Abstract
AIM: To identify the proteins interacting with nucleostemin (NS), thereby gaining an insight into the function of NS.
METHODS: Yeast two-hybrid assay was performed to screen a human placenta cDNA library with the full length of NS as a bait. X-Gal assay and β-galactosidase filter assay were subsequently conducted to check the positive clones and the gene was identified by DNA sequencing. To further confirm the interaction of two proteins, the DNA fragment coding NS and the DNA fragment isolated from the positive clone were inserted into the mammalian expression vector pcDNA3 and pcDNA3-myc, respectively. Then, two plasmids were cotransfected into the COS-7 cells by DEAE-dextron. The total protein from the cotransfected cells was extracted and coimmunoprecipitation and Western blot were performed with suitable antibodies sequentially.
RESULTS: Two positive clones that interacted with NS were obtained from human placenta cDNA library. One was an alpha isoform of human protein phosphatase 2 regulatory subunit B (B56) (PPP2R5A) and the other was a novel gene being highly homologous to the gene associated with spondylo paralysis. The co-immunoprecipitation also showed that NS specifically interacted with PPP2R5A.
CONCLUSION: NS and PPP2R5A interact in yeast and mammalian cells, respectively, which is helpful for addressing the function of NS in cancer development and progression.
Keywords: Nucleostemin, Yeast two-hybrid, Co-IP
INTRODUCTION
There is evidence that cancer originates from cancerous stem cells[1-7]. Nucleostemin (NS) is a novel protein found in the nucleoli of CNS stem cells, embryonic stem cells, and several cancer cell lines, and plays a critical role in controlling the proliferation of stem cells and some cancer cells[7]. NS contains a N-terminal basic domain and two GTP-binding motifs. Mutation analysis indicates that excessive NS, particularly mutant NS that lacks the GTP-regulatory domain, prevents cells from mitosis and causes apoptosis in a p53-dependent manner. The N-terminal basic domain specifies nucleolus localization, p53 interaction, and is required for cell death caused by NS overexpression. To investigate the function of NS in cancer development and progression, yeast two-hybrid assay was used to screen the proteins associated with NS from a human placenta cDNA library, and the interaction between the alpha isoform of human protein phosphatase 2 regulatory subunit B (B56) known as PPP2R5A and NS was identified in yeast and COS-7 cells, thus providing new clues to the functional study of NS as well as related proteins.
MATERIALS AND METHODS
Cell culture and reagents
Monkey Cercopithecus Aethiops COS-7 cells provided by American Type Culture Collection (Manassas, VA, USA) were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen Corporation, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (Sigma Chemical Co., St. Louis, MO, USA), and maintained in a humidified chamber with 50 mL/L CO2 at 37 °C.
Plasmid pGBKT7, matchmaker 3 pretransformed human placenta cDNA library, X-Gal and all other yeast two-hybrid components were purchased from Clontech. Various restriction endonucleases were products of New England Biolabs. T4 DNA ligase was purchased from Promega. The kits for PCR and purification reagents of PCR products were obtained from Qiagen. Antibody against NS or c-myc tag and other regents were all kept in our laboratory.
Plasmid construction
The full length of NS cDNA was cut by BamHI and XhoI restriction endonucleases from pCDNA3-NS, then inserted into the downstream of the Gal4 DNA-binding domain of the bait vector pGBKT7 (Clontech Laboratories) with T4 DNA ligase. The recombinant vector pGBKT7-NS was sequenced and NS protein was in the reading frame. The expression of NS fusion protein with yeast Gal4 DNA-binding domain was checked by Western blot with antibody against NS.
To construct the eukaryotic vector expressing the protein of the positive clone, DNA of the positive clone was digested by restriction endonucleases BamHI and XhoI, and the DNA fragment was inserted into the pCDNA-myc vector.
Screening of clones interacted with NS
The experiments were carried out according to the protocols described in the MATCHMAKER Libraries User Manual (PT3042-1)[8-11]. Briefly, the pGBKT7-NS plasmid was initially introduced into the AH109 yeast strains using a modified lithium acetate protocol and the transformed clones were selected on SD/-Trp plates. The mating between the selected AH109 and human placenta cDNA library was performed and co-transformed clones were selected on SD/-Leu-Trp-His plates and SD/-Leu-Trp-His-Ade pulsing 3AT (DO) plates with X-Gal to detect the transcription of reporter genes (HIS, LEU, TRP, and ADE). Colonies growing at 30 °C and having turned into blue in 8 h were selected as positive clones. Plasmid DNA from the single positive clones was extracted and sequenced with the primer provided with the kit. The clone was identified by DNA sequence and compared with GenBank. To further exclude the false positives, plasmid was transformed into yeast strain AH109 to rule out its self-activation.
In order to further confirm the positive clones in yeast system, experiments were performed in AH109 yeast strain to detect the transcription of LacZ reporter gene according to the protocols. Colony-lift filter assay was used to check the activity of β-galactosidase. Briefly, fresh colonies growing to about 1-3 mm in diameter were transferred completely to a sterile filter and submerged in liquid nitrogen for 10 s and thawed at room temperature, then put on a pre-soaked filter with Z buffer (Na2HPO4·7H2O 16.1 g/L, NaH2PO4·H2O 5.50 g/L, KCl 0.75 g/L, and MgSO4·H2O 0.246 g/L, pH 7.0) containing 0.27 mL β-mercaptoethanol and 1.67 mL X-Gal stock solution (20 mg/mL) in 100 mL volume. The filters were incubated at 30 °C and the colors of colonies were checked periodically.
Co-immunoprecipitation
When COS-7 cells reached 50-70% confluence on the dish, the plasmids expressing NS and interacted protein of NS were cotransfected into COS-7 cells with DEAE-dextron as described[12], and total plasmid DNA was 8 µg/100 mm/dish. After 48 h, the cells were washed with PBS, then scraped and collected by spinning down. The cell pellet was lysed in 0.5 mL HEDL buffer (50 mmol/L HEPES pH 8.0, 150 mmol/L NaCl, 2 mmol/L EDTA, 1% Triton X-100, 10 mmol/L MgCl2, 1 mmol/L Na3VO4, 25 mmol/L NaF, 1 mmol/L PMSF) and shaked for 2 h at 4 °C, centrifuged at 12 000 g for 15 min at 4 °C. The resulting supernatants were immunoprecipitated with various antibodies (mouse anti-c-myc, mouse anti-nucleostemin and mouse non-specific antibodies) respectively as described[12]. SDS-PAGE and Western blot were performed with suitable antibodies and the signal was detected with ECL (Pulilai Co.).
RESULTS
Plasmids construction
The constructed plasmid pGBKT7-NS was identified by restriction endonucleases BamHI/XhoI and DNA sequence analysis. As shown in Figure 1A, there was an expected DNA band about 1.7 kb released from the digested plasmid pGBKT7-NS and the DNA sequence was completely identical with NS in GenBank. Western blot showed that the NS protein fused with yeast Gal4 DNA-binding domain was expressed in yeast (Figure 1B). The recombinant plasmids PPP2R5A/pCDNA3-myc and pCDNA3-NS were identified by restriction endonucleases BamHI/XhoI and DNA sequencing, respectively (Figure 2). As shown in Figure 3, there was an expected band about 2.3 or 1.7 kb released from the digested plasmid and the DNA sequence was completely identical with PPP2R5A or NS in GenBank.
Figure 1.
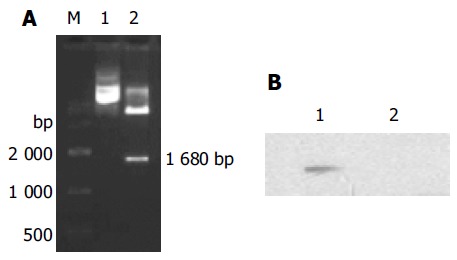
Identification of the recombinant clone of NS and its expression in yeast cell. A: Analysis of pGBKT7-NS with restriction enzyme digestion. M: DNA markers; lane 1: undigested pGBKT7-NS; lane 2: digestion of pGBKT7-NS with BamHI/XhoI and 1 680-bp fragment was released; B: Western blotting analysis of NS expression by pGBKT7-NS in yeast. Total protein from yeast AH109 transferred with pGBKT7-NS was subjected to Western blot and the NS mAb was used to detect the NS protein. Lane 1: total proteins from pGBKT7-NS transferred AH109; lane 2: total proteins from pGBKT7 transferred AH109.
Figure 2.
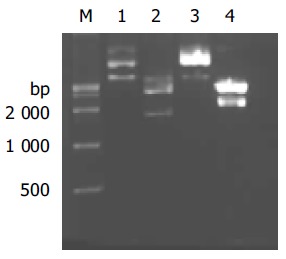
Restriction enzyme analysis of recombinant pcDNA3-NS and pcDNA3-myc-PPP2R5A. M: DNA markers; lane 1: undigested pCDNA3-NS; lane 2: digestion of pCDNA3-NS by BamHI/XhoI and 1 680-bp fragment was released; lane 3: undigested pcDNA3-myc-PPP2R5A; lane 4: digestion of pcDNA3-myc-PPP2R5A with BamHI/XhoI and 2 300 bp fragment was released.
Figure 3.

Co-immunoprecipitation of protein NS and PPP2R5A. Total proteins from cells co-transferred with plasmids pCDNA3-NS and pcDNA3-myc-PPP2R5A were used for immunoprecipitation with antibody against NS or c-myc and antibody against c-myc or NS was used in Western blot. Mouse IgG was used in immunoprecipitation as negative control.
Screening of clones interacted with NS
To rule out the transcription activity of NS, the bait plasmid pGBKT7-NS was transformed into yeast AH109 and the transcription initiation of HIS and MEL1 reporter genes was tested. The results showed that the self-activation of NS was negative.
Amplification of human placenta cDNA library was pre-performed and transformed into the AH109 yeast strain containing pGBKT7-NS plasmids using modified lithium acetate. The transformants of pGBKT7 and pCL1 were used as negative and positive control respectively in the experiment. There were two positive clones, which were subjected to DNA sequence analysis. The results showed that one clone was identical to gene PPP2R5A (GenBank, GI: 30795205), the other was a novel gene being highly homologous to the gene associated with spondylo paralysis. To identify the specificity of the protein interaction, PPP2R5A/pACT2 was co-transformed into yeast cells with plasmids pGBKT7-NS or pGBKT7. The results showed that PPP2R5A/pACT2 interacted with pGBKT7-NS, but not with pGBKT7.
Co-immunoprecipitation
To further testify the two protein interaction in mammalian cells, eukaryotic plasmids expressing NS or PPP2R5A were cotransfected into COS-7 cells and the total protein was extracted and used for immunoprecipitation and Western blot sequentially. The proteins precipitated with NS antibody could be recognized by c-myc tag antibody, and vice versa, indicating the interaction between NS and PPP2R5A in mammalian cell.
DISCUSSION
Yeast two-hybrid assay is an effective method to isolate interacted proteins[8], and can screen all protein-protein interactions in vivo. The proteins obtained by yeast two-hybrid assay are more likely in their native conformations[12]. However, the yeast two-hybrid assay also has its limitations, and false positive clones may occur. In this study, more sensitive and credible yeast strain AH109 was used, which has three reporter genes regulated by different promoters, such as GAL1-HIS3, GAL2-ADE2, and MEL1-LacZ. By using this system, a novel binding protein PPP2R5A of NS was obtained from human placenta cDNA library and its interaction with NS and PPP2R5A was further characterized in mammalian cells using co-immunoprecipitation.
P53 plays an important role in the physiological or pathological processes, including cell growth regulation and cell cycle progress. It was reported that the p53 protein can bind to NS protein by GST pull-down and co-immunoprecipitation, respectively[7]. In our study, a novel NS binding protein was identified as an alpha isoform of human protein phosphatase 2 regulatory subunit B (B56) (PPP2R5A). PPP2R5A belongs to the phosphatase 2A regulatory subunit B family. Phosphatase 2A is one of the four major Ser/Thr phosphatases, and plays an important role in negative control of cell growth and division as well as cell cycle progress[13]. According to Gene Database, PPP2R5A is located in cytoplasm[14-18]. It was found in our study that NS was expressed in cytoplasm and nucleoli (data not shown). NS expression in gastric cancer tissue is higher than that in other gastric tissues (data not shown), suggesting that NS may play a role in the carcinogenesis of gastric cancer. In conclusion, the results of our study help to investigate the functions of NS.
Footnotes
Supported by the National High Technology Research and Development Program of China, No. 200BA711A11A06 and Beijing Science and Technology Project, No. H020220020310
Science Editor Wang XL and Guo SY Language Editor Elsevier HK
References
- 1.Lessard J, Sauvageau G. Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature. 2003;423:255–260. doi: 10.1038/nature01572. [DOI] [PubMed] [Google Scholar]
- 2.Marx J. Cancer research. Mutant stem cells may seed cancer. Science. 2003;301:1308–1310. doi: 10.1126/science.301.5638.1308. [DOI] [PubMed] [Google Scholar]
- 3.Jamieson CH, Ailles LE, Dylla SJ, Muijtjens M, Jones C, Zehnder JL, Gotlib J, Li K, Manz MG, Keating A, et al. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 4.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- 5.Normile D. Cell proliferation. Common control for cancer, stem cells. Science. 2002;298:1869. doi: 10.1126/science.298.5600.1869. [DOI] [PubMed] [Google Scholar]
- 6.Bernardi R, Pandolfi PP. The nucleolus: at the stem of immortality. Nat Med. 2003;9:24–25. doi: 10.1038/nm0103-24. [DOI] [PubMed] [Google Scholar]
- 7.Tsai RY, McKay RD. A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 2002;16:2991–3003. doi: 10.1101/gad.55671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 9.Bartel P, Chien CT, Sternglanz R, Fields S. Elimination of false positives that arise in using the two-hybrid system. Biotechniques. 1993;14:920–924. [PubMed] [Google Scholar]
- 10.Ye Q, Worman HJ. Protein-protein interactions between human nuclear lamins expressed in yeast. Exp Cell Res. 1995;219:292–298. doi: 10.1006/excr.1995.1230. [DOI] [PubMed] [Google Scholar]
- 11.Shou C, Wurmser A, Suen KL, Barbacid M, Feig LA, Ling K. Differential response of the Ras exchange factor, Ras-GRF to tyrosine kinase and G protein mediated signals. Oncogene. 1995;10:1887–1893. [PubMed] [Google Scholar]
- 12.Phizicky EM, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuner R. Identifizierung differenziell exprimieter Gene bei Brust-und Ovarialkarzinomen in den chromosomalen Regionen 1q32-q41 und 11q12-q23. MetaGen Pharmaceuticals GmbH. 2002;23:1. [Google Scholar]
- 14.Sijin L, Ziwei C, Yajun L, Meiyu D, Hongwei Z, Guofa H, Siguo L, Hong G, Zhihong Z, Xiaolei L, et al. The effect of knocking-down nucleostemin gene expression on the in vitro proliferation and in vivo tumorigenesis of HeLa cells. J Exp Clin Cancer Res. 2004;23:529–538. [PubMed] [Google Scholar]
- 15.Tsai RY, McKay RD. A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J Cell Biol. 2005;168:179–184. doi: 10.1083/jcb.200409053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martens E, Stevens I, Janssens V, Vermeesch J, Götz J, Goris J, Van Hoof C. Genomic organisation, chromosomal localisation tissue distribution and developmental regulation of the PR61/B' regulatory subunits of protein phosphatase 2A in mice. J Mol Biol. 2004;336:971–986. doi: 10.1016/j.jmb.2003.12.047. [DOI] [PubMed] [Google Scholar]
- 17.McCright B, Brothman AR, Virshup DM. Assignment of human protein phosphatase 2A regulatory subunit genes b56alpha, b56beta, b56gamma, b56delta, and b56epsilon (PPP2R5A-PPP2R5E), highly expressed in muscle and brain, to chromosome regions 1q41, 11q12, 3p21, 6p21.1, and 7p11.2 --& gt; p12. Genomics. 1996;36:168–170. doi: 10.1006/geno.1996.0438. [DOI] [PubMed] [Google Scholar]
- 18.McCright B, Virshup DM. Identification of a new family of protein phosphatase 2A regulatory subunits. J Biol Chem. 1995;270:26123–26128. doi: 10.1074/jbc.270.44.26123. [DOI] [PubMed] [Google Scholar]


