Summary
Somatic LINE-1 (L1) retrotransposition during neurogenesis is a potential source of genotypic variation among neurons. As a neurogenic niche, the hippocampus supports pronounced L1 activity. However, the basal parameters and biological impact of L1-driven mosaicism remain unclear. Here, we performed single-cell retrotransposon capture sequencing (RC-seq) on individual human hippocampal neurons and glia, as well as cortical neurons. An estimated 13.7 somatic L1 insertions occurred per hippocampal neuron and carried the sequence hallmarks of target-primed reverse transcription. Notably, hippocampal neuron L1 insertions were specifically enriched in transcribed neuronal stem cell enhancers and hippocampus genes, increasing their probability of functional relevance. In addition, bias against intronic L1 insertions sense oriented relative to their host gene was observed, perhaps indicating moderate selection against this configuration in vivo. These experiments demonstrate pervasive L1 mosaicism at genomic loci expressed in hippocampal neurons.
Graphical Abstract
Highlights
-
•
An estimated 13.7 somatic L1 insertions occur per hippocampal neuron, on average
-
•
Target-primed reverse transcription drives somatic L1 retrotransposition
-
•
Somatic L1 insertions sense oriented to introns are depleted in neurons and glia
-
•
Hippocampus genes and enhancers are strikingly enriched for somatic L1 insertions
Somatic genome mosaicism among neurons has the potential to impact brain function. L1 retrotransposons mobilize extensively in hippocampal neurons, preferentially in hippocampally expressed loci, and are depleted from mature neurons when oriented in the most deleterious configuration to host genes, suggesting functional significance.
Introduction
The extent to which the genome of one cell differs from that of any other cell from the same body is unclear. DNA replication errors, mitotic recombination, aneuploidy, and transposable element activity can cause somatic mosaicism during ontogenesis and senescence. In humans, the consequences of somatic mosaicism are most apparent in disease, including cancer and developmental syndromes (Youssoufian and Pyeritz, 2002). The impact of mosaicism among normal cells is relatively undefined beyond the notable exception of V(D)J recombination and somatic hypermutation intrinsic to lymphocyte antigen recognition (Hozumi and Tonegawa, 1976). Reports of retrotransposition (Baillie et al., 2011; Coufal et al., 2009; Evrony et al., 2012; Li et al., 2013; Muotri et al., 2005; Perrat et al., 2013) and other genomic abnormalities (Cai et al., 2014; Gole et al., 2013; McConnell et al., 2013) in animal neurons may therefore be important given that, as for immune cells, mosaicism is a plausible route to neuron functional diversification.
Of approximately 500,000 LINE-1 (L1) copies present in the human genome, only ∼100 members of the L1-Ta and pre-Ta subfamilies remain transposition-competent (Beck et al., 2010; Brouha et al., 2003). L1 mobilization primarily occurs via target primed reverse transcription (TPRT), a process catalyzed in cis by two proteins, ORF1p and ORF2p, translated from the bicistronic 6 kb L1 mRNA. L1 ORF2p encodes endonuclease (EN) and reverse transcriptase (RT) activities essential to L1 retrotransposition and also responsible for trans mobilization of Alu and SVA retrotransposons (Dewannieux et al., 2003; Hancks et al., 2011; Raiz et al., 2012). A typical TPRT-mediated L1 insertion involves a degenerate L1 EN recognition motif (5′-TT/AAAA), an L1 poly-A tail and, crucially, produces target site duplications (TSDs) (Jurka, 1997; Luan et al., 1993). Various host defense mechanisms suppress L1 activity (Beck et al., 2011), including via methylation of the CpG-rich L1 promoter. Neural progenitors and other multipotent cells can nonetheless permit L1 promoter activation (Coufal et al., 2009; Garcia-Perez et al., 2007; Wissing et al., 2012), a pattern accentuated in the hippocampus, likely due to its incorporation of the neurogenic subgranular zone (Baillie et al., 2011; Coufal et al., 2009). This coincidence of neurogenesis, L1 activity, and mosaicism has elicited speculation that L1 mobilization could impact cognitive function rooted in the hippocampus (Richardson et al., 2014).
Despite extensive evidence of somatic retrotransposition in the brain, many fundamental aspects of the phenomenon remain unclear. The rate of L1 mobilization in the neuronal lineage is, for instance, a major unresolved issue. Estimates range from <0.1 to 80 somatic L1 insertions per neuron (Coufal et al., 2009; Evrony et al., 2012). Experiments using engineered L1 reporter systems have shown that L1 mobilization is likely to occur via TPRT in neuronal precursor cells and may be altered by neurological disease (Coufal et al., 2011; Coufal et al., 2009; Muotri et al., 2005; Muotri et al., 2010). However, it is unknown whether endogenous L1 retrotransposition in hippocampal neurons adheres to these predictions. Most importantly, it is unclear whether somatic L1 insertions influence neuronal phenotype or endow carrier neuronal progenitor cells with a selective advantage or disadvantage in vivo. To address these questions, we applied single-cell retrotransposon capture sequencing (RC-seq) to hippocampal neurons and glia, as well as cortical neurons, and found that L1 retrotransposition is a major endogenous driver of somatic mosaicism in the brain.
Results
Pervasive L1 Mobilization in Hippocampal Neurons
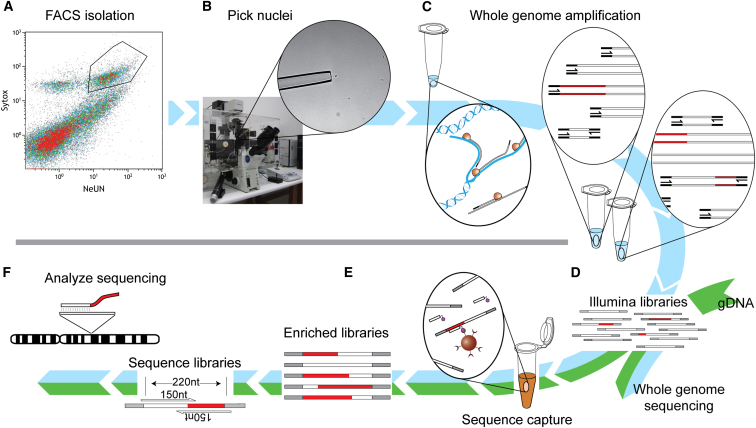
Several biological and technical factors hinder accurate calculation of somatic L1 mobilization frequency using bulk DNA extracted from tissue, as well as subsequent PCR validation and structural characterization of individual somatic L1 insertions (Richardson et al., 2014). We therefore developed a single-cell RC-seq protocol to detect somatic L1 insertions in individual neurons. Briefly, NeuN+ hippocampal nuclei were purified by fluorescence activated cell sorting (FACS) (Figures 1A and S1), with single nuclei isolated using a self-contained microscope and micromanipulator (Figure 1B). Whole-genome amplification (WGA) was achieved through an extensively optimized version of the quasi-linear Multiple Annealing and Looping Based Amplification Cycles (MALBAC) protocol (Zong et al., 2012) and was followed by Illumina library preparation (Figures 1C and 1D). Libraries were then subjected to low-coverage (0.35×) whole-genome sequencing (WGS) as a quality control step to assess amplification bias and, in parallel, hybridized and processed by RC-seq (Figures 1E and 1F).
Figure 1.
Single-Cell RC-Seq Workflow
(A) NeuN+ hippocampal nuclei were first purified by FACS (see also Figure S1).
(B) Nuclei were then picked using a self-contained microscope and micromanipulator.
(C) DNA was extracted from nuclei and subjected to linear WGA, followed by exponential PCR in two separate reactions for each nucleus, using different enzymes.
(D) Exponential WGA products for each nucleus were combined, used to prepare Illumina libraries, and analyzed via WGS to assess genome coverage and possible amplification biases.
(E) Libraries prepared in (D) were enriched via hybridization to L1-Ta LNA probes.
(F) Enriched libraries were sequenced with 2 × 150-mer Illumina reads and analyzed to identify novel L1 integration sites (see also Figure S2).
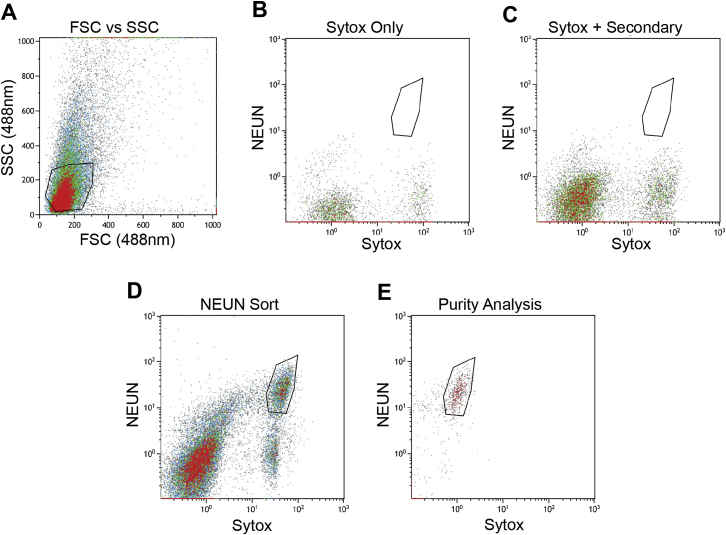
Figure S1.
Identification and Purity Confirmation of NeuN+ Hippocampal Nuclei via Fluorescence Activated Cell Sorting, Related to Figure 1
(A) Events were first gated on forward scatter of cells (FSC) and side scatter of cells (SSC).
(B) A Sytox blue control confirmed clear separation of fluorescent spectra, and an absence of events in the sorting gate.
(C) As for (B), except including a secondary antibody control.
(D) NeuN+/Sytox+ events (in polygonal gate) were sorted into PBS.
(E) A sample of sorted nuclei was re-analyzed by FACS to confirm sort purity. Note: expected photobleaching reduced signal intensities.
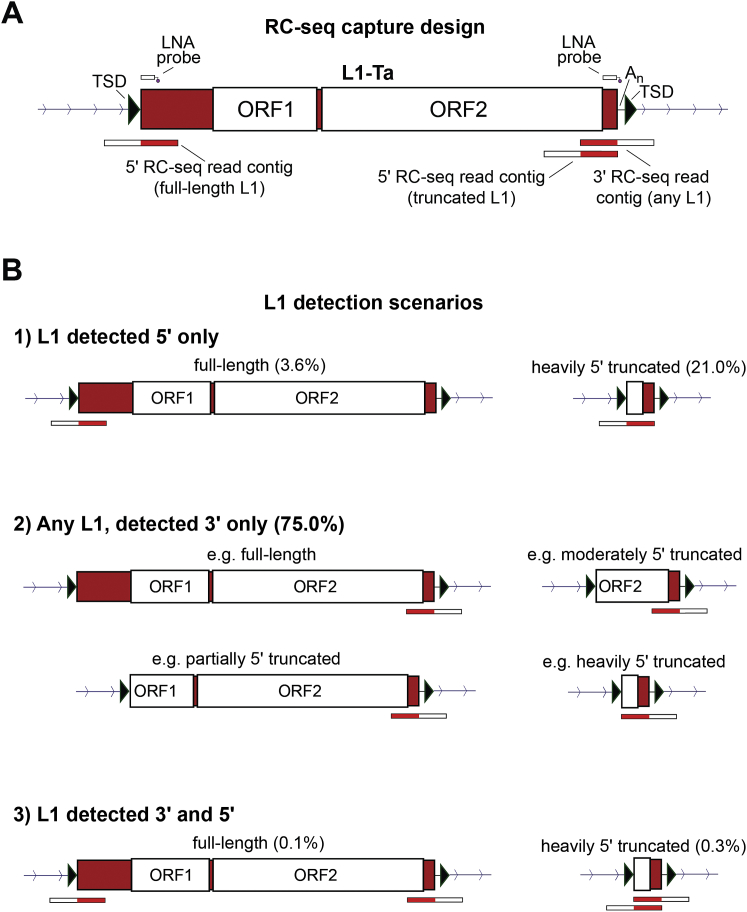
RC-seq utilizes sequence capture to enrich DNA for the junctions between retrotransposon termini and adjacent genomic regions, followed by paired-end sequencing, alignment, and clustering, to reveal L1 insertions absent from the reference genome. Here, we replaced previous RC-seq sequence capture pools (Baillie et al., 2011; Shukla et al., 2013) with two locked nucleic acid (LNA) probes respectively targeting the extreme 5′ and 3′ ends of L1-Ta. These probes capture typical L1 insertions at a 3′ L1-genome junction, and full-length or heavily 5′ truncated L1 insertions at a 5′ L1-genome junction (Figure S2), and delivered a 15-fold improvement in L1 enrichment compared with previous RC-seq applied to brain (Baillie et al., 2011). Assembly of each overlapping read pair into a “contig” enabled computational identification of molecular chimeras and removal of PCR duplicates, and provided single-nucleotide resolution of L1 integration sites by fully spanning L1-genome junctions (Figure S2).
Figure S2.
RC-Seq Capture Design and L1 Insertion Scenarios Detected, Related to Figure 1
(A) A full-length L1-Ta structure indicates the positions of two RC-seq probes designed to detect the 5′ or 3′ L1-genome junction of a given L1 insertion. Three categories of RC-seq reads are therefore generated, namely those that detect: the 5′ L1-genome junction of a full-length L1, the 5′ L1-genome junction of a heavily truncated L1 and the 3′ L1-genome junction of any L1.
(B) L1 detection scenarios as outlined in (A). Insertions are either 1) full-length or heavily truncated and detected at only a 5′ L1-genome junction, 2) of any length and detected at only a 3′ L1-genome junction, 3) full-length or heavily truncated and detected at both L1-genome junctions. Note the percentages given in brackets, indicating the relative occurrence of each scenario in the single-cell RC-seq data presented.
Prior to single-cell RC-seq, we performed deep coverage (∼80×) RC-seq on bulk DNA extracted from the post-mortem hippocampus and matched liver samples of four individuals (identifiers CTRL-36, CTRL-42, CTRL-45, and CTRL-55) without evidence of neurological disease (Table S1). Bulk RC-seq on average detected 97.5% of 960 annotated reference genome L1-Ta copies (Evrony et al., 2012), indicating high assay sensitivity. As expected, we detected ∼210 polymorphic L1-Ta insertions absent from the reference genome, per individual (Tables S1 and S2). This defined the polymorphic (germline) L1-Ta insertion cohort for each individual and provided a positive control for subsequent single-cell RC-seq analyses.
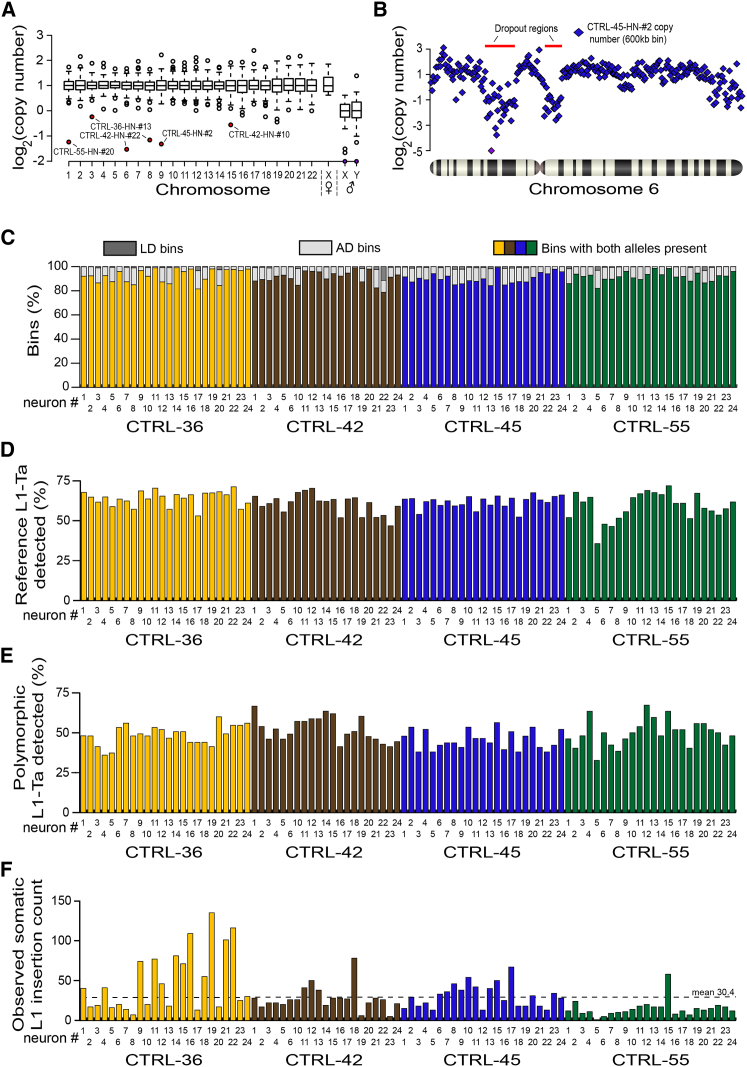
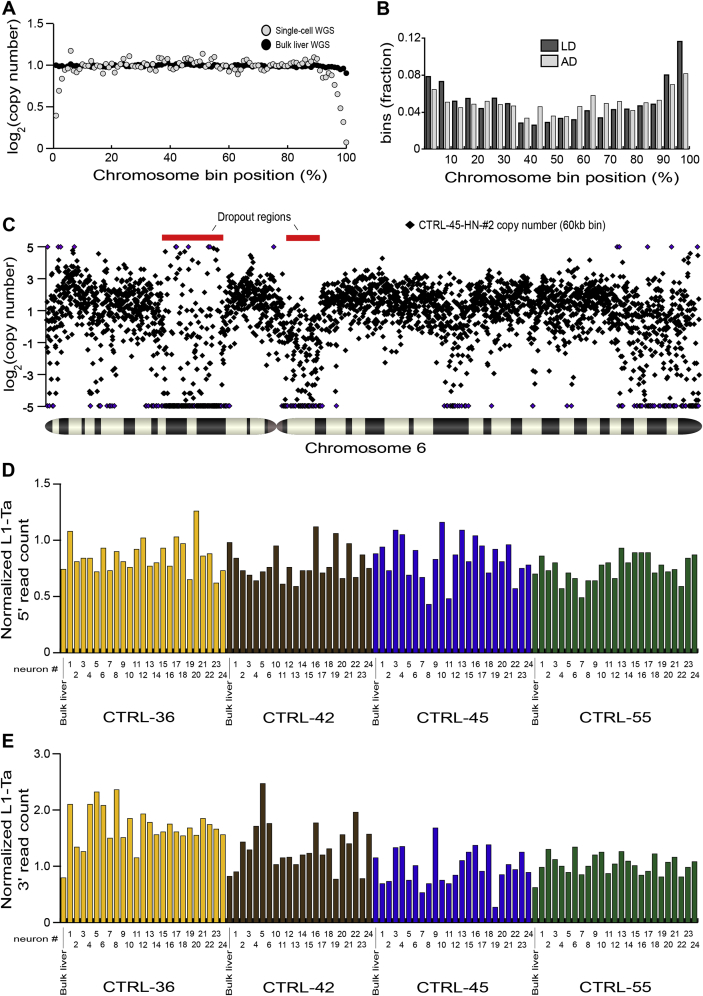
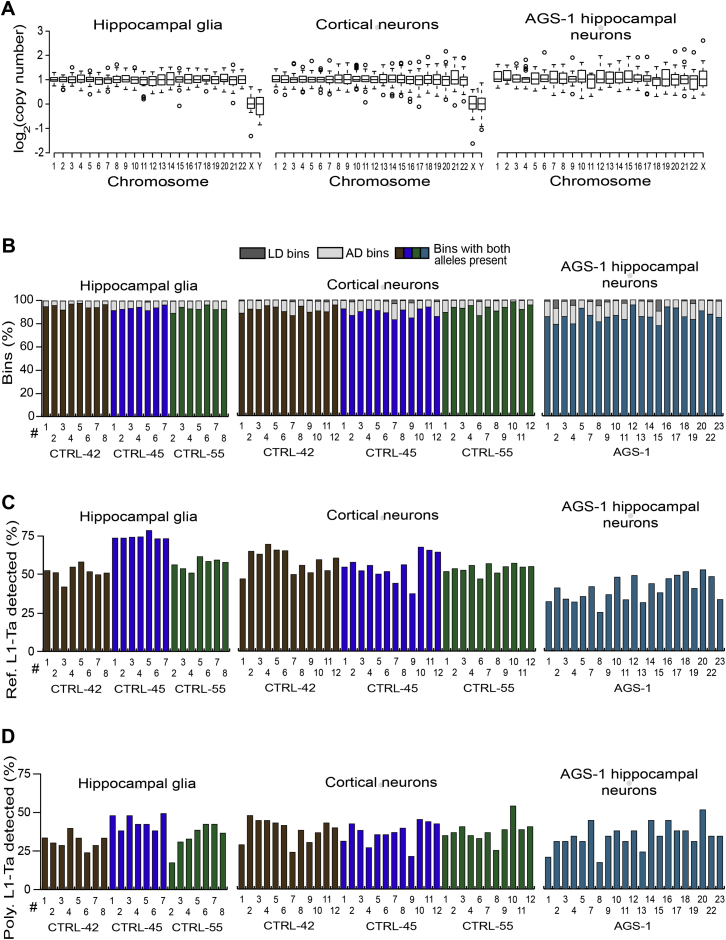
Next, 92 individual neuronal nuclei were isolated from the aforementioned hippocampi, subjected to WGA and analyzed by WGS. Globally, WGS revealed that 4,226/4,232 (99.9%) chromosomes amplified (Figure 2A) with recurring WGA bias largely limited to telomeres (Figures S3, S4A and S4B). Higher-resolution copy-number variation (CNV) analysis based on the division of the genome into adjustable-width “bins” with an average size of ∼600 kb revealed five non-telomeric deletions larger than ∼5 Mb. The largest and third largest of these occurred on chromosome 6 of CTRL-45 hippocampal neuron 2 (CTRL-45-HN-#2) and were 16.2 Mb and 9.4 Mb in length (Figure 2B). An alternative CNV analysis using ∼60 kb bins indicated the presence of numerous subregions in the 16.2 Mb example where chromosomal copy number was ≥2 (Figure S4C), depicting a region of highly variable WGA performance and, arguably, contraindicative of a genuine deletion in vivo. Genome-wide, allelic dropout (AD) and locus dropout (LD) respectively affected 8.0% and 0.7% of bins at 600 kb resolution (Figure 2C, Table S1), indicating efficient amplification across >90% of the genome. Importantly, we optimized WGA parameters to not deplete L1-Ta copies from amplified DNA, with the mean ratio of WGS reads aligned to reference L1-Ta 5′ or 3′ L1-genome junctions at 0.81 and 1.28 of expected values, respectively (Figures S4D and S4E; Table S1). These results show robust WGA for individual neurons, without significant loss of reference genome L1-Ta copies.
Figure 2.
Single-Cell WGS and RC-Seq Analyses of 92 Hippocampal Neurons
(A) Chromosome copy number in each amplified genome, assessed by WGS. Box-and-whisker plots indicate median chromosomal copy number and quartiles across all neurons. Empty circles represent chromosomes with copy number >1.5 IQR from the median. Sex chromosomes for CTRL-36 (female, ♀) and CTRL-42, CTRL-45, and CTRL-55 (male, ♂) are presented separately. Six autosomes, marked in red, had copy number ≤ 1. Two sex chromosomes with log2 copy number < −2 are colored purple.
(B) WGS indicated 16.2 Mb and 9.4 Mb regions of localized AD (indicated by red bars) on chromosome 6 of neuron CTRL-45-HN-#2. Each blue diamond corresponds to a 600 kb “bin”. One bin with log2 copy number < −5 is colored purple.
(C) Percentages of LD (dark gray) and AD (light gray) bins in each neuron, assessed by WGS.
(D) Percentage of reference genome L1-Ta copies detected by single-cell RC-seq in each neuron.
(E) Percentage of polymorphic L1-Ta insertions found in the corresponding bulk RC-seq libraries for each individual and also detected by single-cell RC-seq.
(F) Somatic L1 insertion counts observed in each neuron by single-cell RC-seq.
Note: in (C-F) yellow, brown, blue, and green histogram columns correspond to individuals CTRL-36, CTRL-42, CTRL-45, and CTRL-55, respectively. See also Figures S3 and S4 and Tables S1 and S2.
Figure S3.
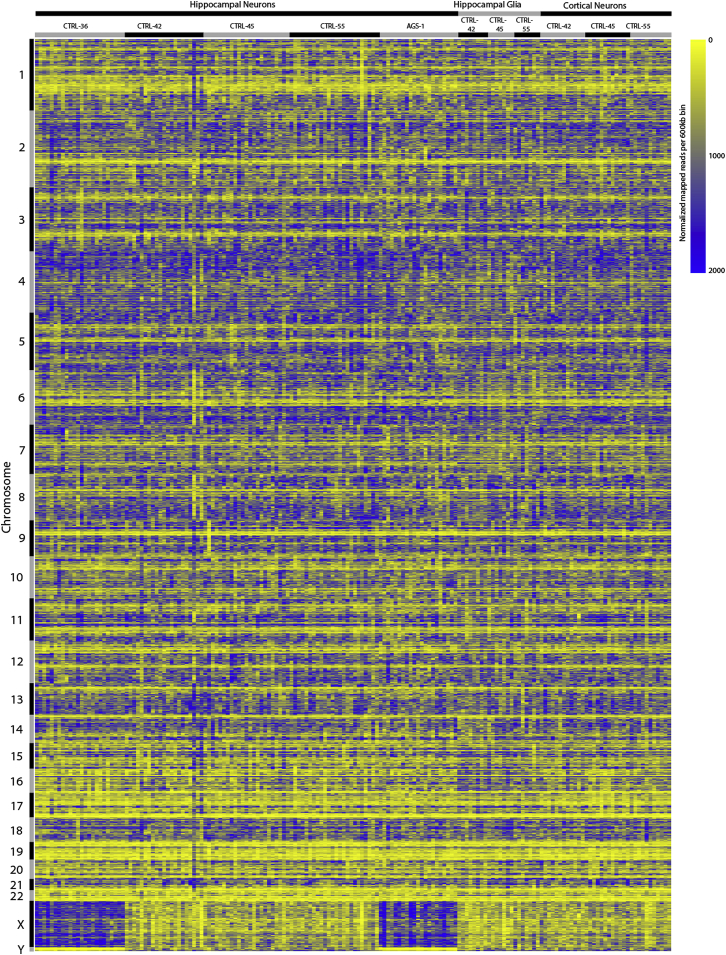
Heatmap Representing Sequence Coverage across the Genome, Related to Figure 2
For each sample, sequence alignments were binned by alignment start position into 600 kb intervals across the human genome, excluding unplaced contigs, extra haplotypes, and the mitochondrial genome. Counts were quantile normalized across all samples before plotting. Chromosomes are indicated on the vertical axis. Sample brain region location (cortex, hippocampus), cell type (glial, neuron) and individual ID are indicated for groups of columns on the horizontal axis. For each individual, single cells are ordered numerically. Note: low and high coverage bins are indicated in yellow and blue, respectively.
Figure S4.
WGS Revealed Limited Amplification Bias in Individual Neuronal Genomes, Related to Figure 2
(A) Median copy number for bins across all single-cell (gray circles) and bulk liver (black circles) WGS libraries, versus the percentile position of bins along the length of their corresponding chromosomes.
(B) Fractions of LD and AD bins versus bin chromosome percentile position. Note: LD and AD fractions are highest at telomeres.
(C) High resolution analysis of two localized AD regions on chromosome 6 of CTRL-45 hippocampal neuron 2 (CTRL-45-HN-#2), also presented at 600 kb resolution in Figure 2B. Copy number is displayed for ∼60 kb bins (black diamonds). Bins with absolute log2(copy number) ≥ 5 are colored in purple. Dropout regions are indicated by red bars.
(D) Observed RC-seq read counts across reference 5′ L1-genome junctions for bulk liver and single-cell WGS libraries, normalized as a ratio to counts obtained by random sampling 107 220 bp sequences from the human reference genome, revealing minimal dropout of L1-genome junctions due to WGA.
(E) As for (D) except at 3′ L1-genome junctions.
Single-cell RC-seq applied to each of the 92 libraries analyzed by WGS detected 61.3% of reference genome L1-Ta copies (Figure 2D, Table S1) and 49.0% of polymorphic L1-Ta insertions in each neuron (Figure 2E), as defined by the earlier bulk RC-seq experiments. The latter figure provided a provisional estimate of assay sensitivity for somatic L1 insertions. A total of 2,782 putative somatic L1-Ta and pre-Ta insertions (Figure 2F, Table S2) were identified in at least one hippocampal neuron, were not detected in any bulk liver RC-seq library or more than one hippocampus by single-cell or bulk RC-seq, and were absent from existing L1 polymorphism databases (Ewing and Kazazian, 2010, 2011; Iskow et al., 2010; Shukla et al., 2013; Wang et al., 2006). Of these insertions, 1,024 (36.8%) and 34 (1.2%) were found in introns and exons, respectively. Twelve (0.4%) somatic L1 insertions were detected at both their 5′ and 3′ L1-genome junctions, 760 (27.3%) at only a 5′ junction, and 2,010 (72.3%) at only a 3′ junction. Notably, nine somatic L1 insertions detected by single-cell RC-seq were also detected and annotated as somatic in the corresponding hippocampus bulk RC-seq library, and 13 were detected by single-cell RC-seq in more than one neuron from the same hippocampus. Of somatic L1 insertions, 98.2% belonged to the L1-Ta subfamily, and 1.8% were annotated as pre-Ta. Although at 5′ L1-genome junctions RC-seq captures only full-length and very heavily truncated L1s (Figure S2), we found 123 full-length L1 insertions, representing 4.4% of all events and including two instances of 5′ transduction. Of those insertions detected at their 3′ L1-genome junction, 151 (7.5%) carried a putative transduced 3′ flanking sequence (Moran et al., 1999). This L1 3′ transduction rate was lower than reported for germline L1 retrotransposition (Goodier et al., 2000), likely due to assay design not encompassing 3′ transductions longer than ∼100 bp, as reported elsewhere (Goodier et al., 2000; Macfarlane et al., 2013).
PCR Validation and Structural Characterization of Somatic L1 Insertions
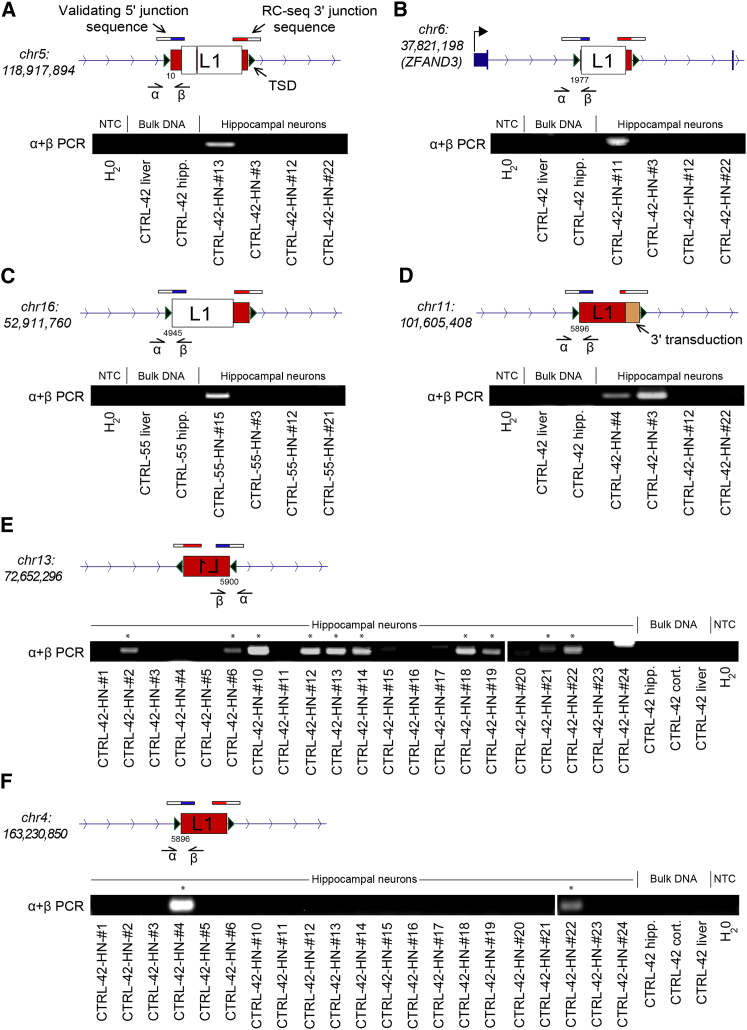
To determine the true positive rate of single-cell RC-seq, we randomly selected 20 somatic L1 insertions detected at only a 3′ L1-genome junction and PCR amplified the opposing 5′ L1-genome junction. This enabled detection of TPRT sequence hallmarks that distinguish WGA artifacts from most genuine L1 integration sites; specifically a TSD, an L1 EN target motif and an L1 poly-A tail (Jurka, 1997; Luan et al., 1993). Through PCR and sequencing, 5′ L1-genome junctions were identified for nine insertions and, when combined with the corresponding 3′ L1-genome junctions described by RC-seq, indicated TSDs and polyA-tails in all cases, and plausible L1 EN motifs for 7/9 (77.8%) examples (Tables S2 and Data S1). PCR validated insertions included full-length (Figure 3A) and variably 5′ truncated (Figures 3B–F) L1s. Intronic L1 insertions were found sense oriented to two genes expressed in brain, ZFAND3 (Figure 3B) and USP33 (Table S2). One L1 insertion incorporated a 3′ transduction and was detected by PCR in two neurons of CTRL-42 (Figure 3D). Further, PCR applied to the full panels of analyzed neurons from each individual revealed that two other L1 insertions were present in 10/21 and 2/21 neurons, respectively (Figures 3E and 3F). Three of the validated L1 insertions generated TSDs >40 bp in length.
Figure 3.
PCR Validation of Somatic L1 Insertions
(A–F) Validated examples from hippocampal neuron single-cell RC-seq data included: (A) a full-length L1 insertion in neuron CTRL-42-HN-#13; (B) a truncated L1 insertion in neuron CTRL-42-HN-#11; (C) a heavily truncated L1 insertion in neuron CTRL-55-HN-#15; and (D) a very heavily truncated L1 insertion yielding a 3′ transduction in neuron CTRL-42-HN-#4, also validated in neuron CTRL-42-HN-#3, and traced to a donor L1-Ta on chromosome 3; (E) a very heavily truncated L1 insertion detected in CTRL-42-HN-#13 and validated in 10/21 CTRL-42 hippocampal neurons tested. Asterisks denote neurons where validation succeeded; (F) a very heavily truncated L1 insertion detected in CTRL-42-HN-#4 and also validated in CTRL-42-HN-#22. Note: in (A–F) the 3′ L1-genome junction was detected by single-cell RC-seq, while the 5′ L1-genome junction was identified by insertion-site PCR (using primers indicated by α and β) and sequencing. Green triangles indicate TSDs. Numbers below the 5′ L1-genome junction indicate the equivalent L1-Ta consensus position. See also Table S2 and Data S1.
These experiments showed that nearly half of somatic L1 insertions detected by single-cell RC-seq at a 3′ L1-genome junction could be confirmed as genuine TPRT-mediated retrotransposition events. By contrast, PCR validation for 10 randomly selected exonic L1 insertions detected at a 5′ L1-genome junction by single-cell RC-seq failed to find the opposing 3′ L1-genome junction in all cases (Table S2). This was consistent with the L1 polyA-tail obstructing PCR amplification of somatic L1 insertion 3′ ends (Baillie et al., 2011) and arguably did not resolve whether L1 insertions detected only at a 5′ L1-genome junction were false positives. Finally, we selected 4 L1 insertions found at both their 5′ and 3′ L1-genome junctions by single-cell RC-seq; all four were confirmed by PCR and presented TPRT hallmarks, including one with a 92 bp TSD (Table S2).
Nearly 75% of somatic L1 insertions found by single-cell RC-seq were detected only at a 3′ L1-genome junction (Figure S2). Given this preponderance, we sought to ascertain why the matching 5′ L1-genome junction could not be identified by PCR for 11/20 selected examples of this type. PCR amplification failure was potentially due to RC-seq false positives, structurally exotic L1 insertions (Gilbert et al., 2005) or, alternatively, WGA inconsistently amplifying the 5′ L1-genome junctions of insertions detected at a 3′ L1-genome junction by single-cell RC-seq. To model the latter possibility, we randomly selected 12 polymorphic L1 insertions detected by bulk RC-seq and confirmed as heterozygous by genotype PCR. We performed PCR using bulk DNA to confirm each insertion was detectable at its 5′ L1-genome junction and then selected 100 random examples in individual neurons where these polymorphic L1s were detected at only a 3′ L1-genome junction by single-cell RC-seq (Table S2). We attempted PCR amplification of the corresponding 5′ L1-genome junction for each neuron, hence recapitulating the validation process for somatic L1 insertions, and confirmed 50/100 examples. This assay indicated the maximum PCR validation rate (50.0%) for somatic L1 insertions detected at only a 3′ L1-genome junction by single-cell RC-seq and, given the validation rate reported above (9/20, 45%), implied a true positive rate potentially as high as 9/10 (90.0%).
L1 Mobilization Frequency in Diverse Neural Cell Populations
Single-cell RC-seq identified mean somatic L1 insertion counts of 48.4, 27.5, 30.5, and 14.8 per hippocampal neuron in CTRL-36, CTRL-42, CTRL-45, and CTRL-55, respectively, yielding an overall mean count of 30.4 (Figure 2F). To estimate the overall true positive mean, we incorporated the PCR validation rate (45.0%) calculated above, leading to a conservative rate calculation of 13.7 somatic L1 insertions per hippocampal neuron. If, more conservatively, only L1 insertions detected at a 3′ L1-genome junction were considered, the true positive mean was 9.9. Conversely, if all L1 insertions were considered, we generously incorporated the maximum PCR validation rate calculated above (90%) and we corrected for assay sensitivity in terms of polymorphic L1 insertions detected (49.0%), the estimated true positive mean was greatly increased to 55.8. Thus, given a true positive mean of 13.7 somatic L1 insertions per neuron, and the detection of at least one event in every neuron (Figure 2F), we concluded that L1 mosaicism was ubiquitous among the hippocampal neurons studied.
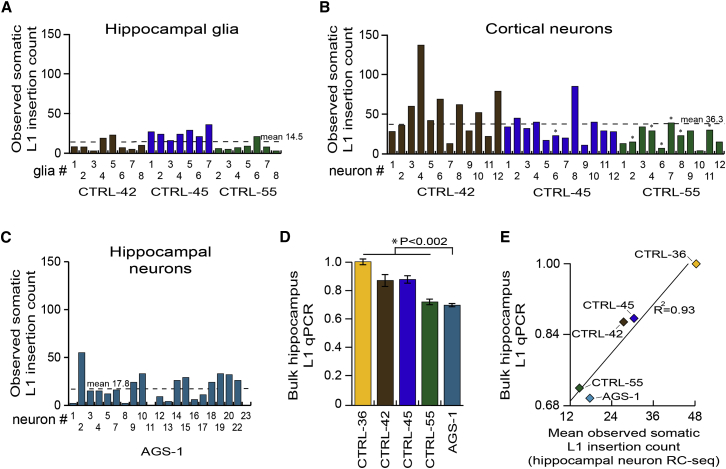
Prior in vitro experiments based on an engineered L1 reporter indicated that glia may support far less L1 mobilization than neurons (Coufal et al., 2009). To evaluate glial lineage endogenous L1 retrotransposition in vivo, we performed single-cell RC-seq upon 22 glial nuclei (NeuN−/Ki67−) isolated from CTRL-42, CTRL-45, and CTRL-55 hippocampi, and detected 316 putative somatic L1 insertions (Figures 4A and S5). This produced a mean true positive estimate of 6.5 insertions per glial cell, based on the PCR validation rate determined for hippocampal neurons (45.0%). This rate was 52.6% lower than the estimated 13.7 insertions for hippocampal neurons, a significant difference (p < 0.005, two-tailed t test, df = 112). Interestingly, four insertions were found in both glial and neuronal cells by single-cell RC-seq, with one of these instances detected at both its 5′ and 3′ L1-genome junctions, revealing a 12 bp TSD (Table S2). We concluded that L1 insertions can arise in proliferating neural stem cells prior to glial or neuronal commitment, while glia otherwise support less L1 mobilization than neurons.
Figure 4.
L1 Mobilization in Diverse Neural Cell Types
(A) Somatic L1 insertion counts observed by single-cell RC-seq applied to hippocampal glia.
(B) As for (A) except for cortical neurons. Seven pyramidal neurons are indicated by an asterisk.
(C) As for (A) except for AGS-1 hippocampal neurons.
(D) L1 qPCR indicated lower L1 copy number in AGS-1 hippocampus versus controls (p < 0.002, two-tailed t test, df = 23). Data represent the mean of 5 technical replicates ± SD.
(E) Mean somatic L1 insertion counts detected by single-cell RC-seq in each hippocampus strongly correlated (R2 = 0.93) with L1 copy number quantified by qPCR (D).
Figure S5.
WGA Quality Control Analyses for Hippocampal Glia, Cortical Neurons and AGS-1 Hippocampal Neurons, Assessed by WGS, Related to Figure 4
(A) Chromosome copy number in each amplified genome. Box-and-whisker plots indicate median chromosome copy number and quartiles across all neurons. No examples of chromosome-wide AD were observed.
(B) Percentages of LD (dark gray) and AD (light gray) bins in each cell.
(C) Percentage of reference genome L1-Ta copies detected by RC-seq in each cell.
(D) Percentage of polymorphic L1-Ta insertions found in the corresponding bulk RC-seq libraries for each individual and also detected by single-cell RC-seq.
A recent single-cell genomic analysis of 300 cortex and caudate nucleus pyramidal neurons elucidated <0.1 somatic L1 insertions per cell, and concluded that L1 was not a major driver of neuronal diversity (Evrony et al., 2012). However, the biological or technical reasons for such disparate results compared with prior data from the hippocampus were unclear. We therefore performed single-cell RC-seq upon 35 NeuN+ nuclei isolated from CTRL-42, CTRL-45 and CTRL-55 cortex tissue, including seven pyramidal neurons, and identified 1,262 putative somatic L1 insertions (Figures 4B and S5). This provided a true positive mean estimate of 16.3 insertions per cortical neuron, a figure higher than hippocampal neurons, but not significantly different. An estimated 10.7 insertions occurred per cortex pyramidal neuron, a rate substantially lower than the remaining cortical neurons but a difference that fell short of statistical significance (p < 0.16, two-tailed t test, df = 33). These data elucidate L1 mosaicism in cortical neurons and exclude a biological explanation for inconsistency with the previous study.
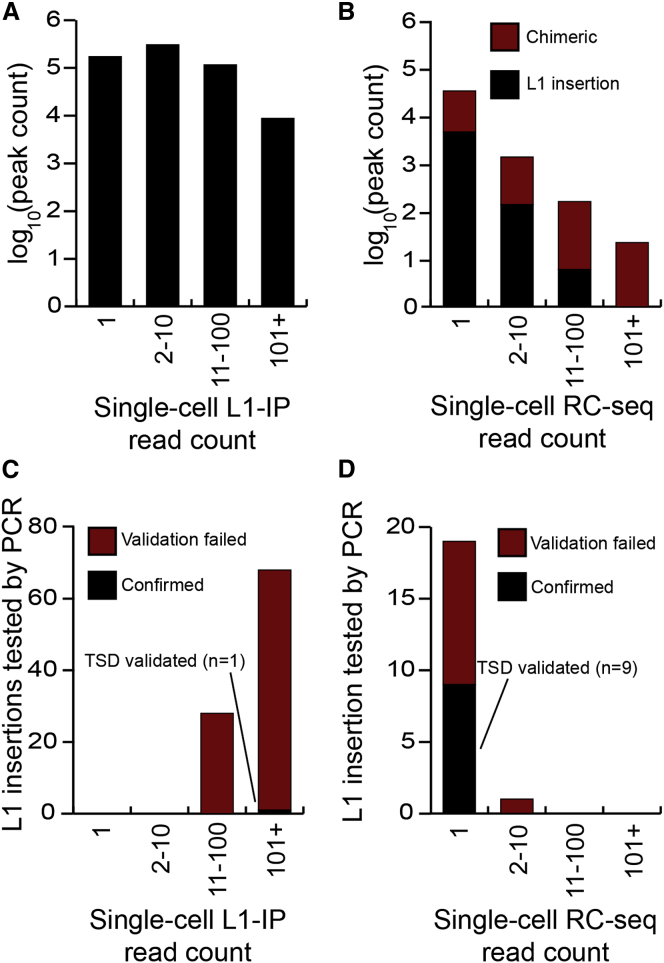
PCR validation including TSD discovery underpins accurate calculation of L1 mobilization frequency and reflects experimental veracity independent of methodology (Richardson et al., 2014). It is therefore notable that, at this stringency, Evrony et al. reported a PCR validation rate of 1/96 and a consequential paucity of L1 activity. Two key technical considerations may explain our discrepant findings. First, RC-seq reads fully span L1-genome junctions (Figure S2), enabling bioinformatic identification of molecular chimeras before PCR validation. The earlier work by contrast followed a design (Ewing and Kazazian, 2010) that typically did not resolve L1-genome junctions, prohibiting computational removal of chimeric reads. Instead, the authors maintained that artifacts, including those generated by WGA and Illumina library preparation, should present lower read depth than genuine L1 insertions, and essentially adhered to the same principle in a very recent study applying WGS to a smaller number of neurons (Evrony et al., 2015). This assumption is crucial as, at least in single-cell RC-seq libraries, putative chimeras are disproportionately likely to amplify efficiently and accrue high read depth (Figures 5A and 5B). Second, Evrony et al. selected candidates for PCR validation effectively as a function of high read count and not at random (Figure 5C). This approach would strongly enrich for artifacts if applied to single-cell RC-seq data (Figure 5B). It follows that, without the capacity to filter artifacts a priori, the previous study resolved numerous molecular chimeras after PCR and capillary sequencing of putative L1 insertions, substantially reducing the reported validation rate. By contrast, we selected PCR validation candidates at random (Figure 5D). These factors plausibly explain why our validation rate of 9/20 (45.0%) was significantly higher than the rate of 1/96 (1.0%) reported by the earlier work (p < 1 × 10−10, chi-square test, df = 1), as well as the disparate estimates of somatic L1 retrotransposition made by each study.
Figure 5.
Single-Cell RC-Seq Efficiently Excludes Molecular Artifacts
(A) Distribution of read “peaks” indicating possible somatic L1 insertions detected by single-neuron L1 insertion profiling (L1-IP) (Evrony et al., 2012).
(B) As for (A), except for all single-cell RC-seq data presented here. Peaks were annotated as chimeric or as likely genuine L1 insertions by sequence analysis of RC-seq reads.
(C) Distribution of read peak height for L1 insertions selected for validation by Evrony et al. The L1 insertion successfully validated by TSD discovery is colored black. The remaining insertions not validated to this standard are colored red.
(D) As for (C), except for L1 insertions detected by single-cell RC-seq and selected at random for validation.
Recent qPCR based estimates of L1 CNV in human tissue, as well as in vitro L1 reporter assays, indicate L1 mobilization may be pronounced in a range of neurodevelopmental and psychiatric diseases (Richardson et al., 2014) including Aicardi-Goutières syndrome (AGS). AGS is a rare, severe neurodevelopmental condition, characterized by mutations in several genes thought to inhibit reverse transcription, including SAMHD1 (Zhao et al., 2013). To address whether SAMHD1 deficiency in AGS patients increases neuronal L1 mobilization, we first applied bulk RC-seq to the post-mortem hippocampus and fibroblasts of an AGS patient (identifier AGS-1) carrying two loss-of-function SAMHD1 mutations. We then performed single-cell RC-seq upon 21 neuronal nuclei from AGS-1 hippocampus and identified 373 putative somatic L1 insertions (Figures 4C and S5), leading to a true positive mean estimate of 8.0 insertions per AGS-1 neuron. This figure was significantly (p < 0.03, two-tailed t test, df = 112) lower than the 13.7 somatic L1 insertions found for control hippocampal neurons. A more significant difference was observed when AGS-1 neurons were compared only with the age (18 years) and gender (female) matched hippocampal neurons of CTRL-36 (p < 0.0001, two-tailed t test, df = 44). As corollary, L1 qPCR also indicated significantly lower (p < 0.002, two-tailed t test, df = 23) L1 copy number in AGS-1 hippocampus versus controls (Figure 4D). Finally, the results of the L1 CNV assay were strongly correlated (R2 = 0.93) with the mean somatic L1 insertion frequencies estimated by single-cell RC-seq (Figure 4E). We therefore concluded that L1 mobilization was unlikely to be elevated in AGS-1 hippocampus.
Somatic L1 Retrotransposition Occurs via TPRT
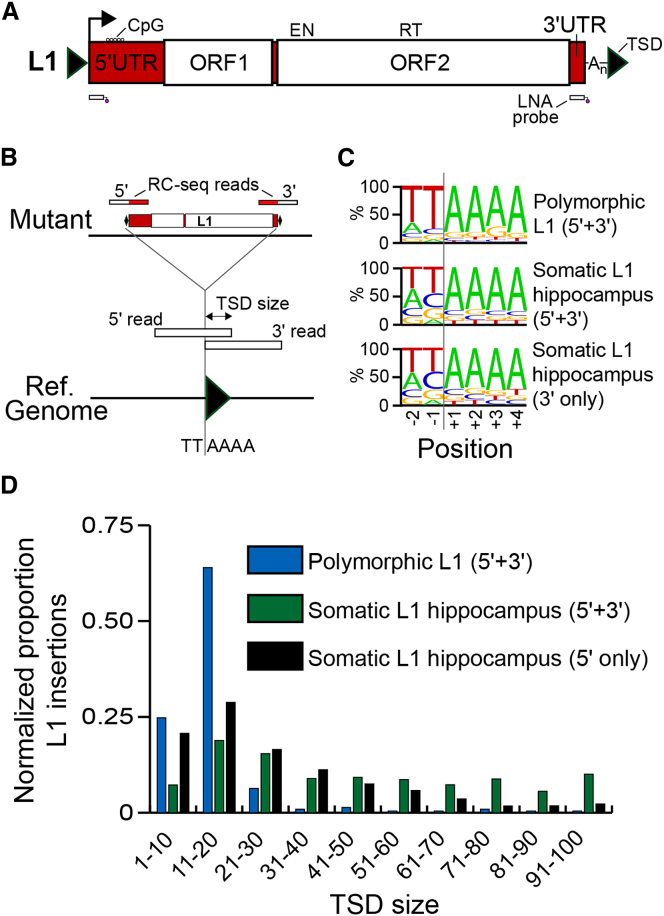
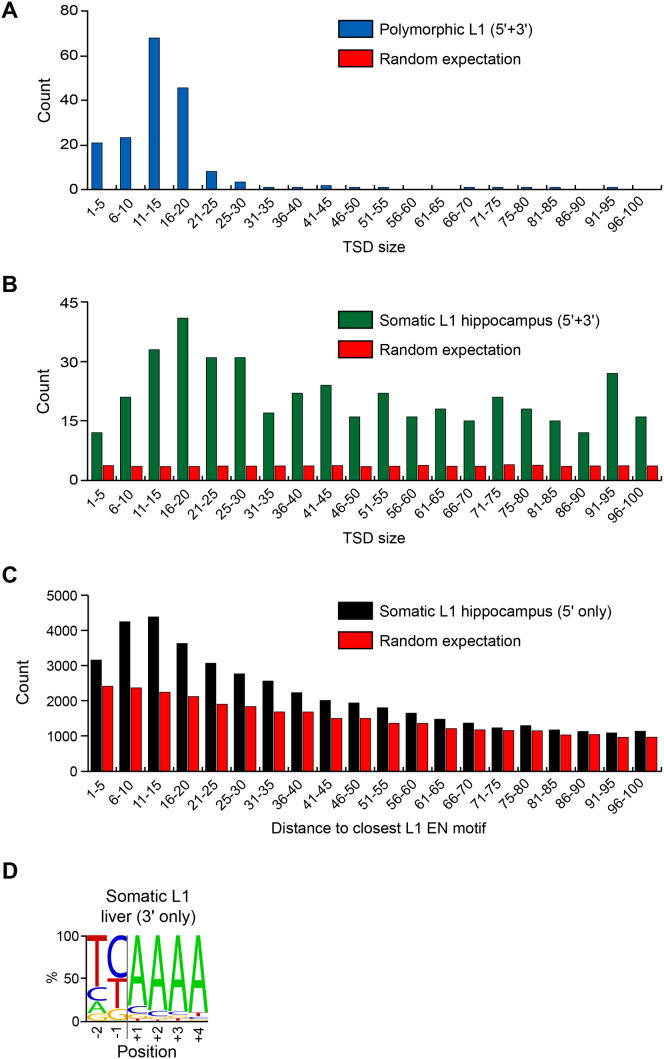
As the 13 total somatic L1 insertions detected by single-cell RC-seq and validated by PCR generally followed the TPRT model, we next assessed whether somatic L1 insertions detected by bulk RC-seq also carried TPRT signatures. RC-seq separately applied to DNA extracted from the four control hippocampus samples elucidated 318,866 putative somatic L1 insertions (Table S1). Again exploiting L1-genome junction resolution by RC-seq reads (Figures 6A and 6B and S2), we found a strong enrichment for the L1 EN motif (Figure 6C), a typical TSD size range of 5–35 nt (Figures 6D and S6) and a median L1 poly-A tail length of 33 nt for somatic L1 integration sites identified by bulk RC-seq. We also identified a substantial group of insertions with TSDs > 40 bp in length (Figure S6). Thus, single-cell RC-seq and RC-seq applied to bulk DNA both elucidated the hallmark sequence features of TPRT-mediated retrotransposition.
Figure 6.
Hallmarks of TPRT Revealed by Bulk RC-Seq
(A) A 6 kb L1-Ta element incorporates 5′ and 3′ UTRs and two ORFs. ORF2p presents EN and RT domains. Methylation of a CpG island present in the 5′ UTR regulates L1 promoter activity. The locations of two capture probes used by RC-seq are indicated below the L1. Note: TSDs and probes are not drawn to scale. See also Figure S2.
(B) TPRT hallmark features, including TSDs and an L1 EN recognition motif, can be identified by RC-seq, including for insertions detected at only a 5′ or 3′ L1-genome junction.
(C) Consensus L1 EN motifs for polymorphic and somatic L1 insertions detected at their 5′ and 3′ L1-genome junctions, and somatic L1 insertions found at only a 3′ L1-genome junction.
(D) Observed TSD size distributions for polymorphic and somatic L1 insertions, normalized to random expectation. See also Figure S6.
Figure S6.
Signatures of L1 Mobilization via TPRT Detected by Hippocampus and Liver Bulk RC-Seq, Related to Figure 6
(A) TSD size distribution for polymorphic L1 insertions detected at their 5′ and 3′ L1-genome junctions.
(B) As for (A), except for hippocampal somatic L1 insertions.
(C) TSD size distribution for hippocampal somatic L1 insertions detected at only a 5′ L1-genome junction.
(D) Consensus L1 EN motif for liver somatic L1 insertions detected at only a 3′ L1-genome junction by RC-seq.
Expected values for (A) and (B) were calculated by randomizing sense and antisense RC-seq read cluster genomic coordinates, to ascertain how many overlapping clusters in the opposing orientation and detecting opposite ends of an L1 insertion were found, using the same bioinformatics process as used for observed clusters. Expected values for (C) were calculated by random sampling of genomic coordinates and searching for the nearest upstream L1 EN motif, again following the same string matching process as for observed values. Note: the corresponding TSD size distribution for liver somatic L1 insertions detected at only their 5′ L1-genome junction contained insufficient data (n = 7) to make a meaningful comparison with hippocampal somatic L1 insertions.
Somatic L1 Insertions Are Enriched in Neurobiology Genes
Substrate DNA chromatinization modulates L1 EN target site nicking in vitro (Cost et al., 2001). As such, dynamic changes to chromatin state during neurogenesis may impact the associated genome-wide pattern of L1 mobilization. An intersection of somatic L1 insertion sites detected by hippocampus bulk RC-seq with RefSeq gene coordinates revealed significant (p < 1.0 × 10−150, Fisher’s exact test, Bonferroni correction) depletion for insertions in exons and promoters versus random sampling and significant (p < 3.8 × 10−10) enrichment for introns versus polymorphic insertions (Table S3). Exons and introns carrying gene ontology (GO) terms relevant to neurobiology were however enriched for somatic L1 insertions (Tables S4 and S5) compared with random sampling performed by gene identifier or by genomic coordinate (p < 4.5 × 10−5 and p < 0.03, respectively, Fisher’s exact test, Benjamini-Hochberg correction). The latter result indicated enrichment for L1 insertions in genes expressed in the brain, despite taking into account that their length is on average >50% greater than that of other genes. By considerable margin, the most enriched GO term found (Table S5) was “regulation of synapse maturation” (p < 1.7 × 10−60, Fisher’s exact test, Benjamini-Hochberg correction). Genome-wide patterns for somatic L1 insertions detected in glia and neurons by single-cell RC-seq typically corroborated those found by bulk RC-seq, including enrichment in introns and depletion from promoters and exons (Table S3) and even stronger enrichment in neurobiology genes annotated by GO term (Tables S4 and S5). Intriguingly, in AGS-1 hippocampal neurons we did not observe enrichment for L1 insertions in neurobiology genes (Table S4), whereas enrichment was observed for control hippocampal neurons, even if each individual was analyzed separately. As a control experiment, from the liver bulk RC-seq data we identified a set of 175 potential liver-specific L1 insertions (see Extended Experimental Procedures) that collectively presented a clear L1 EN consensus motif (Figure S6D) and, owing to the sensitivity of bulk RC-seq, were unlikely to represent incorrectly annotated polymorphic L1 insertions (Table S1). Notably, these liver-specific L1 insertions exhibited no enrichment for neurobiology genes (Table S4). We concluded that somatic L1 retrotransposition in neural cells preferentially occurs into the euchromatic regions of the genome contributing to neurobiology.
Extended Experimental Procedures.
Human Tissue Samples
Snap frozen hippocampus and liver tissue from four post-mortem individuals (IDs: CTRL-36, CTRL-42, CTRL-45, CTRL-55) without neurological disease was provided to P.M.B. by the Edinburgh Sudden Death Brain and Tissue Bank with ethical approval to be used as described in the study (East of Scotland Research Ethics Service, Reference: LR/11/ES/0022). Matched snap frozen frontal cortex tissue was also obtained for CTRL-42, CTRL-45 and CTRL-55. Snap frozen hippocampus tissue and fibroblasts were obtained post-mortem from an AGS patient (ID: AGS-1) in the context of a joint study between M.S.v.d.K and A.V. with ethical approval to be used as described (Children’s National Medical Center IRB, Reference: Pro00000082). Further ethics approvals were provided by the Mater Health Services Human Research Ethics Committee (Reference: 1915A) and the University of Queensland Medical Research Ethics Committee (Reference: 2013001113). Age and gender information for each de-identified individual is provided in Table S1. AGS-1 carried SAMHD1 mutations c.646-647 delAT (p.Met216fs) and c.1223G>C (p.Arg408Pro). From each sample, DNA was extracted using a standard phenol-chloroform protocol. DNA was re-suspended in 75 μl TE, dissolved overnight at 4°C, diluted 10-fold and quantified by Nanodrop and by Qubit HS DNA Fluorometer (Life Technologies Carlsbad, CA).
Purification and Isolation of Neural Cell Nuclei
For each individual, intact nuclei were purified from 5 mg hippocampus tissue and labeled with an anti-NeuN antibody, as described previously (Jiang et al., 2008; Okada et al., 2011). Reagents were pre-chilled and the entire procedure performed on ice. Frozen samples were first gently dounce homogenized for 2 min in 2 ml nuclei extraction buffer (NEB) with 0.1% Triton X-100. Brain homogenates were then filtered through a 40 μm cell strainer and centrifuged at 3,000 rpm for 7 min. Following centrifugation, a block buffer with 10% goat serum was added to the nuclei pellet and incubated for 10 min without disturbing the pellet. To immuno-tag neuronal nuclei, anti-NeuN (Millipore #MAB377) and mouse IgG1 APC (Abcam #ab130786) antibodies were co-incubated for 10 min at 4°C. The nuclei solution was added to the mixed antibodies and incubated for one hour at 4°C. Nuclei were spun down as above and re-suspended in 1X PBS. Prior to FACS, samples including unlabeled controls were stained for 5 min on ice with SYTOX Blue dead cell stain (Life technologies: #S34857) at 1/400 dilution to eliminate debris and collect only intact nuclei (Figure S1). NeuN+ nuclei were sorted with a MOFLO Astrios high speed cell sorter (Beckman Coulter) into 1.5 ml collection tubes and re-analyzed to confirm purity (Figure S1E). Purified nuclei were then picked using an Olympus IX71 inverted microscope, with an Eppendorf TransferMan 2 micromanipulator and Eppendorf Cell Tram. After picking, nuclei were washed in PBS, transferred to individual UV sterilized 0.2 ml PCR tubes and snap frozen. Nuclei were isolated in batches of 24 for amplification. Negative (washed PBS aliquots), positive (gDNA and 1 μl nuclei solution) and no template controls were also included for each batch, with two of each control type included (8 total). To isolate hippocampal glia, we followed the above protocol with the exception that intact nuclei were incubated with anti-NeuN antibody and a conjugated Ki67 antibody (eBioscience#50-5698-80) for one hour at 4°C. Before FACS, samples were again stained with SYTOX Blue for 5 min. Glia were then purified by sorting NeuN–/Ki67– nuclei. NeuN+ cortical nuclei were purified as per hippocampal neurons, instead using matched cortex tissue. Pyramidal neurons were identified as a sorted population of large NeuN+ nuclei, as performed previously (Evrony et al., 2012).
Single-Cell Whole-Genome Amplification
Whole genome amplification (WGA) was performed following extensive optimization of the MALBAC protocol (Zong et al., 2012). Nuclei were denatured at 95°C for 2 min in 15 μl denaturation buffer (20% Bst2.0 buffer, 0.4 μM dNTPs, 0.3 μM Bst primer (GTG AGT GAT GGT TGA GGT CTT GTG GAG NNN NNN NN)), then transferred to an ice slurry. 5 μl Bst2.0 (NEB M0537L) enzyme mix was then added (10% Bst2.0 buffer, 10% Bst2.0) and reactions were placed on an Eppendorf ProCycler S thermal cycler and amplified using the following program: 4°C 10 sec, 1% ramp to 65°C 5 min, then 4 cycles of 94°C 10 sec, 4°C 10 sec, 1% ramp to 65°C 5 min, then 80°C 20 min with a 4°C hold. Individual Bst products were then evenly split into 3 individual PCR reactions. One reaction was performed using Expand Long Range Enzyme Blend (Roche Cat# 04829069001) (0.6X Buffer 2, 5% DMSO, 0.5 μM dNTPs, 0.3 μM PCR primer (GTG AGT GAT GGT TGA GGT CTT GTG GAG), 2.2 units Expand Long Range Enzyme Blend) with the following program: 92°C 2 min, 10 cycles of 92°C 30 sec, 62°C 30 sec, 68°C 6.5 min, then 15 cycles of 92°C 30 sec, 62°C 30 sec, 68°C 6.5 min + 20 sec/cycle, then 68°C 10 min, followed by a 10°C hold. The remaining two reactions were performed with Kapa Robust polymerase (Kapa Biosystems, KK5525) (0.6X BufferA, 1X Enhancer, 0.2 μM dNTPs, 0.3 μM PCR primer (GTG AGT GAT GGT TGA GGT CTT GTG GAG), 2 units Kapa Robust polymerase) with the following program: 94°C 30 sec, 25 cycles of 94°C 30 sec, 62°C 30 sec, 72°C 5 min, then 72°C 5 min, 10°C hold. DNA from PCR amplification was quantified using a Qubit HS dsDNA kit (Invitrogen). Samples were ranked on DNA yield and uniformity of amplification between enzymes. Highest yielding reactions with even amplification across Kapa and Expand reactions were selected, with one of each reaction pooled together using a 1:2 (weight:weight) mix of Expand and Kappa reactions. These pools were then used for Illumina library preparation and later PCR validation experiments.
Illumina Library Construction
Multiplexed Illumina libraries were prepared from bulk input DNA following a previous method (Shukla et al., 2013) with the following modifications: 1 μg input DNA quantified using a Qubit HS DNA Fluorometer was sonicated using a Covaris M220 (Covaris Woburn, Massachusetts) with the following settings: peak power 50, duty factor 20, cycles per burst 200, and time 120 sec. DNA was end repaired, A-tailed, and ligated following the Illumina TruSeq library preparation protocol. Pre-hybridization ligation mediated PCR (LMPCR) consisted of one reaction per library with 50 μl Phusion High-Fidelity Master Mix with HF Buffer (NEB Ipswich, MA), 1 μl of 100 μM TruSeq Forward Primer (TS-F) 5′-AATGATACGGCGACCACCGAGA, 1 μl of 100 μM TruSeq Reverse Primer (TS-R) 5′-CAAGCAGAAGACGGCATACGAG, and molecular grade water to 100 μl. Cycling conditions were: 98°C for 45 sec followed by 8 cycles of: 98°C for 10 sec, 60°C for 30 sec, and 72°C for 30 sec, ending with 72°C for 5 min and held at 4°C until sample clean up. Samples were cleaned with AMpure XP beads (Beckman Coulter Brea, CA) following manufacturer instructions, except using a 1:1.1 ratio of DNA to beads. Samples were eluted in molecular grade water and quantified using the Bioanalyzer DNA 1000 chip (Agilent Technologies Santa Clara, CA) following manufacturer’s instructions.
Single-cell Illumina libraries were prepared as described for bulk input DNA with the following modifications: DNA input consisted of 500 ng of WGA material, with Expand and Kapa PCR products mixed at a 1:2 ratio, and sonication time was reduced to 90 sec to account for shorter fragments generated by WGA. For end repair, A-tailing and ligation half volume reactions were used. LMPCR reaction volumes were also reduced by half. Single-cell libraries were barcoded, pooled, and subjected to Illumina whole genome sequencing (WGS) for QC purposes and copy-number analysis, prior to RC-seq hybridization.
Single-Cell Copy-Number Variation Analysis
We assessed allelic dropout (AD) and locus dropout (LD) in single-cell libraries using low coverage WGS, as well as WGS performed on matched bulk samples. 7/120 single hippocampal neuron libraries (CTRL-42-HN-#7, CTRL-42-HN-#8, CTRL-42-HN-#9, CTRL-45-HN-#14, AGS-1- HN-#6, AGS-1-HN-#21, AGS-1-HN-#24), 1/36 single cortical neuron libraries (CTRL-55-CN-#5) and 2/24 single hippocampal glial cell libraries (CTRL-45-HG-#5, CTRL-55-HG-#1) were excluded from analysis due to low concentration before or after pooling. The remaining 170 libraries were Illumina sequenced (Ambry Genetics, USA; Macrogen, South Korea), generating 1,097,474,654 2x150-mer read pairs (Table S1). Matched bulk WGS for all 5 individuals (liver for controls, fibroblast for AGS-1) were also Illumina sequenced, generating 37,601,911 2x150-mer read pairs. Read pairs were first trimmed from their 5′ and 3′ ends to remove any bases with quality <10, then assembled into contigs using FLASH (Magoč and Salzberg, 2011) and default parameters. Read contigs were then sequentially aligned to amplification primers by BLAST (parameters -m 8 -a 4 -F F, minimum score 22) to trim reads of these sequences, then to the reference genome (hg19) using SOAP2 (Li et al., 2009) (parameters -M 4 -v 2 -r 1 -p 8). Unmapped reads were then aligned to hg19 using LAST (Kiełbasa et al., 2011) (parameters –s 2 –l 11 –d 30 –q 3 –e 30) to allow for soft clip alignments. Only uniquely aligned reads were retained. PCR duplicates were removed if they shared the genomic coordinates of another read. Bulk liver and fibroblast libraries were not subjected to WGA and therefore were not trimmed of amplification primers by BLAST. ∼6.4 M uniquely mapped read pairs were produced per single-cell WGS library (Table S1). On average, WGS generated 0.35X genome coverage for single-cell WGS libraries and 0.49X genome coverage for bulk WGS libraries.
To identify instances of AD and LD, we first divided the genome into 5,000 variable size “bins” with an average size of ∼600 kb, following established methods (Evrony et al., 2012; Navin et al., 2011). This strategy was chosen to avoid false positives due to variation in read mappability across the genome, as demonstrated elsewhere (Navin et al., 2011). Reads mapping to each bin were then counted, with an average of 916 reads per bin in each single-cell library and 1,270 reads per bin in each bulk library. Following established statistical methods (Evrony et al., 2012; McConnell et al., 2013) to correct for GC bias in Illumina sequencing, we divided the bin counts for each single-cell library by the median count of all bins in the same decile of GC content for that library. For each chromosome, copy number was grossly calculated by separately totaling the normalized bin counts in each library and then dividing by the median of the total values for that chromosome. Autosome and female library (CTRL-36, AGS-1) chromosome X total values were then multiplied by two, with male and female sex chromosome data processed separately. For each chromosome, we then divided the normalized bin counts by the median normalized bin count across all libraries for that chromosome, multiplied by 2 for autosomes and female chromosome X to calculate copy number, and calculated the median absolute difference (MAD). Bulk liver libraries were processed separately using the same method. Bins with a normalized count 1.5 MADs or less below the median chromosomal copy number were annotated as AD or LD, defined as a normalized count ≤ 1/2 or ≤ 1/16 the median, respectively. Any bin called as AD or LD in a bulk liver library was not considered for analysis of AD or LD in the matched single-cell libraries, to exclude putative germline structural variants. Male sex chromosomes were considered only for LD. These analyses revealed six examples of chromosome-wide AD (Table S1), where >50% of bins were annotated as AD or LD. To find localized, high-confidence examples of AD or LD, such as those described elsewhere (McConnell et al., 2013), and distinct to underlying amplification variability as shown in Figure S3, we sought genomic segments in each library where >8 bins (∼5 Mb) in a row were found to be AD or LD in only one individual and more than 10 bins away from a telomere, revealing 7 such regions (Table S1), two of which are highlighted in Figure 2B. For these two examples, on chromosome 6 of CTRL-45 hippocampal neuron 2 (CTRL-45-HN-#2), we repeated the above analysis using a genome divided into ∼50,000 variable size bins to achieve a higher resolution (∼60 kb) view of each dropout region (Figure S4C).
During early optimization we noted that WGA in some cases depleted amplified material of L1-Ta 5ʹ and 3ʹ ends. To overcome this, we optimized the Bst and PCR reaction parameters (temperature and cycling conditions) and used combined Expand and Kapa PCR products for library generation, noting that the former kit provided superior genome-wide coverage, and the latter superior detection of L1-Ta insertions (data not shown). In the final method presented here, analysis of WGS reads from single-cell libraries revealed negligible average depletion, or even slight enrichment, for L1-Ta 5′ and 3′ ends versus bulk liver WGS (Figure S4D, Figure S4E, Table S1), where normalization was performed against a set of 107 220 bp sequences randomly sampled from the human reference genome.
RC-Seq Hybridization Reactions
The same single-cell libraries analyzed by WGS were also subjected to RC-seq. First, 20 μl SeqCap Easy Developer Reagent (Roche NimbleGen Madison, WI), 20 μl of 100 μM blocking oligo (AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATC/3 ddC/) analogous to the universal end of the Illumina adaptor, and 20 μl of 100 μM blocking oligo corresponding to the index specific adaptor used for each library (SeqCap EZ HE-Oligo Kits A and B, Roche NimbleGen) was added to each pool. We then added an additional 10 μl of a 100 μM stock of blocking oligo 5′-GTGAGTGATGGTTGAGGTCTTGTGGAG to single-cell pool hybridizations to block the WGA primer. All blocking oligos were 3ʹ terminated with a dideoxy cytosine to prevent priming during post-hybridization LMPCR.
Each tube containing library, developer reagent, and blockers was mixed, spun down briefly, and split into two 1.5 ml tubes of equal volume, with each tube used for separate reactions targeting the 5ʹ and 3ʹ end of an L1-Ta consensus sequence (Dombroski et al., 1993). For each end, a single Locked Nucleic Acid (LNA) (Exiqon Vedbaek, Denmark) probe was identified with high melting temperature and high specificity to L1-Ta. LNA probe LNA-D /5Biosg/CTCCGGT+C+T+ACAGCTC+C+C+AGC targeted the 5ʹ end and LNA-B /5Biosg/AG+A+TGAC+A+C+ATTAGTGGGTGC+A+GCG targeted the 3ʹ end (+ denotes LNA positions within each probe). Probes were biotinylated at their 5ʹ end for capture with strepdavidin beads. Prior to hybridization with these LNA probes, sample pools were dried down in a SpeedVac at 60°C for 30 min. To the dried samples, 7.5 μl of hybridization buffer and 3 μl of Component A (Roche NimbleGen Madison, WI) were added. Samples were vortexed and briefly spun down then placed on a heat block at 95°C for 5 min. Samples were mixed to ensure all DNA was dissolved and then quickly spun. Each halved library pool was then added to a 0.2 ml PCR tube containing 4.5 μl LNA capture probes normalized to 10 μM. Each reaction was mixed by pipetting up and down 10 times. These hybridization cocktails were placed into a pre-heated thermocycler at 95°C for 2 min, then transferred directly to a pre-heated thermocycler at 47°C with heated lid set to 57°C and incubated for 3 days.
Following incubation, hybridization cocktails were washed following the SeqCap SR User’s Guide (Roche NimbleGen) and eluted in 50 μl water. Post-hybridization LMPCR was identical to pre-hybridization LMPCR with two exceptions: two LMPCR reactions were used for each sample, and then combined, and the number of PCR cycles was increased to 10. Reactions were cleaned with a MinElute column (QIAGEN) and eluted in 15 μl EB heated to 60°C. Ampure XP beads were not used for this step due to poor recovery. Samples were quantified with a Qubit HS DNA Fluorometer. L1 5ʹ and 3ʹ capture pools were then combined, with 70% of the final mass of each pool comprising 3ʹ captured DNA and 30% 5ʹ captured DNA. Samples were quantified using a Bioanalyzer DNA 1000 chip. Single-cell capture pools were sequenced with an Illumina MiSeq to verify successful L1-Ta enrichment (data not shown).
Bulk RC-Seq Analysis
Post-enrichment, bulk RC-seq library pools were Illumina sequenced, generating 462,031,340 2x150-mer read pairs (Table S1). Read contig assembly and QC were performed as for WGS. Read contigs were then aligned to hg19 using SOAP2 (Li et al., 2009) (parameters -M 4 -v 2 -r 1 -p 8). As for WGS data, only uniquely aligned reads were retained and PCR duplicates were removed if they shared the genomic coordinates of another read. Unmapped reads were then aligned to a set of potentially active human retrotransposon consensus sequences listed elsewhere (Baillie et al., 2011), using LAST (Kiełbasa et al., 2011) (parameters –s 2 –l 11 –d 30 –q 3 –e 30). Reads aligned at >95% identity to L1-Ta, L1-pre-Ta or L1PA2 families were retained if this alignment spanned ≥33 nt of one contig end. The computational pipeline for alignment, filtering, and clustering then followed an existing strategy (Shukla et al., 2013) including alignment to hg19 with SOAP2 and LAST, with the main exception being that clusters needed to only contain a single aligned read to be reported. This reduction in reporting threshold led us to include an additional step, with reference to our earlier strategy (Shukla et al., 2013), to exclude molecular chimeras by removing any read with an alignment of its non-retrotransposon section plus 5 nt to the genome, rather than 10 nt. We also extracted, in silico, 1 kb from the reference genome extending downstream from an aligned non-retrotransposon section and aligned the full read contig against this region with BLAT and default parameters, to exclude genomic rearrangements (e.g., deletions) involving nearby L1 copies. RC-seq reads processed in this way were used to determine bulk RC-seq sensitivity, as well as L1-Ta enrichment and sequencing depth, with reference to 960 L1-Ta copies annotated in hg19 (Evrony et al., 2012). Sensitivity was also assessed based on how many polymorphic L1-Ta insertions were found per sample. Polymorphic L1-Ta insertions were called as present in an individual if they met a stringent reporting threshold of having at least 8 RC-seq reads in at least one sample from that individual, were detected in both hippocampus and liver (or AGS-1 fibroblast) from that individual, and the read count for at least one sample from that individual exceeded >0.1X the max number of reads in any bulk RC-seq library. L1 insertions found only in one bulk hippocampus library by ≥1 RC-seq reads, and absent from any bulk liver RC-seq library or previous publications (Ewing and Kazazian, 2010, 2011; Iskow et al., 2010; Shukla et al., 2013; Wang et al., 2006), were annotated as somatic hippocampal insertions. Similarly, a control cohort of 175 liver-only L1 insertions were annotated as for hippocampal insertions, instead with a reporting threshold of ≥4 RC-seq reads intended to detect possible somatic L1 mobilization events early in liver organogenesis. Notably, the bulk hippocampus RC-seq libraries detected 97.7% of reference genome L1-Ta copies on average (Table S1), indicating that these annotated liver-only L1 insertions were very unlikely to be mis-annotated polymorphic L1 insertions. Ambiguous RC-seq clusters not annotated as either polymorphic or somatic were discarded.
Single-Cell RC-Seq Analysis
Nine single-cell RC-seq pools (CTRL-36, CTRL-42, CTRL-45, CTRL-55 and AGS-1 hippocampal neurons; CTRL-42, CTRL-45 and CTRL-55 hippocampal glia; CTRL-42, CTRL-45 and CTRL-55 cortical neurons) were Illumina sequenced, generating a total of 3,494,481,909 2x150-mer read pairs (Table S1). Computational analysis then proceeded using the method outlined above for bulk RC-seq, with additional filtering of reads pre- and post-clustering to remove potential artifacts generated by WGA and Illumina library preparation. Pre-clustering, each read was re-aligned to hg19 using BLAST (parameters -m 8 -a 4 -F F, minimum score 22) to find the corresponding best alignments for the non-retrotransposon and retrotransposon segments, genome-wide. Reads corresponding to a 5′ L1-genome junction involving an overlap of non-retrotransposon and retrotransposon segments of >2 bp, or a 3′ L1-genome junction with an overlap of >10 bp, were removed as putative chimeras. Reads corresponding to 3′ L1-genome junction with a non-retrotransposon/retrotransposon segment overlap of 6–10 bp were removed if the proportion of adenosine nucleotides in this overlap was <50%. Removal of chimeric reads at 3′ L1-genome junctions was complicated by the priming of L1 reverse transcription typically requiring hybridization in vivo of the L1 polyA-tail with genomic thymine residues proximal to a site nicked by the L1 EN domain (Feng et al., 1996; Jurka, 1997; Luan et al., 1993), in effect resembling a ‘chimera’ of the L1 3′ end and the host genome at the integration site. The known location of the RC-seq LNA probes, and a known library insert size, allowed us to remove reads indicating 5′ L1-genome junctions >40 nt and <5,700 nt, or >5,930 nt, away from the L1-Ta 5′ end as probable artifacts. Reads indicating truncations of >5 nt from the L1-Ta 3′ end were removed, as a polyA-tail is an established feature of most retrotransposition-competent L1 copies (Moran et al., 1996). Prospective L1 3′ transduced regions were also required to terminate with a polyA-tail of at least 10 bp and 50% adenosine. Any reads where the retrotransposon segment aligned within 5 kb of the non-retrotransposon segment on the genome were also removed, to filter out potential WGA artifacts involving the formation of loops between reference L1-Ta copies and nearby regions of microhomology. Post-filtering, clusters overlapping a RepeatMasker annotated primate-specific L1 insertion (“L1H” and “L1P”) or an Alu were also removed, due to expected difficulty in validating these events, and their high potential to generate WGA artifacts. Polymorphic and somatic L1 insertions were annotated as for bulk RC-seq, with ≥1 single-cell RC-seq reads required to annotate the latter category.
We assessed the sensitivity of single-cell RC-seq by selecting polymorphic insertions detected by at least 40 reads in the corresponding bulk RC-seq, representing a cohort of ‘gold-standard’ variants likely to approximate a true positive rate of 100% based on prior experiments (Shukla et al., 2013), and determined how many of these polymorphic insertions were detected in the corresponding single-cell libraries. Notably, somatic variants are all heterozygous, while some polymorphic variants are homozygous. Thus, using detection of polymorphic L1 insertions as a measure of sensitivity for detection of somatic L1 insertions may underestimate somatic retrotransposition.
Structural Characterization and Validation of Somatic L1 Insertions
Twenty somatic L1 insertions detected by single-cell RC-seq at a 3′ L1-genome junction were selected at random for validation and structural elucidation (Tables S2 and Data S1), each requiring resolution of the corresponding 5′ L1-genome junction by insertion site PCR and sequencing. Insertion site primers, as denoted by an α symbol in Figure 3 and listed in Table S2, were first designed by Primer3, or by hand where a primer was not initially identified within 200 bp of the integration site. As the extent of L1 5′ truncation was unknown, primers oriented antisense to L1 (denoted by a β symbol in Figure 3) were designed approximately every 500 bp through L1-Ta. PCR reactions were then prepared for each possible insertion site / L1 primer combination. PCRs were performed using Invitrogen Platinum Taq DNA Polymerase High Fidelity with the following conditions: 1x High Fidelity PCR buffer, 2 mM MgSO4, 0.2 mM dNTPs, 0.2 μM primers, 7.5U polymerase and 10 ng template DNA. Thermal cycling was performed as follows: initial denaturation at 94°C for 3 min, then 27 cycles of 94°C for 30 sec, 57°C for 30 sec, 68°C for 30 sec, followed by final extension at 68°C for 10 min, then hold at 10°C. PCR products were quantified using a Qubit HS dsDNA kit following the manufacturer’s protocol. Amplicons were then combined in equimolar ratios to give 500 ng of pooled DNA. Illumina libraries were then prepared using the NebNEXT ultra library preparation kit with the following modifications: an initial size selection was performed using 1:0.6 then 1:3 sample to Ampure XP bead ratios. Post-ligation size selection was not performed. Libraries were amplified by 6 cycles of LM-PCR. Library molarity was determined using an Agilent DNA 1000 chip. Concentration was determined using a Qubit HS dsDNA kit. Libraries were sequenced on a MiSeq using a 600 cycle v3 kit. Reads were then interrogated using custom Python scripts to identify sequences corresponding to a full-length or truncated L1 5′ end and also aligning to one of the 20 putative integration sites. 5′ L1-genome junctions identified in this manner were then validated by PCR followed by standard agarose gel electrophoresis using the same amplification conditions as described above. In some instances, nested primers were required, and nested PCR was performed using 27 cycles, then 35 cycles of amplification. Ten L1 insertions detected only at a 5′ L1-genome junction were also tested, although as we have previously encountered (Baillie et al., 2011), PCR amplification of the corresponding 3′ L1-genome junction was extremely challenging, and none of these examples were confirmed (Table S2). Four L1 insertions detected at both their 5′ and 3′ ends were however confirmed by PCR (Table S2).
5′ L1-Genome Junction Amplification Analysis
Polymorphic L1 insertions detected at their 3′ L1-genome junction by single-cell RC-seq from CTRL-42, CTRL-45 and CTRL-55 (four L1s from each individual) were confirmed as heterozygous by empty/filled site PCR using template bulk hippocampus DNA (Table S2). PCR involved the following reagents: 1U MyTaq DNA polymerase (Bioline, Australia), 1x MyTaq Reaction Buffer, 2 μM primers and 10 ng template DNA. Thermal cycling was performed as follows: initial denaturation at 95°C for 1 min, then 35 cycles of 95°C for 30 sec, 57°C for 15 sec, 72°C for 10 sec, followed by final extension at 72°C for 10 min, then hold at 10°C. Insertion site and L1 primers were then combined to amplify the 5′ L1-genome junction for each heterozygous L1 again using bulk hippocampus DNA as template and the same reagent mixture and cycling conditions as for the empty/filled site assay above. Once the 5′ L1-genome junctions were confirmed, targets were re-amplified using the WGA material from ∼8 randomly selected neurons for each insertion where the 3′ L1-genome junction was already identified by single-cell RC-seq. For this PCR, we used the following reagent mixture: 1U MyTaq DNA polymerase, 1x MyTaq Reaction Buffer, 2 μM primers and 50 ng WGA template DNA. Thermal cycling was performed as follows: initial denaturation at 95°C for 1 min, then 37 cycles of 95°C for 30 sec, 57°C for 15 sec, 72°C for 10 sec, followed by final extension at 72°C for 10 min, then hold at 10°C. Twelve heterozygous polymorphic L1s were analyzed across 100 neurons in total, with a 5′ L1-genome junction found in exactly 50 instances (Table S2), indicating that the maximum possible estimated validation rate with TSDs was 50%.
TaqMan qPCR L1 CNV Assay
1 μg DNA from each hippocampus sample was run on a 0.7% agarose gel. Bands ≥10 kb in size were selected by excision to exclude any possible L1 cDNA and subjected to gel purification using a Qiagen gel purification kit (#28606). This high molecular weight genomic DNA was then used as input for qPCR, using the ‘ORF2 #1’ combination first used to assay L1 CNV by Coufal et al. (Coufal et al., 2009), with α-satellite repetitive elements (SATA) as an immobile control. Incorporating minor modifications made by Baillie et al. (Baillie et al., 2011), experiments were performed using Roche TaqMan master mix (#04707494001) and run on a LightCycler 480 (Roche). Quantification included five technical replicates. The ratio of L1 ORF2 to SATA in each hippocampal DNA sample was finally normalized to the signal for CTRL-36.
Insertion Enrichment Analyses
To determine whether somatic L1 insertions were enriched in genes differentially expressed in the same spatiotemporal context, we downloaded single-molecule CAGE data published by FANTOM5 (Forrest et al., 2014), including hippocampus, caudate nucleus, frontal cortex, liver, and heart CAGE experiments. CAGE tag clusters within 1 kb of, and on the same strand as, the annotated 5′ end of RefSeq genes (available from the UCSC Table Browser) were used to compile a table of expression for each gene, with expression normalized to tags-per-million. Pairwise comparisons were then performed between each tissue (as displayed in Figure 7A) to identify genes differentially upregulated in each tissue. Differential expression was defined by edgeR (Robinson et al., 2010), using an exact negative binomial test with Bonferroni correction. Overexpressed genes were ranked by P-value, with the most significant 5% of genes selected as a differentially expressed cohort in each pairwise comparison. For each list, we then calculated enrichment for somatic L1 insertions versus what proportion of insertions were expected based on a random selection of 1x107 genomic points. Significance testing involved two-tailed Fisher’s exact tests, with Benjamini-Hochberg correction.
To assess whether somatic L1 insertions were enriched in enhancer elements, we first downloaded .bed tracks for robust and permissive bidirectionally transcribed enhancers defined by FANTOM5 (Andersson et al., 2014), as well as 47 .bed tracks for cell-type specific enhancer elements. We then intersected the genomic coordinates of L1 insertions with those of each enhancer class, requiring an L1 insertion to be within 100 nt of an enhancer to record an intersection, as for Figure 7C. In Figure 7D, this window was extended to assess how L1 insertion frequency progressively decreased as the distance to the nearest NSC-specific enhancer increased. Observed values were compared with values expected from random sampling using two-tailed Fisher’s exact tests with Bonferroni correction.
Meta-Analysis of L1-IP Data
Single-neuron L1 insertion profiling (L1-IP) reads were downloaded, trimmed, and then aligned as per Evrony et al. (Evrony et al., 2012) using Bowtie version 1.1.1. (Langmead et al., 2009). Reads less than 500 bp apart and on the same strand were clustered into peaks containing a minimum of a single read and annotated using the RepeatMasker track from human genome build hg19 and locations of known non-reference elements. Peaks with consistent orientations and overlapping locations ±500 bp were merged, and counts were summed across samples using BEDTools version 2.20.1-4-gb877b35.
Hippocampal L1 Insertions Prefer Genomic Loci Transcribed in the Hippocampus
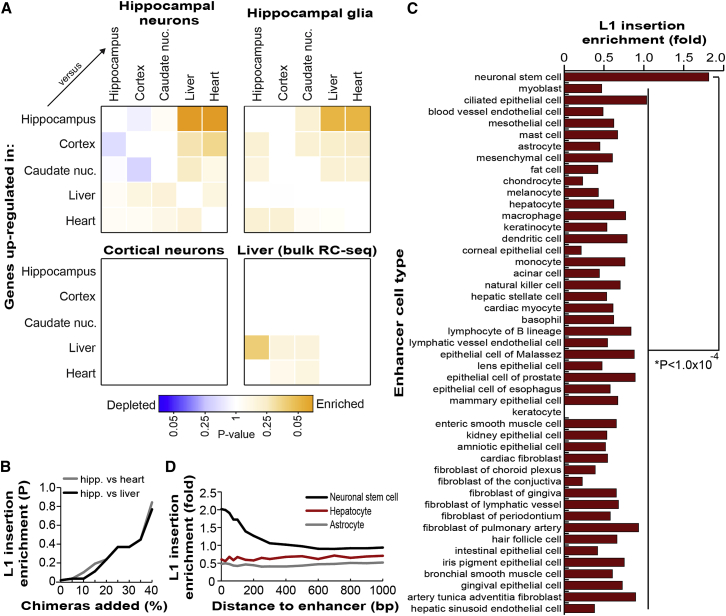
Open chromatin is a typical prerequisite for efficient transcription (Neph et al., 2012). With this in mind, we used single-molecule cap analysis of gene expression (CAGE) transcriptome profiling data from the FANTOM5 consortium (Forrest et al., 2014) to test whether genes strongly transcribed in the hippocampus were specifically enriched for somatic L1 insertions in hippocampal neurons. We first identified genes differentially upregulated in hippocampus, cortex, caudate nucleus, liver, or heart tissue surveyed by CAGE and then intersected these gene lists with the cohort of intragenic somatic L1 insertions detected by single-cell RC-seq applied to hippocampal neurons. Only those genes upregulated in hippocampus versus heart, and hippocampus versus liver, were significantly enriched (p < 0.05, Fisher’s exact test, Benjamini-Hochberg correction) for insertions (Figure 7A, Table S6). Somatic L1 insertions in hippocampal glia were also most enriched in genes upregulated in the hippocampus (p < 0.07). No enrichment was observed for cortical neurons while, intriguingly, the liver-specific L1 insertion cohort exhibited enrichment (p < 0.11) in genes upregulated in liver versus hippocampus (Figure 7A). Finally, we calculated the significance of enrichment for hippocampal neuron L1 insertions in genes upregulated in hippocampus while incrementally introducing putative artifacts described in Figure 5B. We found that statistical significance was no longer achieved once the dataset contained 15% or more artifacts (Figure 7B), hence demonstrating how experimental noise reduced in single-cell RC-seq analyses would otherwise obscure genome-wide enrichment. These experiments altogether reveal context-dependent, preferential L1 mobilization into strongly transcribed loci.
Figure 7.
Genome-Wide Somatic L1 Insertion Patterns
(A) Somatic L1 insertions detected by single-cell RC-seq in hippocampal neurons and glia were enriched in genes differentially upregulated in hippocampus. Liver-specific L1 insertions detected by bulk RC-seq were moderately enriched in genes upregulated in liver. No enrichment was observed for cortical neurons. Color intensity is based on the absolute log2 transformed p value determined by Fisher’s exact test (Benjamini-Hochberg correction) with blue and orange colors representing depletion and enrichment, respectively. Note: in each matrix pairwise comparison, the more highly expressed tissue is on the y axis.
(B) Hippocampal somatic L1 insertions were statistically enriched in genes upregulated in hippocampus versus liver (black) or hippocampus versus heart (gray), as shown in (A). However, as previously filtered molecular chimeras (see Figure 5B) were re-introduced into this dataset, enrichment rapidly became no longer significant.
(C) Of the transcribed cell-type specific enhancers defined by FANTOM5, only those of neuronal stem cells were enriched (observed/expected) for somatic L1 insertions detected by bulk hippocampus RC-seq, compared with other enhancers (p < 1.0 × 10−4, Fisher’s exact test, Bonferroni correction).
(D) Somatic L1 insertion enrichment in neuronal stem cell enhancers (black) extended 500 bp from enhancer boundaries. No enrichment was observed for astrocyte (gray) or hepatocyte (red) enhancers.
Noting that euchromatin is also a signature of active enhancer elements, we intersected our list of somatic L1 insertions detected by hippocampus bulk RC-seq with an extensive FANTOM5 catalog of transcribed constitutive and cell-type specific enhancers defined by histone modifications and CAGE-delineated transcriptional activity (Andersson et al., 2014). Globally, no substantial difference was observed in the rate of L1 insertions in all enhancers versus random expectation. However, of 47 cell-type specific enhancer sets, only neuronal stem cell enhancers were significantly enriched for somatic L1 insertions, compared with random expectation (p < 0.01, Fisher’s exact test, Bonferroni correction) and compared with the union of the remaining 46 cell-type specific enhancer sets (Figure 7C; p < 1.0 × 10−4, Fisher’s exact test). This enrichment was highest for L1 insertions within 100 nt of an enhancer, and was observed up to 500 nt from defined enhancer boundaries (Figure 7D). No enrichment was observed for astrocytes or for other cells not of the neuronal lineage, such as hepatocytes (Figure 7D). The smaller cohorts of somatic L1 insertions detected by single-cell RC-seq and liver bulk RC-seq were insufficient to perform meaningful statistical analyses of L1 insertional preference with regards to enhancers. Nonetheless, hippocampus bulk RC-seq indicated that neuronal stem cell-specific enhancers were the most highly enriched genome functional element in absolute terms (1.8-fold) for somatic L1 insertions. This reinforced the view that L1 mobilization during neurogenesis impacts regulatory and protein-coding loci specifically active in the hippocampus.
A Potential Signature of Neurogenic L1 Selection
De novo germline L1 insertions can be highly deleterious to gene function, and commonly undergo purifying selection (Boissinot et al., 2001; Han et al., 2004). The L1 ORF2 segment of sense oriented intronic L1 insertions particularly hinders RNA polymerase processivity (Han et al., 2004; Lee et al., 2012). Hence, while sense and antisense intronic L1 insertions are assumed to occur with equal frequency in the germline, sense insertions are selected against more strongly and tend to be eliminated from the population. It follows that an estimated 43.3% of recent intronic L1-Ta insertions are sense oriented, versus only 34.1% of fixed L1-Ta insertions and 39.7% of all polymorphic L1-Ta insertions (Ewing and Kazazian, 2010). By contrast, sense oriented intronic L1 insertions are not depleted in tumors (Lee et al., 2012). Among the control individuals examined here, we found that, as expected, 42/101 (41.6%) of intronic, polymorphic germline L1 insertions were sense oriented to their host gene. Surprisingly, 406/1,024 (39.6%) of intronic somatic L1 insertions detected in hippocampal neurons by single-cell RC-seq were also sense oriented, significantly less than the expected 50% (p < 0.0001, exact binomial test). This proportion was 47/136 (34.6%) and 166/503 (33.0%) for glia and cortical neurons, respectively. Adhering to the prevailing germline model of L1 evolutionary selection, we concluded that some somatic L1 insertions may arise sufficiently early in neurogenesis to impact neural progenitor cell fitness, as indicated by a depletion of sense oriented events in mature neurons and glia.
Discussion
Our experiments firmly establish that L1-driven mosaicism pervades the hippocampus and is mediated by TPRT. That we found 13.7 somatic L1 insertions per hippocampal neuron was unexpected given a prior estimate of <0.1 insertions per cortical neuron (Evrony et al., 2012). By discovering here a myriad of L1 insertions in cortical neurons, we exclude a biological explanation for this discrepancy and instead propose that the process by which the earlier work selected insertions for validation led to a significant underestimate of L1 retrotransposition frequency. Indeed, the mobilization rate reported here much more closely resembles an earlier estimate of 80 somatic L1 insertions per brain cell, calculated via L1 qPCR (Coufal et al., 2009).
Beyond this, our data demonstrate that L1 insertions in hippocampal neurons and glia are preferentially found in protein-coding genes highly transcribed in the hippocampus. Transcribed enhancers active in neuronal stem cells are also enriched for somatic L1 insertions, indicating likely L1 perturbation of regulatory elements. L1 insertions in cortical neurons were however not significantly enriched in genes highly transcribed in the cortex. We speculate that this could be due to cortical neurogenesis primarily occurring during fetal development (Spalding et al., 2005), which presents a genome-wide transcriptional profile different to that of the adult cortex. Although L1 mobilization was not increased in AGS-1 hippocampal neurons, the pattern of L1 insertions was prospectively different to that of controls, the reasons for which are presently unclear. The most obvious caveat of this analysis is that, due to the extreme rarity of the disease, only one AGS patient hippocampus was studied. Nonetheless, this experiment serves as a proof-of-principle demonstration that single-cell RC-seq could be used in the future to assess abnormal L1 mobilization in neurological disease. Finally, we noted that somatic L1 insertions in neurons bore substantially longer TSDs on average than polymorphic L1 insertions, corroborated by structural characterization of L1 integration sites found by single-cell RC-seq. Unusually long TSDs have previously been identified using an engineered L1 reporter system in HeLa cells (Gilbert et al., 2005). As also hypothesized in that context, pervasive euchromatinization in neural progenitor cells may promote the formation of long TSDs.
The predominant developmental timing of endogenous L1 mobilization in the brain remains unclear. Although the vast majority of somatic L1 insertions detected by single-cell RC-seq were found in one cell each, a small proportion of L1s were detected in multiple cells, including examples found in both glia and neurons, indicating L1 mobilization in a common multipotent progenitor cell. Three somatic L1 insertions were validated by PCR in multiple neurons, including one example found in nearly 50% of the neurons assayed. Thus, although most L1 insertions may occur in one or a handful of neurons, a substantial number appear to arise during early neurogenesis. Indeed, the signature of potential selection against somatic L1 insertions sense oriented to host gene introns suggests that many retrotransposition events precede terminal neural cell maturation. We speculate that depletion of these events could be explained by preferential L1 integration into neurogenesis genes, thereby impacting the survival or differentiation potential of neural progenitor cells. It also cannot be excluded that somatic L1 integration primarily occurs antisense to host gene introns, though we currently lack a mechanistic explanation for this preference.
Neuronal genome mosaicism may not be restricted to somatic L1 insertions. Alu and SVA retrotransposons trans mobilized by L1 may also contribute mosaic insertions. Other than transposable element activity, recent studies have reported localized and chromosome-wide CNV in normal neurons (Cai et al., 2014; Gole et al., 2013; McConnell et al., 2013). We find no definitive evidence of these events in our data, though it must be noted that our CNV analyses were expressly geared to discern genomic deletions caused by WGA failure or variability. However, it must be noted that we found consistent WGA inefficiency at telomeres, while others have reported that most apparent small genomic deletions occur close to telomeres (McConnell et al., 2013).
L1 mosaicism may also occur outside of the brain, for instance during early embryogenesis (Garcia-Perez et al., 2007; Kano et al., 2009) or, as we previously reported for a single L1 insertion, in the liver (Shukla et al., 2013). However, some cell types present practical and technical challenges not posed by neural cells. For example, hepatocytes are frequently multinucleated and sustain aneuploidy and polyploidy, greatly complicating single-cell genomic analysis. Thus, although the liver-specific L1 insertions detected here by bulk RC-seq consistently bore L1 EN motifs and were enriched in genes differentially upregulated in liver, we were unable to corroborate these findings with single-cell RC-seq or downstream PCR validation. Future methodological advances will therefore likely be required to elucidate L1 mosaicism in the liver, and elsewhere in the body.
The capacity to locate somatic L1 insertions in individual neural cell genomes is a major step toward determining whether mosaicism impacts neurobiological function. Limitations in assaying the transcriptome and genome of the same cell however currently prohibit functional assays of individual somatic L1 insertions. Nonetheless, given the frequency of these events, their mutagenic potential for protein-coding and regulatory regions and an apparent preference for euchromatic DNA linked to neurobiological function, it is not unreasonable to predict that L1-driven somatic mosaicism may alter the functional properties of the brain.
Experimental Procedures
Full protocols can be found in the Extended Experimental Procedures.
Samples
Control tissues were provided by the Edinburgh Sudden Death Brain and Tissue Bank. Tissues were obtained post-mortem from AGS-1 with ethical approval to be used as described. AGS-1 carried SAMHD1 mutations c.646-647 delAT (p.Met216fs) and c.1223G>C (p.Arg408Pro). Patient age and gender information is provided in Table S1.
Single-Cell RC-Seq
NeuN+ (neuronal) and NeuN–/Ki67– (glial) nuclei were isolated via FACS from brain tissue, individually picked under microscope and subjected to linear WGA. Products were split into three exponential PCR reactions utilizing two different kits, and then combined for library preparation and downstream PCR validation. Multiplexed Illumina libraries were pooled and sequenced (2 × 150-mer reads) to assess allelic dropout and L1-genome junction depletion, then hybridized separately to two LNA probes respectively matching the 5ʹ and 3ʹ ends of L1-Ta. Post-enrichment, RC-seq libraries were sequenced (2 × 150-mer reads), computationally processed, filtered to exclude artifacts, and finally used to call polymorphic and somatic L1 insertions.
5′ L1-Genome Junction Validation and Characterization
Twenty somatic L1 insertions detected by single-cell RC-seq at a 3′ L1-genome junction were selected at random for structural characterization by PCR amplification and sequencing of the corresponding 5′ L1-genome junction. For each example, initial PCR template DNA consisted of WGA material from the relevant neuron. As the extent of L1 5′ truncation was unknown, primers oriented antisense to L1 were designed approximately every 500 bp through the L1-Ta consensus and combined with an insertion site primer unique to each locus. 5′ L1-genome junctions were identified by PCR and sequencing and then separately PCR amplified again using WGA material from the selected neuron, WGA material from other single neurons from the same individual, as well as matched bulk DNA. Amplified material was stored and handled separately to bulk DNA.
Author Contributions
K.R.U., D.J.G., J.S.J., S.R.R., F.J.S-L., G.O.B., A.D.E., C.S-P., P.M.B., and G.J.F. designed and performed experiments. K.R.U. optimized single-cell WGA and analyzed RC-seq data. D.J.G. optimized RC-seq capture. J.S.J. isolated nuclei and performed WGA. P.M.B. provided control samples. M.S.v.d.K, and A.V. provided AGS patient samples. G.J.F. led the bioinformatic analyses, wrote the manuscript and directed the study. All authors commented on the manuscript.
Acknowledgments
We thank Haig Kazazian and Ryan Lister for critical reading of the manuscript. We also thank Marianna Bugiani for her work in the autopsy of AGS-1. G.J.F. acknowledges the support of an NHMRC Career Development Fellowship (GNT1045237), NHMRC Project Grants (GNT1042449, GNT1045991, GNT1067983 and GNT1068789), and the EU FP7 under grant agreement No. 259743 underpinning the MODHEP consortium. F.J.S-L. was supported by the Alfonso Martín Escudero Foundation. P.M.B. was supported by a Wellcome Trust Clinical Fellowship (090386/Z/09/Z).
Footnotes
This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Accession Numbers
RC-seq and WGS data are available from the European Nucleotide Archive (ENA) using the identifier PRJEB5239.
Supplemental Information
“Bulk library stats” tab details bulk RC-seq mapping statistics, L1 enrichment and L1-Ta detection rates, as well as counts of polymorphic and somatic L1 insertions detected in each individual. “Single-cell library stats” tab details WGS quality control and single-cell RC-seq results for each individual cell studied.
“Polymorphic L1” tab details polymorphic L1s detected by bulk RC-seq. “Sense start” and “Sense stop” refer to the start and stop positions of an RC-seq read contig arranged in sense to the genome. “Antisense start” and “Antisense stop” refer to a cluster arranged antisense to the genome. RC-seq read counts are provided for each sample. Examples with sense and antisense RC-seq clusters depict simultaneous 5′/3′ detection of an insertion. “Donor sense” records the retrotransposon consensus start position (base 0) of sense insertions (negative values or 0) or the stop position of antisense insertions (positive values). “Donor antisense” records the retrotransposon consensus stop position of sense insertions (positive values) or the start position of antisense insertions (negative values or 0). “Polymorphic L1 PCR” tab details 12 heterozygous polymorphic L1 insertions detected at their 3′ L1-genome junction by single-cell RC-seq and tested by PCR for amplification of the corresponding 5′ L1-genome junction in 100 neurons (see Extended Experimental Procedures). “Somatic L1” tab provides single-cell RC-seq results for all hippocampal neurons, hippocampal glia, and cortical neurons presented. Columns are labeled similar to the “Polymorphic L1” tab, with single-cell RC-seq read counts provided here. A large (>50 MB) table detailing hippocampus and liver somatic L1 insertions is available on request. “Somatic L1 PCR” tab provides detailed information for somatic L1 insertions detected by single-cell RC-seq and characterized by PCR and sequencing. Information provided by validation sequencing is highlighted in green. Characterization includes the capillary sequences of confirmed somatic insertions, TSD sizes, the L1 endonuclease recognition motif and the start position of truncated insertions.
Each datasheet indicates the chromosomal position of an L1 insertion, with first- and second-strand L1 EN cleavage sites indicated by red and black arrows, respectively, and the L1 EN motif found at the first-strand cleavage site underlined in black. Target site duplications are highlighted with blue nucleotides. Numbers at the 5′ L1-genome junction in structure cartoons indicate the length of the L1. See Table S2 for additional information.
References
- Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T., FANTOM Consortium An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillie J.K., Barnett M.W., Upton K.R., Gerhardt D.J., Richmond T.A., De Sapio F., Brennan P.M., Rizzu P., Smith S., Fell M. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C.R., Collier P., Macfarlane C., Malig M., Kidd J.M., Eichler E.E., Badge R.M., Moran J.V. LINE-1 retrotransposition activity in human genomes. Cell. 2010;141:1159–1170. doi: 10.1016/j.cell.2010.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck C.R., Garcia-Perez J.L., Badge R.M., Moran J.V. LINE-1 elements in structural variation and disease. Annu. Rev. Genomics Hum. Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S., Entezam A., Furano A.V. Selection against deleterious LINE-1-containing loci in the human lineage. Mol. Biol. Evol. 2001;18:926–935. doi: 10.1093/oxfordjournals.molbev.a003893. [DOI] [PubMed] [Google Scholar]
- Brouha B., Schustak J., Badge R.M., Lutz-Prigge S., Farley A.H., Moran J.V., Kazazian H.H., Jr. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. USA. 2003;100:5280–5285. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Evrony G.D., Lehmann H.S., Elhosary P.C., Mehta B.K., Poduri A., Walsh C.A. Single-cell, genome-wide sequencing identifies clonal somatic copy-number variation in the human brain. Cell Rep. 2014;8:1280–1289. doi: 10.1016/j.celrep.2014.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost G.J., Golding A., Schlissel M.S., Boeke J.D. Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Res. 2001;29:573–577. doi: 10.1093/nar/29.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal N.G., Garcia-Perez J.L., Peng G.E., Yeo G.W., Mu Y., Lovci M.T., Morell M., O’Shea K.S., Moran J.V., Gage F.H. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal N.G., Garcia-Perez J.L., Peng G.E., Marchetto M.C., Muotri A.R., Mu Y., Carson C.T., Macia A., Moran J.V., Gage F.H. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. USA. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewannieux M., Esnault C., Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 2003;35:41–48. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- Evrony G.D., Cai X., Lee E., Hills L.B., Elhosary P.C., Lehmann H.S., Parker J.J., Atabay K.D., Gilmore E.C., Poduri A. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony G.D., Lee E., Mehta B.K., Benjamini Y., Johnson R.M., Cai X., Yang L., Haseley P., Lehmann H.S., Park P.J., Walsh C.A. Cell lineage analysis in human brain using endogenous retroelements. Neuron. 2015;85:49–59. doi: 10.1016/j.neuron.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing A.D., Kazazian H.H., Jr. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–1270. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing A.D., Kazazian H.H., Jr. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res. 2011;21:985–990. doi: 10.1101/gr.114777.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., Kulakovskiy I.V., Lizio M., Itoh M., FANTOM Consortium and the RIKEN PMI and CLST (DGT) A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Perez J.L., Marchetto M.C., Muotri A.R., Coufal N.G., Gage F.H., O’Shea K.S., Moran J.V. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 2007;16:1569–1577. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- Gilbert N., Lutz S., Morrish T.A., Moran J.V. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 2005;25:7780–7795. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gole J., Gore A., Richards A., Chiu Y.J., Fung H.L., Bushman D., Chiang H.I., Chun J., Lo Y.H., Zhang K. Massively parallel polymerase cloning and genome sequencing of single cells using nanoliter microwells. Nat. Biotechnol. 2013;31:1126–1132. doi: 10.1038/nbt.2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodier J.L., Ostertag E.M., Kazazian H.H., Jr. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum. Mol. Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- Han J.S., Szak S.T., Boeke J.D. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–274. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- Hancks D.C., Goodier J.L., Mandal P.K., Cheung L.E., Kazazian H.H., Jr. Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum. Mol. Genet. 2011;20:3386–3400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hozumi N., Tonegawa S. Evidence for somatic rearrangement of immunoglobulin genes coding for variable and constant regions. Proc. Natl. Acad. Sci. USA. 1976;73:3628–3632. doi: 10.1073/pnas.73.10.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow R.C., McCabe M.T., Mills R.E., Torene S., Pittard W.S., Neuwald A.F., Van Meir E.G., Vertino P.M., Devine S.E. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. USA. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H., Godoy I., Courtney C., Vetter M.R., Gerton G.L., Ostertag E.M., Kazazian H.H., Jr. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E., Iskow R., Yang L., Gokcumen O., Haseley P., Luquette L.J., 3rd, Lohr J.G., Harris C.C., Ding L., Wilson R.K., Cancer Genome Atlas Research Network Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–971. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Prazak L., Chatterjee N., Grüninger S., Krug L., Theodorou D., Dubnau J. Activation of transposable elements during aging and neuronal decline in Drosophila. Nat. Neurosci. 2013;16:529–531. doi: 10.1038/nn.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan D.D., Korman M.H., Jakubczak J.L., Eickbush T.H. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- Macfarlane C.M., Collier P., Rahbari R., Beck C.R., Wagstaff J.F., Igoe S., Moran J.V., Badge R.M. Transduction-specific ATLAS reveals a cohort of highly active L1 retrotransposons in human populations. Hum. Mutat. 2013;34:974–985. doi: 10.1002/humu.22327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell M.J., Lindberg M.R., Brennand K.J., Piper J.C., Voet T., Cowing-Zitron C., Shumilina S., Lasken R.S., Vermeesch J.R., Hall I.M., Gage F.H. Mosaic copy number variation in human neurons. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J.V., DeBerardinis R.J., Kazazian H.H., Jr. Exon shuffling by L1 retrotransposition. Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- Muotri A.R., Chu V.T., Marchetto M.C., Deng W., Moran J.V., Gage F.H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri A.R., Marchetto M.C., Coufal N.G., Oefner R., Yeo G., Nakashima K., Gage F.H. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neph S., Vierstra J., Stergachis A.B., Reynolds A.P., Haugen E., Vernot B., Thurman R.E., John S., Sandstrom R., Johnson A.K. An expansive human regulatory lexicon encoded in transcription factor footprints. Nature. 2012;489:83–90. doi: 10.1038/nature11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrat P.N., DasGupta S., Wang J., Theurkauf W., Weng Z., Rosbash M., Waddell S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science. 2013;340:91–95. doi: 10.1126/science.1231965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raiz J., Damert A., Chira S., Held U., Klawitter S., Hamdorf M., Löwer J., Strätling W.H., Löwer R., Schumann G.G. The non-autonomous retrotransposon SVA is trans-mobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012;40:1666–1683. doi: 10.1093/nar/gkr863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson S.R., Morell S., Faulkner G.J. L1 retrotransposons and somatic mosaicism in the brain. Annu. Rev. Genet. 2014;48:1–27. doi: 10.1146/annurev-genet-120213-092412. [DOI] [PubMed] [Google Scholar]
- Shukla R., Upton K.R., Muñoz-Lopez M., Gerhardt D.J., Fisher M.E., Nguyen T., Brennan P.M., Baillie J.K., Collino A., Ghisletti S. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–111. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spalding K.L., Bhardwaj R.D., Buchholz B.A., Druid H., Frisén J. Retrospective birth dating of cells in humans. Cell. 2005;122:133–143. doi: 10.1016/j.cell.2005.04.028. [DOI] [PubMed] [Google Scholar]
- Wang J., Song L., Grover D., Azrak S., Batzer M.A., Liang P. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Hum. Mutat. 2006;27:323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S., Muñoz-Lopez M., Macia A., Yang Z., Montano M., Collins W., Garcia-Perez J.L., Moran J.V., Greene W.C. Reprogramming somatic cells into iPS cells activates LINE-1 retroelement mobility. Hum. Mol. Genet. 2012;21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youssoufian H., Pyeritz R.E. Mechanisms and consequences of somatic mosaicism in humans. Nat. Rev. Genet. 2002;3:748–758. doi: 10.1038/nrg906. [DOI] [PubMed] [Google Scholar]
- Zhao K., Du J., Han X., Goodier J.L., Li P., Zhou X., Wei W., Evans S.L., Li L., Zhang W. Modulation of LINE-1 and Alu/SVA retrotransposition by Aicardi-Goutières syndrome-related SAMHD1. Cell Rep. 2013;4:1108–1115. doi: 10.1016/j.celrep.2013.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong C., Lu S., Chapman A.R., Xie X.S. Genome-wide detection of single-nucleotide and copy-number variations of a single human cell. Science. 2012;338:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Supplemental References
- Dombroski B.A., Scott A.F., Kazazian H.H., Jr. Two additional potential retrotransposons isolated from a human L1 subfamily that contains an active retrotransposable element. Proc. Natl. Acad. Sci. USA. 1993;90:6513–6517. doi: 10.1073/pnas.90.14.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q., Moran J.V., Kazazian H.H., Jr., Boeke J.D. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–916. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- Jiang Y., Matevossian A., Huang H.S., Straubhaar J., Akbarian S. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiełbasa S.M., Wan R., Sato K., Horton P., Frith M.C. Adaptive seeds tame genomic sequence comparison. Genome Res. 2011;21:487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- Magoč T., Salzberg S.L. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics. 2011;27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J.V., Holmes S.E., Naas T.P., DeBerardinis R.J., Boeke J.D., Kazazian H.H., Jr. High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–927. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- Navin N., Kendall J., Troge J., Andrews P., Rodgers L., McIndoo J., Cook K., Stepansky A., Levy D., Esposito D. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada S., Saiwai H., Kumamaru H., Kubota K., Harada A., Yamaguchi M., Iwamoto Y., Ohkawa Y. Flow cytometric sorting of neuronal and glial nuclei from central nervous system tissue. J. Cell. Physiol. 2011;226:552–558. doi: 10.1002/jcp.22365. [DOI] [PubMed] [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
“Bulk library stats” tab details bulk RC-seq mapping statistics, L1 enrichment and L1-Ta detection rates, as well as counts of polymorphic and somatic L1 insertions detected in each individual. “Single-cell library stats” tab details WGS quality control and single-cell RC-seq results for each individual cell studied.
“Polymorphic L1” tab details polymorphic L1s detected by bulk RC-seq. “Sense start” and “Sense stop” refer to the start and stop positions of an RC-seq read contig arranged in sense to the genome. “Antisense start” and “Antisense stop” refer to a cluster arranged antisense to the genome. RC-seq read counts are provided for each sample. Examples with sense and antisense RC-seq clusters depict simultaneous 5′/3′ detection of an insertion. “Donor sense” records the retrotransposon consensus start position (base 0) of sense insertions (negative values or 0) or the stop position of antisense insertions (positive values). “Donor antisense” records the retrotransposon consensus stop position of sense insertions (positive values) or the start position of antisense insertions (negative values or 0). “Polymorphic L1 PCR” tab details 12 heterozygous polymorphic L1 insertions detected at their 3′ L1-genome junction by single-cell RC-seq and tested by PCR for amplification of the corresponding 5′ L1-genome junction in 100 neurons (see Extended Experimental Procedures). “Somatic L1” tab provides single-cell RC-seq results for all hippocampal neurons, hippocampal glia, and cortical neurons presented. Columns are labeled similar to the “Polymorphic L1” tab, with single-cell RC-seq read counts provided here. A large (>50 MB) table detailing hippocampus and liver somatic L1 insertions is available on request. “Somatic L1 PCR” tab provides detailed information for somatic L1 insertions detected by single-cell RC-seq and characterized by PCR and sequencing. Information provided by validation sequencing is highlighted in green. Characterization includes the capillary sequences of confirmed somatic insertions, TSD sizes, the L1 endonuclease recognition motif and the start position of truncated insertions.
Each datasheet indicates the chromosomal position of an L1 insertion, with first- and second-strand L1 EN cleavage sites indicated by red and black arrows, respectively, and the L1 EN motif found at the first-strand cleavage site underlined in black. Target site duplications are highlighted with blue nucleotides. Numbers at the 5′ L1-genome junction in structure cartoons indicate the length of the L1. See Table S2 for additional information.