Summary
Phosphoinositide-3-kinase (PI3K)-α inhibitors have shown clinical activity in squamous carcinoma (SCC) of head and neck (H&N) bearing PIK3CA mutations or amplification. Studying models of therapeutic resistance we have observed that SCCs cells that become refractory to PI3Kα inhibition maintain PI3K-independent activation of the mammalian target of rapamycin (mTOR). This persistent mTOR activation is mediated by the tyrosine kinase receptor AXL. AXL is overexpressed in resistant tumors from both laboratory models and patients treated with the PI3Kα inhibitor BYL719. AXL dimerizes with and phosphorylates epidermal growth factor receptor (EGFR), resulting in activation of phospholipase Cγ (PLCγ)- protein kinase C (PKC), which in turn activates mTOR. Combined treatment with PI3Kα and either EGFR, AXL, or PKC inhibitors reverts this resistance.
Introduction
H&N and esophageal SCCs are among the most common types of cancer with an annual worldwide incidence rate of approximately 600,000 and 400,000, respectively (Jemal et al., 2011). Despite the efforts to improve their outcome, the 5-year survival rate is only ∼50% in H&N and ∼10% in esophageal cancer (Jemal et al., 2011; Kamangar et al., 2006). Cetuximab, a monoclonal antibody targeting the EGFR, is currently the only approved targeted agent for the therapy of H&NSCC, with only a modest improvement in the overall survival of these patients (Baselga et al., 2005; Bonner et al., 2006; Vermorken et al., 2008). No targeted therapy is currently available for esophageal SCC.
The PI3K pathway plays a key role in the regulation of multiple cellular events, including cell growth, proliferation, cell cycle progression, and survival (Vivanco and Sawyers, 2002). The PI3K family of enzymes is divided into three main classes (classes I to III), with class I being the most often implicated in human cancer (Engelman, 2009; Hennessy et al., 2005). Class I PI3K is comprised of a regulatory subunit (p85), which mediates binding to membrane growth factor receptors, and one of four catalytic subunits (p110α, β, γ or δ), which are responsible for the activity of the enzyme (Engelman, 2009). Activating mutations of PI3KCA have been found in 6 to 20% of H&N and 4–10 % of esophageal SCC, with the hot spot E542K, E545K and H1047R substitutions being the most common (Agrawal et al., 2011; Lui et al., 2013; Song et al., 2014; Stransky et al., 2011). Moreover, increase in PIK3CA copy number has been found in up to 30% of H&N and 40% of esophageal tumors and is associated with poor prognosis (Agrawal et al., 2011; Akagi et al., 2009; Lin et al., 2014; Song et al., 2014; Stransky et al., 2011; Suda et al., 2012).
Tumors with activating alterations in PIK3CA are more responsive to therapy with specific PI3Kα inhibitors (Elkabets et al., 2013; Fritsch et al., 2014; Furet et al., 2013). In the first-in-human clinical trial of the PI3Kα specific inhibitor BYL719 in solid tumors (NCT01387321), we have recently reported that eight patients with H&N tumors harboring PI3KCA mutations had a clinical response to therapy (Juric et al., unpublished results). These are encouraging early results and support the clinical development of these agents in patients with H&N SCC tumors. Given the similarities between H&N and esophageal SCCs, this approach should also be explored in esophageal SCC. However, as with other targeted therapies, we can anticipate that their efficacy will be limited by the development of acquired resistance. In fact, all the responding patients in our clinical trial with BYL719 became eventually refractory to treatment. In this work we investigated the primary and acquired mechanisms of resistance to PI3Kα inhibitors in H&N and esophageal SCC tumors. Our ultimate goal was to design treatment strategies that could prevent or delay the appearance of resistance and that would be potentially applicable to the treatment of patients.
Results
Persistent mTOR activation defines SCC with acquired resistance to BYL719
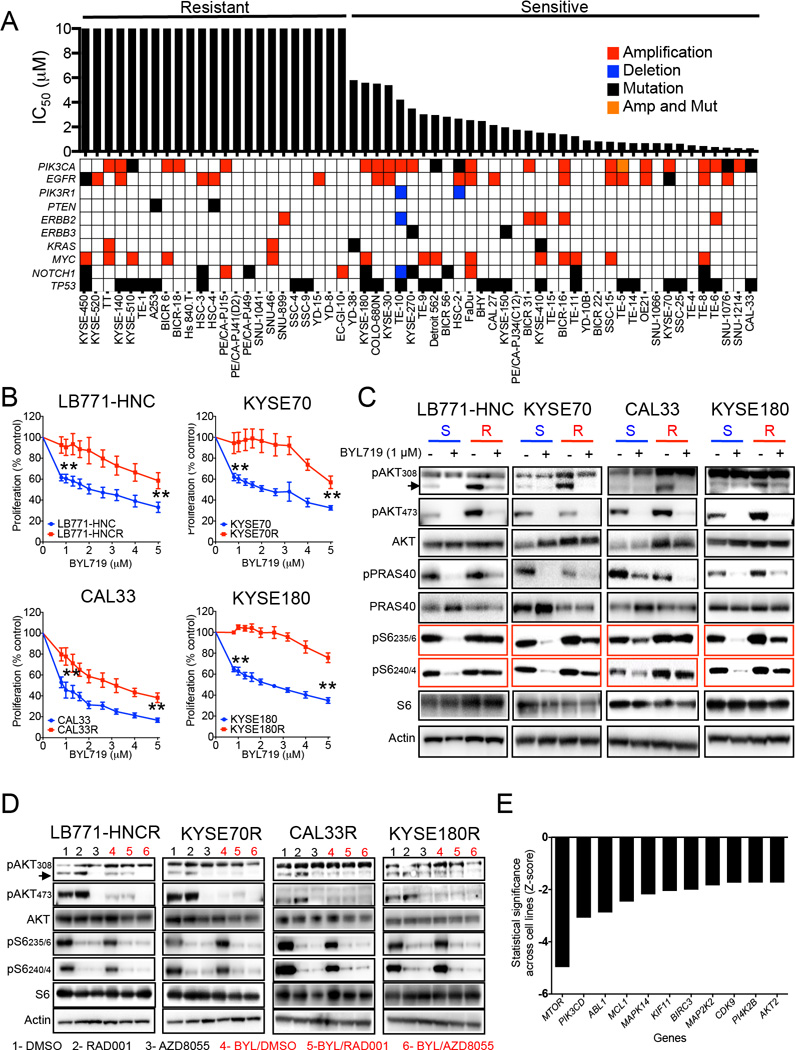
In an initial approach to explore the sensitivity of SCC to the PI3Kα inhibitor BYL719, we measured in a panel of 58 SCC cell lines the median inhibitory concentration (IC50) after five days of treatment. Some cell lines displayed absolute resistance at doses up to 10 µM while the remaining cells showed a gradient of sensitivity to BYL719. The concentration of 10 µM was chosen to define resistance because it is the highest achievable concentration, albeit briefly, in the plasma of patients and therefore likely to be clinically relevant (Juric et al., unpublished results). In terms of PIK3CA status and sensitivity to BYL719, 76% (19/25) of cell lines bearing either mutations or amplification (copy number>4) in PIK3CA were sensitive while only 48% (16/33) of cell lines bearing WT-PIK3CA were sensitive (Figure 1A). PIK3CA mutation/amplification was the only genomic alteration that predicted sensitivity to BYL719 (Figure S1A).
Figure 1. PI3K/AKT-independent mTOR activation in BYL719-resistant cells.
(A) Main genetic features and IC50 for BYL719 of 58 cell lines from H&N and esophageal SCC.
(B) Proliferation (5 days) of four cell lines sensitive to BYL719 in comparison with their resistant counterparts upon increasing doses of BYL719.
(C) Western blot analysis of PI3K/AKT pathway signaling using cell lysates from BYL719-sensitive (S) and resistant (R) cells treated with 1 µM BYL719 for 4 hr and the indicated antibodies.
(D) Western blot with the indicated antibodies of protein lysates from BYL719-resistant cells upon 4 hr of treatment with 100 nM RAD001 or 500 nM AZD8055 with and without 1 µM BYL719.
(E) Ranking of knock-down genes which re-sensitizes resistant cells to BYL719 (Z-score).
Data are presented as means ± SEM. p values were calculated using two-sided Student's t test.
To study the molecular mechanisms by which acquisition of resistance to BYL719 emerges, we selected four sensitive cell lines bearing either amplified or mutated PIK3CA (CAL33, LB771-HNC, KYSE70 and KYSE180) and exposed them over time to increasing concentrations of BYL719 until resistance emerged (Figure 1B; Figure S1B and S1C).
In order to elucidate the underlying mechanisms of resistance, we first analyzed potential differences of pathway inhibition with BYL719 between parental cells and their resistant counterparts. While AKT and its downstream effector PRAS40 were equally suppressed by BYL719 treatment in both parental and resistant cells, mTOR activity was not abolished upon PI3Kα inhibition in resistant cells as indicated by persistent phosphorylation of its downstream ribosomal protein S6 (pS6) in residues 235/6 and 240/4 (Figure 1C).
The addition of the allosteric mTORC1 inhibitor RAD001 abolished the phosphorylation of S6 (Figure 1D) and was sufficient to re-sensitize the resistant cells to BYL719 (Figure S1D). Given that the mTORC2 complex directly activate AKT, treatment with the catalytic mTOR inhibitor AZD8055, which targets both mTORC1 and mTORC2, was very effective in suppressing both AKT and mTOR phosphorylation, resulting in strong antiproliferative activity even when used as a single agent (Figure 1D).
We were able to independently demonstrate the causative role of mTOR in resistance to BYL719 in a synthetic lethality shRNA screen for 134 cancer-related genes performed in three BYL719-resistant cell lines (CAL33R, KYSE180R and the intrinsically resistant HSC3) (Elkabets et al., 2013). We observed that MTOR, which encodes mTOR, was the top gene that when knocked down re-sensitized resistant cells to the antiproliferative activity of BYL719 (Figures 1E; Figure S1E; Table S1).
Taken together, these results show that SCC cells with acquired resistance to BYL719 maintain mTOR activity and that pharmacological or shRNA inhibition of mTOR results in re-sensitization to BYL719.
EGFR activation is sufficient to limit the sensitivity to BYL719 in SCC cells
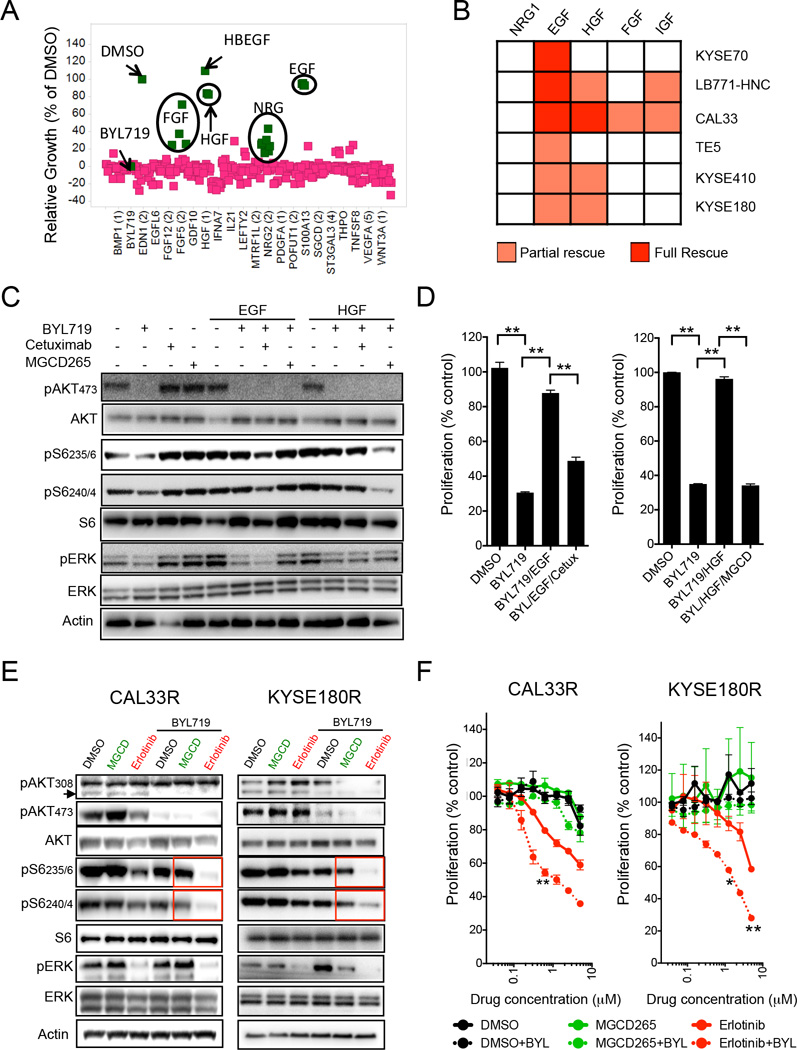
We and others have shown that ligand-mediated activation of receptor tyrosine kinases (RTKs) can rescue the efficacy of a variety of agents against key signaling pathways(Elkabets et al., 2013; Harbinski et al., 2012; Straussman et al., 2012; Wilson et al., 2012). In order to explore whether RTKs activation can sustain mTOR activity in the presence of BYL719 and induce resistance to this agent in our models, we performed a high throughput secretome screen (Harbinski et al., 2012) using the BYL719-sensitive cell lines CAL33 and SNU1214 (Figure 2A; Table S2). Among the over 300 secreted proteins tested, epidermal growth factor (EGF), heparin-binding EGF (HBEGF, both binding EGFR), hepatocyte growth factor (HGF, binding cMET), fibroblast growth factors (FGFs, binding FGF receptors) and neuregulin (NRG, binding HER3) prevented in some degree BYL719-induced growth inhibition. In order to study the reproducibility of these results, we tested whether these ligands could rescue the inhibitory effects of BYL719 in five additional BYL719-sensitive cell lines (1 PIK3CA WT, 2 PIK3CA mutated and 2 PIK3CA amplified). Among the tested recombinant ligands, only EGF and to a lesser extent HGF could either partially or completely prevent BYL719 antiproliferative activity in all cell lines (Figure 2B; Figure S2A and S2B). The same ligands did not prevent BYL719-induced antiproliferative activity in either breast or ovarian cell lines, underscoring their lineage specificity for SCC cells (Figure S2C). The addition of the anti-EGFR antibody cetuximab or the anti-cMET inhibitor MGCD265 opposed to the effects of EGF or HGF, respectively, and suppressed S6 phosphorylation and re-sensitized cells to BYL719 (Figures 2C and 2D; Figure S2D). Of note, in LB771-HNC cells EGF was superior to HGF in maintaining S6 phosphorylation and inducing proliferation in the presence of BYL719 (Figure S2D). Next, we tested whether co-inhibition of EGFR or cMET with BYL719 was sufficient to reduce mTOR activity in cell lines with acquired resistance to BYL719. Treatment with BYL719 in combination with erlotinib, an EGFR kinase inhibitor, but not with MGCD235 or crizotinib, another cMET kinase inhibitor, resulted in suppression of S6 phosphorylation (Figure 2E; Figure S2E). These biochemical effects translated into superior antiproliferative activity in cells where both PI3Kα and EGFR were inhibited, as opposed to lack of significant additive effects when BYL719 was combined with cMET inhibition (Figure 2F). These effects seemed to be mediated mainly by reduction of proliferation (S phase) compared to single agent (Figure S2E).
Figure 2. Role of EGFR in maintaining mTOR activity upon BYL719 treatment.
(A) Results from a secretome screen conducted in CAL33 cells upon treatment for 4 days with 1 µM BYL719. Each square represents the average of three replicates of cells conditioned with the same ligand. Y axis show relative cell growth as a % of DMSO.
(B) Heatmap showing the effects of recombinant ligands in limiting the antiproliferative activity of 1 µM of BYL719 for six days in H&N and esophageal SCC cell lines. Red and orange indicate full and partial rescue from treatment, respectively.
(C) Western blot with the indicated antibodies of CAL33 cell line treated for 4 hr with 1 µM BYL719, 10 µg/ml cetuximab and 1 µM MGCD265 in the presence of 50 ng/ml of EGF or HGF as indicated.
(D) Proliferation (6 days) of CAL33 cells treated as indicated with 1 µM BYL719, 10 µg/ml cetuximab and 1 µM MGCD265 in the presence of 50 ng/ml of EGF or HGF.
(E) Western blot with the indicated antibodies of protein lysates from CAL33R and KYSE180R cells treated with 1 µM BYL719 in combination with either 1 µM MGCD265 or 5 µM erlotinib.
(F) Proliferation (5 days) of resistant cell lines treated with different concentrations of erlotinib or MGCD265 with or without 1 µM BYL719. Significance of erlotinib+BYL versus erlotinib is displayed.
Data are presented as means ± SEM. p values were calculated using two-sided Student's t test.
Based on these results we decided to confirm that activation of EGFR is sufficient to reverse tumor growth inhibition induced by treatment with BYL719 in vivo. Animals bearing CAL33-derived xenografts and implanted with pumps that release recombinant human EGF showed a reduced inhibition of tumor growth upon BYL719 treatment compared to animals with pumps releasing vehicle. Furthermore, as we observed in vitro, treatment with cetuximab re-sensitized the tumors to PI3K inhibition and induced tumor shrinkage (Figure S2F). Pharmacodynamic studies showed that addition of EGF induced phosphorylation of EGFR, activated the mTOR pathway (indicated by pS6) and promoted proliferation (Figure S2G and S2H). The combination of BYL719 and cetuximab prevented EGFR phosphorylation, mTOR activation and resulted in decreased proliferation and induction of cell death (Figure S2G and S2H).
Anti-EGR treatment reverses acquired and intrinsic resistance to PI3Kα inhibition in SCC
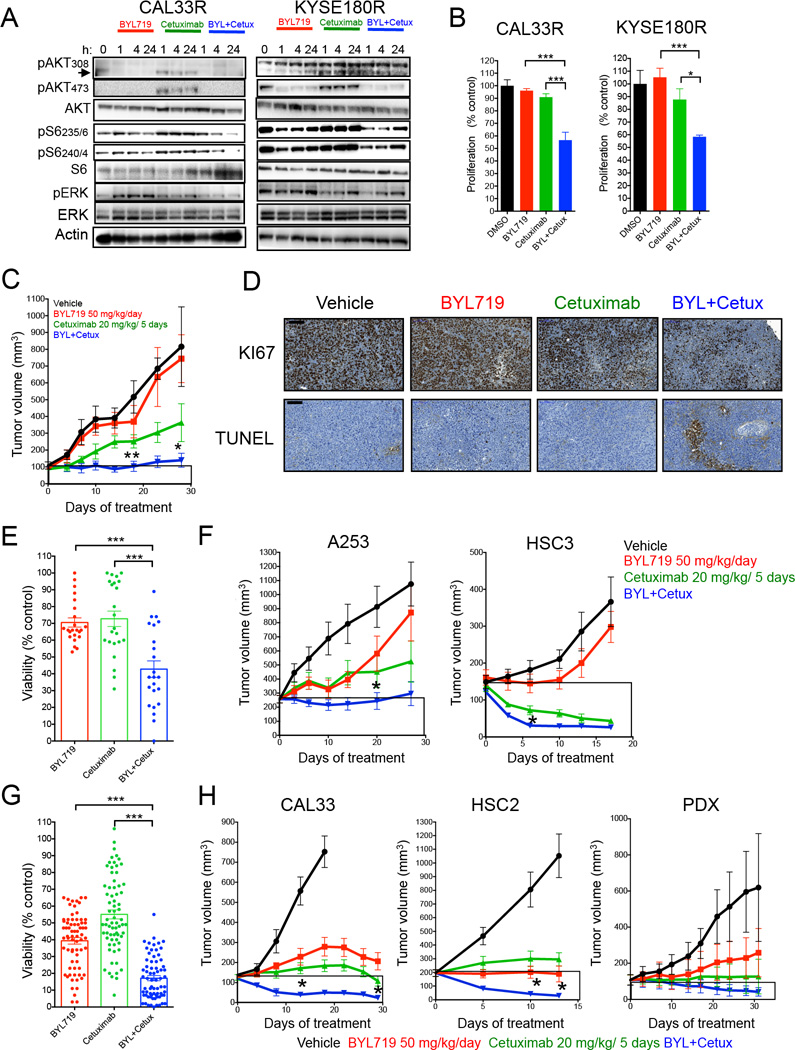
To further characterize the effects of EGFR blockade in reverting resistance, we conducted a series of studies with the anti-EGFR monoclonal antibody cetuximab that prevents ligand binding to the receptor and is approved for the treatment of head and neck SCC. Like the majority of antibodies, cetuximab is devoid of off-target effects even at high doses. In SCC cells with acquired resistance to BYL719, the addition of cetuximab to BYL719 was superior to both agents given alone in reducing S6 phosphorylation (Figure 3A) and in induction of cell death and/or reduction of proliferation (Figure 3B; Figure S3A and S3B). These results were confirmed in vivo with KYSE180R xenografts where, as expected, the tumors were non-responsive to BYL719 and the combination of BYL719 and cetuximab was superior to cetuximab alone (Figure 3C). In these tumors, cetuximab and BYL719 was sufficient to block most of the phosphorylation of S6 (Figure S3C), and this correlated with decreased tumor proliferation and increased cell death (Figure 3D).
Figure 3. Antitumor activity of BYL719 in combination with cetuximab.
(A) Western blot with the indicated antibodies of protein lysates from resistant cells treated as indicated with 1 µM BYL719, 5 µg/ml cetuximab or the combination.
(B) Proliferation (6 days) of resistant cells treated with 1 µM BYL719, 5 µg/ml cetuximab and the combination.
(C) Tumor growth of KYSE180R-derived xenografts treated as indicated (n= 6–10 per arm). Significance of combination versus cetuximab is displayed.
(D) KYSE180R derived xenografts tumors from mice treated as indicated were analyzed for proliferation (Ki67) and apoptosis (TUNEL).
(E) Cell viability (4 days) of 23 cell lines intrinsically resistant to BYL719 treated with 3 µM BYL719, 10 ng/ml cetuximab or the combination.
(F) Tumor growth of A253- and HSC3-derived xenografts (intrinsically resistant to BYL719) treated as indicated (n= 6–10 per arm). Significance of combination versus cetuximab is displayed.
(G) Cell viability (4 days) of 35 cell lines intrinsically sensitive to BYL719 treated with 3 µM BYL719, 10 ng/ml cetuximab or the combination.
(H) Tumor growth of xenograft derived from BYL719-sensitive cell lines treated as indicated (n= 6–10 per arm). For CAL33 significance of combination versus cetuximab is displayed. For HSC2 significance of combination versus BYL719 is displayed.
Data are presented as means ± SEM. p value was calculated using two-sided Student’s t test.
*p<0.05, **p<0.01, ***p<0.005. Scale bar = 100 µm. See also Figure S3 and Table S3.
A similar antitumor activity of the combination of BYL719 and cetuximab was observed also in cells with intrinsic resistance to BYL719, both in vitro (Figure 3E; Table S3) and in vivo (Figure 3F).
Simultaneous suppression of EGFR and PI3Kα resulted in superior activity versus single agent also in cells intrinsically sensitive to BYL719. Despite both BYL719 and cetuximab effectively reducing cell viability, the combination was significantly superior to single agent (Figure 3G; Figure S3D; Table S3). In tumor xenografts, even if treatment with either BYL719 or cetuximab reduced tumor growth in BYL719-sensitive models, including H&N patient-derived xenografts (PDX) available in our laboratory, the combination resulted in greater tumor inhibition and shrinkage (Figures 3H; Figure S3E). Pharmacodynamic analysis of KYSE180 xenografts confirmed that the combination of BYL719 and cetuximab reduced cell proliferation and induced cell death compared to single agent treatment (Figure S3F). Although we cannot exclude a priori that some of the antitumor effects observed with cetuximab were due to antibody-dependent cell cytotoxicity (ADCC), there is ample evidence that the major role that inhibition of receptor activation and signaling play in the antitumor effects of cetuximab (Fan et al., 1993). In those cases where the combination achieved complete tumor remission (11 of 17 tumors for CAL33 and 8 of 20 tumors for KYSE180) the tumors did not recur after therapy completion. This combination therapy showed no appreciable toxicity in mice, as indicated by their unchanged body weights (Figure S3G).
Up-regulation of AXL and its interaction with EGFR are associated with resistance to BYL719
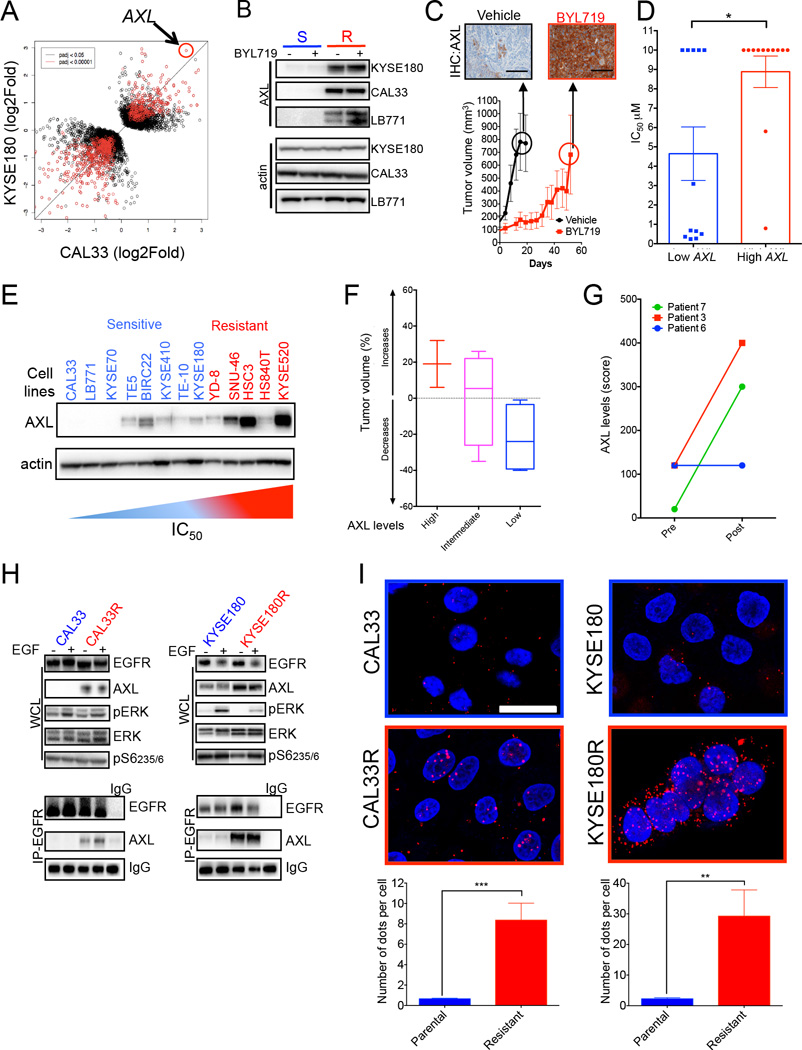
Aiming to investigate the molecular mechanisms leading to EGFR activation during the acquisition of resistance to PI3Kα inhibition, we analyzed the transcriptome of KYSE180 and CAL33 cells using RNA-sequencing and compared it to their resistant counterparts. A differential expression analysis was performed to identify genes that associated with resistance to BYL719. Our analysis was restricted to genes whose expression varied in the same direction in both cell lines upon emergence of resistance. We identified 56 and 153 genes that were upregulated (Log of change>1.2) or down-regulated (Log of change<-1.2), respectively, in both KYSE180R and CAL33R cell lines (Figure 4A; Figure S4A). The most significantly upregulated gene in the resistant cells was AXL in both cell lines (Figures 4A; Figure S4B; Table S4). AXL is a membrane-bound receptor tyrosine kinase that interacts with HER receptors (Meyer et al., 2013; Paccez et al., 2014) and limits the sensitivity to anti-EGFR or anti-HER2 therapies (Byers et al., 2013; Liu et al., 2009; Zhang et al., 2012). Analysis from the TCGA database (Cerami et al., 2012) indicated that high expression of AXL (among the top 10 up-regulated genes) was associated with poor-prognosis in patients with H&N tumors bearing either mutations or amplification of PIK3CA (Figure S4C and S4D) (Cerami et al., 2012). Thus, with our findings, we hypothesized that overexpression of AXL and its interaction with EGFR could play a role in mediating resistance to PI3Kα inhibition.
Figure 4. Up-regulation of AXL in BYL719 resistant tumors, and direct interaction with EGFR.
(A) Dot plot analysis from RNA sequencing of KYSE180 and CAL33 cells compared to their resistant counterparts.
(B) AXL levels in the indicated sensitive (S) and resistant (R) cell lines treated with 1 µM BYL719.
(C) Tumor growth of KYSE180-derived xenografts treated with 50 mg/Kg BYL719 daily. AXL levels in control tumors (vehicle) and in xenografts after treatment with BYL719 for 2 months are shown. Scale bar: 100 µm.
(D) Comparison of IC50 between SCC cells with high and low (1st Vs 4th quartile) AXL mRNA expression (data extracted from the TCGA).
(E) Baseline AXL expression on a panel of BYL719-sensitive and resistant cell lines.
(F) Box plot representation of tumor volume as a function of AXL levels quantified in patient tissue samples by immunohistochemistry. Axl levels are split into three groups from high expressing on the left to low expressing on the right. For each group the tumor volume distribution is represented by its statistics, the median as the middle line, the inter quartile range (25%-75%) as the box and the minimum and maximum as the whisker. (G) Changes in AXL levels following BYL719 treatment in three SCC patients. Pre = pre-treatment samples; Post = samples collected at disease progression upon BYL719 therapy. (H) Western blot with the indicated antibodies of whole cell lysates (WCL) or proteins immunoprecipitated with an anti-EGFR antibody (IP) from the indicated cell lines. EGF (50ng/ml) was supplemented to the culture media 20 minutes prior to lysate collection. (I) Proximity ligation assay (PLA) of AXL and EGFR on parental and resistant cells. Quantification of AXL/EGFR complex was carried out using Image J. Scale bar = 25 µn.
Data are presented as means ± SEM. p value was calculated using two-sided Student's t test.
*p<0.05, **p<0.01, ***p<0.005. See also Figure S4 and Table S4.
A number of additional observations provided support to the role of AXL in mediating resistance to PI3Kα inhibition. First, we confirmed that cells with acquired resistance to BYL719 and xenografts that were initially sensitive but that eventually escaped BYL719 treatment had higher levels of AXL compared to their parental equivalents (Figure 4B and 4C). In addition, when we examined the basal mRNA levels of AXL in our panel of 58 cell lines we observed an inverse correlation between AXL expression and sensitivity to BYL719 (Figure 4D; Table S4). This observation was further validated at the protein level in a smaller cohort of cell lines (Figure 4E). We next investigated the association between AXL expression and resistance to PI3Kα inhibition in tumors from patients that participated in the initial clinical study of BYL719 (Juric et al., unpublished results). We were able to obtain high quality tumor samples from ten SCC patients (9 with H&N cancer and one with cervical cancer) treated with BYL719 (one patient was treated with the combination of BYL719 and the MEK inhibitor MK162) (Table S5). In these samples collected before initiation of treatment, those tumors with lower AXL expression had higher shrinkage upon PI3Kα inhibition (Figure 4F; Figure S4E and S4F; Table S5). Strikingly, the 5 patients with the best responses were the patients with the lowest levels of AXL detected (Table S5). Moreover, in three of these patients we obtained and analyzed also tumor biopsies obtained at time of disease progression to BYL719. Two of the three patients presented a marked increase in AXL levels in the post-treatment sample when compared with the baseline tumor expression, while no changes were appreciated in the third patient (Figure 4G; Figure S4F; Table S6). Of note, the post-treatment specimen that did not experience changes in AXL expression (Patient 6) was obtained from a brain metastasis. Given that BYL719 does not permeate the blood-brain barrier, it is likely that this lesion was never exposed to pharmacologically meaningful concentrations of the drug.
We then examined the interaction of AXL and EGFR in both sensitive and resistant cells. The higher levels of AXL in our resistant models (CAL33R and KYSAE180R) coincided with increased EGFR and AXL interaction as shown by EGFR-AXL co-immunoprecipitation (Figure 4H). This interaction was further validated by in-situ proximity ligation assay, which enables to visualize and quantify the interaction of AXL and EGFR (Figure 4I) (Soderberg et al., 2008). The number of AXL/EGFR complexes, indicated by the red dots, is significantly higher in resistant cell lines compared to the parental counterpart. Over all our data indicate that AXL expression and its interaction with EGFR are associated with resistance to PI3Kα inhibition.
The AXL/EGFR interaction activates mTOR and limits the sensitivity to BYL719
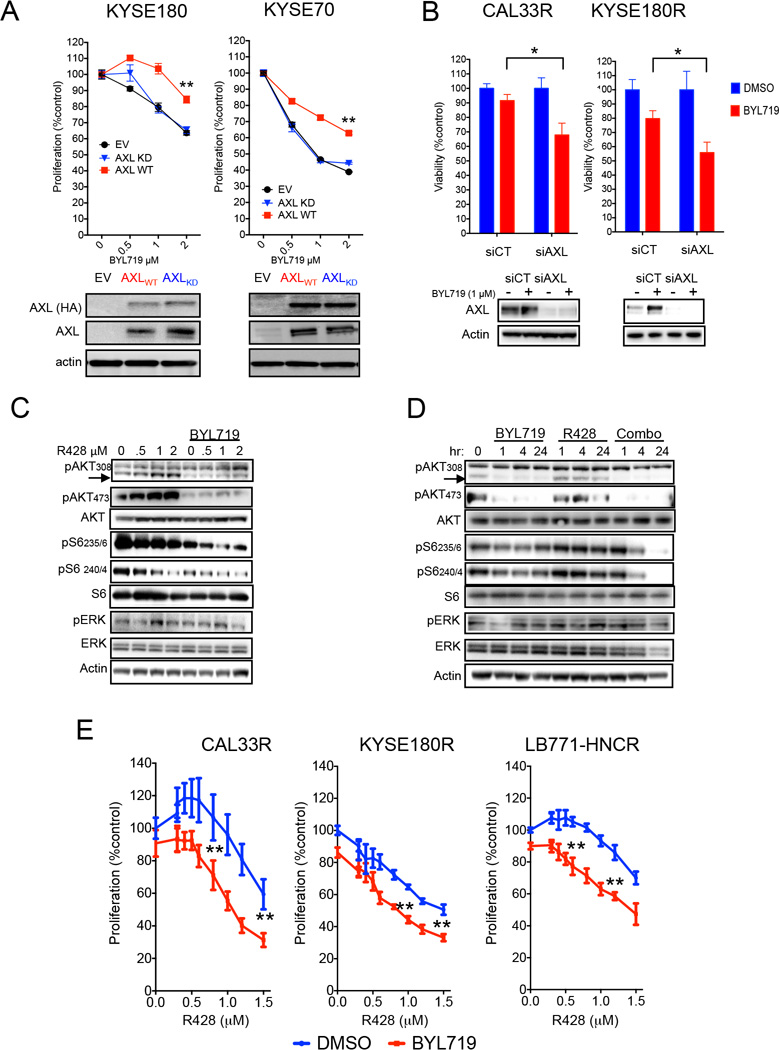
In order to assess whether AXL overexpression plays a causative role in mediating resistance to PI3Kα inhibition, we ectopically transduced this receptor in two BYL719-sensitive cell lines and found that forced expression of AXL was sufficient to limit the sensitivity to BYL719 (Figure 5A). Overexpression of a kinase-dead version of AXL (Zhang et al., 2012) did not affect BYL719 sensitivity, suggesting that the kinase activity of the receptor is required to induce resistance to PI3Kα inhibitors (Figure 5A). Biochemical analysis of these cells indicate that high levels of WT-AXL was sufficient to prevent BYL719-dependent inhibition of mTOR but failed to do so when EGFR expression was concomitantly knocked-down (Figure S5A). Moreover, the inhibition of EGFR was sufficient to re-sensitize WT-AXL-overexpressing cells to BYL719 (Figure S5B). Likewise, knocking-down the expression of AXL in cell lines with acquired resistance to BYL719 re-sensitized the cells to PI3Kα inhibition (Figure 5B). Pharmacological inhibition of AXL by the small molecule ATP competitor R428 (Holland et al., 2010) was sufficient to achieve greater inhibition of pS6 upon BYL719 treatment (Figures 5C and 5D; Figure S5C) and enhanced the antiproliferative activity of BYL719 in resistant cells (Figure 5E; Figure S5D). Co-inhibition of AXL and PI3Kα in BYL719-sensitive cells resulted in further inhibition of pS6 and greater tumor cell growth inhibition (Figure S5E, S5F and S5G), suggesting that even relatively low levels of AXL may be sufficient to partially counteract the activity of BYL719.
Figure 5. AXL/EGFR derives mTOR activation and cause resistance to BYL719.
(A) Cell viability of CAL33 and KYSE70 cells overexpressing either wild type (WT) or kinase dead (KD) AXL upon treatment with BYL719 (upper panel) and western blot showing both endogenous and exogenous expression of AXL (lower panel).
(B) Cell viability (3 days) of KYSE180R and CAL33R cells subjected to AXL knock-down and treated as indicated (upper panel) and western blot showing AXL knockdown (lower panel).
(C) Western blot with the indicated antibodies of cell lysates from KYSE180R cells treated for 4 hr with different concentration of R428 and 1 uM of BYL719.
(D) Western blot with the indicated antibodies of cell lysates from KYSE180R treated with 1 µM BYL719, 1 µM R428 or the combination at different time points.
(E) Cell viability (4 days) of cells with acquired resistance to BYL719 treated with increased concentrations of R428 with or without 1 µM of BYL719.
Data are presented as means ± SEM. p value was calculated using two-sided Student's t test.
*p<0.05, **p<0.01. Scale bar = 100 µm. See also Figure S5 and Table S5 and S6.
Taken together, these results show that BYL719-resistant cells up- display high levels of AXL and the interaction of this receptor with EGFR leads to sustained mTOR activation enabling cells to overcome PI3Kα inhibition.
AXL/EGFR binding activates the PLCγ-PKC signaling cascade
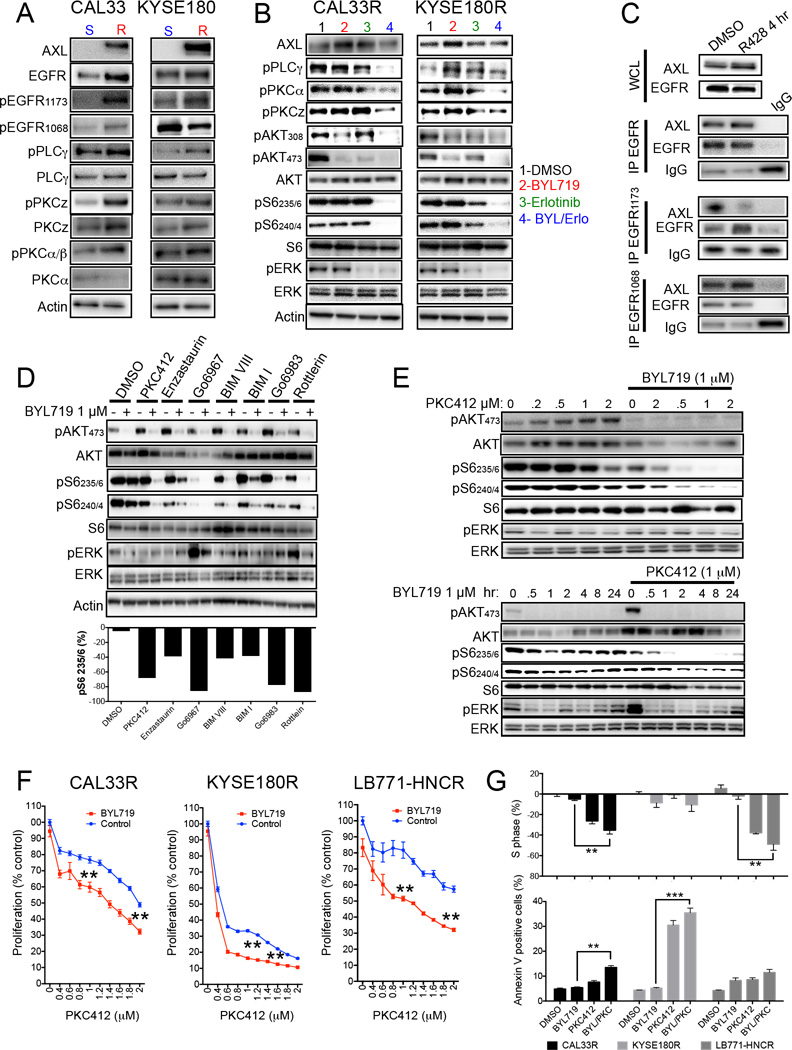
Next we aimed to elucidate the AXL/EGFR downstream signaling pathway(s) that lead(s) to mTOR activation in the presence of PI3Kα blockade. When we investigated the activation status of EGFR in our models, we observed a consistent increase of EGFR phosphorylation at tyrosine 1173 (and not other tyrosine residues such as 1068) associated with AXL overexpression (Figure 6A). Tyrosine1173 in EGFR, when phosphorylated, functions as a docking site for phospholipase Cγ (PLCγ) (Chattopadhyay et al., 1999). PLCγ cleaves phosphatidylinositol 4,5-bisphosphate (PIP2) at the plasma membrane, resulting in the production of the second messengers diacyl glycerol (DAG) and inositol 1,4,5-triphosphate (IP3). DAG activates members of the protein kinase C (PKC) family at the membrane (Rosse et al., 2010). PKC is a family of serine/threonine kinases capable of interacting with a plethora of substrates and ultimately leading to cell proliferation, survival and migration (Disatnik et al., 1994; Mochly-Rosen et al., 2012). PKC has also been shown to activate mTOR in an AKT-independent manner (Fan et al., 2009). Accordingly, the increased EGFR 1173 phosphorylation observed in our cells with acquired resistance to BYL719 was associated with activation of PLCγ and PKCζ the member of the PKC family mostly implicated in mediating EGFR signaling in H&N cancer (Cohen et al., 2006) (Figure 6A). Inhibition of EGFR by erlotinib in the presence of BYL719 was sufficient to block PLCγ/PKC axis in resistant cell lines (Figure 6B).
Figure 6. AXL/EGFR complexes activate mTOR via PLCγ-PKC signaling.
(A) Western blot with the indicated antibodies of protein lysates from the indicated cell lines. S: BYL719-sensitive; R: BYL719-resistant.
(B) Western blot with the indicated antibodies of protein lysates from CAL33R and KYSE180R cells treated for 4 hr with 5 µM of erlotinib, with or without 1 µM BYL719.
(C) Immunoprecipitation (IP) of EGFR1173 and western blot with the indicated antibodies of CAL33R treated for 4 hr with 1 µM R428. WCL: whole cell lysates. IgG: unrelated antibody.
(D) Western blot with the indicated antibodies of protein lysates from KYSE180R cells treated for 4 hr with different PKC inhibitors (0.5 µM PKC412, 1 µM Enzastaurin, 10 µM Go6967, 10 µM BIMVIII, 2 µM BIMI, 10 µM Go6983 and 5 µM Rottlerin) with or without 1 µM BYL719.
(E) Western blot with the indicated antibodies of protein lysates from KYSE180R cells treated with PKC412 and BYL719 as indicated.
(F) Cell viability (4 days) of the indicated cell lines in the presence of 1 µM BYL719 and different concentrations of PKC412.
(G) Analysis of cell cycle (S-phase arrest) and cell survival (annexin V) in BYL719-resistant cells after 48 hr of treatment with 1 µM BYL719, 0.5 µM PKC412 or their combination. Means of two independent experiments performed in duplicate per cell line are shown.
Data are presented as means ± SEM. p value was calculated using two-sided Student’s t test.
**p<0.01. See also Figure S6.
To elucidate the effect of AXL on EGFR phosphorylation, we knocked down AXL in resistant cells and observed a specific reduction in EGFR 1173 phosphorylation (Figure S6A). Similarly, inhibition of AXL by R428 prevented the specific interaction between the residue 1173 of EGFR with AXL, as shown in the immunoprecipitation experiment (Figure 6C; Figure S6B) or by proximity ligation assay (Figure S6C). Furthermore, co-treatment with BYL719 and R428 inhibited PLCγ phosphorylation and cooperated in suppressing S6 phosphorylation in cells with acquired resistance to BYL719 (Figure S6D). These results indicate that AXL can interact with EGFR in a way that leads to the activation of the PLC/PKC axis. To investigate the role of PKC as a mediator of mTOR activation, we treated resistant cells with BYL719 in combination with several inhibitors of PKC. Co-suppression of PI3Kα and PKC invariably resulted in reduction of S6 phosphorylation (Figures 6D and S6E). These effects were dose-dependent and durable in time in both resistant and sensitive cells (Figure 6E; Figure S6F). As a result, combined PI3Kα and PKC inhibition (PKC412) resulted in superior antiproliferative activity compared to single agent treatment in both BYL719-resistant and BYL719-sensitive cells (Figures 6F and 6G; Figure S6G and S6H).
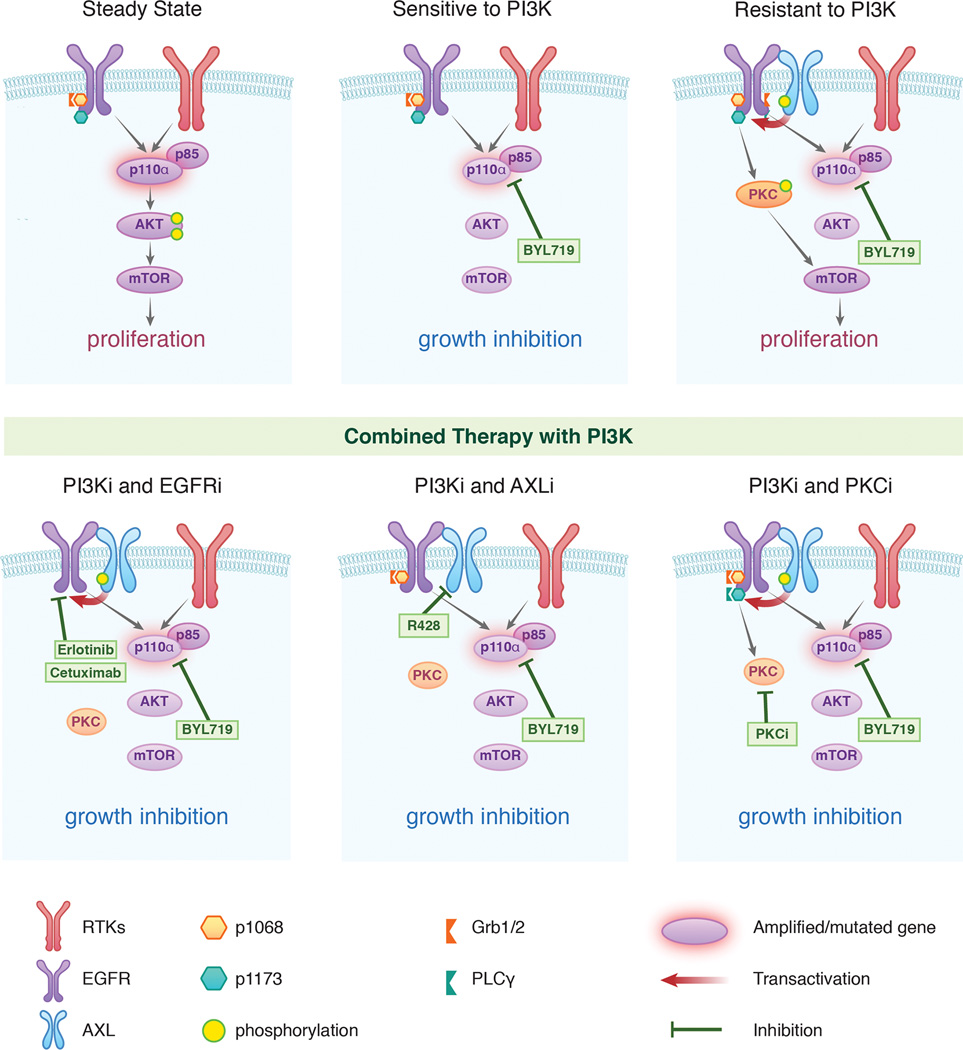
Overall, these data show that up-regulation of AXL and its interaction with EGFR can sustain mTOR activity in the presence of PI3K inhibition. AXL trans-actives EGFR in a ligand independent manner and induces phosphorylation of EGFR on tyrosine 1173. Such event results in PLCγ and PKC activation that, in turn, leads to a PI3K/AKT-independent activation of mTOR (Figure 7).
Figure 7. Scheme summarizing the proposed mechanism by which AXL drives resistance to PI3Kα in SCC.
Up regulation of AXL and its interaction with EGFR leads to PLCγ-PKC activation via phosphorylation of EGFR on tyrosine1173. This results in sustained mTOR activity upon PI3Kα suppression. Combination of PI3Kα inhibition with EGFR, AXL or PKC suppression would prevent this occurrence by blocking the PLCγ/PKC axis resulting in superior antitumor activity compared as monotherapy.
Discussion
In this work we show that H&N and esophageal SCCs escape the antitumor activity of PI3Kα inhibition via up-regulation of AXL and consequent increase in AXL-EGFR interaction that, subsequently, initiates the PLCγ-PKC signaling cascade. The activation of this pathway sustains mTOR activity in the presence of PI3K/AKT inhibition and is prevented by direct inhibition of either EGFR/AXL upstream or PKC downstream activity. We recently reported that residual mTOR activation upon PI3Kα inhibition is sufficient to limit its antitumor activity also in breast cancer (Elkabets et al., 2013), suggesting that this may be a shared feature of PI3Kα resistance. However, we suspect that the mechanisms adopted by cancer cells to bypass PI3K/AKT inhibition and maintain mTOR activation are cancer lineage dependent. AXL is a ubiquitously expressed receptor tyrosine kinase that belongs to the TAM family of receptors (including also TYRO and MER)(Graham et al., 2014). When overexpressed, AXL is capable of transforming fibroblasts and promote xenograft growth of various human cancers (Holland et al., 2005; Janssen et al., 1991). Moreover, expression of AXL has been shown to induce resistance to targeted agents such as the EGFR/HER2 inhibitors lapatinib, erlotinib and cetuximab by re-activating the AKT, ERK and NF-KB pathways in breast, lung and H&N cancers (Brand et al., 2014a; Giles et al., 2013; Liu et al., 2009; Zhang et al., 2012). Here we show that in cell lines, xenograft models and in tumors from SCC patients treated with BYL719, AXL is up-regulated in primary and acquired resistance states.
In our models AXL seems to induce a re-wiring of the EGFR signaling towards the PLCγ-PKC axis by receptor phosphorylation at tyrosine 1173. The specificity of this activation is underscored by the fact that either knockdown or pharmacological inhibition of AXL was sufficient to decrease phosphorylation of this residue in our models. This findings is in accordance with a recent report showing increased phosphorylation of this tyrosine residue in H&N cells with acquired resistance to cetuximab and overexpressing AXL (Brand et al., 2014b). Based on our findings, it is also tempting to speculate that other cancer types characterized by EGFR dependency, such as colon or lung, may adopt a similar mechanism to escape PI3Kα inhibition. Likewise, these tumors may be exquisitely sensitive to dual PI3Kα and EGFR suppression.
It remains to be elucidated whether upfront treatment with dual PI3K and EGFR inhibitors would avoid AXL overexpression and prevent or delay the emergence of resistance. The combination of BYL719 and cetuximab is currently being tested in head and neck cancer patients (Razak et al., unpublished results). Interim results from this pilot study were recently presented and a clinical benefit rate of 28% (4 partial responses and 5 stable diseases out of 32 patients) was reported. However, presence of PIK3CA alterations was not required for patient enrollment (unlike the first-in-man phase I clinical trial (Juric et al., unpublished results), and the status of AXL expression in these tumors has not been reported.
In summary, our study indicates that an AXL-EGFR interaction mediates resistance to PI3Kα inhibition by shifting EGFR signaling toward the PLCγ-PKC axis that maintain mTOR activity in a PI3K/AKT independent fashion. As a result, we propose simultaneous EGFR and PI3Kα blockade as a potential therapeutic strategy to prevent or delay the emergence of resistance to PI3Kα inhibitors in patients with SCC of head and neck and esophagus.
Experimental Procedures
Cell lines and chemical compounds
All other cell lines were purchased from commercial vendors. For the entire list of cell lines see Supplemental Experimental Procedures.
CAL33R, LB771-HNCR, KYSE70R, KYSE180R cells were obtained after chronic exposure to increasing concentrations of BYL719 for ∼8 months. All cells were maintained at 37°C in a humidified atmosphere at 5% CO2. The PI3Kα inhibitor, BYL719, was kindly provided by Novartis. The AXL inhibitor R428, EGFR inhibitor erlotinib, MET inhibitor crizotinib, mTOR inhibitors RAD001 and AZD8055 and PKC inhibitors were purchased from Santa Cruz Chemicals or Selleckchem. All compounds were dissolved in dimethyl sulfoxide (DMSO) for in vitro experiments.
Western blotting and Immunoprecipitation
Cells were washed with ice-cold PBS and scraped into ice-cold RIPA lysis buffer (Cell Signaling) supplemented with phosphatase inhibitor cocktails (Complete Mini, and PhosphoStop (Roche). Lysates were cleared by centrifugation at 13,000 rpm for 10 minutes at 4°C, and supernatants removed and assayed for protein concentration using the Pierce BCA Protein Assay Kit (Thermo Scientific). For immunoprecipitation experiments, cells were lysed using lysis buffer (Cell Signaling #9803) and immunoprecitpiated with anti-EGFR antibodies bind to Protein A magnetic beads (Cell Signaling #8687), after incubation overnight at 4°C. The immunoprecipitate, a sample of the initial whole cell lysate for each condition were then analyzed by western blot.
Thirty-five micrograms of total lysate was resolved on NuPAGE 4–12% Bis-Tris gels (Life Technologies) and electrophoretically transferred to Immobilon transfer membranes (Millipore). Membranes were blocked for 1 hr in 5% nonfat dry milk in TBS-Tween and then hybridized using the primary antibodies in 5% BSA TBS-Tween. Mouse and rabbit horseradish peroxidase (HRP)-conjugated secondary antibodies (1:50,000, Amersham Biosciences) were diluted in 2% nonfat dry milk in TBS-Tween. Protein-antibody complexes were detected by chemiluminescence with SuperSignal West Femto Chemiluminescent Substrate (Thermo Scientific) and images were captured with a G-BOX camera system.
Establishment of tumor xenografts and studies in nude mice
Six-week-old female athymic NU/NU nude mice purchased from Charles River were injected with 1X107 of KYSE180, KYSE180R and CAL33. 5X106 HSC3, 5X106 A253 5X106 HSC2 and 3X106 FaDu subcutaneously in 100 µL culture media/Matrigel (BD Biosciences) 1:5. The patient-derived xenografts belong to Oncotest (http://www.oncotest.com/tumor-models.html) and experiments were conducted at their site (fee for service) in accordance with their Institutional Animal Care and Use Committee.
For cell line-derived xenografts, animals were randomized at a tumor volume of 70 to 120 mm3, approximately 2 to 4 weeks after injection. Animals were randomized to 4–6 groups, with n=8–10 tumors per group. Animals were orally treated daily with BYL719 (25 mg/kg or 50 mg/kg in 0.5% carboxymethylcellulose sodium salt (CMC) (Sigma). Mice were treated every five days with cetuximab (10 mg/kg) via intra-peritoneal injection.
Xenografts were measured with digital caliper, and tumor volumes were determined with the formula: (length × width2) × (π/6). At the end of the experiment, animals were euthanized using CO2 inhalation. Tumor volumes are plotted as means ± SEM.
Mice were maintained and treated in accordance with Institutional Guidelines of Massachusetts General Hospital and Memorial Sloan Kettering Cancer Center. Mice were housed in air-filtered laminar flow cabinets with a 12-hr light cycle and food and water ad libitum. Animal experiments were approved by the Institutional Animal Care and Use Committee from both Massachusetts General Hospital and Memorial Sloan Kettering Cancer Center.
Patients
Tumor response was assessed according to the RECIST criteria. All measurable lesions up to a maximum of 5 lesions per organ and 10 lesions in total, representative of all involved organs, were identified as target lesions and recorded and measured at baseline, and then followed throughout the study. A sum of the longest diameter for all target lesions at baseline was used as a reference by which the objective tumor response was characterized. Pretreatment biopsies were obtained within 2 weeks of starting the study agent. Progression biopsies were performed at the time of disease progression. These procedures were approved by the IRB of Massachusetts General Hospital and by the ethical committee of the Vall d’Hebron University Hospital. Informed consent was obtained from all subjects.
Secreted protein screen
The secretome screen was assessed as previously described (Harbinski et al., 2012). Briefly, secreted protein library cDNAs were reverse transfected into HEK293T cells with FuGENE HD (4:1 ratio transfection reagent to DNA) and incubated for 4 days to allow accumulation of secreted proteins in supernatant. The supernatant was then transferred to CAL33 and SNU-1214 cells, followed by addition of BYL719 to a final concentration of 3 µM. After 96 hr, growth was measured using CellTiter-Glo.
Statistical analysis
Two-way t-test was done using GraphPad Prism (GraphPad Software). Error bars represent the SEM. *p<0.05, **p<0.01, ***p<0.005. All the in vitro experiments were repeated at least three times. Some in vivo experiments were run in duplicate with at least n=6 for each treatment arm.
Supplementary Material
Significance.
The therapeutic options for patients with H&N and esophageal SCC are limited and their survival is poor. These tumors have frequent mutations and amplifications of PIK3CA, which encodes the p110α subunit of PI3Kα. Clinical activity of PI3Kα inhibitors under clinical development in these tumors has been reported. However, as with many other targeted therapies, clinical resistance eventually develops and dissecting the underlying mechanisms driving resistance could lead to improved treatment approaches. Here, we show that the activity of these compounds may be limited by the presence of AXL that, interacting with EGFR, initiates a survival mechanism resulting in drug resistance. Our work provides the basis to explore dual PI3Kα and EGFR blockade in patients with H&N and esophageal SCC.
Acknowledgment
We would like to acknowledge the MSKCC Genomics Core Laboratory and Caitlin J. Jones and Mono Pirun from the MSKCC Bioinformatics Core for assistance in performing and analyzing the RNA sequencing experiments. The Molecular Cytology Core Facility; Yarilin Dmitry and Turkekul Mesruh from MSKCC for sample preparation and staining of IHC, Sho Fujisawa and Yevgeniy Romin for confocal microscope analysis.
Funding
This work was funded by the Cycle for Survival (to J.B. and M.S.), Banco Bilbao Vizcaya Argentaria (BBVA) Foundation (Tumor Biomarker Research Program) and a Molecular Cytology Core Facility-Core grant (P30 CA0087748). M.E. is an International Sephardic Education Foundation postdoctoral fellow. D.J. is funded by a National Institute of Health Training Grant (T32 CA-71345-15). T.G.B. is funded by the NIH Director’s Award DP2 CA174497.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Number
The RNAseq count data have been deposited in the GEO database under accession numbers GSE61515.
Author contributions
M.E., M.S., and J.B. designed the research. M.E., E.P., Q.S., S.B, N.M., J.J.Y., R.D., Y.C., H.W., J.L. performed the experiments. D.J., A.O.B and J.R. provided the human tissue samples and clinical data. A.H., Q.S., A.T. performed the secreted protein screening. M.L. performed the shRNA screen analysis. R.A.P. performed the RNAseq analysis. J.M.G., D.A.K., R.R. and C.T. processed the human tissue samples and performed the IHC staining for AXL. R.A.G. scored and quantified IHC experiments. T.G.B. and N.R. assisted with research design. M.E. and M.S. prepared the figures. M.E., M.S., and J.B. wrote the manuscript.
Conflict of interest
J.B., D.J. and N.R. have consulted for Novartis Pharmaceuticals. A.H., Q.S., J.J.Y., R.D., Y.C., A.T., H.W., J.L., and M.L. are full-time employees of Novartis Pharmaceuticals.
Data and materials availability
BYL719 was obtained with a material transfer agreement (MTA) from Novartis Pharmaceuticals. Requests for BYL719 will be accommodated upon execution of an MTA with Novartis.
References
- Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akagi I, Miyashita M, Makino H, Nomura T, Hagiwara N, Takahashi K, Cho K, Mishima T, Ishibashi O, Ushijima T, et al. Overexpression of PIK3CA is associated with lymph node metastasis in esophageal squamous cell carcinoma. Int J Oncol. 2009;34:767–775. doi: 10.3892/ijo_00000202. [DOI] [PubMed] [Google Scholar]
- Baselga J, Trigo JM, Bourhis J, Tortochaux J, Cortes-Funes H, Hitt R, Gascon P, Amellal N, Harstrick A, Eckardt A. Phase II multicenter study of the antiepidermal growth factor receptor monoclonal antibody cetuximab in combination with platinum-based chemotherapy in patients with platinum-refractory metastatic and/or recurrent squamous cell carcinoma of the head and neck. J Clin Oncol. 2005;23:5568–5577. doi: 10.1200/JCO.2005.07.119. [DOI] [PubMed] [Google Scholar]
- Bonner JA, Harari PM, Giralt J, Azarnia N, Shin DM, Cohen RB, Jones CU, Sur R, Raben D, Jassem J, et al. Radiotherapy plus cetuximab for squamous-cell carcinoma of the head and neck. N Engl J Med. 2006;354:567–578. doi: 10.1056/NEJMoa053422. [DOI] [PubMed] [Google Scholar]
- Brand TM, Iida M, Stein AP, Corrigan KL, Braverman CM, Luthar N, Toulany M, Gill PS, Salgia R, Kimple RJ, Wheeler DL. AXL mediates resistance to cetuximab therapy. Cancer Res. 2014a;74:5152–5164. doi: 10.1158/0008-5472.CAN-14-0294. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Byers LA, Diao L, Wang J, Saintigny P, Girard L, Peyton M, Shen L, Fan Y, Giri U, Tumula PK, et al. An epithelial-mesenchymal transition gene signature predicts resistance to EGFR and PI3K inhibitors and identifies Axl as a therapeutic target for overcoming EGFR inhibitor resistance. Clin Cancer Res. 2013;19:279–290. doi: 10.1158/1078-0432.CCR-12-1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattopadhyay A, Vecchi M, Ji Q, Mernaugh R, Carpenter G. The role of individual SH2 domains in mediating association of phospholipase C-gamma1 with the activated EGF receptor. J Biol Chem. 1999;274:26091–26097. doi: 10.1074/jbc.274.37.26091. [DOI] [PubMed] [Google Scholar]
- Cohen EE, Lingen MW, Zhu B, Zhu H, Straza MW, Pierce C, Martin LE, Rosner MR. Protein kinase C zeta mediates epidermal growth factor-induced growth of head and neck tumor cells by regulating mitogen-activated protein kinase. Cancer Res. 2006;66:6296–6303. doi: 10.1158/0008-5472.CAN-05-3139. [DOI] [PubMed] [Google Scholar]
- Disatnik MH, Hernandez-Sotomayor SM, Jones G, Carpenter G, Mochly-Rosen D. Phospholipase C-gamma 1 binding to intracellular receptors for activated protein kinase C. Proc Natl Acad Sci U S A. 1994;91:559–563. doi: 10.1073/pnas.91.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkabets M, Vora S, Juric D, Morse N, Mino-Kenudson M, Muranen T, Tao J, Campos AB, Rodon J, Ibrahim YH, et al. mTORC1 Inhibition Is Required for Sensitivity to PI3K p110alpha Inhibitors in PIK3CA-Mutant Breast Cancer. Sci Transl Med. 2013;5 doi: 10.1126/scitranslmed.3005747. 196ra199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9:550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- Fan QW, Cheng C, Knight ZA, Haas-Kogan D, Stokoe D, James CD, McCormick F, Shokat KM, Weiss WA. EGFR signals to mTOR through PKC and independently of Akt in glioma. Science signaling. 2009;2 doi: 10.1126/scisignal.2000014. ra4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z, Masui H, Altas I, Mendelsohn J. Blockade of epidermal growth factor receptor function by bivalent and monovalent fragments of 225 anti-epidermal growth factor receptor monoclonal antibodies. Cancer Res. 1993;53:4322–4328. [PubMed] [Google Scholar]
- Fritsch C, Huang A, Chatenay-Rivauday C, Schnell C, Reddy A, Liu M, Kauffmann A, Guthy D, Erdmann D, De Pover A, et al. Characterization of the novel and specific PI3Kalpha inhibitor NVP-BYL719 and development of the patient stratification strategy for clinical trials. Mol Cancer Ther. 2014;13:1117–1129. doi: 10.1158/1535-7163.MCT-13-0865. [DOI] [PubMed] [Google Scholar]
- Furet P, Guagnano V, Fairhurst RA, Imbach-Weese P, Bruce I, Knapp M, Fritsch C, Blasco F, Blanz J, Aichholz R, et al. Discovery of NVP-BYL719 a potent and selective phosphatidylinositol-3 kinase alpha inhibitor selected for clinical evaluation. Bioorg Med Chem Lett. 2013;23:3741–3748. doi: 10.1016/j.bmcl.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Giles KM, Kalinowski FC, Candy PA, Epis MR, Zhang PM, Redfern AD, Stuart LM, Goodall GJ, Leedman PJ. Axl mediates acquired resistance of head and neck cancer cells to the epidermal growth factor receptor inhibitor erlotinib. Mol Cancer Ther. 2013;12:2541–2558. doi: 10.1158/1535-7163.MCT-13-0170. [DOI] [PubMed] [Google Scholar]
- Graham DK, DeRyckere D, Davies KD, Earp HS. The TAM family: phosphatidylserine-sensing receptor tyrosine kinases gone awry in cancer. Nat Rev Cancer. 2014;14:769–785. doi: 10.1038/nrc3847. [DOI] [PubMed] [Google Scholar]
- Harbinski F, Craig VJ, Sanghavi S, Jeffery D, Liu L, Sheppard KA, Wagner S, Stamm C, Buness A, Chatenay-Rivauday C, et al. Rescue screens with secreted proteins reveal compensatory potential of receptor tyrosine kinases in driving cancer growth. Cancer Discov. 2012;2:948–959. doi: 10.1158/2159-8290.CD-12-0237. [DOI] [PubMed] [Google Scholar]
- Hennessy BT, Smith DL, Ram PT, Lu Y, Mills GB. Exploiting the PI3K/AKT pathway for cancer drug discovery. Nature reviews Drug discovery. 2005;4:988–1004. doi: 10.1038/nrd1902. [DOI] [PubMed] [Google Scholar]
- Holland SJ, Pan A, Franci C, Hu Y, Chang B, Li W, Duan M, Torneros A, Yu J, Heckrodt TJ, et al. R428, a selective small molecule inhibitor of Axl kinase, blocks tumor spread and prolongs survival in models of metastatic breast cancer. Cancer Res. 2010;70:1544–1554. doi: 10.1158/0008-5472.CAN-09-2997. [DOI] [PubMed] [Google Scholar]
- Holland SJ, Powell MJ, Franci C, Chan EW, Friera AM, Atchison RE, McLaughlin J, Swift SE, Pali ES, Yam G, et al. Multiple roles for the receptor tyrosine kinase axl in tumor formation. Cancer Res. 2005;65:9294–9303. doi: 10.1158/0008-5472.CAN-05-0993. [DOI] [PubMed] [Google Scholar]
- Janssen JW, Schulz AS, Steenvoorden AC, Schmidberger M, Strehl S, Ambros PF, Bartram CR. A novel putative tyrosine kinase receptor with oncogenic potential. Oncogene. 1991;6:2113–2120. [PubMed] [Google Scholar]
- Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA-Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- Kamangar F, Dores GM, Anderson WF. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol. 2006;24:2137–2150. doi: 10.1200/JCO.2005.05.2308. [DOI] [PubMed] [Google Scholar]
- Lin DC, Hao JJ, Nagata Y, Xu L, Shang L, Meng X, Sato Y, Okuno Y, Varela AM, Ding LW, et al. Genomic and molecular characterization of esophageal squamous cell carcinoma. Nature Genet. 2014;46:467–473. doi: 10.1038/ng.2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Greger J, Shi H, Liu Y, Greshock J, Annan R, Halsey W, Sathe GM, Martin AM, Gilmer TM. Novel mechanism of lapatinib resistance in HER2-positive breast tumor cells: activation of AXL. Cancer Res. 2009;69:6871–6878. doi: 10.1158/0008-5472.CAN-08-4490. [DOI] [PubMed] [Google Scholar]
- Lui VW, Hedberg ML, Li H, Vangara BS, Pendleton K, Zeng Y, Lu Y, Zhang Q, Du Y, Gilbert BR, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3:761–769. doi: 10.1158/2159-8290.CD-13-0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer AS, Miller MA, Gertler FB, Lauffenburger DA. The receptor AXL diversifies EGFR signaling and limits the response to EGFR-targeted inhibitors in triple-negative breast cancer cells. Sci Signal. 2013;6:ra66. doi: 10.1126/scisignal.2004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D, Das K, Grimes KV. Protein kinase C, an elusive therapeutic target? Nat. Rev. Drug. Discov. 2012;11:937–957. doi: 10.1038/nrd3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paccez JD, Vogelsang M, Parker MI, Zerbini LF. The receptor tyrosine kinase Axl in cancer: biological functions and therapeutic implications. Int. J. Cancer. 2014;134:1024–1033. doi: 10.1002/ijc.28246. [DOI] [PubMed] [Google Scholar]
- Rosse C, Linch M, Kermorgant S, Cameron AJ, Boeckeler K, Parker PJ. PKC and the control of localized signal dynamics. Nat. Rev. Mol. Cell. Biol. 2010;11:103–112. doi: 10.1038/nrm2847. [DOI] [PubMed] [Google Scholar]
- Soderberg O, Leuchowius KJ, Gullberg M, Jarvius M, Weibrecht I, Larsson LG, Landegren U. Characterizing proteins and their interactions in cells and tissues using the in situ proximity ligation assay. Methods. 2008;45:227–232. doi: 10.1016/j.ymeth.2008.06.014. [DOI] [PubMed] [Google Scholar]
- Song Y, Li L, Ou Y, Gao Z, Li E, Li X, Zhang W, Wang J, Xu L, Zhou Y, et al. Identification of genomic alterations in oesophageal squamous cell cancer. Nature. 2014;509:91–95. doi: 10.1038/nature13176. [DOI] [PubMed] [Google Scholar]
- Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence MS, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straussman R, Morikawa T, Shee K, Barzily-Rokni M, Qian ZR, Du J, Davis A, Mongare MM, Gould J, Frederick DT, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487:500–504. doi: 10.1038/nature11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suda T, Hama T, Kondo S, Yuza Y, Yoshikawa M, Urashima M, Kato T, Moriyama H. Copy number amplification of the PIK3CA gene is associated with poor prognosis in non-lymph node metastatic head and neck squamous cell carcinoma. BMC Cancer. 2012;12:416. doi: 10.1186/1471-2407-12-416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermorken JB, Mesia R, Rivera F, Remenar E, Kawecki A, Rottey S, Erfan J, Zabolotnyy D, Kienzer HR, Cupissol D, et al. Platinum-based chemotherapy plus cetuximab in head and neck cancer. N. Engl. J. Med. 2008;359:1116–1127. doi: 10.1056/NEJMoa0802656. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Wilson TR, Fridlyand J, Yan Y, Penuel E, Burton L, Chan E, Peng J, Lin E, Wang Y, Sosman J, et al. Widespread potential for growth-factor-driven resistance to anticancer kinase inhibitors. Nature. 2012;487:505–509. doi: 10.1038/nature11249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, Abdel-Rahman M, Wang X, Levine AD, Rho JK, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–860. doi: 10.1038/ng.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.