Nat. Methods. 2006;3:369–75. doi: 10.1038/nmeth873. [DOI] [PubMed] [Google Scholar]
- 54.Miller JS, Stevens KR, Yang MT, Baker BM, Nguyen DHT, et al. Rapid casting of patterned vascular networks for perfusable engineered three-dimensional tissues. Nat. Mater. 2012;11:768–74. doi: 10.1038/nmat3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lim SH, Mao HQ. Electrospun scaffolds for stem cell engineering. Adv. Drug Deliv. Rev. 2009;61:1084–96. doi: 10.1016/j.addr.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 56.Katz JS, Burdick JA. Light-responsive biomaterials: development and applications. Macromol. Biosci. 2010;10:339–48. doi: 10.1002/mabi.200900297. [DOI] [PubMed] [Google Scholar]
- 57.Li CY, Wood DK, Hsu CM, Bhatia SN. DNA-templated assembly of droplet-derived PEG microtissues. Lab Chip. 2011;11:2967–75. doi: 10.1039/c1lc20318e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yeh J, Ling Y, Karp JM, Gantz J, Chandawarker A, et al. Micromolding of shape-controlled, harvestable hydrogels. Biomaterials. 2006;27:5391–98. doi: 10.1016/j.biomaterials.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 59.Malda J, Visser J, Melchels FP, Juengst T, Hennink WE, et al. 25th anniversary article: engineering hydrogels for biofabrication. Adv. Mater. 2013;25:5011–28. doi: 10.1002/adma.201302042. [DOI] [PubMed] [Google Scholar]
- 60.Baranski JD, Chaturvedi RR, Stevens KR, Eyckmans J, Carvalho B, et al. Geometric control of vascular networks to enhance engineered tissue integration and function. Proc. Natl. Acad. Sci. USA. 2013;110:7586–91. doi: 10.1073/pnas.1217796110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim K, Ohashi K, Utoh R, Kano K, Okano T. Preserved liver-specific functions of hepatocytes in 3D co-culture with endothelial cell sheets. Biomaterials. 2012;33:1406–13. doi: 10.1016/j.biomaterials.2011.10.084. [DOI] [PubMed] [Google Scholar]
- 61a.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008;26:120–26. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 62.Zinchenko YS, Schrum LW, Clemens M, Coger RN. Hepatocyte and Kupffer cells co-cultured on micropatterned surfaces to optimize hepatocyte function. Tissue Eng. 2006;12:751–61. doi: 10.1089/ten.2006.12.751. [DOI] [PubMed] [Google Scholar]
- 63.Ganesan LP, Mohanty S, Kim J, Clark KR, Robinson JM, Anderson CL. Rapid and efficient clearance of blood-borne virus by liver sinusoidal endothelium. PLoS Pathog. 2011;7:e1002281. doi: 10.1371/journal.ppat.1002281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cormier EG, Durso RJ, Tsamis F, Boussemart L, Manix C, et al. L-SIGN (CD209L) and DC-SIGN (0209) mediate transinfection of liver cells by hepatitis C virus. Proc. Natl. Acad. Sci. USA. 2004;101:14067–72. doi: 10.1073/pnas.0405695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dolganiuc A, Norkina O, Kodys K, Catalano D, Bakis G, et al. Viral and host factors induce macrophage activation and loss of Toll-like receptor tolerance in chronic HCV infection. Gastroenterology. 2007;133:1627–36. doi: 10.1053/j.gastro.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu J, Meng Z, Jiang M, Pei R, Trippler M, et al. Hepatitis B virus suppresses Toll-like receptor–mediated innate immune responses in murine parenchymal and nonparenchymal liver cells. Hepatology. 2009;49:1132–40. doi: 10.1002/hep.22751. [DOI] [PubMed] [Google Scholar]
- 67.Choi NW, Cabodi M, Held B, Gleghorn JP, Bonassar LJ, Stroock AD. Microfluidic scaffolds for tissue engineering. Nat. Mater. 2007;6:908–15. doi: 10.1038/nmat2022. [DOI] [PubMed] [Google Scholar]
- 68.Tomei AA, Siegert S, Britschgi MR, Luther SA, Swartz MA. Fluid flow regulates stromal cell organization and CCL21 expression in a tissue-engineered lymph node microenvironment. J. Immunol. 2009;183:4273–83. doi: 10.4049/jimmunol.0900835. [DOI] [PubMed] [Google Scholar]
- 69.Murphy BR, Whitehead SS. Immune response to dengue virus and prospects for a vaccine. Annu. Rev. Immunol. 2011;29:587–619. doi: 10.1146/annurev-immunol-031210-101315. [DOI] [PubMed] [Google Scholar]
- 70.Phillips JE, Burns KL, Le Doux JM, Guldberg RE, Garcia AJ. Engineering graded tissue interfaces. Proc. Natl. Acad. Sci. USA. 2008;105:12170–75. doi: 10.1073/pnas.0801988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Glorieux S, Bachert C, Favoreel HW, Vandekerckhove AP, Steukers L, et al. Herpes simplex virus type 1 penetrates the basement membrane in human nasal respiratory mucosa. PLoS ONE. 2011;6:e22160. doi: 10.1371/journal.pone.0022160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hogk I, Rupp S, Burger-Kentischer A. 3D-tissue model for herpes simplex virus-1 infections. Methods Mol. Biol. 2013;1064:239–51. doi: 10.1007/978-1-62703-601-6_17. [DOI] [PubMed] [Google Scholar]
- 73.Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, et al. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 2006;80:7469–80. doi: 10.1128/JVI.02677-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ehre C, Worthington EN, Liesman RM, Grubb BR, Barbier D, et al. Overexpressing mouse model demonstrates the protective role of Muc5ac in the lungs. Proc. Natl. Acad. Sci. USA. 2012;109:16528–33. doi: 10.1073/pnas.1206552109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wyckoff J, Wang WG, Lin EY, Wang YR, Pixley F, et al. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–29. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 76.Kidd ME, Shin S, Shea LD. Fibrin hydrogels for lentiviral gene delivery in vitro and in vivo. J. Controlled Release. 2012;157:80–85. doi: 10.1016/j.jconrel.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thomas AM, Shea LD. Polysaccharide-modified scaffolds for controlled lentivirus delivery in vitro and after spinal cord injury. J. Controlled Release. 2013;170:421–29. doi: 10.1016/j.jconrel.2013.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Castleberry S, Wang M, Hammond PT. Nanolayered siRNA dressing for sustained localized knockdown. ACS Nano. 2013;7:5251–61. doi: 10.1021/nn401011n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jindal R, Patel SJ, Yarmush ML. Tissue-engineered model for real-time monitoring of liver inflammation. Tissue Eng. C. 2011;17:113–22. doi: 10.1089/ten.tec.2009.0782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smith LE, Smallwood R, Macneil S. A comparison of imaging methodologies for 3D tissue engineering. Microsc. Res. Tech. 2010;73:1123–33. doi: 10.1002/jemt.20859. [DOI] [PubMed] [Google Scholar]
- 81.Hong CS, Fellows W, Niranjan A, Alber S, Watkins S, et al. Ectopic matrix metalloproteinase-9 expression in human brain tumor cells enhances oncolytic HSV vector infection. Gene Ther. 2010;17:1200–5. doi: 10.1038/gt.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81a.Valyi-Nagy K, Dosa S, Kovacs SK, Bacsa S, Voros A, et al. Identification of virus resistant tumor cell subpopulations in three-dimensional uveal melanoma cultures. Cancer Gene Ther. 2010;17:223–34. doi: 10.1038/cgt.2009.73. [DOI] [PubMed] [Google Scholar]
- 82.Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21:745–54. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim HJ, Huh D, Hamilton G, Ingber DE. Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow. Lab Chip. 2012;12:2165–74. doi: 10.1039/c2lc40074j. [DOI] [PubMed] [Google Scholar]
- 84.Nichols JE, Cortiella J, Lee J, Niles JA, Cuddihy M, et al. In vitro analog of human bone marrow from 3D scaffolds with biomimetic inverted colloidal crystal geometry. Biomaterials. 2009;30:1071–79. doi: 10.1016/j.biomaterials.2008.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hadziyannis SJ, Vassilopoulos D, Hadziyannis E. The natural course of chronic hepatitis B virus infection and its management. Antivir. Agents. 2013;67:247–91. doi: 10.1016/B978-0-12-405880-4.00007-X. [DOI] [PubMed] [Google Scholar]
- 86.Yamane D, McGivern DR, Masaki T, Lemon SM. Liver injury and disease pathogenesis in chronic hepatitis C. Curr. Top. Microbiol. Immunol. 2013;369:263–88. doi: 10.1007/978-3-642-27340-7_11. [DOI] [PubMed] [Google Scholar]
- 87.Wedemeyer H, Pischke S, Manns MP. Pathogenesis and treatment of hepatitis E virus infection. Gastroenterology. 2012;142:1388–97. doi: 10.1053/j.gastro.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 88.Martin A, Lemon SM. Hepatitis A virus: from discovery to vaccines. Hepatology. 2006;43:S164–72. doi: 10.1002/hep.21052. [DOI] [PubMed] [Google Scholar]
- 89.Pavio N, Meng XJ, Renou C. Zoonotic hepatitis E: animal reservoirs and emerging risks. Vet. Res. 2010;41:46. doi: 10.1051/vetres/2010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.von Weizsäcker F, Roggendorf M, editors. Models of Viral Hepatitis. Karger; Basel: 2005. [Google Scholar]
- 91.Jopling CL, Yi MK, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific microRNA. Science. 2005;309:1577–81. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 92.Tang H, McLachlan A. Transcriptional regulation of hepatitis B virus by nuclear hormone receptors is a critical determinant of viral tropism. Proc. Natl. Acad. Sci. USA. 2001;98:1841–46. doi: 10.1073/pnas.041479698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Konduru K, Kaplan GG. Stable growth of wild-type hepatitis A virus in cell culture. J. Virol. 2006;80:1352–60. doi: 10.1128/JVI.80.3.1352-1360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–26. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 95.Gottwein JM, Bukh J. Cutting the Gordian knot—development and biological relevance of hepatitis C virus cell culture systems. Adv. Virus Res. 2008;71:51–133. doi: 10.1016/S0065-3527(08)00002-X. [DOI] [PubMed] [Google Scholar]
- 96.Lohmann V, Korner F, Koch JO, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–13. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 97.Saeed M, Scheel TKH, Gottwein JM, Marukian S, Dustin LB, et al. Efficient replication of genotype 3a and 4a hepatitis C Virus replicons in human hepatoma cells. Antimicrob. Agents Chemother. 2012;56:5365–73. doi: 10.1128/AAC.01256-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Bartosch B, Dubuisson J, Cosset FL. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 2003;197:633–42. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pietschmann T, Kaul A, Koutsoudakis G, Shavinskaya A, Kallis S, et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA. 2006;103:7408–13. doi: 10.1073/pnas.0504877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Tanaka T, Takahashi M, Kusano E, Okamoto H. Development and evaluation of an efficient cell-culture system for hepatitis E virus. J. Gen. Virol. 2007;88:903–11. doi: 10.1099/vir.0.82535-0. [DOI] [PubMed] [Google Scholar]
- 101.Tanaka T, Takahashi M, Takahashi H, Ichiyama K, Hoshino Y, et al. Development and characterization of a genotype 4 hepatitis E virus cell culture system using a HE-JF5/15F strain recovered from a fulminant hepatitis patient. J. Clin. Microbiol. 2009;47:1906–10. doi: 10.1128/JCM.00629-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Emerson SU, Zhang M, Meng XJ, Nguyen H, St Claire M, et al. Recombinant hepatitis E virus genomes infectious for primates: importance of capping and discovery of a cis-reactive element. Proc. Natl. Acad. Sci. USA. 2001;98:15270–75. doi: 10.1073/pnas.251555098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Huang YW, Haqshenas G, Kasorndorkbua C, Halbur PG, Emerson SU, Meng XJ. Capped RNA transcripts of full-length cDNA clones of swine hepatitis E virus are replication competent when transfected into Huh7 cells and infectious when intrahepatically inoculated into pigs. J. Virol. 2005;79:1552–58. doi: 10.1128/JVI.79.3.1552-1558.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sells MA, Chen ML, Acs G. Production of hepatitis B virus particles in Hep G2 cells transfected with cloned hepatitis B virus DNA. Proc. Natl. Acad. Sci. USA. 1987;84:1005–9. doi: 10.1073/pnas.84.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Paran N, Geiger B, Shaul Y. HBV infection of cell culture: evidence for multivalent and cooperative attachment. EMBO J. 2001;20:4443–53. doi: 10.1093/emboj/20.16.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ge D, Fellay J, Thompson AJ, Simon JS, Shianna KV, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 107.Thomas DL, Thio CL, Martin MP, Qi Y, Ge D, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Aizaki H, Nagamori S, Matsuda M, Kawakami H, Hashimoto O, et al. Production and release of infectious hepatitis C virus from human liver cell cultures in the three-dimensional radial-flow bioreactor. Virology. 2003;314:16–25. doi: 10.1016/s0042-6822(03)00383-0. [DOI] [PubMed] [Google Scholar]
- 109.Aly HH, Shimotohno K, Hijikata M. 3D cultured immortalized human hepatocytes useful to develop drugs for blood-borne HCV. Biochem. Biophys. Res. Commun. 2009;379:330–34. doi: 10.1016/j.bbrc.2008.12.054. [DOI] [PubMed] [Google Scholar]
- 110.Aly HH, Qi Y, Atsuzawa K, Usuda N, Takada Y, et al. Strain-dependent viral dynamics and virus-cell interactions in a novel in vitro system supporting the life cycle of blood-borne hepatitis C virus. Hepatology. 2009;50:689–96. doi: 10.1002/hep.23034. [DOI] [PubMed] [Google Scholar]
- 111.Mizumoto H, Ishihara K, Nakazawa K, Ijima H, Funatsu K, Kajiwara T. New culture technique for hepatocyte organoid formation and long-term maintenance of liver-specific functions. Tissue Eng. C. 2008;14:167–75. doi: 10.1089/ten.tec.2007.0373. [DOI] [PubMed] [Google Scholar]
- 112.Ndongo-Thiam N, Berthillon P, Errazuriz E, Bordes I, De Sequeira S, et al. Long-term propagation of serum hepatitis C virus (HCV) with production of enveloped HCV particles in human HepaRG hepatocytes. Hepatology. 2011;54:406–17. doi: 10.1002/hep.24386. [DOI] [PubMed] [Google Scholar]
- 113.Narbus CM, Israelow B, Sourisseau M, Michta ML, Hopcraft SE, et al. HepG2 Cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J. Virol. 2011;85:12087–92. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Sainz B, Jr, TenCate V, Uprichard SL. Three-dimensional Huh7 cell culture system for the study of hepatitis C virus infection. Virol. J. 2009;6:103. doi: 10.1186/1743-422X-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sainz B, Jr, Chisari FV. Production of infectious hepatitis C virus by well-differentiated, growth-arrested human hepatoma–derived cells. J. Virol. 2006;80:10253–57. doi: 10.1128/JVI.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Murakami K, Ishii K, Ishihara Y, Yoshizaki S, Tanaka K, et al. Production of infectious hepatitis C virus particles in three-dimensional cultures of the cell line carrying the genome-length dicistronic viral RNA of genotype 1b. Virology. 2006;351:381–92. doi: 10.1016/j.virol.2006.03.038. [DOI] [PubMed] [Google Scholar]
- 117.Molina-Jimenez F, Benedicto I, Dao Thi VL, Gondar V, Lavillette D, et al. Matrigel-embedded 3D culture of Huh-7 cells as a hepatocyte-like polarized system to study hepatitis C virus cycle. Virology. 2012;425:31–39. doi: 10.1016/j.virol.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 117a.Cho NJ, Elazar M, Xiong AM, Lee W, Chiao E, et al. Viral infection of human progenitor and liver-derived cells encapsulated in three-dimensional PEG-based hydrogel. Biomed. Mater. 2009;4:011001. doi: 10.1088/1748-6041/25/1/011001. [DOI] [PubMed] [Google Scholar]
- 118.Rumin S, Berthillon P, Tanaka E, Kiyosawa K, Trabaud MA, et al. Dynamic analysis of hepatitis C virus replication and quasispecies selection in long-term cultures of adult human hepatocytes infected in vitro. J. Gen. Virol. 1999;80:3007–18. doi: 10.1099/0022-1317-80-11-3007. [DOI] [PubMed] [Google Scholar]
- 119.Steenbergen RHG, Joyce MA, Thomas BS, Jones D, Law J, et al. Human serum leads to differentiation of human hepatoma cells, restoration of very-low-density lipoprotein secretion, and a 1000-fold increase in HCV Japanese fulminant hepatitis type 1 titers. Hepatology. 2013;58:1907–17. doi: 10.1002/hep.26566. [DOI] [PubMed] [Google Scholar]
- 120.Berto A, Van der Poel WHM, Hakze-van der Honing R, Martelli F, La Ragione RM, et al. Replication of hepatitis E virus in three-dimensional cell culture. J. Virol. Methods. 2013;187:327–32. doi: 10.1016/j.jviromet.2012.10.017. [DOI] [PubMed] [Google Scholar]
- 121.Rogée S, Talbot N, Caperna T, Bouquet J, Barnaud E, Pavio N. New models of hepatitis E virus replication in human and porcine hepatocyte cell lines. J. Gen. Virol. 2013;94:549–58. doi: 10.1099/vir.0.049858-0. [DOI] [PubMed] [Google Scholar]
- 122.Blank CA, Anderson DA, Beard M, Lemon SM. Infection of polarized cultures of human intestinal epithelial cells with hepatitis A virus: vectorial release of progeny virions through apical cellular membranes. J. Virol. 2000;74:6476–84. doi: 10.1128/jvi.74.14.6476-6484.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Snooks MJ, Bhat P, Mackenzie J, Counihan NA, Vaughan N, Anderson DA. Vectorial entry and release of hepatitis A virus in polarized human hepatocytes. J. Virol. 2008;82:8733–42. doi: 10.1128/JVI.00219-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446:801–5. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 125.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457:882–86. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Sourisseau M, Michta ML, Zony C, Israelow B, Hopcraft SE, et al. Temporal analysis of hepatitis C virus cell entry with occludin directed blocking antibodies. PLoS Pathog. 2013;9:e1003244. doi: 10.1371/journal.ppat.1003244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Schulze A, Mills K, Weiss TS, Urban S. Hepatocyte polarization is essential for the productive entry of the hepatitis B virus. Hepatology. 2012;55:373–83. doi: 10.1002/hep.24707. [DOI] [PubMed] [Google Scholar]
- 128.Mee CJ, Grove J, Harris HJ, Hu K, Balfe P, McKeating JA. Effect of cell polarization on hepatitis C virus entry. J. Virol. 2008;82:461–70. doi: 10.1128/JVI.01894-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Mee CJ, Farquhar MJ, Harris HJ, Hu K, Ramma W, et al. Hepatitis C virus infection reduces hepatocellular polarity in a vascular endothelial growth factor–dependent manner. Gastroenterology. 2010;138:1134–42. doi: 10.1053/j.gastro.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Timpe JM, Stamataki Z, Jennings A, Hu K, Farquhar MJ, et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology. 2008;47:17–24. doi: 10.1002/hep.21959. [DOI] [PubMed] [Google Scholar]
- 131.Gripon P, Rumin S, Urban S, Le Seyec J, Glaise D, et al. Infection of a human hepatoma cell line by hepatitis B virus. Proc. Natl. Acad. Sci. USA. 2002;99:15655–60. doi: 10.1073/pnas.232137699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yan H, Zhong G, Xu G, He W, Jing Z, et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife. 2012;1:e00049. doi: 10.7554/eLife.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yi N, Lempp FA, Mehrle S, Nkongolo S, Kaufman C, et al. Hepatitis B and D viruses exploit sodium taurocholate co-transporting polypeptide for species-specific entry into hepatocytes. Gastroenterology. 2013;146:1070–83. doi: 10.1053/j.gastro.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 134.Quasdorff M, Hoesel M, Odenthal M, Zedler U, Bohne F, et al. A concerted action of HNF4α and HNF1α links hepatitis B virus replication to hepatocyte differentiation. Cell. Microbiol. 2008;10:1478–90. doi: 10.1111/j.1462-5822.2008.01141.x. [DOI] [PubMed] [Google Scholar]
- 135.Deleted in proof
- 136.Bhatia SN, Balis UJ, Yarmush ML, Toner M. Effect of cell-cell interactions in preservation of cellular phenotype: cocultivation of hepatocytes and nonparenchymal cells. FASEB J. 1999;13:1883–900. doi: 10.1096/fasebj.13.14.1883. [DOI] [PubMed] [Google Scholar]
- 137.Iacovacci S, Manzin A, Barca S, Sargiacomo M, Serafino A, et al. Molecular characterization and dynamics of hepatitis C virus replication in human fetal hepatocytes infected in vitro. Hepatology. 1997;26:1328–37. doi: 10.1002/hep.510260535. [DOI] [PubMed] [Google Scholar]
- 138.Lin M, Chen Q, Yang LY, Li WY, Cao XB, et al. Hepatitis B virus infection and replication in primarily cultured human fetal hepatocytes. World J. Gastroenterol. 2007;13:1027–31. doi: 10.3748/wjg.v13.i7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Fournier C, Sureau C, Coste J, Ducos J, Pageaux G, et al. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J. Gen. Virol. 1998;79:2367–74. doi: 10.1099/0022-1317-79-10-2367. [DOI] [PubMed] [Google Scholar]
- 140.Schulze-Bergkamen H, Untergasser A, Dax A, Vogel H, Buchler P, et al. Primary human hepatocytes—a valuable tool for investigation of apoptosis and hepatitis B virus infection. J. Hepatol. 2003;38:736–44. doi: 10.1016/s0168-8278(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 141.Lanford RE, Sureau C, Jacob JR, White R, Fuerst TR. Demonstration of in vitro infection of chimpanzee hepatocytes with hepatitis C virus using strand-specific RT/PCR. Virology. 1994;202:606–14. doi: 10.1006/viro.1994.1381. [DOI] [PubMed] [Google Scholar]
- 142.Tam AW, White R, Yarbough PO, Murphy BJ, McAtee CP, et al. In vitro infection and replication of hepatitis E virus in primary cynomolgus macaque hepatocytes. Virology. 1997;238:94–102. doi: 10.1006/viro.1997.8817. [DOI] [PubMed] [Google Scholar]
- 143.Zhao XP, Tang ZY, Klumpp B, Wolff-Vorbeck G, Barth H, et al. Primary hepatocytes of Tupaia belangeri as a potential model for hepatitis C virus infection. J. Clin. Investig. 2002;109:221–32. doi: 10.1172/JCI13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.von Weizsäcker F, Kock J, MacNelly S, Ren S, Blum HE, Nassal M. The tupaia model for the study of hepatitis B virus: direct infection and HBV genome transduction of primary tupaia hepatocytes. Methods Mol. Med. 2004;96:153–61. doi: 10.1385/1-59259-670-3:153. [DOI] [PubMed] [Google Scholar]
- 145.Gripon P, Diot C, Theze N, Fourel I, Loreal O, et al. Hepatitis B virus infection of adult human hepatocytes cultured in the presence of dimethyl sulfoxide. J. Virol. 1988;62:4136–43. doi: 10.1128/jvi.62.11.4136-4143.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Buck M. Direct infection and replication of naturally occurring hepatitis C virus genotypes 1, 2, 3 and 4 in normal human hepatocyte cultures. PLoS ONE. 2008;3:e2660. doi: 10.1371/journal.pone.0002660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Agnello V, Abel G, Elfahal M, Knight GB, Zhang QX. Hepatitis C virus and other Flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA. 1999;96:12766–71. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011;17:589–95. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Nahmias Y, Casali M, Barbe L, Berthiaume F, Yarmush ML. Liver endothelial cells promote LDL-R expression and the uptake of HCV-like particles in primary rat and human hepatocytes. Hepatology. 2006;43:257–65. doi: 10.1002/hep.21016. [DOI] [PubMed] [Google Scholar]
- 150.Banaudha K, Orenstein JM, Korolnek T, St Laurent GC, III, Wakita T, Kumar A. Primary hepatocyte culture supports hepatitis C virus replication: a model for infection-associated hepatocarcinogenesis. Hepatology. 2010;51:1922–32. doi: 10.1002/hep.23616. [DOI] [PubMed] [Google Scholar]
- 151.Zhou M, Zhao F, Li J, Cheng Z, Tian X, et al. Long-term maintenance of human fetal hepatocytes and prolonged susceptibility to HBV infection by co-culture with nonparenchymal cells. J. Virol. Methods. 2014;195:185–93. doi: 10.1016/j.jviromet.2013.10.010. [DOI] [PubMed] [Google Scholar]
- 151a.Harding MJ, Lepus CM, Gibson TF, Shepherd BR, Gerber SA, et al. An implantable vascularized protein gel construct that supports human fetal hepatoblast survival and infection by hepatitis C virus in mice. PLoS ONE. 2010;5:e9987. doi: 10.1371/journal.pone.0009987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Ploss A, Khetani SR, Jones CT, Syder AJ, Trehan K, et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc. Natl. Acad. Sci. USA. 2010;107:3141–45. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jones CT, Catanese MT, Law LMJ, Khetani SR, Syder AJ, et al. Real-time imaging of hepatitis C virus infection using a fluorescent cell-based reporter system. Nat. Biotechnol. 2010;28:167–71. doi: 10.1038/nbt.1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.March S, Ng S, Velmurugan S, Galstian A, Shan J, et al. A microscale human liver platform that supports the hepatic stages of Plasmodium falciparum and vivax. Cell Host Microbe. 2013;14:104–15. doi: 10.1016/j.chom.2013.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154a.Farkas D, Tannenbaum SR. In vitro methods to study chemically induced hepatotoxicity: a literature review. Curr. Drug Metab. 2005;6:111–25. doi: 10.2174/1389200053586118. [DOI] [PubMed] [Google Scholar]
- 155.Lagaye S, Shen H, Saunier B, Nascimbeni M, Gaston J, et al. Efficient replication of primary or culture hepatitis C virus isolates in human liver slices: a relevant ex vivo model of liver infection. Hepatology. 2012;56:861–72. doi: 10.1002/hep.25738. [DOI] [PubMed] [Google Scholar]
- 156.Yoshida T, Takayama K, Kondoh M, Sakurai F, Tani H, et al. Use of human hepatocyte-like cells derived from induced pluripotent stem cells as a model for hepatocytes in hepatitis C virus infection. Biochem. Biophys. Res. Commun. 2011;416:119–24. doi: 10.1016/j.bbrc.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 157.Wu X, Robotham JM, Lee E, Dalton S, Kneteman NM, et al. Productive hepatitis C virus infection of stem cell–derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2012;8:e1002617. doi: 10.1371/journal.ppat.1002617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Schwartz RE, Trehan K, Andrus L, Sheahan TP, Ploss A, et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 2012;109:2544–48. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Roelandt P, Obeid S, Paeshuyse J, Vanhove J, Van Lommel A, et al. Human pluripotent stem cell–derived hepatocytes support complete replication of hepatitis C virus. J. Hepatol. 2012;57:246–51. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 159a.Schwartz RE, Fleming HE, Khetani SR, Bhatia SN. Pluripotent stem cell–derived hepatocyte-like cells. Biotechnol. Adv. 2014;32:504–13. doi: 10.1016/j.biotechadv.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159b.Shan J, Schwartz RE, Ross NT, Logan DJ, Duncan SA, et al. Identification of small molecules for human hepatocyte expansion and iPS differentiation. Nat. Chem. Biol. 2013;9:514–20. doi: 10.1038/nchembio.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159c.Vallier L. Heps with pep: direct reprogramming into human hepatocytes. Cell Stem Cell. 2014;3:267–69. doi: 10.1016/j.stem.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 159d.Zhu S, Rezyani M, Harbell J, Mattis AN, Wolfe AR, et al. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature. 2014;508:93–97. doi: 10.1038/nature13020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Deleted in proof
- 161.Deleted in proof
- 162.Legrand N, Ploss A, Balling R, Becker PD, Borsotti C, et al. Humanized mice for modeling human infectious disease: challenges, progress, and outlook. Cell Host Microbe. 2009;6:5–9. doi: 10.1016/j.chom.2009.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bukh J. Animal models for the study of hepatitis C virus infection and related liver disease. Gastroenterology. 2012;142:1279–87. doi: 10.1053/j.gastro.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 164.Dorner M, Horwitz JA, Robbins JB, Barry WT, Feng Q, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–11. doi: 10.1038/nature10168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dorner M, Horwitz JA, Donovan BM, Labitt RN, Budell WC, et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237–41. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bitzegeio J, Bankwitz D, Hueging K, Haid S, Brohm C, et al. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 2010;6:e1000978. doi: 10.1371/journal.ppat.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Flaim CJ, Chien S, Bhatia SN. An extracellular matrix microarray for probing cellular differentiation. Nat. Methods. 2005;2:119–25. doi: 10.1038/nmeth736. [DOI] [PubMed] [Google Scholar]
- 168.Flaim CJ, Teng D, Chien S, Bhatia SN. Combinatorial signaling microenvironments for studying stem cell fate. Stem Cells Dev. 2008;17:29–39. doi: 10.1089/scd.2007.0085. [DOI] [PubMed] [Google Scholar]
- 169.Lee MY, Kumar RA, Sukumaran SM, Hogg MG, Clark DS, Dordick JS. Three-dimensional cellular microarray for high-throughput toxicology assays. Proc. Natl. Acad. Sci. USA. 2008;105:59–63. doi: 10.1073/pnas.0708756105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Deiss F, Mazzeo A, Hong E, Ingber DE, Derda R, Whitesides GM. Platform for high-throughput testing of the effect of soluble compounds on 3D cell cultures. Anal. Chem. 2013;85:8085–94. doi: 10.1021/ac400161j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mali P, Yang L, Esvelt KM, Aach J, Guell M, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–26. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat. Biotechnol. 2011;29:731–34. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Patel SJ, King KR, Casali M, Yarmush ML. DNA-triggered innate immune responses are propagated by gap junction communication. Proc. Natl. Acad. Sci. USA. 2009;106:12867–72. doi: 10.1073/pnas.0809292106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kwong GA, von Maltzahn G, Murugappan G, Abudayyeh O, Mo S, et al. Mass-encoded synthetic biomarkers for multiplexed urinary monitoring of disease. Nat. Biotechnol. 2013;31:63–70. doi: 10.1038/nbt.2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Miyata T, Uragami T, Nakamae K. Biomolecule-sensitive hydrogels. Adv. Drug Deliv. Rev. 2002;54:79–98. doi: 10.1016/s0169-409x(01)00241-1. [DOI] [PubMed] [Google Scholar]
- 176.Emmert-Buck MR, Bonner RF, Smith PD, Chuaqui RF, Zhuang ZP, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 177.Kalantari M, Garcia-Carranca A, Morales-Vazquez CD, Zuna R, Perez Montiel D, et al. Laser capture microdissection of cervical human papillomavirus infections: copy number of the virus in cancerous and normal tissue and heterogeneous DNA methylation. Virology. 2009;390:261–67. doi: 10.1016/j.virol.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Lovatt D, Ruble BK, Lee J, Dueck H, Kim TK, et al. Transcriptome in vivo analysis (TIVA) of spatially defined single cells in live tissue. Nat. Methods. 2014;11:190–96. doi: 10.1038/nmeth.2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Kloxin AM, Tibbitt MW, Anseth KS. Synthesis of photodegradable hydrogels as dynamically tunable cell culture platforms. Nat. Protoc. 2010;5:1867–87. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Li CY, Wood DK, Huang JH, Bhatia SN. Flow-based pipeline for systematic modulation and analysis of 3D tumor microenvironments. Lab Chip. 2013;13:1969–78. doi: 10.1039/c3lc41300d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Tan JKH, Watanabe T. Artificial engineering of secondary lymphoid organs. Adv. Immunol. 2010;105:131–57. doi: 10.1016/S0065-2776(10)05005-4. [DOI] [PubMed] [Google Scholar]
- 182.Breiner KM, Schaller H, Knolle PA. Endothelial cell–mediated uptake of a hepatitis B virus: a new concept of liver targeting of hepatotropic microorganisms. Hepatology. 2001;34:803–8. doi: 10.1053/jhep.2001.27810. [DOI] [PubMed] [Google Scholar]
- 183.Dalrymple NA, Mackow ER. Endothelial cells elicit immune-enhancing responses to dengue virus infection. J. Virol. 2012;86:6408–15. doi: 10.1128/JVI.00213-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Gardner JP, Durso RJ, Arrigale RR, Donovan GP, Maddon PJ, et al. L-SIGN (CD 209L) is a liver-specific capture receptor for hepatitis C virus. Proc. Natl. Acad. Sci. USA. 2003;100:4498–503. doi: 10.1073/pnas.0831128100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Tarran R, Button B, Picher M, Paradiso AM, Ribeiro CM, et al. Normal and cystic fibrosis airway surface liquid homeostasis—the effects of phasic shear stress and viral infections. J. Biol. Chem. 2005;280:35751–59. doi: 10.1074/jbc.M505832200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Shi ZD, Tarbell JM. Fluid flow mechanotransduction in vascular smooth muscle cells and fibroblasts. Ann. Biomed. Eng. 2011;39:1608–19. doi: 10.1007/s10439-011-0309-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.DuRose JB, Li J, Chien S, Spector DH. Infection of vascular endothelial cells with human cytomegalovirus under fluid shear stress reveals preferential entry and spread of virus in flow conditions simulating atheroprone regions of the artery. J. Virol. 2012;86:13745–55. doi: 10.1128/JVI.02244-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Mueller AJ, Filipe-Santos O, Eberl G, Aebischer T, Spaeth GF, Bousso P. CD4+ T cells rely on a cytokine gradient to control intracellular pathogens beyond sites of antigen presentation. Immunity. 2012;37:147–57. doi: 10.1016/j.immuni.2012.05.015. [DOI] [PubMed] [Google Scholar]
- 189.Xu N, Zhang ZF, Wang L, Gao B, Pang DW, et al. A microfluidic platform for real-time and in situ monitoring of virus infection process. Biomicrofluidics. 2012;6:34122. doi: 10.1063/1.4756793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Zhu Y, Warrick JW, Haubert K, Beebe DJ, Yin J. Infection on a chip: a microscale platform for simple and sensitive cell-based virus assays. Biomed. Microdevices. 2009;11:565–70. doi: 10.1007/s10544-008-9263-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Akin D, Li HB, Bashir R. Real-time virus trapping and fluorescent imaging in microfluidic devices. Nano Lett. 2004;4:257–59. [Google Scholar]
- 192.Costello DA, Millet JK, Hsia CY, Whittaker GR, Daniel S. Single particle assay of coronavirus membrane fusion with proteinaceous receptor-embedded supported bilayers. Biomaterials. 2013;34:7895–904. doi: 10.1016/j.biomaterials.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Schudel BR, Harmon B, Abhyankar VV, Pruitt BW, Negrete OA, Singh AK. Microfluidic platforms for RNA interference screening of virus-host interactions. Lab Chip. 2013;13:811–17. doi: 10.1039/c2lc41165b. [DOI] [PubMed] [Google Scholar]
- 194.Algar WR, Prasuhn DE, Stewart MH, Jennings TL, Blanco-Canosa JB, et al. The controlled display of biomolecules on nanoparticles: a challenge suited to bioorthogonal chemistry. Bioconjug. Chem. 2011;22:825–58. doi: 10.1021/bc200065z. [DOI] [PubMed] [Google Scholar]
- 195.Gregory A, Stenzel MH. Complex polymer architectures via RAFT polymerization: from fundamental process to extending the scope using click chemistry and nature’s building blocks. Prog. Polymer Sci. 2012;37:38–105. [Google Scholar]
- 196.Wang WW, Khetani SR, Krzyzewski S, Duignan DB, Obach RS. Assessment of a micropatterned hepatocyte coculture system to generate major human excretory and circulating drug metabolites. Drug Metab. Dispos. 2010;38:1900–5. doi: 10.1124/dmd.110.034876. [DOI] [PubMed] [Google Scholar]
- 197.Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. Reproducible, ultra high-throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS ONE. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Lovett M, Lee K, Edwards A, Kaplan DL. Vascularization strategies for tissue engineering. Tissue Eng. B. 2009;15:353–70. doi: 10.1089/ten.teb.2009.0085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Morinet F, Casetti L, Francois JH, Capron C, Pillet S. Oxygen tension level and human viral infections. Virology. 2013;444:31–36. doi: 10.1016/j.virol.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 200.Hanjaya-Putra D, Bose V, Shen YI, Yee J, et al. Controlled activation of morphogenesis to generate a functional human microvasculature in a synthetic matrix. Blood. 2011;118:804–15. doi: 10.1182/blood-2010-12-327338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Samuel R, Daheron L, Liao S, Vardam T, Kamoun WS, et al. Generation of functionally competent and durable engineered blood vessels from human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA. 2013;110:12774–79. doi: 10.1073/pnas.1310675110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Wu W, Hansen CJ, Aragon AM, Geubelle PH, White SR, Lewis JA. Direct-write assembly of biomimetic microvascular networks for efficient fluid transport. Soft Matter. 2010;6:739–42. [Google Scholar]
- 203.Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, et al. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 2006;80:5308–20. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Schoggins JW, Dorner M, Feulner M, Imanaka N, Murphy MY, et al. Dengue reporter viruses reveal viral dynamics in interferon receptor–deficient mice and sensitivity to interferon effectors in vitro. Proc. Natl. Acad. Sci. USA. 2012;109:14610–15. doi: 10.1073/pnas.1212379109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Tanimizu N, Miyajima A, Mostov KE. Liver progenitor cells develop cholangiocyte-type epithelial polarity in three-dimensional culture. Mol. Biol. Cell. 2007;18:1472–79. doi: 10.1091/mbc.E06-09-0848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Si-Tayeb K, Noto FK, Nagaoka M, Li J, Battle MA, et al. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology. 2010;51:297–305. doi: 10.1002/hep.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Goldman O, Han S, Sourrisseau M, Dziedzic N, Hamou W, et al. KDR identifies a conserved human and murine hepatic progenitor and instructs early liver development. Cell Stem Cell. 2013;12:748–60. doi: 10.1016/j.stem.2013.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Discher DE, Mooney DJ, Zandstra PW. Growth factors, matrices, and forces combine and control stem cells. Science. 2009;324:1673–77. doi: 10.1126/science.1171643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Fernandes TG, Kwon SJ, Bale SS, Lee MY, Diogo MM, et al. Three-dimensional cell culture microarray for high-throughput studies of stem cell fate. Biotechnol. Bioeng. 2010;106:106–18. doi: 10.1002/bit.22661. [DOI] [PMC free article] [PubMed] [Google Scholar]