Highlights
-
•
AFM measured the equivalent diameters of LDL (∼22.7 nm) and HDL (∼14.4 nm) in PBS.
-
•
AFM detected the mean Young’s moduli of LDL (∼0.39 GPa) and HDL (∼0.47 GPa) in PBS.
-
•
AFM detected the mean adhesive forces of LDL (∼0.19 nN) and HDL (∼0.15 nN) in PBS.
-
•
AFM detected much larger sizes of Young’s moduli and adhesive forces of LDL/HDL in air.
Abbreviations: AFM, atomic force microscopy; CAD, coronary artery disease; HDL, high-density lipoprotein; LDL, low-density lipoprotein
Keywords: Low-density lipoprotein (LDL), High-density lipoprotein (HDL), Atomic force microscopy (AFM), Young’s modulus, Adhesive force
Abstract
The size and biomechanical properties of lipoproteins are tightly correlated with their structures/functions. While atomic force microscopy (AFM) has been used to image lipoproteins the force measurement of these nano-sized particles is missing. We detected that the sizes of LDL and HDL in liquid are close to the commonly known values. The Young’s modulus of LDL or HDL is ∼0.4 GPa which is similar to that of some viral capsids or nanovesicles but greatly larger than that of various liposomes. The adhesive force of LDL or HDL is small (∼200 pN). The comparison of AFM detection in air and liquid was also performed which is currently lacking. Our data may provide useful information for better understanding and AFM detection of lipoproteins.
1. Introduction
Low-density lipoprotein (LDL) and high-density lipoprotein (HDL) are the two most intriguing classes of circulating lipoproteins in the plasma of vertebrates, mainly involved in the transport of water insoluble molecules (e.g. cholesterol, triacylglycerols, phospholipids, etc.) to and from peripheral cells, respectively. They are also the most commonly accepted indicators of risk for coronary artery disease (CAD). LDL and HDL are complex, nanometer-sized particles consisting of phospholipids, cholesterol, cholesterol esters, triglycerols, as well as different apolipoproteins (mainly apoB-100 for LDL and apoA-I for HDL). Since the size and biomechanical properties of lipoprotein particles are relevant to their structures/functions and the risk of diseases (for instance, smaller or softer or more adhesive lipoproteins may be more readily to adhere to other molecules/cells or to pass through intercellular gaps; the oxidation of lipoproteins with different sizes or biomechanical properties may be different), it is vital to characterize these properties of lipoproteins.
While the sizes of LDL and HDL have been extensively studied by different techniques including gradient gel electrophoresis (GGE) [1], high-performance gel-filtration chromatography [2], dynamic light scattering (DLS) [3], transmission electron microscopy (TEM), cryo-electron microscopy [4], nuclear magnetic resonance (NMR) [5], and others [6], the biomechanical properties of individual LDL and HDL particles have been less investigated until now.
Atomic force microscopy (AFM) is a powerful tool for nanoscale imaging in air or liquid and has been widely applied in measuring circulating lipoproteins [7,8], reconstituted lipoproteins or nanolipoproteins [9–11], apolipoproteins in phospholipid monolayer [12,13], and even apolipoproteins [14] either in air or in liquid. Considering that many AFM studies of lipoproteins were performed under a non-physiological condition (in air), a comparative study of AFM detection in air and in liquid is needed which has been lacking. Thanks to its force measurement function, AFM is also an ideal technique for measuring the biomechanical properties of biological samples with different sizes ranging from small molecules to cells or even tissues. Unfortunately, according to our knowledge, no AFM studies on the biomechanical properties of plasma lipoproteins have so far been reported although there are many AFM studies on the changes in biomechanical properties of the cells treated by native or modified lipoproteins [15,16].
In this study, we utilized AFM to image the two lipoprotein classes (human plasma LDL and HDL) and, for the first time, measure their biomechanical properties (adhesive force and Young’s modulus). At the same time, the comparison of the morphological/biomechanical properties detected by AFM in air and in liquid was also performed.
2. Materials and methods
2.1. Reagents
LDL and HDL were purchased from Yiyuan Biotechnologies (Guangzhou, China). N,N-diisopropylethylamine (DIPEA), aminopropyltriethoxysilane (APTES), and glutaraldehyde were from TCI (Shanghai, China; or Tokyo, Japan for APTES).
The purchased LDL or HDL solutions contained EDTA which is widely used to prevent lipoproteins from oxidation during preparation and storage. To certify that the purchased LDL and HDL were not oxidized, agarose gel electrophoresis was performed on 0.5% agarose gel in sodium barbital buffer at 55 V for 1 h (Fig. 1). The fresh serum has two bands (α and β). LDL and HDL correspond to the β and α band, respectively, indicating that the LDL and HDL used in this study were native and not oxidized.
Fig. 1.
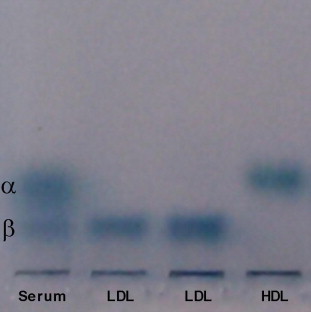
Agarose gel electrophoresis of lipoproteins. Lane 1: fresh human serum; lanes 2 and 3: purchased LDL; lane 4: purchased HDL. The gel was stained with Sudan Black B. The staining of the origins of all lanes was probably due to dye aggregation.
2.2. Sample preparations
For AFM detection in air, LDL and HDL were directly deposited on freshly cleaved mica overnight, rinsed with double distilled water twice, air-dried at room temperature for more than 1 h, and then immediately subjected to AFM imaging and force measurements in air. For AFM detection in liquid, the mica was functionalized as previously described [17]. Briefly, freshly cleaved mica sheets were functionalized with 30 μl of N,N-diisopropylethylamine (DIPEA) and 50 μl of aminopropyltriethoxysilane (APTES), followed by incubation with 0.2% glutaraldehyde (GD) to make GD-APTES-mica. The entire procedure was performed in a dessicator and ultra pure argon was used to remove air and moisture. Then, LDL and HDL were deposited on the GD-APTES-mica, rinsed with double distilled water twice, incubated with l-glycine, rinsed with ultra pure water again, and immediately subjected to AFM imaging and force measurements in PBS. For force measurements, a lipoprotein monolayer was made by depositing a relatively higher concentration of LDL or HDL on the mica to avoid the interference of bare mica surface with the force measurements as little as possible.
2.3. Atomic force microscopy (AFM)
The experiments were performed using an Asylum MFP-3D-SA AFM (Asylum Research, USA) equipped with a scanner of 90 μm × 90 μm × 15 μm in tapping mode for imaging or in contact mode for force measurement. The data were acquired using silicon nitride tips (AppNano, USA) with an end radius of 10 nm and a spring constant of ∼0.04 N/m. After AFM force measurement (obtaining force-vs-distance curves), the Young’s modulus and adhesive force data were directly extracted by the instrument-equipped software (Igor Pro 6.31). To obtain the Young’s modulus, the retrace curve is fitted automatically by the software using the Hertz model. Prior to AFM detection of lipoproteins, the imaging and force measurement of the surfaces of the bare mica and GD-APTES-mica have been performed in air and liquid (data not shown) to exclude the possibility that the detected particles are not lipoproteins.
2.4. Statistical analysis
All values are expressed as mean ± SD (more than 100 particles were measured for each group). Statistic analyses were performed using Student t test to determine the significance between different groups. A value of P < 0.05 was considered statistically significant.
3. Results
3.1. The size, adhesive force, and Young’s modulus of LDL detected by AFM in air
Fig. 2 shows the data from AFM detection of LDL in air. The representative 2-dimentional (2-D) and 3-D topographical images and a cross-section height profile were shown in Fig. 2A–C, respectively. Obviously, besides the particles of ∼20–40 nm in diameter, there are many relatively large particles with a diameter of ∼40–80 nm. Moreover, the boundaries of many clustered particles cannot be distinguished from one another (Fig. 2A–C). Quantification determined that the average diameter and height of the LDL particles detected in air are 45.8 ± 19.6 nm (full width at half maximum, FWHM) and 8.5 ± 1.8 nm, respectively. The equivalent diameter of LDL as a sphere with the calculated volume from AFM detection in air is 26.4 nm. Fig. 2D and E show the representatives of the adhesive force and Young’s modulus mapping of LDL particles, respectively. The average adhesive force and Young’s modulus of LDL particles detected by AFM in air are 24.1 ± 0.7 nN and 36.6 ± 4.4 GPa, respectively.
Fig. 2.
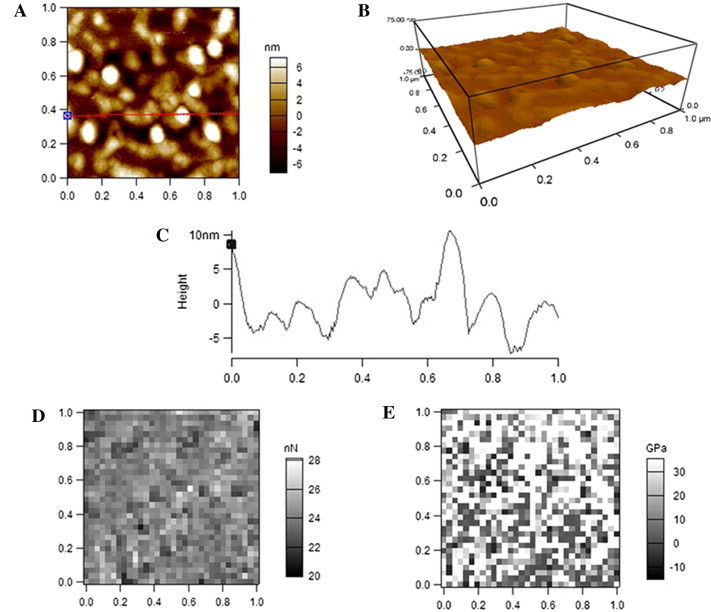
AFM detection of LDL particles in air. (A) A representative AFM 2-dimentional (2-D) topographical image. (B) The corresponding 3-D image. (C) A cross-section height profile across the indicated line in A. (D) Representative adhesive force mapping. (E) Representative Young’s modulus mapping. Scan size: 1 μm × 1 μm. Resolution: (A, B) 256 pixel × 256 pixel; (D, E) 32 pixel × 32 pixel.
3.2. The size, adhesive force, and Young’s modulus of HDL detected by AFM in air
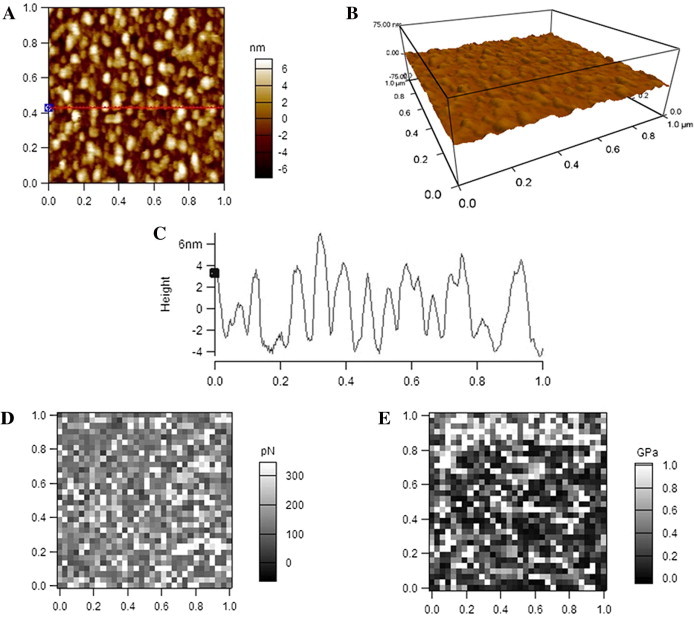
Fig. 3 shows the data from AFM detection of HDL in air, also displaying the 2-D (Fig. 3A), 3-D (Fig. 3B) topographical images, cross-section height profile (Fig. 3C), adhesive force mapping (Fig. 3D), and Young’s modulus mapping (Fig. 3E), respectively. The boundaries of many clustered particles also cannot be distinguished from one another (Fig. 3A–C). The average diameter and height of HDL particles detected by AFM in air are 23.7 ± 6.9 nm and 2.2 ± 0.4 nm, respectively, which are quite smaller than those of LDL particles. The equivalent diameter of HDL as a sphere with the calculated volume from AFM detection in air is 12.0 nm. The average adhesive force of HDL particles detected by AFM in air is 50.5 ± 1.5 nN, which is significantly larger than that of LDL particles detected in air; the average Young’s modulus of HDL detected by AFM in air is 22.6 ± 2.7 GPa, which is significantly smaller than that of LDL particles detected in air.
Fig. 3.
AFM detection of HDL particles in air. (A) A representative AFM 2-D topographical image. (B) The corresponding 3-D image. (C) A cross-section height profile across the indicated line in A. (D) Representative adhesive force mapping. (E) Representative Young’s modulus mapping. Scan size: 1 μm × 1 μm. Resolution: (A, B) 256 pixel × 256 pixel; (D, E) 32 pixel × 32 pixel.
3.3. The size, adhesive force, and Young’s modulus of LDL detected by AFM in liquid
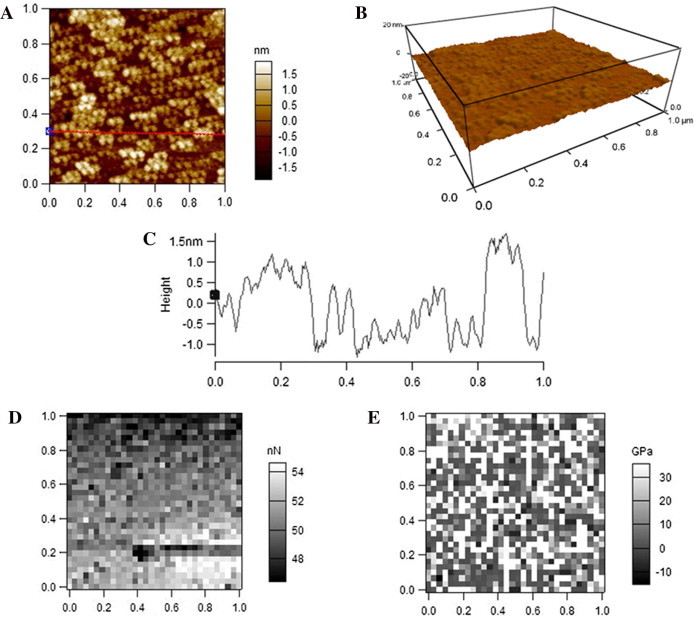
When imaged by AFM in PBS, visually and obviously, LDL particles are much smaller but a little higher than those imaged in air (Fig. 2A–C) as showed by the topographical images (Fig. 4A and B) and the height profile (Fig. 4C). Moreover, the boundaries of most particles can be distinguished clearly from one another (Fig. 4A–C). The average diameter and height of LDL particles detected in PBS are 28.9 ± 9.2 nm (FWHM) and 8.7 ± 2.0 nm, respectively. The equivalent diameter of LDL as a sphere with the calculated volume from AFM observation in PBS is 22.6 nm. Moreover, compared with the data from AFM detection in air (Fig. 2D and E), both the adhesive force (Fig. 4D) and Young’s modulus (Fig. 4E) of LDL particles detected in PBS dramatically dropped. The average adhesive force and Young’s modulus of the AFM-measured LDL particles in PBS are 0.19 ± 0.12 nN and 0.39 ± 0.15 GPa, respectively.
Fig. 4.
AFM detection of LDL particles in PBS. (A) A representative AFM 2-D topographical image. (B) The corresponding 3-D image. (C) A cross-section height profile across the indicated line in A. (D) Representative adhesive force mapping. (E) Representative Young’s modulus mapping. Scan size: 1 μm × 1 μm. Resolution: (A, B) 256 pixel × 256 pixel; (D, E) 32 pixel × 32 pixel.
3.4. The size, adhesive force, and Young’s modulus of HDL detected by AFM in liquid
Similarly, topographical images (Fig. 5A and B) and height profile (Fig. 5C) shows that HDL particles imaged by AFM in PBS are slightly smaller but significantly higher than those imaged in air (Fig. 3A–C). The boundaries of most particles also can be distinguished clearly from one another (Fig. 5A–C). Quantification analysis shows that the average diameter and height of HDL particles detected in PBS are 21.5 ± 6.5 nm (FWHM) and 4.1 ± 0.9 nm, respectively. The equivalent diameter of HDL as a sphere with the calculated volume from AFM detection in air is 14.4 nm. The adhesive force (0.15 ± 0.13 nN) and Young’s modulus (0.47 ± 0.14 GPa) of HDL particles detected in PBS (Fig. 5D and E) are also dramatically smaller than those of HDL particles detected in air (Fig. 3D and E).
Fig. 5.
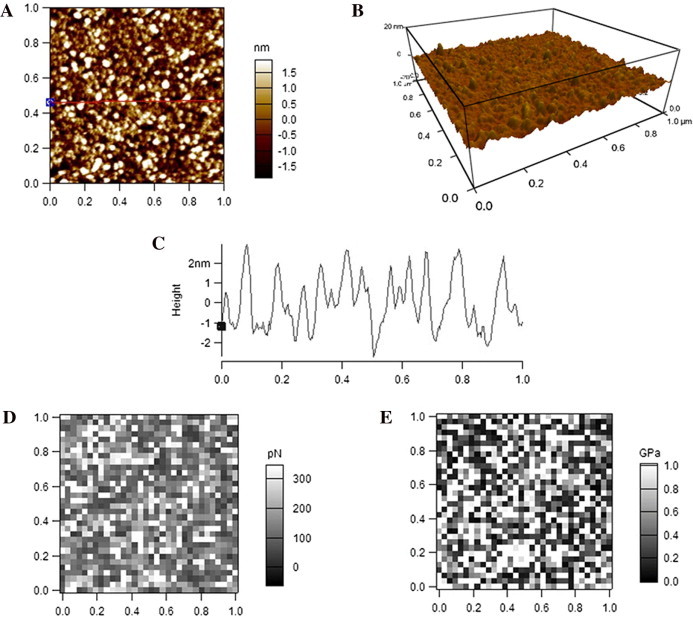
AFM detection of HDL particles in PBS. (A) A representative AFM 2-D topographical image. (B) The corresponding 3-D image. (C) A cross-section height profile across the indicated line in A. (D) Representative adhesive force mapping. (E) Representative Young’s modulus mapping. Scan size: 1 μm × 1 μm. Resolution: (A, B) 256 pixel × 256 pixel; (D, E) 32 pixel × 32 pixel.
4. Discussion
4.1. The size and biomechanical properties (Young’s modulus and adhesive force) of LDL and HDL detected by AFM in liquid
In Table 1, we summarize the AFM-measured sizes of LDL and HDL obtained by us and previously reported by others. In our study, the average diameter and height of LDL measured in PBS are 28.9 ± 9.2 nm (FWHM; the equivalent diameter of a sphere is ∼22.7 nm) and 8.7 ± 2.0 nm, respectively, which are similar to the data in previous reports [7,18–21] but closer to the value determined by NMR spectroscopy (∼18–23 nm) [22]. The average diameter and height of HDL measured in PBS are 21.5 ± 6.5 nm (FWHM; the equivalent diameter of a sphere is ∼14.4 nm) and 4.1 ± 0.9 nm, respectively, which are smaller than the data in previous reports [23–25] and closer to the value determined by NMR spectroscopy (∼7–14 nm) [22]. Moreover, our data of the average diameter of LDL/HDL have relatively large standard deviations (9.2 nm for LDL and 6.5 nm for HDL), implying the existence of various LDL/HDL subclasses with different sizes which also can be directly observed in the AFM topographical images (Fig. 4A for LDL and Fig. 5A for HDL).
Table 1.
A brief summary for the AFM-measured sizes of LDL and HDL.
| Lipoprotein | Diameter (nm) | Height (nm) | In air or liquid | Refs. |
|---|---|---|---|---|
| LDL | 26 ± 3a | 17 ± 1 | In liquid | [18] |
| 26b or 17c | 7.5 | In liquid | [7] | |
| 23 ± 3a or ∼17c | 10 ± 2 | In liquid | [20] | |
| 20.6 ± 1.9c (lbLDL) | NA | In liquid | [21] | |
| 16.2 ± 1.4c (sdLDL) | NA | In liquid | [21] | |
| 28.9 ± 9.2a or 22.6c | 8.7 ± 2.0 | In liquid | Current work | |
| 55–65a | 2.5–3 | In air | [19] | |
| 45.8 ± 19.6a or 26.4c | 8.5 ± 1.8 | In air | Current work | |
| HDL | 30 ± 10a | ∼5 | In liquid | [23] |
| >33a (rHDL) | 5.5 ± 0.4 | In liquid | [24] | |
| 22b or 15c | 6.3 | In liquid | [7] | |
| 21.5 ± 6.5a or 14.4c | 4.1 ± 0.9 | In liquid | Current work | |
| 8–20a | 2–3 | In air | [25] | |
| 23.7 ± 6.9a or 12.0c | 2.2 ± 0.4 | In air | Current work | |
Abbreviations: lbLDL: large buoyant LDL; sdLDL: small dense LDL; rHDL: reconstituted HDL with 2-4 apoA-I molecules per rHDL; NA: not available.
Other reports might also image LDL or HDL in air or liquid using AFM but did not clearly determine the size of these particles.
The diameter determined from the full width at half maximum (FWHM).
The diameter determined from the average area contribution per lipoprotein.
The equivalent diameter of a sphere with the calculated volume from observation.
According to our knowledge, it is the first study to detect the biomechanical properties (Young’s modulus and adhesive force) of plasma lipoproteins using AFM. We determinate that the Young’s moduli of LDL and HDL in PBS were 0.39 ± 0.15 GPa and 0.47 ± 0.14 GPa, respectively. Our data are slightly larger than the Young’s moduli of some viral capsids with similar sizes (∼22–28 nm in diameter), e.g. ∼0.26–0.37 GPa for hepatitis B virus (HBV, an animal virus) [26,27] and 0.14–0.28 GPa for cowpea chlorotic mottle virus (CCMV, a plant virus) [28–30], as well as that (∼0.2–0.3 GPa) of the plasma/inner membrane nanovesicles (∼80 nm in diameter) derived from yeast cells [31], but 4- to 40-fold larger than the Young’s moduli (0.01–0.1 GPa) of various liposomes and polymersomes (∼100–200 nm in diameter) [32,33]. The small sizes and unique structures (a highly hydrophobic core surrounded by a monolayer of phospholipids, cholesterol, and apolipoproteins) of LDL/HDL may be responsible for it. HDL (0.47 ± 0.14 GPa) is stiffer than LDL (0.39 ± 0.15 GPa) probably due to the smaller size of HDL. It has been reported that the stiffness of liposomes increase with decreasing liposome diameter [32]. Moreover, with the decrease of the particle size, the protein components of a particle will potentially contribute more to its higher stiffness since proteins generally have relatively high Young’s moduli (⩾0.5 GPa) [34]. The content of cholesterol in lipoproteins might contribute to their stiffness. More in-depth research will be needed to clarify it.
We also determinate that the adhesive forces of LDL and HDL in PBS were 0.19 ± 0.12 nN and 0.15 ± 0.13 nN, respectively. The adhesive force is probably derived from the electrostatic and hydrophobic interactions between AFM tips and lipoprotein particles (the apolipoproteins and the exposed lipids, respectively). The pI values of LDL and HDL are ∼5.5 [35,36] and ∼5.0 [25], respectively. At pH ∼7.4 (under physiological condition or in PBS), both LDL and HDL are negatively charged. Even then, the electrostatic interactions also can occur between the negatively charged AFM tip and positively charged domains on apolipoproteins therefore causing a relatively small adhesive force (e.g. ∼200 pN for LDL and HDL). Considering the pI value of LDL is a little higher than that of HDL, LDL potentially has more positively charged domains than HDL. It might be responsible for the slightly stronger adhesive force of LDL (0.19 ± 0.12 nN) than HDL (0.15 ± 0.13 nN).
Obtaining the biomechanical properties of lipoproteins will help to the better understanding of their structures and functions. Moreover, to leave from or access to the peripheral cells, circulating lipoproteins have to pass through the intercellular gaps or extracellular matrix. During these or other processes including the modification of lipoproteins, the size and/or biomechanical properties (e.g. stiffness and adhesive force) of lipoproteins may play vital roles. More in-depth studies on the detection/comparison of the biomechanical properties of various lipoprotein types or subclasses as well as modified lipoproteins will be needed in the future.
4.2. Comparison of AFM detection in air and in liquid
Since AFM detection in air was performed in many studies on lipoproteins and other nano-sized particles, it is necessary to certify the quality or fidelity of the data from detection in air by comparing with the data from detection in liquid. We found that AFM detection in air acquires much worst quality or fidelity of the morphological and biomechanical properties of lipoproteins than detection in liquid.
First, LDL and HDL particles detected in air were significantly lower than those detected in liquid (8.5 ± 1.8 nm vs. 8.7 ± 2.0 nm in height for LDL, 2.2 ± 0.4 nm vs. 4.1 ± 0.9 nm in height for HDL). The previous studies also reported a quite low height of LDL (2.5–3 nm) or HDL (2–3 nm) particles detected by AFM in air (Table 1). Second, the particles detected by AFM in air have much larger average diameters and standard deviations than those detected in liquid (mean diameters: 45.8 nm vs. 28.9 nm in FWHM or 26.4 nm vs. 22.6 nm in the equivalent diameter of a sphere for LDL and 23.7 nm vs. 21.5 in FWHM for HDL; standard deviations: 19.6 nm vs. 9.2 nm for LDL and 6.9 nm vs. 6.5 nm for HDL).
The size enlargement of the particles detected in air might be due to the formation of water film while air drying. For the dispersed individual particles, the water film covering or surrounding a particle will be recognized as a part of the particle by AFM therefore misleading an enlarged contour; for the clustered particles, the water in the spaces among these particles will also be imaged by AFM, therefore two or more particles clustered together will be regarded as one large particle by AFM. This effect of water film can be observed directly in the topographical images (Figs. 2A and 3A) and the cross-section height profiles (Figs. 2C and 3C). This is the reason why the boundaries of many clustered LDL and HDL particles detected in air cannot be distinguished from one another.
More important evidence of the existence of the water film is the dramatic changes in the mechanical properties of the particles. In PBS, the Young’s moduli and adhesive forces of LDL and HDL were relatively small (∼0.4 GPa and ∼200 pN, respectively). When detected in air, their Young’s moduli and adhesive forces increased ∼50- to 200-fold (∼20–40 GPa and ∼20–50 nN, respectively). It was caused by the surface tension or viscosity of water film during the approach or retraction of the tips to or from the particle surfaces. Predictably, the AFM detection of other biochemical or mechanical properties of lipoproteins in air will probably be influenced dramatically by water film.
Therefore, due to the formation of a water film on samples, AFM detection in air will significantly affect the quality or fidelity of the data on the properties of nano-sized particles (e.g. the morphological and mechanical properties of plasma lipoproteins). The comparison of the morphological or mechanical data detected in air (especially the latter) among different types of particles (e.g. LDL and HDL) is insignificant since the situation of water film or the distribution of particles on the substrate but not the particles’ own properties greatly determine the values. To acquire better quality or fidelity of the data, AFM detection in liquid or avoidance of the formation of water film will be required.
Author contributions
Y.C. conceived and designed the project, C.G., M.A. and Z.L. acquired the data, Y.C., C.G., M.A. and Z.L. analyzed and interpreted the data, Y.C. wrote the paper.
Acknowledgements
This work was supported by the grants from the National Natural Science Foundation of China (31260205) and the Scientific Research Foundation for Returned Overseas Chinese Scholar of State Education Ministry.
References
- 1.Hirano T., Ito Y., Yoshino G. Measurement of small dense low-density lipoprotein particles. J. Atheroscler. Thromb. 2005;12:67–72. doi: 10.5551/jat.12.67. [DOI] [PubMed] [Google Scholar]
- 2.Scheffer P.G., Bakker S.J.L., Heine R.J., Teerlink T. Measurement of low-density lipoprotein particle size by high-performance gel-filtration chromatography. Clin. Chem. 1997;43:1904–1912. [PubMed] [Google Scholar]
- 3.Sakurai T., Trirongjitmoah S., Nishibata Y., Namita T., Tsuji M., Hui S.P., Jin S., Shimizu K., Chiba H. Measurement of lipoprotein particle sizes using dynamic light scattering. Ann. Clin. Biochem. 2010;47:476–481. doi: 10.1258/acb.2010.010100. [DOI] [PubMed] [Google Scholar]
- 4.van Antwerpen R., Chen G.C., Pullinger C.R., Kane J.P., LaBelle M., Krauss R.M., Luna-Chavez C., Forte T.M., Gilkey J.C. Cryo-electron microscopy of low density lipoprotein and reconstituted discoidal high density lipoprotein: imaging of the apolipoprotein moiety. J. Lipid Res. 1997;38:659–669. [PubMed] [Google Scholar]
- 5.Otvos J.D., Jeyarajah E.J., Bennett D.W., Krauss R.M. Development of a proton nuclear magnetic resonance spectroscopic method for determining plasma lipoprotein concentrations and subspecies distributions from a single, rapid measurement. Clin. Chem. 1992;38:1632–1638. [PubMed] [Google Scholar]
- 6.Williams P.T., Zhao X.Q., Marcovina S.M., Otvos J.D., Brown B.G., Krauss R.M. Comparison of four methods of analysis of lipoprotein particle subfractions for their association with angiographic progression of coronary artery disease. Atherosclerosis. 2014;233:713–720. doi: 10.1016/j.atherosclerosis.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Legleiter J., DeMattos R.B., Holtzman D.M., Kowalewski T. In situ AFM studies of astrocyte-secreted apolipoprotein E- and J-containing lipoproteins. J. Colloid Interface Sci. 2004;278:96–106. doi: 10.1016/j.jcis.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 8.Faustino A.F., Carvalho F.A., Martins I.C., Castanho M.A., Mohana-Borges R., Almeida F.C., Da Poian A.T., Santos N.C. Dengue virus capsid protein interacts specifically with very low-density lipoproteins. Nanomedicine. 2014;10:247–255. doi: 10.1016/j.nano.2013.06.004. [DOI] [PubMed] [Google Scholar]
- 9.Blanchette C.D., Law R., Benner W.H., Pesavento J.B., Cappuccio J.A., Walsworth V., Kuhn E.A., Corzett M., Chromy B.A., Segelke B.W., Coleman M.A., Bench G., Hoeprich P.D., Sulchek T.A. Quantifying size distributions of nanolipoprotein particles with single-particle analysis and molecular dynamic simulations. J. Lipid Res. 2008;49:1420–1430. doi: 10.1194/jlr.M700586-JLR200. [DOI] [PubMed] [Google Scholar]
- 10.Blanchette C.D., Cappuccio J.A., Kuhn E.A., Segelke B.W., Benner W.H., Chromy B.A., Coleman M.A., Bench G., Hoeprich P.D., Sulchek T.A. Atomic force microscopy differentiates discrete size distributions between membrane protein containing and empty nanolipoprotein particles. Biochim. Biophys. Acta. 2009;1788:724–731. doi: 10.1016/j.bbamem.2008.11.019. [DOI] [PubMed] [Google Scholar]
- 11.Blanchette C.D., Segelke B.W., Fischer N., Corzett M.H., Kuhn E.A., Cappuccio J.A., Benner W.H., Coleman M.A., Chromy B.A., Bench G., Hoeprich P.D., Sulchek T.A. Characterization and purification of polydisperse reconstituted lipoproteins and nanolipoprotein particles. Int. J. Mol. Sci. 2009;10:2958–2971. doi: 10.3390/ijms10072958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chieze L., Bolanos-Garcia V.M., Pinot M., Desbat B., Renault A., Beaufils S., Vie V. Fluid and condensed ApoA-I/phospholipid monolayers provide insights into ApoA-I membrane insertion. J. Mol. Biol. 2011;410:60–76. doi: 10.1016/j.jmb.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 13.Chieze L., Bolanos-Garcia V.M., Le Caer G., Renault A., Vie V., Beaufils S. Difference in lipid packing sensitivity of exchangeable apolipoproteins apoA-I and apoA-II: an important determinant for their distinctive role in lipid metabolism. Biochim. Biophys. Acta. 1818;2012:2732–2741. doi: 10.1016/j.bbamem.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 14.Winford S., Tobin M., Gross E. Surface-induced assembly of apolipoprotein A-I: implications for symmetry-driven non-cooperative clustering. J. Cryst. Growth. 2012;343:38–44. [Google Scholar]
- 15.Shao W., Jin H., Huang J., Qiu B., Xia R., Deng Z., Cai J., Chen Y. AFM investigation on Ox-LDL-induced changes in cell spreading and cell-surface adhesion property of endothelial cells. Scanning. 2013;35:119–126. doi: 10.1002/sca.21040. [DOI] [PubMed] [Google Scholar]
- 16.Chouinard J.A., Grenier G., Khalil A., Vermette P. Oxidized-LDL induce morphological changes and increase stiffness of endothelial cells. Exp. Cell Res. 2008;314:3007–3016. doi: 10.1016/j.yexcr.2008.07.020. [DOI] [PubMed] [Google Scholar]
- 17.Lohr D., Bash R., Wang H., Yodh J., Lindsay S. Using atomic force microscopy to study chromatin structure and nucleosome remodeling. Methods. 2007;41:333–341. doi: 10.1016/j.ymeth.2006.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang J., Mou J., Yuan J.Y., Shao Z. The effect of deformation on the lateral resolution of atomic force microscopy. J. Microsc. 1996;182:106–113. doi: 10.1046/j.1365-2818.1996.140422.x. [DOI] [PubMed] [Google Scholar]
- 19.Ho C.H., Britt D.W., Hlady V. Human low density lipoprotein and human serum albumin adsorption onto model surfaces studied by total internal reflection fluorescence and scanning force microscopy. J. Mol. Recognit. 1996;9:444–455. doi: 10.1002/(sici)1099-1352(199634/12)9:5/6<444::aid-jmr281>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 20.Chouinard J.A., Khalil A., Vermette P. Method of imaging low density lipoproteins by atomic force microscopy. Microsc. Res. Tech. 2007;70:904–907. doi: 10.1002/jemt.20492. [DOI] [PubMed] [Google Scholar]
- 21.Sakurai T., Takeda S., Takahashi J.Y., Takahashi Y., Wada N., Trirongjitmoah S., Namita T., Jin S., Ikuta A., Furumaki H., Hui S.P., Fuda H., Fujikawa M., Shimizu K., Chiba H. Measurement of single low-density lipoprotein particles by atomic force microscopy. Ann. Clin. Biochem. 2013;50:564–570. doi: 10.1177/0004563213481586. [DOI] [PubMed] [Google Scholar]
- 22.Jeyarajah E.J., Cromwell W.C., Otvos J.D. Lipoprotein particle analysis by nuclear magnetic resonance spectroscopy. Clin. Lab. Med. 2006;26:847–870. doi: 10.1016/j.cll.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 23.Feng M., Morales A.B., Beugeling T., Bantjes A., vanderWerf K., Gosselink G., deGrooth B., Greve J. Adsorption of high density lipoproteins (HDL) on solid surfaces. J. Colloid Interface Sci. 1996;177:364–371. [Google Scholar]
- 24.Carlson J.W., Jonas A., Sligar S.G. Imaging and manipulation of high-density lipoproteins. Biophys. J. 1997;73:1184–1189. doi: 10.1016/S0006-3495(97)78150-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vainikka K., Chen J., Metso J., Jauhiainen M., Riekkola M.L. Coating of open tubular capillaries with discoidal and spherical high-density lipoprotein particles in electrochromatography. Electrophoresis. 2007;28:2267–2274. doi: 10.1002/elps.200600766. [DOI] [PubMed] [Google Scholar]
- 26.Uetrecht C., Versluis C., Watts N.R., Roos W.H., Wuite G.J., Wingfield P.T., Steven A.C., Heck A.J. High-resolution mass spectrometry of viral assemblies: molecular composition and stability of dimorphic hepatitis B virus capsids. Proc. Natl. Acad. Sci. USA. 2008;105:9216–9220. doi: 10.1073/pnas.0800406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roos W.H., Gibbons M.M., Arkhipov A., Uetrecht C., Watts N.R., Wingfield P.T., Steven A.C., Heck A.J., Schulten K., Klug W.S., Wuite G.J. Squeezing protein shells: how continuum elastic models, molecular dynamics simulations, and experiments coalesce at the nanoscale. Biophys. J. 2010;99:1175–1181. doi: 10.1016/j.bpj.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michel J.P., Ivanovska I.L., Gibbons M.M., Klug W.S., Knobler C.M., Wuite G.J., Schmidt C.F. Nanoindentation studies of full and empty viral capsids and the effects of capsid protein mutations on elasticity and strength. Proc. Natl. Acad. Sci. USA. 2006;103:6184–6189. doi: 10.1073/pnas.0601744103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gibbons M.M., Klug W.S. Nonlinear finite-element analysis of nanoindentation of viral capsids. Phys. Rev. E. 2007;75:031901. doi: 10.1103/PhysRevE.75.031901. [DOI] [PubMed] [Google Scholar]
- 30.Gibbons M.M., Klug W.S. Influence of nonuniform geometry on nanoindentation of viral capsids. Biophys. J. 2008;95:3640–3649. doi: 10.1529/biophysj.108.136176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Calo A., Reguera D., Oncins G., Persuy M.A., Sanz G., Lobasso S., Corcelli A., Pajot-Augy E., Gomila G. Force measurements on natural membrane nanovesicles reveal a composition-independent, high Young’s modulus. Nanoscale. 2014;6:2275–2285. doi: 10.1039/c3nr05107b. [DOI] [PubMed] [Google Scholar]
- 32.Li S., Eghiaian F., Sieben C., Herrmann A., Schaap I.A. Bending and puncturing the influenza lipid envelope. Biophys. J. 2011;100:637–645. doi: 10.1016/j.bpj.2010.12.3701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaskiewicz K., Makowski M., Kappl M., Landfester K., Kroeger A. Mechanical properties of poly(dimethylsiloxane)-block-poly(2-methyloxazoline) polymersomes probed by atomic force microscopy. Langmuir. 2012;28:12629–12636. doi: 10.1021/la301608k. [DOI] [PubMed] [Google Scholar]
- 34.Kurland N.E., Drira Z., Yadavalli V.K. Measurement of nanomechanical properties of biomolecules using atomic force microscopy. Micron. 2012;43:116–128. doi: 10.1016/j.micron.2011.07.017. [DOI] [PubMed] [Google Scholar]
- 35.Ghosh S., Basu M.K., Schweppe J.S. Agarose gel electrophoresis of serum lipoproteins: determination of true mobility, isoelectric point, and molecular size. Anal. Biochem. 1972;50:592–601. doi: 10.1016/0003-2697(72)90070-x. [DOI] [PubMed] [Google Scholar]
- 36.Hurt-Camejo E., Camejo G., Rosengren B., Lopez F., Wiklund O., Bondjers G. Differential uptake of proteoglycan-selected subfractions of low density lipoprotein by human macrophages. J. Lipid Res. 1990;31:1387–1398. [PubMed] [Google Scholar]


