Abstract
Endosomal entrapment is known to be a major bottleneck to successful cytoplasmic delivery of nucleic acids (NAs) using cationic liposome-NA nanoparticles (NPs). Quantitative measurements of distributions of NPs within early endosomes (EEs) have proven difficult due to the sub-resolution size and short lifetime of wildtype EEs. In this study we used Rab5-GFP, a member of the large family of GTPases which cycles between the plasma membrane and early endosomes to fluorescently label early endosomes. Using fluorescence microscopy and quantitative image analysis of cells expressing Rab5-GFP, we found that at early time points (t < 1 h), only a fraction (≈35%) of RGD-tagged NPs (which target cell surface integrins) colocalize with wildtype EEs, independent of the NP’s membrane charge density. In comparison, a GTP-hydrolysis deficient mutant, Rab5-Q79L, which extends the size and lifetime of EEs yielding giant early endosomes (GEEs), enabled us to resolve and localize individual NPs found within the GEE lumen. Remarkably, nearly all intracellular NPs are found to be trapped within GEEs implying little or no escape at early time points. The observed small degree of colocalization of NPs and wildtype Rab5 is consistent with recycling of Rab5-GDP to the plasma membrane and not indicative of NP escape from EEs. Taken together, our results show that endosomal escape of PEGylated nanoparticles occurs downstream of EEs i.e., from late endosomes/lysosomes. Our studies also suggest that Rab5-Q79L could be used in a robust imaging assay which allows for direct visualization of NP interactions with the luminal membrane of early endosomes.
1. Introduction
Synthetic nucleic acid carriers whether—lipid-, dendrimer- or polymer -based—are promising candidates for the treatment of various disease [1–14]. Relative to viral vectors, synthetic vectors show low immunogenic response and are generally considered safer [15–17]. Furthermore, synthetic vector/nucleic acid complexes such as cationic liposome–DNA (CL–DNA) complexes are not limited by the finite capsid size of viral vectors and can deliver large genetic constructs, including entire genes (exons and introns) and regulatory sequences [18]. Surface functionalization of liposomes and lipid-based delivery systems, typically through PEGylation (PEG; polyethylene-glycol) with PEG-lipids, is required to achieve extended circulation times in vivo [19–21]. However, PEGylation of CL–DNA nanoparticles (NPs) typically reduces their transfection efficiency (TE; a measure of exogenous gene expression) by presenting barriers to cell attachment and endosomal escape [21–23]. One common approach to improve NP internalization is to use a targeting or cell penetrating peptide at the distal end of the PEG-lipid. An added benefit of targeted vectors is that the selective delivery of payload to the proper tissue or cell type can reduce side effects and improve efficacy [24–27]. Although a large library of tissue or cell targeting peptides is being developed [28,29], relatively little is known about how targeting peptides alter the endocytosis and intracellular trafficking of drugs or nanoparticles.
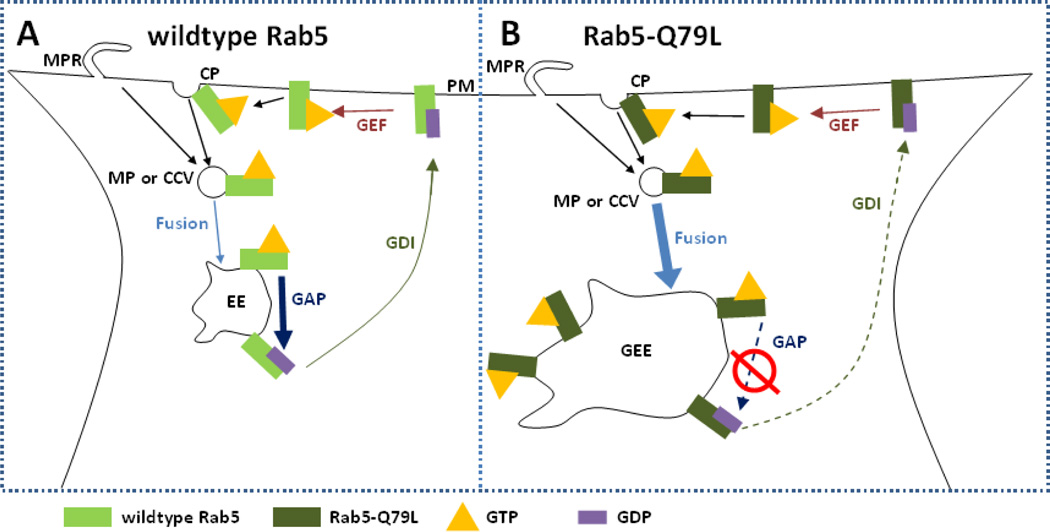
To elucidate the uptake and intracellular behavior of RGD-tagged CL–DNA NPs, we used fluorescence microscopy and automated particle colocalization with both wildtype Rab5-GFP and Rab5-Q79L-GFP, a very slowly hydrolyzing mutant, to measure colocalization of NPs and early endosomes (EEs) in fixed mammalian cells. Rab5, a member of the Rab family of GTPases that coordinate intracellular vesicle budding, trafficking and fusion [30], plays a dominate role in the formation and function of early endosomes [30–32]. Fig. 1 A shows a typical cycle of wildtype Rab5 during the endosomal process. Initially, Rab5 accumulates at the sites of clathrin-coated pits or macropinocytic ruffles where it recruits the necessary proteins for endosomal budding from the plasma membrane [33–35]. In the GTP-bound form, Rab5 interacts with effectors which mediate homotypic fusion of other GTP-Rab5 containing endocytic vesicles [36, 37]. Upon GTP hydrolysis, GDP-bound Rab5 will complex with guanosine nucleotide disassociation inhibitor (GDI) which facilitates transport back to the plasma membrane [38]. The GDP-bound form of Rab5 cannot mediate fusion and is considered inactive [36]. EEs gradually lose Rab5 as GTP hydrolysis continues and they simultaneously accumulate Rab7 signifying the maturation of the early endosome into a late endosome [39]. The point mutation Q79L hinders GTP hydrolysis activity of Rab5 (labeled Rab5-Q79L), which increases the ratio of membrane bound GTP-Rab5 to cytosolic GDP-Rab5 [36]. When Rab5 is unable to efficiently hydrolyze GTP early endosomes continuously fuse and form giant early endosomes (GEEs) [40]. In contrast to EEs, GEEs are longer lived and spatially resolvable. Although the mutant Rab5-Q79L alters the maturation process of the early endosomes from what is found in the wildtype case, our findings show that Rab5-Q79L is a useful tool for understanding the intracellular pathway of peptide-tagged PEGylated CL–DNA NPs.
Figure 1.
Schematic showing the cycle of wildtype Rab5 and mutant Rab5-Q79L. (A) In the case of wildtype Rab5, GTP-bound Rab5 is recruited to the lumen side of clathrin pits (CP) or macropinocytotic ruffles (MPR). Once the macropinosome (MP) or clathrin-coated vesicle (CCV) has pinched off from the plasma membrane (PM), GTP-bound Rab5 mediates homotypic fusion with other GTP-bound Rab5-labeled vesicles forming the early endosome (EE). On the surface of the EE, GTPase activating protein (GAP) stimulates hydrolysis of GTP to GDP. GDP-bound Rab5 complexes with guanosine nucleotide disassociation inhibitor (GDI) and undergoes transport to the PM. At the PM, guanine nucleotide exchange factor (GEF) promotes exchange of GDP to GTP which completes the cycle. (B) In the case of Rab5-Q79L, GTP hydrolysis through GAP is severely reduced, resulting in two major phenotypic changes from the widltype case. First, due to fusion of MPs and CCVs being mediated by GTP-bound Rab5, reduced GTP hydrolysis increases the number of fusion events, leading to the formation of giant early endosomes (GEE) that contain an abundance of Rab5-Q79L on the membrane. Second, loss of Rab5 from the EE through GTP hydrolysis is necessary for maturation of EEs into late endosomes, thus the reduced GTP hydrolysis of Rab5-Q79L delays maturation and extends the lifetime of EEs.
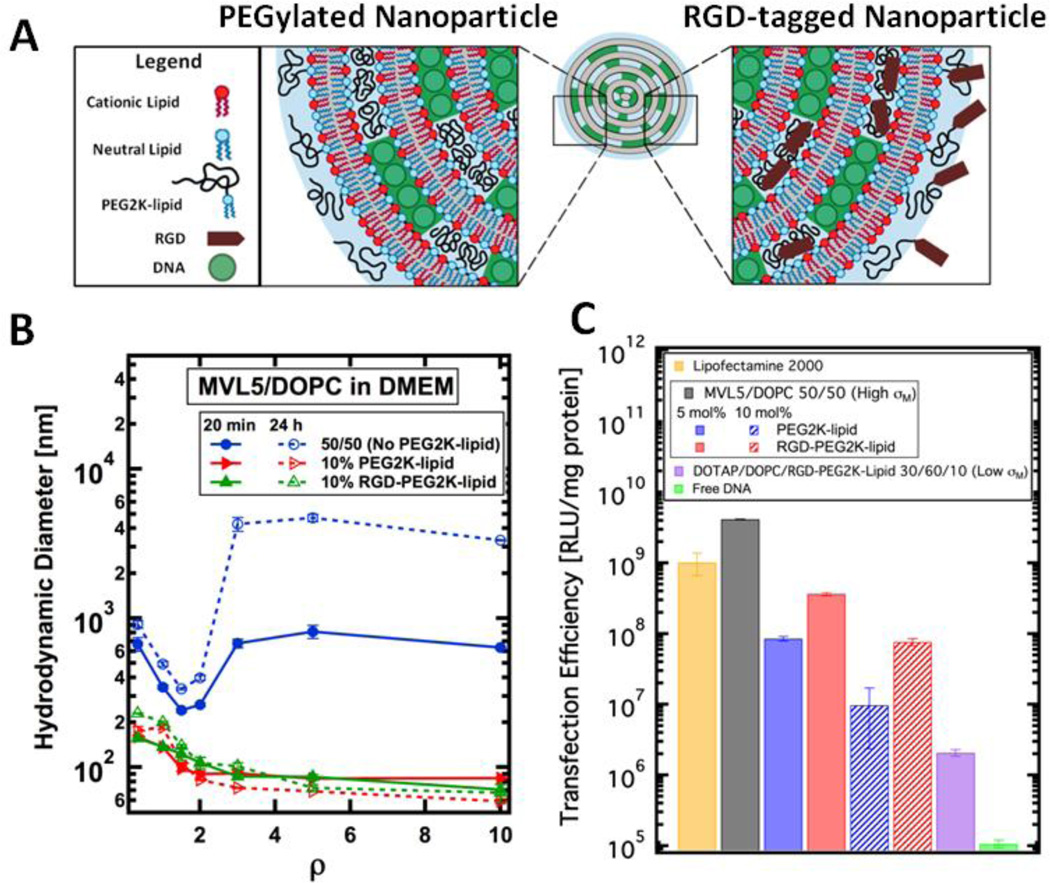
We prepared PEGylated CL–DNA nanoparticles presenting a linear RGD peptide (GRGDSP) at the distal end of the PEG2000-lipid (PEG2K-lipid). The RGD motif is common in extracellular matrix proteins and specifically binds to integrins; plasma membrane-bound receptors which are commonly over-expressed in cancer cells [41,42]. Our CL–DNA nanoparticles consisted of MVL5 (a pentavalent cationic lipid), a neutral lipid (DOPC), and a PEG2K-lipid with or without RGD (Fig. 2 A). CL–DNA complexes based on MVL5 show remarkably high transfection efficiency when compared to monovalent lipids (e.g. DOTAP) [43]. Furthermore, MVL5 has also found applications in gene silencing due to its lower toxicity as well as high transfection in the presence of serum [44–46]. Currently, a cornucopia of literature exists for conventional lipoplexes lacking PEG-lipid [8, 43–50] but we want to emphasize that our vector is a self-assembled PEGylated CL–DNA nanoparticle. Conventional lipoplexes show a large polydispersity of sizes (from nanometers to microns) but the use of PEG-lipids leads to spontaneous formation of equilibrium CL–DNA nanoparticles with a smaller (≈ 100 nm) and narrower size distribution [22, 23, 51–55]. Furthermore, previous work has shown that the grafted PEG polymer significantly alters the NP’s interaction with biological membranes which effects cellular uptake and endsomal escape [52]. We confirmed that complexes lacking PEG2K-lipid (i.e., composed only of MVL5/DOPC and plasmid DNA) aggregate in the presence of cell media but the addition of PEG2K-lipid leads to the formation of sub-200 nm sterically-stabilized nanoparticles (NPs). We compared the TE of MVL5/DOPC complexes lacking PEGylation with that of PEGylated MVL5/DOPC NPs both with and without RGD-tagging. Relative to the control (no PEGylation), PEGylation reduces TE and RGD-tagging partially recovers TE. Quantitative particle localization, measured via fluorescent live-cell imaging, revealed that PEGylated CL–DNA NPs were internalized more efficiently upon RGD-tagging. Both types of NPs (with and without RGD-tagging) showed perinuclear accumulation, indicating endosomal entrapment and motor-based transport of endosomes. To further probe the endocytic pathway of RGD-tagged CL–DNA NPs, we used Rab5-GFP constructs to fluorescently label early endosomes. We used a semi-automated image processing routine to count the number of intracellular particles found inside and outside of early endosomes. Although the TE of RGD-tagged NPs strongly increases with membrane charge density, their uptake and colocalization with Rab5-labeled EEs was independent of membrane charge density. Surprisingly, the fraction of NPs colocalizing with wildtype EEs at early times points (< 1 h) was relatively small. While this observation could suggest another mechanism of entry or efficient escape from early endosomes, our experiments using the mutant Rab5-Q79L-GFP, show that upon extending the lifetime of EEs (by inhibiting GTP hydrolysis), all intracellular NPs were localized inside EEs or GEEs. This suggests that the lack of colocalization of PEGylated NPs with wildtype Rab5-GFP is merely a result of the short half-life of wildtype early endosomes and that endosomal escape occurs later in the endocytic pathway, for example, at the late endosome/lysosome or recycling stage.
Figure 2.
Size and transfection efficiency (TE) of MVL5-based CL–DNA complexes and NPs. (A) Schematic showing the internal nano-structure and functionalized surface of a PEGylated and RGD-tagged CL–DNA nanoparticle. (B) Dynamic light scattering shows that CL–DNA complexes lacking PEGylation form micron-sized aggregates when complexed in the cell culture medium DMEM. (blue curve). PEGylation with 10 mol% PEG2K-lipid or RGD-PEG2K-lipid induces the formation of sub-200 nm, sterically-stabilized nanoparticles (red and green curves). (C) MVL5/DOPC complexes show remarkably high TE, outperforming the commercial reagent Lipofectamine® 2000. As the concentration of PEG2K-lipid increases (blue bars), TE decreases. RGD-tagging of MVL5-based NPs partially recovers TE (red bars) relative to the PEGylated NPs lacking RGD. Low-σM NPs contain 30/60/10 DOTAP/DOPC/RGD-PEG2Klipid by mol% and show low TE.
2. Materials and Methods
2. 1 Materials
DOTAP, DOPC and DOPE-PEG2000 were purchased from Avanti Polar lipids as chloroform solutions. MVL5 was synthesized as described previously [39]. The fluorescent lipids TRITC-DHPE and Texas Red-DHPE (Invitrogen, Carlsbad, California) have excitation and emission maxima of 555/589 nm and 580/615 nm, respectively. The RGD-PEG2K-lipid contained a GRGDSP sequence covalently bound to the distal end of PEG2K. It was custom synthesized on solid phase using Fmoc-amino acids and a lipid-PEG2K acid. The pGL3-control vector coding the Luciferase gene (Promega, Fitchburg, Wisconsin) was propagated in E. coli, and purified using a Qiagen Plasmid Mega Prep Kit. The GFP-Rab5-Q79L plasmid was a gift from the Weimbs lab (UCSB) and propagated and purified as decribed above for pGL3. For cell imaging studies, the pGL3 vector was labeled using the Mirus Bio Label IT Nucleic Acid Labeling Kit with Cy5 (excitation/emission maximum: 649 nm/670 nm) according to the manufacturer’s protocol. For labeling of early endosomes, the CellLights Early Endosome-GFP BacMam 2.0 (Life Technologies, Carlsbad, California) reagent was used according to manufacturer’s protocol.
2.2 Liposome Preparation
Lipid solutions in 3:1 chloroform/methanol were combined at the desired molar ratio of lipid in glass vials. The RGD-PEG2K-lipid was dissolved in a 1:1 solution of deionized water/acetonitrile. For complexes or nanoparticles containing MVL5, liposomes were prepared using MVL5/DOPC/PEG2K-lipid at 50/50–x/x where x is 0, 5 or 10. DOTAP-based complexes were formed with liposomes composed of DOTAP/DOPC/PEG2K-lipid at 30/60/10 molar ratio. Liposomes used in live or fixed cell imaging contained 0.5 wt% (of total lipid) TRITC-DHPE-lipid or Texas Red-DHPE-lipid label. After mixing, the lipid solutions in organic solvent were dried, first by a stream of nitrogen and then in a vacuum for 12 h. The appropriate amount of sterile, high resistivity (18.2 MΩ cm) water to achieve a final concentration of 1 mM lipid was then added to the dried lipid films, and the resulting mixtures were incubated at 37 °C for 16 h to form liposomes. Following this incubation, the liposome solutions were sonicated using a tip sonicator to form small unilamellar vesicles. When these unilamellar vesicles are mixed with DNA they spontaneously self-assemble into multilamellar nanoparticles [56].
2.3 Cell Culture and Transfection
Mouse L-cells (ATCC number: CCL-1) were cultured in Dubelco’s Modified Eagle Medium supplemented with 5% fetal bovine serum (HyClone, Logan, Utah) and 1% Penicillin/Streptomyocin (Invitrogen, Carlsbad, California). Cells were kept at 37 °C in a humidified atmosphere containing 5% CO2 and were reseeded every 72 h to maintain subconfluency. For transfection studies, cells were seeded in 24-wellplates (2.5×105 cells/well) such that confluency at transfection was 60–80%. CL–DNA complexes were formed by diluting 1 µg of DNA and the appropriate amount of liposome solution to 250 µL each with Optimem (Invitrogen, Carlsbad, California) and mixing. Complexes were incubated for 20 minutes at room temperature before addition to cells. Cells were washed once with PBS and then incubated with 200 µL of complex suspension (0.4 µg of DNA per well) for 6 h. After 6 h, the transfection medium was removed, and cells were rinsed once with PBS and then incubated in supplemented DMEM for 18 h. Cells were harvested in 150 µL of Passive Lysis Buffer (Promega, Fitchburg, Wisconsin) and subjected to one freeze-thaw cycle. Luciferase expression was measured using a Perkin-Elmer 1420 Victor3 V multilabel counter following the assay manufacturer’s (Promega, Fitchburg, Wisconsin) instructions. TE results are normalized to total cellular protein as measured by a Bradford Assay (BioRad, Hercules, California). Data points represent an average of two measurements with error bars showing the standard deviation. All experiments were repeated at least two times to ensure reproducibility.
2. 4 Dynamic Light Scattering
The size of CL–DNA complexes and nanoparticles was measured using a Malvern Nanosizer ZS. CL–DNA particles were prepared in light-scattering vials in a similar manner to the transfection experiment; DNA and lipid were diluted in equal volumes before mixing. A total of 2 µg of DNA and the appropriate amount of liposome dispersion (to achieve the desired lipid/DNA charge ratio ρ) were mixed in 1 mL of DMEM and incubated at room temperature for 20 minutes. Plots show the z-average diameter. All data points are the average of two measurements performed on the same sample. Error bars show the standard deviation.
2.5 Live-Cell Imaging and Particle Localization
Live-cell imaging was performed using dual-labeled (see Materials) complexes at ρ=10. The concentration of CL–DNA NPs was the same as that used in the transfection assay. Cells were grown to 60% confluency on poly(L-lysine)-coated coverslips (22 mm) and maintained at 37 °C using a Harvard Warner flow chamber (Model #P2 and RC21-B Harvard Apparatus, Holliston, Massachusetts). Images were taken on a Nikon Diaphot 300 using a Nikon 1.4 NA 60× Plan Apo DIC objective (Nikon, Tokyo, Japan) and Sensicam QE CCD (PCO AG, Kelheim, Germany). Brightfield images were captured at a magnification of 60 × in differential interference contract (DIC) mode. Fluorescent images are composed of two merged channels where one channel shows lipid (TRITC-DHPE) and the other DNA (Cy5). Images were analyzed using a MATLAB (The Mathworks, Natick, MA) routine that measured the intracellular spatial distribution of fluorescently labeled DNA. Data points and error bars represent the average and standard deviation of 10 to 20 representative cells. The Matlab routine first locates the cell boundary and nuclear membrane using the DIC image. Next, all intracellular fluorescent particles are located by fitting a 2D Gaussian to all fluorescent spots contained within the cell boundary. Finally, the closest distance to the nuclear membrane is measured and recorded for each intracellular particle. Our Matlab code uses scripts inspired by [57].
2.6 Endosome Labeling Assay and Particle Colocalization
L-cells were seeded on poly-L-lysine coated coverslips in 6-well plates such that the confluency was 60 – 80% 24 hours after seeding. For wildtype Rab5 studies, at 24 hours post seeding, 15 µL of Invitrogen’s CellLight Early Endosome-GFP Marker (a viral transduction system) was added to cells and incubated overnight. For mutant Rab5-Q79L studies, at 24 hours post seeding, 4 µg of Rab5-Q79L-GFP plasmid was complexed with 10 µl of Lipofectamine 2000 according the manufacturers protocol, the cell medium was changed to OptiMEM and complexes were incubated with cells for 4–6 hours. The medium was then aspirated, the cells were washed with PBS, supplemented DMEM was added and the cells were incubated overnight. After overnight incubation in serum-containing DMEM, cells were further serum starved for 24 hours to synchronize cells and reduce cell-to-cell variability as well as minimize any lingering effects from the Rab-GFP vectors. From this point forward, the protocol used for wildtype and mutant Rab5 is identical.
The appropriate amount of fluorescently labeled liposome solution was added to a solution of 0.1 µg of Cy5-labeled plasmid such that the final charge ratio was ρ=10 in a total OptiMEM volume of 50 µL. The resulting dual labeled NP solution was incubated at room temperature for 20 minutes. Cells were removed from the incubator, washed with PBS and 2 mL of ice-cold OptiMEM were added to the cells. Dual labeled particles were then added and the cells were incubated at 4 °C for 1 hour. In summary, for colocalization studies, a 2 ml solution containing 0.1 µg of Cy5-labeled DNA and appropriate lipid was added to each well. This cold incubation allows particles to bind to the plasma membrane while endocytosis is thermally inhibited. After 1 hour of cold incubation cells were transferred to 37 °C and 5% CO2 for either 30 or 60 minutes, as noted with the presented data. After warm incubation, cells were washed three times with ice-cold PBS containing 50 U/mL of heparin to remove extracellular complexes [58]. After heparin washing, cells were fixed with 3.7% formaldehyde for 15 minutes, washed three times using room temperature PBS, and mounted using Invitrogen AntiFade media containing DAPI. Fixed cells were then imaged using an Olympus (Tokyo, Japan) DSU equipped with a 100 × UPlanSApo objective, Hamamatsu (Hamamatsu, Japan) ImagEM CCD camera and Metamorph (Nashville, Tennessee) software. Representative cells were chosen and imaged at z steps of 250 nm. Z-stacks of the fluorescent channels were deconvolved using the ImageJ plugin Iterative Deconvolve 3D. Image processing consisted of Background Subtraction with a 20 pixel rolling ball radius as well as the smooth filter which improves image clarity. Z-stacks were then overlayed and Z-projected for analysis. Finally, an automated Matlab routine was used to measure the number of colocalized fluorescent signals within each cell. The routine locates all intracellular particles in each fluorescent channel, yielding a list of coordinates for each channel e.g.; {(x_red1, y_red1), (x_red2, …)}, {(x_green1, y_green1), …} where the red channel corresponds to lipid, the green channel to endosomes and the blue channel to DNA. If the distance between particles in different channels was below a chosen threshold (typically 3 pixels ~ 500 nm) then the particle was identified as being either (a) a trapped complex (red, blue, green) or (b) a free complex (red, blue). Similar colocalization routines have been previously reported in studies of synthetic vectors. [59, 60]
3 Results
In this study complexes consisting of cationic lipid, neutral lipid, PEG2000-lipid with (RGD-PEG2K-lipid) or without (PEG2K-lipid) RGD at the distal end of PEG and plasmid DNA (Fig. 2 A) are referred to as nanoparticles (NPs) due to their stable sub-100 nm size. CL–DNA complexes (i.e. lipoplexes) without the PEG-lipid component do not form NPs as their size increases with time in DMEM or physiological buffer (see Fig. 2B) conditions [61–63]. In contrast to lipid NPs reported elsewhere in the literature [64], our CL–DNA NPs are simply formed by mixing liposomes and DNA and allowing the components to assemble into their equilibrium structure. In addition to varying the coverage of PEG2K (by varying the mol% of PEG2K-lipid) we also investigated the effect of membrane charge density. CL–DNA NPs with high membrane charge density (referred to as High-σM) were composed of MVL5/DOPC/PEG2K-lipid at 50/50-x/x mol % where x = 0, 5, 10. Low membrane charge density CL–DNA NPs (referred to as Low-σM) were composed of DOTAP/DOPC/PEG2K-lipid at 30/60/10 mol %. When referred to as RGD-tagged, the PEG2K-lipid presented a GRGDSP-OH peptide at the distal end of the PEG2K. The charge ratio (ratio of positive lipid charges to negative DNA charges) ρ was set at 10 for all data shown except for dynamic light scattering where the charge ratio is indicated on the x axis.
3.1 Size and Transfection Efficiency of CL–DNA Complexes and PEGylated Nanoparticles
The diameter of MVL5/DOPC complexes with no PEG2K-lipid, 10 mol% PEG2K-lipid and 10 mol% RGD-PEG2K-lipid in cell culture media (DMEM) was measured using dynamic light scattering (Fig. 2 B). MVL5/DOPC complexes lacking PEGylation form large, micron-sized aggregates for all ρ > 2 which continue to aggregate for up to 24 hours post complex formation. MVL5/DOPC complexes containing 10 mol% PEG2K-lipid or RGD-PEG2K-lipid, form stable sub-200 nm NPs which are resistant to aggregation at all ρ and for at least 24 hours after formation.
Using a luciferase assay, we measured the TE of High-σM MVL5/DOPC complexes at different PEG2K or RGD surface coverage (Fig. 2 C). MVL5/DOPC complexes without PEGylation showed high TE, even outperforming the benchmark commercial lipofection reagent Lipofectamine2000. PEGylation with 5 or 10 mol% PEG2K-lipid reduces TE by 1 and 2 orders of magnitude, respectively. RGD-tagging of PEGylated NPs partially recovers TE (to a level in between PEGylated NPs and bare CL–DNA complexes). Low-σM RGD-tagged NPs composed of DOTAP/DOPC/RGD-PEG2K-lipid at a molar ratio of 30/60/10 showed low TE (≈106 RLU/mg protein), nearly two orders of magnitude lower than MVL5/DOPC NPs at the same RGD-PEG2K grafting density and only one order of magnitude higher than the naked DNA control.
3.2 Live Cell Imaging and Particle Localization
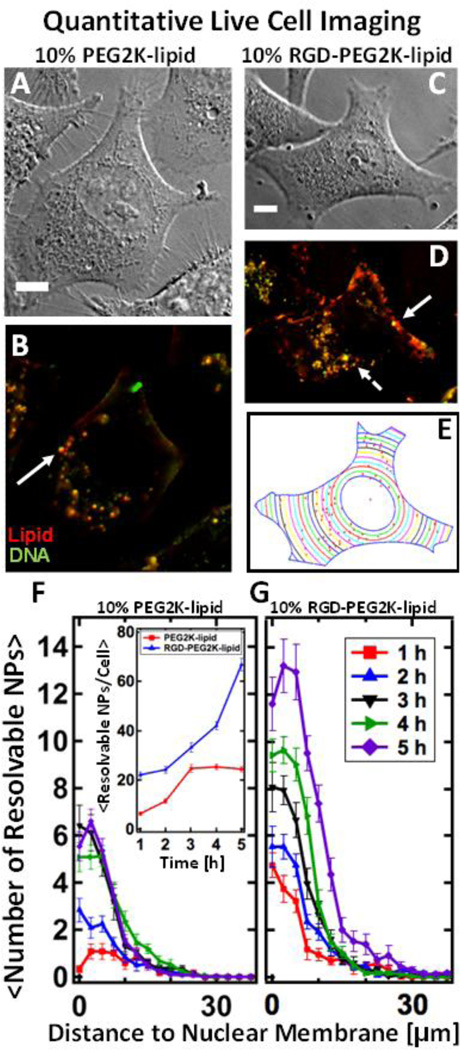
Live cell imaging using differential interference contrast (DIC, Fig. 3 A and C) and fluorescence microscopy (Fig. 3 B and D) allowed direct observation of binding, uptake and intracellular localization. Quantitative imaging allows us to measure NP uptake while easily differentiating between NPs bound to plasma membrane versus NPs internalized within cells. PEGylated NPs lacking RGD adhere to the plasma membrane via nonspecific electrostatic-mediated adhesion, undergo internalization and accumulate in the perinuclear region. The solid white arrow marks a large, spatially resolvable fluorescent spot which we interpret as an endosome containing multiple NPs (Fig. 3 B and D). When the NPs contain RGD-PEG2K-lipid, a larger number of intracellular particles adhere to cells and undergo internalization (Fig. 3 D). Furthermore, along with large resolvable endosomes containing multiple NPs, small resolution-limited spots are visible (dashed arrows). To quantify intracellular particle localization we used a semi-automated particle localization routine (see Methods: Live Cell Imaging and Particle Localization) which measures the average number of fluorescent particles found inside cells at a given distance to the nuclear membrane (Fig. 3 G and H). Fig. 3 E displays the method for the case of the cell imaged in DIC (Fig. 3 C) and fluorescent (Fig. 3 D) mode. For both NPs with and without RGD-tagging, particles accumulate in the perinuclear region. Furthermore, by counting the total number of distinct fluorescent objects per cell (Fig. 3 G, inset) we confirm quantitatively that RGD-tagged NPs are internalized much more efficiently.
Figure 3.
Live-cell imaging of fluorescently-labeled PEGylated MVL5-based nanoparticles with and without RGD-tagging. NPs containing multivalent lipid (MVL5/DOPC/(PEG2K-lipid or RGD-PEG2K-lipid) at 50/40/10 mol% at ρ=10) are internalized in live L-cells. (Red – TRITC-DHPE (lipid), Green – Cy5 (DNA), Yellow – Nanoparticle composed of complexed lipid and DNA) (A, B) DIC and fluorescent images of Lcells after 4 hours of incubation with PEGylated NPs which lack RGD. (C, D) DIC and fluorescent images of L-cells after 4 hours of incubation with RGD-tagged NPs. Relative to NPs lacking RGD (A, B), RGDtagging causes a higher number of NPs to bind to the plasma membrane as well as internalize. Large, bright fluorescent particles (solid arrows) are likely endosomes containing multiple NPs which are spatially irresolvable. The dashed arrow points to a tiny resolution-limited spot which is believed to be a single NP. (E) A MatLab routine was used to determine the boundary and nuclear location of the cell in (C). Using (D), the routine locates all intracellular particles (red spots in (E)) and measures their distance to the nucleus. (F, G) The results of the localization routine for NPs without and with RGD respectively. By defining the nuclear membrane as a reference point, we can average the localization results over 20 cells. Spatial localization shows that while both types of MVL5 NPs (with and without RGD-tagging) are uptaken and trafficked to the perinuclear region, approximately three times as many RGD-tagged particles are found in cells after 5 hours of incubation (a standard incubation time for a TE assay). Inset shows total NPs per cell. All scale bars are 10 µm.
3.3 Fixed-Cell Imaging with Rab5-GFP and Rab5-Q79L-GFP
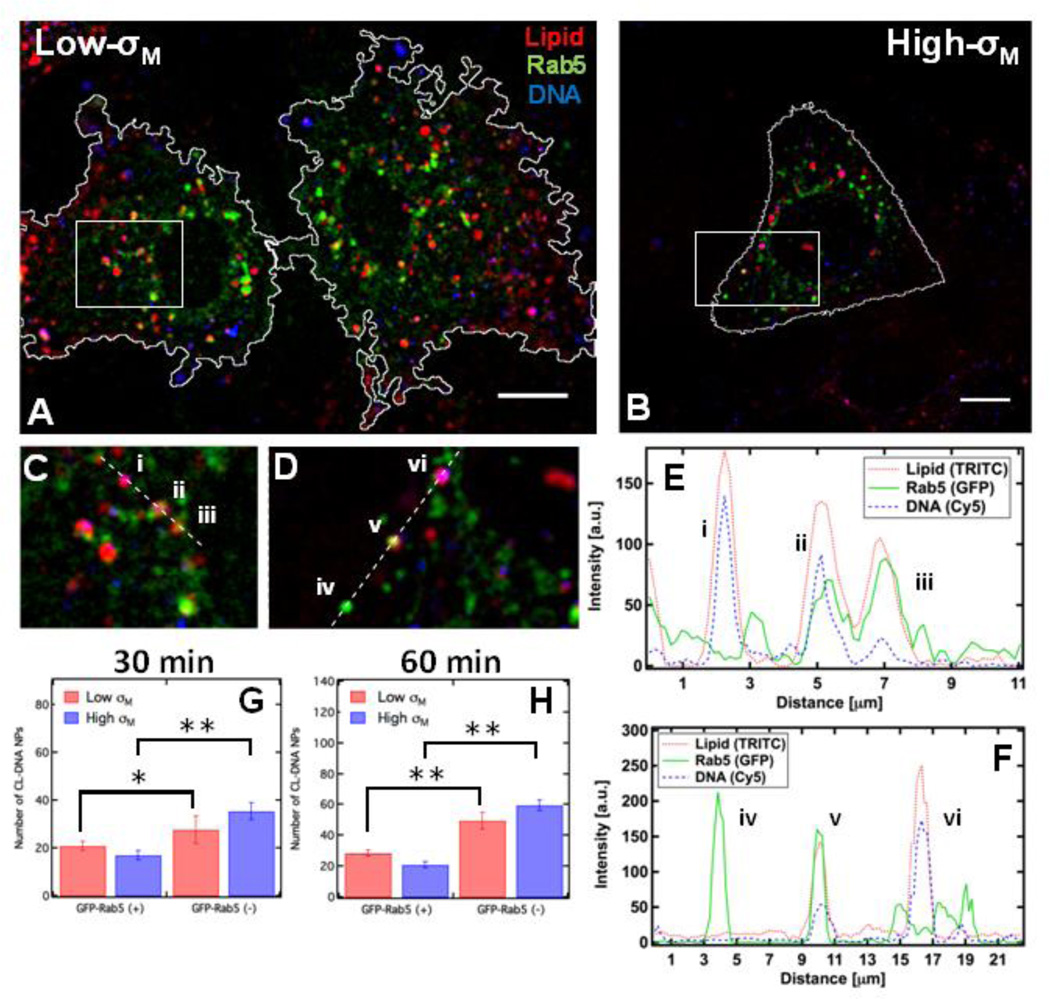
L-cells expressing wildtype Rab5-GFP were incubated with fluorescently labeled RGD-tagged CL–DNA NPs and fixed after 30 minutes (Fig. 4 A–D). To visualize only particles in similar endocytic stages, we employed a two-step incubation. Briefly, cells were incubated at 4 °C in the presence of NPs, allowing NPs to settle and bind to the outside of cells while endocytosis remained thermally inhibited (Fig. S1). After 1 hour of incubation with NPs at 4 °C, cells were incubated at 37 °C for the designated time e.g.; 30 or 60 minutes and extracellular NPs were removed via cold heparin washing. Intensity profiles (Fig. 4 E and F) of intracellular sections show a variety of colocalized signals e.g.; NPs inside early endosomes (EEs) (Fig. 4 C (ii, iii) and Fig. 2 D (v)), NPs lacking Rab5-GFP colocalization (Fig. 4 C (i) and Fig. 4 D (vi)), and EEs void of NPs (Fig. 4 D (iv)). To quantitatively measure what fraction of intracellular NPs colocalize with Rab5-labeled EE, we used a semi-automated MatLab routine which measured the number of intracellular particles found inside (GFP-Rab5(+)) and outside (GFP-Rab5(−)) of early endosomes (see Methods: Endosome Labeling and Particle Colocalization). Fig. 4 G and H shows the results of the NP-EE colocalization routine at 30 and 60 minutes. The total number (NTotal = NGFP(+) + NGFP(−)) of intracellular NPs increases with time and is effectively the same for both membrane charge densities. Furthermore, a statistically significant larger number of NPs are found outside of EEs at both time points. Although the number of Low-σM NPs inside EEs slightly increases from 30 to 60 minutes, the number of High-σM NPs colocalized with Rab5 slightly decreases with time. We found that for both High- and Low-σM NPs, the number of particles lacking Rab5 colocalization significantly increases from 30 to 60 minutes.
Figure 4.
CL-DNA NPs colocalize with Rab5-GFP-labeled endosomes. (A, B) Fluorescent micrographs and cell outlines of wildtype Rab5-GFP expressing L-cells that have been incubated with fluorescent RGDtagged NPs. The three overlayed channels represent lipid (red), DNA (blue) and Rab5-GFP (green). In (A) Low-σM refers to NPs with DOTAP/PC/RGD-PEG2K-lipid at a molar ratio of 30/60/10. The cells in (B) were incubated with High-σM NPs composed of MVL5/PC/RGD-PEG2K-lipid at a molar ratio of 50/45/5. (C, D) High magnification of boxed regions from (A, B) show intracellular complexes with (ii, iii, v) and without (i, vi) Rab5 colocalization. (E, F) Intensity profiles (dashed lines in C, D) showing signals from all three fluorescent channels. Early endosomes lacking NPs are also observed (iv). (G, H) Quantitative colocalization shows that a statistically significant difference in the number of NPs found with (+) and without (−) Rab5-GFP at 30 and 60 minutes respectively. Although an increase in the number of NPs lacking colocalization with Rab5-GFP is observed from 30 to 60 minutes. The number of intracellular NPs colocalized with Rab5-GFP does not significantly change. All scale bars are 10 µm. Student t-test for colocalization * P < 0.025, ** P < 0.001.
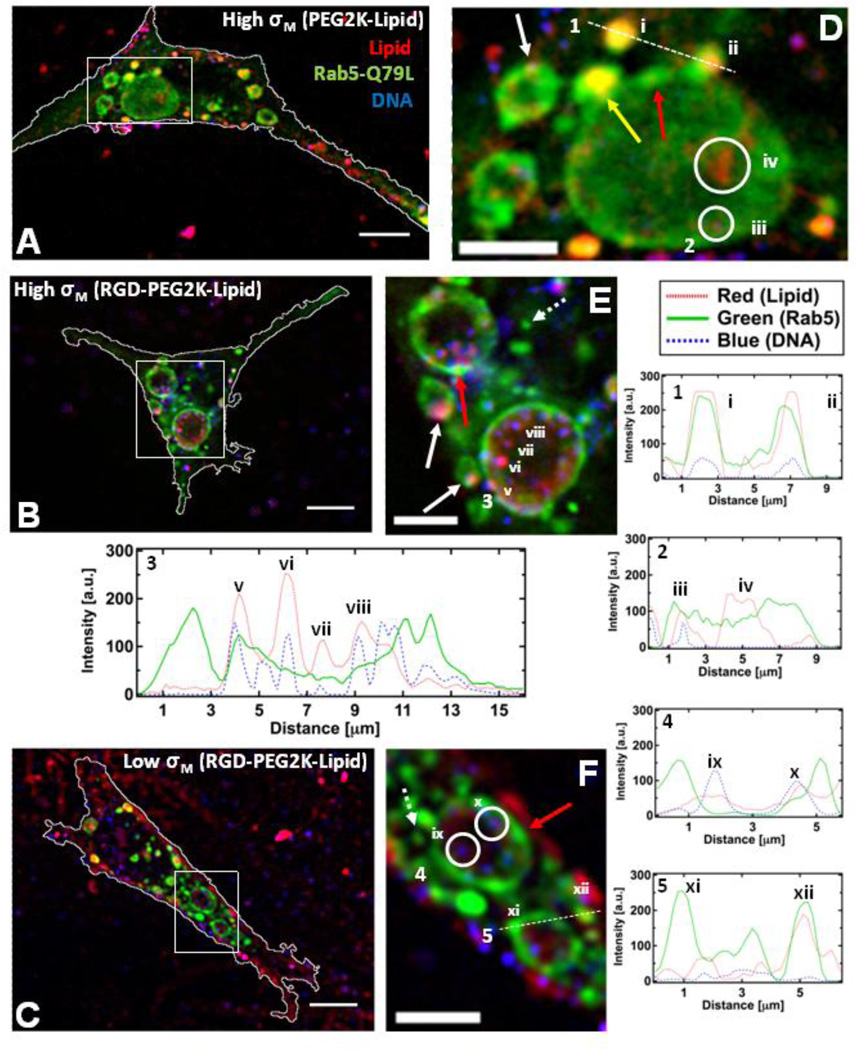
Fig. 5 A–F contains micrographs and corresponding cropped regions of cells expressing mutant Rab5-Q79L-GFP that have been incubated with High-σM NPs that lack RGD-PEG2K-lipid (Fig. 5 A and B) as well as High- and Low-σM NPs with RGD-PEG2K-lipid (Fig. 5 C and D and Fig. 5 E and F respectively). All cells shown were fixed after 1 hour of incubation at 4 °C followed by 1 hour of incubation at 37 °C. Cells expressing Rab5-Q79L show giant (> 3 µm) early endosomes (GEEs) as well as a few EEs with similar fluorescence intensity to those observed with wildtype Rab5 (dashed arrows in Fig. 5 E and F, see Fig. S2 for micrographs and cropped regions without markings). GEEs show non-uniform GFP fluorescence around their perimeter due to regions of the membrane rich in Rab5-GFP (red arrows in Fig. 5 D, E and F and Fig. 5 F (xi)) suggesting recent fusion of EEs with GEEs. Furthermore, the yellow arrow and (ii) in Fig. 5 D highlight examples where EEs containing a NP are undergoing fusion with a GEE. Visual inspection shows that in the case of NPs lacking the RGD motif we see fewer NPs colocalized with GEEs (Fig. 5 D (i, ii, iii)) relative to those that are RGD-tagged (see numerous NPs in magnified images Fig. 5 E and F). The cationic liposome uptake observed in the case of NPs lacking RGD (Fig. 5 D (iv)) arises because without the RGD-targeting moiety cationic charge is the main mechanism of cellular adhesion and uptake where cationic liposomes, which coexist with NPs for ρ > 1 [62, 63], are more cationic compared to NPs containing charge neutralizing DNA. Significantly, the creation of GEEs in cells expressing Rab5-Q79L has allowed us to spatially resolve individual RGD-tagged NPs within GEEs (Fig. 3 E (v, vi, vii, viii) and Fig. 5 F (ix and x) and corresponding intensity profiles). This NP resolvability has led to images, which strongly hint at RGD-tagged NPs adhering to GEE membranes in the case of High-σM (Fig. 5 E (v) and white arrows). Similar to what was observed with NPs lacking RGD, we also find examples of cationic liposomes inside EEs (Fig. 5 F (xii)). Finally, overall inspection of all images confirms that nearly all intracellular NPs, regardless of σM or RGD-tagging are found within GFP-labeled GEEs.
Figure 5.
Colocalization of CL-DNA NPs and giant early endosomes (GEE). Rab5-Q79L inhibits endosome maturation, showing large (>5 µm) endosomes with spatially resolvable NPs. Nearly all intracellular CLDNA particles are found within Rab5-Q79L-GFP labeled endosomes. (Red – TRITC (Lipid), Green – GFP (Endosomes), Blue – Cy5 (DNA)) (A, B, C) Fluorescent micrographs and cell boundaries (white outlines) of L-cells after 1 hour of incubation with (A) PEGylated High-σM NPs (MVL5/DOPC/PEG2K-lipid at a molar ratio of 50/40/10), (B) RGD-tagged High-σM NPs (MVL5/DOPC/RGD-PEG2K-lipid at a molar ratio 50/45/5) and (C) RGD-tagged Low-σM NPs (DOTAP/DOPC/RGD-PEG2K-lipid at a molar ratio of 30/60/10), (D, E, F) High magnification of boxed regions in (A, B, C). (1–5) Intensity profiles of labeled scans from high magnification regions. Intensity profile 1 and 2 show NPs (i, ii, iii) and liposomes (iv) found in EEs and GEEs. Intensity profile 3 shows clear evidence of individually resolvable NPs (v, vi, vii, viii) inside the lumen of the GEE. Intensity profile 4 shows two dim NPs (ix, x) inside a GEE. Intensity profile 5 shows GFP-rich region of the GEE membrane (xi) and a cationic liposome within an EE (xii).The yellow arrow in (D) points to a EE containing a NP fusing with a GEE. Red arrows in (D, E, F) show GFP-rich regions of the GEE membrane. Dashed arrows in (E, F) show smaller EEs similar to what is observed with wildtype Rab5. Solid white arrows in (E) point to clear examples of NPs adhering to the GEE membrane. Scale bars in (A, B, C) and (D, E, F) are 10 µm and 5 µm respectively.
4 Discussion
Although PEGylation of lipid-based NPs significantly improves circulation times in vivo, numerous studies have found that PEGylated NPs suffer from reduced cellular uptake and inefficient endosomal escape [23, 52]. RGD-tagging of PEGylated NPs improves cellular uptake, but when comparing High- and Low-σM RGD-tagged NPs which differed in TE by nearly 2 orders of magnitude (Fig. 2 C), we found that the uptake and fraction contained in early endosomes at early timepoints were within error (Fig. 4 G and H).
Our first aim was to confirm that PEGylation of MVL5-based CL–DNA complexes yields sterically stabilized NPs (Fig. 2). One interesting feature in the DLS data is that contrary to CL–DNA complexes formed with monovalent lipids [63, 65, 66], MVL5-based complexes are not electrostatically stabilized by overcharging at high ρ where they formed micron-sized aggregates for all ρ > 2. This weak colloidal stability suggests that steric stabilization through surface functionalization is essential for the use of MVL5 complexes in vivo, where serum or plasma are high ionic strength buffers (≈150mM 1:1 NaCl), which screen electrostatic repulsion. The addition of 10 mol% PEG2K-lipid or RGD-PEG2K-lipid (where the PEG polymer is in the brush regime [67]) induces the formation of sub-200 nm, sterically stabilized nanoparticles which retain colloidal stability for up to 24 hours after formation (Fig. 2 B). Taken together with the live cell imaging results, DLS data shows that PEGylation at 10 mol% screens the attractive van der Waals force between NPs but our live cell imaging shows that the attractive electrostatic interaction between High-σM NPs and the anionic plasma membrane is not fully screened at full polymer coverage (Fig. 3).
PEGylation has been previously reported to give rise to a steric-based repulsive force between NPs and cells [52,68], so as PEG2K-lipid mol% increases, both adhesion to the plasma membrane and fusion to the endosomal membrane upon NP uptake become energetically unfavorable, resulting in low TE [22, 23, 52]. One strategy to recover the lost adhesion to the plasma membrane is covalent attachment of a ligand to the distal end of the PEG2K-lipid, allowing NPs to specifically bind to cells via a ligand-receptor interaction. Although the TE of non-PEGylated MVL5/DOPC complexes is remarkably high, the addition of 5 and 10 mol% PEG2K-lipid significantly reduces TE (Fig. 2 C). Previously it was found that uptake of Low-σM NPs was dramatically enhanced by addition of RGD ligand, implying integrin receptor mediated uptake [52]. Here we find a similar result where RGD further enhances uptake of High- σM NPs (Fig. 3). Although RGD-tagged CL–DNA NPs can undergo internalization in the absence of electrostatic-mediated adhesion to the plasma membrane [52], endosomal escape requires sufficient electrostatic attraction between the anionic endosomal membrane and cationic NP. This steric-induced inefficient endosomal escape explains why RGD-tagged NPs cannot transfect as efficiently as CL–DNA complexes lacking PEGylation. In fact previous work has shown that the endosomal escape of non-PEGylated CL–DNA complexes is strongly dependent on σM [47], where high σM promotes electrostatically mediated fusion of the endosomal membrane and outer bilayer of the CL–DNA complex, leading to endosomal escape. Our PEGylated NPs are formed at high-σM but the steric repulsion of the RGD-PEG2K motifs present on the surface of the NP inhibit endosomal escape by giving rise to a polymer brush-induced repulsive steric force between the endosomal membrane and NP. More evidence for weak endosomal escape of PEGylated NPs with and without RGD-tagging can be found in the live cell imaging data; our NPs lack the required machinery to directly interact with motor proteins, implying that the observed perinuclear accumulation of RGD-tagged NPs is a result of active trafficking of intra-endosomal NPs (Fig. 3). Finally, recent literature has shown that targeted vectors with high internalization can still show low TE due to weak endosomal escape [52].
Our next aim was to measure the propensity of NPs to escape endosomes, where we used Rab5-GFP to allow endogenous labeling of early endosomes [70]. Rab5 is involved in both clathirin-mediated endocytosis and macropinocytosis [71], which are the main uptake pathways of the adenovirus [72], a viral vector which also presents RGD motifs on its surface (i.e. as part of the penton base of the viral capsid [73]). Furthermore, colocalization of fluorescently labeled adenovirus particles with Rab5-GFP has been previously reported [74]. While previous studies [75–77] have shown colocalization of synthetic nucleic acid vectors and Rab5-GFP labeled endosomes, our quantitative colocalization imaging of RGD-tagged NPs and Rab5-GFP and comparisons to hydrolysis-deficient mutant Rab5-Q79L provides direct visual evidence that nanoparticles do not escape early endosomes. Interestingly, despite an extensive incubation protocol, only a small but significant fraction of both Low and High-σM NPs were found to colocalize with wildtype EEs. The High- and Low-σM NPs used in our Rab5 studies differed in TE by nearly two orders of magnitude while showing a relatively small difference in uptake and colocalization with Rab5 endosomes.
Rab5-EEs have been previously shown to have a half life of 10–15 minutes [36] implying that at 30 minutes post internalization NPs should be further along the endocytic pathway. Although this line of evidence (the short lifetime of EEs) should suggest that no NP-EE colocalization should occur at later time points (60 minutes), Fig. 4 H shows statistically significant colocalization of NPs with EEs at 60 minutes. Although our incubation protocol was designed to synchronize internalization of NPs, the increase in total uptake from 30 to 60 minutes in combination with NP-EE colocalization at 60 minutes suggests that once bound to the plasma membrane NPs cannot all simultaneously internalize. This results in individual endocytic events occurring over a range of times after activation of endocytosis upon warming the cells from 4 °C to 37 °C. The live cell images in Fig. 3 support this conclusion as they show plasma membrane bound NPs at up to 4 hours of incubation. Regarding NPs lacking EE colocalization we see an increase from 30 to 60 minutes (GFP-Rab5(−) in Fig. 4 G and H). We interpret this as RGD-tagged NPs internalizing throughout the warm incubation, briefly colocalizing with Rab5 EEs and accumulating in later stage endocytic vesicles lacking Rab5. In fact, recent literature [78] has shown that cationic lipid-NA NPs undergo slower internalization kinetics relative to particles with similar size and ligands, which is in a agreement with our model that at any given early time point, a fraction of NPs are surface bound, in EEs or further along the endocytic pathway. In summary, our wildtype Rab5 results show that (1) once bound to the plasma membrane there is a broad distribution of time constants for NPs to undergo internalization, (2) upon internalization NPs pass through EEs on the scale of 30 minutes and accumulate in later endocytic stages and (3) initial internalization and EE colocalization do not depend on σM for RGD-tagged NPs.
Cell imaging with the mutant Rab5-Q79L has allowed us to unambiguously determine whether the NPs observed not to be colocalized with EEs have escaped early endosomes into the cytoplasm, internalized through a Rab5-independent mechanism, or progressed further along the endocytic pathway. Rab5-Q79L slows maturation of early endosomes due to very slow GTP hydrolysis activity, causing early endosomes to continuously fuse and grow into GEEs with internal NPs which are spatially resolvable. Nearly all fluorescent NPs are found to directly colocalize with or within GFP-Rab5-Q79L labeled GEEs (Fig. 5). The high level of colocalization of NPs with GEEs rules out the possibility of RGD-tagged NPs using non-Rab5 dependent pathways or efficiently escaping EEs. More importantly, the Rab5-Q79L data strongly suggests that the relatively low amounts of colocalization observed in wildtype Rab5-GFP is a result of the short half life of wildtype early endosomes because increasing the lifetime of the early endosome (i.e. with Rab5-Q79L which inhibits EE maturation) increases the colocalization of NPs with mutant Rab5. With wildtype Rab5, a large fraction of intracellular NPs do not colocalize with early endosomes but taken together with the observation of that all particles colocalize with mutant Rab5-Q79L we conclude that the fraction of NPs which escape endosomes do so during the later stages of endocytosis. Our model is in agreement with recent work using EM to directly measure lipid-siRNA access to the cytoplasm, where the authors propose that endosomal escape only occurs after NPs have resided within cells for longer than 1.5 hours [78]. Furthermore, our conclusion is also in agreement with another recent literature report [79] which showed that polyplexes and conventional lipoplexes (e.g.; lacking PEG-lipid) that internalize via clathrin-mediated endocytosis show low transfection efficiency unless they are rerouted (via protein kinase A inhibition) to another late stage endosome that lacks the familiar components of the wildtype late endosome pathway. These authors, in agreement with our study, concluded through indirect evidence that escape from early endosomes is unlikely. There has also been a recent report in the literature that did directly visualize release of nucleic acid into the cytoplasm at short time points (5 minutes) which is in stark contrast to our results with Rab5-Q79L where we see no evidence of escape even after 1 hour of incubation [80]. Two major differences between this report and our findings lie in the vector and the cargo. First, our vector contains PEG-lipids yield which have been previously shown to inhibit endosomal escape [52] as opposed to conventional lipoplexes which are more efficient carriers of nucleic acid and could be escaping from early endosomes. Second, the early release of cargo was observed for small (20 bp) oligonucleotides via a hypothesized pore forming mechanism. Our NPs carry plasmid which we speculate would not be able to escape endosomes via pores as efficiently as small oligonucleotides.
Interestingly, the mutant and wildtype Rab5 data both show that early endosome colocalization is not indicative of high or low TE. NPs used in the Rab studies differed in TE by nearly two orders of magnitude but show similar uptake, similar wildtype Rab5 colocalization and finally similar mutant Rab5-colocalization at early time points. In summary, the combination of wildtype and mutant Rab5 data provides direct evidence that (1) RGD-tagged CL-DNA NPs use a Rab5-dependent pathway for cellular uptake, (2) colocalization of PEGylated NPs, or lack thereof, with Rab5-GFP does not depend on σM and is not indicative of high or low TE and (3) the major bottlenecks to high TE for RGD-tagged CL-DNA NPs are downstream of uptake and early endosomes.
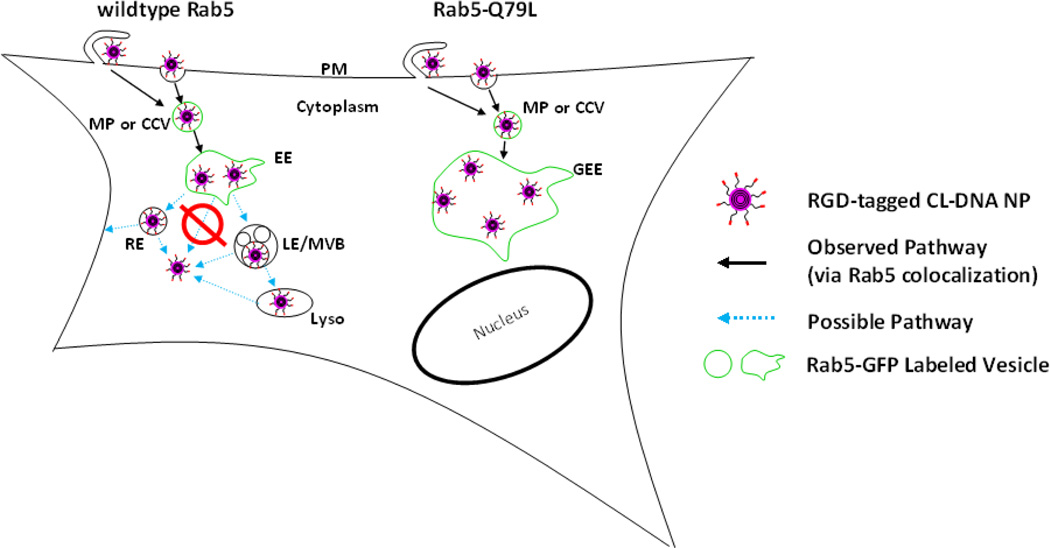
Fig. 6 contains a schematic which summarizes our findings. The observed pathways of RGD-tagged CL–DNA NPs are shown with solid black arrows. NPs adhere to the plasma membrane where they undergo endocytosis through either macropinocytosis or clathrin-mediated endocytosis. Upon internalization, NPs are trafficked in macropinosomes or clathrin coated vesicles (which contain Rab5) to early endosomes. From the early endosome, in the case of wildtype Rab5, we highlight possible pathways of RGD-tagged CL–DNA NPs (blue, dashed arrows), which include but are not limited to recycling endosomes or late endosomes/multivesicular bodies. From these later endocytic stages we know that a fraction of our particles escape the endosomal pathway due to moderate to high levels of gene expression in our luciferase assay. In the case of Rab5-Q79L, NPs (whether RGD-tagged or not) are trafficked to large, swollen GEEs where the vast majority remain even up to 1 hour post incubation. Finally, our observation that NPs do not escape from GEEs at early time points is strong evidence that NPs cannot escape from short-lived wildtype EEs (red symbol, Fig. 6). One possible explanation for our observation that NPs were not observed to escape GEEs is that the formation of recycling endosomes through fission of early endosomes may be a crucial step for endosomal escape. It has been previously shown that Rab5-Q79L significantly reduces (by up to 60%) recycling, suggesting that the growth of GEEs is due to both increased fusion between EEs as well as reduced fission (e.g.; formation of recycling endosomes) [36]. Previous work strongly suggests that fission is not the dominant means of endosomal escape. First, a previous report has shown that using a PEG-lipid capable of cleavage at low pH leads to improved transfection efficiency due to improved endosomal escape, suggesting that endosomal escape of PEGylated NPs occurs at low pH endocytic stages which are downstream of early endosomes [23]. Second, recent work has shown that the membrane charge density mediates endosomal escape which strongly suggests that escape occurs through fusion or pore forming process. If escape could occur through fission events we would not expect endosomal escape to strongly depend on membrane charge density, as has been previously reported [52].
Figure 6.
Schematic Showing Known and Possible Pathways of RGD-tagged CL-DNA NPs. Legend: PM: plasma membrane, MP: macropinosome, CCV: clathrin-coated vesicle, (G)EE: (giant) early endosome, RE: recycling endosome, LE: late endosome, MVB: multivesicular body, Lyso: lysosome. In both cases (wildtype or mutant Rab5) NPs bind to the plasma membrane and undergo endocytosis via CCV or MPs. In the case of wildtype Rab5, NPs initially colocalize with Rab5-GFP labeled vesicles and then proceed to two possible pathways e.g.; recycling or degradative. From these later stages, a fraction of NPs escape from the endosomal pathway as evidenced by measured gene expression. In the case of mutant Rab5-Q79L-GFP, at short time scales (≈1 hr) an accumulation of CL-DNA NPs are found in early endosomes and no other pathways are explored. Two distinct observations suggest that NPs do not escape wildtype early endosomes: (1) NPs were found to not escape GEEs and (2) colocalization with EEs is not membrane charge density but gene expression (TE) which requires endosomal escape is strongly dependent on membrane charge density.
5 Conclusion
PEGylation of MVL5-based CL–DNA complexes promotes the formation of sterically-stabilized nanoparticles while RGD-tagging of the PEGylated nanoparticles improves cellular uptake and TE. Previous works shows that the uptake of PEGylated NPs lacking RGD-tagging strongly depends on membrane charge density [52]. Comparing our results with those found in the literature for NPs containing univalent cationic lipids, we note that MVL5-based NPs show improved cellular uptake and TE when compared to NPs based on the monovalent lipid DOTAP, consistent with the hypothesis that high membrane charge density improves endosomal escape [52]. To further elucidate early events in the endocytic pathway of RGD-tagged NPs we used Rab5-GFP for endogenous labeling of early endosomes. We find that only a minority of intracellular NPs colocalize with Rab5-GFP at early time points (t < 1 h). A brief survey of the literature [75–77, 81–83] shows that low levels of colocalization of synthetic vectors and Rab5-GFP endosomes is frequently observed. We conclude that this observation is a result of the inherent short life time of wildtype EE. Low levels of colocalization with wildtype Rab5-GFP could imply efficient escape from the early endosome or an alternative internalization mechanism, but our data using mutant Rab5-Q79L shows a significant fraction of intracellular NPs colocalizing with relatively long-lived giant endosomes (resulting from self-fusion of early endosomes) suggesting that escape from early endosomes is a rare event. This hypothesis is further substantiated by the observation that although fluorescence microscopy shows that total uptake and colocalization of NPs with early endosomes does not depend on σM, transfection efficiency (a measure of exogenous gene expression), which requires endosomal escape, strongly depends on σM implying that PEGylated NPs do not escape from EEs. Future studies using other GTPase deficient Rab mutants will allow for a better understanding of lipid-NA intracellular pathways as well as optimization of vectors for improved efficiency. Finally, development of NPs has been hampered by the lack of a robust imaging platform to directly visualize the dynamics and motion of NPs trapped in endosomes. Thus, Rab5-Q79L could allow for optical assessment of early endosomal membrane penetrating peptides of different NPs with the goal of designing membrane-penetrating NPs that escape early endosomes.
Highlights.
-
-
GTP hydrolysis deficient Rab5 slows endsosome maturation and increases their size.
-
-
Small fraction of nanoparticles colocalizes with wildtype Rab5 labeled endosomes.
-
-
Nearly all intracellular nanoparticles colocalized with mutant early endosomes.
-
-
Absence of Rab5-nanoparticle colocalization does not indicate endosomal escape.
-
-
Endosomal escape occurs downstream of early endosomes.
Acknowledgements
This work was supported by NIH GM-59288 (transfection efficiency and colocalization studies with Rab GTPases) and NSF DMR-1401784 (nanoparticle imaging and automated image analysis). CLC was supported by NSRRC and Academia Sinica, Taiwan. DLS characterization was performed using the Central Facilities of the Materials Research Laboratory at UCSB which are supported by the MRSEC Program of the NSF under award no. DMR-1121053; a member of the NSF-funded Materials Research Facilities Network (www.mrfn.org). BFBS was supported by a Marie Curie International Outgoing Fellowship within the EU Seventh Framework Programme for Research and Technological Development (2007–2013), under grant agreement no. PIOF-GA-2009-252701. The Rab5-Q79L-GFP plasmid was a gift from the Weimbs Lab (UCSB). We thank Adrian Giovannone and Thomas Weimbs for critical reading of the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
RNM and CRS designed the experiments. RNM performed microscopy, DLS, and data analysis. RNM, KKE and CRS wrote the manuscript. CLC performed the transfection experiments. KKE designed and synthesized RGD-PEG2K-lipid and MVL5. BFBS, KSL and CLC made critical comments on the manuscript.
References
- 1.Yin H, Kanasty RL, Eltoukhy AA, Vegas AJ, Dorkin JR, Anderson DG. Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 2014;15:541–555. doi: 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- 2.Guo X, Huang L. Recent Advances in Nonviral Vectors for Gene Delivery. Acc. Chem. Res. 2012;45:971–979. doi: 10.1021/ar200151m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Salcher E, Wagner E. Chemically Programmed Polymers for Targeted DNA and siRNA Transfection. Top. Curr. Chem. 2010;296:227–249. doi: 10.1007/128_2010_69. [DOI] [PubMed] [Google Scholar]
- 4.Sioud M. Therapeutic siRNAs. Trends Pharmacol. Sci. 2004;25:22–28. doi: 10.1016/j.tips.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Caplen NJ. RNAi as a gene therapy approach. Expert Opin. Biol. Ther. 2003;3:575–586. doi: 10.1517/14712598.3.4.575. [DOI] [PubMed] [Google Scholar]
- 6.Yin JY, Ma ZY, Selliah N, Shivers DK, Cron RQ, Finkel TH. Effective gene suppression using small interfering RNA in hard-to-transfect human T cells. J. Immunol. Methods. 2006;312:1–11. doi: 10.1016/j.jim.2006.01.023. [DOI] [PubMed] [Google Scholar]
- 7.Bielke W, Erbacher C, editors. Topics in Current Chemistry, Vol. 296: Nucleic Acid Transfection. Heidelberg: Springer; 2010. [PubMed] [Google Scholar]
- 8.Ewert KK, Zidovska A, Ahmad A, Bouxsein NF, Evans HM, McAllister CS, Samuel CE, Safinya CR. Cationic Lipid-Nucleic Acid Complexes for Gene Delivery and Silencing: Pathways and Mechanisms for Plasmid DNA and siRNA. Top. Curr. Chem. 2010;296:191–226. doi: 10.1007/128_2010_70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Safinya CR, Ewert KK, Majzoub RN, Leal C. Cationic liposome–nucleic acid complexes for gene delivery and gene silencing. New J. Chem. 2014;38:5164–5172. doi: 10.1039/C4NJ01314J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang L, Hung M-C, Wagner E. Non-Viral Vectors for Gene therapy, Part I, Advances in Genetics. Vol. 53. San Diego: Elsevier; 2005. [Google Scholar]
- 11.Ewert KK, Ahmad A, Evans HM, Safinya CR. Cationic lipid-DNA complexes for non-viral gene therapy: relating supramolecular structure to cellular pathways. Expert Opin. Biol. Ther. 2005;5:33–53. doi: 10.1517/14712598.5.1.33. [DOI] [PubMed] [Google Scholar]
- 12.Ewert KK, Evans HM, Ahmad A, Slack NL, Lin AJ, Martin-Herranz A, Safinya CR. Lipoplex Structures and their Distinct Cellular Pathways. In: Huang L, Hung M-C, Wagner E, editors. Non-Viral Vectors for Gene therapy, Part I, Advances in Genetics. Vol. 53. San Diego: Elsevier; 2005. [Google Scholar]
- 13.Ewert KK, Slack NL, Ahmad A, Evans HM, Lin AJ, Samuel CE, Safinya CR. Cationic lipid-DNA complexes for gene therapy: Understanding the relationship between complex structure and gene delivery pathways at the molecular level. Curr. Med. Chem. 2004;11:133–149. doi: 10.2174/0929867043456160. [DOI] [PubMed] [Google Scholar]
- 14.Li SD, Huang L. Gene therapy progress and prospects: non-viral gene therapy by systemic delivery. Gene Ther. 2006;13:1313–1319. doi: 10.1038/sj.gt.3302838. [DOI] [PubMed] [Google Scholar]
- 15.Yi Y, Hahm SH, Lee KH. Retroviral Gene Therapy: Safety Issues and Possible Solutions. Curr. Gene Ther. 2005;5:25–35. doi: 10.2174/1566523052997514. [DOI] [PubMed] [Google Scholar]
- 16.Young L, Mautner V. The promise and potential hazards of adenovirus gene therapy. Gut. 2001;48:733–736. doi: 10.1136/gut.48.5.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raper S, Chirmule N, Lee F, Wivel N, Bagg A, Gao GP, Wilson JM, Batshaw ML. Fatal systemic inflammatory response syndrome in a ornithine transcarbamylase deficient patient following adenoviral gene transfer. Mol. Genet. Metab. 2003;80:148–158. doi: 10.1016/j.ymgme.2003.08.016. [DOI] [PubMed] [Google Scholar]
- 18.Willard HF. Artificial chromosomes coming to life. Science. 2000;290:1308. doi: 10.1126/science.290.5495.1308. [DOI] [PubMed] [Google Scholar]
- 19.Plank C, Mechtler K, Szoka FC, Jr, Wagner E. Activation of the complement system by synthetic DNA complexes: a potential barrier for intravenous gene delivery. Hum. Gene Ther. 1996;7:1437–1446. doi: 10.1089/hum.1996.7.12-1437. [DOI] [PubMed] [Google Scholar]
- 20.Fenske DB, MacLachlan I, Cullis PR. Stabilized plasmid-lipid particles: A systemic gene therapy vector. Methods Enzymol. 2002;346:36–71. doi: 10.1016/s0076-6879(02)46048-x. [DOI] [PubMed] [Google Scholar]
- 21.Immordino ML, Dosio F, Cattel L. Stealth liposomes: review of the basic science, rationale and clinical applications, existing and potential. Int. J. Nanomedicine. 2006;1:297–315. [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Herranz A, Ahmad A, Evans HM, Ewert KK, Schulze U, Safinya CR. Surface functionalized cationic lipid–DNA complexes for gene delivery: PEGylated lamellar complexes exhibit distinct DNA–DNA interaction regimes. Biophys. J. 2004;86:1160–1168. doi: 10.1016/S0006-3495(04)74190-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chan CL, Majzoub RN, Shirazi RS, Ewert KK, Chen YJ, Liang KS, Safinya CR. Endosomal Escape and Transfection Efficiency of PEGylated Cationic Liposome–DNA Complexes Prepared with an Acid-Labile PEG-Lipid. Biomaterials. 2012;33:4928–4935. doi: 10.1016/j.biomaterials.2012.03.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langer R. Drug delivery and Targeting. Nature. 1998;392:S5–S10. [PubMed] [Google Scholar]
- 25.Ruoslahti E, Bhatia SN, Sailor MJ. Targeting of drugs and nanoparticles to tumors. J. Cell Biol. 2010;188:759–768. doi: 10.1083/jcb.200910104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Torchilin VP. Recent advances with liposomes as pharmaceutical carriers. Nat. Rev. Drug Discov. 2005;4:145–160. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 27.Koo OM, Rubinstein I, Onyuksel H. Role of nanotechnology in targeted drug delivery and imaging: a concise review. Nanomed.-Nanotechnol. 2005;1:193–212. doi: 10.1016/j.nano.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 28.Teesalu T, Sugahara KN, Ruoslahti E. Mapping of vascular ZIP codes by phage display. Methods Enzymol. 2012;503:35–56. doi: 10.1016/B978-0-12-396962-0.00002-1. [DOI] [PubMed] [Google Scholar]
- 29.Sugahara KN, Teesalu T, Karmali PP, Kotamraju VR, Agemy L, Girard OM, Hanahan D, Mattrey RF, Ruoslahti E. Tissue-penetrating delivery of compounds and nanoparticles into tumors. Cancer Cell. 2009;8:510–520. doi: 10.1016/j.ccr.2009.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zerial M, McBride H. Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Bio. 2001;2:107–117. doi: 10.1038/35052055. [DOI] [PubMed] [Google Scholar]
- 31.Gorvel JP, Chavrier P, Zerial M, Gruenberg J. rab5 constrols early endosome fusion in vitro. Cell. 1991;64:915–925. doi: 10.1016/0092-8674(91)90316-q. [DOI] [PubMed] [Google Scholar]
- 32.Rubino M, Miaczynska M, Lippe R, Zerial M. Selective membrane recruitment of EEA1 suggests a role in directional transport of clathrin-coated vesicles to early endosomes. J. Biol. Chem. 2000;275:3745–3748. doi: 10.1074/jbc.275.6.3745. [DOI] [PubMed] [Google Scholar]
- 33.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase Rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 34.Houiuchi H, Giner A, Hoflack B, Zerial M. A GDP/GTP exchange-stimulatory activity for the Rab5-RabGDI complex on clathri-coated vesicles from bovine brain. J. Biol. Chem. 1995;270:11257–11262. doi: 10.1074/jbc.270.19.11257. [DOI] [PubMed] [Google Scholar]
- 35.Lanzetti L, Palamidessi A, Areces L, Scita G, Di Fiorre PP. Rab5 is a signaling GTPase involved in actin remodeling by receptor tyrosine kinases. Nature. 2004;429:309–314. doi: 10.1038/nature02542. [DOI] [PubMed] [Google Scholar]
- 36.Stenmark H, Parton RG, Steele-Mortimer O, Lutcke A, Gruenberg J, Zerial M. Inhibition of rab5 GTPase activity stimulates membrane fusion in endocytosis. EMBO J. 1994;13:1287–1296. doi: 10.1002/j.1460-2075.1994.tb06381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- 38.Ullrich O, Stenmark H, Alexandrov K, Huber LA, Kaibuchi K, Sasaki T, Takai Y, Zerial M. Rab GDP-dissociation inbitor as a general regulator for the membrane association of Rab Proteins. J. Biol. Chem. 1993;268:18143–18150. [PubMed] [Google Scholar]
- 39.Rink J, Ghigo E, Kalaidzidis Y, Zerial M. Rab conversion as a mechanism of progression from early to late endosomes. Cell. 2005;122:735–749. doi: 10.1016/j.cell.2005.06.043. [DOI] [PubMed] [Google Scholar]
- 40.Roberts RL, Barbieri MA, Pryse KM, Chua M, Morisaki JH, Stahl PD. Endosome fusion in living cells overexpressing GFP-Rab5. J. Cell Sci. 1999;112:3667–3675. doi: 10.1242/jcs.112.21.3667. [DOI] [PubMed] [Google Scholar]
- 41.Ruoslati E. RGD and other recognition sequences for integrins. Annu. Rev. Cell Dev. Biol. 1996;12:697–715. doi: 10.1146/annurev.cellbio.12.1.697. [DOI] [PubMed] [Google Scholar]
- 42.Mizejewski GJ. Role of integrins in cancer: survey of expression patterns. Proc. Soc. Exp. Biol. Med. 1999;222:124–138. doi: 10.1177/153537029922200203. [DOI] [PubMed] [Google Scholar]
- 43.Ewert KK, Ahmad A, Evans HM, Schmidt HW, Safinya CR. Efficient synthesis and cell-transfection properties of a new multivalent cationic lipid for nonviral gene delivery. J. Med. Chem. 2002;45:5023–5029. doi: 10.1021/jm020233w. [DOI] [PubMed] [Google Scholar]
- 44.Ahamd A, Evans HM, Ewert K, George CX, Samuel CE, Safinya CR. New multivalent cationic lipids reveal bell curve for transfection efficiency versus membrane charge density: lipid-DNA complexes for gene delivery. J. Gene Med. 2005;7:739–748. doi: 10.1002/jgm.717. [DOI] [PubMed] [Google Scholar]
- 45.Bouxsein NF, McAllister CS, Ewert KK, Samuel CE, Safinya CR. Structure and gene silencing activities of monovalent and pentavalent cationic lipid vectors complexed with siRNA. Biochemistry. 2007;46:4785–4792. doi: 10.1021/bi062138l. [DOI] [PubMed] [Google Scholar]
- 46.Chan CL, Ewert KK, Majzoub RN, Hwu Y-K, Liang KS, Leal C, Safinya CR. Optimizing cationic and neutral lipids for efficient gene delivery at high serum content. J. Gene Med. 2014;16:84–96. doi: 10.1002/jgm.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin AJ, Slack NL, Ahmad A, George CX, Samuel CE, Safinya CR. Three-dimensional imaging of lipid gene-carriers: Membrane charge density controls universal transfection behavior in lamellar cationic liposome-DNA complexes. Biophys. J. 2003;84:3307–3316. doi: 10.1016/S0006-3495(03)70055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zuhorn IS, Kalicharan R, Hoekstra D. Lipoplex-mediated Transfection of Mammalian Cells Occurs through the Cholesterol-dependent Clathrin-mediated Pathway of Endocytosis. J. Biol. Chem. 2002;277:18021–18028. doi: 10.1074/jbc.M111257200. [DOI] [PubMed] [Google Scholar]
- 49.Zuhorn IS, Kalicharan R, Hoekstra D. Gene transfer by Means of Lipo- and Polyplexes: Role of Clathrin and Caveolae-Mediated Endocytosis. J. Liposome Res. 2006;16:237–247. doi: 10.1080/08982100600848819. [DOI] [PubMed] [Google Scholar]
- 50.Rehman Z, Sjollema KA, Kuipers J, Hoekstra D, Zuhorn IS. Nonviral gene delivery vectors use syndecan-dependent transport mechanisms in filopodia to reach the cell surface. ACS Nano. 2012;6:7521–7532. doi: 10.1021/nn3028562. [DOI] [PubMed] [Google Scholar]
- 51.Garbuzenko O, Barenholz Y, Priev A. Effect of grafted PEG on liposome size and on compressibility and packing of lipid bilayer. Chem. Phys. Lipids. 2005;135:117–129. doi: 10.1016/j.chemphyslip.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 52.Majzoub RN, Chan CL, Ewert KK, Silva BFB, Liang KS, Jacovetty EL, Carragher B, Potter CS, Safinya CR. Uptake and transfection efficiency of PEGylated cationic liposome-DNA complexes with and without RGD-tagging. Biomaterials. 2014;35:4996–5005. doi: 10.1016/j.biomaterials.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gjetting T, Arildsen NS, Christensen CL, Poulsen TT, Roth JA, Handlos VN, Poulsen HS. In vitro and in vivo effects of polyethylene glycol (PEG)-modified lipid in DOTAP/cholesterol-mediated gene transfection. Int. J. Nanomedice. 2010;9:371–383. doi: 10.2147/ijn.s10462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sadzuka Y, Kishi K, Hirota S, Sonobe T. Effect of polyethyleneglycol (PEG) chain on cell uptake of PEG-modified liposomes. J. Liposome Res. 2003;13:157–172. doi: 10.1081/lpr-120020318. [DOI] [PubMed] [Google Scholar]
- 55.Adil MM, Erdman ZZ, Kokkoli E. Transfection Mechanisms of Polyplexes, Lipoplexes and Stealth Liposomes in α5β1 Integrin Bearing DLD-1 Colorectal Cancer Cells. Langmuir. 2014;30:3802–3810. doi: 10.1021/la5001396. [DOI] [PubMed] [Google Scholar]
- 56.Silva BFB, Majzoub RN, Chan C-L, Li Y, Olsson U, Safinya CR. PEGylated Cationic Liposome–DNA Complexation in Brine is Pathway-Dependent. Biochim. Biophys. Acta – Biomembranes. 2014;1838:398–412. doi: 10.1016/j.bbamem.2013.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Crocker JC, Grier DG. Methods of Digital Video Microscopy for Colloidal Studies. J. Colloid Interf. Sci. 1996;159:298–310. [Google Scholar]
- 58.Iwasa A, Akia H, Khalil I, Kogure K, Futaki S, Harashima H. Cellular uptake and subsequent intracellular trafficking of R8-liposomes introduced at low temperature. BBA-Biomembranes. 2006;1758:713–720. doi: 10.1016/j.bbamem.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 59.Hama S, Akita H, Ito R, Mizuguchi H, Hayakawa T, Harashima H. Quantitative Comparison of Intracellular Trafficking and Nuclear Transcription between Adenoviral and Lipoplex Systems. Mol. Ther. 2006;13:786–794. doi: 10.1016/j.ymthe.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 60.El-Sayed A, Khalil IA, Kogure K, Futaki S, Harashima H. Octaarginine- and Octalysine-modified Nanoparticles Have Different Modes of Endosomal Escape. J. Biol. Chem. 2008;283:23450–23461. doi: 10.1074/jbc.M709387200. [DOI] [PubMed] [Google Scholar]
- 61.Rädler JO, Koltover I, Salditt T, Safinya CR. Structure of DNA-cationic liposome complexes: DNA intercalation in multilamellar membranes in distinct interhelical packing regimes. Science. 1997;275:810–814. doi: 10.1126/science.275.5301.810. [DOI] [PubMed] [Google Scholar]
- 62.Koltover I, Salditt T, Rädler JO, Safinya CR. An inverted hexagonal phase of cationic liposome-DNA complexes related to DNA release and delivery. Science. 1998;281:78–81. doi: 10.1126/science.281.5373.78. [DOI] [PubMed] [Google Scholar]
- 63.Koltover I, Salditt T, Safinya CR. Phase diagram, stability, and overcharging of lamellar cationic lipid–DNA self-assembled complexes. Biophys. J. 1999;77:915–924. doi: 10.1016/S0006-3495(99)76942-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laouini A, Jaafar-Maalej C, Limayem-Blouza I, Sfar S, Charcosset C, Fessi H. Preparation, characterization and applications of liposomes: state of the art. J. Colloid Sci. Biotech. 2012;1:147–168. [Google Scholar]
- 65.Eastman SJ, Siegel C, Tousignant J, Smith AE, Cheng SH, Scheule RK. Biophysical characterization of cationic lipid:DNA complexes. Biochim. Biophys. Acta. 1997;1325:41–62. doi: 10.1016/s0005-2736(96)00242-8. [DOI] [PubMed] [Google Scholar]
- 66.Trinh MU, Ralston J, Fornasiero D. Characterisation and stability of lipid-DNA complexes. Colloids Surf. B. Biointerfaces. 2008;67:85–91. doi: 10.1016/j.colsurfb.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 67.Kuhl TL, Leckband DE, Lasic DD, Israelachvili JN. Modulation of interaction forces between bilayers exposing short-chained ethylene oxide headgroups. Biophys J. 1995;66:1479–1488. doi: 10.1016/S0006-3495(94)80938-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Israelachvili J. Intermolecular and surface forces. London: Academic Press; 1992. p. 31. [Google Scholar]
- 69.Lin AJ, Slack NL, Ahmad A, George CX, Samuel CE, Safinya CR. Three-dimensional imaging of lipid gene-carriers: Membrane charge density controls universal transfection behavior in lamellar cationic liposome–DNA complexes. Biophys. J. 2003;84:3307–3316. doi: 10.1016/S0006-3495(03)70055-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bucci C, Parton RG, Mather IH, Stunnenberg H, Simons K, Hoflack B, Zerial M. The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell. 1992;70:715–728. doi: 10.1016/0092-8674(92)90306-w. [DOI] [PubMed] [Google Scholar]
- 71.Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Bio. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- 72.Meier O, Boucke K, Hammer SV, Keller S, Stidwill RP, Hemmi S, Greber UF. Adenovirus triggers macropinocytosis and endosomal leakage together with its clathrin-mediated uptake. J. Cell Bio. 2002;158:1119–1131. doi: 10.1083/jcb.200112067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Neumann R, Chroboczek J, Jacrot B. Determination of the nucleotide sequence for the penton-base gene of human adenovirus type 5. Gene. 1988;69:153–157. doi: 10.1016/0378-1119(88)90389-7. [DOI] [PubMed] [Google Scholar]
- 74.Rauma T, Tuukkanen J, Bergelson JM, Denning G, Hautala T. rab5 GTPase Regulates Adenovirus Endocytosis. J. Virol. 1999;73:9664–9668. doi: 10.1128/jvi.73.11.9664-9668.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wong AW, Scales SJ, Reilly DE. DNA Internalized via Caveloae Requires Microtubule-dependent, Rab7-independent Transport to the Late endocytic pathway for Delivery to the Nucleus. J. Bio. Chem. 2007;282:22953–22963. doi: 10.1074/jbc.M611015200. [DOI] [PubMed] [Google Scholar]
- 76.Billiet L, Gomez JP, Berchel M, Jaffres PA, Le Gall T, Montier T, Bertrand E, Cheradame H, Geugan P, Pitard B, Benvegnu T, Lehn T, Pichon C, Midoux P. Gene transfer by chemical vectors, and endocytosis routes of polyplexes, lipoplexes and lipopolyplexes in a myoblast cell line. Biomaterials. 2012;33:2980–2990. doi: 10.1016/j.biomaterials.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 77.Perche F, Gosset D, Mevel M, Miramon ML, Yaouanc J-J, Pichon C, Benvegnu T, Jaffres PA, Midoux P. Selective gene delivery in dendritic cells with mannosylated and histidylated lipopolyplexes. J. Drug Target. 2011;19:315–325. doi: 10.3109/1061186X.2010.504262. [DOI] [PubMed] [Google Scholar]
- 78.Gilleron J, Querbes W, Zeigerer A, Borodovsky A, Marisco G, Schubert U, Manygoats K, Seifert S, Andree C, Stoter M, Epstein-Barash H, Zhang L, Koteliansky V, Fitzgerald K, Fava E, Bickle M, Kalaidzidis Y, Akinc A, Maier M, Zerial M. Image-based analysis of lipid nanoparticle-mediated siRNA delivery, intracellular trafficking and endosomal escape. Nat. Biotechnol. 2013;31:638–646. doi: 10.1038/nbt.2612. [DOI] [PubMed] [Google Scholar]
- 79.Rehman Z, Hoekstra D, Zuhorn IS. Protein kinase A inhibition modulates the intracellular routing of gene delivery vehicles in HeLa cells, leading to productive transfection. J. Control. Release. 2011;156:76–84. doi: 10.1016/j.jconrel.2011.07.015. [DOI] [PubMed] [Google Scholar]
- 80.Rehman Z, Hoekstra D, Zuhorn IS. Mechanism of Polyplex- and Lipoplex-Mediated Delivery of Nucleic Acids: Real-Time Visualization of Transient Membrane Destabilization without Endosomal Lysis. ACS Nano. 2013;7:3767–3777. doi: 10.1021/nn3049494. [DOI] [PubMed] [Google Scholar]
- 81.Sandin P, Fitzpatrick L, Simpson J, Dawson K. High-Speed Imaging of Rab Family GTPases Reveals Rare Events in Nanoparticle Trafficking in Living Cells. ACS Nano. 2012;6:1513–1521. doi: 10.1021/nn204448x. [DOI] [PubMed] [Google Scholar]
- 82.Song Q, Yao L, Huang M, Hu Q, Lu Q, Qu B, Qi H, Rong Z, Jiang X, Gao X, Chen J, Chen H. Mechanisms of transcellular transport of wheat germ agglutinin-functionalized polymeric nanoparticles in Caco-2 cells. Biomaterials. 2012;33:6769–6782. doi: 10.1016/j.biomaterials.2012.05.066. [DOI] [PubMed] [Google Scholar]
- 83.Vercauteren D, Deschout H, Remaut K, Engbersen JFJ, Jones AT, Demeester J, De Smedt SC, Braeckmans K. Dynamic colocalization microscopy to characterize intracellular trafficking of nanomedicines. ACS Nano. 2011;5:7874–7884. doi: 10.1021/nn2020858. [DOI] [PubMed] [Google Scholar]