Summary
MicroRNA (miR)-155 has been implicated in regulating inflammatory responses and tumorigenesis, but its precise role in linking inflammation and cancer has remained elusive. Here, we identify a connection between miR-155 and Notch signaling in this context. Loss of Notch signaling in the bone marrow (BM) niche alters hematopoietic homeostasis and leads to lethal myeloproliferative-like disease. Mechanistically, Notch signaling represses miR-155 expression by promoting binding of RBPJ to the miR-155 promoter. Loss of Notch/RBPJ-signaling upregulates miR-155 in BM endothelial cells, leading to miR-155-mediated targeting of the NF-κB inhibitor κB-Ras1, NF-κB activation and increased proinflammatory cytokine production. Deletion of miR-155 in the stroma of RBPJ-/- mice prevented the development of myeloproliferative-like disease and cytokine induction. Analysis of BM from patients carrying myeloproliferative neoplasia also revealed elevated expression of miR-155. Thus, the Notch/miR155/kB-Ras1/NF-kB axis regulates the inflammatory state of the BM niche and affects the development of myeloproliferative disorders.
Introduction
Notch signaling plays an essential role in regulating normal and abnormal hematopoietic stem and progenitor cell development and functions. While Notch's cell-autonomous role in this process is well established, its non-cell autonomous role remains poorly understood. Specifically, the cellular and molecular mechanism(s) by which Notch loss-of-function regulates the integrity of the BM niche is poorly defined. Here, we used a conditional knock-out model of RBPJ, a non-redundant downstream effector of the canonical Notch signaling cascade, to determine the contribution of Notch signaling to the non-cell autonomous regulation of hematopoiesis.
Notch genes encode large, highly conserved type 1 transmembrane receptors, which are activated through cell-cell contact by binding to one of their ligands on neighboring cells (Artavanis-Tsakonas et al., 1999). Notch binding and activation is regulated at multiple steps by molecules that control endocytosis, O-fucosylation and proteolytic cleavage, leading to the release of the Notch intracellular domain (NICD) and its translocation to the nucleus (De Strooper et al., 1999). Following ligand activation, Notch signalling can be distinguished into canonical and non-canonical pathways on the basis of whether NICD interacts with a CSL transcription factor (CBF1/RBP-J, Su(H), Lag-1) (Kopan and Ilagan, 2009). In mice, the CSL factor is known as RBPJk (recombination signal binding protein for immunoglobulin kappa J region) and functions as a transcriptional repressor. Canonical Notch signalling involves NICD binding to RBPJ and converting it from a repressor to an activator, resulting in the transcription of Notch-dependent genes which can influence the developmental and differentiation programs (Davis and Turner, 2001). Evidences of NICD binding to RBPJ maintaining a repressor status have been recently reported and involve dislocation and recruitment of co-activators and co-repressors, respectively (Sakano et al., 2010; Tiberi et al., 2012).
Although the precise mechanism(s) involved in the regulation of hematopoiesis via the non-cell-autonomous Notch signaling cascade remain unclear, recent studies have begun to shed some insight into this process (Kim et al., 2008; Yao et al., 2011; Yoda et al., 2011, Klinakis et al, 2011). While informative, the genetic models used in these studies involved deletion of genes that affect global Notch signaling, both CSL-dependent and CSL-independent Notch signaling, and regulate other molecules/effectors in addition to Notch (Pruessmeyer and Ludwig, 2009;De Strooper, 2005), thus, preventing a clear understanding of the specific downstream mechanisms.
In this study, we show that RBPJ functions as a transcriptional repressor on the promoter of the microRNA miR-155. miR-155 is encoded from the B cell integration cluster locus and is upregulated in cancer and in inflammation (Tili et al., 2013). Loss of canonical Notch signaling induces direct upregulation of miR-155 expression on BM stromal and endothelial cells and causes significant alterations of hematopoiesis. Constitutive miR-155 up-regulation due to loss of RBPJ transcriptional repression induces NF-κB activation and a global state of inflammation in the BM niche, leading to an uncontrolled expansion of myeloid cells and to the development of a myeloproliferative-like disease. Our results demonstrate a connection between Notch signaling, miR-155 and NF-κB and suggest a critical role for this pathway in maintaining hematopoietic homeostasis and linking inflammation and cancer.
Results
RBPJ deletion in the BM microenvironment disrupts hematopoietic homeostasis and induces a non-cell autonomous myeloproliferative-like disease
Inhibition of RBPJ transcriptional activity by deletion of its DNA binding motif results in the complete loss of signaling via all Notch receptors (Han et al., 2002). This RBPJ knock-out model has been successfully used to unveil the role of Notch in the lymphoid compartment; however, the effects of RBPJ deletion on myeloid cells were not investigated. RBPJ was conditionally deleted in the hematopoietic system by injecting mice with pIpC, which induces MX1-Cre expression in hematopoietic (CD45+) as well as in stromal cells (CD45-) of the BM (Figure S1A-B). Analysis of stem and progenitor pools within the BM, spleen and peripheral blood (PB) of mice lacking RBPJ revealed a significant increase in the frequency and absolute number of phenotypically defined primitive lineage negative Kit+ Sca-1+ (LSK) cells, including long-term HSCs (LT-HSC), of common myeloid progenitors (CMP) and of granulocyte-macrophage progenitors (GMP; Figure 1A, D and S1C-F). These increases were reflected as expansion of immature myeloid (Gr1-Mac1+ cells) and neutrophils in the BM, spleen and PB of RBPJ-/- mice relative to controls (Figure 1G-H, left panels and Fig.7G). The number of megakaryocyteerythroid progenitors (MEP), red blood cells (RBC), hemoglobin (Hb), hematocrit (HCT) and platelets (PLT) were reduced in mice lacking RBPJ (Figure S1G, Q). RBPJ-/- mice developed significant splenomegaly (Figure 1C) and histologic analysis confirmed myeloid expansion in the BM and spleen, and myeloid cell infiltrates in liver and other parenchymal organs. Overall, BM cellularity was significantly increased (Figure S1H). Leukocyte counts and differentials showed a progressive increase of myeloid cells in the PB of RBPJ-/- mice compared to controls: from 2 fold increase at week 4, to 7 fold at week 20 after pIpC (Figure 1H and S1N-O). As reported in a previous study (Han et al., 2002), loss of Notch signaling in the lymphoid compartment was characterized by reduction of T-cells and relative expansion of B-cells (Figure S1P, left graph). Interestingly, B-cells were increased in the PB, but not in the BM and spleen of RBPJ-/- mice (data not shown). RBPJ-/- mice failed to live past 27 weeks and died of a myeloproliferative disease (Fig. 1I). In contrast, mice haplo-insufficient for RBPJ+/- did not develop disease, showing comparable survival to control animals up to 40 weeks of follow-up.
Figure 1. Cell-autonomous and non-cell autonomous impact of RBPJ deletion in the hematopoietic compartment.
(A) % of LSK cells in BM and spleen of RBPJ-/- and RBPJ+/+ mice at 15-20 wks after pIpC (n=8-12).
(B) LSK cells in BM and spleen of WT (CD45.1) recipient mice transplanted with CD45.2+ RBPJ-/- or RBPJ+/+ BM donor cells at 30wks post-transplant (left), and of RBPJ-/- and RBPJ+/+ recipient mice transplanted with WT donor cells at 8wks after RBPJ deletion in the microenvironment (right). Results are expressed as fold change of % (n=3-10).
(C) Bar graph summarizes average spleen weight from RBPJ-/-, RBPJ+/+, RBPJ-/- and RBPJ+/+ recipient mice (n=6-9).
(D) % of CMP and GMP in BM and spleen of RBPJ-/- and RBPJ+/+ mice (n=8-12). Total CMP and GMP numbers/femur in: (E) WT (CD45.1) recipients transplanted with RBPJ-/- or RBPJ+/+ BM cells; 30wks post-transplant (n=3-5) or (F) RBPJ-/- and RBPJ+/+ recipients transplanted with WT CD45.1+ BM cells; 6-8wks after pIpC (n=6).
(G) Dot Blots indicate Gr1/Mac1 expression in PB, spleen and BM. Bar graphs summarize % of Gr1+/Mac1+ cells over whole population; 3 independent experiments (n=6-14).
(H) Neutrophil counts in PB of RBPJ-/- and RBPJ+/+ mice (left), WT recipients of RBPJ-/- and RBPJ+/+ donor cells (middle) and of RBPJ-/- and RBPJ+/+ recipients (right).
(I) Kaplan-Meyer survival curve of RBPJ-/-, RBPJ+/-, RBPJ+/+ mice (n=7-14; p<0.01), WT mice recipients of RBPJ-/- BM cells, and of RBPJ-/- or RBPJ+/+ mice recipients of WT cells (n=9-11; p<0.01).
All results are shown as mean±SEM; *p<0.05, **p<0.01, ***p<0.005 unless otherwise indicated. See also Figure S1.
Figure 7. Loss of miR-155 rescues the RBPJ-/- phenotype.
(A) Neutrophil counts in PB of RBPJ+/+miR-155+/+, RBPJ-/-miR-155+/+ (KO) and RBPJ-/- miR-155-/- (DKO) mice up to 16wks from pIpC induction (n=3-8).
(B) % of BM CMP and GMP subsets in the Lin- population of RBPJ+/+miR-155+/+, RBPJ-/-miR-155+/+ (KO) and RBPJ-/- miR-155-/- (DKO) mice at 20wks after induction (mean±SD; n= 3-8).
(C-E) RBPJ+/+miR-155+/+, RBPJ-/-miR-155+/+ (KO) and RBPJ-/-miR-155-/- (DKO) were induced with pIpC and after 2 weeks were transplanted with WT BM cells. At week 10 post-transplant, when level of engraftment was > 90%, data were collected:
(C) % of Gr1+/Mac1+ in PB; (mean±SD; n= 3-7).
(D) Representative spleens. Bar graph indicates spleen weight (n=3-6).
(E) % of CMP and GMP in the BM (mean±SD; n= 3-8).
(F) κB-Ras1, G-CSF and TNFα expression by qRT-PCR in sorted BM CD45+, EC, MSC and cultured OB cells from RBPJ-/- and DKO mice. Fold change (n=3-6).
(G) Wright/Gimsa staining of BM cytospins and histological (H&E) analyses of spleen. Scale bar for BM cytospin, 20μm. Scale bar for spleen, 500μm.
(H) miR-155 expression by qRT-PCR in MPN patients. Dot scatter plot indicates relative expression of miR-155 in the BM of 85 MF patients and 10 age-matched healthy donors.
All results express as mean±SEM; *p<0.05, **p<0.01, ***p<0.005, unless otherwise indicated. See also Figure S6.
To assess the relative contribution of cell-autonomous and non-cell-autonomous mechanisms, we performed reciprocal transplantation studies. BM cells from RBPJ+/+ and RBPJ-/- expressing the CD45.2 allele were transplanted into wild-type (WT) mice expressing the CD45.1 allele. Similarly to the RBPJ-/-mice, the LSK pool was significantly increased in WT mice reconstituted with RBPJ-/- donor cells compared to controls (Figure 1B, left side). However, there was no difference in the representation of GMP, CMP and MEP subsets in RBPJ-/- and RBPJ+/+ donor populations (Figure 1E and S1I). Mice reconstituted with RBPJ-/- donor cells did not show myeloid cell expansion and exhibited normal levels of Gr1+Mac1+ neutrophils in the BM, SP and PB (Figure 1G-H, center panels). These mice did not display alterations in RBC, Hb, HCT or PLT (Figure S1Q), nor did they develop splenomegaly, and their survival was similar to the control cohort (Figure 1I). As previously reported, mice reconstituted with RBPJ-/- donor cells presented an overall decrease in T-cells accompanied by a relative increase in B-cells (data not shown). The leukocyte differential count in the recipients of RBPJ-/- donor cells did not show neutrophilia or lymphocytosis (Figure S1N), indicating that the effects observed in RBPJ-/- mice were not solely mediated by a cell-autonomous mechanism.
To examine the contribution of the microenvironment to the myeloproliferative-like disease, we transplanted WT BM cells into RBPJ-/- or RBPJ+/+ recipients. Comparative analysis of hematopoietic subsets in the BM and spleen of RBPJ-/- recipients revealed that the loss of RBPJ in the BM microenvironment had a lower impact on the LSK pool in RBPJ-/- recipients (Figures 1B, right side and S1J) than in parental RBPJ-/- mice or WT mice transplanted with RBPJ-/- cells, demonstrating a cell-autonomous contribution of RBPJ on HSC homeostasis. However, similar to the RBPJ-/- parental animals, RBPJ-/- recipient mice transplanted with WT cells showed a significant increase in the frequencies and absolute numbers of CMP and GMP, in both BM and spleen (Figure 1F and S1L). These changes were associated with a marked decrease of MEP, RBC, Hb, and HCT (Figure S1K, Q), and a significant increase in circulating LSK cells (Figure S1J). RBPJ-/- recipients showed a rapid and progressive increase in neutrophils and Gr1+/Mac1+ cells in the PB, spleen and BM (Figure 1G-H, right panel) as well as increased BM cellularity (Figure S1M). RBPJ-/- recipients developed marked splenomegaly (Figure 1C, 7D), hypercellular BM with myeloid cell expansion and myeloid infiltrates in the parenchymal organs (data not shown). A complete leukocyte count and differential in the PB of RBPJ-/- recipients indicated a significant increase of myeloid cells (3 fold) compared to RBPJ+/+ recipients, whereas lymphoid cells showed a kinetic of engraftment similar to the controls, and a relative decrease in frequency in favor of myeloid cells (Figure 1SN-P). Thus, the loss of RBPJ in the microenvironment resulted in a lethal myeloproliferative-like disease, which evolved rapidly following BM transplant, as indicated by the shorter survival of RBPJ-/- recipients compared to RBPJ-/- mice (100% of the mice were dead by week 10; Figure 1I).
These data demonstrate that loss of Notch/RBPJ canonical pathway is not sufficient to induce a cell-autonomous myeloid disease. Furthermore, they clearly show that loss of RBPJ in the microenvironment is necessary and sufficient to induce a lethal myeloproliferative-like disease in a non-cell autonomous manner.
RBPJ deficiency in the BM niche favors myeloid cell expansion and mobilization by upregulating expression of G-CSF
Given that Mx1-Cre is expressed in the stromal cells of multiple organs (Schneider et al., 2003), we questioned whether the BM microenvironment was a critical site for disease initiation of RBPJ-/- mice. RBPJ-/- and RBPJ+/+ littermates were transplanted with equal number of BM cells harvested from Lys-EGFP reporter mice (wild-type RBPJ), in which EGFP expression is driven by the lysozyme promoter (Faust et al., 2000). As lysozyme expression is specific to myeloid cells (Miyamoto et al., 2002), this strategy provided optimal visualization of the myeloid compartment by intravital fluorescent microscopy (IVFM). The BM niche was imaged at 24 hours and at weeks 2, 4 and 6 after transplantation (Figure 2A). Homing at 24 hours was similar in both conditions; however, by week 2, Lys-EGFP cells transplanted in RBPJ-/- recipients had underwent a more rapid and robust expansion then Lys-EGFP cells transplanted in RBPJ+/+ mice (Figure 2A-B). At weeks 4 and 6 after transplantation, the content of Lys-EGFP cells in the BM of RBPJ-/- recipients remained higher but not substantially different from RBPJ+/+ recipients, as Lys-EGFP cells had also expanded in the RBPJ+/+ recipients (Figure 2B). However, in RBPJ-/- recipients the Lys-EGFP cell expansion in the BM was associated with a greater output of neutrophils and Gr1+/Mac1+ cells in the PB and progressive enlargement of the spleen (Figure 2D-E). Significant increase in the proportion of myeloid cells in the BM of RBPJ-/- recipients was observed again at advanced stages of disease (Figure 1G).
Figure 2. The RBPJ-/- BM niche favors myeloid expansion and trafficking.
(A) Intravital imaging of mouse calvarium. RBPJ-/- mice were induced with pIpC 4wks prior transplantation of BM cells from Lys-EGFP donor mice; imaging was performed at 24hrs and wks 2, 4 and 6 post-transplant. Series of 60 μm Z-stacks were collected from 6 regions of calvarium BM in each mouse. Images of 1 representative region of 6 at wks 2 and 6 in RBPJ-/- and RBPJ+/+ recipients (myeloid cells: green; vasculature; red). Cells were detected automatically and numbers were computed in each region. Scale bar, 50μm.
(B) Upper graph: number of Lys-EGFP+ cells/region homed to BM at 24 hrs: 32±6 cells in RBPJ-/- vs. 31 ±8 cells in RBPJ+/+ recipient mice (n=12 regions; 2 independent experiments; p=NS). Lower graph: kinetics of engraftment of Lys-EGFP cells into RBPJ-/- and RBPJ+/+ recipients over time (mean±SEM of cells in each region; n=12-18 regions; 2-3 mice; *p=0.019).
(C) Lys-EGFP expression on CMP, GMP and LSK at 2wks post-transplant in a representative experiment. Values show % of cells expressing high levels of Lys-EGFP within each specific subset (n=2).
(D) Kinetics of neutrophil mobilization into PB in RBPJ-/- and RBPJ+/+ recipients following transplantation of Lys-EGFP cells into RBPJ-/- recipients (mean±SEM; n=4-13; *p<0.005).
(E) Spleen size in RBPJ-/- recipients during time following transplantation of Lys-EGFP cells (mean±SD; n=4-6; *p<0.05; ***p<0.005).
Lys-EGFP is increasingly expressed from CMP to GMP to mature neutrophils, allowing measurement of the kinetics of differentiation by fluorescent intensity. As expected, Lys-EGFP expression was low in LSK cells in both groups, while analysis of CMP and GMP revealed a higher number of high Lys-EGFP expressing cells in RBPJ-/- recipients than in RBPJ+/+ recipients, suggesting a more rapid rate of myeloid cell differentiation (Figure 2C). These data indicate that the RBPJ-/- BM microenvironment is a primary site of myeloid cell expansion, which is accompanied by rapid differentiation and mobilization of myeloid cells in the PB and spleen.
Analysis of serum cytokines by cytokine arrays revealed a significant increase in the levels of several inflammatory cytokines in RBPJ-/- mice (Figure S2A), in particular G-CSF, a driver of myeloid cell proliferation and differentiation. Increased G-CSF levels were confirmed in the BM milieu and in the serum of RBPJ-/- mice by ELISA (Figure 3A-B). Heterozygous RBPJ+/- mice, which did not develop disease, displayed G-CSF levels just above the baseline of normal mice. To determine the contribution of G-CSF to the myeloproliferative-like disease, we treated RBPJ-/- mice with anti-G-CSF neutralizing antibodies. Anti-G-CSF treatment resulted in the normalization of Gr1+/Mac1+ cells in the PB (Figure 3C), in a significant reduction in the spleen size (Figure 3D), a notable decrease in the proportion of CMPs and a significant reduction in level of GMPs (Figure 3E). These data suggest that G-CSF is a major but not the sole contributor to the myeloproliferative-like disease occurring in RBPJ-/- mice. TNFα levels were also consistently elevated in the absence of RBPJ (Figure S2A-C), suggesting that TNFα, a cytokine involved in myeloid progenitor homeostasis (Walkley et al., 2007), may play a synergistic role with G-CSF in promoting myeloid cell expansion.
Figure 3. The RBPJ-/- BM niche secrets increased levels of G-CSF.
(A) G-CSF levels (pg/mL) by ELISA in fresh BM eluted from femurs of RBPJ-/- or RBPJ+/+ recipients 6wks after pIpC induction (n=3).
(B) Serum G-CSF-levels (pg/mL) by ELISA in RBPJ+/+, RBPJ+/-, RBPJ-/- and RBPJ-/- recipient mice 6wks after pIpC induction (n=4).
(C) % of Gr1+/Mac1+ cells in PB of RBPJ-/- and RBPJ+/+ mice treated with anti-G-CSF neutralizing Ab or vehicle control (n=3).
(D) Spleens from RBPJ+/+ and RBPJ-/- mice treated with G-CSF Ab or vehicle control; representative experiment (n=3).
(E) % of CMP (left) and GMP subsets (right) in spleen of RBPJ-/- and RBPJ+/+ mice treated with anti-G-CSF Ab or vehicle control (n=3).
(F) RBPJ and (G) Notch target gene Hes1 expression by qRT-PCR in BM stroma from RBPJ-/- or RBPJ+/+ mice (n=3).
(H) G-CSF and TNFα levels by cytokine array in BM-derived stroma supernatant from RBPJ-/- and RBPJ+/+ mice. Results are expressed as pixel density (n=3).
(I) Left: number of WT Lin- progenitors from 48hrs co-culture on BM stroma from RBPJ-/- or RBPJ+/+ mice (n=7). Right: acquisition of myeloid markers by Lin- progenitor cells; results are expressed as % of Gr1+/Mac1+ cells (n=4; p=0.09).
All results express as mean±SEM; *p<0.05, **p<0.01, ***p<0.005, unless otherwise indicated. See also Figure S2.
Analysis of cytokines in the supernatant of RBPJ-/- BM-derived stromal cells, validated for loss of RBPJ and Notch signaling (Figure 3F-G), demonstrated a pro-inflammatory profile similar to the one observed in the serum (Figure S2B). These experiments confirmed a significant increase in the levels of G-CSF and TNFα in RBPJ-/- stromal cells (Figure 3H). Accordingly, RBPJ-/- BM-derived stromal cells showed a greater ability to support myeloid cell expansion and differentiation of BM Lin- progenitors in co-culture experiments (Figure 3I). These results demonstrate that RBPJ deletion in the BM stromal cells induces pro-inflammatory cytokines, generating a BM niche exceedingly favorable to myeloid progenitor cell expansion.
Contribution of RBPJ-/- BM endothelial cells to the myeloproliferative-like disease
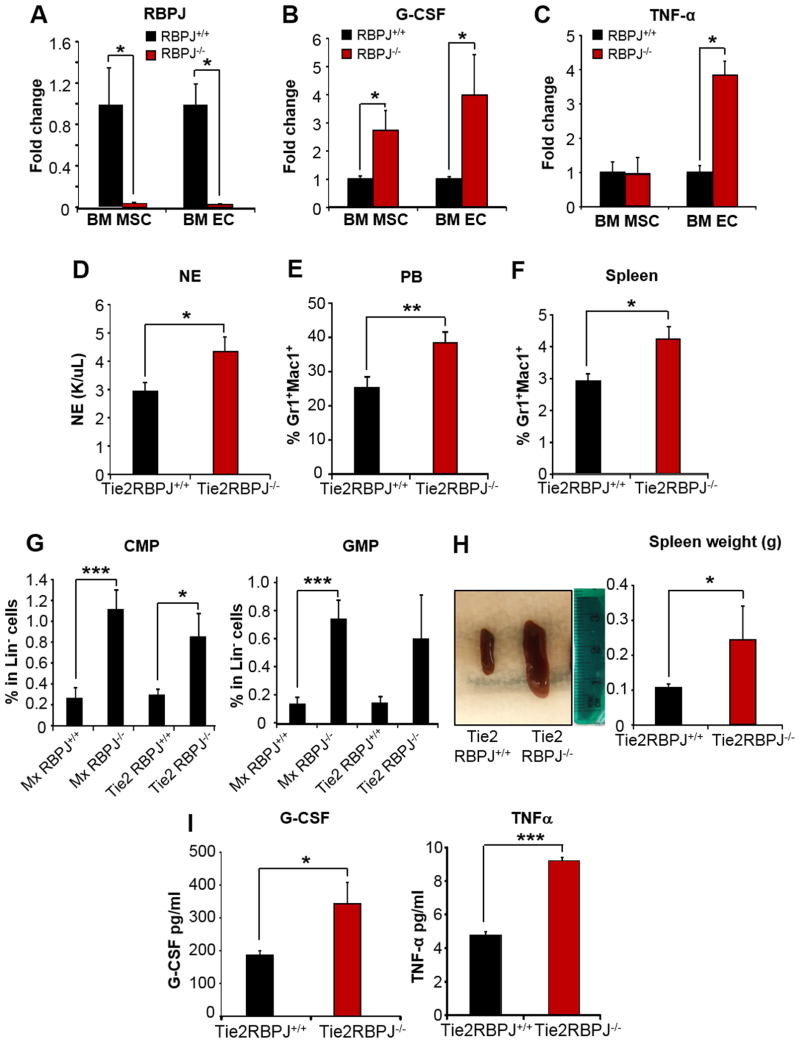
Next, we sought to determine which specific stromal cell type(s) in the BM were responsible for the increased cytokine production in RBPJ deficient mice. Co-culture experiments of Lin- progenitor cells with RBPJ-/- derived osteoblasts (OB) or characterization of hematopoiesis in mice with RBPJ deletion in osteocytes did not show impact on the myeloid compartment, ruling out a major role of mature bone cells in the myeloid expansion (Figure S3A-E). In contrast, sorted BM CD45-CD105+CD31- cells (enriched in mesenchymal like-cells; BM MSC) and BM CD45-CD105+CD31+ VE-cadherin+ cells (enriched in endothelial cells; BM EC) demonstrated higher levels of G-CSF and TNFα in association with RBPJ deletion (Figure 4A-C). Given the significantly higher expression of G-CSF and TNFα in BM ECs of RBPJ-/- mice and the known role these cells play in producing pro-inflammatory cytokines, we sought to further investigate the role of the BM endothelial niche. To this end, we crossed RBPJ-/- mice with the tamoxifen-inducible Tie2CreER mice. In these mice, the Cre recombinase is expressed under the control of the Tie2 promoter in which specific regulatory elements were modified to restrict the expression of Cre to ECs, with negligible expression in the hematopoietic system (Forde et al., 2002). Tamoxifen-induced Tie2CreER+/RBPJlox/lox and Tie2CreER-/RBPJlox/lox mice (referred as Tie2RBPJ-/- and Tie2RBPJ+/+ mice) were analyzed at 6 months post-induction. In these mice, the average reduction of RBPJ on sorted BM EC was approximately 30%. Despite the incomplete deletion of EC RBPJ, analysis of the hematopoietic compartment of Tie2RBPJ-/- mice demonstrated a significant increase in neutrophils and Gr1+/Mac1+ cells in the PB (Figure 4D-E), increased Gr1+/Mac1+, CMP and GMPs cells in the spleen (Figure 4F-G) and splenomegaly (Figure 4H) compared to Tie2RBPJ+/+ mice. These changes were associated with increased levels of G-CSF and TNFα in Tie2RBPJ-/- mice serum (Figure 4I). All parameters of myeloid cell expansion were similarly increased in Tie2RBPJ-/- mice as those observed in Mx1RBPJ-/- mice (i.e. CMP and GMP; Figure 4G); however these changes occurred less rapidly in the Tie2RBPJ-/- background. Thus, deletion of RBPJ in ECs is sufficient to induce myeloid cell expansion. However, the contribution of other stromal cell types, including MSCs, may be important for the development of a full blown and rapid myeloproliferative-like disease.
Figure 4. Contribution of the RBPJ-/- BM endothelial niche to the myeloid expansion.
(A) RBPJ (B) G-CSF and (C) TNFα transcript levels by qRT-PCR in sorted CD45-CD105+C31- (MSC) and CD45-CD105+C31+ (EC) cells from BM of RBPJ+/+ or RBPJ-/- mice. Fold change (n=3).
(D) Neutrophil counts and (E) % of Gr1+/Mac1+ cells in PB (F) % of Gr1+/Mac1+ cells and (G) % of CMP and GMP in spleens of Tie2-RBPJ-/- and Tie2RBPJ+/+ mice at 6 months after tamoxifen induction compared to Mx-RBPJ-/- mice induced with pIpC (n=6-7).
(H) Spleens from Tie2-RBPJ-/- and Tie2RBPJ+/+ mice in representative experiment. Bar graph: spleen weights (n=6-7).
(I) G-CSF and TNFα levels (pg/mL) by ELISA in serum of Tie2-RBPJ-/- and Tie2RBPJ+/+ mice 6 months after tamoxifen induction (n=6-7).
All results express as mean±SEM; *p<0.05, **p<0.01, ***p<0.005. See also Figure S3.
Loss of RBPJ transcriptional repression induces up-regulation of miR-155
The coordinated upregulation of multiple inflammatory cytokines upon deletion of RBPJ suggests the involvement of master regulators triggering a proinflammatory circuitry. Likely candidates for such function are microRNAs. Analysis of the expression of several microRNAs implicated in inflammation showed a significant increase in the expression of miR-155 in RBPJ-/- BM stromal cells (Figure 5A). No significant changes in the expression of miR-196b, miR-210, miR-125a and miR-146a were noted under similar conditions (Figure S4A). Increased expression of miR-155 in RBPJ-/- stroma was further validated using BM sorted populations enriched in MSCs and ECs, as well as in ECs derived from Tie2RBPJ-/- mice (Figure 5B). miR-155 expression was also up-regulated in Lin- RBPJ-/- hematopoietic cells, but not in bulk CD45+ cells (Figure S4B and S5D). To determine the molecular link between RBPJ and miR-155 in vitro, murine microvascular endothelial cells (here indicated as EC) and BM stromal cell lines Raw264.7 (Raw) and OP9, were transfected with shRNAs directed towards RBPJ (shRBPJ) (Figure S4C). Expression of the shRBPJ upregulated miR-155 expression in ECs, Raw cells and in OP9 cell lines (Figures 5C-F, S4D-E). The upregulated miR-155 was functional, as shown by the target luciferase assay (Figure S4F). As RBPJ is a transcriptional repressor, we hypothesized that RBPJ associates with the miR-155 promoter and suppresses its transcription. In silico analysis of the BIC/miR-155 promoter revealed the presence of two putative RBPJ binding sites (Jarriault et al., 1995) at -1792 and at -1703 nt upstream of the start site (Figure 5G). Chromatin immunoprecipitation analysis (ChIP) of the promoter using primers (Table S1) for these two regions confirmed binding of endogenous RBPJ to the endogenous miR-155 promoter in cells derived from RBPJ+/+ mice but not RBPJ-/- mice (Figure 5H, left panel). Similarly, binding of RBPJ to miR-155 promoter was observed in EC and Raw cells and was abrogated following the expression of shRBPJ (Figure 5H, middle and right panels). Notch was also found in the complex when RBPJ was present (Figure S4G). To further confirm the repressor activity of RBPJ on the miR-155 promoter, we cloned the 2kb pre-miR-155 promoter region containing the two RBPJ binding sites into a luciferase reporter construct. Consistent with a repressor function of RBPJ, miR-155 transcriptional activation was increased 2 to 3 fold following RBPJ repression (Figure 5I). miR-155 expression was also increased following treatment of Raw cells with the γ-secretase inhibitor (GSI; Figure 5I), which inhibits the generation of NICD and ultimately prevents the formation of the NICD/RBPJ complex on the promoter. Conversely, overexpression of NICD decreased miR-155 expression in Raw cells and in T-cells (Figure S4H-I). As RBPJ has been shown to associate with the histone deacetylase complex (HDAC) (Kao et al., 1998), and HDAC2 binding is observed at the miR-155 promoter in wild-type Raw cells (Figure S4J), we investigated whether RBPJ regulated miR-155 repression through HDAC function. Treatment with a pan HDAC inhibitor led to a 3 fold increase in the expression of miR-155, similar to the effect induced by deletion of RBPJ (Figure 5I).
Figure 5. RBPJ functions as repressor on miR-155 promoter.
miR-155 levels by qRT-PCR in (A) BM stroma cells from RBPJ+/+ or RBPJ-/- mice (n=4) and (B) BM sorted CD45-CD105+C31- (MSC) or CD45-CD105+C31+ (BM EC) cells from RBPJ+/+, RBPJ-/- mice (n=3); CD45-CD105+C31+ cells (n=6) include also BM cells sorted from Tie2Cre+RBPJ-/- and Tie2Cre+RBPJ+/+ mice. Fold change.
(C) RBPJ and (D) miR-155 transcripts levels by qRT-PCR in EC cells treated with shRBPJ. Fold change (mean±SD; n=2-3).
(E) RBPJ and (F) miR-155 transcripts levels by qRT-PCR in Raw cells treated with shRBPJ. Fold change (n=3).
(G) Scheme of the RBPJ binding sites in the pre-miR-155/BIC promoter.
(H) ChIP assay of total BM from RBPJ-/- or RBPJ+/+ mice and EC and Raw cells transfected with shRBPJ or scrambled shRNA. IP were conducted with no Ab, an irrelevant Ab, or Abs directed to RBPJ, followed by PCR of the miR-155 promoter. 3 independent experiments/each.
(I) miR-155 luciferase assay. Raw cells expressing the shRBPJ or treated with HDAC inhibitor (1μM) or with GSI (1μM) were transfected with the miR-155 promoter region containing the RBPJ binding sites. Results are expressed as fold change in RLU (n=4; 2 independent experiments).
All results express as mean±SEM; *p<0.05, **p<0.01, ***p<0.005. See also Figure S4.
miR-155 induces G-CSF and TNFα expression via NF-κB activation
Given the strong correlation between miR-155 upregulation and increased G-CSF and TNFα levels, we hypothesized that miR-155 regulates G-CSF and TNFα production by BM stromal cells. Transfection of shRBPJ in ECs and Raw cells resulted in increased G-CSF and TNFα expression and protein levels (Figure 6A-B and S5A) and enhanced their ability to promote myeloid cell expansion in co-culture (Figure 6C). Similarly, overexpression of miR-155 in Raw cells resulted in rapid up-regulation of G-CSF and TNFα (Figure 6D). Consistently, treatment of RBPJ knocked down cells with an anti-miR-155 LNA inhibited shRBPJ-mediated induction of G-CSF and TNFα (Figure 6E and S5B). Thus, loss of RBPJ augments G-CSF and TNFα levels by directly up-regulating the expression of miR-155. To identify the molecular link between miR-155 and inflammatory cytokines, we used the microRNA target predictions program https://cm.jefferson.edu/rna22v1.0-musmusculus/ (Miranda et al., 2006) and searched for putative miR-155 targets whose inhibition might result in the transcriptional activation of G-CSF and TNFα. Among the predicted targets, the NF-κB inhibitor κB-Ras1 (also known as Nkiras1) that inhibits IkB-β phosphorylation (Fenwick C, Science 2000), was found to have miR-155 target sequences at its 3′UTR. Given that the inflammatory profile observed in RBPJ-/- stromal cells has a NF-κB signature (Figure S2) and that the transcription of G-CSF and TNFα is regulated principally by NF-κB, we tested the hypothesis that miR-155 up-regulation increased NF-κB activation by decreasing its inhibition. κB-Ras1 protein levels were noticeably reduced in RPBJ-/- mice (Figure S5C) and κB-Ras1 expression was significantly decreased in RPBJ-/- BM stroma (Figure 6F). In Raw cells, κB-Ras1 expression was significantly reduced by repressing RBPJ or by overexpressing miR-155 (Figure 6G). Importantly, κB-Ras1 downregulation in these cells was antagonized by miR-155 LNA (Figure 6H). Next, we cloned the κB-Ras1 3′UTR into the pMIR-RL vector and confirmed that κB-Ras1 is a bona fide miR-155 target, as shown by decreased luciferase activity in response to miR-155 overexpression (Figure 6I). Targeting of κB-Ras1 by shRNA (shκB-Ras1) increased expression of G-CSF and TNFα in both Raw and EC cells (Figure 6J-K). κB-Ras1 inhibition correlated with increased transcriptional activation of NF-κB in Raw shRBPJ and Raw MSCV-miR155 expressing cells (Figure 6L) and inhibition of NF-κB by the NF-κB inhibitor DMAPT (dimethylamino-parthenolide) (Neelakantan et al., 2009), resulted in a significant decrease in the expression of G-CSF and TNFα (Figure 6M). Finally, analysis of miR-155, κB-Ras1, G-CSF and TNFα expression, showed that sorted BM EC and MSC from RBPJ-/-mice had the highest expression of miR-155, correlating with the lowest expression of κB-Ras1 and higher levels of G-CSF and TNFα, compared to purified osteoblasts and sorted CD45+ cells (Figure S5D).
Figure 6. Inhibition of κB-Ras1 by miR-155 upregulates G-CSF and TNFα via NF-κB.
(A) EC cells and (B) Raw cells transfected with shRBPJ or scrambled shRNA (sc) were analyzed for G-CSF and TNFα expression by qRT-PCR. Fold change (n=3-5).
(C) Total number WT BM Lin- progenitors co-cultured for 48 hrs with Raw cells transduced with shRBPJ or scrambled shRNA (n=4-6).
(D) G-CSF and TNFα transcript levels by qRT-PCR in Raw cells transduced with MSCV/GFP or MSCV-miR-155/GFP. Fold change (n=5).
(E) G-CSF transcript levels by qRT-PCR in Raw cells transfected with shRBPJ and or scrambled shRNA, in the presence of 75nM LNA control or LNA anti-miR155. Representative experiment express in fold change. κB-Ras1 transcript levels by qRT-PCR in: (F) BM stroma cells from RBPJ+/+ or RBPJ-/- mice. Fold change (n=4), (G) Raw cells transfected with shRBPJ and or scrambled shRNA (sc) or transduced with MSCV/GFP (Co) or MSCV-miR-155/GFP (miR155). Fold change (n=4-6), and (H) Raw cells controls (Co) or transduced with MSCV-miR-155/GFP (miR155) in the presence of 75nM LNA control or LNA anti-miR-155 (n=3-6).
(I) κB-Ras1 3′UTR Luciferase assay in Raw cells transduced with MSCV/GFP or MSCV-miR-155/GFP. Fold change RLU (n=3).
(J) κB-Ras1 transcript levels by qRT-PCR in Raw and EC cells transfected with shκB-Ras1 or scrambled shRNA. Fold change (n=3-4).
(K) Raw and EC cells transfected with shκB-Ras1 or scrambled shRNA were analyzed for G-CSF and TNFα expression by qRT-PCR. NFκB Luciferase assay (right panel) in EC cells transfected with shκB-Ras1 or scrambled shRNA. Fold change (n=3-4).
(L) NFkB Luciferase assay in Raw cells transfected with shRBPJ and or scrambled shRNA (sc) or transduced with MSCV/GFP (Co) or MSCV-miR155/GFP. Fold change RLU (n=4-6).
(M) G-CSF and TNFα transcripts by qRT-PCR in Raw cells transfected with shRBPJ or scrambled shRNA (sc), in culture for 24 hrs with or without 5μM NFkB inhibitor DAMPT. Fold change (n=3).
All results express as mean±SD; *p<0.05, **p<0.01, ***p<0.005. See also Figure S5.
Taken together, our results demonstrate that, following Notch/RBPJ loss of function, miR-155 up-regulates NF-κB activation by targeting one of its inhibitors, κB-Ras1, leading to increased expression of G-CSF and TNFα. Furthermore, these results suggest that this axis may be cell context-dependent and likely to be more active in BM EC and MSC.
miR-155 is required for the development of RBPJ-dependent myeloproliferative-like disease and is upregulated in the BM of MPN patients
To examine the impact of miR-155 deletion on RBPJ dependent MPN development, we generated Mx1RBPJlox/lox/miR-155-/- (DKO) and Mx1RBPJlox/lox/miR-155+/+ (KO) littermates. Consistent with our previous results, deletion of RBPJ induced a lethal myeloproliferative-like disease and KO mice died within 20 weeks of pIpC treatment. In contrast, all DKO mice remained viable at this same time point. Comparative analysis of the hematopoietic compartment in these mice showed that neutrophils increased dramatically in the PB of KO mice, while DKO mice exhibited normal levels of PB neutrophils (Figure 7A), lacked the development of splenomegaly (Figure S6A) and showed only a modest increase in CMP and GMPs in the BM compared to KO mice (Figure 7B). Importantly, G-CSF levels in the serum of DKO recipients were similar to those seen in healthy controls (Figure S6B).
To prove that miR-155 knock down in the stroma is critical for myeloid cell expansion, we transplanted WT BM cells into lethally irradiated DKO and KO mice. As anticipated, KO recipients developed disease, whereas deletion of miR-155 in the stroma of DKO recipients correlated with only a modest increase in Gr1+/Mac1+ cells in the PB (Figure 7C), prevented the development of splenomegaly and the increase of myeloid progenitors (CMP and GMP) in the BM (Figure 7D-E). Histologic analysis of the DKO recipients showed absence of the myeloid expansion with normal myeloid/erythroid ratio in the BM and intact structure of the spleen (Figure 7G). A significant decrease of κB-Ras1 expression was observed in sorted KO BM EC and MSC compared to WT cells (Figure S5D). Conversely, the levels of κB-Ras1 were increased or restored to normal in sorted DKO BM EC and MSC compared to KO cells (Figure 7F). The normalization of κB-Ras1 expression in DKO cells correlated with the normalization of G-CSF and TNFα expression.
Taken together, these experiments show a key role for miR-155 in sustaining the inflammatory reaction driving the lethal myeloproliferative-like disease triggered by the loss of RBPJ. To determine the potential clinical relevance of our observations, we examined whether miR-155 was also altered in patients with myeloproliferative neoplasia (MPN) characterized by a high inflammatory component, such as myelofibrosis (MF). Analysis of 85 MF patients revealed a significant increase in the expression of miR-155 in total BM cells compared to normal control patients (Figure 7H), whereas an increase was not detected in mononuclear cells of the PB (data not shown). Collectively, this observation suggests that miR-155 may play a role in the human disease and may represent a potential therapeutic target for the control of myeloid cell expansion.
Discussion
Evidence supporting a non-cell-autonomous role for Notch signaling in the regulation of hematopoiesis has recently emerged; however, the cellular and molecular mechanism(s) by which Notch regulates the integrity of the BM niche are still poorly understood. By using a Notch/RBPJ loss-of-function model we demonstrated that RBPJ functions as a transcriptional repressor of the microRNA miR-155: a microRNA involved in inflammation and frequently up-regulated in leukemia cells and solid tumors. miR155 up-regulation due to loss of the Notch/RBPJ axis increased NF-κB activation by targeted inhibition of κB-Ras1 and induced a persistent pro-inflammatory state of the BM niche leading to a myeloproliferative-like disease.
Global (canonical and non-canonical) loss of Notch signaling, due to deletion of Mindbomb (Kim et al., 2008b), ADAM10 (Yoda et al., 2011) or Pofut1 (Yao et al., 2011) has been correlated with the development of a myeloproliferative-like syndrome driven by both cell-autonomous and non-cell autonomous mechanisms, whereas deletion of Nicastrin has been shown to lead instead to a cell-autonomous chronic myelo-monocytic leukemia (CMML)-like disease (Klinakis et al., 2011). Discrepancies among these models may be due to the contribution of other targets regulated by ADAM10 (Pruessmeyer and Ludwig, 2009), Pofut1 and the Nicastrin γ-secretase complex (De Strooper, 2005), which acting in combination with global Notch loss-of-function may account for the variable involvement of the microenvironment. However, the molecular mechanisms linking loss of Notch signaling with altered myelopoiesis were not defined in these models.
Our study clearly shows that loss of RBPJ, a non-redundant downstream effector of canonical Notch signaling, is necessary and sufficient to induce a lethal myeloproliferative-like disease driven by the microenvironment. RBPJ-/- stem and progenitor cells were not capable of inducing myeloid expansion or a myeloproliferative disease in a cell-autonomous manner. This effect was further confirmed by using the Lys-CreRBPJlox/lox model, in which the RBPJ deletion occurs only in the myeloid progenitors starting at the CMP stage (Figure S3F-G). Of note, our studies clearly uncouple the direct, cell autonomous role of Notch/RBPJ signaling on HSC homeostasis from its non-cell autonomous impact on myeloid progenitors. Interestingly, lymphocytosis was observed only in the parental RBPJ-/-mice but not in the BM chimeras, suggesting that this process requires both cell autonomous and non-cell autonomous mechanisms; further studies are warranted to understand the impact of RBPJ loss on this process.
Here, we unveil a previously unrecognized link between RBPJ-dependent Notch signaling and miR-155. RBPJ represses transcription by binding to specific sequence motifs in the promoter of target genes and by recruiting co-repressors such SMRT and SHARP (Oswald et al., 2005) and the HDAC complex (Kao et al., 1998). Our study shows that RBPJ binds to the miR-155 promoter and that either deletion of RBPJ DNA binding domain or loss of full-length RBPJ causes miR-155 up-regulation. Importantly, our data show that RBPJ-mediated repression of miR-155 is NICD-dependent, suggesting the possibility that Notch may also function as a transcriptional repressor. This is intriguing considering that it is well established that NICD converts RBPJ from a transcriptional repressor to a transcriptional activator (Kopan and Ilagan, 2009). However, precedents of Notch signaling as repressor have been reported and involved dislocation and recruitment of coactivators and corepressors, respectively (Sakano et al., 2010; Tiberi et al., 2012). Alternatively, it is posible that the association of NICD to RBPJ, which can enhance its binding to a specific promoter (Krejci and Bray, 2007; Castel et al., 2013), may result in RBPJ repressor function on the miR-155 promoter; however, whether the presence of NICD on the miR-155 promoter enhances RBPJ repressor activity directly or indirectly needs to be further investigated. Collectively, these data support the possibility that miR-155 expression may be physiologically regulated by tuning of the Notch signaling and may indicated a more general mechanism of miR-155 regulation through repression. This hypothesis is supported by a recent study reporting inhibition of miR-155 by transcriptional repression in breast cancer cells, where mutation in the tumor suppressor BRCA1 gene abolishes its repressive function on the miR-155 promoter, resulting in increased miR-155 expression in breast cancer cells and in tumor progression (Chang et al., 2011).
miR-155 plays a broad role in inflammation and in the regulation of the immune-system and it is induced by a wide range of inflammatory factors including LPS/TLR responses. Although increased expression of miR-155 has been positively associated with increased cytokine production, in particular G-CSF and TNFα (Tili et al., 2009; Worm et al., 2009) the molecular mechanism(s) by which miR-155 enhances the expression of these cytokines were not investigated. Our results show that miR-155 increases NF-κB activation by targeting one of its negative regulators, κB-Ras1 (Fenwick et al., 2000). κB-Ras1 (also known as Nkiras1) is a small protein with similarity to Ras-like small GTPases that functions as a negative modulator of NF-κB by binding to NF-κB/IkB complexes and preventing lκBβ and IκBα phosphorylation by IKK (Chen et al., 2004). κB-Ras1 is ubiquitously expressed and its levels inversely correlate with poor prognosis in tumors. While prior studies have suggested an important role for NF-κB in regulating miR-155 expression (Baltimore et al., 2008), our findings reveal that miR-155 can reciprocally upregulate NF-κB activity and amply the inflammatory response by inducing NF-κB-dependent cytokines, in particular G-CSF and TNFα.
A previous report demonstrated a cell intrinsic role for miR-155 in driving myeloproliferative-like disease in a murine transduction/transplantation model (O'Connell et al., 2008). Our findings clearly demonstrate a non-cell autonomous mechanism by which miR-155 overexpression contributes to a myeloproliferative-like disease in RBPJ deficient mice. Interestingly, while RBPJ-/- progenitors exhibit miR-155 up-regulation, transplantation of these cells in WT recipients did not lead to a myeloid expansion. This discrepancy is likely due to the differential expression of miR-155 in the two murine models as well as to differences in the cell types producing this micro-RNA. As discussed by Tili and Croce (Tili et al., 2009), the differential impact of miR-155 on its targets can depend significantly on the level of its expression. Thus, in contrast to models of forced expression, resulting in a ∼30 fold increase of miR-155 on hematopoietic cells compared to normal levels (O'Connell et al., 2008), the miR-155 levels reached in our model (3-5 fold increase) were likely not enough to drive a cell-autonomous MPN, but were sufficient to drive cytokine production in cells of the BM microenvironment specialized to generate an inflammatory response, such as endothelial cells. Our studies point to cell context differences and a higher ability of BM EC and MSC to activate the miR-155/NF-κB/G-CSF/TNFα pathway compared to hematopoietic cells and osteoblasts. Indeed, we show that RBPJ deletion in endothelial cells is sufficient to drive a myeloproliferative-like disease. A relevant role of ECs in this process is not surprising, given that stem cells and progenitors localize adjacent to sinusoids in the BM and that ECs are poised to regulate the hematopoietic response during inflammation through production of pro-inflammatory cytokines (Ding et al., 2012; Fernandez et al., 2008)
Both G-CSF and TNFα are commonly upregulated upon TLR-mediated stimulation during emergency granulopoiesis and following infections (Hirai et al., 2006; Ueda et al., 2005). Their increased expression is also a common feature of several animal models of myeloproliferative-like disease (Dumortier et al., 2010; Fulzele et al., 2013; Walkley et al., 2007; Yoda et al., 2011) and it is observed in patients with MPN (Tefferi et al., 2011). Our discovery of the Notch/miR155/NF-κB/cytokines axis suggests the hypothesis that Notch signaling may contribute to hematopoietic homeostasis by regulating the level of the inflammatory state in the BM niche. In support of this idea, a recent report noted physiologic downregulation of Notch signaling in response to LPS (Kim et al., 2008a); furthermore epigenetic silencing of Notch in myeloid malignancies has been documented (Lobry et al., 2013). Thus, while transitory inhibition of Notch signaling in the BM microenvironment may trigger a physiologic inflammatory circuit characterized by miR-155/NF-κB/cytokine induction driving myeloid cell expansion in response to BM stress (such as infection or acute inflammation), continuous inhibition of Notch signaling due to persistence of inflammatory feed-back loops or/and epigenetic mechanisms, may contribute to the development or progression of a myeloproliferative disorder.
Consistent with our observations demonstrating an essential role for Notch/RBPJ in regulating miR-155 and driving a myeloproliferative-like disease in a murine model, we observed a significant overexpression of miR-155 in Myelofibrosis (MF) patients' BM samples (enriched in stromal cellular components), but not in the PB. Relevant to these findings is the notion that NF-κB plays an essential role in multiple myeloid malignancies and that a comprehensive cytokine profile on serum derived from MF patients demonstrated a cytokine profile very similar to that observed in our mouse model of RBPJ deficiency (Tefferi et al., 2011). Collectively these findings provide a strong link between miR-155/NF-κB and MPN and the rationale to explore this molecular axis to target inflammation and disease progression in these diseases.
Experimental Procedures
Mice
RBPJlox/lox mice were obtained from T. Honjo; Tie2CreER mice were obtained from B. Arnold (European Mouse Mutant Archive; Monterotondo, Italy); miR-155-/- and Lys-Cre mice were obtained from Jackson Laboratories; Lys-EGFP mice were obtained from Dr. T. Graf; DMP1Cre/RBPJ-/- mice were obtained from Dr. T.Bellido. Mx1-Cre recombinase was induced by pIpC 200μg i.p., 3 times at 2-day interval (wk1) plus one single dose (wk2). Efficiency of RBPJ excision was assessed by qRT-PCR (primers in Table S1); Tie2CreER expression was induced by tamoxifen 100mg/kg body for 4-5 weeks, twice/day by gavage. Animal experiments were performed using protocols approved by the IUSM Animal Care and Use Committee.
Cells and cell culture
Raw264.7 and OP9 cells were cultured in DMEM and α-MEM with 10%FBS at 37°C. Cells were transfected with pLKO.1-shRBPJ construct (Thermo) or pLKO.1 vector; or with psiLvU6-shκB-Ras1 or control vector (Genecopoeia) using Lipofectamine 2000 reagent (Life Technologies); or with the retroviral vectors MSCV-GFP, MSCV-ICN/GFP or MGP M155 (MSCV-miR-155, Addgene). BM stromal cells were generated from long bones (tibias, femurs) pre-incubated with 1% collagenase. Total BM cells were flushed and cultured in α-MEM with 20% FBS. Non-adherent cells were removed after 2 days of culture, and adherent cells were expanded for 1wk prior sorting to purify CD45- BM stromal cells by CD45 microbeads (Miltenyi Biotec).
Mouse lung microvascular endothelial cells (Kuhlencordt et al., 2004) were provided by I. Petrache. Cells were grown in DMEM with 20% FBS. Osteoblasts were obtained by long bones as described (Chitteti et al, 2010).
Co-cultures: Lin- cells were purified from femur's BM using the lineage-cell depletion kit (Miltenyi), plated at a density of 5×104/ml on a confluent BM stromal cell layer, and cultured for 48 hrs in IMDM (Gibco, Invitrogen) containing 10% FBS, 50ng/mL SCF and 50ng/mL IL-3 (Miltenyi). Raw cells were treated with 1 μM GSI (EMD Calbiochem), 1 μM HDAC inhibitor (Vorinostat) or 5μM DMAPT for 24 or 48 hours. LNA treatment: cells were transfected with 75nM LNA scramble or anti-miR-155 (Exiqon Company) using lipofectamine 2000 (Invitrogen). Cells were collected for analysis at d2 and d4 post-transfection.
Patients Cells
Primary patients' BM aspirates were obtained from MPN patients with MF seen at the University of Texas MD Anderson Cancer Center, and normal BM cells samples were obtained from Stem Cell Technologies (Vancouver, Canada). BM cells were fractionated using Ficoll Hypaque 1077 (Sigma-Aldrich).
Flow cytometry and cell sorting
BM, spleen and PB cell suspensions were labeled with monoclonal antibodies (Abs) used to define distinct hematopoietic subsets (Table S2). Cells were acquired on a LSRII (BD) and events (0.5 to 5×106//staining) collected and analyzed using Flowjo. CD45-CD31+CD105+ and CD45-CD31-CD105+ cells were sorted in the Reflection3 cell sorter from total BM cells after staining with CD45-APC, CD31-FITC and CD105-PE .
Bone Marrow Transplantation
Total BM cells (5 × 106) from pIpC-induced Mx-Cre+ RBPJlox/lox or control Mx-Cre− littermates (CD45.2) were transplanted by tail vein injection into lethally irradiated B6-SJL (CD45.1) recipients. Total BM cells (3 × 106) from CD45.1 WT or Lys-EGFP mice were transplanted into irradiated, pIpC-induced Mx-Cre+ RBPJlox/lox or control Mx-Cre− recipient mice.
Histology: Tissues were fixed in Z-fix buffer, paraffin-embedded, sectioned, and stained with hematoxylin-eosin.
G-CSF Ab in vivo
Mx1Cre+ and Mx1Cre- RBPJlox/lox 6 to 8wk-old mice were induced with pIpC and after 6 wks were treated with anti-G-CSF neutralizing Ab (R&D # AB-414-NA) 10 μg/d i.p. for 2 cycles of 12 days.
Cytokine array and ELISA
BM milieu was obtained by flashing 1 femur with 1ml of Milliplex reagent; serum was collected from PB; BM-stroma supernatants at 72hrs collection were used. Cytokine levels were measured using a mouse cytokine array (R&D) and Luminex MAP assays (MILLIPLEX MAP Mouse Cytokine/Chemokine, Millipore # MCYTOMAG-70K-09).
Quantitative RT-PCR and microRNA quantification
RNA was purified with the RNeasy Mini Kit (QIAGEN) and cDNA prepared using the Superscript II Kit (Invitrogen). qRT-PCR was performed using SYBR green II Brilliant (Stratagene); Primers are shown in Table S1.
Total RNA from BM sorted cells, BM-derived stroma and Raw cells were extracted with RNAqueous Micro Kit (Ambion). MicroRNA expression was assessed by qRT-PCR (Applied Biosystems) using microRNA-specific TaqMan MicroRNA Assays (mmu-miR-155, U6snRNA). BM from healthy donors and PMF patients was extracted using Trizol (Invitrogen) and 10ng of total RNA from each sample was transcribed using the Taqman MicroRNA reverse Transcriptase kit on ABI 7900HT FAST platform (Applied Biosystems). miR-155 and control gene RUN6B (Applied Biosystems) expression were analyzed by qRT-PCR, using Taqman probes.
Cloning of miR-155 and promoter, κB-Ras1 3′UTR, gene reporter assay and ChIP analysis
The mouse miR-155 promoter region (-1842 to -18) containing the RBPJ binding sites was amplified by PCR from C57BL/6 mouse spleen genomic DNA (primers in table S1). Cloning was performed into the site between XhoI and HindIII of the pGL4.10 luciferase reporter vector (Promega) and confirmed by sequencing. A Dual-Luciferase Assay (Promega) was performed to quantify miR-155 promoter activity. Raw cells were co-transfected with pGL4.10/miR-155Luc and the CMV/Renilla luc control plasmid. The mouse κB-Ras1 3′UTR region (+779 to +4592) containing two miR155 binding sites was amplified by PCR from C57BL/6 mouse genomic DNA (primers in table S1). Cloning was performed into the site between SacI and PmeI of the pmiR-RL vector. Raw cells were co-transfected with the pmiR-RL vector and the CMV/Renilla luc control plasmid to determine targeting of the κB-Ras1 3′UTR by miR155. miR-155 levels and function were also measured in cells by co-transfecting a firefly luciferase reporter containing a binding site for miR-155 at 3′UTR downstream of the luciferase gene (Signosis) and the control Renilla plasmid. Luminescence was measured 24-48hrs after transfection, using a TD-20/20 luminometer (Turner Biosystems).
Chromatin preparations from RBPJ-/- and RBPJ+/+ BM cells, or Raw transfected with shRBPJ or scrambled shRNA, were cross-linked with endogenous DNA and immunoprecipitated with anti-RBPJ (Santa Cruz, 28713)(Barbarulo et al., 2011), anti-Notch1 (Santa Cruz, 6014-R), anti-HDAC2(Cell Signaling, 2545S). As negative controls IgG or irrelevant antibodies (anti-Jagged1; Santa Cruz, 8303) were used; anti-PolII was used as positive control. Recovered DNA was analyzed for miR-155 and GAPDH (control) promoter fragments by qRT-PCR using specific primers (TableS1).
Imaging acquisition and image analysis
Two-photon images of calvarium BM were collected using Olympus XLUMPLFL 20xW, NA 0.95 objective. Z-stacks were collected through the depth of tissue (60 μm Z-stacks) from 6 regions of calvarium BM, at step size settings of 1 μm and 512x512 pixels frame size. Projection images were created using MetaMorph imaging software (Molecular 135 Devices). Volume rendering software Voxx was used to create 3D reconstructions of BM.
Statistical Analysis
Equality of distributions for matched pairs of observations was tested using the t test. Survival was calculated using Kaplan-Meier calculations. P<0.05 was considered to be statistically significant.
Supplementary Material
Highlights.
Notch/RBPJ functions as a transcriptional repressor of miR-155
miR-155 targets the NF-kB inhibitor kB-Ras1 increasing NF-kB activation
Loss of endothelial Notch/RBPJ signaling increases NF-kB activation via miR155
Persistent miR155/NF-kB activation in the BM niche drives myeloproliferation
Acknowledgments
This work was supported by grant NHLBI R01 HL068256 (N.C) the ITRAC program at Indiana University Simon Cancer Center (N.C.), the RSFG at IUPUI (NC), the Indiana Center for Excellence in Molecular Hematology (NIDDK P30 DK090948) and NHLBI HL55716 (E.F.S.).
Footnotes
None of the authors of this manuscript has any financial conflict of interest that might be construed to influence the results or interpretation of the results reported in this communication.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- Barbarulo A, Grazioli P, Campese AF, Bellavia D, Di Mario G, Pelullo M, Ciuffetta A, Colantoni S, Vacca A, Frati L, et al. Notch3 and canonical NF-kappaB signaling pathways cooperatively regulate Foxp3 transcription. J Immunol. 2011;186:6199–6206. doi: 10.4049/jimmunol.1002136. [DOI] [PubMed] [Google Scholar]
- Castel D, Mourikis P, Bartels SJ, Brinkman AB, Tajbakhsh S, Stunnenberg HG. Dynamic binding of RBPJ is determined by Notch signaling status. Genes Dev. 2013;27:1059–1071. doi: 10.1101/gad.211912.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Wang RH, Akagi K, Kim KA, Martin BK, Cavallone L, Haines DC, Basik M, Mai P, Poggi E, et al. Kathleen Cunningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) Tumor suppressor BRCA1 epigenetically controls oncogenic microRNA-155. Nat Med. 2011;17:1275–1282. doi: 10.1038/nm.2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Vallee S, Wu J, Vu D, Sondek J, Ghosh G. Inhibition of NF-kappaB activity by IkappaBbeta in association wtih kappaB-Ras. Mol cell Biol. 2004;24:3048–3056. doi: 10.1128/MCB.24.7.3048-3056.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitteti BR, Cheng YH, Streicher DA, Rodriguez-Rodriguez S, Carlesso N, Srour EF, Kacena MA. Osteoblast lineage cells expressing high levels of Runx2 enhance hematopoietic progenitor cell proliferation and function. J Cell Biochem. 2010;111:284–294. doi: 10.1002/jcb.22694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Turner DL. Vertebrate hairy and Enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- De Strooper B. Nicastrin: gatekeeper of the gamma-secretase complex. Cell. 2005;122:318–320. doi: 10.1016/j.cell.2005.07.021. [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Ding L, Saunders TL, Enikolopov G, Morrison SJ. Endothelial and perivascular cells maintain haematopoietic stem cells. Nature. 2012;481:457–462. doi: 10.1038/nature10783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumortier A, Durham AD, Di Piazza M, Vauclair S, Koch U, Ferrand G, Ferrero I, Demehri S, Song LL, Farr AG, et al. Atopic dermatitis-like disease and associated lethal myeloproliferative disorder arise from loss of Notch signaling in the murine skin. PloS one. 2010;5:e9258. doi: 10.1371/journal.pone.0009258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96:719–726. [PubMed] [Google Scholar]
- Fenwick C, Na SY, Voll RE, Zhong H, Im SY, Lee JW, Ghosh S. A subclass of Ras proteins that regulate the degradation of IkappaB. Science. 2000;287:869–873. doi: 10.1126/science.287.5454.869. [DOI] [PubMed] [Google Scholar]
- Fernandez L, Rodriguez S, Huang H, Chora A, Fernandes J, Mumaw C, Cruz E, Pollok K, Cristina F, Price JE, et al. Tumor necrosis factor-alpha and endothelial cells modulate Notch signaling in the bone marrow microenvironment during inflammation. Exp Hematol. 2008;36:545–558. doi: 10.1016/j.exphem.2007.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forde A, Constien R, Grone HJ, Hammerling G, Arnold B. Temporal Cre-mediated recombination exclusively in endothelial cells using Tie2 regulatory elements. Genesis. 2002;33:191–197. doi: 10.1002/gene.10117. [DOI] [PubMed] [Google Scholar]
- Fulzele K, Krause DS, Panaroni C, Saini V, Barry KJ, Liu X, Lotinun S, Baron R, Bonewald L, Feng JQ, et al. Myelopoiesis is regulated by osteocytes through Gsalpha-dependent signaling. Blood. 2013;121:930–939. doi: 10.1182/blood-2012-06-437160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. International immunology. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- Hirai H, Zhang P, Dayaram T, Hetherington CJ, Mizuno S, Imanishi J, Akashi K, Tenen DG. C/EBPbeta is required for ‘emergency’ granulopoiesis. Nat Immunol. 2006;7:732–739. doi: 10.1038/ni1354. [DOI] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Park JH, Mo JS, Ann EJ, Han SO, Baek SH, Kim KJ, Im SY, Park JW, Choi EJ, et al. Downregulation by lipopolysaccharide of Notch signaling, via nitric oxide. Journal of cell science. 2008a;121:1466–1476. doi: 10.1242/jcs.019018. [DOI] [PubMed] [Google Scholar]
- Kim YW, Koo BK, Jeong HW, Yoon MJ, Song R, Shin J, Jeong DC, Kim SH, Kong YY. Defective Notch activation in microenvironment leads to myeloproliferative disease. Blood. 2008b;112:4628–4638. doi: 10.1182/blood-2008-03-148999. [DOI] [PubMed] [Google Scholar]
- Klinakis A, Lobry C, Abdel-Wahab O, Oh P, Haeno H, Buonamici S, van De Walle I, Cathelin S, Trimarchi T, Araldi E, et al. A novel tumour-suppressor function for the Notch pathway in myeloid leukaemia. Nature. 2011;473:230–233. doi: 10.1038/nature09999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krejci A, Bray S. Notch activation stimulates trannsient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 2007;21:1322–1327. doi: 10.1101/gad.424607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlencordt PJ, Rosel E, Gerszten RE, Morales-Ruiz M, Dombkowski D, Atkinson WJ, Han F, Preffer F, Rosenzweig A, Sessa WC, et al. Role of endothelial nitric oxide synthase in endothelial activation: insights from eNOS knockout endothelial cells. American journal of physiology Cell physiology. 2004;286:C1195–1202. doi: 10.1152/ajpcell.00546.2002. [DOI] [PubMed] [Google Scholar]
- Lobry C, Ntziachristos P, Ndiaye-Lobry D, Oh P, Cimmino L, Zhu N, Araldi E, Hu W, Freund J, Abdel-Wahab O, et al. Notch pathway activation targets AML-initiating cell homeostasis and differentiation. J Exp Med. 2013;210:301–319. doi: 10.1084/jem.20121484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Miyamoto T, Iwasaki H, Reizis B, Ye M, Graf T, Weissman IL, Akashi K. Myeloid or lymphoid promiscuity as a critical step in hematopoietic lineage commitment. Developmental cell. 2002;3:137–147. doi: 10.1016/s1534-5807(02)00201-0. [DOI] [PubMed] [Google Scholar]
- Neelakantan S, Nasim S, Guzman ML, Jordan CT, Crooks PA. Aminoparthenolides as novel anti-leukemic agents: Discovery of the NF-kappaB inhibitor, DMAPT (LC-1) Bioorganic & medicinal chemistry letters. 2009;19:4346–4349. doi: 10.1016/j.bmcl.2009.05.092. [DOI] [PubMed] [Google Scholar]
- O'Connell RM, Rao DS, Chaudhuri AA, Boldin MP, Taganov KD, Nicoll J, Paquette RL, Baltimore D. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald F, Winkler M, Cao Y, Astrahantseff K, Bourteele S, Knochel W, Borggrefe T. RBP-Jkappa/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol Cell Biol. 2005;25:10379–10390. doi: 10.1128/MCB.25.23.10379-10390.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruessmeyer J, Ludwig A. The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Seminars in cell & developmental biology. 2009;20:164–174. doi: 10.1016/j.semcdb.2008.09.005. [DOI] [PubMed] [Google Scholar]
- Sakano D, Kato A, Parikh N, McKnight K, Terry D, Stefanovic B, Kato Y. BCL6 canalizes Notch-dependent transcription, excluding Mastermind-like1 from selected target genes during left-right patterning. Developmental cell. 2010;18:450–462. doi: 10.1016/j.devcel.2009.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider A, Zhang Y, Guan Y, Davis LS, Breyer MD. Differential, inducible gene targeting in renal epithelia, vascular endothelium, and viscera of Mx1Cre mice. Am J Physiol Renal Physiol. 2003;284:F411–F417. doi: 10.1152/ajprenal.00235.2002. [DOI] [PubMed] [Google Scholar]
- Tefferi A, Vaidya R, Caramazza D, Finke C, Lasho T, Pardanani A. Circulating interleukin (IL)-8, IL-2R, IL-12, and IL-15 levels are independently prognostic in primary myelofibrosis: a comprehensive cytokine profiling study. J Clin Oncol. 2011;29:1356–1363. doi: 10.1200/JCO.2010.32.9490. [DOI] [PubMed] [Google Scholar]
- Tiberi L, van den Ameele J, Dimidschstein J, Piccirilli J, Gall D, Herpoel A, Bilheu A, Bonnefont J, Iacovino M, Kyba M, et al. BCL6 controls neurogenesis through Sirt1-dependent epigenetic repression of selective Notch targets. Nature neuroscience. 2012;15:1627–1635. doi: 10.1038/nn.3264. [DOI] [PubMed] [Google Scholar]
- Tili E, Croce CM, Michaille JJ. miR-155: on the crosstalk between inflammation and cancer. Int Rev Immunol. 2009;28:264–284. doi: 10.1080/08830180903093796. [DOI] [PubMed] [Google Scholar]
- Tili E, Michaille JJ, Croce CM. MicroRNAs play a central role in molecular dysfunctions linking inflammation with cancer. Immunol Rev. 2013;253:167–184. doi: 10.1111/imr.12050. [DOI] [PubMed] [Google Scholar]
- Ueda Y, Kondo M, Kelsoe G. Inflammation and the reciprocal production of granulocytes and lymphocytes in bone marrow. J Exp Med. 2005;201:1771–1780. doi: 10.1084/jem.20041419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkley CR, Olsen GH, Dworkin S, Fabb SA, Swann J, McArthur GA, Westmoreland SV, Chambon P, Scadden DT, Purton LE. A microenvironment-induced myeloproliferative syndrome caused by retinoic acid receptor gamma deficiency. Cell. 2007;129:1097–1110. doi: 10.1016/j.cell.2007.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worm J, Stenvang J, Petri A, Frederiksen KS, Obad S, Elmen J, Hedtjarn M, Straarup EM, Hansen JB, Kauppinen S. Silencing of microRNA-155 in mice during acute inflammatory response leads to derepression of c/ebp Beta and down-regulation of G-CSF. Nucleic Acids Res. 2009;37:5784–5792. doi: 10.1093/nar/gkp577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao D, Huang Y, Huang X, Wang W, Yan Q, Wei L, Xin W, Gerson S, Stanley P, Lowe JB, et al. Protein O-fucosyltransferase 1 (Pofut1) regulates lymphoid and myeloid homeostasis through modulation of Notch receptor ligand interactions. Blood. 2011;117:5652–5662. doi: 10.1182/blood-2010-12-326074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kimura T, Tohmonda T, Uchikawa S, Koba T, Takito J, Morioka H, Matsumoto M, Link DC, Chiba K, et al. Dual functions of cell-autonomous and non-cell-autonomous ADAM10 activity in granulopoiesis. Blood. 2011;118:6939–6942. doi: 10.1182/blood-2011-06-357210. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.