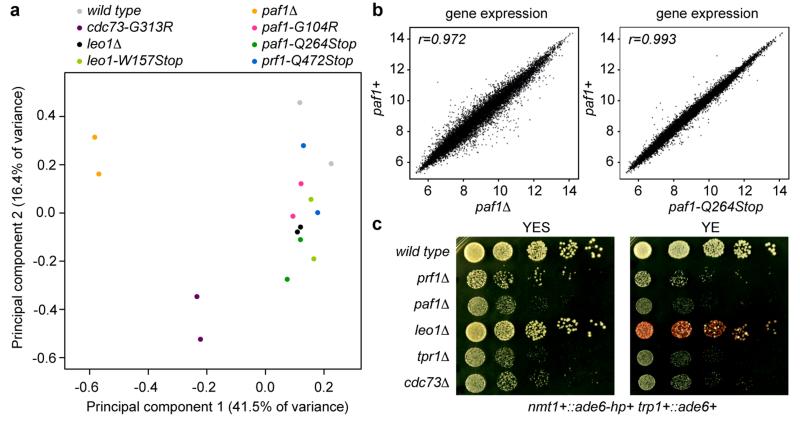
Extended Data Figure 8. Effect of Paf1C mutations on global gene expression and silencing.
a, The impact of the Paf1C mutations on genome expression was assessed by hybridizing total RNA to whole-genome tiling arrays. The parental wild type strain, all Paf1C point mutations discovered in the screen, and full deletions of the paf1+ and leo1+ genes were included in the analysis. In order to compare the genome-wide expression profiles of the mutants with the wild type strain, a Principal Component Analysis (PCA) was performed on the data obtained for two biological replicates of each strain. Principal Component (PC) 1 and 2 explained 41.5% and 16.4% of the variance between samples and were selected for visualization, revealing that cdc73-G313R and paf1Δ cells are most different from wild type cells. All the other mutants clustered together in a group of samples that also includes wild type, demonstrating that RNA steady-state levels are only minimally affected in these mutants. Note that leo1Δ is more similar to wild type than paf1Δ, as well as that paf1Δ clusters separately from the Paf1C point mutants. b, Pairwise comparisons of gene expression between wild type and paf1 mutant strains. c, leo1Δ cells have no growth defect but are susceptible for de novo formation of heterochromatin by siRNAs acting in trans. These results suggest that Leo1 might be a bona fide repressor of small RNA-mediated heterochromatin formation.