Abstract
The use of paramagnetic constraints in protein NMR is an active area of research because of the benefits of long-range distance measurements (>10 Å). One of the main issues in successful execution is the incorporation of a paramagnetic metal ion into diamagnetic proteins. The most common metal ion tags are relatively long aliphatic chains attached to the side chain of a selected cysteine residue with a chelating group at the end where it can undergo substantial internal motions, decreasing the accuracy of the method. An attractive alternative approach is to incorporate an unnatural amino acid (UAA) that binds metal ions at a specific site on the protein using the methods of molecular biology. Here we describe the successful incorporation of the unnatural amino acid 2-amino-3-(8-hydroxyquinolin-3-yl) propanoic acid (HQA) into two different membrane proteins by heterologous expression in E. coli. Fluorescence and NMR experiments demonstrate complete replacement of the natural amino acid with HQA and stable metal chelation by the mutated proteins. Evidence of site-specific intra- and inter-molecular PREs by NMR in micelle solutions sets the stage for the use of HQA incorporation in solid-state NMR structure determinations of membrane proteins in phospholipid bilayers.
Keywords: membrane protein, micelle, unnatural amino acid, UAA, PRE, protein structure, HQA, CXCR1, p7
Introduction
Many analogies can be drawn between the earliest protein NMR studies and present day studies of proteins in biological supramolecular assemblies, such as membrane proteins, amyloid fibrils, chromatin, etc. Both were severely limited by resolution and sensitivity of the spectra, and the ability to interpret the data in terms of the three-dimensional structures of the proteins, which has always been the major goal of protein NMR spectroscopy. Starting with the initial NMR spectrum of a protein in solution (Saunders et al. 1957), which consisted of four broad, overlapping signals, it was clear that additional steps were needed to extract the underlying spectroscopic and structural information. Some gains resulted from increasing the 1H resonance frequency and the introduction of signal averaging of the continuous wave signals (Meadows et al. 1967). Nonetheless, the early demonstrations of protein NMR were limited to a few residues whose resonances could be resolved in the spectra of the most favorable globular proteins available in large quantities at the time, for example ribonuclease and lysozyme. The introduction of isotopic labeling of proteins was a major step forward in attaining both improved spectral resolution and resonance assignments (Markley et al. 1968) but was restricted to selected examples, such as Staphylococcal nuclease because heterologous expression of proteins was not yet feasible. The only available spectral parameters were the isotropic chemical shift frequencies, which varied among amino acid residues because of the differences in environment resulting from protein folding, and resonance line widths that reflected protein dynamics. The first spectra of membrane proteins were not reported for another ten years (Cross and Opella 1979; Cross and Opella 1980; Hagen et al. 1978; Hagen et al. 1979).
In the early 1970s the addition of directly bound paramagnetic species made a substantial difference in the prospects for NMR of proteins. Two similar, parallel paths were introduced. McConnell and coworkers (Morrisett et al. 1973; Wien et al. 1972) exploited the stable paramagnetic center of 1-oxyl-2,2,6,6,-tetramethyl-4-piperidinyl (TEMPO) to covalently label lysozyme and then measure the broadening effects on resonances of bound ligands. The goal then, as now, was to make direct distance measurements between the electron spin-label and nuclei at covalently bonded sites on the protein. However, this was not possible because the studies were limited by the experiments being performed at the relatively low field strength corresponding to a 1H resonance frequency of 100 MHz and other technical issues. Around the same time, Campbell and coworkers (Campbell et al. 1973) were able to convincingly demonstrate the ability of a lanthanide ion (Gd3+) bound to lysozyme to selectively broaden resonances from residues proximate to the binding site, aided in large part by performing the experiments at the significantly higher 1H resonance frequency of 270 MHz. These early experiments that exploited the broadening effects of paramagnetic species, whether TEMPO-containing spin labels or lanthanide ions bound to protein ligands, were key predecessors for the current activity in paramagnetic NMR of proteins.
Solution NMR of proteins has advanced significantly over the past forty years based on improvements in instrumentation, implementation of new spectroscopic techniques, and the application of sophisticated computational methods to both the processing of experimental data and structure calculations. Nonetheless, limitations remain for structure determination of several important classes of proteins, especially large proteins in complexes in aqueous solution and membrane proteins in various detergent/lipid environments. Both classes of proteins present difficulties for the resolution of individual resonances that result from both the number of overlapping resonances and the broad line widths of the resonances associated with slowly reorienting proteins. Even as these problems have been incrementally addressed, there remains the problem of resolving and assigning a sufficient number of 1H/1H NOEs for structure determination with conventional approaches that measure inter-proton distances of <5 Å. Membrane proteins with multiple trans-membrane helices have the additional problem of identifying the correct alignment and relative positioning of the helices, which is difficult to do with only measurements of short-range distances. Further progress in NMR spectroscopy of these classes of proteins would be greatly aided by the ability to measure relatively long-range (>10 Å) distances. Such long-range distance constraints have many benefits, including improving the resolution of protein structure determination, defining the overall folding topology, and identifying residues in binding sites. They are especially advantageous in offering a method for positioning multiple trans-membrane helices in membrane proteins (Chen et al. 2011; Ganguly et al. 2011).
This Perspective focuses on membrane proteins, in particular the use of paramagnetic metals attached by tags to otherwise diamagnetic proteins. Our primary research interest is in structure determination of membrane proteins in their native environment of phospholipid bilayers (Opella 2013) (Radoicic et al. 2014). However, on the path towards this goal, aspects of membrane protein sample preparation and, to some extent, experimental methods are first worked out with micelle (Mesleh et al. 2003), bicelle (Son et al. 2012), or nanodisc (Park et al. 2011a) samples that are tractable for solution NMR. This is the situation for the examples described here, as we demonstrate the applicability of the genetically incorporated unnatural amino acid (UAA) 1-amino-3-(8-hydroxyquionolin-3-yl)-propanoic acid (HQA) (Figure 1C) (Lee et al. 2009) as a metal-binding tag to enable the use of paramagnetic ions to provide long-range paramagnetic relaxation enhancements that serve as intra- and inter- molecular distance measurements in membrane proteins.
Figure 1.
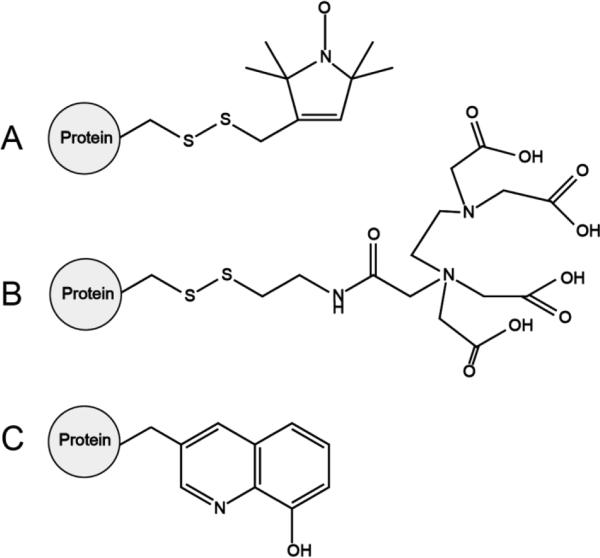
Paramagnetic probes attached to a protein for site-specific incorporation of protein labeling. (A) Nitroxide spin label. (B) Ethylenediaminetetraacetic acid (EDTA). (C) 2-amino-3-(8-hydroxyquinolin-3-yl)propanoic acid (HQA). Nitroxide spin labels and EDTA tags are mobile, containing multiple rotatable bonds and are incorporated by chemical reaction at a surface exposed cysteine residue, while HQA is approximately the size of tryptophan and is incorporated genetically at any position in the protein of interest.
Paramagnetic protein NMR
The unpaired electron on a paramagnetic ion has spectroscopic effects that are several orders of magnitude larger than those of the spin S=1/2 nuclei (1H, 13C, 15N) that are commonly observed in NMR studies of proteins (Liu et al. 2014a; Otting 2010) (Bertini et al. 2008) (Clore 2013) (Sengupta et al. 2013) (Knight et al. 2013) (Ganguly et al. 2011) (Zhuang et al. 2008). Briefly, paramagnetic ions induce three effects on the diamagnetic spectra of proteins, only two of which are generally observed and incorporated into the experiments - paramagnetic relaxation enhancements (PREs) and pseudocontact shifts (PCSs). In addition, there are through-bond contact shifts; however, because they only occur in close proximity to the metal ions, their effects are typically overwhelmed by strong PRE broadening of the resonances. In general, both PREs and PCSs are widely used in paramagnetic NMR studies of proteins. Their occurrence and properties can be controlled by the selection of the metal ions, the ligands, and other factors. The magnetic susceptibility tensor is a key parameter. The two metals used here, Mn2+ and Gd3+, have isotropic magnetic susceptibility tensors and therefore induce only PRE effects. Nitroxide spin labels also only induce PRE effects (McConnell and McFarland 1970). Many other metals, especially the lanthanides (with the exception of gadolinium) and Co2+ have highly anisotropic susceptibility tensors (Abragam and Bleaney 2012). This gives rise to PCSs, which can result in large changes in chemical shifts at distant sites.
There are other beneficial effects of adding paramagnetic metals to the samples in a controlled manner. They can reduce the longitudinal relaxation times so that data can be acquired much more quickly (Inubushi and Becker 1983; Parthasarathy et al. 2013; Ullrich et al. 2014; Ward et al. 2014), and they can weakly align proteins in solution (Prestegard et al. 2000; Tolman et al. 1995), providing an alternative to conventional alignment media for the measurement of residual dipolar couplings (RDCs).
Much of the groundwork for the use of paramagnetic ions in protein NMR was laid by Bertini and coworkers on metalloproteins, which contain a natural paramagnetic center (Balayssac et al. 2007; Bertini et al. 2008). For convenience, in appropriate cases, the metal ion can be exchanged for one with more favorable properties for the studies of interest. As an example, Bertini and Pintacuda have used metal ions in structural studies of superoxide dismutase (Knight et al. 2012). They were able to measure many 15N and 13C PREs based on the high-resolution two-dimensional heteronuclear correlation ‘fingerprint’ spectrum of the protein. These paramagnetic constraints significantly reduced the RMSD of the calculated protein structure.
However, most proteins, especially the membrane proteins of interest, are not metalloproteins. Because of the advantages of introducing a paramagnetic ion, there has been a great deal of activity in the design and implementation of tags that attach a paramagnetic ion to proteins. There are three main approaches to specifically attaching a paramagnetic metal to a protein. One is to attach residues corresponding to a natural or engineered metal binding site to the C- or N- terminus or a loop of the protein. The second is to attach a chemical linker to a reactive site, almost universally a surface cysteine side chain, which can bind a metal ion. The third is to incorporate an unnatural metal-binding amino acid into the sequence at a specific location. The incorporation of an unnatural metal-binding amino acid into membrane proteins is the principal subject of this Perspective.
In the first case, twelve amino acid residues corresponding to an “EF-hand” calcium-binding site were added to the N-terminus of the membrane protein Vpu from HIV-1 (Ma and Opella 2000). This provided a covalently attached lanthanide ion binding site. The protein itself was not altered by the added residues, as evidenced by a lack of perturbation of the chemical shifts, and it was possible to observe long-range paramagnetic effects in the spectra. In addition, the added lanthanide served to weakly align the protein for measurement of residual dipolar couplings. Imperiali and Schwalbe designed seventeen residue lanthanide binding tags (LBTs) for proteins with improved properties over native calcium binding sites (Wohnert et al. 2003). Subsequently, they inserted LBTs into protein loops (Barthelmes et al. 2011), which were shown to give complementary results. In a similar vein, Gaponenko et al (Gaponenko et al. 2000) fused zinc fingers to the N- and C- termini of a protein and demonstrated that they could be substituted with paramagnetic cobalt and manganese.
Most paramagnetic NMR studies of proteins have placed the metal ion on the surface of the protein with a covalent tag (Keizers and Ubbink 2011). Generally this has been done through a linkage to a selected cysteine side chain. In many cases this requires the removal of competitive reactive sites through mutation. It has also become a very active area of research with the development of linkers to two cysteine residues to reduce the local dynamics of the metal ion in order to increase the precision of the measurements. There have also been examples where other protein functional groups are involved in the chelation for the same reason. As a result of this activity, this area has been the subject of a number of reviews, including by Ubbink (Hass and Ubbink 2014) and Otting (Otting 2010). The tables and figures of these reviews provide a thorough listing of the wide variety of tags that have been utilized to tag proteins with paramagnetic ions. Figure 3 of the review by Otting (Otting 2010) is particularly helpful in understanding and planning paramagnetic NMR experiments. It shows the relative paramagnetism and asymmetry of the magnetic susceptibility tensors in a way that facilitates direct comparisons.
Figure 3.
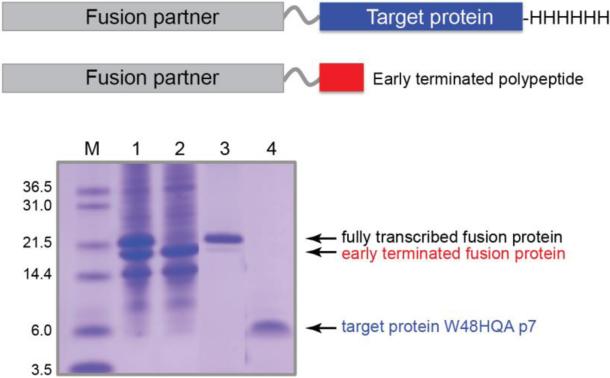
Schematic drawings of the proteins prepared by using two expression vectors shown in Figure 2 (top) and a SDS-PAGE showing the purification of a HQA-incorporated membrane protein p7 (bottom): lane 1, inclusion bodies solubilized in detergents; lane 2, flow through of the Ni-affinity chromatography; lane 3, elution from the Ni-affinity chromatography; lane 4, HPLC pure W48HQA p7 after CNBr cleavage. Note that the His-tag attached to the C-terminus of the target protein facilitates the separation of the fully transcribed proteins from the early-terminated proteins using Ni-affinity chromatography.
Using covalent paramagnetic tags, Otting and coworkers have utilized pseudocontact shifts in their studies (Yagi et al. 2013). They discussed how the assignment problem caused by the large shifts could be addressed. One way is to take advantage of the shifts for two bonded nuclei, e.g., 1H and 15N, which shift along parallel frequencies from the same metal ion. Fast exchange between diamagnetic and paramagnetic species enables titration of the protein and the resulting paramagnetic ion-induced chemical shifts. Pseudocontact shifts can be used as constraints for calculations of protein structures. With the use of multiple tags, significant improvements in the precision of structures result from the measurement of PCSs.
Ubbink and coworkers have also focused on the use of PCSs in the structure determination of protein complexes (Liu et al. 2014a) Their results have been facilitated by the development of metal binding tags. The magnetic properties of lanthanides are relatively insensitive to variations in the coordination. However, since many tags are more flexible than a typical chemical coordination site, it is essential to restrict their dynamics. This has been achieved with bulky tags, including peptide tags as discussed above, two-point attached tags (Liu et al. 2014b), and tags that interact with protein side chains (Su et al. 2008). With dynamically restricted lanthanide tags, significant PCSs can be observed over very large distances, perhaps beyond 100 Å (Hass and Ubbink 2014).
The groups of Jaroniec and Huber have used a variety of tagged mutants of the protein GB1 as a model polycrystalline protein (Jaroniec 2012; Sengupta et al. 2013) (Li et al. 2013). Jaroniec and coworkers observed substantial PRE effects in well resolved magic angle spinning spectra by comparing samples with and without free electrons on the TEMPO moiety. Significantly, some of the strong signals in the control sample are missing in the corresponding spectrum of the spin-labeled protein. A key feature of their experimental samples is that the spin-labeled protein was diluted with unlabeled protein to enable the intramolecular effects of interest to be separated from any intermolecular effects of nearby protein molecules. Although clearly beneficial in certain applications, the use of nitroxides as relaxation agents suffers from a major draw-back, namely the presence of large transverse PREs, which lead to severely attenuated signal intensities for numerous residues; this precludes quantitative PRE and distance measurements. They overcame this problem by using tags containing a more rapidly relaxing paramagnetic center, namely Cu2+ (Nadaud et al. 2009), that does not elicit significant paramagnetic shifts due to its effectively isotropic magnetic susceptibility tensor. The longitudinal PREs could be used to determine the global fold of GB1. They were able to calculate the backbone fold of GB1 based solely on PREs using only six mutants.
Paramagnetic NMR has been applied to a number of membrane proteins. Building on the structure of the Anabaena sensory rhodopsin (ASR) (Wang et al. 2013) Ladizhansky and co-workers employed nitroxide spin labels and PRE data to map the oligomerization interface of the receptor (Wang et al. 2012). Gottstein et al (Gottstein et al. 2012) have summarized the alpha helical membrane proteins whose structures have been determined with the aid of PREs. Other notable studies of membrane proteins using paramagnetic NMR include: protein-lipid interactions of outer membrane protein X (OmpX) and DHPC using various nitroxide spin labels (Hilty et al. 2004), improvements of the outer membrane protein A (OmpA) backbone structure via ‘parallel spin labeling’ (Liang et al. 2006), characterization of the Influenza M2 proton channel (Su et al. 2012), topology determination in DPC micelles of SCO3063 and YbdK, two bacterial histidine kinase membrane proteins (Yeo et al. 2010), DsbB (Tang et al. 2011), structural studies of the M. tuberculosis RV1761C protein (Page et al. 2009), and the characterization of an 80 residue region of the GPCR Ste2p using PREs in solution NMR (Cohen et al. 2011).
Perhaps the most powerful approach to introducing tags into proteins is unnatural amino acid incorporation (Liu and Schultz 2010; Noren et al. 1989). This very promising method for paramagnetic NMR will be discussed in more depth in the following section along with a sampling of recent studies performed in our laboratory using membrane proteins and unnatural amino acid incorporation of a novel paramagnetic tag. The best currently available metal-binding unnatural amino acid, HQA, is shown in Figure 1C, which we used in our studies.
Experimental section
Synthesis of HQA
The synthesis of 2-amino-3-(8-hydroxyquinolin-3-yl)propanoic acid dihydrochloride (HQA) was carried out as previously reported (Lee et al. 2009) with a few minor changes. The synthesis of the first intermediate, 8-methoxy-3-methylquinoline, was carried out according to Patent DE 3719014 C2, and all silica flash column chromatography solvent systems used ethyl acetate and hexanes rather than dichloromethane.
Protein preparation
Interleukin-8 (IL-81-66), a monomeric construct with the 6 C-terminal residues of IL-8 removed, was expressed and purified as previously described (Rajarathnam et al. 1997). For 1TM-CXCR1 and p7 constructs, an original pET31b(+) vector (Novagen, www.novagen.com) was modified by including a thrombin cleavage site with a 6-Gly linker between the KSI fusion partner and the polypeptide of interest to facilitate enzymatic cleavage (Figure 2B). They were purified and refolded as described previously for similar constructs of the proteins (Casagrande et al. 2011) (Cook et al. 2011) .
Figure 2.
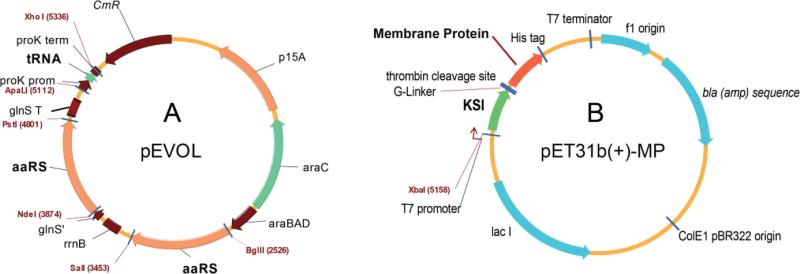
Vector maps for unnatural amino acid incorporation into proteins in E. coli. (A) The pEVOL vector optimized for efficient expression of orthogonal pairs of tRNA/aminoacyl-tRNA synthetase (Young et al. 2010). (B) Modified pET31b(+) vector for HQA-incorporated membrane protein expression as a KSI-fusion protein containing a thrombin cleavage site between KSI and the target membrane protein.
For expression of HQA-incorporated p7 and 1TM-CXCR1 constructs, the pEVOL vector (chloramphenicol resistant, arabinose inducible) encoding the orthogonal tRNA/aminoacyl-tRNA synthetase pair specific for HQA (Young et al. 2010) (Figure 2A) and the modified pET31b(+) vector (ampicillin resistant, isopropyl β-D-1-thiogalactopyranoside (IPTG) inducible) encoding the target mutant were co-transformed into BL21(DE3) cells. Uniformly 15N-labeled HQA-incorporated 1TM-CXCR1 and p7 constructs were prepared using a media exchange method (Marley et al. 2001; Venditti et al. 2012): 500 mL Luria-Bertani (LB) media with chloramphenicol/carbenicillin was inoculated with 10% overnight LB starter culture and grown in a shaker/incubator at 37°C. At OD600 ~0.4 the cells were induced with a final concentration of 0.02% L-arabinose and grown for two hours at 37°C. They were then spun down by centrifugation at 1350 × g. The cell pellets were gently re-suspended in 500 mL minimal media with 15N-labeled ammonium sulfate as the sole nitrogen source, induced with 0.02% final concentration of L-arabinose and grown at 37°C for one more hour following which 1 mM HQA and 100 μM IPTG were added. Cells were grown for four hours, after which they were harvested via centrifugation at 6200 × g and stored at −80°C prior to further purification. Purification and refolding procedures of HQA-incorporated 1TM-CXCR1 and p7 constructs were identical to those of their wild-type counterparts.
Fluorescence experiments
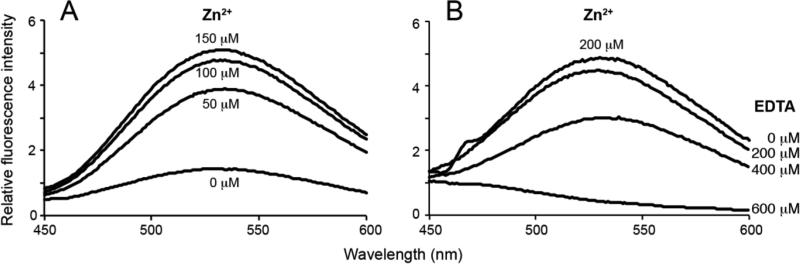
The fluorescence experiments were performed on a FluoroMax-4 spectrofluorometer (Horiba Scientific, New Jersey, NY) at room temperature with excitation at 400 nm. Fluorescence of the wild-type and HQA-incorporated proteins at 20-50 μM (200 μL) was measured in 300 mM SDS or 150 mM DPC detergent micelles, 20 mM HEPES (pH 7.0) and different concentrations of ZnCl2 (0, 50, 100, 150, and 200 μM). Fluorescence quenching experiments were performed on Zn2+ -bound W48HQA p7 by titrating EDTA to final concentrations of 0, 200, 400, and 600 μM.
NMR experiments
NMR samples were prepared by dissolving the lyophilized proteins in a solution containing 150 mM DHPC, 20 mM HEPES, pH 7.0 at a final protein concentration of 50 μM −100 μM. For the PRE experiments, aliquots of stock solutions of 10 mM or 100 mM MnCl2 were added to the protein samples at final Mn2+ concentrations of 0.1 mM for 1TM CXCR1 samples and 0.5 mM for p7 samples. For the IL-8 and 1TM CXCR1 binding experiments, unlabeled W10HQA 1TM CXCR1 was complexed with uniformly 15N-labeled IL-8 in a 1:1 molar ratio. The NMR experiments were performed on a Bruker Avance 600 MHz spectrometer equipped with 5-mm triple-resonance cryoprobe with z-axis gradient. One-dimensional 15N-edited 1H NMR and 1H-15N HSQC (Bodenhausen and Ruben 1980) NMR spectra were obtained at 50°C for the 1TM-CXCR1 and p7 samples, and at 40°C for IL-8-containing samples.
Results and discussion
Metal-chelating Unnatural amino acid
Although the use of fusions with cysteines and metal-binding peptides for site-specific paramagnetic labeling of proteins has helped probe protein structure determination, analysis of protein-ligand binding, and descriptions of molecular dynamics, their use is limited by their bulk, local dynamics, and potential sample heterogeneity. Large conformational spaces for the paramagnetic center lead to observed PRE measurements whose properties may be influenced by the flexibility of the probe itself. Rigid paramagnetic probes have been introduced into proteins to counter this problem (Liu et al. 2014c; Loh et al. 2013), but the location of anchoring cysteine residues in the protein limit their placement and their often-bulky structure can also affect native protein structure. The incorporation of unnatural amino acids (UAAs) addresses both the issues of bulk and local dynamics and allows for homogeneous protein samples. They can be placed anywhere in the protein without being limited by disulfide bonds, cysteine residues, and attachments to the protein termini (Otting 2010). In addition, a paramagnetic metal ion can be introduced anywhere in the protein with minimal protein structure perturbation (Jones et al. 2010) due to a single amino acid substitution with a UAA with high metal affinity in the protein sequence.
Site-specific genetic incorporation of HQA involves an orthogonal aminoacyl-tRNA/tRNA synthetase pair specific to the HQA that reads the amber TAG codon (Lee et al. 2009). Under normal circumstances, E. coli would recognize TAG as a stop codon. With the addition of the orthogonal pair, however, HQA is added to the growing protein chain. The orthogonal tRNA synthetase is not recognized by endogenous tRNA/amino acid and vice versa (Noren et al. 1989). Of the more than 100 unnatural amino acids incorporated successfully, a small fraction is useful for NMR. These include isotopically labeled p-methoxy-phenylalanine and its fluorinated analogs (Cellitti et al. 2008; Deiters et al. 2005), photocaged unnatural amino acids for site-specific labeling (Jones et al. 2010), metal-chelating unnatural amino acids (Lee et al. 2009; Loh et al. 2013; Nguyen et al. 2011), and an amino acid that ligates with a lanthanide tag (Liu and Schultz 2010). Incorporation of isotopically labeled amino acids at specific residues have helped identify ligand binding sites and conformational changes of large proteins (Deiters et al. 2005). Aside from these, to date, only five unnatural amino acids that have been successfully incorporated into proteins can be used as paramagnetic probes. Three are metal chelating and one is used to ligate with a lanthanide tag (Jones et al. 2010). Protein engineering with unnatural amino acids has been discussed (Zhang et al. 2013), and it is possible for site-directed spin labeling of a genetically encoded unnatural amino acid (Fleissner et al. 2009) (Schmidt et al. 2014).
The use of lanthanides for PRE studies are of particular interest because of their varying magnetic properties; different lanthanides can be incorporated into the same metal-chelation site. Previous lanthanide tags could only be non-covalently bound to the N- or C- termini of a protein, resulting in flexibility of the tag and poor measurements. While the lanthanide-p-azido-L-phenylalanine (AzF) fusion allowed for placement of the tag independent of the location of other residues in the protein, there are limitations that hinder its widespread application (Loh et al. 2013). The lanthanide tag is also limited by the need for known structural data for tether optimization. Using metal-chelating unnatural amino acids, on the other hand, provides a straightforward way to introduce a paramagnetic ion for PRE measurements (Nguyen et al. 2011). For example, the UAA bipyridylalanine (BpyAla) was successfully incorporated into the West Nile virus NS2B-NS3 protease and bound to cobalt (II) to obtain PCS measurements. Of the three metal-chelating unnatural amino acids, only HQA chelates to lanthanides (Jones et al. 2010; Lee et al. 2009), making it the best candidate for PRE studies. Moreover, HQA is the smallest of the three UAA, making it a favorable replacement for the similarly sized aromatic canonical amino acids, in particular tryptophan. It is also fluorescently active, allowing for easy detection of metal-bound protein.
Successful efforts to obtain NMR spectra of HQA-incorporated protein have not been reported previously (Jones et al. 2010). Here we show by solution NMR the successful incorporation of HQA into two membrane proteins: the cytoplasmic N-terminal region of the G-protein coupled receptor (GPCR) CXCR1 and the second transmembrane helix of viroporin p7 from the hepatitis C virus. We were also able to successfully obtain NMR data showing specific binding of a paramagnetic ion to the HQA-incorporated proteins, offering powerful insight to protein-ligand binding and protein structure.
Genetic incorporation of HQA into membrane proteins
Genetic incorporation of the unnatural amino acid 2-amino-3-(8-hydroxyquinolin-3-yl) propanoic acid (HQA) was done via amber codon suppression as demonstrated by Schultz and co-workers (Lee et al. 2009). For the 1TM-CXCR1 construct the tryptophan at position 10 in the amino acid sequence was chosen as the site of mutation due to its location in the binding region of the receptor and its ligand, interleukin-8 (IL-8). For the p7 construct, the tryptophan at residue 48 in the sequence was chosen as the site of mutation due to its location in the second transmembrane helix, with the primary goal to obtain intramolecular PRE data.
The pEVOL vector encoding the orthogonal tRNA/aminoacyl-tRNA synthetase pair for HQA (Young et al. 2010) and the modified pET31b(+) vector encoding our target mutant (Figure 2) were co-expressed in minimal media and we were able to obtain high yields of mutant, HQA incorporated, membrane protein for both the 1TM-CXCR1 and p7 constructs. Growth and incorporation protocols for both the orthogonal pair and target genes were optimized and included testing various growth temperatures, induction times and concentrations, as well as optimizing the growth media itself (data not shown). Following growth and induction both the truncated and incorporated forms of the protein were produced, as can be seen in Figure 3, lane 1. The truncated form, which is the result of the presence of the amber stop codon at the mutation site, was also observed in HQA-incorporated 1TM-CXCR1 expression (data not shown). Due to the presence of a C-terminal histidine tag, which is only present in the full-length mutant and not in the truncated form (Figure 3, top), the two could be successfully separated via Ni-immobilized metal affinity chromatography (IMAC). Further purification by HPLC resulted in high yields of pure protein (Figure 3).
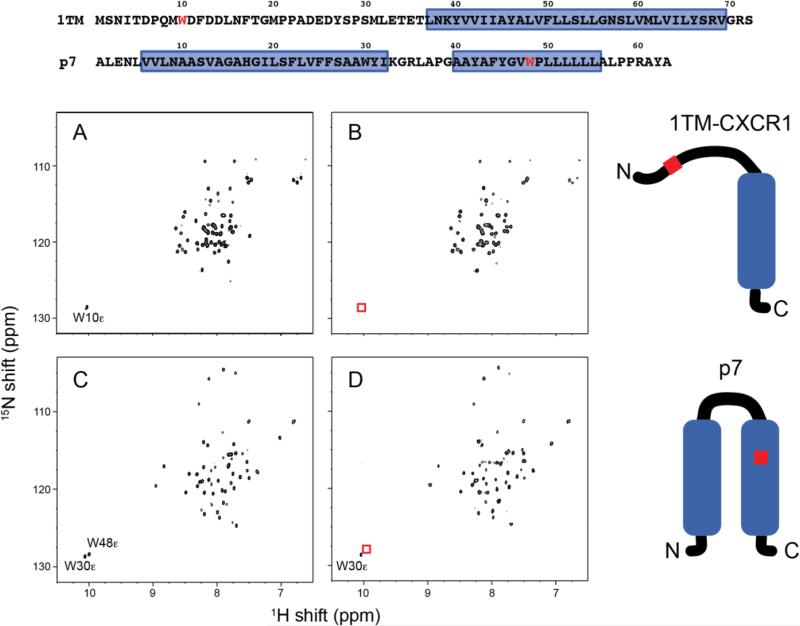
In the 15N HSQC spectra obtained at 600 MHz for both W10HQA 1TM-CXCR1 and W48HQA p7 in DHPC micelles at pH 7 (Figure 4) it was observed that the resonances from the tryptophan indole nitrogen sites, which were mutated at positions 10 and 48, respectively, were noticeably absent in the spectra of the mutant proteins, indicating that the unnatural amino acid was successfully incorporated into the proteins and at the correct positions. Furthermore, the overlap between the spectra of the wild-type and mutant proteins is very significant for both mutants (Figure 4) indicating that incorporation of the unnatural amino acid did not cause significant chemical shift perturbations in the spectra, and that the native structures and conformations are retained.
Figure 4.
Comparison of 1H-15N HSQC spectra of uniformly 15N-labeled membrane protein constructs in DHPC micelles. (A) Wild-type 1TM-CXCR1. (B) W10HQA 1TM-CXCR1. (C) Wild-type p7. (D) W48HQA p7. Absence of Trp indole NHε signals shown in red boxes indicates complete incorporation of unnatural amino acid HQA into residue Trp 10 in 1TM-CXCR1 and into residue Trp 48 in p7. The mutation site is colored in red in the amino acid sequences and schematic drawings of proteins. Note that the amide chemical shifts for wild-type and HQA-incorporated proteins are identical except in the vicinity of the mutated site.
Fluorescence induced by HQA-metal chelate
8-hydroxyquinoline forms chelate compounds with many metal ions including lanthanides, and Cd2+, Mg2+, and Zn2+ form a strong fluorescent complex with 8-hydroxyquinoline (Soroka et al. 1987). In order to investigate site-specific fluorescence from binding of metal ions to HQA-incorporated membrane proteins in detergent micelles, W10HQA 1TM-CXCR1 and W48HQA p7 proteins were titrated with Zn2+ ions and fluorescence was measured (Figure 5). No fluorescence of the HQA-incorporated proteins was observed in the absence of Zn2+, but the fluorescence increased with increasing the concentrations of Zn2+, and was saturated at about 5:1 (Zn2+:HQA) molar ratio. As a control, the fluorescence of the wild-type proteins was examined; however they showed no fluorescence in the presence of 200 μM Zn2+ (data not shown).
Figure 5.
Fluorescence spectra of HQA-incorporated membrane proteins in detergent micelles. (A) W10HQA 1TM-CXCR1 complexed with varying concentrations of ZnCl2. (B) W48HQA p7 complexed with 200 μM ZnCl2 and with varying concentrations of EDTA. Protein concentration was approximately 50 μM.
In figure 5B, complete quenching of fluorescence of Zn2+-HQA chelates was observed at a 3:1 (EDTA:HQA-Zn2+) molar ratio, suggesting that Zn2+ forms stable chelates with HQA in the transmembrane region of p7 in micelles. These results prove that genetic incorporation of the unnatural amino acid HQA into membrane proteins can serve as a powerful site-specific biophysical probe for studies of the structures and dynamics of membrane proteins in membrane environments.
Intramolecular PREs by HQA-metal chelate
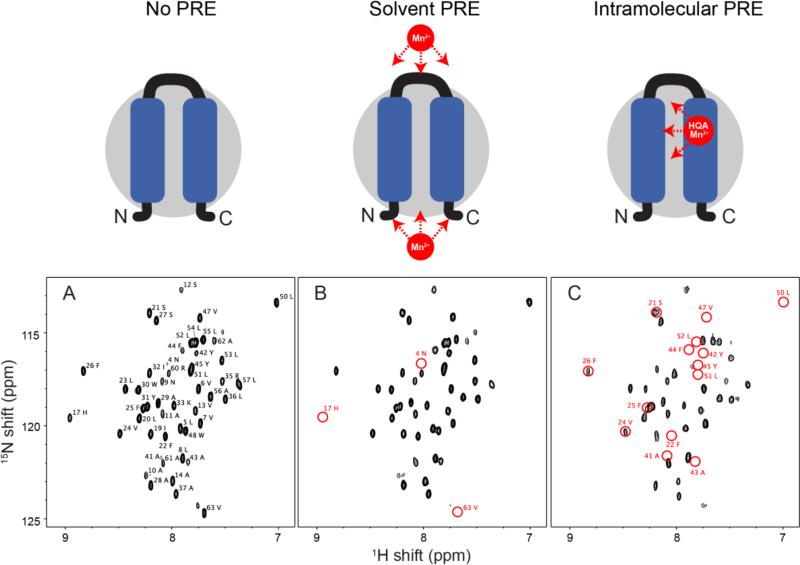
In Figure 6, the comparison of HSQC spectra of p7 constructs demonstrates the different PRE effects on the wild-type and HQA-incorporated p7 samples. All of the p7 resonances, except for the first three N-terminal residues, were observed and assigned in DHPC micelles at pH 7 (Figure 6A). When paramagnetic Mn2+ ions were titrated to the solution containing wild-type p7, several resonances (residues 4, 17, 18, and 63) were significantly broadened, indicating these residues are located in the solvent accessible regions of the molecular surface of p7 in detergent micelles (Figure 6B). When Mn2+ ions were titrated in the W48HQA p7 sample at an approximately 5:1 (Mn2+:W48HQA p7) molar ratio, many more signals were broadened or disappeared compared to the wild-type p7 (Figure 6C). This result suggests that Mn2+ can specifically bind to the HQA located in the second transmembrane helix of p7 in detergent micelles, and therefore yields intramolecular PREs. Many of the missing resonances (residues 41-47 and residues 50-52) are near the mutated position of the residue 48 as expected. However, it is noteworthy that the residues 21, 22, 24-26 were also significantly broadened or disappeared, indicating that these residues, which are located in the first transmembrane helix are in close proximity to residue 48 which is located in the second transmembrane helix of p7. These intramolecular PREs are very helpful in structure determination of membrane proteins, since they provide long-range distance restraints that are very challenging to obtain by conventional NOE experiments.
Figure 6.
Expanded region of the 1H-15N HSQC spectra of uniformly 15N-labeled p7 in DHPC micelles. (A) Wild-type p7 alone. (B) Wild-type p7 in the presence of 0.5 mM MnCl2. (C) W48HQA p7 in the presence of 0.5 mM MnCl2. The resonances broadened by solvent PREs or intramolecular PREs are indicated by red circles. Note that glycine residues 15, 18, 34, and 46 do not lie in the expanded region.
1TM-CXCR1 consists of the ligand binding N-terminal domain, which is mobile and solvent accessible, and the first transmembrane helix of CXCR1, which is buried in the membrane environment (Park et al. 2011c). Therefore, the resonances of the N-terminal domain of wild-type 1TM-CXCR1 and several C-terminal residues were broadened beyond detection in the presence of Mn2+ or Gd3+, while only 7 residues near the HQA mutation site of W10HQA 1TM-CXCR1 were broadened by the titration of paramagnetic ions due to a site-specific HQA and metal ion interactions at approximately 1:1 (Zn2+:HQA) molar ratio (data not shown). Although successful efforts to obtain NMR spectra of HQA-incorporated protein have not been reported previously due to metal mediated protein oligomerization (Jones et al. 2010), we do not observe any evidence of oligomerization or aggregation of HQA-incorporated membrane proteins in detergent micelles. Optimization of experimental conditions was straightforward.
Intermolecular PREs by HQA-metal chelate
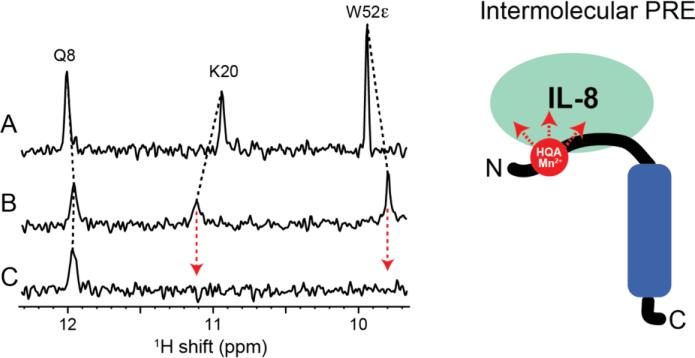
The N-terminal domain of the chemokine receptor CXCR1 contains the major binding site of the ligand IL-8 (Park et al. 2011b). Figure 7 shows that the HQA in 1TM-CXCR1 serves as an excellent probe for intermolecular PREs, providing crucial long-range distance restraints for structure determination of the IL-8 and 1TM-CXCR1 complex. The binding sites of IL-8, identified by chemical shift perturbation upon interaction with W10HQA 1TM-CXCR1 (Figure 7B), were identical to those of the IL-8 interaction with wild-type 1TM-CXCR1. Adding Mn2+ ions to the complex selectively broadened the residues of the binding sites (e.g. residues K20 and W52ε) beyond detection because of the site specific intermolecular PREs (Figure 7C), providing the distance restraints between IL-8 and 1TM-CXCR1 that are essential for structure determination of the ligand-receptor complex.
Figure 7.
Expanded region of the 15N-edited 1H spectra of uniformly 15N-labeled IL-8 in DHPC micelles. (A) IL-8 alone. (B) IL-8 bound to unlabeled W10HQA 1TM-CXCR1. (C) IL-8 bound to unlabeled W10HQA 1TM-CXCR1 in the presence of MnCl2. Close proximity between residues K20 and W52ε of IL-8 and the paramagnetic HQA-Mn2+ group at residue 10 of CXCR1 is shown by observation of select intermolecular PREs.
Conclusions
The research described here is a prelude to the use of paramagnetic tags on membrane proteins in phospholipid bilayers. This involves the use of solid-state NMR because the membrane proteins are immobilized by their interactions with the phospholipids in the bilayer environment. Thus, significant background comes from prior solid-state NMR studies of paramagnetically tagged proteins.
The in vivo incorporation of unnatural amino acids into proteins is a well-established technique requiring an orthogonal tRNA/aminoacyl-tRNA synthetase pair specific for the unnatural amino acid that is incorporated at a position encoded by a TAG amber codon. Recently developed metal-chelating unnatural amino acid 2-amino-3-(8-hydroxyquinolin-3-yl)propanoic acid (HQA) forms highly stable complexes with various transition metal ions and lanthanides, serving as an excellent probe for paramagnetic relaxation enhancement NMR experiments. Optimization of the expression of orthogonal aminoacyl-tRNA synthetases/suppressor tRNA pairs from the pEVOL vector and target proteins from the KSI-fusion expression system provided high yields of HQA-incorporated proteins, essentially equivalent to those for wild-type proteins. NMR spectral comparisons of the wild-type and HQA-incorporated mutants demonstrate complete incorporation of HQA into the membrane proteins. Zn2+-HQA induced fluorescence confirms a stable metal chelation.
Acknowledgments
We thank Dr. P. G. Schultz for providing the pEVOL vector, Ms. G. Kang in the laboratory of Dr. J. Kim for assistance with the fluorescence measurements, and Dr. M. Burkhart for access to his laboratory for the organic synthesis of HQA. The research was supported by grants P41EB002031, R01EB005161, R01GM099986, and R01GM066978 from the National Institutes of Health. It utilized the Biotechnology Resource Center for NMR Molecular Imaging of Proteins at the University of California, San Diego.
References
- Abragam A, Bleaney . Electron Paramagnetic Resonance of Transition Ions. Oxford University Press; 2012. [Google Scholar]
- Balayssac S, Bertini I, Lelli M, Luchinat C, Maletta M. Paramagnetic ions provide structural restraints in solid-state NMR of proteins. J Am Chem Soc. 2007;129:2218–2219. doi: 10.1021/ja068105a. doi:10.1021/ja068105a. [DOI] [PubMed] [Google Scholar]
- Barthelmes K, et al. Engineering encodable lanthanide-binding tags into loop regions of proteins. J Am Chem Soc. 2011;133:808–819. doi: 10.1021/ja104983t. doi:10.1021/ja104983t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I, Luchinat C, Parigi G, Pierattelli R. Perspectives in paramagnetic NMR of metalloproteins. Dalton transactions. 2008:3782–3790. doi: 10.1039/b719526e. doi:10.1039/b719526e. [DOI] [PubMed] [Google Scholar]
- Bodenhausen G, Ruben DJ. Natural abundance nitrogen-15 NMR by enhanced heteronuclear spectroscopy. Chemical Physics Letters. 1980;69:185–189. doi: http://dx.doi.org/10.1016/0009-2614(80)80041-8. [Google Scholar]
- Campbell ID, Dobson CM, Williams RJ, Xavier AV. The determination of the structure of proteins in solution: lysozyme. Annals of the New York Academy of Sciences. 1973;222:163–174. doi: 10.1111/j.1749-6632.1973.tb15259.x. [DOI] [PubMed] [Google Scholar]
- Casagrande F, Maier K, Kiefer H, Opella SJ, Park SH. Expression and Purification of G-Protein-Coupled Receptors for Nuclear Magnetic Resonance Structural Studies. In: Robinson AS, editor. Production of Membrane Proteins: Strategies for Expression and Isolation. Wiley-VCH Verlag GmbH & Co. KGaA; Weinheim, Germany: 2011. pp. 297–316. doi:10.1002/9783527634521.ch11. [Google Scholar]
- Cellitti SE, et al. In vivo incorporation of unnatural amino acids to probe structure, dynamics, and ligand binding in a large protein by nuclear magnetic resonance spectroscopy. J Am Chem Soc. 2008;130:9268–9281. doi: 10.1021/ja801602q. doi:10.1021/ja801602q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, et al. Optimal mutation sites for PRE data collection and membrane protein structure prediction. Structure. 2011;19:484–495. doi: 10.1016/j.str.2011.02.002. doi:10.1016/j.str.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G. Seeing the invisible by paramagnetic and diamagnetic. NMR Biochem Soc Trans. 2013;41:1343–1354. doi: 10.1042/BST20130232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen LS, Arshava B, Neumoin A, Becker JM, Guntert P, Zerbe O, Naider F. Comparative NMR analysis of an 80-residue G protein-coupled receptor fragment in two membrane mimetic environments. Biochimica et biophysica acta. 2011;1808:2674–2684. doi: 10.1016/j.bbamem.2011.07.011. doi:10.1016/j.bbamem.2011.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook GA, Stefer S, Opella SJ. Expression and purification of the membrane protein p7 from hepatitis C virus. Biopolymers. 2011;96:32–40. doi: 10.1002/bip.21453. doi:10.1002/bip.21453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross TA, Opella SJ. NMR of fd coat protein. Journal of supramolecular structure. 1979;11:139–145. doi: 10.1002/jss.400110204. doi:10.1002/jss.400110204. [DOI] [PubMed] [Google Scholar]
- Cross TA, Opella SJ. Structural properties of fd coat protein in sodium dodecyl sulfate micelles. Biochemical and biophysical research communications. 1980;92:478–484. doi: 10.1016/0006-291x(80)90358-7. doi:0006-291X(80)90358-7 [pii] [DOI] [PubMed] [Google Scholar]
- Deiters A, Geierstanger BH, Schultz PG. Site-specific in vivo labeling of proteins for NMR studies. Chembiochem : a European journal of chemical biology. 2005;6:55–58. doi: 10.1002/cbic.200400319. doi:10.1002/cbic.200400319. [DOI] [PubMed] [Google Scholar]
- Fleissner MR, et al. Site-directed spin labeling of a genetically encoded unnatural amino acid. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:21637–21642. doi: 10.1073/pnas.0912009106. doi:10.1073/pnas.0912009106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S, Weiner BE, Meiler J. Membrane protein structure determination using paramagnetic tags. Structure. 2011;19:441–443. doi: 10.1016/j.str.2011.03.008. doi:10.1016/j.str.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaponenko V, Dvoretsky A, Walsby C, Hoffman BM, Rosevear PR. Calculation of z-coordinates and orientational restraints using a metal binding tag. Biochemistry. 2000;39:15217–15224. doi: 10.1021/bi001381w. [DOI] [PubMed] [Google Scholar]
- Gottstein D, Reckel S, Dotsch V, Guntert P. Requirements on paramagnetic relaxation enhancement data for membrane protein structure determination by NMR. Structure. 2012;20:1019–1027. doi: 10.1016/j.str.2012.03.010. doi:10.1016/j.str.2012.03.010. [DOI] [PubMed] [Google Scholar]
- Hagen DS, Weiner JH, Sykes BD. Fluorotyrosine M13 coat protein: fluorine-19 nuclear magnetic resonance study of the motional properties of an integral membrane protein in phospholipid vesicles. Biochemistry. 1978;17:3860–3866. doi: 10.1021/bi00611a028. [DOI] [PubMed] [Google Scholar]
- Hagen DS, Weiner JH, Sykes BD. Investigation of solvent accessibility of the fluorotyrosyl residues of M13 coat protein in deoxycholate micelles and phospholipid vesicles. Biochemistry. 1979;18:2007–2012. doi: 10.1021/bi00577a026. [DOI] [PubMed] [Google Scholar]
- Hass MA, Ubbink M. Structure determination of protein-protein complexes with long-range anisotropic paramagnetic NMR restraints. Current opinion in structural biology. 2014;24:45–53. doi: 10.1016/j.sbi.2013.11.010. doi:10.1016/j.sbi.2013.11.010. [DOI] [PubMed] [Google Scholar]
- Hilty C, Wider G, Fernandez C, Wuthrich K. Membrane protein-lipid interactions in mixed micelles studied by NMR spectroscopy with the use of paramagnetic reagents. Chembiochem : a European journal of chemical biology. 2004;5:467–473. doi: 10.1002/cbic.200300815. doi:10.1002/cbic.200300815. [DOI] [PubMed] [Google Scholar]
- Inubushi T, Becker E. Efficient detection of paramagnetically shifted NMR resonances by optimizing the WEFT pulse sequence. Journal of magnetic resonance. 1983;51:128–133. [Google Scholar]
- Jaroniec CP. Solid-state nuclear magnetic resonance structural studies of proteins using paramagnetic probes Solid state nuclear magnetic resonance. 2012;43-44:1–13. doi: 10.1016/j.ssnmr.2012.02.007. doi:10.1016/j.ssnmr.2012.02.007. [DOI] [PubMed] [Google Scholar]
- Jones DH, et al. Site-specific labeling of proteins with NMR-active unnatural amino acids. Journal of biomolecular NMR. 2010;46:89–100. doi: 10.1007/s10858-009-9365-4. doi:10.1007/s10858-009-9365-4. [DOI] [PubMed] [Google Scholar]
- Keizers PH, Ubbink M. Paramagnetic tagging for protein structure and dynamics analysis Progress in nuclear magnetic resonance spectroscopy. 2011;58:88–96. doi: 10.1016/j.pnmrs.2010.08.001. doi:10.1016/j.pnmrs.2010.08.001. [DOI] [PubMed] [Google Scholar]
- Knight MJ, Felli IC, Pierattelli R, Emsley L, Pintacuda G. Magic angle spinning NMR of paramagnetic proteins. Accounts of chemical research. 2013;46:2108–2116. doi: 10.1021/ar300349y. doi:10.1021/ar300349y. [DOI] [PubMed] [Google Scholar]
- Knight MJ, et al. Structure and backbone dynamics of a microcrystalline metalloprotein by solid-state NMR. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:11095–11100. doi: 10.1073/pnas.1204515109. doi:10.1073/pnas.1204515109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HS, Spraggon G, Schultz PG, Wang F. Genetic incorporation of a metal-ion chelating amino acid into proteins as a biophysical probe. J Am Chem Soc. 2009;131:2481–2483. doi: 10.1021/ja808340b. doi:10.1021/ja808340b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Pilla K, Li Q, Zhanag A, Su XC, Huber T, Yang J. Magic angle spinning NMR structure determination of proteins from pseudocontact shifts. J Am Chem Soc. 2013;135:8294–8303. doi: 10.1021/ja4021149. [DOI] [PubMed] [Google Scholar]
- Liang B, Bushweller JH, Tamm LK. Site-directed parallel spin-labeling and paramagnetic relaxation enhancement in structure determination of membrane proteins by solution NMR spectroscopy. J Am Chem Soc. 2006;128:4389–4397. doi: 10.1021/ja0574825. doi:10.1021/ja0574825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annual review of biochemistry. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. doi:10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- Liu W-M, Overhand M, Ubbink M. The application of paramagnetic lanthanoid ions in NMR spectroscopy on proteins. Coordination Chemistry Reviews. 2014a;273–274:2–12. doi: http://dx.doi.org/10.1016/j.ccr.2013.10.018. [Google Scholar]
- Liu WM, et al. A two-armed lanthanoid-chelating paramagnetic NMR probe linked to proteins via thioether linkages. Chemistry. 2014b;20:6256–6258. doi: 10.1002/chem.201400257. doi:10.1002/chem.201400257. [DOI] [PubMed] [Google Scholar]
- Liu Z, Gong Z, Guo DC, Zhang WP, Tang C. Subtle dynamics of holo glutamine binding protein revealed with a rigid paramagnetic probe. Biochemistry. 2014c;53:1403–1409. doi: 10.1021/bi4015715. doi:10.1021/bi4015715. [DOI] [PubMed] [Google Scholar]
- Loh CT, Ozawa K, Tuck KL, Barlow N, Huber T, Otting G, Graham B. Lanthanide tags for site-specific ligation to an unnatural amino acid and generation of pseudocontact shifts in proteins. Bioconjugate chemistry. 2013;24:260–268. doi: 10.1021/bc300631z. doi:10.1021/bc300631z. [DOI] [PubMed] [Google Scholar]
- Ma C, Opella SJ. Lanthanide ions bind specifically to an added “EF-hand” and orient a membrane protein in micelles for solution NMR spectroscopy. Journal of magnetic resonance. 2000;146:381–384. doi: 10.1006/jmre.2000.2172. doi:10.1006/jmre.2000.2172. [DOI] [PubMed] [Google Scholar]
- Markley JL, Putter I, Jardetzky O. High-resolution nuclear magnetic resonance spectra of selectively deuterated staphylococcal nuclease. Science. 1968;161:1249–1251. doi: 10.1126/science.161.3847.1249. [DOI] [PubMed] [Google Scholar]
- Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. Journal of biomolecular NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- McConnell HM, McFarland BG. Physics and chemistry of spin labels. Quarterly reviews of biophysics. 1970;3:91–136. doi: 10.1017/s003358350000442x. [DOI] [PubMed] [Google Scholar]
- Meadows DH, Markley JL, Cohen JS, Jardetzky O. Nuclear magnetic resonance studies of the structure and binding sites of enzymes. I. Histidine residues Proceedings of the National Academy of Sciences of the United States of America. 1967;58:1307–1313. doi: 10.1073/pnas.58.4.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesleh MF, Lee S, Veglia G, Thiriot DS, Marassi FM, Opella SJ. Dipolar waves map the structure and topology of helices in membrane proteins. J Am Chem Soc. 2003;125:8928–8935. doi: 10.1021/ja034211q. doi:10.1021/ja034211q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisett JD, Wien RW, McConnell HM. The use of spin labels for measuring distances in biological systems. Annals of the New York Academy of Sciences. 1973;222:149–162. doi: 10.1111/j.1749-6632.1973.tb15258.x. [DOI] [PubMed] [Google Scholar]
- Nadaud PS, Helmus JJ, Kall SL, Jaroniec CP. Paramagnetic ions enable tuning of nuclear relaxation rates and provide long-range structural restraints in solid-state NMR of proteins. J Am Chem Soc. 2009;131:8108–8120. doi: 10.1021/ja900224z. doi:10.1021/ja900224z. [DOI] [PubMed] [Google Scholar]
- Nguyen TH, Ozawa K, Stanton-Cook M, Barrow R, Huber T, Otting G. Generation of pseudocontact shifts in protein NMR spectra with a genetically encoded cobalt(II)-binding amino acid. Angewandte Chemie. 2011;50:692–694. doi: 10.1002/anie.201005672. doi:10.1002/anie.201005672. [DOI] [PubMed] [Google Scholar]
- Noren CJ, Anthony-Cahill SJ, Griffith MC, Schultz PG. A general method for site-specific incorporation of unnatural amino acids into proteins. Science. 1989;244:182–188. doi: 10.1126/science.2649980. [DOI] [PubMed] [Google Scholar]
- Opella SJ. Structure determination of membrane proteins in their native phospholipid bilayer environment by rotationally aligned solid-state NMR spectroscopy. Accounts of chemical research. 2013;46:2145–2153. doi: 10.1021/ar400067z. doi:10.1021/ar400067z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otting G. Protein NMR using paramagnetic ions. Annual review of biophysics. 2010;39:387–405. doi: 10.1146/annurev.biophys.093008.131321. doi:10.1146/annurev.biophys.093008.131321. [DOI] [PubMed] [Google Scholar]
- Page RC, Lee S, Moore JD, Opella SJ, Cross TA. Backbone structure of a small helical integral membrane protein: A unique structural characterization. Protein science : a publication of the Protein Society. 2009;18:134–146. doi: 10.1002/pro.24. doi:10.1002/pro.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Berkamp S, Cook GA, Chan MK, Viadiu H, Opella SJ. Nanodiscs versus macrodiscs for NMR of membrane proteins. Biochemistry. 2011a;50:8983–8985. doi: 10.1021/bi201289c. doi:10.1021/bi201289c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Casagrande F, Cho L, Albrecht L, Opella SJ. Interactions of interleukin-8 with the human chemokine receptor CXCR1 in phospholipid bilayers by NMR spectroscopy. Journal of molecular biology. 2011b;414:194–203. doi: 10.1016/j.jmb.2011.08.025. doi:10.1016/j.jmb.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Casagrande F, Das BB, Albrecht L, Chu M, Opella SJ. Local and global dynamics of the G protein-coupled receptor CXCR1. Biochemistry. 2011c;50:2371–2380. doi: 10.1021/bi101568j. doi:10.1021/bi101568j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthasarathy S, Nishiyama Y, Ishii Y. Sensitivity and resolution enhanced solid-state NMR for paramagnetic systems and biomolecules under very fast magic angle spinning. Accounts of chemical research. 2013;46:2127–2135. doi: 10.1021/ar4000482. doi:10.1021/ar4000482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestegard JH, al-Hashimi HM, Tolman JR. NMR structures of biomolecules using field oriented media and residual dipolar couplings. Quarterly reviews of biophysics. 2000;33:371–424. doi: 10.1017/s0033583500003656. [DOI] [PubMed] [Google Scholar]
- Radoicic J, Lu GJ, Opella SJ. NMR structures of membrane proteins in phospholipid bilayers. Quarterly reviews of biophysics. 2014;47:249–283. doi: 10.1017/S0033583514000080. doi:10.1017/S0033583514000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajarathnam K, Kay CM, Clark-Lewis I, Sykes BD. Characterization of quaternary structure of interleukin-8 and functional implications. Methods in enzymology. 1997;287:89–105. doi: 10.1016/s0076-6879(97)87009-7. [DOI] [PubMed] [Google Scholar]
- Saunders M, Wishnia A, Kirkwood JG. The nuclear magnetic resonance spectrum of ribonuclease. J Am Chem Soc. 1957;79:3289–3290. doi:10.1021/ja01569a083. [Google Scholar]
- Schmidt M, Borbas J, Drescher J, Summerer D. A genetically encoded spin label for electron paramagnetic resonance distance measurements. J Am Chem Soc. 2014;136:138–141. doi: 10.1021/ja411535q. [DOI] [PubMed] [Google Scholar]
- Sengupta I, Nadaud PS, Jaroniec CP. Protein structure determination with paramagnetic solid-state NMR spectroscopy. Accounts of chemical research. 2013;46:2117–2126. doi: 10.1021/ar300360q. doi:10.1021/ar300360q. [DOI] [PubMed] [Google Scholar]
- Son WS, et al. ‘q-Titration’ of long-chain and short-chain lipids differentiates between structured and mobile residues of membrane proteins studied in bicelles by solution NMR spectroscopy. Journal of magnetic resonance. 2012;214:111–118. doi: 10.1016/j.jmr.2011.10.011. doi:10.1016/j.jmr.2011.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soroka K, Vithanage RS, Phillips DA, Walker B, Dasgupta PK. Fluorescence properties of metal complexes of 8-hydroxyquinoline-5-sulfonic acid and chromatographic applications. Analytical Chemistry. 1987;59:629–636. doi:10.1021/ac00131a019. [Google Scholar]
- Su XC, et al. A dipicolinic acid tag for rigid lanthanide tagging of proteins and paramagnetic NMR spectroscopy. J Am Chem Soc. 2008;130:10486–10487. doi: 10.1021/ja803741f. doi:10.1021/ja803741f. [DOI] [PubMed] [Google Scholar]
- Su Y, Hu F, Hong M. Paramagnetic Cu(II) for probing membrane protein structure and function: inhibition mechanism of the influenza M2 proton channel. J Am Chem Soc. 2012;134:8693–8702. doi: 10.1021/ja3026328. doi:10.1021/ja3026328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M, Berthold DA, Rienstra CM. Solid-State NMR of a Large Membrane Protein by Paramagnetic Relaxation Enhancement. The journal of physical chemistry letters. 2011;2:1836–1841. doi: 10.1021/jz200768r. doi:10.1021/jz200768r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolman JR, Flanagan J, Kennedy M, Prestegard JH. Nuclear magnetic dipole interactions in filed-oriented proteins: Information for structure determination in solution. Proceedings of the National Academy of Sciences of the United States of America. 1995;92:9279–9283. doi: 10.1073/pnas.92.20.9279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich SJ, Holper S, Glaubitz C. Paramagnetic doping of a 7TM membrane protein in lipid bilayers by Gd(3)(+)-complexes for solid-state NMR spectroscopy. Journal of biomolecular NMR. 2014;58:27–35. doi: 10.1007/s10858-013-9800-4. doi:10.1007/s10858-013-9800-4. [DOI] [PubMed] [Google Scholar]
- Venditti V, Fawzi N, Clore G. An efficient protocol for incorporation of an unnatural amino acid in perdeuterated recombinant proteins using glucose-based media. Journal of biomolecular NMR. 2012;52:191–195. doi: 10.1007/s10858-012-9606-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Munro RA, Kim SY, Jung KH, Brown LS, Ladizhansky V. Paramagnetic relaxation enhancement reveals oligomerization interface of a membrane protein. J Am Chem Soc. 2012;134:16995–16998. doi: 10.1021/ja308310z. doi:10.1021/ja308310z. [DOI] [PubMed] [Google Scholar]
- Wang S, et al. Solid-state NMR spectroscopy structure determination of a lipid-embedded heptahelical membrane protein. Nature methods. 2013;10:1007–1012. doi: 10.1038/nmeth.2635. doi:10.1038/nmeth.2635. [DOI] [PubMed] [Google Scholar]
- Ward ME, Wang S, Krishnamurthy S, Hutchins H, Fey M, Brown LS, Ladizhansky V. High-resolution paramagnetically enhanced solid-state NMR spectroscopy of membrane proteins at fast magic angle spinning. Journal of biomolecular NMR. 2014;58:37–47. doi: 10.1007/s10858-013-9802-2. doi:10.1007/s10858-013-9802-2. [DOI] [PubMed] [Google Scholar]
- Wien RW, Morrisett JD, McConnell HM. Spin-label-induced nuclear relaxation. Distances between bound saccharides, histidine-15, and tryptophan-123 on lysozyme in solution. Biochemistry. 1972;11:3707–3716. doi: 10.1021/bi00770a008. doi:10.1021/bi00770a008. [DOI] [PubMed] [Google Scholar]
- Wohnert J, Franz KJ, Nitz M, Imperiali B, Schwalbe H. Protein alignment by a coexpressed lanthanide-binding tag for the measurement of residual dipolar couplings. J Am Chem Soc. 2003;125:13338–13339. doi: 10.1021/ja036022d. doi:10.1021/ja036022d. [DOI] [PubMed] [Google Scholar]
- Yagi H, Pilla KB, Maleckis A, Graham B, Huber T, Otting G. Three-dimensional protein fold determination from backbone amide pseudocontact shifts generated by lanthanide tags at multiple sites. Structure. 2013;21:883–890. doi: 10.1016/j.str.2013.04.001. doi:10.1016/j.str.2013.04.001. [DOI] [PubMed] [Google Scholar]
- Yeo KJ, et al. Rapid exploration of the folding topology of helical membrane proteins using paramagnetic perturbation. Protein science : a publication of the Protein Society. 2010;19:2409–2417. doi: 10.1002/pro.521. doi:10.1002/pro.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TS, Ahmad I, Yin JA, Schultz PG. An enhanced system for unnatural amino acid mutagenesis in E. coli. Journal of molecular biology. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. doi:10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- Zhang WH, Otting G, Jackson CJ. Protein engineering with unnatural amino acids. Current opinion in structural biology. 2013;23:581–587. doi: 10.1016/j.sbi.2013.06.009. doi:10.1016/j.sbi.2013.06.009. [DOI] [PubMed] [Google Scholar]
- Zhuang T, Lee H-S, Imperiali B, Prestegard JH. Structure determination of a galectin-3-carbohydrate complex using paramagnetism-based NMR constraints. Protein Science. 2008;17:1220–11231. doi: 10.1110/ps.034561.108. [DOI] [PMC free article] [PubMed] [Google Scholar]