Abstract
Objectives
Growing evidence suggests that mutations in the connection domain of the HIV-1 reverse transcriptase (RT) can contribute to viral resistance to RT inhibitors. This work was designed to determine the effects of a novel mutation, D404N, in the connection subdomain of RT of HIV-1 CRF08_BC subtype on drug resistance, viral replication capacity (RC) and RT activity.
Methods
Mutation D404N, alone or together with the other reported mutations, was introduced into an HIV-1 CRF08_BC subtype infectious clone by site-directed mutagenesis. Viral susceptibility to nine RT inhibitors, viral RC and the DNA polymerase activity of viral RT of the constructed virus mutants were investigated. A modelling study using the server SWISS-MODEL was conducted to explore the possible structure-related drug resistance mechanism of the mutation D404N.
Results
Single mutations D404N and H221Y conferred low-level resistance to nevirapine, efavirenz, rilpivirine and zidovudine. Double mutations Y181C/D404N and Y181C/H221Y significantly reduced susceptibility to NNRTIs. The most pronounced resistance to NNRTIs was observed with the triple mutation Y181C/D404N/H221Y. Virus containing D404N as the only mutation displayed ∼50% RC compared with the WT virus. The modelling study suggested that the D404N mutation might abolish the hydrogen bonds between residues 404 and K30 in p51 or K431 in p66, leading to impaired RT subunit structure and enhanced drug resistance.
Conclusions
These results indicate that D404N is a novel NNRTI-associated mutation in the HIV-1 subtype CRF08_BC and provides information valuable for the monitoring of clinical RTI resistance.
Keywords: drug susceptibility, replication capacity, modelling
Introduction
The HAART regimen, containing three or more different drugs, such as two NRTIs and a PI, or two NRTIs and a NNRTI, or other combinations (http://aids.about.com/od/hivaidsletterh/g/haartdef.htm), has been widely used as the first-line regimen for HIV-1 treatment.1 NRTIs, including zidovudine, lamivudine, abacavir sulphate, didanosine, stavudine, zalcitabine, tenofovir disoproxil fumarate and emtricitabine, are structurally similar to the natural deoxynucleoside but lack the 3′-hydroxyl group, which is necessary for DNA strand elongation. Therefore, they can cause DNA chain termination by competing with normal nucleotides after activation through intracellular phosphorylation.2 NNRTIs, including nevirapine, delavirdine and efavirenz (first generation) and etravirine and rilpivirine (second generation) are non-competitive inhibitors. They bind to a hydrophobic pocket that is close to but distinct from the polymerase active site, and induce a conformational change resulting in inhibition of dNTP binding.3
Although HAART has dramatically reduced the morbidity and mortality of HIV infection, the emergence of drug-resistance mutations inevitably leads to virological failure and progress to AIDS in the majority of infected patients.4 Therefore, identifying and understanding HIV-1 drug-resistance mutations can help in the development of effective antiretroviral drugs for newly infected patients or patients in whom treatment has failed.5 Currently, most genotypic analyses for drug resistance mutations are restricted to residues 1–300 in reverse transcriptase (RT),6,7 including the well-studied NRTI resistance mutations M41L, D67N, K70R, Y115F, Q151M, M184V and T210W, as well as the NNRTI resistance-related mutations L100I, K101E/P, K103N/S, V106A/M, E138A/G/K/Q, Y181C/I/V, Y188L/C/H, G190A/S/E and M230L (http://hivdb.stanford.edu/DR/). However, recent reports indicate that several residues in the connection subdomain (residues 319–426) of RT could also induce resistance to NNRTIs and even NRTIs through direct or indirect mechanisms.1,6,8,9 The connection subdomain not only acts as a tether between the polymerase and the RNase H subdomain, but also plays a role in nucleic acid substrate interaction and RT inter-subunit interaction.10 The most relevant mutations are N348I, T369I/V, Y318F/W and A376S.6 E312Q, G335C/D, N348I, A360I/V, V365I and A376S were observed to confer resistance to zidovudine by decreasing RNase H activity.7,11,12
In China, the HIV-1 epidemic is driven by the wide dissemination of subtypes CRF01_AE, B′, CRF07_BC and CRF08_BC.13 The last two subtypes used to circulate predominantly in intravenous drug users, whereas in recent years they have been widely transmitted sexually.14,15 This indicates that subtypes CRF07_BC and CRF08_BC have begun to be transmitted from high-risk groups to the general population. Although free antiretroviral treatment has been offered by the Chinese government since 2003, the emergence of drug-resistance mutations has become a considerable hindrance to effective HIV-1 management.15 One report showed that drug resistance had reached 30.3% in Henan, 26.6% in Anhui and 19.2% in Hubei province.15 However, drug-resistance data in China are still very limited and most studies have focused on the first 300 amino acids in RT only.16–18 To date, no studies have been conducted on the connection subdomain of HIV-1 CRF08_BC RT in patients in whom treatment has failed.
In our previous selection culture assay using one clinical isolate of HIV-1 subtype CRF08_BC,19 we identified a novel mutation, D404N, in RT together with another reported mutation, H221Y, which evolved after the appearance of the primary mutation Y181C under increasing nevirapine pressure. The selected viruses containing these accumulated mutations were >1000-fold less susceptible to nevirapine than the WT virus.19 Interestingly, the addition of D404N/H221Y enabled the virus to be 10-fold more resistant to nevirapine than those without these two mutations.19
This study was designed to determine whether the novel mutation D404N plays a role in resistance to RT inhibitors and to evaluate RT polymerase activity and viral replication capacity (RC). The interaction between D404N and H221Y or Y181C was further investigated from the aspects of drug susceptibility and viral RC. A molecular modelling study was performed to explore the possible structural mechanisms. Besides, since E404 mainly emerges in the WT virus of subtype B and was not induced in our previous study,19 we also tested whether D404E influences viral phenotypic susceptibility, viral RC and RT activity in the CRF08_BC subtype.
Materials and methods
Cells, plasmids and drugs
We obtained 293FT cells from ATCC. TZM-bl cells were obtained from Dr John C. Kappes, Dr Xiaoyun Wu and Tranzyme Inc. through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH. The WT infectious plasmid pBRGX was constructed and confirmed from a clinical isolate of CRF08_BC subtype in our previous study.20 Drugs, including four NNRTIs (nevirapine, efavirenz, etravirine and rilpivirine) and five NRTIs (zidovudine, abacavir sulphate, lamivudine, emtricitabine and tenofovir disoproxil fumarate), were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH.
Site-directed mutagenesis and virus production
Eight mutation patterns in RT, including single (D404N, D404E, Y181C and H221Y), double (Y181C/D404N, Y181C/H221Y and D404N/H221Y) and triple (Y181C/D404N/H221Y) mutations, were introduced into the pBRGX plasmid by site-directed mutagenesis as described previously21 using AccuPrime™ Taq DNA Polymerase High Fidelity (Invitrogen). Details of the amino acid substitutions are listed in Table 1. The presence of mutations in the plasmids was confirmed by DNA sequencing. Mutation-specific primers and primers for amplifying RT during positive clone identification were designed and are illustrated in Table 2. Constructed mutants and WT plasmids were transfected into 293FT cells in six-well plates uisng Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Seventy-two hours after transfection, the viruses were collected, aliquoted and stored in a −80°C freezer for future use. Viral titre was determined by measuring p24 with a Vironostika kit (bioMérieux, France) and determining the TCID50 in TZM-bl cells as described previously.22 Only those with p24 production >50 mg/L were subjected to phenotypic drug resistance assay and analysis of RC and RT activity.
Table 1.
Amino acid substitutions in HXB2a and pBRGXb
| Codon site in RT | Position in HXB2 | Codon in HXB2 (amino acid) | Position in pBRGX | Codon in BRGX (amino acid) | Mutant codon (amino acid) |
|---|---|---|---|---|---|
| 181 | 3090–3092 | TAT (Y) | 3061–3063 | TAT (Y) | TGT (C) |
| 221 | 3210–3212 | CAT (H) | 3181–3183 | CAT (H) | TAT (Y) |
| 404 | 3759–3761 | GAG (E) | 3730–3732 | GAC (D) | AAC (N) |
| 404 | 3730–3732 | GAC (D) | GAG (E) |
Table 2.
Primers for site-directed mutagenesis and RT amplification
| Amino acid substitution/name | Genetic mutation | Sequence (5′–3′) |
|---|---|---|
| Primers for site-directed mutagenesis | ||
| Y181C_F | A3062G | TATCTGTCAATACATGGATGACTTGTATGTAGGATCTG |
| Y181C_R | A3062G | TATTGACAGATAACTATGTCTGGATTTTGTTTTC |
| H221Y_F | C3181T | AAGAAATATCAGAAAGAACCTCCATTTCTTTGGATGGGGTATG |
| H221Y_R | C3181T | TTCTGATATTTCTTGTCTGGTGTGGTAAATCCCCACTTTAACAG |
| D404N_F | G3730A | TGGACAAACTGGTGGCAAGCCACCTGGATTC |
| D404N_R | G3730A | ACCAGTTTGTCCACCATGTCTCCCATGTTTCTT |
| D404E_F | C3732G | TGGACAGAGTGGTGGCAAGCCACCTGGATTC |
| D404E_R | C3732G | ACCACTCTGTCCACCATGTCTCCCATGTTTCTT |
| Primers for RT amplification | ||
| HI1395 | ATGAAGAGGCTGCAGAATGGG | |
| HI4635 | GGATTCTACTACTCCCTGACTTTGG | |
Mutated nucleic acids in the primer sequences are shown in bold.
Phenotypic drug susceptibility of the virus mutants
The susceptibility of the mutant viruses to NNRTIs (nevirapine, efavirenz, etravirine and rilpivirine) and NRTIs (zidovudine, abacavir sulphate, lamivudine, emtricitabine and tenofovir disoproxil fumarate) was measured in a single-cycle cell-culture-based phenotypic assay as described previously.19,22 In brief, TZM-bl cells (1 × 104/well) were seeded into a 96-well flat-bottom plate. On the next day, mutant and WT viruses equal to 200–400 blue focus units (BFUs) in serially diluted drugs were added to each well of the plate in triplicate. Wells infected with equal amounts of virus but without drugs were included as virus controls. The plates were incubated at 37°C in a 5% CO2 incubator for 48 h. After the cells had been fixed and stained, the number of blue cells was calculated using ImmunoSpot 3.2 software after being imaged with an ELIspot plate reader (Cellular Technology Limited, C.T.L.). IC50 values were determined using SigmaPlot 12.0 software (four-parameter logistic analysis). Susceptibility to drugs was expressed as the fold change in IC50 of mutant virus compared with the WT reference virus. As in our previous report,22 drug resistance levels were divided into four groups according to the fold change in IC50 for the mutant virus relative to WT, i.e. high-level resistance (>20-fold), moderate-level resistance (4- to 20-fold), low-level resistance (2- to 4-fold) and susceptible (0- to 2-fold).
Determination of RC of the virus mutants
The RC of the mutant and WT viruses was determined in a modified single-cycle cell-culture-based assay as described previously.8,23,24 Briefly, TZM-bl cells (1 × 104/well) were seeded in a 96-well plate. On the next day, serially diluted mutant and WT viruses were applied to TZM-bl cells in triplicate for each dilution. The subsequent steps were the same as described above for the phenotypic assays. RC was determined by normalizing the number of blue cells per millilitre to the corresponding p24 concentration (mg/L) for each mutant virus. Final relative RC was expressed as a percentage of the value for the WT reference virus, calculated as follows: RC of test virus (%) = (blue cell number/ng p24test virus/blue cell number/ng p24WT reference virus) × 100.
RT activity assay
The effect of D404N on RT activity was assessed with an RT assay using a colorimetric kit (Roche, USA) as described previously with some modifications.21 Briefly, 750 μL of viral supernatant was ultracentrifuged at 4°C for 2 h at 100 000 g and the viral pellets were resuspended in 75 μL of lysis buffer. Viral lysate was serially diluted 4-fold in lysis buffer and the subsequent procedures were carried out according to the manufacturer's instructions. Each sample was detected in duplicate. Viral RT activity was quantified and normalized by the input amount of p24.
Modelling study
The models of HIV-1 CRF08_BC RT (1–560 amino acids) (WT containing D404 and mutant containing N404) were built by homology modelling with HIV-1 RT, nevirapine and RNA/DNA hybrid crystal structure 4PUO25 using the server SWISS-MODEL26 with default parameters. The model was further analysed using Discovery Studio 3.1 visualizer (Accelrys), and the Ramachandran plot was examined to ensure that the structure of the model was not in the unfavourable regions.
Statistical analysis
The data were evaluated for statistical significance with an unpaired Student's t-test using Prism (version 5.0, GraphPad, Inc.). Values of P < 0.05 were considered to represent a statistically significant difference. Results are given as the mean ± SD of triplicate independent experiments.
Results
Low-level resistance induced by single mutations
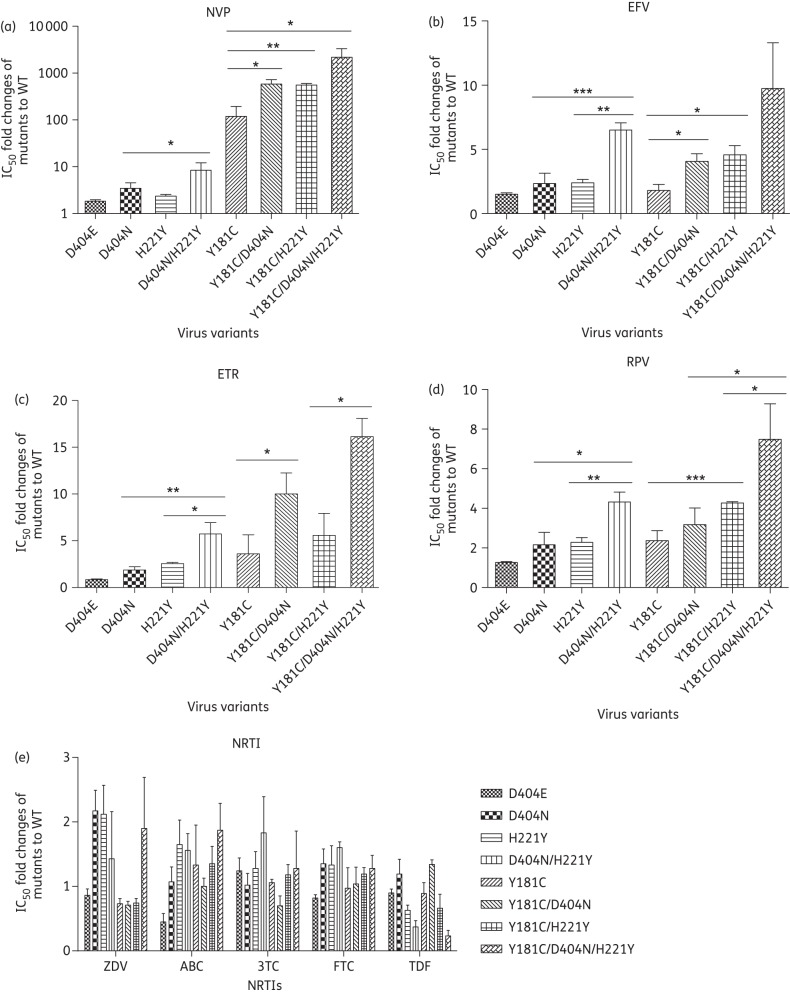
Figure 1 shows that the mutation D404N conferred low-level resistance to nevirapine (3.5-fold), efavirenz (2.3-fold) and rilpivirine (2.2-fold) but did not affect susceptibility to etravirine (1.7-fold). Viruses with the single mutation H221Y were cross-resistant to nevirapine (2.4-fold), efavirenz (2.4-fold), etravirine (2.6-fold) and rilpivirine (2.3-fold) (Figure 1a–d). Mutants with both D404N and H221Y showed low-level resistance to zidovudine (2.2- and 2.1-fold) but were still susceptible to all the other NRTIs, including abacavir sulphate, lamivudine, emtricitabine and tenofovir disoproxil fumarate (Figure 1e). Additionally, viruses containing the single mutation Y181C displayed high-level resistance to nevirapine (119.4-fold) and low-level resistance to etravirine (3.6-fold) and rilpivirine (2.7-fold) (Figure 1a, c and d). No resistance to efavirenz or NRTIs was observed in the case of Y181C (Figure 1b and e). However, D404E did not confer resistance to all the NNRTIs and NRTIs compared with WT (<2-fold) (Figure 1a–e).
Figure 1.
Phenotypic analysis of the susceptibility of virus variants to RT inhibitors: (a) nevirapine (NVP), (b) efavirenz (EFV), (c) etravirine (ETR), (d) rilpivirine (RPV) and (e) NRTIs (ZDV, zidovudine; ABC, abacavir sulphate; 3TC, lamivudine; FTC, emtricitabine; TDF, tenofovir disoproxil fumarate). The fold changes in IC50 for virus mutants compared with WT are shown. TZM-bl cells were infected with virus variants and treated with serially diluted RT inhibitors. At 48 h post-infection, the number of blue cells, representing HIV infection, was calculated. The IC50 was estimated from the standard curve plotted by the percentage of reduction of blue cell numbers and serial concentrations of inhibitors. *P < 0.05; **P < 0.01; ***P < 0.001.
Moderate to strong resistance induced by double mutations
To investigate the interaction between component mutations of double mutations with respect to drug susceptibility, viruses carrying D404N/H221Y or Y181C/D404N or Y181C/H221Y were generated and tested for susceptibility to both NRTIs and NNRTIs. Results showed that the double mutation D404N/H221Y rendered the virus resistant to all the approved NNRTIs at a moderate level. Compared with H221Y, the addition of D404N significantly enhanced viral resistance to efavirenz (6.5-fold), etravirine (5.7-fold) and rilpivirine (4.3-fold) (Figure 1b–d). In addition, D404N in combination with H221Y also increased viral resistance to nevirapine (8.4-fold) (Figure 1a), though this increase was not statistically different from that induced by H221Y alone. Similarly, compared with D404N, the addition of H221Y significantly reduced viral susceptibility to nevirapine, efavirenz, etravirine and rilpivirine (Figure 1a–d). For the five NRTIs (zidovudine, abacavir sulphate, lamivudine, emtricitabine and tenofovir disoproxil fumarate), the virus with D404N/H221Y in RT showed no resistance compared with the WT reference virus (Figure 1e).
Interestingly, when D404N or H221Y was introduced in addition to the primary mutation Y181C, mutant viruses showed a sharp decrease in susceptibility to nevirapine (583.5-fold for Y181C/D404N and 555.0-fold for Y181C/H221Y) (Figure 1a) as well as moderate resistance to etravirine (10- and 5.6-fold), efavirenz (4.1- and 4.6-fold) and rilpivirine (3.2- and 4.8-fold) (Figure 1b–d). Compared with Y181C, the addition of D404N significantly increased resistance to nevirapine, efavirenz and etravirine (Figure 1a–c). Likewise, the addition of H221Y also significantly enhanced viral resistance to nevirapine, efavirenz and rilpivirine compared with Y181C alone (Figure 1a–c). The combination of D404N or H221Y with Y181C did not influence viral susceptibility to the NRTIs (Figure 1e).
High-level cross-resistance to NNRTIs induced by multiple mutations
We further examined the resistance to reverse transcription inhibitors of viruses carrying multiple mutations (D404N, H221Y and Y181C). Our results demonstrated >2000-fold increase in resistance to nevirapine when these three mutations co-existed. A moderate level of resistance was also observed for three other NNRTIs (efavirenz, 9.7-fold; etravirine, 16.1-fold; rilpivirine, 7.5-fold) (Figure 1b–d) relative to the WT reference virus. Of note, compared with Y181C/H221Y, the addition of D404N significantly reduced viral susceptibility to etravirine and rilpivirine (Figure 1c and d). Compared with Y181C/D404N, H221Y also significantly increased viral resistance to rilpivirine (Figure 1d). However, the combination of the three mutations had no influence on the resistance to NRTIs (Figure 1e).
D404N and H221Y impaired viral RC
To investigate the effects of D404N and H221Y on viral RC and the interaction between them and Y181C, single-cycle cell-culture-based assays were performed. As shown in Figure 2, the RC of the virus containing a single D404N mutation was decreased by ∼50% in comparison with that of the WT virus. The single H221Y mutant displayed the lowest RC (30.95%). The combination of D404N and H221Y had a similar impact on viral RC (48.52% of WT) compared with either mutation alone. The above results indicate that the presence of D404N, H221Y or D404N/H221Y was clearly able to impair viral RC. The virus with the single mutation D404E showed similar RC to WT (Figure 2).
Figure 2.
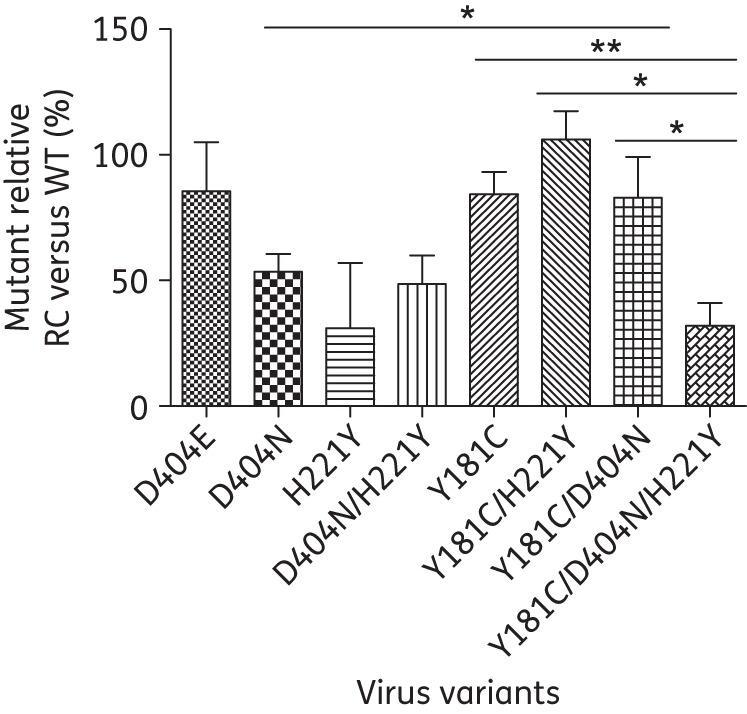
RC of HIV-1 virus variants. TZM-bl cells were infected with virus variants without drugs. At 48 h post-infection, numbers of blue cells, representing HIV infection, were calculated and normalized by the amount of p24. The relative RC of virus variants was determined by the ratio of the blue cell number normalized by input p24 amount (BFU/p24) of mutant virus compared with that of WT virus (%). *P < 0.05; **P < 0.01.
Impact of Y181C mutation on viral RC
The RC of the virus with Y181C alone was close to that of the WT virus (84.26% of WT) (Figure 2). The addition of D404N or H221Y to Y181C did not further suppress the replication of virus compared with Y181C alone. This indicated that Y181C could completely compensate for the impaired RC of D404N or H221Y. However, when D404N was added to Y181C/H221Y, viral RC was significantly impaired compared with Y181C/H221Y, dropping from 106.17% to 46.86% relative to WT (Figure 2). Likewise, the addition of H221Y to Y181C/D404N also led to a significant decrease in viral RC in comparison with Y181C/D404N (Figure 2). In addition, viruses with Y181C/D404N/H221Y displayed significantly decreased RC compared with those having Y181C alone. Thus, both D404N and H221Y are deleterious mutations for viral replication and their co-existence results in irreversible RC loss.
RT activity assay
The effects of these mutations on the polymerase activity of RT were detected using a colorimetric kit. The template/primer included in the colorimetric kit was a hybrid of poly(A)/oligo(dT)15 and the included nucleotides were labelled with digoxigenin and biotin in an optimized ratio. The signal to be detected was from the synthesized minus-strand cDNA and the activity determined by this kit was the RNA-dependent DNA polymerase activity of RT. Results showed that D404N conferred 2.6-fold and D404E 1.6-fold RT polymerase activity relative to that of WT. No statistical difference was observed between other mutant viruses and WT reference virus (Figure 3).
Figure 3.
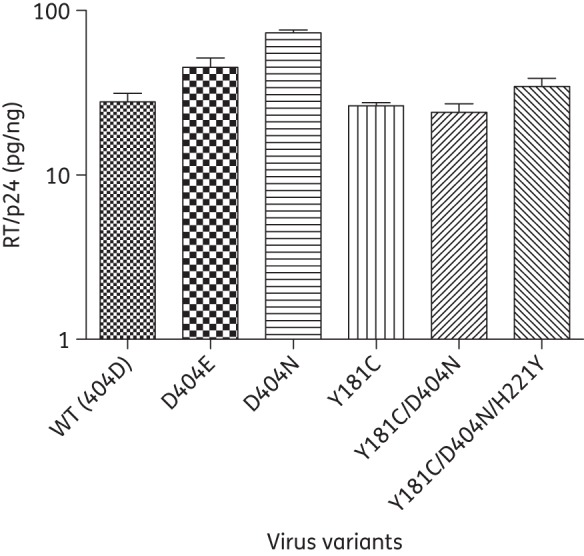
RNA-dependent DNA polymerase activity of RT. The polymerase activity of virus variants and WT virus was tested by a colorimetric RT assay. Viral particles in the supernatant were concentrated by ultracentrifugation. Final RT activity was normalized by the amount of p24. Results are expressed as mean ± SD of two independent experiments.
Molecular modelling of RT of HIV-1 CRF08_BC
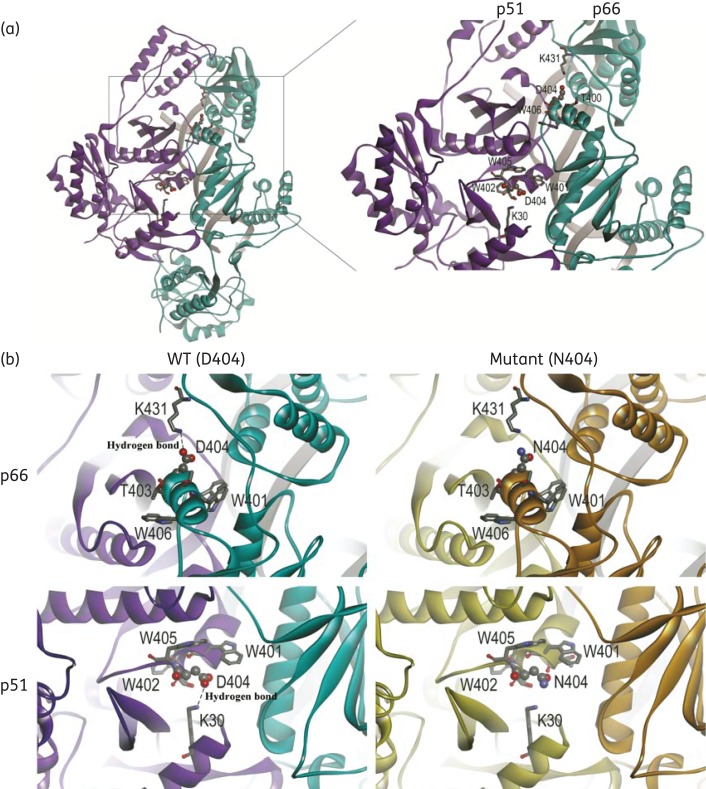
NNRTI-related resistance is always induced by RT conformational changes resulting from the emergence of mutations such as Y181C and Y188C. To test whether structural changes could be induced by the novel mutation D404N, the HIV-1 CRF08_BC RT model was built by homology modelling with HIV-1 RT, nevirapine and the RNA/DNA hybrid crystal structure (4PUO) through the server SWISS-MODEL. As shown in Figure 4(a), residue D404 is adjacent to K30 in p51 and K431 in the p66 subunit. Since aspartic acid (D) is a negatively charged amino acid and lysine (K) is a positively charged amino acid, it is apparent in Figure 4(b) that the negatively charged oxygen in the carboxylate radical group (red) in the side chain of D404 is in close proximity to the positively charged hydrogen in the amine group (blue) of K30 and K431. Therefore, a hydrogen bond can form between D404 and K30 or K431. When D404 is replaced with N404, the negatively charged O− will be replaced by neutral NH2 and the hydrogen bonds will be abolished. The residues within 4 Å (hydrogen bond distance) from residue 404 were highlighted and mainly restricted to the helix of 400–406.
Figure 4.
D404N RT models. (a) The models are shown as ribbon diagrams. Subunits of p66 and p51 are coloured cyan and purple, respectively. Residue 404 and its vicinal amino acids on both subunits are highlighted. The atoms of residue 404 are labelled with balls and those of lysine 30 and 431 with sticks. The DNA/RNA hybrid is shown in tube format, coloured grey. (b) Subunits of p66 and p51 in WT RT with D404 are coloured cyan and purple, respectively, and those in mutant RT with N404 are coloured copper and khaki, respectively. Red, oxygen atom; blue, nitrogen atom; grey, carbon atom. The figures were produced using DS visualizer (Accelrys).
Discussion
The current study characterized a novel NNRTI-related mutation D404N in RT, which was selected previously by nevirapine during in vitro cell culture.19 D404N not only conferred low-level resistance to nevirapine, efavirenz, rilpivirine and zidovudine when it was the only mutation, but also increased resistance to all NNRTIs when it was combined with H221Y or Y181C. Viruses with D404N or H221Y displayed impaired RC that could be compensated by Y181C. The combination of D404N, H221Y and Y181C significantly reduced susceptibility to NNRTIs, accompanied with a great loss of viral RC.
Here, D404N is described as a novel NNRTI-associated mutation because it has not been reported to be associated with NNRTI resistance in the Stanford HIV Drug Resistance Database (http://hivdb.stanford.edu) or by the International AIDS Society in the USA.27 Although D404N has not been observed in patients infected with HIV-1 subtype CRF08_BC virus, it has indeed been found in HIV-1 subtype C viruses from the sera of treatment-failure patients receiving an HAART regimen containing nevirapine and efavirenz, but not treatment-naive patients.1 In addition, E404N was also found in one patient infected by HIV-1 subtype B with reduced susceptibility to delavirdine, efavirenz or nevirapine, while E404D was detected in four patients with reduced susceptibility to delavirdine, efavirenz, nevirapine or zidovudine.8 Besides, according to the HIV Drug Resistance Database of Stanford University (http://hivdb.stanford.edu/cgi-bin/RTPosMutSummary.cgi), the substitution E404D showed a significantly higher frequency (>10%) than E404N (<2%) in ART-experienced patients. The higher frequency of E404D compared with E404N in subtype B might be explained by a codon difference. A single G-to-C transition at the third position of the codon (E404D, GAG to GAC) has a much higher chance of occurring than a two-nucleotide substitution (E404N, GAG to AAC). For HIV-1 subtypes CRF08_BC and C, however, mutation D404N requires only a single G-to-A transition (the most common RT error) at the first position of codon 404 (GAC to AAC). Since the viral genome of CRF08_BC subtype contains two short fragments with GAG (residues 17–176) and RT regions (residues 102–200) of subtype B′ inserted in the backbone of subtype C,19 the connection subdomain sequence of subtype CRF08_BC is close to that of subtype C. In this regard, the frequency of occurrence of 404N in subtypes CRF08_BC and C might be expected to be similar to and greater than, respectively, that in subtype B. D404E did not confer resistance to all the RT inhibitors, which is similar to what was found for E404 in WT of HIV subtype B and might explain why this D404E mutation did not occur in our previous selection cultures.
Previously, the emergence of D404N/H221Y in the Y181C mutant was found to significantly increase viral resistance to nevirapine.19 To further confirm whether it was due to the effect of a single D404N or H221Y mutation or to the synergetic impact of both mutations and whether it was related to the presence of Y181C, we showed in this report that a single D404N mutation conferred low-level resistance to nevirapine, efavirenz and rilpivirine, but not to etravirine. This is similar to what was found for another mutation, A400T, in the connection domain of RT. One recent study reported that A400T reduced resistance to nevirapine 5-fold and efavirenz 2-fold but did not affect etravirine resistance.28 Additionally, the N348I substitution in the connection domain was associated with resistance to nevirapine,29 while other mutations, such as E312Q, Y318F, T369I, T369V, A376S and I393L, showed low-level resistance to nevirapine but had little or no effect on efavirenz and etravirine.6 Furthermore, we found marginal resistance to zidovudine in the virus containing D404N, which was also reported for other amino acid substitutions, such as E312Q, G335C/D, N348I, A360I/V, V365I and A376S.7 The functions of single Y181C and H221Y mutations (Figure 1) in drug resistance were also investigated and the results were consistent with other studies.30–33
Next, we performed drug susceptibility assays to further confirm the influence of double (D404N/H221Y, Y181C/D404N and Y181C/H221Y) and triple (Y181C/D404N/H221Y) mutations. The addition of D404N to H221Y enabled the viruses to significantly reduce susceptibility to efavirenz, etravirine and rilpivirine, which indicated that there was synergism in resistance to NNRTIs between D404N and H221Y. When D404N or H221Y coexisted with Y181C in RT of HIV-1 subtype CRF08_BC, viral resistance to nevirapine was dramatically increased (>500-fold). Also observed were a significantly higher level of resistance to etravirine (12.6-fold for Y181C/D404N; 7.3-fold for Y181C/H221Y) and slightly increased resistance to efavirenz and rilpivirine. These results indicated that D404N and H221Y could enhance the resistance of Y181C to nevirapine and etravirine. Some studies found that the Y181C and H221Y mutations were significantly correlated with resistance to nevirapine and efavirenz, with an association P value of 7.2E-56.32,34 Therefore, this pathway warrants further analysis in clinical drug resistance mutation tests. Of note, the addition of D404N to Y181C/H221Y significantly increased resistance to etravirine and rilpivirine compared with the combination Y181C/H221Y. Lower susceptibility to nevirapine and efavirenz was observed for Y181C/D404N/H221Y compared with Y181C/H221Y, but the difference was not significant. Highest resistance to all the NNRTIs was conferred by the accumulation of these three mutations (Y181C/D404N/H221Y).
After confirming the effect of the D404N mutation on resistance to RT inhibitors, we also characterized its impact on viral RC using a single-cycle cell-culture assay in the absence of RT inhibitors. Viral RC refers to the ability of the virus to adapt and reproduce in a defined environment.35 It is different from RT activity, which refers to the whole reverse transcription process from the viral single-strand RNA genome to double-strand DNA, including RNA- and DNA-dependent DNA polymerase activity and RNase H activity.36 RC depends on multiple viral and host factors such as the events involved in the virus life cycle and selective factors such as drugs. Reverse transcription by viral RT is an important event during viral replication. Emergence of mutations in RT during therapy potentially affects viral replication. Therefore, we compared mutant viral RC with that of WT in the absence of drugs to confirm whether the mutation has any impact on viral replication. Our results showed that the single mutation D404N significantly reduced viral RC relative to that of the WT virus. This is similar to what was found for some NNRTI-related mutations, such as V106A, G190E and P236L, which were reported to confer reduced RC, possibly resulting from impaired RNase H activity.37,38 The RC of viruses containing mutation Y181C was 84.26% of that of WT, consistent with the result of another study in which the single mutation Y181C was also observed to slightly reduce viral RC (∼1.5-fold) in the backbone of pNL4-3 (infectious clone of subtype B).23 Here, the RC and RT activity of D404E were similar to those of WT, which indicates that 404E does not influence viral replication.
Although D404N alone impaired viral RC approximately by half, with the addition of Y181C to D404N the RC was restored to 82.91%. This suggests that Y181C is a compensatory mutation to D404N. It was reported that NNRTI-resistant HIV-1 viruses tend to persist in patients over long periods of time after interruption of NNRTI therapy and have a higher possibility of transmission into new hosts than PI-resistant viruses because the NNRTI-resistant viruses have small to moderate impact on viral fitness.39 Therefore, although the addition of D404N to Y181C increased resistance to NNRTIs, it did not affect viral RC, which could enable the virus to persist or readily transmit to a new host.
Interestingly, although no influence of the combination of Y181C with H221Y on viral RC was detected, the addition of D404N to Y181C/H221Y resulted in a significant loss of viral RC. In general, viral fitness decreases as the number of mutations increases.40 Our results confirmed that the strong cross-resistance to NNRTIs induced by the triple mutation Y181C/D404N/H221Y occurred at the price of a severe impairment in RC. We also found that a single H221Y substitution significantly reduced the RC to 30.95% of the WT value. However, if H221Y co-existed with Y181C, viral RC was restored to approximately the WT level and was 1.3-fold higher than that occurring with the single Y181C substitution. A similar result was reported by another group.41
Results in this report might be ascribed to conformational changes induced by D404N. According to our high-resolution crystal structure (Figure 4) of the complexes of HIV-1 RT, nevirapine and RNA/DNA hybrid, residue 404 locates in the helix αL (residues 394–404) on the interface42 and close to K30 in p51 and K431 in the p66 subunit. The change from negatively charged D404 to hydrophilic polar N404 might abolish the hydrogen bonds between D404 and K30 or K431. K30 is in the finger subdomain of p51, while K431 is in the RNase H subdomain in the p66 subunit. The structure between the connection and the finger subdomain in p51, as well as the structure between the connection and the RNase H subdomain in the p66 subunit, might be impaired and result in reduction of susceptibility to RT inhibitors. This structural change might also influence the RNase H activity of RT and affect viral RC. In our study, Y181C also did not produce significant loss of polymerase activity, which is consistent with the results of other studies.43,44
In summary, we identified and characterized a novel NNRTI-associated mutation, D404N, in the connection subdomain in the RT of HIV-1 subtype CRF08_BC. This is important because it is very possible for the mutation D404N to emerge in subtype CRF08_BC because it has been found in patients infected with HIV-1 subtype C and the connection subdomains of these two subtypes are very similar. However, the emergence frequency of the mutation D404N in patients infected with HIV-1 CRF08_BC subtype still needs to be demonstrated in more clinical studies; besides nevirapine and efavirenz, D404N also reduced susceptibility to the second-generation NNRTI rilpivirine by itself or in the presence of other mutations, such as Y181C and H221Y. In addition, this mutation could confer low-level resistance to zidovudine. Thus, the presence of this mutation alone or in combination with Y181C might potentially lead to failure of HAART therapy including NNRTI; because Y181C could compensate for the replication loss of D404N, viruses with these two mutations might persist in the host for a long time and be transmitted to a new host. CRF08_BC has become a prevalent subtype in China and studies of novel drug-resistance mutations in this subtype have become increasingly important. The results of this study are valuable because they facilitate the development of novel RT inhibitors and the prevention of therapy failure.
Funding
This work was supported in part by grants from the AIDS Trust Fund of Hong Kong Special Administrative Region of the People's Republic of China.
Transparency declarations
None to declare.
Disclaimer
The results and conclusions expressed in this article are views of the authors alone and do not necessarily reflect the opinions of the Department of Health and Human Services. The funders had no role in study design, data collection and analysis, the decision to publish or preparation of the manuscript.
Acknowledgements
We are highly appreciative of the resources made available by the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH; these include TZM-bl from Dr John C. Kappes, Dr Xiaoyun Wu and Tranzyme Inc., and the RT inhibitors zidovudine, lamivudine, abacavir, emtricitabine tenofovir, nevirapine, efavirenz, etravirine and rilpivirine.
References
- 1.Brehm JH, Koontz DL, Wallis CL, et al. Frequent emergence of N348I in HIV-1 subtype C reverse transcriptase with failure of initial therapy reduces susceptibility to reverse-transcriptase inhibitors. Clin Infect Dis 2012; 55: 737–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Menendez-Arias L. Mechanisms of resistance to nucleoside analogue inhibitors of HIV-1 reverse transcriptase. Virus Res 2008; 134: 124–46. [DOI] [PubMed] [Google Scholar]
- 3.Cullen MD, Ho WC, Bauman JD, et al. Crystallographic study of a novel subnanomolar inhibitor provides insight on the binding interactions of alkenyldiarylmethanes with human immunodeficiency virus-1 reverse transcriptase. J Med Chem 2009; 52: 6467–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delviks-Frankenberry KA, Nikolenko GN, Pathak VK. The “connection” between HIV drug resistance and RNase H. Viruses 2010; 2: 1476–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang MW, Shafer RW. HIV-1 antiretroviral resistance: scientific principles and clinical applications. Drugs 2012; 72: e1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Menendez-Arias L, Betancor G, Matamoros T. HIV-1 reverse transcriptase connection subdomain mutations involved in resistance to approved non-nucleoside inhibitors. Antiviral Res 2011; 92: 139–49. [DOI] [PubMed] [Google Scholar]
- 7.Nikolenko GN, Delviks-Frankenberry KA, Palmer S, et al. Mutations in the connection domain of HIV-1 reverse transcriptase increase 3′-azido-3′-deoxythymidine resistance. Proc Natl Acad Sci USA 2007; 104: 317–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta S, Fransen S, Paxinos EE, et al. Combinations of mutations in the connection domain of human immunodeficiency virus type 1 reverse transcriptase: assessing the impact on nucleoside and nonnucleoside reverse transcriptase inhibitor resistance. Antimicrob Agents Chemother 2010; 54: 1973–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hachiya A, Kodama EN, Sarafianos SG, et al. Amino acid mutation N348I in the connection subdomain of human immunodeficiency virus type 1 reverse transcriptase confers multiclass resistance to nucleoside and nonnucleoside reverse transcriptase inhibitors. J Virol 2008; 82: 3261–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sluis-Cremer N, Temiz NA, Bahar I. Conformational changes in HIV-1 reverse transcriptase induced by nonnucleoside reverse transcriptase inhibitor binding. Curr HIV Res 2004; 2: 323–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delviks-Frankenberry KA, Nikolenko GN, Boyer PL, et al. HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc Natl Acad Sci USA 2008; 105: 10943–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nikolenko GN, Palmer S, Maldarelli F, et al. Mechanism for nucleoside analog-mediated abrogation of HIV-1 replication: balance between RNase H activity and nucleotide excision. Proc Natl Acad Sci USA 2005; 102: 2093–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liao L, Xing H, Shang H, et al. The prevalence of transmitted antiretroviral drug resistance in treatment-naive HIV-infected individuals in China. J Acquir Immune Defic Syndr 2010; 53 Suppl 1: S10–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu L, Jia M, Ma Y, et al. The changing face of HIV in China. Nature 2008; 455: 609–11. [DOI] [PubMed] [Google Scholar]
- 15.Ouyang Y, Shao Y, Ma L. HIV-1 CRF_BC recombinants infection in China: molecular epidemic and characterizations. Curr HIV Res 2012; 10: 151–61. [DOI] [PubMed] [Google Scholar]
- 16.Sui HS, Gui T, Jia L, et al. Different frequencies of drug resistance mutations among HIV-1 subtypes circulating in China: a comprehensive study. PLoS One 2014; 9: e91803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen M, Ma Y, Duan S, et al. Genetic diversity and drug resistance among newly diagnosed and antiretroviral treatment-naive HIV-infected individuals in western Yunnan: a hot area of viral recombination in China. BMC Infect Dis 2012; 12: 382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li H, Zhong M, Guo W, et al. Prevalence and mutation patterns of HIV drug resistance from 2010 to 2011 among ART-failure individuals in the Yunnan Province, China. PLoS One 2013; 8: e72630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu H, Zhang XM, Zhang HJ, et al. In vitro selection of HIV-1 CRF08_BC variants resistant to reverse transcriptase inhibitors. AIDS Res Hum Retro 2014; Epub ahead of print Dec 2014; doi: 10.1089/AID.2013.0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Q, Zhang X, Wu H, et al. Parental LTRs are important in a construct of a stable and efficient replication-competent infectious molecular clone of HIV-1 CRF08_BC. PLoS One 2012; 7: e31233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang HJ, Wang YX, Wu H, et al. The Y271 and I274 amino acids in reverse transcriptase of human immunodeficiency virus-1 are critical to protein stability. PLoS One 2009; 4: e6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Zhang HJ, Zhang XM, et al. Identification of drug resistant mutations in HIV-1 CRF07_BC variants selected by nevirapine in vitro. PLoS One 2012; 7: e44333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Asahchop EL, Wainberg MA, Oliveira M, et al. Distinct resistance patterns to etravirine and rilpivirine in viruses containing nonnucleoside reverse transcriptase inhibitor mutations at baseline. AIDS 2013; 27: 879–87. [DOI] [PubMed] [Google Scholar]
- 24.Xu HT, Quan Y, Schader SM, et al. The M230L nonnucleoside reverse transcriptase inhibitor resistance mutation in HIV-1 reverse transcriptase impairs enzymatic function and viral replicative capacity. Antimicrob Agents Chemother 2010; 54: 2401–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Das K, Martinez SE, Bandwar RP, et al. Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage. Nucleic Acids Res 2014; 42: 8125–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold K, Bordoli L, Kopp J, et al. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 2006; 22: 195–201. [DOI] [PubMed] [Google Scholar]
- 27.Johnson VA, Calvez V, Gunthard HF, et al. Update of the drug resistance mutations in HIV-1: March 2013. Top Antivir Med 2013; 21: 6–14. [PMC free article] [PubMed] [Google Scholar]
- 28.Wright DW, Deuzing IP, Flandre P, et al. A polymorphism at position 400 in the connection subdomain of HIV-1 reverse transcriptase affects sensitivity to NNRTIs and RNaseH activity. PLoS One 2013; 8: e74078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yap SH, Sheen CW, Fahey J, et al. N348I in the connection domain of HIV-1 reverse transcriptase confers zidovudine and nevirapine resistance. PLoS Med 2007; 4: e335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Azijn H, Tirry I, Vingerhoets J, et al. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob Agents Chemother 2010; 54: 718–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anta L, Llibre JM, Poveda E, et al. Rilpivirine resistance mutations in HIV patients failing non-nucleoside reverse transcriptase inhibitor-based therapies. AIDS 2013; 27: 81–5. [DOI] [PubMed] [Google Scholar]
- 32.Reuman EC, Rhee SY, Holmes SP, et al. Constrained patterns of covariation and clustering of HIV-1 non-nucleoside reverse transcriptase inhibitor resistance mutations. J Antimicrob Chemother 2010; 65: 1477–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asahchop EL, Oliveira M, Wainberg MA, et al. Characterization of the E138K resistance mutation in HIV-1 reverse transcriptase conferring susceptibility to etravirine in B and non-B HIV-1 subtypes. Antimicrob Agents Chemother 2011; 55: 600–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ceccherini-Silberstein F, Svicher V, Sing T, et al. Characterization and structural analysis of novel mutations in human immunodeficiency virus type 1 reverse transcriptase involved in the regulation of resistance to nonnucleoside inhibitors. J Virol 2007; 81: 11507–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.De Luca A. The impact of resistance on viral fitness and its clinical implications. In: Geretti AM, ed. Antiretroviral Resistance in Clinical Practice. London: Mediscript, 2006. Chapter 12. http://www.ncbi.nlm.nih.gov/books/NBK2244/. [PubMed] [Google Scholar]
- 36.Esposito F, Corona A, Tramontano E. HIV-1 reverse transcriptase still remains a new drug target: structure, function, classical inhibitors, and new inhibitors with innovative mechanisms of actions. Mol Biol Int 2012; 2012: 586401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W, Wrin T, Gamarnik A, et al. Reverse transcriptase mutations that confer non-nucleoside reverse transcriptase inhibitor resistance may also impair replication capacity. Antivir Ther 2002; 7: S79 (Abstract 72). [Google Scholar]
- 38.Martinez-Picado J, Martinez MA. HIV-1 reverse transcriptase inhibitor resistance mutations and fitness: a view from the clinic and ex vivo. Virus Res 2008; 134: 104–23. [DOI] [PubMed] [Google Scholar]
- 39.de Bethune MP. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection: a review of the last 20 years (1989–2009). Antiviral Res 2010; 85: 75–90. [DOI] [PubMed] [Google Scholar]
- 40.Collins JA, Thompson MG, Paintsil E, et al. Competitive fitness of nevirapine-resistant human immunodeficiency virus type 1 mutants. J Virol 2004; 78: 603–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yin LL, Han XX, Zhao B, et al. Impact of HIV-1 reverse transcriptase H221Y mutation on replication capacity of Y181C mutant viral strain. Chin J Microb Immunol 2013; 33: 423–8. [Google Scholar]
- 42.Lapkouski M, Tian L, Miller JT, et al. Complexes of HIV-1 RT, NNRTI and RNA/DNA hybrid reveal a structure compatible with RNA degradation. Nat Struct Mol Biol 2013; 20: 230–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Archer RH, Dykes C, Gerondelis P, et al. Mutants of human immunodeficiency virus type 1 (HIV-1) reverse transcriptase resistant to nonnucleoside reverse transcriptase inhibitors demonstrate altered rates of RNase H cleavage that correlate with HIV-1 replication fitness in cell culture. J Virol 2000; 74: 8390–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kroeger Smith MB, Michejda CJ, Hughes SH, et al. Molecular modeling of HIV-1 reverse transcriptase drug-resistant mutant strains: implications for the mechanism of polymerase action. Protein Eng 1997; 10: 1379–83. [DOI] [PubMed] [Google Scholar]