Abstract
AIM: To investigate the regulation of phosphatase and tensin homolog deleted on chromosome ten (PTEN) gene expression in human hepatocellular carcinoma (HCC) cell lines.
METHODS: The mRNA and protein levels of PTEN were detected by Northern blot and Western blot in HCC cell lines, respectively. Plasmids containing different fragments of PTEN promoter with Luciferase reporter were constructed and transiently transfected into HCC cell lines to study the promoter activity. DNA analysis and RT-PCR were performed to detect the mutation of PTEN promoter and PTEN cDNA.
RESULTS: Either protein or mRNA levels of PTEN in L02 cells (as a control) were significantly higher than that in HCC cell lines. The profile of PTEN promoter activity in 8 cell lines was closely correlated with levels of PTEN mRNA and PTEN protein. Furthermore, the sequence analysis of 8 cells lines showed no mutation in the region of PTEN promoter and PTEN cDNA.
CONCLUSION: PTEN expression is down-regulated in HCC cell lines probably due to loss of activity of PTEN promoter.
Keywords: Phosphatase, Tensin homolog, Hepatocellular carcinoma
INTRODUCTION
The tumor suppressor gene phosphatase and tensin homolog deleted on chromosome ten (PTEN), mutated in a wide range of human cancers[1,2], encodes a protein containing 403 amino acids with phospholipid and protein phosphatase activity[3-6]. Consequently, PTEN inhibits the generation of phosphatidylinositol 3,4,5-trisphosphate (PIP3)[7] and then blocks the activation of proto-oncogene PKB/Akt[8,9]. The loss of PTEN in human tumors leads to an increase in PI(3,4,5)P3 and the uncontrolled stimulation of growth and survival signals[10]. PTEN also dephosphorylated focal adhesion kinase because of its tyrosine phosphatase activity[11,12], which might lead to the inactivation of Ras/mitogen-activated protein kinase(MAPK) pathway[12-15]. It is well known that both pathways mentioned above are intimately involved in control of cell growth and survival, so PTEN appears to impinge on cell proliferation, adhesion, cell migration, and cell invasion[14,15]. Moreover, germline mutations in PTEN cause Cowden disease, which is characterized by the formation of multiple hamartomas and increased susceptibility to skin, thyroid, and breast tumors[16]. Together, these findings suggest that loss of PTEN activity sensitizes cells to malignant transformation and PTEN is an important protein to regulate various physiological pathways. Despite extensive characterization of PTEN mutations in human cancers and relatively good understanding of the molecular roles of PTEN in the control of cellular processes, little is known about modes of PTEN regulation. Recently, scientists have paid more attention to the regulation of PTEN expression. It was reported that the transcription of PTEN could be regulated by p53 and Sp1[17,18]. In addition, 5’-untranslated region (5-UTR) of PTEN gene was responsible for constitutive PTEN expression in mice[18]. Salvesen et al[19] found that PTEN promoter methylation was relatively frequent in endometrial carcinoma. Till now, the regulation of PTEN expression is still unclear especially in HCC cells. It is well known that the regulation of gene expression is a multi-step process in eukaryotes, and the transcriptional regulation plays a important role in it. So, we attempted to study the transcriptional regulation of PTEN expression in HCC cell lines.
MATERIALS AND METHODS
Cell culture
Human hepatocellular carcinoma (HCC) cell lines (SMMC-7721, BEL-7402, BEL-7404, and BEL-7405) and human liver immortal cell line L02, purchased from Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China), were routinely maintained in RPMI 1640 (Gibco BRL, USA) supplemented with 100 mL/L fetal bovine serum (HyClone, USA) at 37 °C in a humidified atmosphere containing 50 mL/L CO2 in air. HepG2 (human hepatoblastoma) was obtained from American Type Culture Collection (ATCC). HCC cell lines MHCC-97H and MHCC-97L kindly provided by Liver Cancer Institute of Zhongshan Hospital, Fudan University (Shanghai, China), were maintained in Dulbecco’s modified Eagle’s medium (Gibco BRL, USA) supplemented with 100 mL/L fetal bovine serum (HyClone, USA) at 37 °C in a humidified atmosphere containing 50 mL/L CO2 in air.
Western blot analysis
After being grown into confluence, cells were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed in cold lysis buffer (50 mmol/L HEPES, pH 7.5, 150 mmol/L NaCl, 1.5 mmol/L MgCl2, 1 mmol/L EDTA, 0.2 mmol/L EGTA, 10 mL/L NP-40, 100 g/L glycerol, 1 mmol/L dithiothreitol, 1 mmol/L phenylmethylsulfonyl fluoride, 20 mmol/L sodium fluoride, 5 mmol/L sodium orthovanadate, 10 g/L aprotinin, 10 g/L leupeptin, 2 g/L pepstatin, and 1 mmol/L benzamidine). Lysates were incubated for 20 min on ice and centrifugated at 12 000 g for 20 min. The supernatants were collected and protein concentration was determined by Lowry protein assay. Cell lysates were electrophoresed by SDS-PAGE and then transferred onto polyvinylidene fluoride (PVDF) membrane. The membranes were blocked with 50 g/L nonfat dry milk in PBST (PBS, 0.5 mL/L Tween-20) for 4 h at room temperature and incubated overnight at 4 °C with a mAb against human PTEN (Santa Cruz, CA, USA), followed by incubation with HRP-conjugated secondary antibody at room temperature for 3 h. Antibody binding was detected by enhanced chemiluminescence (ECL).
Northern blot analysis
Total RNA was isolated using the TRIzol reagent (Invitrogen, CA, USA) according to the manufacturer’s directions. The 20 μg RNA was electrophoresed on a 12 g/L agarose/formaldehyde gel and blotted onto a nylon membrane (Schleicher & Schuell, Germany) by capillary transfer. Hybridization was performed in 0.2 mol/L Na2HPO4/NaH2PO4 (pH 7.2), 1 mmol/L EDTA, 10 g/L BSA, 70 g/L SDS and 150 mL/L formamide at 50 °C, and the filters were washed extensively with 40 mmol/L Na2HPO4/NaH2PO4 (pH 7.2), 1 mmol/L EDTA, 10 g/L SDS at 65 °C. A 1.2 kb DNA fragment representing the entire coding region of PTEN was used as a probe and was labeled by Prime-a-Gene labeling system (Promega, Madison, WI, USA). Air-dried blots were autoradiographed onto Kodak film (Eastman Kodak, Rochester, NY, USA) and the RNA signal was detected using an ImageMaster VDS system (Pharmacia Biotech, San Francisco, CA, USA) and normalized against the signal for β-actin using ImageMaster TotalLab 1D software.
Isolation of 5’-flanking and promoter region of PTEN gene
Based on the published sequence of PTEN (accession number AF067844), a 2.7 kb DNA fragment of PTEN containing 5’-flanking region, 5’-untranslated region (5’-UTR) and full-length of PTEN promoter region were obtained by PCR using primers 5’-GATAGATCTGGGTG-GGGTGCGGGGTAGGAGTGC-3’ and 5’-GAGAAG-CTTGCTGCGGCGGCTGCTGGATGGTTG-3’. The fragment was subcloned into the luciferase reporter plasmid pGL3-basic (Promega, Madison, WI, USA) which was digested twice with BglII and HindIII restriction enzymes. Positive clones, pGL3-2768, from 8 cell lines were identified by restriction enzymes digest and DNA sequencing, and aligned with the GenBank databases.
Reporter gene plasmids constructions
Several specific primers containing BglII and HindIII restriction enzyme sites (listed below) were designed to amplify serial deletion fragment of PTEN using the clone pGL3-2768 (-2 927/-160) as a template.
5’-CGGAGATCTGTGTTTGGATGTGGGTGCTTTT-3’ (-2 403),
5’-GCTAGATCTTCATTTAGATAGGTGCCCTTTGG-3’ (-1 794),
5’-GAGAGATCTGCGTGGTCACCTGGTCCTT-3’ (-1 389),
5’-CTGAGATCTCTCAGTAGAGCCTGCGGCTTGG-3’ (-1 118),
5’-GGCAGATCTGCGGTGATGTGGCGGGACTCTT-3’ (-916),
5’-GCGAGATCTCGCGACTGCGCTCAGTTCTCTCCT-3’ (-858),
5’-CCGAAGCTTGGCCTCGCCTCACAGCGGCTCAACT-3’ (-778),
5’-CAGAGATCTGGTCTGAGTCGCCTGTCACCATTT-3’ (-458),
5’-GAGAAGCTTGCTGCGGCGGCTGCTGGATGGTTG-3’ (-160).
Fragments of 2 234 (-2 403/-160), 1 635 (-1 794/-160), 1 230 (-1 389/-160), 1 526 (-2 403/-778), 1 016 (-1 794/-778), 612 (-1 389/-778), 341 (-1 118/-778), 139 (-916/-778), 81 (-858/-778) and 299 (-458/-160) were cloned into the vector pGL3-basic at the BglII and HindIII sites to individually generate pGL3-2234, pGL3-1635, pGL3-1230, pGL3-1526, pGL3-1016, pGL3-612, pGL3-341, pGL3-139, pGL3-81, pGL3-299, respectively. These constructs were sequenced and aligned with the GenBank databases.
Transient transfections and luciferase activity assays
Cells were seeded into 6-well plates at a density of 150 000 cells per well 1 day before transfection. The transfection was performed with the Lipofectamine 2000 transfection reagent (Invitrogen, CA, USA) according to the manufacturer’s guidelines. Typically, 3 µg of pGL3 vector and 1 μg of pGFP-β-Gal (a gift from Houyan Song, Department of Molecular Genetics, Shanghai Medical College, Fudan University, Shanghai, China) were used per well. After 48 h, cells were lysed with lysis buffer (Promega, Madison, WI, USA). The mixtures were centrifugated at 12 000 g for 15 s at 4 °C, and the supernatant was preserved at -70 °C. Activities of firefly luciferases were measured in a luminometer Lumat LB 9507 using the luciferase assay system (Promega, Madison, WI, USA). β-Gal activity was measured by β-galactosidase enzyme assay system (Promega, Madison, WI, USA). Promoter activity was quantified by calculating the ratio of firefly luciferase activity/β-gal activity of the same sample. Transfection efficiency was determined through the positive cells with green fluorescence from the green fluorescence protein (GFP) under fluorescent microscope. All the luciferase assays were carried out at least in triplicate, and the experiments were repeated thrice.
RT-PCR and DNA sequencing of PTEN cDNA and PTEN promoter
Total RNA was isolated from cell lines using the TRIzol RNA isolation kit (Invitrogen, CA, USA) according to the manufacturer’s protocols. After synthesis of first strand cDNA using oligo-d(T)12-18 primer and moloney murine leukemia virus (M-MuLV) reverse transcriptase (Promega, Madison, WI, USA), PTEN cDNA was amplified using PCR with pyrococcus furiosus (Pfu) DNA-polymerase (Promega, Madison, WI, USA). The primer sequences were as follows: upper primer, 5’-ACAGGC-TCCCAGACATGACA-3’ and lower primer, 5’-TCAG-ACTTTTGTAATTTGTGTATG-3’. PCR amplification was carried out for 30 cycles under denaturing-annealing-extension conditions of 94 °C for 30 s, 60 °C for 1 min and 72 °C for 1 min, respectively. The PCR product was cloned into the T vector and was identified by DNA sequencing of at least three independent clones and aligned with the GenBank databases.
Statistical analysis
F test was used for statistical analysis.
RESULTS
PTEN protein and mRNA expression in 8 cell lines
Most mammalian cells containing the wild-type PTEN gene expressed detectable levels of PTEN mRNA and protein under normal growth conditions[17]. L02, a human liver immortal cell line, was used as a control in the present study. There was one 55 ku PTEN protein detected with various levels in 8 cell lines by Western blot analysis (Figure 1). The protein level of PTEN in L02 cells was the highest among the 8 cell lines, whereas the PTEN protein in HepG2 cells was almost undetectable. Simultaneously, Northern blot analysis showed a major 2.5-kb transcript and a lower abundance 5.0-kb transcript of PTEN mRNA in all 8 cell lines, which was consistent with previous reports[20] (Figure 2). The total mRNA of PTEN was calculated in both 5.0-kb and 2.5-kb transcripts. The mRNA level of PTEN was much higher in L02 cells than the other 7 HCC cell lines, especially in HepG2 cells (Figure 2). The mRNA level of PTEN in L02 cells was over five-folds than in HepG2 cells. However, the profile of PTEN protein level in each of 8 cell lines closely parallelized with its PTEN mRNA.
Figure 1.
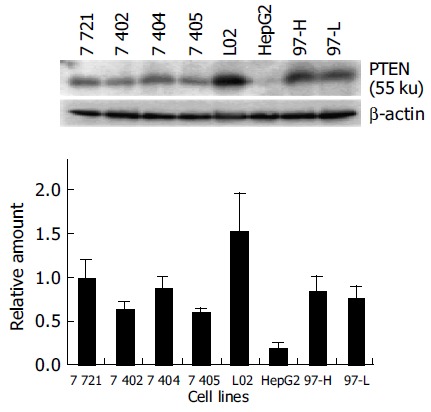
Western blot analysis for PTEN protein expression in 8 cell lines under normal growth conditions.
Figure 2.
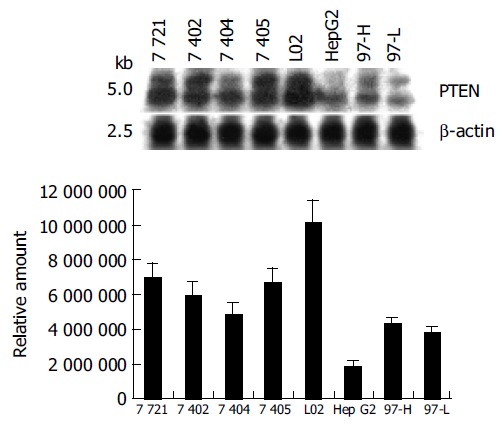
Northern blot analysis of PTEN mRNA expression in 8 cell lines under normal growth conditions.
Absence of PTEN promoter and PTEN cDNA mutation in 8 cell lines
Deletions or mutations of PTEN encoding gene are associated with a variety of human cancers[12]. Furthermore, decreased expression of PTEN was associated with advanced glioma, melanoma, and prostate cancer, implicating losses of PTEN by mutation involved in tumor progression[21-23]. To investigate whether the deletion or mutation exists in PTEN gene, leading to the lost expression of PTEN in HCC cell lines, we analyzed the sequence of PTEN cDNA and PTEN promoter (-2 927/-160 bp) in 8 cell lines. We found no mutation in PTEN cDNA and PTEN promoter region of 8 cell lines (data not shown), indicating that the different levels of PTEN mRNA and protein in 8 cell lines were not caused by the mutation of PTEN cDNA and promoter region. It might be related to PTEN transcriptional or post-transcriptional regulation.
Core region of PTEN promoter identified in SMMC-7721 and L02 cell lines
It is well known that promoter plays the most important role in gene transcription. In an attempt to analyze the function of PTEN promoter, we isolated a DNA fragment containing 5-flanking region and the 5-untranslated region (5-UTR) from PTEN gene, and performed a series of promoter deletion. Eleven fragments of PTEN gene promoter were constructed into pGl3-basic with luciferase reporter (Figure 3 A) and were transiently transfected into L02 and SMMC-7721 cell lines. It was found that the profiles of luciferase activities of various plasmids were the same in the two cell lines (Figures 3B and C). The 612-bp fragment (-1 389/-778) was sufficient to induce maximum luciferase activity in L02 and SMMC-7721 cell lines. The plasmid pGL3-2768 (-2 927/-160), which contained full-length promoter, appeared to have lower activity than that of pGL3-612 (-1 389/-778), indicating that the suppressor elements or special DNA structure may exist in double ends, especially in 3 end fragment (-777/-160). The 341 bp fragment (-1 118/-778) possessed over 60% of the promoter activity of the 612 bp fragment. The 81-bp fragment (-858/-778), however, appeared to be insufficient for inducing transcriptional activation of PTEN. The 299-bp downstream fragment (-458/-160) possessed no significant promoter activity in L02 and SMMC-7721 cells. No further increase in activity was observed when longer fragments than 612 bp were transfected, which was consistent with previous report[24]. Taken together, these data showed that the 612-bp fragment (-1 389/-778) had optimal promoter activity and the core region of PTEN promoter was located within the -1 118 to -778 region.
Figure 3.
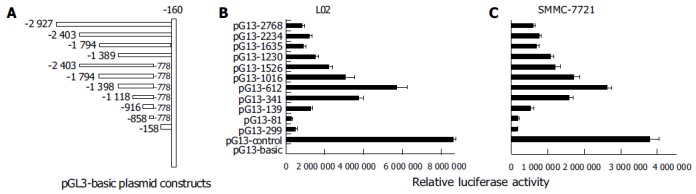
PTEN promoter deletion analysis in SMMC-7721 and L02 cell lines. A: Schematic representation of serial deletion constructs of human PTEN genomic locus; B: promoter activity of serial deletion constructs of PTEN in L02 cell lines; C: promoter activity of serial deletion constructs of PTEN in SMMC-7721 cell lines.
Different activities of PTEN promoter in 8 cell lines
When transfecting the plasmid pGl3-612 that possessed maximum PTEN promoter activity into the 8 cell lines, we found the activity of pGl3-612 in SMMC-7721 and L02 cells were much higher than that in others (Figure 4), while the activity of pGl3-612 in HepG2 cells was the lowest, only 17% of that in L02 cells. The activities of pGl3-612 in BEL-7402, BEL-7404, BEL-7405, MHCC-97H and MHCC-97L were 55%, 65%, 62%, 45%, and 39% of that in L02 cells, respectively. The profile of PTEN promoter activity in 8 cell lines was mostly parallelized to the profiles of PTEN protein and PTEN mRNA. These results demonstrated that the changes of PTEN protein and PTEN mRNA in 8 cell lines might result from the function of PTEN promoter.
Figure 4.
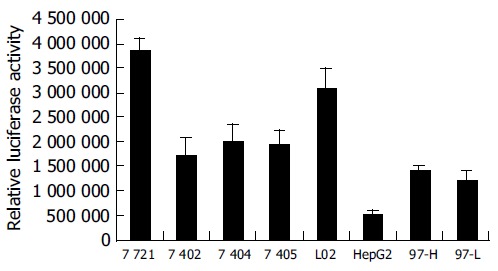
Activity of PTEN promoter in 8 cell lines.
DISCUSSION
Since the isolation of PTEN/MMAC1/TEP1 (acronyms for phosphatase and tensin homolog[2], mutated in multiple advanced cancers[1], and TGF-β (transforming growth factor-β)-regulated and epithelial cell-enriched phosphatase[25] as a candidate tumor suppressor gene, hundreds of reports have been published focusing on its structure and function, as well as on mutations that cause human diseases[26]. Mutations of the PTEN gene arise during cancer progression in a remarkable variety of cancers, including brain, prostate, breast, endometrial cancers and melanoma[26]. The frequency of PTEN mutations observed in endometrial tumors[26], malignant glial tumors[25], malignant melanoma cell lines[26] and metastatic prostate carcinomas[27] was about 45%, 24%, 40%, and 10%, respectively. In addition, germline mutations in the PTEN gene have been associated with Cowden syndrome and a significantly increased risk of certain tumors, including cancer of the breast and thyroid[21,28]. These data further support that PTEN is a tumor suppressor gene.
The structure of PTEN contains a phosphatase domain that has a structure resembling tyrosine phosphatase and a C2 domain appears to bind PTEN to the plasma membrane, which might orientate the catalytic domain appropriately for interactions with phosphatidylinositol 3,4,5-trisphosphate (PIP3) and other potential substrates[29]. A PDZ binding motif in the tail might also play a role in altering the balance of PTEN effects on potential downstream signaling targets such as Akt[16]. PTEN is also known to be critically important both during embryonic development and in mature organisms as a tumor suppressor[30-32]. Studies of PTEN functions have provided a novel insight into the regulation of apoptosis, migration and tumor progression. PTEN appears to serve as a hub or switchpoint linking complex signaling pathways[33,34].
HCC presents a major health threat in South-East Asia, especially in China. It ranks the third among all malignancies both in incidence and mortality in China and accounts for approximately 42.5% of the total incidence worldwide[35]. As a tumor suppressor gene, PTEN expression is downregulated in tumors and tumor cell lines by genetic and epigenetic mechanisms[26]. Therefore, it is very important to study the regulation of PTEN expression in human HCC cell lines. In this study, the results of Western blot analysis demonstrated that the protein level of PTEN in L02 cells was the highest among 8 cell lines, whereas there was almost undetectable PTEN protein expression in HepG2 cells. Northern blot analysis showed that the profile of PTEN mRNA in 8 cell lines almost parallelized to the profile of PTEN protein, indicating that the variation of PTEN protein was mostly dependent on the change of PTEN mRNA. Moreover, deletions or mutations of PTEN gene are associated with a variety of human cancers[12]. Does such deletion or mutation of PTEN gene exist in HCC cell lines, causing the cut down of PTEN expression in HCC cell lines? The sequence analysis of PTEN cDNA and PTEN promoter region showed no mutations in these HCC cell lines. Hence the downregulation of PTEN expression in HCC cell lines probably existed in transcriptional or post-transcriptional levels.
The deletion analysis of PTEN promoter showed that the fragment of 612 bp (-1 389/-778) could produce maximum promoter activity in 8 cell lines and the core region of PTEN promoter was within the 341 bp (-1 118/-778) fragment. The full-length fragment possessing low activity indicated that the double ends of the 612 bp fragment contained suppressive elements or special structures. We used the Genomatix Suite/MatInspector software[36] to analyze the potential binding sites in PTEN core promoter region and its downstream DNA sequence (-1 118/-160 bp), and found a variety of binding sites for p53, NF-kappaB, Ap2, MAZ, Sp1, E4F and Egr-1. Particularly, there were five MAZ binding sites in core promoter region (-1 118/-778) area and 11 Egr-1 in its downstream area (-779/-160). Our results suggested that these two transcription factors might play an important role in control of PTEN expression. After the transfection of pGL3-612 (-1 389/-778), which could produce maximum promoter activity into 8 cell lines, we found the profile of PTEN promoter activity was almost parallelized with the profiles of PTEN mRNA and PTEN protein in 8 cell lines. Furthermore, a recent study reported that the PTEN had no internal ribosome entry site (IRES) that could mediate cap-independent initiation of translation[18]. Taken together, we conclude that the downregulation of PTEN expression in 7 HCC cell lines may not be responsible for the mutation of PTEN, but mainly contribute to the loss of PTEN promoter activity.
ACKNOWLEDGMENT
We gratefully acknowledge the kind support of Dr. Sui-Quan Wang in our research for the using of luminometer Lumat LB 9507.
Footnotes
Supported by the National Natural Science Foundation of China, No. 39970338; Shanghai Municipal Government Science and Technology Committee, No. 00JC14042; and Innovation fund of Fudan University, CQF109801
Science Editor Kumar M and Ma JY Language Editor Elsevier HK
References
- 1.Steck PA, Pershouse MA, Jasser SA, Yung WK, Lin H, Ligon AH, Langford LA, Baumgard ML, Hattier T, Davis T, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 2.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang SI, Puc J, Miliaresis C, Rodgers L, McCombie R, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 3.Maehama T, Taylor GS, Dixon JE. PTEN and myotubularin: novel phosphoinositide phosphatases. Annu Rev Biochem. 2001;70:247–279. doi: 10.1146/annurev.biochem.70.1.247. [DOI] [PubMed] [Google Scholar]
- 4.Simpson L, Parsons R. PTEN: life as a tumor suppressor. Exp Cell Res. 2001;264:29–41. doi: 10.1006/excr.2000.5130. [DOI] [PubMed] [Google Scholar]
- 5.Waite KA, Eng C. Protean PTEN: form and function. Am J Hum Genet. 2002;70:829–844. doi: 10.1086/340026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leslie NR, Downes CP. PTEN: The down side of PI 3-kinase signalling. Cell Signal. 2002;14:285–295. doi: 10.1016/s0898-6568(01)00234-0. [DOI] [PubMed] [Google Scholar]
- 7.Myers MP, Pass I, Batty IH, Van der Kaay J, Stolarov JP, Hemmings BA, Wigler MH, Downes CP, Tonks NK. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maehama T, Dixon JE. PTEN: a tumour suppressor that functions as a phospholipid phosphatase. Trends Cell Biol. 1999;9:125–128. doi: 10.1016/s0962-8924(99)01519-6. [DOI] [PubMed] [Google Scholar]
- 9.Stambolic V, Suzuki A, de la Pompa JL, Brothers GM, Mirtsos C, Sasaki T, Ruland J, Penninger JM, Siderovski DP, Mak TW. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell. 1998;95:29–39. doi: 10.1016/s0092-8674(00)81780-8. [DOI] [PubMed] [Google Scholar]
- 10.Furnari FB, Lin H, Huang HS, Cavenee WK. Growth suppression of glioma cells by PTEN requires a functional phosphatase catalytic domain. Proc Natl Acad Sci USA. 1997;94:12479–12484. doi: 10.1073/pnas.94.23.12479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li DM, Sun H. PTEN/MMAC1/TEP1 suppresses the tumorigenicity and induces G1 cell cycle arrest in human glioblastoma cells. Proc Natl Acad Sci USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myers MP, Stolarov JP, Eng C, Li J, Wang SI, Wigler MH, Parsons R, Tonks NK. P-TEN, the tumor suppressor from human chromosome 10q23, is a dual-specificity phosphatase. Proc Natl Acad Sci USA. 1997;94:9052–9057. doi: 10.1073/pnas.94.17.9052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gu J, Tamura M, Yamada KM. Tumor suppressor PTEN inhibits integrin- and growth factor-mediated mitogen-activated protein (MAP) kinase signaling pathways. J Cell Biol. 1998;143:1375–1383. doi: 10.1083/jcb.143.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tamura M, Gu J, Matsumoto K, Aota S, Parsons R, Yamada KM. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science. 1998;280:1614–1617. doi: 10.1126/science.280.5369.1614. [DOI] [PubMed] [Google Scholar]
- 15.Tamura M, Gu J, Takino T, Yamada KM. Tumor suppressor PTEN inhibition of cell invasion, migration, and growth: differential involvement of focal adhesion kinase and p130Cas. Cancer Res. 1999;59:442–449. [PubMed] [Google Scholar]
- 16.Liaw D, Marsh DJ, Li J, Dahia PL, Wang SI, Zheng Z, Bose S, Call KM, Tsou HC, Peacocke M, et al. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 17.Stambolic V, MacPherson D, Sas D, Lin Y, Snow B, Jang Y, Benchimol S, Mak TW. Regulation of PTEN transcription by p53. Mol Cell. 2001;8:317–325. doi: 10.1016/s1097-2765(01)00323-9. [DOI] [PubMed] [Google Scholar]
- 18.Han B, Dong Z, Liu Y, Chen Q, Hashimoto K, Zhang JT. Regulation of constitutive expression of mouse PTEN by the 5'-untranslated region. Oncogene. 2003;22:5325–5337. doi: 10.1038/sj.onc.1206783. [DOI] [PubMed] [Google Scholar]
- 19.Salvesen HB, MacDonald N, Ryan A, Jacobs IJ, Lynch ED, Akslen LA, Das S. PTEN methylation is associated with advanced stage and microsatellite instability in endometrial carcinoma. Int J Cancer. 2001;91:22–26. doi: 10.1002/1097-0215(20010101)91:1<22::aid-ijc1002>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 20.Wu RC, Blumenthal M, Li X, Schönthal AH. Loss of cellular adhesion to matrix induces p53-independent expression of PTEN tumor suppressor. BMC Mol Biol. 2002;3:11. doi: 10.1186/1471-2199-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guldberg P, thor Straten P, Birck A, Ahrenkiel V, Kirkin AF, Zeuthen J. Disruption of the MMAC1/PTEN gene by deletion or mutation is a frequent event in malignant melanoma. Cancer Res. 1997;57:3660–3663. [PubMed] [Google Scholar]
- 22.Rasheed BK, Stenzel TT, McLendon RE, Parsons R, Friedman AH, Friedman HS, Bigner DD, Bigner SH. PTEN gene mutations are seen in high-grade but not in low-grade gliomas. Cancer Res. 1997;57:4187–4190. [PubMed] [Google Scholar]
- 23.Suzuki H, Freije D, Nusskern DR, Okami K, Cairns P, Sidransky D, Isaacs WB, Bova GS. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 1998;58:204–209. [PubMed] [Google Scholar]
- 24.Sheng X, Koul D, Liu JL, Liu TJ, Yung WK. Promoter analysis of tumor suppressor gene PTEN: identification of minimum promoter region. Biochem Biophys Res Commun. 2002;292:422–426. doi: 10.1006/bbrc.2002.6662. [DOI] [PubMed] [Google Scholar]
- 25.Li DM, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 26.Ali IU, Schriml LM, Dean M. Mutational spectra of PTEN/MMAC1 gene: a tumor suppressor with lipid phosphatase activity. J Natl Cancer Inst. 1999;91:1922–1932. doi: 10.1093/jnci/91.22.1922. [DOI] [PubMed] [Google Scholar]
- 27.Liu W, James CD, Frederick L, Alderete BE, Jenkins RB. PTEN/MMAC1 mutations and EGFR amplification in glioblastomas. Cancer Res. 1997;57:5254–5257. [PubMed] [Google Scholar]
- 28.Cairns P, Okami K, Halachmi S, Halachmi N, Esteller M, Herman JG, Jen J, Isaacs WB, Bova GS, Sidransky D. Frequent inactivation of PTEN/MMAC1 in primary prostate cancer. Cancer Res. 1997;57:4997–5000. [PubMed] [Google Scholar]
- 29.Lee JO, Yang H, Georgescu MM, Di Cristofano A, Maehama T, Shi Y, Dixon JE, Pandolfi P, Pavletich NP. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 30.Nelen MR, van Staveren WC, Peeters EA, Hassel MB, Gorlin RJ, Hamm H, Lindboe CF, Fryns JP, Sijmons RH, Woods DG, et al. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum Mol Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 31.Leslie NR, Gray A, Pass I, Orchiston EA, Downes CP. Analysis of the cellular functions of PTEN using catalytic domain and C-terminal mutations: differential effects of C-terminal deletion on signalling pathways downstream of phosphoinositide 3-kinase. Biochem J. 2000;346 Pt 3:827–833. [PMC free article] [PubMed] [Google Scholar]
- 32.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 33.Suzuki A, de la Pompa JL, Stambolic V, Elia AJ, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, et al. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 34.Yamada KM, Araki M. Tumor suppressor PTEN: modulator of cell signaling, growth, migration and apoptosis. J Cell Sci. 2001;114:2375–2382. doi: 10.1242/jcs.114.13.2375. [DOI] [PubMed] [Google Scholar]
- 35.Yu J, Ni M, Xu J, Zhang H, Gao B, Gu J, Chen J, Zhang L, Wu M, Zhen S, et al. Methylation profiling of twenty promoter-CpG islands of genes which may contribute to hepatocellular carcinogenesis. BMC Cancer. 2002;2:29. doi: 10.1186/1471-2407-2-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]


