Abstract
Lipoxygenases (LOX) regulate inflammation through the production of a variety of molecules whose specific downstream effects are not entirely understood due to the complexity of the inflammation pathway. The generation of these biomolecules can potentially be inhibited and/or allosterically regulated by small synthetic molecules. The current work describes the first mass spectrometric, high throughput method for identifying small molecule LOX inhibitors and LOX allosteric effectors, which change the substrate preference of human lipoxygenase enzymes. Using a volatile buffer and an acid-labile detergent, enzymatic products can be directly detected using liquid chromatography-mass spectrometry (HPLC-MS), without the need of organic extraction. The method also reduces the required enzyme concentration compared to traditional UV absorbance methods by approximately 30-fold, allowing accurate binding affinity measurements for inhibitors with nanomolar affinity. The procedure was validated using known LOX inhibitors and the allosteric effector, 13(S)-hydroxy-9Z,11E-octadecadienoic acid (13-HODE).
Keywords: Human 15-lipoxygenase-1, human 15-lipoxygenase-2, high-throughput assay, inhibitor, allostery, mass spectroscopy
1.1 Introductory Statement
The emergence of mass spectrometry (MS) based enzyme assays has rapidly increased in recent years, due in large part to advances in both MS instrument hardware and software capabilities.[1] HPLC-MS provides an increasingly convenient and economical means for direct detection of enzymatic products, and thus offers a viable alternative to more limited detection methods. A number of exciting HPLC-MS based inhibition assays have been described recently, [2–4] however, no HPLC-MS based inhibitor assays compatible with lipoxygenase (LOX) enzymes and their hydrophobic substrates/inhibitors have currently been reported.
Lipoxygenase enzymes oxygenate 1,4-cis,cis-pentadiene-containing polyunsaturated fatty acids to produce signaling molecules which regulate inflammation. Human 15-lipoxygenase-1 (15-LOX-1 or 12/15-LOX) is implicated in atherogenesis,[5] diabetes,[6] Alzheimer’s disease,[7] breast cancer,[8] and stroke.[9] Human 15-lipoxygenase-2 (15-LOX-2) is implicated in breast,[10] prostate,[11] and renal cancers,[12] as well as atherosclerosis.[13] Identifying inhibitors to these and other human LOXs has been an area of continued research. [14–20] The common method for discovering LOX inhibitors is the UV absorbance based assay, however, this method requires high nanomolar concentrations of LOX to produce measurable enzymatic rates. Thus, this method limits the lower range of inhibitor potency to greater than 10 nM, as has already been observed.[14] The high sensitivity of mass spectrometry provides a means for measuring product formation in smaller amounts than the UV method, potentially allowing inhibitor measurements for more potent compounds. However, LOX product measurements by mass spectrometry are impeded by the requirement of detergents to inhibit chemical aggregates and prevent promiscuous inhibition.[21] To circumvent this HPLC-MS compatibility problem, the acid cleavable detergent, sodium 3-(4-(1,1-bis(hexyloxy)ethyl)pyridinium-1-yl)propane-1-sulfonate (PPS), is utilized, which degrades before injection onto the HPLC-MS. The use of PPS, coupled with a volatile buffer, provides a reaction condition which does not require organic extraction, yields high sensitivity measurements of LOX reaction products and is fully compatible with a high-throughput, direct injection HPLC-MS process.
In addition to high sensitivity, mass spectrometry based product detection also provides the ability to differentiate unique enzymatic products. This opens the possibility of studying substrate specificity changes in addition to enzymatic inhibition, providing a means to determine if an inhibitor is binding the allosteric site of LOX. LOXs make particularly interesting targets for allosteric regulation as their various isozymes can act on multiple substrates producing an array of products, whose ratios have unique biological effects. [22,23] Previous competitive substrate experiments have shown that the allosteric sites of both 15-LOX-1 and 15-LOX-2 bind specific LOX products to regulate substrate preference,[24,25] however, non-native allosteric molecules have not been identified. The ability to identify small molecules capable of modulating substrate preference of LOX enzymes would be a valuable tool for probing the downstream biological effects of specific LOX product ratios on inflammatory related diseases. Unfortunately, current methods for measuring LOX substrate preference require large-scale reactions and time-consuming HPLC analysis, and are thus inadequate for compound screening.
In the current work, we utilize high sensitivity HPLC-MS analysis, coupled with HPLC-MS compatible enzyme reaction conditions, to provide fast and reliable inhibitor data for a set of 15-LOX-1 tight binding inhibitors whose exact potency was previously unknown.[14] Furthermore, the reported method opens the possibility of analyzing multiple reaction products simultaneously, potentially leading to the discovery of allosteric effector molecules of LOX. Finally, this novel method is amenable to miniaturization, allowing for adaptation to a 96-well format, with reagent requirements and analysis times compatible with high throughput (HTP) screening.
1.2 Materials and Methods
1.2.1 Materials
All commercial fatty acids were purchased from Nu Chek Prep, Inc. (MN, USA) and were further re-purified using a Higgins HAISIL (5 µm, 250 × 10 mm) C-18 column. An isocratic elution of 85% A (99.9% methanol and 0.1% acetic acid): 15% B (99.9% water and 0.1% acetic acid) was used to purify all the fatty acids. Post purification, the fatty acids were stored at −80 °C for a maximum of 6 months. LOX products 15(S)-hydroxy-5Z,8Z,11Z,13E-eicosatetraenoic acid (15-HETE), 13(S)-hydroxy-9Z,11E-octadecadienoic acid (13-HODE) and d30-13-HODE (fully deuterated 13-HODE) were made and purified as previously described. [26,27] NDGA was purchased from Sigma/Aldrich Chemicals. The inhibitors were obtained from the NIH Molecular Libraries Small Molecule Repository: (https://mli.nih.gov/mli/compound-repository/). All other chemicals were reagent grade or better and were used without further purification.
1.2.2 Overexpression and Purification of Lipoxygenases
15-LOX-1 and 15-LOX-2 were expressed as N-terminal His6-tagged proteins and were purified via immobilized metal affinity chromatography using an Ni-NTA resin.[18] The protein purity was evaluated by SDS-PAGE analysis and was found to be greater than 90%.
1.2.3 Lipoxygenase Inhibitor Affinity Assay
LOX turnover reactions were performed at 3 ml volume in a stirred reaction vessel. Assay buffer consisting of 50 mM ammonium bicarbonate adjusted to pH 7.5 with 10 µM arachidonic acid (AA), 0.01% PPS acid cleavable detergent (Figure 1) and 0.2 mg/ml BSA. Various concentrations of inhibitor were added in 10 µl of DMSO or vehicle control. The reaction was initiated by adding 0.6 nM 15-LOX-1, and quenched at <20% turnover (500 sec) by addition of 200 µM glacial acetic acid. 6 µM 13-HODE was added as an internal standard. 10 µl of each sample, without extraction, was injected on to a Thermo LTQ HPLC-MS equipped with an electrospray ionization (ESI) source set to and negative ions [M–H] mode. A Thermo Aquasil (3 µm, 30 mm × 2.1 mm) C-18 reverse phase column, held at ambient temperature (20° C), was used to separate buffer salts and protein from fatty acids. A 5 min gradient was run at a flow rate of 200 µl/min, beginning with 70% water/30% acetonitrile and ending with 10% water, 90% acetonitrle, followed by 100% acetonitrile for 3 minutes. All solvents contained 0.1% formic acid. The first minute of each run was diverted to waste. The operating conditions were determined as follows: sheath gas flow rate, 35 arb. units; auxiliary gas flow rate, 10 arb. units; sweep gas flow rate, 0.0 arb. units; spray voltage, 5.0 kV; capillary temp, 275° C; capillary voltage, −34 V; tube lens voltage, −118.66 V All masses between 290 and 340 m/z were scanned, with LOX products eluding between 5 and 6 minutes. Products were identified based on their MS/MS fragmentation pattern (LipidMaps.org). 15-HETE was identified by its parent mass of 319.5 m/z (negative ion) and key fragments 219, 175, and 121 m/z. 13-HODE was identified by its parent mass of 295.5 m/z (negative ion) and key fragment 195 m/z. Between each sample the column was washed with 50% acetonitrile/50% isopriponol followed by 90% water/10% methanol. Percent inhibition was calculated for each run by normalizing the 15-HETE peak area to the 13-HODE internal standard and comparing to the 15-HETE/13-HODE ratio in control samples. The control reaction was done in parallel and the percent inhibition was determined by comparison of the product area of inhibited and non-inhibited samples. Over the time of the entire experiment, the area of the non-inhibited controls only varied by 7%. A standard curve was performed to verify that products were being detected within their linear range, with all concentrations run in triplicate. Peak areas were linear for all concentrations tested, yielding a standard curve with a coefficient of determination (R2) of 0.998 for 15-HETE (Figure S1). In addition, the area of the LOX products were compared with and without the assay matrix and found to be within 90% of eachother. Inhibitor assays were performed by measuring enzyme inhibition at 5 inhibitor concentrations in triplicate. The Kiapp values were determined by plotting the fractional velocity as a function of the inhibitor concentration, followed by a quadratic fit using the Morrison equation.[28] This was required because the Kiapp value approached the total active enzyme concentration ([E]), indicating that these are tight binding inhibitors and therefore hyperbolic fitting of the data was inappropriate. This treatment is based on defining the Kiapp in terms of bound and free concentrations of inhibitor and LOX, without any assumptions regarding the amount of free component depletion due to binary complex formation, as described before.[14] To determine the average Kiapp and the associated error, the enzyme concentration in the Morrison equation was varied from the maximal active enzyme concentration as measured by the metal content ([E]) to 0.01 nM. The subsequent Kiapp values were averaged and the standard deviation determined. NDGA was used as a control inhibitor to compare UV absorbance and HPLC-MS methods. A UV based IC50 was performed as previously described[14] using ammonium bicarbonate buffer and PPS in 3 ml reaction volume with approximately 20 nM of 15-LOX-1. Aliquots were taken directly from UV cuvettes, quenched and run in HPLC-MS assay. IC50 values for NDGA were determined by fitting data to a hyperbolic equation.
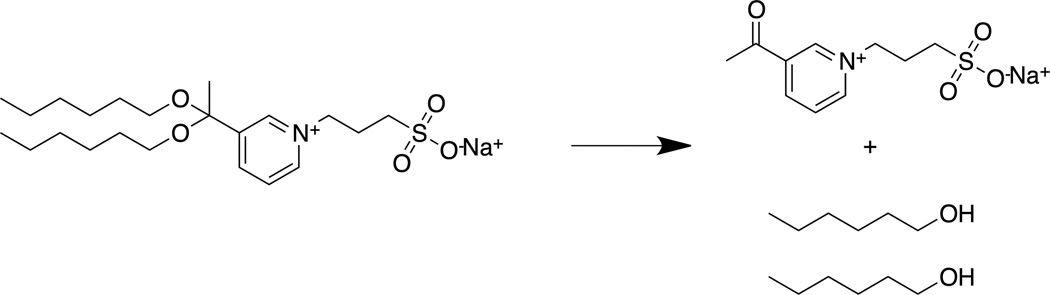
Figure 1. PPS Cleavage Mechanism.
In the presence of acid, PPS is cleaved to yield 3-acetyl-1-(3-sulfopropyl)pyridinium and hexanol.
1.2.4 96 Well Allosteric Effector Assay
Turnover reactions were performed in 96-well plates, with 100 µl total volume using assay buffer consisting of 50 mM ammonium bicarbonate adjusted to pH 7.5 with 0.01% PPS acid cleavable detergent. To verify the allosteric effect on the substrate ratio change, d30-13-HODE at 0, 10 and 20 µM was used. Effector compound was diluted in 25 µl of assay buffer before adding to assay plate. Approximately 700 ng (150 nM in 100 µl) 15-LOX-2 protein in 25 µl assay buffer was added by multichannel pipette and allowed to incubate with the effector for 30 seconds. The reaction was initiated by adding 50 µl of assay buffer containing 16 µM γ-Linolenic acid (GLA) and 4 µM AA and mixed with a multichannel pipette. The reaction was quenched between 5 and 10% total turnover (60 sec) by adding 50 µl MeOH containing 6% glacial acetic acid to cleave PPS detergent, and 500 µM tris(2-carboxyethyl)phosphine (TCEP) to reduce LOX products. Samples were incubated for 30 minutes to allow PPS cleavage to occur, then samples were neutralized by adding 50 µl of 80% MeOH, 20% 4 M NaOH, to facilitate detection of products using negative mode. Addition of methanol to samples increased detected product, presumably by minimizing product adhesion to wells. Well plates were covered with silicon sealing mats and kept at 4° C during analysis to limit evaporation. Twenty µl of each sample was injected on to the HPLC-MS, using conditions identical to those described above. Products were identified based on their MS/MS fragmentation pattern. 15-HETE was identified as described above, while 13(S)-hydroxy-6Z,9Z,11E-octadecatrienoic acid (13-HOTrE(γ)) was identified by its parent mass of 293.4 m/z (negative ion), and its key fragments at 231 and 193 m/z. The ratio of 15-HETE to 13-HOTrE(γ) peak areas was utilized to determine the substrate preference for each individual sample. Typical chromatograms for m/z of 319.5 (15-HETE) and 293.4 (13-HOTrE(γ)) can be found in supplemental data (Figures S2 and S3). A standard curve was performed to verify that products were being detected within their linear range (Figure S4). Standards were run in triplicate and standard curves had R2 values of 0.999 for 15-HETE and 0.996 for 13-HOTrE(γ). An internal standard is not utilized since we are observing the 15-HETE/13-HOTrE(γ) ratio and any detector variations, such as ionization suppression, are reflected in both product areas. In addition, the area of the LOX products were compared with and without the assay matrix and found to be within 90% of eachother.
1.3 Results and Disccussion
1.3.1 Lipoxygenase Inhibitor Affinity Assay
The advent of effective high throughput screening methods for the discovery of human LOX inhibitors has lead to the publication of several highly potent compounds against many of the human LOX isozymes.[14,19,20,29] Ranking the potency of these inhibitors is vital to understanding structure/activity relationships and thus optimizing inhibitor binding. However, several compounds were previously discovered with IC50 values below the limit of the UV LOX assay (IC50 <10 nM).[14] The current HPLC-MS assay is designed to use enzyme concentrations below those for the UV LOX assay, providing the ability to determine greater inhibitor potencies than previously possible.[14]
HPLC-MS is a powerful tool for identifying and quantitating analytes at low concentrations, but several key hurdles must be addressed for its application to an LOX inhibition assay. Ammonium bicarbonate was used due to its wide buffer range (pH 6.8 to 11.3) and volatility, which limits the introduction of salt. A detergent is required due to the hydrophobic nature of the LOX substrates and inhibitors, which tend to aggregate. However, this presents a problem for HPLC-MS detection, as traditional detergents are not compatible with mass spectrometry. To remedy this problem, the acid cleavable detergent PPS was used. The mechanism for PPS cleavage is shown in Figure 1 and is achieved when the reaction is quenched and incubated with acetic acid. To maximize analyte signal and reduce error, the aqueous reducing agent, TCEP was added with the acetic acid, this reduces the peroxide to an alcohol, which eliminates decomposition into the oxo-lipid product and results in a single mass peak (the alcohol). It should be noted that reduction is required because the hydroperoxide product (e.g. 15(S)-hydroperoxy-5Z,8Z,11Z,13E-eicosatetraenoic acid (15-HpETE)), can degrade to the alcohol (15-HETE) and ketone during ionization, thus hampering its detection. Finally, samples are adjusted back to neutral pH, to facilitate negative ion detection. A standard curve was performed to ensure linearity over the concentrations of interest (Figure S1).
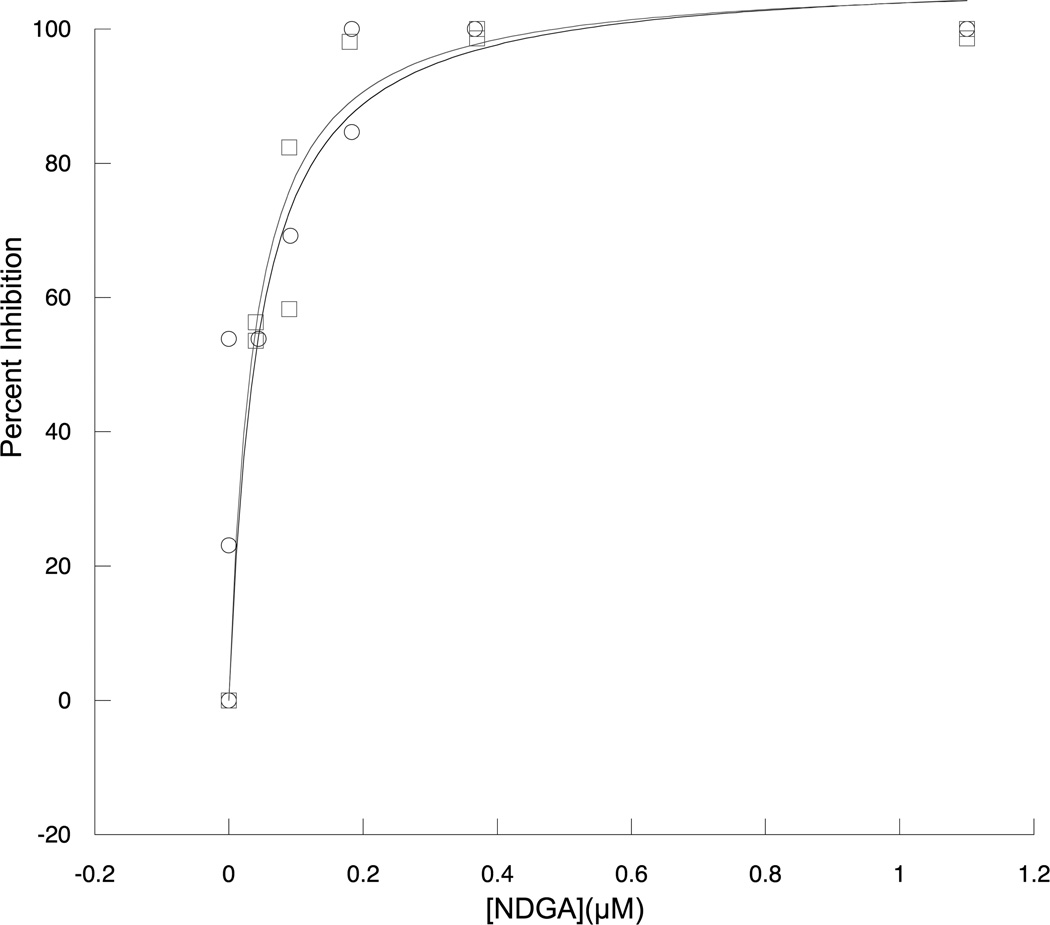
To probe the accuracy of the reported inhibitor assay, the established 15-LOX-1 inhibitor NDGA was used. The HPLC-MS assay was performed with split samples of the UV LOX assay, quenched at appropriate total turnover (<20%). Saturation curves for inhibition of NDGA to 15-LOX-1 obtained using UV and HPLC-MS methods are shown in Figure 2. Data was fit to the simple hyperbolic equation to yield the IC50 values reported in Figure 3 and indicate excellent agreement between the two methods. The IC50 values for NDGA are within the range of published values.[15] To ensure that detector variability had a minimal effect on product measurements, the standard deviation of the internal standard peak area was determined for each day the assay was run. Day to day variations were observed due to instrumental drift (less than 25%), but this drift did not significantly affect the daily slope of the standard curve, nor the determined IC50 values.
Figure 2. Comparison of UV and HPLC-MS based IC50 measurements.
Inhibition of 15-LOX-1 by NDGA was measured with both UV and HPLC-MS based assays. Black circles represent data recorded by UV assay, grey squares represent HPLC-MS assay data. The absolute value of the standard deviation in HPLC-MS assay data does not exceed 15% error for any inhibitor concentration.
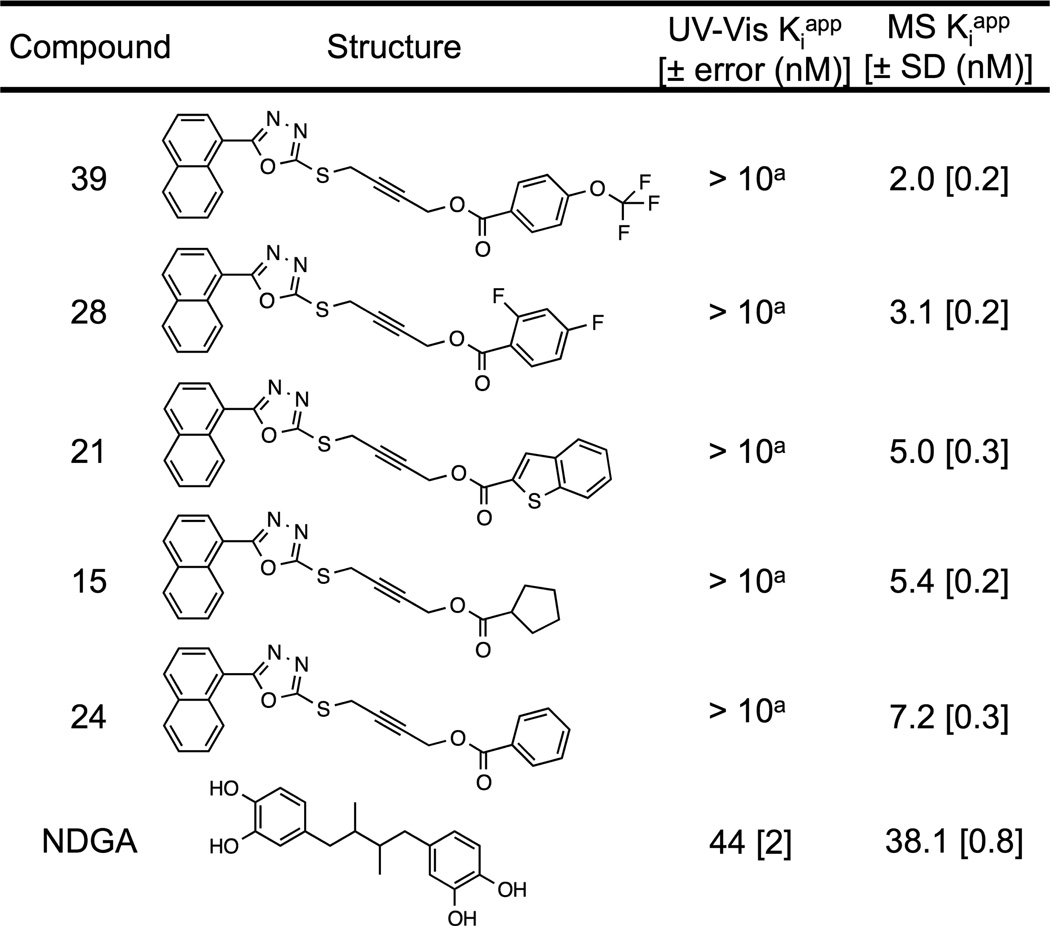
Figure 3. 15-LOX-1 IC50 measurements for tight binding inhibitors.
Binding affinities for tight binding 15-LOX-1 inhibitors as measured by HPLC-MS and UV based methods. a Binding affinities measured by the UV LOX assay were published previously.[14]
Previously published nanomolar active 15-LOX-1 inhibitors possessed binding affinities too great to be measured by traditional UV-Vis methodology.[14] Due to the approximately 30-fold lower enzyme concentration required for the current HPLC-MS assay, five compounds were chosen from the inhibitor series reported by Rai et. al. and their Kiapp values determined (Figure 3). The measured Kiapp values for the inhibitors still approached the concentration of LOX, despite the significant decrease in required enzyme concentration. This precluded the use of hyperbolic data fitting and instead, the Kiapp values were determined using the Morrison equation.[28] The Kiapp values are listed for both the HPLC-MS assay and the UV assay (Figure 3) and it is apparent that the Kiapp values for the new assay are more accurate than the approximate values published previously (IC50 < 10 nM). Inhibition plots for these compounds can be found in the supplemental data (Figure S5). In addition, the data demonstrate that the affinity of the inhibitor series increases with greater hydrophobicity, in agreement with the previous conclusion, however not to the degree that was expected.[14] None of these inhibitors have lower than nanomolar potency. This novel HPLC-MS method lowers the potency limit for inhibitors to the low nanomolar scale (approximately 0.3 nM) and allows for a wider range of the most promising LOX inhibitors for cellular studies.
1.3.2 96-Well Allosteric Effector Assay
Allosteric regulation occurs when enzyme activity is affected by binding of a regulator to a site other than the active site. This change in enzyme activity is the result of changes in active site due to conformational changes facilitated by effector binding. Allosteric regulation can either be positive (allosteric activator) or negative (allosteric inhibitor). To identify changes in active site topography, two substrates with different affinity for the active site are allowed to compete for turnover and the ratio of product formation by each substrate is determined. If the addition of an effector changes this substrate specificity ratio, it indicates that the effector is binding to an allosteric site. This allosteric regulation of substrate specificity has been previously observed for both 15-LOX-1 and 15-LOX-2 with the addition of specific LOX products, indicating auto-regulation.[24–26,30] However, the HTP screening for additional allosteric effector molecules is not possible in the previously published HTP screens because of the difficulty of measuring the change in product ratios.[29,31–34] The current HPLC-MS LOX assay overcomes these challenges due to its rapid nature and its ability to measure product ratios. To ensure a linear response to each of the LOX products, standard curves were performed with each assay. Typical standard curve data can be seen in Figure S4. Each analyte concentration was also tested at multiple fixed concentrations of the other analyte to ensure no interference in detection was observed (data not shown).
To test the validity of the current assay for determining the allosteric effect, changes in the substrate preference ratio of 15-LOX-2 with AA and GLA were measured. d30-13-HODE was used as an external allosteric effector molecule. The perdeuterated product was required in order to differentiate it from the AA and GLA products, as previously published.[24–26] The substrate preference ratio is measured below 20% conversion to product, to ensure a reliable ratio measurement. Due to a high preference for AA under the current assay conditions, a 1:4 ratio of AA to GLA was used to maintain a low overall percent turnover while still having enough of each product for accurate measurements on HPLC-MS. A total substrate concentration of 20 µM was chosen to ensure maximal product detection and minimal in-situ product generation, which could elicit an auto-feedback allosteric mechanism.
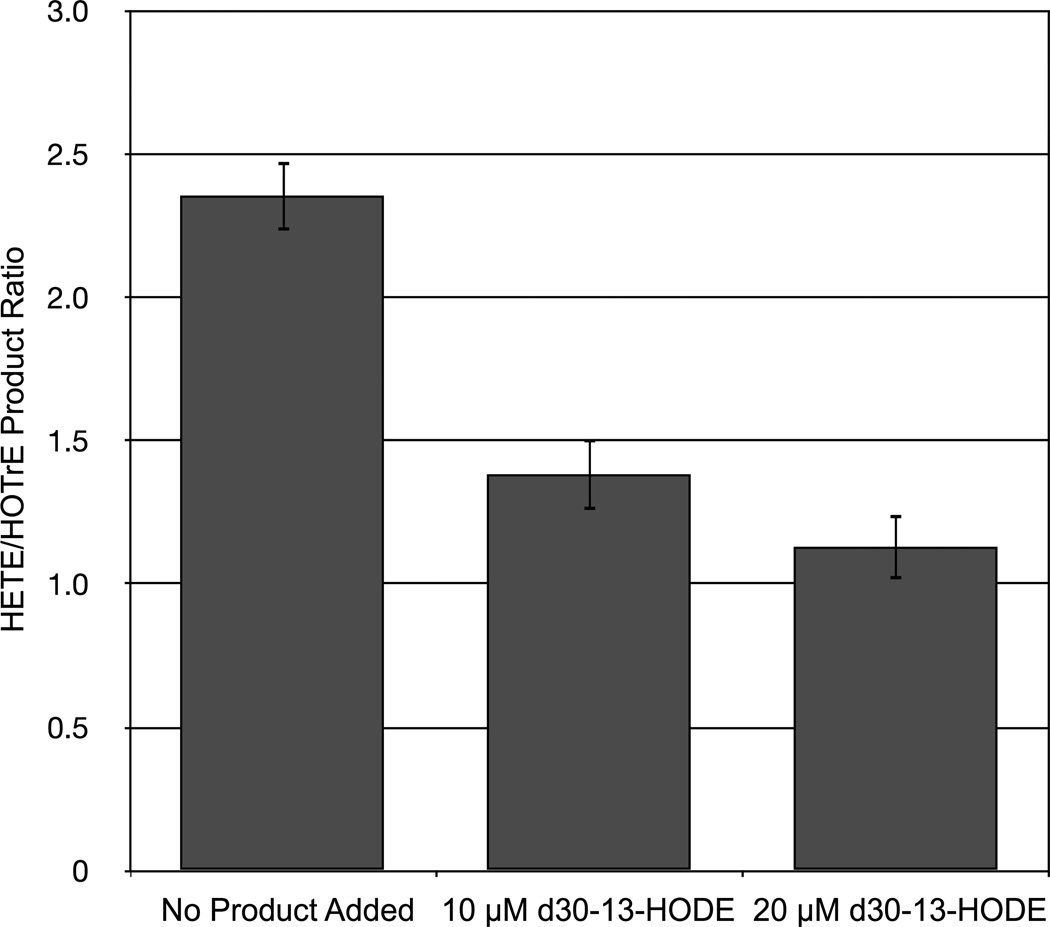
The effect of d30-13-HODE addition on substrate preference is summarized in Figure 4. Using 15-LOX-2 with its substrates AA and GLA, the AA/GLA ratio was determined to be 2.3 ± 0.1. Using the known allosteric effector d30-13-HODE, we were able to titrate a change in substrate preference, eliciting an AA/GLA ratio of 1.4 ± 0.1 with 10 µM d30-13-HODE and ultimately achieving a ratio of 1.1 ± 0.1 with 20 µM d30-13-HODE added. Based on the standard curve for AA and GLA products, the addition of d30-13-HODE changed the AA/GLA ratio by reducing AA catalysis, but not affecting GLA catalysis significantly (data not shown). These data indicate that the current method successfully detects allosteric regulation of 15-LOX-2 at biologically relevant concentrations in a HTP manner. It should be noted that the specific substrate preference ratio observed does not match our previous steady-state kinetic study with AA/GLA and 15-LOX-2.[25] This is most likely due to the variation in conditions between the two assays, and does not affect the ability of the assay to detect allosteric effector molecules.
Figure 4. 15-LOX-2 Allosteric effect of d30-13-HODE.
The ratio of 15-HETE/13-HOTrE(γ), with increasing concentration of the LO allosteric effector, d30-13-HODE. The observed peak area of 15-HETE was divided by the area of 13-HOTrE(γ) to yield peak area ratio. The error bars reflect standard deviation of 8 reactions and as observed, the absolute value of the standard deviation does not change with increasing d30-13-HODE concentration.
To Summarize, the acid cleavable detergent, PPS, is utilized to adequately solvate the hydrophobic LOX substrates and inhibitors, while allowing for usage under the LC-MS conditions due to its cleavage degradation. The hydrophilic reducing agent, TCEP, allows for direct detection of LOX reaction products in a volatile buffer system via HPLC-MS, thus allowing for a HTP screen in 96-well plates. Using this novel system, the amount of LOX needed was also reduced by 30-fold, which allowed for the IC50 determination of low nanomolar inhibitors. As this method represents the first HTP screen capable of identifying allosteric effector molecules of human LOX enzymes, it opens the possibility of identifying drug-like molecules capable of changing the substrate specificity of LOX isozymes. Given the varying biological effects of LOX products, novel allosteric effector molecules could be valuable tools in helping understand the complex and synergistic role of LOX product ratios in the cell.
Supplementary Material
Acknowledgments
The authors wish to thank Qiangli Zhang for HPLC-MS advice and coaching and the NIH Molecular Libraries Small Molecule Repository for providing compounds. This work was supported by the National Institutes of Health, GM56062 and S10-RR20939 (MS Equipment grant).
Abbreviations
- LOX
lipoxygenase
- 15-LOX-1
human reticulocyte 15-lipoxygenase-1
- 15-LOX-2
human epithelial 15-lipoxygenase-2
- NDGA
nordihydroguaiaretic acid
- AA
arachidonic acid
- GLA
γ-Linolenic acid
- 13-HOTrE(γ)
13(S)-hydroxy-6Z,9Z,11E-octadecatrienoic acid
- 13-HODE
13(S)-hydroxy-9Z,11E-octadecadienoic acid
- 15-HETE
15(S)-hydroxy-5Z,8Z,11Z,13E-eicosatetraenoic acid
- d30-13-HODE
fully deuterated 13-HODE
- PPS
sodium 3-(4-(1,1-bis(hexyloxy)ethyl)pyridinium-1-yl)propane-1-sulfonate
- TCEP
tris(2-carboxyethyl)phosphine
- HPLC-MS
Liquid chromatography–mass spectrometry
- R2
coefficient of determination
- HTP
high throughput
- [E]
total active enzyme concentration
- IC50
inhibitor constant at 50% inhibition
- Kiapp
apparent inhibition constant when [E] >> Kiapp
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Greis KD. Mass spectrometry for enzyme assays and inhibitor screening: an emerging application in pharmaceutical research. Mass Spectrometry Reviews. 2007;26:324–339. doi: 10.1002/mas.20127. [DOI] [PubMed] [Google Scholar]
- 2.Hou G, Zhang R, Pi Z, Song F, Liu Z, Liu S. A new method for screening aldose reductase inhibitors using ultrahigh performance liquid chromatography-tandem mass spectrometry. Anal. Methods. 2014;6:7681–7688. [Google Scholar]
- 3.Gurard-Levin ZA, Scholle MD, Eisenberg AH, Mrksich M. High-throughput screening of small molecule libraries using SAMDI mass spectrometry. ACS Comb Sci. 2011;13:347–350. doi: 10.1021/co2000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holt TG, Choi BK, Geoghagen NS, Jensen KK, Luo Q, LaMarr WA, et al. Label-free high-throughput screening via mass spectrometry: a single cystathionine quantitative method for multiple applications. Assay Drug Dev Technol. 2009;7:495–506. doi: 10.1089/adt.2009.0200. [DOI] [PubMed] [Google Scholar]
- 5.Kuhn H, Walther M, Kuban RJ. Mammalian arachidonate 15-lipoxygenases structure, function, and biological implications. Prostaglandins Other Lipid Mediat. 2002;68–69:263–290. doi: 10.1016/s0090-6980(02)00035-7. [DOI] [PubMed] [Google Scholar]
- 6.Obrosova IG, Stavniichuk R, Drel VR, Shevalye H, Vareniuk I, Nadler JL, et al. Different roles of 12/15-lipoxygenase in diabetic large and small fiber peripheral and autonomic neuropathies. Am. J. Pathol. 2010;177:1436–1447. doi: 10.2353/ajpath.2010.100178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Succol F, Praticò D. A role for 12/15 lipoxygenase in the amyloid beta precursor protein metabolism. J. Neurochem. 2007;103:380–387. doi: 10.1111/j.1471-4159.2007.04742.x. [DOI] [PubMed] [Google Scholar]
- 8.O'Flaherty JT, Wooten RE, Samuel MP, Thomas MJ, Levine EA, Case LD, et al. Fatty acid metabolites in rapidly proliferating breast cancer. PLoS ONE. 2013;8:e63076. doi: 10.1371/journal.pone.0063076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yigitkanli K, Pekcec A, Karatas H, Pallast S, Mandeville E, Joshi N, et al. Inhibition of 12/15-lipoxygenase as therapeutic strategy to treat stroke. Ann. Neurol. 2013;73:129–135. doi: 10.1002/ana.23734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang WG, Watkins G, Douglas-Jones A, Mansel RE. Reduction of isoforms of 15- lipoxygenase (15-LOX)-1 and 15-LOX-2 in human breast cancer. Prostaglandins Leukot Essent Fatty Acids. 2006;74:235–245. doi: 10.1016/j.plefa.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 11.Feng Y, Bai X, Yang Q, Wu H, Wang D. Downregulation of 15-lipoxygenase 2 by glucocorticoid receptor in prostate cancer cells. Int. J. Oncol. 2010;36:1541–1549. doi: 10.3892/ijo_00000641. [DOI] [PubMed] [Google Scholar]
- 12.Daurkin I, Eruslanov E, Stoffs T, Perrin GQ, Algood C, Gilbert SM, et al. Tumorassociated macrophages mediate immunosuppression in the renal cancer microenvironment by activating the 15-lipoxygenase-2 pathway. Cancer Res. 2011;71:6400–6409. doi: 10.1158/0008-5472.CAN-11-1261. [DOI] [PubMed] [Google Scholar]
- 13.Gertow K, Nobili E, Folkersen L, Newman JW, Pedersen TL, Ekstrand J, et al. 12- and 15-lipoxygenases in human carotid atherosclerotic lesions: associations with cerebrovascular symptoms. Atherosclerosis. 2011;215:411–416. doi: 10.1016/j.atherosclerosis.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Rai G, Kenyon V, Jadhav A, Schultz L, Armstrong M, Jameson JB, et al. Discovery of potent and selective inhibitors of human reticulocyte 15-lipoxygenase-1. J Med Chem. 2010;53:7392–7404. doi: 10.1021/jm1008852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Whitman S, Gezginci M, Timmermann BN, Holman TR. Structure-activity relationship studies of nordihydroguaiaretic acid inhibitors toward soybean, 12-human, and 15-human lipoxygenase. J Med Chem. 2002;45:2659–2661. doi: 10.1021/jm0201262. [DOI] [PubMed] [Google Scholar]
- 16.Segraves EN, Shah RR, Segraves NL, Johnson TA, Whitman S, Sui JK, et al. Probing the activity differences of simple and complex brominated aryl compounds against 15-soybean, 15-human, and 12-human lipoxygenase. J Med Chem. 2004;47:4060–4065. doi: 10.1021/jm049872s. [DOI] [PubMed] [Google Scholar]
- 17.Cichewicz RH, Kenyon VA, Whitman S, Morales NM, Arguello JF, Holman TR, et al. Redox inactivation of human 15-lipoxygenase by marine-derived meroditerpenes and synthetic chromanes: archetypes for a unique class of selective and recyclable inhibitors. Journal of the American Chemical Society. 2004;126:14910–14920. doi: 10.1021/ja046082z. [DOI] [PubMed] [Google Scholar]
- 18.Amagata T, Whitman S, Johnson TA, Stessman CC, Loo CP, Lobkovsky E, et al. Exploring Sponge-Derived Terpenoids for Their Potency and Selectivity against 12- Human, 15-Human, and 15-Soybean Lipoxygenases. J Nat Prod. 2003;66:230–235. doi: 10.1021/np020462l. [DOI] [PubMed] [Google Scholar]
- 19.Kenyon V, Rai G, Jadhav A, Schultz L, Armstrong M, Jameson JB, et al. Discovery of potent and selective inhibitors of human platelet-type 12- lipoxygenase. J Med Chem. 2011;54:5485–5497. doi: 10.1021/jm2005089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jameson II JB, Kantz A, Schultz L, Kalyanaraman C, Jacobson MP, Maloney DJ, et al. A High Throughput Screen Identifies Potent and Selective Inhibitors to Human Epithelial 15-Lipoxygenase-2. PLoS ONE. 2014;9:e104094. doi: 10.1371/journal.pone.0104094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McGovern SL, Helfand BT, Feng B, Shoichet BK. A specific mechanism of nonspecific inhibition. J Med Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 22.Pasqualini ME, Heyd VL, Manzo P, Eynard AR. Association between E-cadherin expression by human colon, bladder and breast cancer cells and the 13-HODE: 15-HETE ratio. A possible role of their metastatic potential. Prostaglandins Leukot Essent Fatty Acids. 2003;68:9–16. doi: 10.1016/s0952-3278(02)00230-2. [DOI] [PubMed] [Google Scholar]
- 23.O'Flaherty JT, Hu Y, Wooten RE, Horita DA, Samuel MP, Thomas MJ, et al. 15- lipoxygenase metabolites of docosahexaenoic acid inhibit prostate cancer cell proliferation and survival. PLoS ONE. 2012;7:e45480. doi: 10.1371/journal.pone.0045480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wecksler AT, Kenyon V, Deschamps JD, Holman TR. Substrate specificity changes for human reticulocyte and epithelial 15-lipoxygenases reveal allosteric product regulation. Biochemistry. 2008;47:7364–7375. doi: 10.1021/bi800550n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Joshi N, Hoobler EK, Perry S, Diaz G, Fox B, Holman TR. Kinetic and Structural Investigations into the Allosteric and pH Effect on the Substrate Specificity of Human Epithelial 15-Lipoxygenase-2. Biochemistry. 2013 doi: 10.1021/bi4010649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wecksler AT, Kenyon V, Garcia NK, Deschamps JD, van der Donk WA, Holman TR. Kinetic and structural investigations of the allosteric site in human epithelial 15-lipoxygenase-2. Biochemistry. 2009;48:8721–8730. doi: 10.1021/bi9009242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lewis ER, Johansen E, Holman TR. Large competitive kinetic isotope effects in human 15-lipoxygenase catalysis measured by a novel HPLC method. Journal of the American Chemical Society. 1999;121:1395–1396. [Google Scholar]
- 28.Morrison JF. Kinetics of the reversible inhibition of enzyme-catalysed reactions by tight-binding inhibitors. Biochim Biophys Acta. 1969;185:269–286. doi: 10.1016/0005-2744(69)90420-3. [DOI] [PubMed] [Google Scholar]
- 29.Deschamps JD, Gautschi JT, Whitman S, Johnson TA, Gassner NC, Crews P, et al. Discovery of platelet-type 12-human lipoxygenase selective inhibitors by highthroughput screening of structurally diverse libraries. Bioorg Med Chem. 2007;15:6900–6908. doi: 10.1016/j.bmc.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mogul R, Johansen E, Holman TR. Oleyl sulfate reveals allosteric inhibition of soybean lipoxygenase-1 and human 15-lipoxygenase. Biochemistry. 2000;39:4801–4807. doi: 10.1021/bi992805t. [DOI] [PubMed] [Google Scholar]
- 31.Waslidge NB, Hayes DJ. A colorimetric method for the determination of lipoxygenase activity suitable for use in a high throughput assay format. Anal Biochem. 1995;231:354–358. doi: 10.1006/abio.1995.0063. [DOI] [PubMed] [Google Scholar]
- 32.Pufahl RA, Kasten TP, Hills R, Gierse JK, Reitz BA, Weinberg RA, et al. Development of a fluorescence-based enzyme assay of human 5-lipoxygenase. Anal Biochem. 2007;364:204–212. doi: 10.1016/j.ab.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 33.Nair DG, Funk CD. A cell-based assay for screening lipoxygenase inhibitors. Prostaglandins Other Lipid Mediat. 2009;90:98–104. doi: 10.1016/j.prostaglandins.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 34.Dahlström M, Forsström D, Johannesson M, Huque-Andersson Y, Björk M, Silfverplatz E, et al. Development of a fluorescent intensity assay amenable for highthroughput screening for determining 15-lipoxygenase activity. J Biomol Screen. 2010;15:671–679. doi: 10.1177/1087057110373383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.