Abstract
Filamentous growth is one of the key features of pathogenic fungi during the early infectious phase. The pseudohyphal development of yeast Saccharomyces cerevisiae shares similar characteristics with hyphae elongation in pathogenic fungi. The expression of FLO11 is essential for adhesive growth and filament formation in yeast and is governed by a multilayered transcriptional network. Here we discovered a role for the histone acetyltransferase general control nonderepressible 5 (Gcn5) in regulating FLO11-mediated pseudohyphal growth. The expression patterns of FLO11 were distinct in haploid and diploid yeast under amino acid starvation induced by 3-amino-1,2,4-triazole (3AT). In diploids, FLO11 expression was substantially induced at a very early stage of pseudohyphal development and decreased quickly, but in haploids, it was gradually induced. Furthermore, the transcription factor Gcn4 was recruited to the Sfl1-Flo8 toggle sites at the FLO11 promoter under 3AT treatment. Moreover, the histone acetylase activity of Gcn5 was required for FLO11 induction. Finally, Gcn5 functioned as a negative regulator of the noncoding RNA ICR1, which is known to suppress FLO11 expression. Gcn5 plays an important role in the regulatory network of FLO11 expression via Gcn4 by downregulating ICR1 expression, which derepresses FLO11 for promoting pseudohyphal development.
1. Introduction
Fungi can alternate their cellular morphology between unicellular yeast and multicellular hyphae forms in response to environmental stimuli, a process known as dimorphic switching. This phenomenon is intimately linked to pathogens of animals or plants and their pathogenicity [1, 2]. The pathogenic fungus Candida albicans switches from the usual unicellular yeast-like form to a multicellular invasive-filamentous form when it infects host cells [3]. The budding yeast Saccharomyces cerevisiae can switch between different morphological forms under various stress conditions. By doing so, yeast features differential growth modes according to adaptive needs to confer cell protection and enhance dissemination and substrate colonization [4, 5].
In S. cerevisiae, cell-cell and cell-surface adhesion are required for many developmental processes including mating [6], haploid invasive growth [7, 8], diploid pseudohyphal development [9, 10], and biofilm formation [11, 12]. Each of these events is initiated by distinct signals that are coupled to the expression of specific cell surface proteins by corresponding signaling pathways (for reviews, see [13, 14]). For example, in response to nutrient stress such as nitrogen starvation, budding yeast develops two types of cell forms: adhesive/invasive growth for haploids and pseudohyphae for diploids [8, 9]. For both types of cell growth, yeast features increased cell length, change in polarity, and augmented cell-cell adhesion; as in pathogens, this kind of morphological development is essential for host-cell attachment, virulence, and tissue invasiveness [15].
The flocculin protein, Flo11, plays a central role in pseudohyphal growth and biofilm formation [11, 16–20]. In yeast cells with the FLO11 deleted (flo11Δ), diploids do not form pseudohyphae and haploids lose agar invasiveness [17]. Unlike most genes in yeast, the upstream region and promoter of FLO11 consist of an unusually long sequence spanning more than 3 kb and harboring binding sites for transcription factors involved in mitogen-activated protein kinase (MAPK) and protein kinase A (PKA) signaling pathways and the general control nonderepressible (GCN) response pathway [16, 21–26]. Genetic analyses and β-galactosidase reporter assays demonstrated that several transcriptional factors, including repressors and activators, coordinately regulate FLO11 expression [24, 27–30]. Ste12 and Tec1 are nuclear transcription factors in the MAPK signaling cascade activating FLO11 expression under nitrogen starvation [31]. The protein kinase Tpk2 in the cAMP-dependent signaling pathway is required for transcriptional activation of FLO11 expression mediated by the transcriptional factors Flo8 and Gcn4 [21, 25, 26, 32]. GCN pathway is involved in both morphogenesis and biofilm formation in response to amino acid deficiency. The sensor kinase Gcn2 regulates expression of the transcriptional activator Gcn4 under conditions of amino acid deprivation, which consequently activates FLO11 expression and pseudohyphal growth [21, 33].
Chromatin is composed of histone octamers (histones H2A, H2B, H3, and H4) that associate with DNA in nucleosomes. These structures can both repress and activate transcription and other genomic processes, such as cell progression, DNA replication, recombination, and repair, depending on the positioning of nucleosomes relative to binding sites for activators and repressors [34, 35]. A key event in the regulation of eukaryotic gene expression is the posttranslational modification of nucleosomal histones, which converts chromosomes into transcriptionally active or inactive chromatin. A gene-specific and time-dependent order of events is believed to link chromatin structure modifications and transcription activation. Chromatin-modifying enzymes mark histone residues and change nucleosome conformation, which allows the transcriptional machinery to transcribe or repress genes [36, 37].
Many epigenetically regulated genes including FLO11 can be considered transcriptional switches because they have two inheritable expression states, “ON” and “OFF” [38]. FLO11 is generally silent in the yeast form unless transcriptional activation is induced by various environmental stresses for subsequent promotion of invasive growth in haploid cells and filamentous growth in diploid yeast. FLO11 expression is repressed by the histone deacetylase (HDAC) Hda1, and importantly the Hda1-mediated gene silencing of several FLO genes depends on subtelomeric chromatin position [39]. However, a histone deacetylase complex containing Rpd3L plays a seemly paradoxical role in activating instead of repressing FLO11 expression [40]. In addition, the chromatin-remodeling complexes Swi/Snf and Rsc can regulate FLO11 expression [41, 42].
Two long noncoding RNAs (ncRNAs), ICR1 (interfering Crick RNA) and PWR1 (promoting Watson RNA), are transcribed by RNA polymerase II in an opposite direction within an overlapped sequence at the upstream promoter region of FLO11 [43, 44]. Rpd3L acts with two transcription factors, Flo8 (an activator) and Sfl1 (a repressor), to regulate the mutually exclusive transcription of PWR1 and ICR1. Sfl1 suppresses PWR1 transcription by competing with Flo8 to bind the “toggle” sites at the PWR1 promoter and then induces ICR1 expression, which then suppresses FLO11 transcription via a promoter exclusion mechanism. When Rpd3L binds to the FLO11 promoter, Sfl1 is excluded from the PWR1 promoter, which leads to Flo8 associating with the toggle sites to activate PWR1 and concomitantly blocks ICR1 transcription. Consequently, Rpd3L and Flo8 activate FLO11 expression by regulating the reciprocal transcription of a pair of cis-interfering ncRNAs [43, 45].
Gcn5 is a histone acetyltransferase (HAT) that functions as a coactivator in transcriptional regulation and mainly contributes to transcriptional activation and substrate specificity combined with other histone modifiers [46, 47]. Gcn5 modifies several of the aminoterminal lysine residues of histones with acetylation and is the catalytic subunit for three chromatin-modifying complexes, ADA, SAGA, and SLIK/SALSA, to regulate a wide arrange of genes both positively and negatively [48, 49]. The histone acetylation activity of Gcn5 is required for Gcn4 to act as an efficient transcription factor at gene promoters [50]. Gcn4 recruits Gcn5 and its multisubunit complexes to the promoters of a specific subset of genes, such as HIS3, for transcriptional activation [51, 52]. Although Gcn5 was found involved in FLO11 expression under amino acid starvation, such regulation was observed only in haploid strains [42].
The mechanism to control the precisely regulated switch from repression to activation of FLO11 represents a critical question in yeast dimorphism. In this study, we demonstrate that Gcn4 and Gcn5 are new components in controlling FLO11 expression and pseudohyphal development in diploid yeast under amino acid starvation by regulating ncRNA ICR1 transcription at the promoter of FLO11.
2. Materials and Methods
2.1. Plasmids and Yeast Strains
The yeast strains used in this study were all in a S. cerevisiae Σ1278b genetic background and are described in Table 1. Standard yeast culture medium was prepared as described previously for invasive growth in YPD (Bacto Peptone; 4% Bacto Peptone, 2% yeast extract, 2% peptone, and 2% glucose) and for filamentous growth in SLAD (synthetic medium with a low concentration of ammonium (6.7 g/L yeast nitrogen base without amino acids and ammonium sulfate, 2% glucose, and 2% washed Bacto Agar)) [8, 9]. Amino acid starvation medium was prepared by supplementing minimal media (1.7 g/L yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose) with 10 mM 3-amino-1,2,4-triazole (3AT) [47]. Yeast transformation involved the lithium acetate method and all yeast manipulation followed standard methods [53, 54]. The plasmids carrying MATa1 (pRS315-MATa1) or pRS414-MATαAT in haploids were constructed as described previously to generate pseudodiploid strains [55]. The construction of Gcn5 mutants was performed as described previously [47]. The vector pRS316 was used to generate a wild type GCN5 (pRS316+GCN5-WT) and the single amino acid substitution of Glu to Gln (E173Q; pRS316-yGCN5-E173Q) representing functional Gcn5 and a catalytically dead mutant of GCN5, respectively.
Table 1.
Yeast strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| SLY16 | MATa, ura3–52 leu2∷hisG his3∷hisG trp1∷hisG | Laboratory of G. Fink |
|
| ||
| SLY17 | MATα, ura3–52 leu2∷hisG his3∷hisG trp1∷hisG | Laboratory of G. Fink |
|
| ||
| SLY134 | MATa, ura3–52 leu2∷hisG his3∷hisG flo11Δ∷HIS3 | [39] |
|
| ||
| SLY506 | MATa, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG gcn5Δ∷HIS3 | This study |
|
| ||
| SLY507 | MATa, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG snf1Δ∷TRP1 | This study |
|
| ||
| SLY549 | MATα, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG gcn5Δ∷HIS3 | This study |
|
| ||
| SLY551 | MATa, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG gcn5Δ∷HIS3 + [pRS316-GCN5] | This study |
|
| ||
| SLY553 | MATa, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG gcn5Δ∷HIS3 + [pRS316] | This study |
|
| ||
| SLY555 | MATa, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG gcn5Δ∷HIS3 + [pRS316-gcn5-E173Q] | This study |
|
| ||
| SLY565 | MATa, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG + [pRS415-MATα2] | This study |
|
| ||
| SLY569 | MATα, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG + [pRS315-MATa1] | This study |
|
| ||
| SLY570 | MATα, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG gcn5Δ∷HIS3 + [pRS315-MATa1] | This study |
|
| ||
| SLY594 | MATa, ura3–52 leu2∷hisG his3∷hisG trp1∷hsiG gcn5Δ∷HIS3 + [pRS415-MATα2] | This study |
|
| ||
| SLY678 | MATα, ura3–52 leu2∷hisG trp1∷hisG GCN4-2Flag∷TRP1 | This study |
|
| ||
| SLY848 | MATa/α, ura3–52/ura3–52 leu2∷hisG/leu2∷hisGhis3∷hisG/HIS3 GCN4-2Flag∷TRP1/trp1∷hisG | This study |
|
| ||
| SLY1062 | MATa/α, ura3–52/ura3–52leu2∷hisG/leu2∷hisGhis3∷hisG/his3∷hisG trp1∷hisG/trp1∷hisGgcn5Δ∷HIS3/gcn5Δ∷HIS3 + [pRS316-GCN5] | This study |
|
| ||
| SLY1063 | MATa/α, ura3–52/ura3–52leu2∷hisG/leu2∷hisGhis3∷hisG/his3∷hisG trp1∷hisG/trp1∷hisGgcn5Δ∷HIS3/gcn5Δ∷HIS3 + [pRS316] | This study |
|
| ||
| SLY1064 | MATa/α, ura3–52/ura3–52leu2∷hisG/leu2∷hisGhis3∷hisG/his3∷hisG trp1∷hisG/trp1∷hisGgcn5Δ∷HIS3/gcn5Δ∷HIS3 + [pRS316-gcn5-E173Q] | This study |
2.2. Invasive Growth and Pseudohyphal Formation Assays
Measurement of invasive growth of yeast cells was as described [55]. Briefly, a 3 mL YPD liquid culture was grown until it reached OD600 0.8–1 and then 5 μL was plated into two YPD agar plates and incubated for 4 d at 30°C. One of the plates was used for measuring “total growth” and the other for “invasive growth” remaining on the agar media after two washes as described. Total growth was measured by excision of the yeast cells together with agar and liquification and quantification of cells by OD600. The second plate was washed with 9 mL distilled water by use of a microtiter plate shaker at 5 rpm for 5 min each time. The cell number was measured as described for the total growth. The analysis of pseudohyphal filament formation was as described [9]. Yeast cells were streaked on SLAD plates to score phenotype after growth at 30°C for 6 d. Plates were photographed by use of a digital camera attached to a stereomicroscope (ZEISS) and the number of colonies forming filaments was counted.
2.3. RNA Preparation, Reverse Transcription, and Real-Time qPCR
Total RNA was extracted from yeast cells using a total RNA Mini Kit (Geneaid) as described [56, 57]. To remove potential contamination of genomic DNA, total purified RNA was incubated with RQ1 (Promega) DNase at 37°C for 40 min and then inactivated at 65°C for 10 min. Reverse transcription involved a cDNA synthesis kit (Epicentre) in an RNase-free environment. RNA underwent initial incubation at 65°C for 5 min with oligo-dT. Then, the reaction mixture including buffer, dNTPs, DTT, ribonuclease inhibitor, and MMLV reverse transcriptase was incubated at 37°C for 60 min and then inactivated at 85°C for 5 min. cDNA prepared from triplicate biological samples was used for PCR analysis. Real-time quantitative PCR (qPCR) involved use of Stratagene Mx3000p (Agilent Technologies) and Kapa SYBR Fast Universal qPCR Kit (Kapa Biosystems). The primers used in this study are available upon request.
2.4. Chromatin-Immunoprecipitation (ChIP) Assay
ChIP assay was performed as described previously [56, 57]. Briefly, the haploid and diploid Gcn4-Flag cells were fixed with formaldehyde for 2 h. To maintain a proper size of fragmented chromatin, we optimized the sonication parameters to shear DNA to an average size of about 200 bp corresponding to 1 or 2 nucleosomes. Chromatin solution containing 1 mg whole cell extract was immunoprecipitated (IP) with anti-Flag M2 (Sigma-Aldrich) antibody and purified with protein G sepharose (Millipore). The precipitated DNA was analyzed by real-time qPCR. The signal for each gene primer pair in the immunoprecipitation was normalized to that of the input and then divided by the control vector to determine the fold change. Quantification of data, indicated by mean ± SD, was based on the number of independent biological and/or experimental replicates described in the figure legends.
3. Results and Discussion
3.1. Gcn5 Is Involved in FLO11 Expression and Pseudohyphal Development in S. cerevisiae
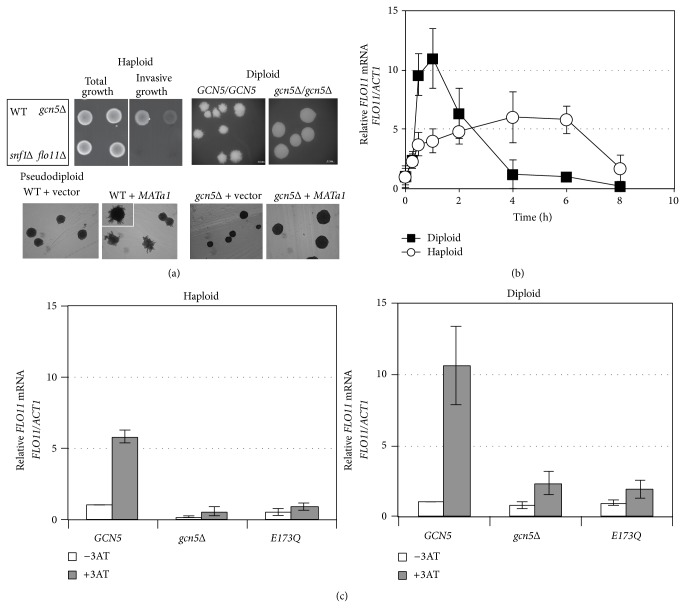
To investigate the role of histone modifiers in pseudohyphal development by controlling FLO11 gene expression in yeast S. cerevisiae, we examined the phenotype of haploid and diploid strains with deletions of GCN5 (gcn5Δ) and SNF1 (snf1Δ), which encode histone acetyltransferases and histone kinase, respectively. In haploids, wild type (WT), but not gcn5Δ, snf1Δ, and flo11Δ, remained on agar plate medium after being washed with water, so GCN5 and SNF1 are required for invasive growth (Figure 1(a)). In diploids, the homozygous mutant gcn5Δ/gcn5Δ did not develop the pseudohyphae shown in the WT (Figure 1(a)). In addition, we constructed a pseudodiploid by introducing a single MATa1 plasmid to the α haploids [55] and found that the pseudohyphal development was suppressed in gcn5Δ (Figure 1(a)). These observations convincingly support the essential role of the histone acetyltransferase Gcn5 in haploid invasive and diploid filamentous growth.
Figure 1.
Gcn5 is involved in pseudohyphal development and FLO11 expression. (a) Gcn5 is required for invasive growth in haploids and filamentous growth in diploids. Haploid cells of wild type (WT) yeast and deletion mutants (gcn5Δ, snf1Δ, and flo11Δ) were grown on YPD agar medium for 4 d at 30°C before analyzing invasive growth (haploid, top left panel). Cells grown on agar surface before (total growth) and remaining after being rinsed with water (invasive growth) are shown. The WT (GCN5/GCN5) and homozygous gcn5Δ diploids (gcn5Δ/gcn5Δ) were grown on nitrogen starvation agar medium (SLAD) for 6 d at 30°C before filamentous growth was recorded (diploid, top right panel). Shows filamentous growth of colonies on SLAD agar medium for diploid WT (GCN5 + vector) and gcn5Δ (gcn5Δ + vector) and pseudodiploid WT (GCN5 + Mat a1) and gcn5Δ (gcn5Δ + Mat a1) (pseudodiploid, bottom panel). Scale bar: 0.5 mm. (b) The expression patterns of FLO11 in haploid and diploid yeast in response to 3AT. The isogeneic haploid and diploid strains were grown at 30°C in minimal medium to 0.8 OD600 (time 0) and induced with 10 mM 3AT. Total RNA was prepared from samples collected at the times indicated. Quantitative RT-PCR (RT-qPCR) analysis of the mRNA levels of FLO11 normalized to that of ACT1. (c) The histone acetyltransferase activity of Gcn5 is required for induced FLO11 expression by 3AT. Total RNA was prepared from samples collected at 2 h (haploid) or 1 h (diploid) after the addition of 3AT. The gcn5 deletion haploid (left panel) or diploid (right panel) strains carrying a wild type GCN5 (GCN5), vector (gcn5Δ), or a catalytic dead mutant (E173Q) were grown in repressed (−3AT) or induced condition (+3AT, 10 mM). RT-qPCR analysis of the mRNA levels of FLO11 normalized to that of ACT1 and compared with the WT (GCN5) without 3AT. Data are mean ± SD from 3 biological repeats ((b)-(c)).
FLO11 is required for both haploid invasive and diploid filamentous growth, generally considered to be mediated by distinct pathways [17, 39]. We examined the induction of FLO11 expression in both haploids and diploids under an amino acid starvation condition induced by 3AT. In a time course experiment, the induction patterns of FLO11 mRNA expression in diploids and haploids differed. In haploids, FLO11 transcription was activated gradually upon 3AT treatment, with approximately 8-fold enhancement 4 h after induction followed by a decrease after 6 h (Figure 1(b)). In diploids, FLO11 expression was quickly induced by 10-fold in <30 min and reached approximately 12-fold enhancement by 1 h and then was reduced to the basal level at 4 h after 3AT induction (Figure 1(b)). This was an unexpected observation, because FLO11 was previously not considered to be expressed in diploids and tetraploids [58, 59] and the induction of FLO11 by nitrogen starvation was found much lower in diploids than haploids [21]. This discrepancy may result from different time courses in sampling.
3.2. HAT Activity of Gcn5 Is Required for FLO11 Expression Induced by Nitrogen Starvation in Dimorphic Growth
Gcn5 was found to regulate FLO11 expression under amino acid starvation in haploid yeast [42]. However, whether Gcn5-mediated FLO11 expression is involved in dimorphic growth was unclear. We previously showed that a mutation (E173Q) in the catalytic domain of Gcn5 disrupted its HAT activity and resulted in poor growth under nitrogen starvation [47]. To ascertain whether Gcn5 and/or its HAT activity is essential for transcriptional regulation of FLO11 with amino acid starvation, we first measured FLO11 transcripts in deletion (gcn5Δ) and catalytically dead (E173Q) Gcn5 strains with and without 3AT. 3AT-induced FLO11 expression was significantly impaired in gcn5Δ in both haploids and diploids at 2 and 1 h after treatment, respectively (Figure 1(c)). Furthermore, HAT activity of Gcn5 was critically important for the full induction of FLO11 by 3AT (Figure 1(c)). Therefore, Gcn5 as an HAT is required for transcriptional activation of FLO11 in both haploids and diploids under amino acid starvation.
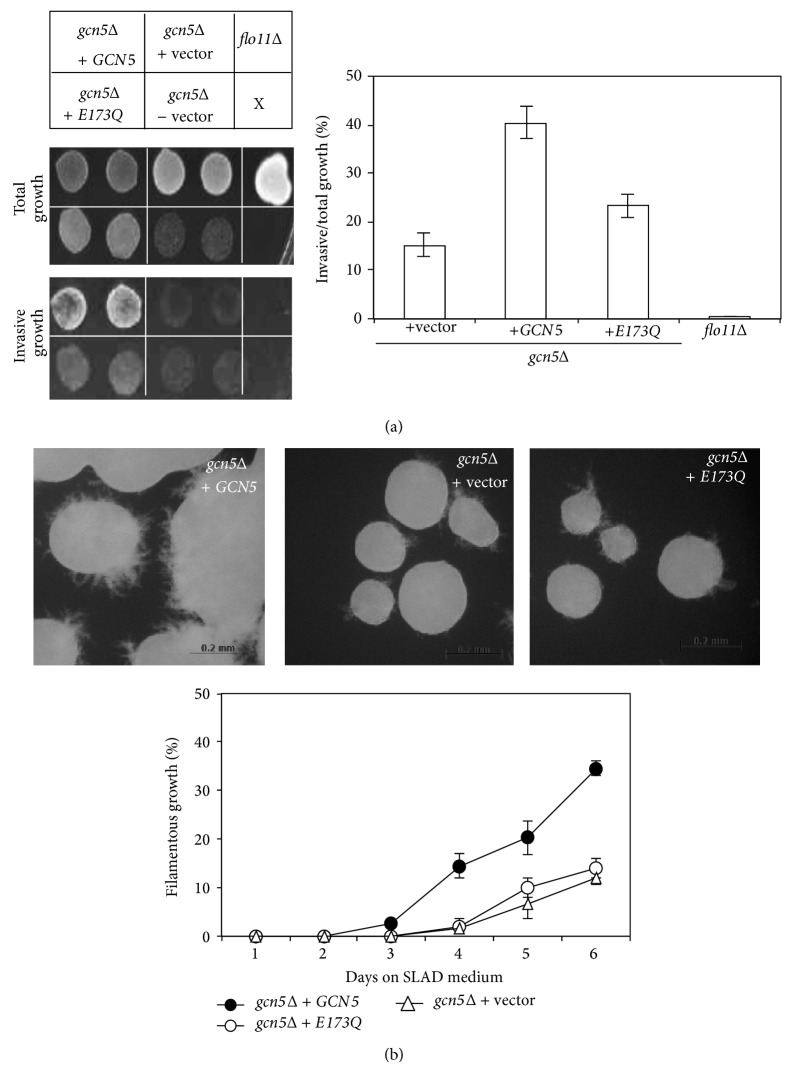
We next examined the association of Gcn5-dependent FLO11 expression and yeast dimorphism. We performed phenotypic analysis with the same strains used for measuring FLO11 transcripts in Figure 1(c). The FLO11-deletion strain (flo11Δ) was used as a control for complete loss of haploid invasive growth. Haploid flo11Δ cells were completely washed away from the surface of YPD agar medium (Figure 2(a), flo11Δ). The presence of a plasmid containing a functional GCN5 in gcn5Δ (gcn5Δ + GCN5) restored about 40% of the cells remaining on the agar surface; less than 15% and approximately 22% cells remained in gcn5Δ (gcn5Δ + vector) and gcn5Δ transformed with the catalytic dead version of Gcn5 (gcn5Δ + E173Q), respectively (Figure 2(a)). In diploids, filamentous development was substantially impaired in gcn5Δ (gcn5Δ + vector) and E173Q (gcn5Δ + E173Q) strains; approximately 10~12% of the population showed pseudohyphal growth after 6 d on SLAD agar medium (Figure 2(b)). The defective phenotype was rescued in part by introducing a copy of GCN5 to the gcn5Δ (gcn5Δ + GCN5). These results suggest that Gcn5 plays a regulatory role in both haploid invasive and diploid filamentous growth in yeast. Induction of FLO11 expression by amino acid starvation (with 3AT) has distinct patterns in haploids and diploids. Gcn5 is required for optimal FLO11 induction and its HAT activity is important for both FLO11 expression and dimorphic growth in yeast.
Figure 2.
The histone acetyltransferase activity of Gcn5 is required for pseudohyphal development. (a) The histone acetyltransferase activity of Gcn5 is required for haploid invasive growth. The gcn5 deletion haploid (gcn5Δ + vector) strain carrying a wild type GCN5 (gcn5Δ + GCN5) or a catalytic dead Gcn5 (gcn5Δ + E173Q) was grown on YPD agar plate for 4 d at 30°C before invasive growth was scored. The flo11Δ was used as a control for complete loss of invasive growth. The quantification of invasiveness in each strain was measured by cells remaining on agar before (total growth) and after being washed (invasive growth) and is shown on the right. Data are mean ± SD from 5 measurements in 10 biological repeats. (b) Histone acetyltransferase activity of Gcn5 is required for diploid filamentous growth. The indicated diploid strains were grown on nitrogen starvation agar medium (SLAD) at 30°C and the filamentous growth of colonies was recorded daily for 6 d. The quantification of filament formation was described in Section 2. Data are mean ± SD from measurements of filamentous colonies in 10 biological repeats. Scale bar: 0.2 mm.
3.3. Transcription Factor Gcn4 Binds to the FLO11 Promoter in Response to Amino Acid Starvation
Gene-specific and time-dependent orders of events are believed to link chromatin structure modifications and transcription activation. The SAGA complex containing Gcn5 plays a key role in activating genes that allow yeast to metabolize galactose as a carbon source. Similar to carbohydrate starvation, amino acid starvation is sensed by yeast cells trough diverse pathways [21, 33]. GCN4 was identified by expression microarray analysis as a positive regulator involved in such nutrient stresses [60]. In addition to being a metabolic regulator, Gcn4 is involved in regulating FLO11 expression [21, 61, 62]. Gcn5 was recruited to gene promoters via the transcription activator Gcn4 under conditions of amino acid starvation [51, 63]. Thus, Gcn4 is likely involved in the Gcn5-mediated FLO11 induction under amino acid starvation condition.
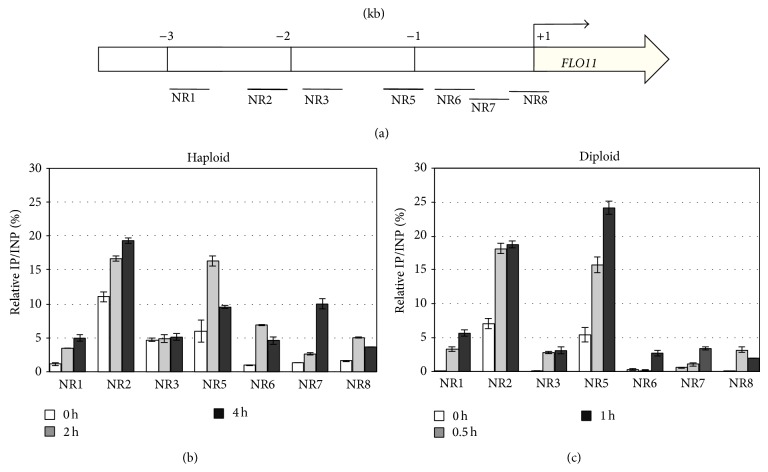
We postulated that the “ordered recruitment” of Gcn4-Gcn5 might take place at the FLO11 promoter, similar to that observed at the GAL1 and INO1 promoters [57, 64]. We tested this possibility by using ChIP assay to analyze Gcn4 occupancy at the upstream region of the FLO11 promoter. We designed specific primer pairs to detect the ChIP-DNA in different regions at the FLO11 promoter (Figure 3(a)). Because of the distinct FLO11 expression patterns shown in Figure 1(b), we performed Gcn4-ChIP experiments at 2 and 4 h for haploids (Figure 3(b)) and 0.5 and 1 h for diploids (Figure 3(c)) with 3AT treatment. In the absence of 3AT (time = 0 h), the chromatin association of Gcn4 at the FLO11 promoter was barely detectable in diploids (Figure 3(b)) and was insignificant in haploids (Figure 3(c)) except at NR2 and NR5, which suggests a very low level of Gcn4 occupancy before 3AT induction. Gcn4 occupancy was enhanced with 3AT treatment in both haploids and diploids (Figures 3(b) and 3(c)), which indicates a direct recruitment of Gcn4 to the FLO11 promoter at the very early stage of amino acid starvation. The most notably enhanced Gcn4 occupancy was at NR2 and NR5 in both haploids and diploids, approximately 2.2 (NR2) and 1.2 kb (NR5) upstream of the initiation start site of FLO11 (Figure 3(a)).
Figure 3.
The transcription factor Gcn4 is recruited to the FLO11 promoter during the course of 3AT induction. (a) A schematic representation of the amplicon locations at the upstream region of FLO11 promoter with primer sets for chromatin immunoprecipitation analysis (ChIP-qPCR). +1 indicates the transcription start site of FLO11. Cells from haploids (b) and diploids (c) were collected at the indicated times to measure the chromatin association of Gcn4 at the FLO11 promoter region (NR1~NR8, noncoding RNA) during 3AT induction. Association of Gcn4-Flag (a chromosome-integrated copy of GCN4 tagged with a Flag epitope) at the NR sites was detected by ChIP and anti-Flag antibody. Data are mean ± SD from three biological repeats and normalized to input (IP/INP). The fold changes of amplified genomic DNA relative to ACT1 are indicated (internal control of qPCR).
Our results agree with findings that the sites for a coordinated regulation of FLO11 transcription by Gcn4 and Hac1 are mapped to approximately 1 and 2 kb upstream of the ATG of FLO11 by FLO11::lacZ promoter assay [61, 62]. Here, we further demonstrate the direct-chromatin association of Gcn4 to specific binding sequences at the FLO11 promoter. Interestingly, the Gcn4 binding region overlaps with the toggle sites of Flo8 and Sfl1 at the FLO11 promoter [43, 45], whereby Flo8 and Sfl1 reciprocally control the switches of FLO11 transcription depending on the Rpd3L complex. Gcn4 may cooperate with Flo8 on 3AT activation to induce FLO11 expression and supports a role of cross talk between transcription factors to restrict chromatin accessibility for activators and transcriptional machinery.
3.4. Gcn5 Regulates Transcription of ncRNA ICR1 and FLO11 in Response to Amino Acid Starvation
Two cis-interfering long noncoding RNAs encoded by PWR1 and ICR1 are located in the intergenic region upstream of the FLO11 promoter [43, 45]. We have shown that Gcn5 is required for induced FLO11 expression (Figure 1) and Gcn4 occupancy at the specific regions of the FLO11 promoter is enriched by 3AT (Figure 3). Gcn4 has been shown to recruit Gcn5 to promoter regions of several genes involved in amino acid biosynthesis, such as HIS3 [51], to regulate gene expression. Interestingly, the binding sites of Gcn4 at the FLO11 promoter overlap with the toggle site of Sfl1 and Flo8 for expression of the PWR1 and ICR1.
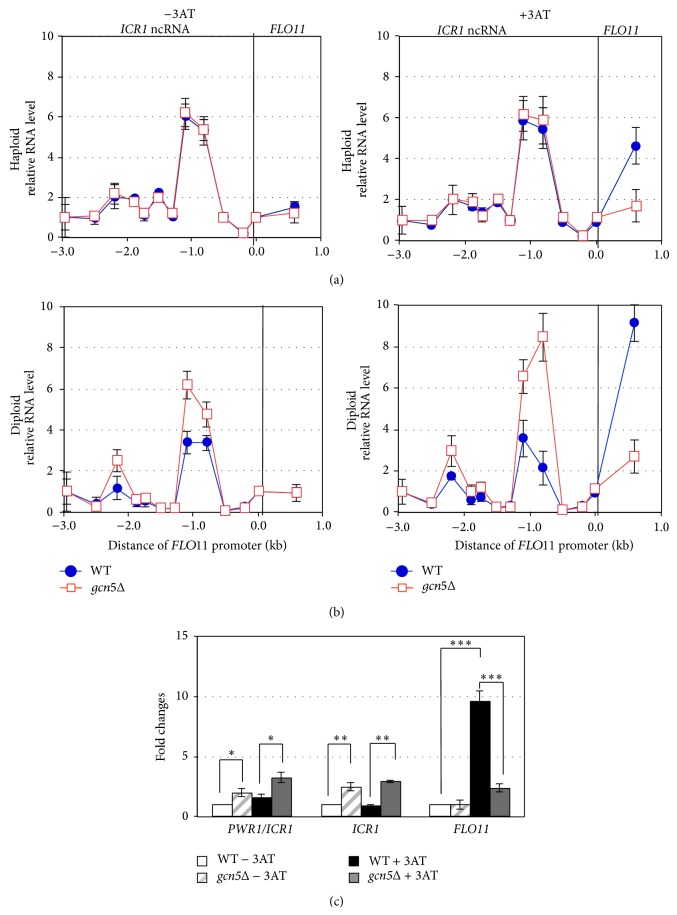
We hypothesized that Gcn5 is involved in transcription of the ncRNA ICR1 to causally regulate FLO11 expression. We examined the transcript levels of ICR1 and FLO11 in response to 3AT by using a set of strand-specific tiling primers spanning 3 kb upstream and 0.5 kb downstream of the transcriptional start site of FLO11 (Figure 4(a)). ICR1 transcript abundance was enriched at approximately 2.2 and 0.8~1.1 kb upstream of the FLO11 transcriptional start site in both haploids and diploids (Figures 4(a) and 4(b)). These two regions with accumulated ICR1 transcripts are fairly colocalized with the preferred Gcn4 binding sites at NR2 and NR5 (Figure 3(a)). Our results agree with previous findings [43]. Furthermore, we found an inverse association of ICR1 and FLO11 transcription in our assay. In haploids, the expression level and pattern of ICR1 were nearly indistinguishable with or without 3AT and in the WT and gcn5Δ; however, FLO11 was significantly induced by 3AT in the WT but not gcn5Δ (Figure 4(a)). Therefore, Gcn5 is essential for the 3AT-dependent FLO11 induction and may be dispensable for ICR1 expression in haploids. In addition, ICR1 expression may be independent of 3AT treatment.
Figure 4.
Gcn5 regulates transcription of ICR1 and FLO11 in response to 3AT. (a) A schematic representation of the transcription initiation sites and direction of ncRNAs ICR1 and PWR1 at the FLO11 promoter (adapted from [43]). (b) RT-qPCR analysis of expression of ncRNA ICR1 and FLO11. Haploid (two top panels) or diploid (two bottom panels) wild type (WT) and gcn5Δ strains were treated with or without 10 mM 3AT. Cells were cultured to early log phase in minimal medium and induced by 3AT for 2 h (haploids) or 1 h (diploids). In total, 12 sets of strand-specific primers tiled from −15 bp to −3 kb upstream of the FLO11 promoter. The transcript levels were normalized to that of ACT1. The signal at −3 kb of the FLO11 promoter in wild type was set to 1. (c) Quantification of transcript levels of ICR1, ICR1/PWR1, and FLO11 in diploids treated with or without 3AT. Data are derived from Figure 4(b) (two bottom panels) and the transcript levels at specified locations are indicated in parentheses. Data are mean ± SD from 3 biological repeats. ∗∗∗ P < 1E − 10; ∗∗ P < 1E − 5; ∗ P < 1E − 2 by Student's t-test.
WT and gcn5Δ diploids differed in the expression of ICR1 and FLO11. With and without 3AT, the ICR1 level was about 2-fold higher in gcn5Δ yeast than the WT, which suggests that Gcn5 represses ICR1 expression regardless of 3AT in diploids (Figure 4(b)). In agreement with these results (Figures 1(c) and 2(b)), FLO11 expression was significantly induced by amino acid starvation to 10-fold in WT but only twofold in gcn5Δ (Figure 4(b)), so Gcn5 is the major regulator of 3AT-induced FLO11 expression, with a minor Gcn5-independent induction in diploids. These results are consistent with the phenotypic observation in Figure 2(b) that pseudohyphal development is lost in gcn5Δ. Gcn5 may exert a differential control for FLO11 induction by 3AT in haploids and diploids (Figures 4(a) and 4(b)) and the repression of ICR1 transcription by Gcn5 in diploids may contribute to elevated FLO11 expression.
3.5. Gcn5 Is a Negative Regulator of ncRNA ICR1 Transcription in Diploid Yeast
To examine whether Gcn5 is involved in a promoter occlusion mechanism of ncRNAs to regulate FLO11 expression in response to 3AT, we further compared the expression profiles of PWR1, ICR1, and FLO11 in diploids (Figure 4(c)). In WT diploids under normal nutritional conditions, both ICR1 and FLO11 were expressed at low levels, whereas nutrient stress induced by 3AT greatly increased FLO11 level, by 10-fold (P = 4.7E − 15) (Figures 4(b) and 4(c)). In gcn5Δ diploids, the basal levels of ICR1 transcripts were higher than those of the WT (P = 2.4E − 9) regardless of 3AT induction (P = 2.3E − 8) (Figures 4(b) and 4(c)), so ICR1 expression by Gcn5 is independent of 3AT. However, with 3AT, FLO11 expression was reduced in gcn5Δ diploids (Figure 4(c); P = 2E − 12), so Gcn5 is a major regulator of FLO11 induction by 3AT. Furthermore, we investigated the association of gene expression changes in ncRNAs and FLO11 by correlation analysis. Interestingly, we found a significant inverse correlation between the expression of ICR1 and FLO11 in gcn5Δ with (R 2 = −0.4, P < 1E − 10) and without (R 2 = −0.72, P < 1E − 10) 3AT induction (Figure 4(c)). Our results suggest a role for Gcn5 in activating FLO11 expression in response to amino acid starvation in diploids by negatively regulating the expression of ncRNA ICR1.
3.6. Cell-Type-Specific Regulation of Gcn5 on ncRNA ICR1 Transcription
The functional link between chromatin structure and transcription processes is the “histone code,” whereby the various covalent modifications on histone tails define special patterns and affect chromatin-associated proteins at specific loci [65]. Such epigenetic controls were reported to regulate variegated FLO11 expression [38, 45]. In this study, we explored the establishment and function of Gcn5-dependent histone acetylation in transcriptional regulation on FLO11. We demonstrate both common and distinct features of Gcn5-mediated transcriptional regulation of FLO11 and ncRNA ICR1 in haploid and diploid yeast under nutrient starvation conditions. Our findings add a new layer of transcriptional control of the regulatory circuit of FLO11 expression in dimorphic growth in response to stress conditions.
In haploids, Gcn5 is not likely involved in the expression of ncRNAs ICR1 and PWR1 but is still required for induced FLO11 expression by 3AT (Figure 4(a)). However, in diploids, Gcn5 suppresses ICR1 expression and significantly enhances the induction of FLO11 responding to amino acid starvation (Figure 4(b)). We propose that Gcn5 is involved in different regulatory complexes and/or mechanisms in haploids and diploids. Gcn5 may simply function as a transcriptional activator of FLO11 in haploid yeast responding to nutrient stress. In diploids, Gcn5 may be recruited by a distinct group of transcriptional factors to modify histones for regulating the mutually exclusive transcription of ncRNAs PWR1 and ICR1 at the FLO11 promoter and acts with additional unknown factors mediated by nutrient stress signal to upregulate FLO11 expression.
In budding yeast, three cell types, including haploids of opposite mating type (MATa and MATα) and diploids (MATa/α), have distinct developmental phenotypes such as mating, meiosis, and budding patterns that are directly attributable to their different genotypes at the mating-type locus [66]. Mating loci (a1 and α2) regulate the expression of cell-type-specific genes as well as that of the INITIATION MEIOSIS genes such as IME1, IME2, and IME4. The histone acetyltransferase Gcn5 may participate in regulating the reciprocal expression of ncRNAs ICR1 and PWR1 via a mating-type specific mode during amino acid starvation. To test this possibility, we analyzed the filamentous phenotype of gcn5Δ and the FLO11 expression in a pseudodiploid strain (Figure 1(a)). The filamentous phenotype in the pseudodiploid was suppressed by gcn5Δ, but the induced FLO11 expression by 3AT in the pseudodiploid was similar to that in haploids (data not shown), so cell-type-specific factors other than mating loci may collaborate with Gcn5 to regulate ncRNA expression.
Alternatively, chromatin structure may play a role to regulate Gcn5-associated cell-type-specific event. For example, in sporulation induced by nutrient starvation, a1/α2 heterodimer suppresses expression of two ncRNAs, IRT1 (IME1 regulatory transcript 1) and IME4-AS, in diploids but not in haploids. IRT1 is located at the IME1 promoter and IME4-AS is an antisense ncRNA located at the coding region of IME4. In haploids, IME4 is repressed by IME4-AS, and transcription of IRT1 represses IME1 by establishing a repressive chromatin state. In contrast, the a1/α2 dimer inhibits the expression of IRT1 and IME4-AS in diploid cells, which then activates IME1 and IME4 and allows diploid cells to enter into sporulation process. Such regulation requires chromatin modifiers including Set2 (histone methyltransferase) and Set3 (histone deacetylase) complexes to establish repressive chromatin structures [67, 68]. Thus, Gcn5 may functionally associate with Set2 and Set3 to regulate ncRNAs ICR1 and PWR1 expression in a cell-type-specific manner during nutrient starvation.
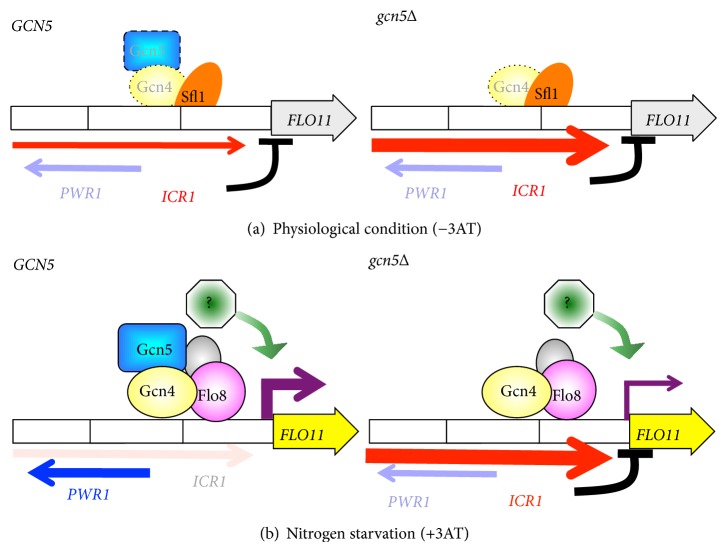
We propose a model to hypothesize the new role of Gcn5 in regulating FLO11 expression in diploids responding to nutrient stress (Figure 5). Under physiological conditions, the Sfl1 repressor associates at the FLO11 promoter to inhibit transcription of the ncRNA PWR1, which leads to permissive transcription of the ncRNA ICR1 [43, 45]. In both haploids and diploids, the net FLO11 transcription remains minimal because of the negative control from the actively transcribed ncRNA ICR1 by an antisense orientation at the promoter region of FLO11. DNA association of the transcription activator Gcn4 near the toggle sites of Sfl1 and Flo8 may have a limited effect on regulating transcription of the ncRNAs PWR1 and ICR1 probably because of low protein concentration and/or less competition for binding to DNA. Marginal upregulation of PWR1, probably by Gcn4-Gcn5, results in slightly reduced ICR1 expression, which may not be sufficient to derepress FLO11 without a stress signal (Figure 5(a)). With 3AT to induce amino acid starvation, Sfl1 is dissociated from the toggle sites, and in turn, the activator Flo8 binds to DNA and activates PWR1 transcription. Gcn4 recruits Gcn5 substantially to the Flo8-Sfl1 toggling sites to further activate PWR1 leading to suppressing ICR1, which resets the transcriptional module for activation of FLO11 responding to nutrient stress (Figure 5(b)). A Gcn5-independent factor may be required for the full induction of FLO11. Our model presents Gcn4-Gcn5 as new regulators involved in the FLO11 induction in diploid yeast responding to nutrient stress by participating in the coordinated regulatory circuit consisting of Sfl1, Flo8, Rpd3L-containing complex, and the ncRNAs ICR1 and PWR1.
Figure 5.
A model of Gcn5-mediated transcriptional regulation of ncRNA ICR1 and FLO11 in diploid yeast. (a) Under physiological conditions (−3AT), the Sfl1 (a repressor, oval in orange) associates with the FLO11 promoter to inhibit the transcription of the ncRNA PWR1, which leads to enhanced transcription of another ncRNA, ICR1, for repressed FLO11 transcription. The reciprocal interference between PWR1 and ICR1 transcription and suppression of FLO11 expression by active ICR1 transcription are adapted from [43]. Without 3AT induction, Gcn5 may be recruited by a marginal level of Gcn4 bound at the FLO11 promoter (left panel). Because deletion of GCN5 enhances ICR1 expression twofold (right panel), Gcn5 may play a role in regulating transcription of the PWR1 and ICR1 independent of 3AT. However, the net transcription of FLO11 is minimal in the WT without 3AT induction. (b) In nutrient stress such as amino acid starvation induced by 3AT (+3AT), a substantial level of Gcn4 and Gcn5 is recruited to the FLO11 promoter. Because of overlapped binding sequences, Gcn4-Gcn5 may collaborate with Flo8 (an activator, circle in purple) to compete with Sfl1 for binding to the toggling sites and activate PWR1 expression to block ICR1 transcription, which then derepresses the expression of FLO11 (left panel). Deletion of GCN5 enhances ICR1 expression and then reduces FLO11 transcription (right panel). Since gcn5Δ diploid yeast still maintains twofold induction of FLO11, a Gcn5-independent factor (octagon in green) is likely involved in FLO11 expression in response to 3AT induction.
4. Conclusions
This study reveals that the histone acetyltransferase Gcn5 is required for pseudohyphal development in the budding yeast S. cerevisiae. We found distinct expression patterns of FLO11 in haploid and diploid yeast responding to amino acid starvation induced by 3AT. Deletion of FLO11 or GCN5 resulted in loss of invasive growth in haploids and filamentous formation in diploids. ChIP analysis revealed for the first time an association of Gcn4 with the FLO11 promoter region that overlaps with the binding sites of Sfl1 and Flo8 at a critical location to mediate the mutually exclusive transcription of two long ncRNAs, ICR1 and PWR1. Gcn5 is required for full induction of FLO11 expression in diploid yeast under nutrient stress conditions. Finally, Gcn5 inhibits transcription of ICR1, which then depresses FLO11 for basal and induced expression. Our research on the mechanistic role of Gcn5 in regulating FLO11 expression and pseudohyphal development suggests potential therapeutic targets for treatment of fungal diseases.
Acknowledgments
This work was supported by grants from the National Science Council (NSC 95-2311-B-001-077-MY3 to Wan-Sheng Lo) and Institute of Plant and Microbial Biology (IPMB), Academia Sinica, Taiwan (to Wan-Sheng Lo and Long-Chi Wang). The authors thank the assistance of the Cell Biology Core Facilities at IPMB and Ms. Laura Smales for editing the paper.
Conflict of Interests
The authors declare that there is no conflict of interests regarding the publication of this paper.
References
- 1.Borges-Walmsley M. I., Walmsley A. R. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends in Microbiology. 2000;8(3):133–141. doi: 10.1016/s0966-842x(00)01698-x. [DOI] [PubMed] [Google Scholar]
- 2.Csank C., Haynes K. Candida glabrata displays pseudohyphal growth. FEMS Microbiology Letters. 2000;189(1):115–120. doi: 10.1016/s0378-1097(00)00241-x. [DOI] [PubMed] [Google Scholar]
- 3.Lo H. J., Köhler J. R., Didomenico B., Loebenberg D., Cacciapuoti A., Fink G. R. Nonfilamentous C. albicans mutants are avirulent. Cell. 1997;90(5):939–949. doi: 10.1016/s0092-8674(00)80358-x. [DOI] [PubMed] [Google Scholar]
- 4.Lengeler K. B., Davidson R. C., D'Souza C., et al. Signal transduction cascades regulating fungal development and virulence. Microbiology and Molecular Biology Reviews. 2000;64(4):746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brückner S., Mösch H.-U. Choosing the right lifestyle: adhesion and development in Saccharomyces cerevisiae . FEMS Microbiology Reviews. 2012;36(1):25–58. doi: 10.1111/j.1574-6976.2011.00275.x. [DOI] [PubMed] [Google Scholar]
- 6.Cappellaro C., Baldermann C., Rachel R., Tanner W. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: cell wall attachment and active sites of a- and α-agglutinin. EMBO Journal. 1994;13(20):4737–4744. doi: 10.1002/j.1460-2075.1994.tb06799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo B., Styles C. A., Feng Q., Fink G. R. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(22):12158–12163. doi: 10.1073/pnas.220420397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roberts R. L., Fink G. R. Elements of a single map kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes & Development. 1994;8(24):2974–2985. doi: 10.1101/gad.8.24.2974. [DOI] [PubMed] [Google Scholar]
- 9.Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell. 1992;68(6):1077–1090. doi: 10.1016/0092-8674(92)90079-r. [DOI] [PubMed] [Google Scholar]
- 10.Mösch H.-U., Roberts R. L., Fink G. R. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae . Proceedings of the National Academy of Sciences of the United States of America. 1996;93(11):5352–5356. doi: 10.1073/pnas.93.11.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reynolds T. B., Fink G. R. Bakers' yeast, a model for fungal biofilm formation. Science. 2001;291(5505):878–881. doi: 10.1126/science.291.5505.878. [DOI] [PubMed] [Google Scholar]
- 12.Verstrepen K. J., Derdelinckx G., Verachtert H., Delvaux F. R. Yeast flocculation: what brewers should know. Applied Microbiology and Biotechnology. 2003;61(3):197–205. doi: 10.1007/s00253-002-1200-8. [DOI] [PubMed] [Google Scholar]
- 13.Banuett F. Signalling in the yeasts: an informational cascade with links to the filamentous fungi. Microbiology and Molecular Biology Reviews. 1998;62(2):249–274. doi: 10.1128/mmbr.62.2.249-274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gancedo J. M. Control of pseudohyphae formation in Saccharomyces cerevisiae . FEMS Microbiology Reviews. 2001;25(1):107–123. doi: 10.1016/s0168-6445(00)00056-5. [DOI] [PubMed] [Google Scholar]
- 15.Cullen P. J., Sprague G. F., Jr. The regulation of filamentous growth in yeast. Genetics. 2012;190(1):23–49. doi: 10.1534/genetics.111.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rupp S., Summers E., Lo H.-J., Madhani H., Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. The EMBO Journal. 1999;18(5):1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo W.-S., Dranginis A. M. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae . Molecular Biology of the Cell. 1998;9(1):161–171. doi: 10.1091/mbc.9.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo W.-S., Dranginis A. M. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. Journal of Bacteriology. 1996;178(24):7144–7151. doi: 10.1128/jb.178.24.7144-7151.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lambrechts M. G., Bauer F. F., Marmur J., Pretorius I. S. MUC1, a mucin-like protein that is regulated by MSS10, is critical for pseudohyphal differentiation in yeast. Proceedings of the National Academy of Sciences of the United States of America. 1996;93(16):8419–8424. doi: 10.1073/pnas.93.16.8419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Karunanithi S., Joshi J., Chavel C., Birkaya B., Grell L., Cullen P. J. Regulation of mat responses by a differentiation MAPK pathway in Saccharomyces cerevisiae . PLoS ONE. 2012;7(4) doi: 10.1371/journal.pone.0032294.e32294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braus G. H., Grundmann O., Brückner S., Mösch H.-U. Amino acid starvation and Gcn4p regulate adhesive growth and FLO11 gene expression in Saccharomyces cerevisiae . Molecular Biology of the Cell. 2003;14(10):4272–4284. doi: 10.1091/mbc.e03-01-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vinod P. K. U., Venkatesh K. V. A steady state model for the transcriptional regulation of filamentous growth in Saccharomyces cerevisiae . In Silico Biology. 2008;8(3-4):207–222. [PubMed] [Google Scholar]
- 23.Vinod P. K., Sengupta N., Bhat P. J., Venkatesh K. V. Integration of global signaling pathways, cAMP-PKA, MAPK and TOR in the regulation of FLO11 . PLoS ONE. 2008;3(2) doi: 10.1371/journal.pone.0001663.e1663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Dyk D., Pretorius I. S., Bauer F. F. Mss11p is a central element of the regulatory network that controls FLO11 expression and invasive growth in Saccharomyces cerevisiae . Genetics. 2005;169(1):91–106. doi: 10.1534/genetics.104.033704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Robertson L. S., Fink G. R. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(23):13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pan X., Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae . Molecular and Cellular Biology. 1999;19(7):4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuchin S., Vyas V. K., Carlson M. Snf1 protein kinase and the repressors Nrg1 and Nrg2 regulate FLO11, haploid invasive growth, and diploid pseudohyphal differentiation. Molecular and Cellular Biology. 2002;22(12):3994–4000. doi: 10.1128/mcb.22.12.3994-4000.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Conlan R. S., Tzamarias D. Sfl1 functions via the co-repressor Ssn6-Tup1 and the cAMP-dependent protein kinase Tpk2. The Journal of Molecular Biology. 2001;309(5):1007–1015. doi: 10.1006/jmbi.2001.4742. [DOI] [PubMed] [Google Scholar]
- 29.Kim H. Y., Lee S. B., Kang H. S., Oh G. T., Kim T. Two distinct domains of Flo8 activator mediates its role in transcriptional activation and the physical interaction with Mss11. Biochemical and Biophysical Research Communications. 2014;449:202–207. doi: 10.1016/j.bbrc.2014.04.161. [DOI] [PubMed] [Google Scholar]
- 30.Fichtner L., Schulze F., Braus G. H. Differential Flo8p-dependent regulation of FLO1 and FLO11 for cell-cell and cell-substrate adherence of S. cerevisiae S288c. Molecular Microbiology. 2007;66(5):1276–1289. doi: 10.1111/j.1365-2958.2007.06014.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Köhler T., Wesche S., Taheri N., Braus G. H., Mösch H.-U. Dual role of the Saccharomyces cerevisiae TEA/ATTS family transcription factor Tec1p in regulation of gene expression and cellular development. Eukaryotic Cell. 2002;1(5):673–686. doi: 10.1128/ec.1.5.673-686.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pan X., Heitman J. Protein kinase A operates a molecular switch that governs yeast pseudohyphal differentiation. Molecular and Cellular Biology. 2002;22(12):3981–3993. doi: 10.1128/MCB.22.12.3981-3993.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valerius O., Kleinschmidt M., Rachfall N., et al. The Saccharomyces homolog of mammalian RACK1, Cpc2/Asc1p, is required for FLO11-dependent adhesive growth and dimorphism. Molecular and Cellular Proteomics. 2007;6(11):1968–1979. doi: 10.1074/mcp.m700184-mcp200. [DOI] [PubMed] [Google Scholar]
- 34.Felsenfeld G., McGhee J. D. Structure of the 30 nm chromatin fiber. Cell. 1986;44(3):375–377. doi: 10.1016/0092-8674(86)90456-3. [DOI] [PubMed] [Google Scholar]
- 35.Felsenfeld G., Emerson B. M., Jackson P. D., Lewis C. D., Nickol J. M. Chromatin structure near transcriptionally active genes. Progress in Clinical and Biological Research. 1986;218:63–74. [PubMed] [Google Scholar]
- 36.Li B., Carey M., Workman J. L. The role of chromatin during transcription. Cell. 2007;128(4):707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 37.Cosma M. P. Ordered recruitment: gene-specific mechanism of transcription activation. Molecular Cell. 2002;10(2):227–236. doi: 10.1016/s1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 38.Octavio L. M., Gedeon K., Maheshri N. Epigenetic and conventional regulation is distributed among activators of FLO11 allowing tuning of population-level heterogeneity in its expression. PLoS Genetics. 2009;5(10) doi: 10.1371/journal.pgen.1000673.e1000673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Halme A., Bumgarner S., Styles C., Fink G. R. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell. 2004;116(3):405–415. doi: 10.1016/s0092-8674(04)00118-7. [DOI] [PubMed] [Google Scholar]
- 40.van Mulders S. E., Christianen E., Saerens S. M. G., et al. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae . FEMS Yeast Research. 2009;9(2):178–190. doi: 10.1111/j.1567-1364.2008.00462.x. [DOI] [PubMed] [Google Scholar]
- 41.Barrales R. R., Jimenez J., Ibeas J. I. Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae . Genetics. 2008;178(1):145–156. doi: 10.1534/genetics.107.081315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fischer C., Valerius O., Rupprecht H., Dumkow M., Krappmann S., Braus G. H. Posttranscriptional regulation of FLO11 upon amino acid starvation in Saccharomyces cerevisiae . FEMS Yeast Research. 2008;8(2):225–236. doi: 10.1111/j.1567-1364.2007.00331.x. [DOI] [PubMed] [Google Scholar]
- 43.Bumgarner S. L., Dowell R. D., Grisafi P., Gifford D. K., Fink G. R. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(43):18321–18326. doi: 10.1073/pnas.0909641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Martens J. A., Wu P. Y. J., Winston F. Regulation of an intergenic transcript controls adjacent gene transcription in Saccharomyces cerevisiae . Genes and Development. 2005;19(22):2695–2704. doi: 10.1101/gad.1367605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bumgarner S. L., Neuert G., Voight B. F., et al. Single-cell analysis reveals that noncoding RNAs contribute to clonal heterogeneity by modulating transcription factor recruitment. Molecular Cell. 2012;45(4):470–482. doi: 10.1016/j.molcel.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clements A., Poux A. N., Lo W.-S., Pillus L., Berger S. L., Marmorstein R. Structural basis for histone and phosphohistone binding by the GCN5 histone acetyltransferase. Molecular Cell. 2003;12(2):461–473. doi: 10.1016/S1097-2765(03)00288-0. [DOI] [PubMed] [Google Scholar]
- 47.Lo W.-S., Trievel R. C., Rojas J. R., et al. Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Molecular Cell. 2000;5(6):917–926. doi: 10.1016/S1097-2765(00)80257-9. [DOI] [PubMed] [Google Scholar]
- 48.Grant P. A., Duggan L., Côté J., et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an ada complex and the saga (spt/ada) complex. Genes & Development. 1997;11(13):1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 49.Sterner D. E., Belotserkovskaya R., Berger S. L. SALSA, a variant of yeast SAGA, contains truncated Spt7, which correlates with activated transcription. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(18):11622–11627. doi: 10.1073/pnas.182021199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Georgakopoulos T., Thireos G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO Journal. 1992;11(11):4145–4152. doi: 10.1002/j.1460-2075.1992.tb05507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuo M. H., vom Baur E., Struhl K., Allis C. D. Gcn4 activator targets Gcn5 histone acetyltransferase to specific promoters independently of transcription. Molecular Cell. 2000;6(6):1309–1320. doi: 10.1016/s1097-2765(00)00129-5. [DOI] [PubMed] [Google Scholar]
- 52.Qiu H., Hu C., Zhang F., et al. Interdependent recruitment of SAGA and Srb mediator by transcriptional activator Gcn4p. Molecular and Cellular Biology. 2005;25(9):3461–3474. doi: 10.1128/mcb.25.9.3461-3474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rothstein R. J. One-step gene disruption in yeast. Methods in Enzymology. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- 54.Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae . Genetics. 1989;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lo W.-S., Raitses E. I., Dranginis A. M. Development of pseudohyphae by embedded haploid and diploid yeast. Current Genetics. 1997;32(3):197–202. doi: 10.1007/s002940050266. [DOI] [PubMed] [Google Scholar]
- 56.Liang C.-Y., Wang L.-C., Lo W.-S. Dissociation of the H3K36 demethylase Rph1 from chromatin mediates derepression of environmental stress-response genes under genotoxic stress in Saccharomyces cerevisiae . Molecular Biology of the Cell. 2013;24(20):3251–3262. doi: 10.1091/mbc.e12-11-0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lo W. S., Gamache E. R., Henry K. W., Yang D., Pillus L., Berger S. L. Histone H3 phosphorylation can promote TBP recruitment through distinct promoter-specific mechanisms. The EMBO Journal. 2005;24(5):997–1008. doi: 10.1038/sj.emboj.7600577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Galitski T., Saldanha A. J., Styles C. A., Lander E. S., Fink G. R. Ploidy regulation of gene expression. Science. 1999;285(5425):251–254. doi: 10.1126/science.285.5425.251. [DOI] [PubMed] [Google Scholar]
- 59.Wu C.-Y., Rolfe P. A., Gifford D. K., Fink G. R. Control of transcription by cell size. PLoS Biology. 2010;8(11) doi: 10.1371/journal.pbio.1000523.e1000523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hinnebusch A. G. Translational regulation of yeast GCN4. A window on factors that control initiator-tRNA binding to the ribosome. The Journal of Biological Chemistry. 1997;272(35):21661–21664. doi: 10.1074/jbc.272.35.21661. [DOI] [PubMed] [Google Scholar]
- 61.Herzog B., Popova B., Jakobshagen A., Shahpasandzadeh H., Braus G. H. Mutual cross talk between the regulators Hac1 of the unfolded protein response and Gcn4 of the general amino acid control of Saccharomyces cerevisiae . Eukaryotic Cell. 2013;12(8):1142–1154. doi: 10.1128/ec.00123-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Herzog B., Streckfuss-Bömeke K., Braus G. H. A feedback circuit between transcriptional activation and self-destruction of Gcn4 separates its metabolic and morphogenic response in diploid yeasts. Journal of Molecular Biology. 2011;405(4):909–925. doi: 10.1016/j.jmb.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 63.Dudley A. M., Gansheroff L. J., Winston F. Specific components of the SAGA complex are required for Gcn4- and Gcr1- mediated activation of the his4-912delta promoter in Saccharomyces cerevisiae. Genetics. 1999;151(4):1365–1378. doi: 10.1093/genetics/151.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lo W.-S., Duggan L., Emre N. C. T., et al. Snf1—a histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293(5532):1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- 65.Strahl B. D., Allis C. D. The language of covalent histone modifications. Nature. 2000;403(6765):41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 66.Siliciano P. G., Tatchell K. Transcription and regulatory signals at the mating type locus in yeast. Cell. 1984;37(3):969–978. doi: 10.1016/0092-8674(84)90431-8. [DOI] [PubMed] [Google Scholar]
- 67.Gelfand B., Mead J., Bruning A., et al. Regulated antisense transcription controls expression of cell-type-specific genes in yeast. Molecular and Cellular Biology. 2011;31(8):1701–1709. doi: 10.1128/mcb.01071-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Werven F. J., Neuert G., Hendrick N., et al. Transcription of two long noncoding RNAs mediates mating-type control of gametogenesis in budding yeast. Cell. 2012;150(6):1170–1181. doi: 10.1016/j.cell.2012.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]