Abstract
S-Adenosyl-L-methionine, an important biological cofactor, exists in two chiral forms, (S,S)- and (R,S)-, only the former of which is biologically active. Herein, we develop a chromatographic method to obtain pure (S,S)-AdoMet using a single C18 column.
Keywords: S-Adenosyl-L-methionine, Chiral resolution, Diastereoisomer, High-performance liquid chromatography, Radioactive, Separation
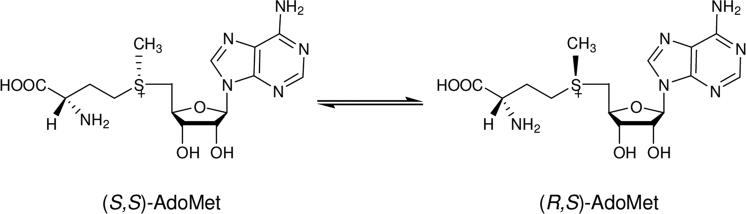
S-Adenosyl-L-methionine[1], usually abbreviated as AdoMet, SAM or SAMe, is distributed ubiquitously throughout all organisms. It is a key metabolite and has various biological functions, acting as the most common and principal methyl donor in the cell[2]. AdoMet has two notable chiral centers: one is the chiral carbon at the α amino position and the other is the sulfonium sulfur. The chiral sulfur center exists in two enantiomeric forms, making AdoMet a diastereomer: (S,S)-AdoMet and (R,S)-AdoMet. The (S,S) configuration is the only biosynthesized form, and the biologically active form for almost all AdoMet dependent methyltransferases[3, 4]; in fact, a homocysteine methyltransferase has been reported to repair unactivated (R,S)-AdoMet [5]. Once isolated or prepared, (S,S)-AdoMet spontaneously converts to the inactive (R,S)-AdoMet, with a racemization rate around 10−16 s−1 under physiological conditions [6, 7](Figure 1). Due to this property, commercial AdoMet samples contain up to 20-30% of the inactive (R,S)- form[7, 8]; even in-house enzymatically synthesized AdoMet includes some (R,S)- product [9, 10]. In our hands, we see 5-10% (R,S)-AdoMet immediately after overnight enzymatic incubation, and this level is expected to increase until sample utilization. Past publications have been largely focused on the analytical studies of AdoMet quantification and stability using NMR, mass spectrometry, capillary electrophoresis, chromatographic, colorimetric and radioactive approaches[3, 7, 8, 11-15]. There is a lack of appropriate preparative protocols to isolate the pure (S,S)-AdoMet in a simple and efficient way. Usage of the mixture of (S,S)- and (R,S)-AdoMet can lead to artifacts in the study of enzymes, leading, for example to overestimation of the Km for AdoMet and underestimation of the kcat. The difficulty of obtaining a complete separation of S,S and R,S diastereoisomers has also led to different conclusions regarding the level of AdoMet in plasma from immunocompromised adults[16, 17]. Here, we report an efficient method to separate the (S,S)- and (R,S)-AdoMet using common elution buffers and a single C18 column.
Figure 1.
Racemization of S-adenosyl- L-methionine (AdoMet)
AdoMet was synthesized biologically from adenosine triphosphate (ATP) and methionine with methionine adenosyltransferase (AdoMet synthetase), using a similar procedure to that outlined by Walsby et al. [10], with minor modifications. A typical 10 mL reaction mixture contained 100 mM Tris (pH=8), 1 mM EDTA, 50 mM KCl, 26 mM MgCl2, 13 mM ATP, 10 mM methionine, 8% β-mercaptoethanol and 1 mL of AdoMet synthetase lysate[18]. The reactions were left stirring vigorously overnight at 37°C in the dark. After approximately 16 h, the reactions were quenched with 1 ml of 1.0 M HCl and then centrifuged at 4°C at 15,000×g for 20 min. The lysates were purified via FPLC (BioRad) using a UNO S-6 (BioRad) cation exchange column. A linear gradient from 0-1 M HCl was run through the column, from which AdoMet was eluted at approximately 0.4 to 0.6 M HCl as a distinct peak. The eluant was collected and dried by lyophilization or by rotary evaporation in an ice-water bath, with a yield of ca. 40%. Due to the common usage of radioactively labeled AdoMet in enzyme assays, tritium [methyl-3H]-AdoMet (specific activity: 80 Ci/mmol) was also purchased from American Radiolabeled Chemicals Inc., United States and analyzed.
In order to obtain pure, fully active (S,S)-AdoMet, reversed phase chromatography was carried out using a commonly used C18 reversed phase analytical column (Kinetex™ 5 μm, 100 Å, 250×4.6 mm from Phenomenex). High-performance liquid chromatography (HPLC) utilized either a PerkinElmer Series 200 system for non- radioactively labeled AdoMet or a composite system with Beckman 110B pumps and a Hitachi 655A UV-visible detector for the radioactively labeled AdoMet. Separations were carried out at room temperature unless specified. HPLC flow rate is 1 ml/min with absorbance monitored at 257 nm. Three elution buffers are used here: Buffer 1, 50 mM ammonium formate buffer (pH=4.0); Buffer 2, 50 mM ammonium acetate buffer (pH=5.4) with 1% TFA and Buffer 3, 10 mM Phosphate buffer (pH=6.8). Between runs, the column is washed with acetonitrile: water (7:3, v/v) and then pre-equilibrated with the appropriate elution buffer for 10-15min.
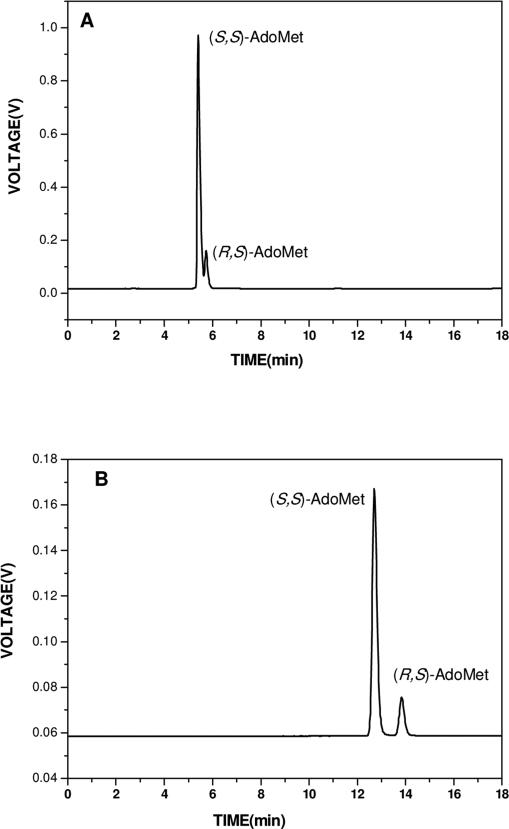
The first step to obtain pure (S,S)-AdoMet is to apply Buffer 1 as the eluant, resulting in retention times of 5.4 min for (S,S)-AdoMet and 5.7 min for (R,S)-AdoMet (Figure 2A) [14]. This step allows baseline separation of (S,S)- and (R,S)-diastereoisomers only if extremely small amounts of AdoMet are injected into the column, making it suitable for analytical purposes [14]. The small retention time difference (18 seconds) between (S,S)- and (R,S)- makes it impossible to separate diastereoisomers using preparative or semi-preparative levels of AdoMet. However, for commercial samples, Step 1 achieves the buffer exchange necessary for purification of AdoMet, especially the radioactively labeled samples, which are stored in sulfuric acid/ethanol buffer [typical condition, sulfuric acid (pH 2.0): ethanol (9:1)]. If these are loaded directly onto the C18 column equilibrated with Buffer 2 (Step 2), AdoMet is found to elute across the entire profile. Step 1 is not necessary if the AdoMet sample has been prepared “in house” and stored in water. Depending on the initial AdoMet storage buffer, the retention time might change, but the overall profile is robust and reproducible.
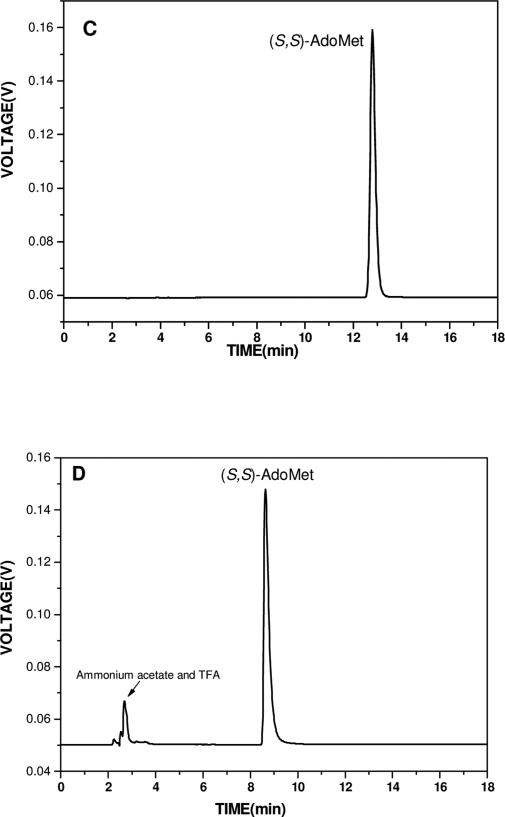
Figure 2.
HPLC elution profiles for different steps. Flow rate, 1 ml/min; isocratic elution; detection wavelength, 257 nm. (A) Step 1, elution buffer 1, 50mM ammonium formate buffer (pH=4.0); (B) Step 2, elution buffer 2, 50mM ammonium acetate buffer (pH=5.4) with 1% TFA; (C) (S,S)- AdoMet re-purified according to Step2; (D) Step 3, elution buffer 3, 10mM Phosphate buffer (pH=6.8). As discussed in the text, the early eluting peak is ammonium acetate and TFA.
The concentrated sample from Step 1 is loaded onto the same HPLC column pre-equilibrated with Buffer 2. In order to reduce any chemical breakdown of AdoMet and conversion of (S,S)-AdoMet to (R,S)-AdoMet, the eluant is pre-cooled to 4°C and stored in an ice bucket before and during the procedures performed in Steps 2 and 3. (S,S)-AdoMet and (R,S)-AdoMet elute at 12.7 min and 13.8 min, respectively (Figure 2B), with baseline separation. Pure (S,S)-AdoMet is collected from Step 2, concentrated at 4 °C and re-injected onto the same column with Buffer 2 (Step 2). From Figure 2C, it is clear that only an (S,S)-AdoMet peak is present, confirming the purity of the bioactive (S,S)-AdoMet. Since Buffer 2 contains ammonium acetate and especially TFA, which are likely to affect subsequent biological assays, a final step was introduced to remove these buffer components from the sample solution.
Due to the polar property of AdoMet, we first attempted cation exchange chromatography. While this procedure allowed removal of the ammonium acetate and TFA, it introduced high concentrations of salt and hence an elevated ionic strength for the final buffer. In light of this, we returned to the C18 column used in previous steps, applying Buffer 3 as the eluant to eliminate ammonium acetate and TFA. These conditions wash the ammonium acetate and TFA off the column between 2-4 min (1st peak in Figure 2D) followed by (S,S)-AdoMet at 8.6 min (Figure 2D). The removal of ammonium acetate and TFA was confirmed by a control HPLC profile using the same concentration of ammonium acetate and TFA in the absence of AdoMet. After elution, H3PO4 was added immediately to bring the pH of the (S,S)-AdoMet sample to around 3. Pure (S,S)-AdoMet sample can be aliquoted and stored in -20 °C or -80°C. We point out that the eluant buffer in Step 3 can be replaced by other buffers (for example, 10 mM HEPES PH=6.8) as long as they have a pH ≥ 6. This offers considerable flexibility for biological studies.
We have presented a straight forward HPLC method for the preparation of pure bioactive (S,S)- AdoMet using a C18 column in three steps. Considering that radioactively labeled AdoMet is frequently employed in biology, we carried out the purification of [methyl-3H]-AdoMet using the protocol developed here. Around 60 μCi of (S,S)- [methyl-3H]-AdoMet was obtained using the procedures outlined above. The purity of the resulting (S,S)- [methyl-3H]-AdoMet was tested by reaction with excess dopamine (5 mM) catalyzed by 50 μM catechol-O-methyltransferase (COMT) in 100 mM phosphate buffer pH=7.4 with 5 mM MgCl2, 4 mM DTT for 30 min [19]. Analysis of the product mixture by HPLC showed that no inactive (R,S)-[methyl-3H]-AdoMet was present in the reaction mixture. Storage of the (S,S)-[methyl-3H]-AdoMet in acidic buffer (pH ≅ 3) at -20 °C preserved the sample purity for at least one month.
In conclusion, a protocol for separation of (S,S)- AdoMet has been established using reversed phase chromatography. This will enable reliable access to pure AdoMet, eliminating artifacts that may arise from the presence of chemical and enantiomeric impurities.
Acknowledgments
The authors are grateful for funding of this laboratory by the National Institutes of Health (NIH) to J.P.K. (GM025765 and GM039296).
Abbreviations used
- AdoMet
S-Adenosyl-L-methionine
- ATP
adenosine triphosphate
- COMT
catechol-O-methyltransferase
- HPLC
high-performance liquid chromatography
- NMR
nuclear magnetic resonance
- TFA
trifluoroacetic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cantoni GL. The nature of the active methyl donor formed enzymatically from l-methionine and adenosinetriphosphate. Journal of the American Chemical Society. 1952;74:2942–2943. [Google Scholar]
- 2.Lu SC, Mato JM. S-adenosylmethionine in liver health, injury, and cancer. Physiological Reviews. 2012;92:1515–1542. doi: 10.1152/physrev.00047.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinci CR, Clarke SG. Recognition of age-damaged (R,S)-adenosyl-L-methionine by two methyltransferases in the yeast Saccharomyces cervisiae. Journal of Biological Chemistry. 2007;282:8604–8612. doi: 10.1074/jbc.M610029200. [DOI] [PubMed] [Google Scholar]
- 4.Vinci CR, Clarke SG. Homocysteine Methyltransferases Mht1 and Sam4 Prevent the Accumulation of Age-damaged (R,S)-AdoMet in the Yeast Saccharomyces cerevisiae. Journal of Biological Chemistry. 2010;285:20526–20531. doi: 10.1074/jbc.M110.113076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Linster CL, Van Schaftingen E, Hanson AD. Metabolite damage and its repair or pre-emption. Nature Chemical Biology. 2013;9:72–80. doi: 10.1038/nchembio.1141. [DOI] [PubMed] [Google Scholar]
- 6.Wu SE, Huskey WP, Borchardt RT, Schowen RL. Chiral instability at sulfur of s-adenosylmethionine. Biochemistry. 1983;22:2828–2832. doi: 10.1021/bi00281a009. [DOI] [PubMed] [Google Scholar]
- 7.Hoffman JL. Chromatographic analysis of the chiral and covalent instability of s-adenosyl-l-methionine. Biochemistry. 1986;25:4444–4449. doi: 10.1021/bi00363a041. [DOI] [PubMed] [Google Scholar]
- 8.Beaudouin C, Haurat G, Laffitte JA, Renaud B. The presence of (+)-s-adenosyl-l-methionine in the rat-brain and its lack of effect on phenylethanolamine n-methyltransferase activity. Journal of Neurochemistry. 1993;61:928–935. doi: 10.1111/j.1471-4159.1993.tb03604.x. [DOI] [PubMed] [Google Scholar]
- 9.McCarthy DL, Capitani G, Feng L, Gruetter MG, Kirsch JF. Glutamate 47 in 1-aminocyclopropane-1-carboxylate synthase is a major specificity determinant. Biochemistry. 2001;40:12276–12284. doi: 10.1021/bi011050z. [DOI] [PubMed] [Google Scholar]
- 10.Walsby CJ, Hong W, Broderick WE, Cheek J, Ortillo D, Broderick JB, Hoffman BM. Electron-nuclear double resonance spectroscopic evidence that S-adenosylmethionine binds in contact with the catalytically active [4Fe-4S](+) cluster of pyruvate formate-lyase activating enzyme. Journal of the American Chemical Society. 2002;124:3143–3151. doi: 10.1021/ja012034s. [DOI] [PubMed] [Google Scholar]
- 11.Cannon LM, Butler FN, Wan W, Zhou ZS. A stereospecific colorimetric assay for (S,S)-adenosylmethionine quantification based on thiopurine methyltransferase-catalyzed thiol methylation. Analytical Biochemistry. 2002;308:358–363. doi: 10.1016/s0003-2697(02)00267-1. [DOI] [PubMed] [Google Scholar]
- 12.Iwig DF, Booker SJ. Insight into the polar reactivity of the onium chalcogen analogues of S-adenosyl-L-methionine. Biochemistry. 2004;43:13496–13509. doi: 10.1021/bi048693+. [DOI] [PubMed] [Google Scholar]
- 13.Desiderio C, Cavallaro RA, De Rossi A, D'Anselmi F, Fuso A, Scarpa S. Evaluation of chemical and diastereoisomeric stability of S-adenosylmethionine in aqueous solution by capillary electrophoresis. Journal of Pharmaceutical and Biomedical Analysis. 2005;38:449–456. doi: 10.1016/j.jpba.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 14.Cataldi TRI, Bianco G, Abate S, Mattia D. Analysis of S-adenosylmethionine and related sulfur metabolites in bacterial isolates of Pseudomonas aeruginosa (BAA-47) by liquid chromatography/electrospray ionization coupled to a hybrid linear quadrupole ion trap and Fourier transform ion cyclotron resonance mass spectrometry. Rapid Communications in Mass Spectrometry. 2009;23:3465–3477. doi: 10.1002/rcm.4274. [DOI] [PubMed] [Google Scholar]
- 15.Farrar CE, Siu KKW, Howell PL, Jarrett JT. Biotin Synthase Exhibits Burst Kinetics and Multiple Turnovers in the Absence of Inhibition by Products and Product-Related Biomolecules. Biochemistry. 2010;49:9985–9996. doi: 10.1021/bi101023c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skelly M, Merali S, Clarkson AB. Pneumocystis pneumonia and S-adenosylmethionine plasma levels. Journal of Infection. 2011;62:490–492. doi: 10.1016/j.jinf.2011.04.012. [DOI] [PubMed] [Google Scholar]
- 17.de Boer MGJ, van Zelst B, Gelinck LBS, Kroon FP, de Jonge R. Pneumocystis pneumonia and measurement of S-adenosylmethionine plasma levels. Journal of Infection. 2011;62:493–495. [Google Scholar]
- 18.A clone containing the AdoMet synthetase overproducing strain DM22 (pK8) was generously donated by Prof. Joan B. Broderick from Montana State University. In brief, DM22 (pK8) was grown at 37°C overnight in LB media containing 50 μg/mL oxytetracycline. The cells were harvested via centrifugation and reconstituted in 4ml/1g wet cell of 100 mM Tris, 1mM EDTA, pH 8. Lysozyme (Sigma, powder) was added to the mixture for a final concentration of 50 μg/mL and left to incubate at room temperature for 30 min. Phenylmethylsulfonyl fluoride (0.1 mM) was added to the solution and the cells lysed via sonication in an ice bath. The cell lysates were cleared via centrifugation and the supernatants stored at −80°C until further use. Typically, 1ml AdoMet synthetase lysate can convert 100μmol of substrate to product in 12 hours.
- 19.Zhang J, Klinman JP. Enzymatic Methyl Transfer: Role of an Active Site Residue in Generating Active Site Compaction That Correlates with Catalytic Efficiency. Journal of the American Chemical Society. 2011;133:17134–17137. doi: 10.1021/ja207467d. [DOI] [PMC free article] [PubMed] [Google Scholar]