Abstract
Objectives
Although founder viruses in primary HIV-1 infections (PHIs) typically use the CCR5 coreceptor (R5-tropic), 3%–19% of subjects also harbour CXCR4-using viruses (X4-tropic), making tropism determination before CCR5 antagonist usage mandatory. Genotypic methods can be used to accurately determine HIV-1 tropism in chronically infected patients.
Methods
We compared the results of genotypic methods [geno2pheno, PSSMx4r5 including a novel nucleotide-input version (ntPSSM) and distant segments (ds)Kernel] to predict coreceptor usage in a cohort of 67 PHIs. Specimens with discrepant results were phenotypically tested after cloning the V3 gene region into proviral backbones. Recombinant viruses were used to infect U87 indicator cell lines bearing CD4 and either CCR5 or CXCR4.
Results
Geno2pheno10%, PSSMx4r5 and (ds)Kernel gave identical predictions in 85% of cases. Geno2pheno10% predicted the presence of CXCR4 viruses in 18% of patients. Two patients were predicted to carry X4-tropic viruses by all algorithms and X4-tropic viruses were detected in at least one of the recombinant AD8 or NL4-3 backbone-based assays. Ten samples resulted in discordant predictions with at least one algorithm. Full concordance between tropism prediction by using population sequencing and phenotypic assays was observed only with ntPSSM. Geno2pheno prediction and the phenotypic assay gave the same results in a minority of ‘discordant’ patients.
Conclusions
Compared with both PSSMx4r5 versions, (ds)Kernel and our phenotypic assay, geno2pheno10% overestimated the frequency of X4-tropic viruses (18% versus 3%). ntPSSM was able to detect one additional X4 virus compared with (ds)Kernel that was confirmed with the phenotypic assay.
Keywords: HIV tropism, maraviroc, primary HIV-1 infection, X4-tropic, CCR5-tropic
Introduction
Depending on the affinity for the CCR5 or CXCR4 coreceptor, HIV strains can be classified either as R5-tropic or as X4-tropic variants or as R5X4 dual-tropic if both coreceptors can be used.1 Current guidelines require viral tropism determination before CCR5 antagonist (maraviroc) initiation, both in treatment-experienced and in treatment-naive patients.2,3
Although the majority of strains observed during primary HIV infection (PHI) are found to be exclusively R5-tropic,4,5 X4/R5X4 viruses continue to be detected during HIV-1 PHI with frequencies of 3%–17.2%, depending on the assay used to measure HIV-1 tropism.5,6 Indeed, viral tropism can be determined both through phenotypic and genotypic assays. Phenotypic assays are expensive and time consuming and for these reasons they have been progressively replaced by genotypic assays. These tests predict viral coreceptor tropism based on sequence analysis of the V3 region.7 Currently, geno2pheno,8–10 PSSMx4r5 (PSSM)11,12 and distant segments (ds)Kernel13 are the most frequently used bioinformatics tools for coreceptor prediction in routine laboratories. Here we report a new version of PSSM that allows direct uploading of nucleotide sequences (including those with mixed bases) to perform tropism prediction on all permutated combinations (ntPSSM).
Recent data from our group showed that in PHI a high level of discrepancy is observed between the genotypic assays.14 Thus, in this novel paper, for the first time in the setting of PHI, we analysed those V3 loop sequences that were predicted to belong to CXCR4-using variants by any of the genotypic methods available and compared these results with an in-house phenotypic assay using fully replication competent viruses.
Materials and methods
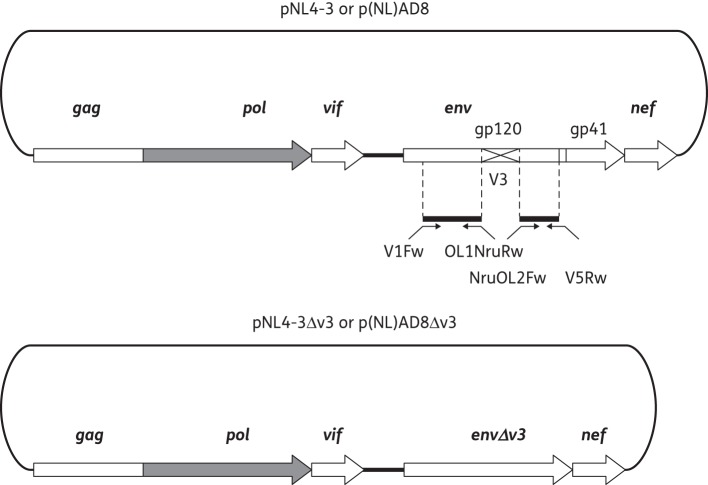
The study included 67 patients with PHI, after approval by the San Raffaele Ethics Committee and patients' informed consent. PHI was defined on the basis of positive plasma HIV RNA and a negative/indeterminate western blot assay. Samples were collected 21 (10–31) days after diagnosis [CD4 count 401 (311–489) cells/mm3 and HIV RNA 5.08 (4.73–5.66) log10 copies/mL]. Viral RNA from the first available plasma sample was extracted, reverse transcribed, PCR amplified, sequenced directly and checked for contamination as described previously.15,16 For all V3 loop sequences, including each cloned variant, HIV-1 coreceptor usage was determined using the following bioinformatics predictors: the geno2pheno algorithm available at http://coreceptor.bioinf.mpi-inf.mpg.de/ adopting a false positive rate (FPR) of 10%; webPSSM (position scoring specific matrix) and ntPSSM available at http://indra.mullins.microbiol.washington.edu/webpssm/; and (ds)Kernel available at http://genome.ulaval.ca/hiv-dskernel. The phenotypic assay used in the text is an improved version of our previously published assays.17–19 Indeed, the NruI cloning site that was introduced into the proviral backbones does not lead to additional artificial amino acid changes. To reduce potential biases due to the contribution of non-V3 regions to coreceptor tropism, two backbones were generated based on R5- or X4-tropic laboratory proviral clones (Figure 1). V3 sequences were then amplified by nested PCR from the gp120 amplicons or from molecular clones NL4-3 (X4 phenotype) and AD8 (R5 phenotype), and cloned into the NruI-digested vectors p(NL)AD8ΔV3 or pNL4-3ΔV320 and used to transform XL1-Blue electrocompetent cells. An aliquot of the transformed culture was plated onto LB agar while the remaining liquid culture was grown overnight. Plasmid DNA from the bulk culture and individual clones were then purified. U87.CD4.CCR5 and U87.CD4.CXCR4 cells (NIH AIDS Reagent Program) were plated in 24-well plates (Nunc) at a density of 40 000 cells/well and independently transfected 24 h later with 500 ng of each recombinant plasmid carrying a patient's V3 region in both NLgp120 or AD8gp120 vectors (X-tremeGENE 9, Roche). Alternatively, an aliquot of the bulk ligation-derived miniprep was used to transform indicator cell lines to evaluate the population for the presence of minor variants not detected by clonal analysis. Cultures were examined daily for syncytia formation until day 9 (see Supplementary Methods available at JAC Online).
Figure 1.
Construction of ΔV3 proviral backbone vectors. Construction of p(NL)AD8ΔV3 and pNLΔV3 vectors used to generate replicating recombinant viruses following insertion of patient-derived V3 sequences. Two ΔV3 vectors were generated starting from two proviral laboratory backbones: pNL4-3 (X4-tropic) or p(NL)AD8 (R5-tropic). For further details see the Supplementary Methods available at JAC Online.
Results
Geno2pheno10%, PSSM and (ds)Kernel gave identical predictions in 57 (85%) cases. In one otherwise discordant case, (ds)Kernel could not be used due to the complexity of the population sequencing result. Fifty-five population-based sequences were predicted to encode R5 viruses with all algorithms. Two patients were predicted to carry X4-tropic viruses by all algorithms. Geno2pheno10% predicted the presence of CXCR4 viruses in 12 (18%) patients. Ten samples, all from subtype B viruses, resulted in discordant predictions with at least one algorithm. From the sequences amplified from these 10 patients, V3 loops were all predicted to be derived from X4 viruses by geno2pheno10%, whereas both (ds)Kernel and PSSM predicted R5 tropism. Interestingly, ntPSSM identified one additional X4 variant (later confirmed by phenotypic analysis) present in the mixed viral population of Patient 5 (Table 1). V3 amplification was successful in 91% (11/12) of the cases evaluated. Ten to 20 clones were sequenced for each patient. Only in two patients (Patients 5 and 12) with mixed bases detected by population sequencing, did more than one clone have to be phenotypically (and genotypically) tested. When using the NL4-3 backbone, V3 loop sequences from Patients 1 (R5 in the AD8 context) and 4 (dual-tropic in the AD8 context) did not result in replication-competent virus, suggesting that an R5 background was necessary and that the analysis of the whole envelope would more closely reflect in vivo tropism. Phenotypic assays performed with the mixed populations of recombinant viruses were concordant with the results obtained by clonal phenotypic assays except for Patient 5, in whom a cloned X4-tropic variant was not detected. The two V3 loops predicted to belong to X4 viruses by all of the genotypic algorithms resulted in X4-tropic viruses in at least one of the recombinant AD8 or NL4-3 backbone-based assays. Using the clonal analysis, we found that these patients carried R5X4 dual-tropic viruses rather than a mixture of solely R5- and X4-tropic variants. One patient (Patient 5) harboured a dual-tropic virus that was correctly predicted only by ntPSSM and geno2pheno. The remaining seven patients harboured R5-tropic viruses (Patients 1, 6, 7, 8, 9, 11 and 12).
Table 1.
Genotypic and phenotypic results
| Genotypic assay |
Phenotype (AD8-bb) |
Phenotype (NL-bb) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patient | G2PH | G2PH FPR% | PSSM | ntPSSMa | (ds)KERNEL | V3 amino acid sequence | U87-CCR5 | U87-CXCR4 | U87-CCR5 | U87-CXCR4 |
| 1 | CXCR4 | 6.8 | CCR5 | CCR5 | CCR5 | CTRPNNNTRKSVHIGPGSAIYTTDIIGDIRKAHC | +++ | − | − | − |
| CTRPNNNTRKSVHIGPGSAIYTTDIIGDIRKAHC | +++ | − | − | − | ||||||
| 2 | CXCR4 | 5 | CCR5 | CCR5 | CCR5 | CTRPGNNTRKGIHMGPGRAFYTTEDIIGDIRQAHC | − | − | − | − |
| CTRPGNNTRKGIHMGPGRAFYTTEDIIGDIRQAHC | − | − | − | − | ||||||
| 3 | CXCR4 | 1.1 | CXCR4 | CXCR4 | CCR5/CXCR4 | CTRPYNNRRQRIHIGPGRAFYATGNILGDIRKAHC | + | +++ | ++ | + |
| CXCR4 | 1.7 | CXCR4 | CXCR4 | CCR5/CXCR4 | CTRPYNNRRQRIHIGPGRAFYATGNILGDIRKAHC | + | +++ | − | ++ | |
| 4 | CXCR4 | 0.2 | CXCR4 | CXCR4 | CCR5/CXCR4 | CTRPNNNTRRRISIGPGRAFYAREQIIGDIRQAHC | + | ++ | − | − |
| CTRPNNNTRRRISIGPGRAFYAREQIIGDIRQAHC | ++ | ++ | − | − | ||||||
| 5 | CXCR4 | 8.7 | CCR5 | CCR5/CXCR4 | CCR5 | CTRPNNNTRKSIQLGPGRAFYTTGK/T/E/AIIGDIRKAHC | +++ | − | +++ | − |
| CXCR4 | 3.9 | CXCR4 | CXCR4 | CCR5 | CTRPNNNTRKSIQLGPGRAFYTTGKIIGDIRKAHC | +++ | ++ | +++ | ++ | |
| CCR5 | 15 | CCR5 | CCR5 | CCR5 | CTRPNNNTRKSIQLGPGRAFYTTGAIIGDIRKAHC | +++ | − | +++ | − | |
| 6 | CXCR4 | 7 | CCR5 | CCR5 | CCR5 | CTRPSNNTRRSIHIGPGRAFYATGVLIGDPRKAHC | ++ | − | ++ | − |
| CTRPSNNTRRSIHIGPGRAFYATGVLIGDPRKAHC | +++ | − | +++ | − | ||||||
| 7 | CXCR4 | 3.8 | CCR5 | CCR5 | CCR5 | CTRPNNNTRKSISIGPGRAFYAHRDVIGDIRQAHC | +++ | − | ++ | − |
| CTRPNNNTRKSISIGPGRAFYAHRDVIGDIRQAHC | +++ | − | +++ | − | ||||||
| 8 | CXCR4 | 9.9 | CCR5 | CCR5 | CCR5 | CTRPNNNTRRSIHIGPGRAVYTTGEIIGDIRQAHC | +++ | − | +++ | − |
| CTRPNNNTRRSIHIGPGRAVYTTGEIIGDIRQAHC | +++ | − | +++ | − | ||||||
| 9 | CXCR4 | 9.8 | CCR5 | CCR5 | CCR5 | CTRPNNNTRRSIHVGPGGAIYTAGNIIGDIRQAHC | +++ | − | ++ | − |
| CTRPNNNTRRSIHVGPGGAIYTAGNIIGDIRQAHC | +++ | − | ++ | − | ||||||
| 10 | CXCR4 | 6.4 | CCR5 | CCR5 | ND | CTRPN/HNNT/RRK/RSI/LN/HI/LGPGK/RAFYTTGDIIGDIRQ/RAH/P/Y/SC | ND | ND | ND | ND |
| 11 | CXCR4 | 8.2 | CCR5 | CCR5 | CCR5 | CTRPNNNTRKGIHTAWGRAFYVTGDIIGDIRQAHC | +++ | − | + | − |
| CTRPNNNTRKGIHTAWGRAFYVTGDIIGDIRQAHC | +++ | − | ++ | − | ||||||
| 12 | CXCR4 | 2.6 | CCR5 | CCR5 | CCR5 | CARPGNNTRKGIHIGPGK/RAFYTTGIIGDIRKAHC | − | − | +++ | − |
| CXCR4 | 2.6 | CCR5 | CCR5 | CCR5 | CARPGNNTRKGIHIGPGRAFYTTGIIGDIRKAHC | + | − | +++ | − | |
| CXCR4 | 2.7 | CCR5 | CCR5 | CCR5 | CARPGNNTRKGIHIGPGKAFYTTGIIGDIRKAHC | ++ | − | ND | ND | |
| V3 NL | CXCR4 | 0.5 | CXCR4 | CXCR4 | CXCR4 | CTRPNNNTRKSIRIQRGPGRAFVTIGKIGNMRQAHC | − | − | − | ++ |
| V3 AD8 | CCR5 | 35 | CCR5 | CCR5 | CCR5 | CTRPNNNTRKSIHIGPGRAFYTTGDIIGDIRQAHC | ++ | − | ++ | − |
G2PH, geno2pheno; FPR, false positive rate; bb, backbone; ND, not done.
Number of syncytia is semi-quantitatively indicated (+, very low; ++, moderate; +++, high). Minus sign (−) indicates absence of syncytia. For each patient, the V3 loop sequences resulting from population sequencing (in bold) and from each clone (in italics) are reported.
All of the clones derived from Patient 2 were incapable of replicating in vitro. Amplification with extensively degenerate primers in Patient 10 gave rise only to defective V3 sequences that could not be further evaluated. In agreement with previous data, control viruses obtained by cloning the NL4-3 V3 sequence into the AD8 backbone were non-functional, while the chimeric virus obtained by cloning the AD8 V3 sequence into the NL4-3 backbone maintained the same coreceptor usage as the donor virus.19
aVersion of ntPSSM implemented for acceptance of nucleotide sequences, including ambiguous bases, as well as amino acid sequences.
If only the detection of CXCR4-using strains (X4- or dual-tropic) is considered, concordance between the phenotypic result and the genotype inferred by PSSM and (ds)Kernel increased to almost 100%. Full concordance between tropism prediction by using population sequencing and phenotypic assays was observed only with ntPSSM. Geno2pheno10% prediction and the phenotypic assay gave the same results in only 20% of ‘discordant’ patients (30% including Patient 5). However, if a lower FPR (such as 5) is selected, geno2pheno predicted X4 tropism in only four (6%) ‘discordant’ patients (namely Patients 3, 4, 7 and 12).
Discussion
Both the US Department of Health and Human Services and the European AIDS Clinical Society antiretroviral guidelines consider the use of maraviroc as an acceptable or alternative regimen for antiretroviral-naive patients.2,3 In our phenotypic assay, we found 3% CXCR4-bearing viruses (2/67) [or 4.5% (3/67) considering Patient 5 as carrying an X4 virus], in agreement with other studies based on phenotypic evaluations in PHI. Only the V3 loop of the entire envelope was cloned into the proviral vectors and this may have impaired in some cases the recovery of the replication capacity or the real tropism of all recombinant viruses even if both R5- and X4-tropic backbones were generated to limit this issue.19 Moreover, even if recombinant viruses are allowed to replicate for several days in indicator cell lines, we cannot exclude for the population analysis of tropism that the presence of minority variants could be missed (such as for Patient 5). Considering our results, for geno2pheno, an FPR of 10%, as currently suggested by European guidelines, seems too high for correct coreceptor prediction in PHI. In fact, in seven patients for which geno2pheno10% predicted the presence of CXCR4 variants, only R5-tropic clonal variants were detected.
The different results obtained with the three algorithms may be due to the different statistical models used for predictions and partially to how they handle insertions, deletions and ambiguous positions. Because geno2pheno and the latest version of webPSSM (referred to as ntPSSM) allow the input of nucleotide sequences for tropism prediction, permutations, alignment and translation are performed automatically and positions with different amino acids can be scored independently. A common drawback of both geno2pheno and ntPSSM is that when all possible amino acid combinations are inferred from a mixed nucleotide population, they could infer tropism for sequences that do not really exist. In the context of PHI, a mono-/oligoclonal viral population is mostly detected, but this will be a greater problem in chronically infected patients where complex quasispecies are present and where deep-sequencing technologies may be more suited to characterize the viral population.
In conclusion, since geno2pheno10% X4 overestimation will preclude CCR5 antagonist usage in patients that could benefit from this treatment in the early stage of HIV infection, and despite the low number of samples tested in the present work, other genotypic tools such as ntPSSM and Kernel should be used in this particular category of subjects. Alternatively, a lower and more specific geno2pheno cut-off should be selected, i.e. an FPR of 5%. Indeed, decreasing the FPR to 5% in our study increased the concordance with the phenotypic assay from 20% to 80% and shifted the frequency of CXCR4-tropic virus to 6% (Table 1).
Funding
This study was partially funded by the Italian Ministry of Health, by the Italian Ministry of University and Research and by the Functional Profiling and Computational Biology Core of the Seattle Center for AIDS Research (NIH P30 AI027757).
Transparency declarations
None to declare.
Supplementary data
Supplementary Methods are available at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
These data were presented in part at the VI Italian Conference on AIDS and Retroviruses, Rome, 25–27 May 2014, OC-70.
References
- 1.Berger EA, Doms RW, Fenyo EM, et al. A new classification for HIV-1. Nature 1998; 391: 240. [DOI] [PubMed] [Google Scholar]
- 2.Vandekerckhove LP, Wensing AM, Kaiser R, et al. European guidelines on the clinical management of HIV-1 tropism testing. Lancet Infect Dis 2011; 11: 394–407. [DOI] [PubMed] [Google Scholar]
- 3.Thompson MA, Aberg JA, Hoy JF, et al. Antiretroviral treatment of adult HIV infection: 2012 recommendations of the International Antiviral Society-USA panel. JAMA 2012; 308: 387–402. [DOI] [PubMed] [Google Scholar]
- 4.Roos MT, Lange JM, de Goede RE, et al. Viral phenotype and immune response in primary human immunodeficiency virus type 1 infection. J Infect Dis 1992; 165: 427–32. [DOI] [PubMed] [Google Scholar]
- 5.Zhu T, Mo H, Wang N, et al. Genotypic and phenotypic characterization of HIV-1 patients with primary infection. Science 1993; 261: 1179–81. [DOI] [PubMed] [Google Scholar]
- 6.Raymond S, Delobel P, Mavigner M, et al. Correlation between genotypic predictions based on V3 sequences and phenotypic determination of HIV-1 tropism. AIDS 2008; 22: F11–6. [DOI] [PubMed] [Google Scholar]
- 7.Cardozo T, Kimura T, Philpott S, et al. Structural basis for coreceptor selectivity by the HIV type 1 V3 loop. AIDS Res Hum Retroviruses 2007; 23: 415–26. [DOI] [PubMed] [Google Scholar]
- 8.Lengauer T, Sander O, Sierra S, et al. Bioinformatics prediction of HIV coreceptor usage. Nat Biotechnol 2007; 25: 1407–10. [DOI] [PubMed] [Google Scholar]
- 9.Sing T, Low AJ, Beerenwinkel N, et al. Predicting HIV coreceptor usage on the basis of genetic and clinical covariates. Antivir Ther 2007; 12: 1097–106. [PubMed] [Google Scholar]
- 10.Sander O, Sing T, Sommer I, et al. Structural descriptors of gp120 V3 loop for the prediction of HIV-1 coreceptor usage. PLoS Comput Biol 2007; 3: e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen MA, van 't Wout AB. Predicting HIV-1 coreceptor usage with sequence analysis. AIDS Rev 2003; 5: 104–12. [PubMed] [Google Scholar]
- 12.Mullins JI, Jensen MA. Evolutionary dynamics of HIV-1 and the control of AIDS. Curr Top Microbiol Immunol 2006; 299: 171–92. [DOI] [PubMed] [Google Scholar]
- 13.Boisvert S, Marchand M, Laviolette F, et al. HIV-1 coreceptor usage prediction without multiple alignments: an application of string kernels. Retrovirology 2008; 5: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nozza S, Canducci F, Galli L, et al. Viral tropism by geno2pheno as a tool for predicting CD4 decrease in HIV-1-infected naive patients with high CD4 counts. J Antimicrob Chemother 2012; 67: 1224–7. [DOI] [PubMed] [Google Scholar]
- 15.Svicher V, D'Arrigo R, Alteri C, et al. Performance of genotypic tropism testing in clinical practice using the enhanced sensitivity version of Trofile as reference assay: results from the OSCAR Study Group. New Microbiol 2010; 33: 195–206. [PubMed] [Google Scholar]
- 16.Boeri E, Canducci F, Grasso MA, et al. Phylogenetic internal control for HIV-1 genotypic antiretroviral testing. New Microbiol 2004; 27 (Suppl 1): 105–9. [PubMed] [Google Scholar]
- 17.Canducci F, Ceresola ER, Saita D, et al. In vitro phenotypes to elvitegravir and dolutegravir in primary macrophages and lymphocytes of clonal recombinant viral variants selected in patients failing raltegravir. J Antimicrob Chemother 2013; 68: 2525–32. [DOI] [PubMed] [Google Scholar]
- 18.Canducci F, Marinozzi MC, Sampaolo M, et al. Genotypic/phenotypic patterns of HIV-1 integrase resistance to raltegravir. J Antimicrob Chemother 2010; 65: 425–33. [DOI] [PubMed] [Google Scholar]
- 19.Bagnarelli P, Fiorelli L, Vecchi M, et al. Analysis of the functional relationship between V3 loop and gp120 context with regard to human immunodeficiency virus coreceptor usage using naturally selected sequences and different viral backbones. Virology 2003; 307: 328–40. [DOI] [PubMed] [Google Scholar]
- 20.Canducci F, Marinozzi MC, Sampaolo M, et al. Dynamic features of the selective pressure on the human immunodeficiency virus type 1 (HIV-1) gp120 CD4-binding site in a group of long term non progressor (LTNP) subjects. Retrovirology 2009; 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.