Abstract
Pseudomonas putida is an uncommon opportunistic pathogen, usually susceptible to antimicrobial agents. Data concerning resistance to antimicrobial agents in clinical P. putida isolates are limited. To the best of our knowledge we report for the first time the isolation of NDM-1-producing multidrug-resistant P. putida from a case of acute gastroenteritis. The isolate showed resistance to a wide range of antimicrobials, including fluoroquinolones, third-generation cephalosporins and carbapenems. The isolate also exhibited multiple mutations in the quinolone resistance determining region and showed the presence of qepA, blaTEM, blaOXA1 and blaOXA7 genes. The present study highlights the importance of looking for the relatively rare aetiological agents in clinical samples that do not yield common pathogens.
Keywords: 16S rRNA, diarrhoea, multidrug resistance, mutation, NDM-1, Pseudomonas putida
Introduction
Pseudomonas putida, a non-fermenting Gram-negative bacillus belonging to the fluorescent group of the genus Pseudomonas is frequently found in the environment, along with other non-fermenting Gram-negative organisms. Previously thought to be of low pathogenicity [1], there are increasing reports of their emergence as opportunistic human pathogens causing bacteraemia and sepsis in neonatal, neutropenic and cancer patients, as well as in people with urinary tract infections [2,3]. Most P. putida are susceptible to antimicrobial agents such as fluoroquinolones, aminoglycosides and carbapenems [4]. In the present study, we report the isolation of an NDM-1-producing multidrug-resistant P. putida from a 2-month-old female child admitted to a tertiary-care hospital with acute gastroenteritis in Belgaum, South India. To the best of our knowledge this is the first report of isolation of NDM-1-producing multidrug-resistant P. putida causing acute gastroenteritis.
A female child aged 2 months was admitted to the gastroenteritis ward of a tertiary-care hospital in Belgaum, Karnataka, South India, with symptoms of acute gastroenteritis on 21 June 2013. She had had watery diarrhoea for 3 days along with vomiting for 5 days, showed signs of acute dehydration and had fever of 38.9°C. The fever was intermittent in nature, associated with chills and rigor. The patient was lethargic, restless and had sunken eyes. The patient was put on rehydration therapy, a stool sample was collected for laboratory investigations before administration of any antibiotic at the hospital, and the patient was later empirically treated for acute diarrhoea with oral ciprofloxacin (50 mg) twice daily and metronidazole (25 mg) thrice daily. She recovered and was discharged on 26 June 2013.
The stool sample was processed for isolation and identification of common enteric bacterial pathogens, which include diarrheagenic Escherichia coli, Shigella sp., Salmonella sp., Vibrio sp. following WHO 1987, and was subjected to ELISA for rotavirus, RT-PCR for identification of common viral pathogens like norovirus, astrovirus and sapovirus [5]. The sample was also subjected to routine microscopy for detection of various parasites like Ascaris lumbricoides, Giardia lamblia, Trichuris trichiura, hook worm and Entamoeba histolytica. The isolate was subjected to identification based on an automated microbial identification system, (Vitec2 Compact; bioMérieux, Marcy l'Etoile, France) which was also used for carrying out Antibiotic sensitivity testing (AST) as per CLSI norms [6]. The identity of the isolate was also confirmed by genotypic-based method of 16S rRNA gene sequencing as defined earlier [7]. The rabbit ileal loop test was carried out for the isolate essentially as described by Koley et al [8]. The volume of the accumulated fluid in millilitres and the length of the loop in centimetres were measured, and the extent of the fluid accumulation was expressed as mL/cm. The ileal loop test was performed with positive and negative controls being Vibrio cholerae O1 (N16961) and phosphate-buffered saline, respectively.
The isolate was further screened for any mutation in the quinolone resistance determining region following an earlier described protocol (Table 1) [9]. Presence of plasmid-mediated quinolone resistance determinants was screened following standard conditions (Table 1) [10]. The isolate was also subjected to PCR for detection of the presence of various β-lactam resistance genes (blaTEM, blaSHV, blaCTX-M-3, blaOXA1 and blaOXA7) following techniques reported previously (Table 1) [10]. Presence of the NDM-1 gene was determined by PCR using published primers (Table 1) as described earlier [11]. All PCR products were subjected to nucleotide sequencing in an automatic sequencer (ABI 3130; Applied Biosystems, Foster City, CA, USA) following the manufacturer's instructions. Contig sequences were aligned and edited with SeqScape v2.7 (Applied Biosystems) and compared in BLAST of the NCBI database.
Table 1.
Details of the primers, amplification temperature and amplicon size used in the study
| Sl No. | Gene | Oligonucleotide sequence (5′–3′) | Amplification temp (°C) | Amplicon size (bp) |
|---|---|---|---|---|
| 1 | NDM-1 | ACCGCCTGGACCGATGACCA | 58°C | 264 |
| GCCAAAGTTGGGCGCGGTTG | ||||
| 2 | gyrA | GACGGCCTGAAGCCGGTGCAC | 64°C | 417 |
| GCCCACGGCGATACCGCTGGA | ||||
| 3 | gyrB | AGTACTTCGCCGACTTCCT | 739 | |
| TACAGGCGCGACAGGCGCTT | ||||
| 4 | parC | TCTACGCCATGAGCGAACTGG | 262 | |
| AGCAGCACCTCGGAA TAGCG | ||||
| 6 | qnrA | ATTTCTCACGCCAGGATTTG | 64°C | 516 |
| GATCGGCAAAGGTTAGGTCA | ||||
| 7 | qnrB | GATCGTGAAAGCCAGAAAGG | 476 | |
| ATGAGCAACGATGCCTGGTA | ||||
| 8 | qnrC | GGGTTGTACATTTATTGAATCG | 307 | |
| CACCTACCCATTTATTTTCA | ||||
| 9 | qnrS | GCAAGTTCATTGAACAGGGT | 428 | |
| TCTAAACCGTCGAGTTCGGCG | ||||
| 10 | aac (6′)-Ib-cr | TTGCGATGCTCTATGAGTGGCTA | 55°C | 482 |
| CTCGAATGCCTGGCGTGTTT | ||||
| 11 | qepA | AACTGCTTGAGCCCGTAGAT | 596 | |
| GTCTACGCCATGGACCTCAC | ||||
| 12 | blaTEM | GAGTATTCAACATTTTCGT | 50°C | 857 |
| ACCAATGCTTAATCAGTGA | ||||
| 13 | blaSHV | TCGCCTGTGTATTATCTCCC | 768 | |
| CGCAGATAAATCACCACAATG | ||||
| 14 | blaCTX-M-3 | AATCACTGCGTCAGTTCAC | 701 | |
| TTTATCCCCCACAACCCAG | ||||
| 15 | blaOXA1 | GCAGCGCCAGTGCATCAAC | 198 | |
| CCGCATCAAATGCCATAAGTG | ||||
| 16 | blaOXA7 | AGTTCTCTGCCGAAGCC | 591 | |
| TCTCAACCCAACCAACCC |
Culture on thiosulphate citrate bile salt sucrose agar and Hektoen enteric agar plates did not yield any isolate whereas on McConkey agar, a pure culture of non-lactose-fermenting colonies appeared. The isolate was identified as P. putida by an automated microbial identification system, which was confirmed by 16s rRNA sequence analysis. The sample was negative for all other bacteria, viruses and parasites tested. Growth of the organism as sole pathogen in the culture media in the absence of any other viral, bacterial or parasitic organism indicates the colonization of the gut of the patient by P. putida, probably through suppression of normal microbiota. The P. putida isolate showed resistance to a wide range of antimicrobials, including fluoroquinolones, third-generation cephalosporins and carbapenems, according to CLSI breakpoint [6]. The isolate showed a MIC (mg/L) of 32 for Ampicillin-sulbactam (SAM), ≥128 for Ticarcillin (TIC), ≥128 for Piperacillin (PIP), ≥64 for Ceftazidime (CAZ), ≥64 for Ceftriaxone (CRO), ≥64 for Cefepime (FEP), ≥16 for Imipenem (IMP), ≥16 for Meropenem (MEM), ≥16 for Amikacin (AMK), ≥32 for Gentamicin (GEN), ≥16 for Tobramycin (TOB), ≥4 for Ciprofloxacin (CIP), ≥8 for Levofloxacin (LVX), ≥16 for Tetracycline (TET), ≥8 for Tigecycline (TGC) and ≥320 for Co-trimoxazole (CoT).
The isolate was tested and found positive for the production of extended spectrum β-lactamase using the combination disc test using ceftazidime-clavulanic acid (30/10 μg) and ceftriaxone -clavulanic acid (30/10 μg) [10]. The 16S rRNA gene sequence of this isolate was also compared with other sequences submitted to the NCBI GenBank to understand its genetic relationship with other P. putida by neighbour-joining phylogenetic analysis using MEGA 5.2 software [12], which showed 100% similarity to P. putida isolated from various other parts of the world (Fig. 1). The isolate resulted in moderate fluid accumulation (0.35 mL/cm), which was higher than the negative control (0.05 mL/cm) and lower than the positive control (1.2 cm/mL).
Fig. 1.
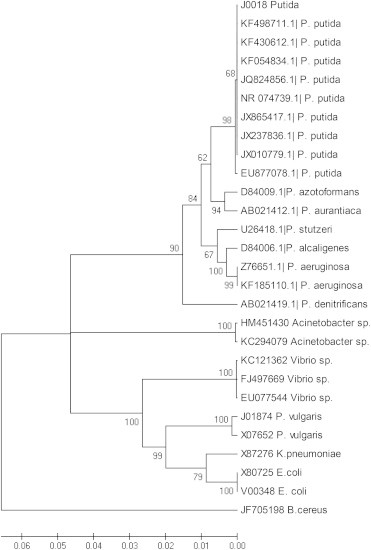
Phylogenetic analysis based on the 16s rRNA gene sequence of the Pseudomonas putida (J0018) isolate.
The isolate exhibited amino acid substitution of T83I and S136A in gyrA; E469D in gyrB; and T105P, V124A and S136A in parC. It also showed the presence of qepA gene. The isolate further showed the presence of blaTEM, blaOXA1 and blaOXA7 genes. The isolate harboured the NDM-1 gene and the partial sequence of the gene showed 100% nucleotide/amino acid identity with those reported from other organisms isolated from various other parts of India and elsewhere in the world.
Since the aetiological agent in this case was resistant to the antibiotic used for therapy, it appears that the disease was self-limiting and resolved on its own within 9 days of onset, and the infant recovered as a result.
This is likely to be the first report of the presence of NDM-1 in P. putida. Studies on antimicrobial resistance in P. putida are scarce. In accordance with one such earlier report [9] we detected multiple mutations in the quinolone resistance determining region of the fluoroquinolone-resistant P. putida. OXA1 and OXA7, which code for oxacillinases, are plasmid-mediated enzymes and were detected in an ampicillin-resistant isolate of Escherichia coli [13]. QepA, a plasmid-mediated efflux pump found in a clinical isolate of E. coli from Japan, is known to elevate levels of resistance to several clinically important FQs, such as ciprofloxacin, norfloxacin and veterinary enrofloxacin [14]. The spread of the opportunistic pathogens carrying NDM-1 gene seems to have become a major global health threat. Reports of microbes previously regarded as non-pathogenic, causing acute diarrhoea in humans is a cause of concern [15]. Although in the present study P. putida was isolated as sole pathogen in the culture media, further in-depth studies are required to understand the potential of this bacterium to cause gastroenteritis as an independent pathogen. The presence of plasmid-mediated resistance determinants in P. putida, which is a well-established bioremediation agent [16,17], adds to the worry as these potent genes may spread to other susceptible bacteria, making them highly resistant. The data highlight the fact that the overuse of antibiotics that are excreted by patients and so find their way into hospital and community wastewater systems provides an environmental selection pressure for the emergence and persistence of multidrug-resistant and pan-drug-resistant bacteria [18]. NDM-1-positive strains can destroy carbapenem antibiotics such as meropenem, imipenem, doripenem and ertapenem by breaking down the carbapenem groups of antibiotics, which have been serving as the basis for the treatment of antibiotic-resistant bacterial infections. Therefore, the spread of the NDM-1 gene to potentially non-pathogenic microorganisms now becomes potentially a major global health threat [11].
Nevertheless, the distinction between causality of microbial resistance and the rate of spread of resistance must be recognized if we are to create a true solution to the problem of antibiotic resistance [19]. If these underlying resistance genes are transferred into otherwise susceptible bacteria, this can lead to therapeutic failure as a consequence of antimicrobial resistance, which may pose a real threat in the near future for the developing countries where antibiotic misuse is common. Emergence of newer mechanisms of resistance under heavy use of antibiotics in resource-poor countries is likely to complicate diagnosis for lesser known pathogens and their therapeutic management. Now is the time for government and health officials to stop the blame game and act together towards a possible solution by undertaking various programmes of antibiotic surveillance and awareness among clinicians, veterinarians and the general public, otherwise we will be headed towards a post-antibiotic era as recently mentioned by WHO (http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf). Although further studies are required to establish or confirm the role of this microorganism as an independent pathogen of gastroenteritis, our report also highlights the importance of looking for the relatively rare aetiological agents in clinical samples that do not yield common pathogens.
Our study once again highlights the problem of transfer of multidrug-resistant genes, including the NDM-1 gene, to these otherwise non-pathogenic bacteria, which calls for serious and rapid implementation of strict antibiotic use policies. There is an urgent need to address the lack of effective treatments to meet the increasing public health burden caused by multidrug-resistant bacteria, in particular against newly emerging pathogens that were previously known to be non-pathogenic.
Nucleotide sequence accession numbers
The sequences of the P. putida have been deposited in the GenBank database under accession numbers, KJ437624 (NDM-1), KJ437625 (blaOXA-7), KJ437626 (qepA), KJ437627 (16s rRNA), KJ437628 (gyrA), KJ437629 (gyrB) and KJ437630 (parC).
Ethics approval
The study was approved by Institutional Ethics Committee of RMRC, Belgaum and KLE University, Belgaum.
Funding
The study was supported by the internal funds of the Regional Medical Research Centre, (ICMR) Belgaum and partially supported by the infrastructure developed for the conduct of ICMR funded extramural task force project ‘National Hospital Based Rotavirus Surveillance Network’ (5/8-1(189)/TF/2011-12 ECD dated 06/12/2012).
Conflicts of interests
The authors declare that they have no conflict of interest.
Acknowledgements
The authors are grateful to the Indian Council of Medical Research for providing intramural funds for the study and the Biomedical Informatics Centre, RMRC, Belgaum for help with data analysis. The authors also acknowledge the services rendered by the Medical Officers (project) Dr Ishrat Madarkar and Dr Nikita Vernekar and the ward boys in the paediatric department of KLES Hospital, Belgaum. The authors are also thankful to European Society of Clinical Microbiology and Infectious Diseases for bearing the publication expenses.
References
- 1.Graevenitz A.V., Weinstein J. Pathogenic significance of Pseudomonas fluorescens and Pseudomonas putida. Yale J Biol Med. 1971;44:265–273. [PMC free article] [PubMed] [Google Scholar]
- 2.Lombardi G., Luzzaro F., Docquier J.D., Riccio M.L., Perilli M., Colì A. Nosocomial infections caused by multidrug-resistant isolates of Pseudomonas putida producing VIM-1 metallo-β-lactamase. J Clin Microbiol. 2002;40:4051–4055. doi: 10.1128/JCM.40.11.4051-4055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Docquier J.D., Riccio M.L., Mugnaioli C., Luzzaro F., Endimiani A., Toniolo A. IMP-12, a new plasmid encoded metallo-β-lactamase from a Pseudomonas putida clinical isolate. Antimicrob Agents Chemother. 2003;47:1522–1528. doi: 10.1128/AAC.47.5.1522-1528.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fass R.J., Barnishan J., Solomon M.C., Ayers L.W. In vitro activities of quinolones, β-lactams, tobramycin, and trimethoprim–sulfamethoxazole against non-fermentative gram-negative bacilli. Antimicrob Agents Chemother. 1996;40:1412–1418. doi: 10.1128/aac.40.6.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yan H., Yagyu F., Okitsu S., Nishio O., Ushijima H. Detection of norovirus (GI, GII), sapovirus, and astrovirus in fecal samples using reverse transcription single-round multiplex PCR. J Virol Meth. 2003;114:37–44. doi: 10.1016/j.jviromet.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 6.Clinical and Laboratory Standards Institute . Clinical and Laboratory Standards Institute; Wayne, PA, USA: 2012. Performance standards for antimicrobial susceptibility testing; Twenty Second Informational Supplement. M100–S22. [Google Scholar]
- 7.Anzai Y., Kim H., Park J.Y., Wakabayashi H., Oyaizu H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int J Syst Evol Microbiol. 2000;50:1563–1589. doi: 10.1099/00207713-50-4-1563. [DOI] [PubMed] [Google Scholar]
- 8.Koley H., Mitra R., Basu A., Mukhopadhyay A.K., Saha P.K., Ramakrishna B.S. Response of wild-type mutants of Vibrio cholerae O1 possessing different combinations of virulence genes in the ligated rabbit ileal loop and in Ussing chambers: evidence for the presence of additional secretogen. J Med Microbiol. 1999;48:51–57. doi: 10.1099/00222615-48-1-51. [DOI] [PubMed] [Google Scholar]
- 9.Horii T., Muramatsu H., Iinuma Y. Mechanisms of resistance to fluoroquinolones and carbapenems in Pseudomonas putida. J Antimicrob Chemother. 2005;56:643–647. doi: 10.1093/jac/dki254. [DOI] [PubMed] [Google Scholar]
- 10.Bhattacharya D., Bhattacharya H., Thamizhmani R., Sayi D.S., Reesu R., Anwesh M. Shigellosis in Bay of Bengal Islands, India: clinical and seasonal patterns, surveillance of antibiotic susceptibility patterns and molecular characterization of multi-drug resistant Shigella strains isolated during a 6-year period, 2006 to 2011. Eur J Clin Res Infect Dis. 2014;33:157–170. doi: 10.1007/s10096-013-1937-2. [DOI] [PubMed] [Google Scholar]
- 11.Bhattacharya D., Thamizhmani R., Bhattacharjee H., Sayi D.S., Muruganandam N., Roy S. Emergence of New Delhi metallo-β-lactamase 1 (NDM-1) producing and multidrug resistant uropathogens causing urinary tract infections in Andaman Islands, India. Microb Drug Resist. 2013;19:457–462. doi: 10.1089/mdr.2013.0070. [DOI] [PubMed] [Google Scholar]
- 12.Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Medeiros A.A., Cohenford M., Jacoby G.A. Five novel plasmid determined β-lactamases. Antimicrob Agents Chemother. 1985;27:715–719. doi: 10.1128/aac.27.5.715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamane K., Wachino J., Suzuki S., Kimura K., Shibata N., Kato H. New plasmid-mediated Fluoroquinolone efflux pump, QepA, found in an Escherichia coli clinical isolate. Antimicrob Agents Chemother. 2007;51:3354–3360. doi: 10.1128/AAC.00339-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dey S., Bhattacharya D., Roy S., Nadgir S.D., Patil A., Kholkute S.D. Shewanella algae in acute gastroenteritis. Indian J Med Microbiol. 2015;33:172–175. doi: 10.4103/0255-0857.148442. [DOI] [PubMed] [Google Scholar]
- 16.Gomes N.C., Kosheleva I.A., Abraham W.R., Smalla K. Effects of the inoculant strain Pseudomonas putida KT2442 (pNF142) and of naphthalene contamination on the soil bacterial community. FEMS Microbiol Ecol. 2005;54:21–33. doi: 10.1016/j.femsec.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Validov S., Kamilova F., Qi S., Stephan D., Wang J.J., Makarova N. Selection of bacteria able to control Fusariumoxysporum f. Sp. Radicis-lycopersici in stonewool substrate. J Appl Microbiol. 2007;102:461–471. doi: 10.1111/j.1365-2672.2006.03083.x. [DOI] [PubMed] [Google Scholar]
- 18.Walsh T.R., Toleman M.A. The emergence of pan-resistant Gram-negative pathogens merits a rapid global political response. J Antimicrob Chemother. 2012;67:1–3. doi: 10.1093/jac/dkr378. [DOI] [PubMed] [Google Scholar]
- 19.Spellberg B., Guidos R., Gilbert D., Bradley J., Boucher H.W., Scheld W.M. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]


