Summary
Drugs of abuse activate the mesolimbic dopaminergic pathway. Genetic variations in the dopaminergic system may contribute to vulnerability to drug addiction. Several processes are shared between cocaine and heroin addictions but some neurobiological mechanisms and environmental influence may be specific. This study examined the association of 98 single nucleotide polymorphisms (SNPs) in 13 dopamine pathway-related genes with heroin and/or cocaine addiction in a sample of 801 African Americans (315 subjects with heroin addiction (OD), with or without cocaine (CD) or alcohol addiction (AD), 279 subjects with CD, with or without AD, and 207 controls). Single-marker analyses provided nominally significant evidence for associations of 24 SNPs in DRD1, ANKK1/DRD2, DRD3, DRD5, DBH, DDC, COMT and CSNK1E with OD and/or CD. A DRD2 7-SNPs haplotype block that includes SNPs rs1075650 and rs2283265, which were shown to alter D2S/D2L splicing, was indicated in both addictions. The Met allele of the COMT Val158Met functional variant was associated with protection from OD. None of the signals remained significant after correction for multiple testing. The study results are in accordance with the results of several previous studies, including our report of association of DRD1 SNP rs5326 with OD in a smaller sample from this cohort. The findings suggest the presence of an overlap in the genetic liability for heroin and cocaine addictions, as well as shared and distinct liability for OD in subjects of African and European descent.
Keywords: dopaminergic pathway; polymorphism; heroin addiction; cocaine addiction; African Americans; DRD1, DRD2, COMT Val158Met, rs5326, rs1075650, rs2283265
INTRODUCTION
Drug addiction is a chronic brain disease that is the product of genetic, environmental and drug-induced factors (Kreek et al., 2012). The genetic determinants likely include interactions among multiple neurochemical systems, including the dopaminergic pathway. Drugs of abuse activate the mesolimbic dopaminergic pathway and dopamine (DA) has been implicated in all stages of drug addiction (Di Chiara et al., 2004). Cocaine inhibits DA reuptake by blocking the dopamine transporter thus directly increasing DA synaptic levels. Heroin increases DA levels indirectly by inhibiting GABA neurons in the ventral tegmental area (VTA). Twin studies indicated that a large proportion of variance in the vulnerability for drug dependence is heritable (Kendler et al., 2000). Although cocaine and heroin addiction share psychological processes and neurobiological substrates, they differ in their behavioral, psychological and neurobiological mechanisms and have unique environmental influence s (Badiani et al., 2011; Peters et al., 2013). In addition, many individuals are addicted to both drugs.
This study examined the association of variations in dopamine pathway-related genes with heroin and/or cocaine addiction in African Americans. Although there is a similar risk to develop drug addiction among different ancestral populations, differences in linkage disequilibrium (LD) and allele frequencies may suggest the existence of some distinct genetic risk factors. African Americans are significantly underrepresented in association studies and this study should help to address this gap.
The genes selected for this study are encoding DA receptors (DRD1-5), a DA transporter (DAT1, SLC6A3), enzymes involved in DA synthesis and degradation (TH, DBH, DDC and COMT) and regulators of DA signaling (ANKK1, CSNK1E and PPP1R1B). Dopamine is the endogenous ligand for two classes of G protein-coupled dopamine receptors; the D-like receptors (D1 and D5) and the D2-like receptors (D2, D3, and D4). Tyrosine hydroxylase (TH) produces L-DOPA that is in turn catalyzed to dopamine by DOPA decarboxylase (DDC). Dopamine is degraded by catechol-O-methyltransferase (COMT), which is a modulator of prefrontal dopaminergic tone. The dopamine transporter (DAT1, SLC6A3) mediates the synaptic reuptake of dopamine. Dopamine beta-hydroxylase (DBH) converts dopamine to norepinephrine thus regulating the ratio of the two neurotransmitters. DARPP-32 (PPP1R1B, dopamine- and cAMP-regulated phosphoprotein, 32 kDa) is a key regulatory molecule in the dopaminergic signaling pathway. The casein kinase 1 epsilon isoform (CSNK1E) is a serine/threonine-selective phosphotransferase that regulates dopaminergic signaling through DARPP-32. ANKK1 is located in close proximity to DRD2, and there is high LD between SNPs in both genes. The TaqIA polymorphism (rs1800497) is located in the coding region of ANKK1. There is a potential relationship between ANKK1 and the dopaminergic system (Hoenicka et al., 2010).
Several association studies of DA-related genes with heroin or cocaine addiction have been reported. ANKK1/DRD2 SNPs were associated with heroin or cocaine addiction/abuse in different populations (Al-Eitan et al., 2012; Clarke et al., 2014; Jacobs et al., 2013a; Lawford et al., 2000; Li et al., 2006; Nelson et al., 2013; Perez de los Cobos et al., 2007; Shahmoradgoli Najafabadi et al., 2005; Vereczkei et al., 2013; Xu et al., 2004). In addition, a DRD1 SNPs were associated with heroin abuse (Jacobs et al., 2013a,b), a DBH SNP was associated with progressive OD in Han Chinese (Xie et al., 2013), and CSNK1E SNPs were associated with OD in subjects with European descent (Levran et al., 2008) and Han Chinese (Wang et al., 2014). COMT variants have been associated with cocaine addiction in African Americans (AA) (Lohoff et al., 2008), and cocaine-induced paranoia in European Americans (EA) and AA (Ittiwut et al., 2011). Two association studies of cocaine dependence in a Spanish Caucasian sample that included dopaminergic genes reported nominal significant association of SNPs in DBH, SLC6A3 and TH (Fernandez-Castillo et al., 2010; Fernandez-Castillo et al., 2013). A recent GWAS of cocaine dependence in EA and AA did not identify association with SNPs in DA-related genes (Gelernter et al., 2013).
This study examined the association of variations in 13 dopamine pathway-related genes with heroin and/or cocaine addiction in a sample of 801 American subjects of predominantly African ancestry. The study is an extension of our previous studies in this population with a larger sample, modified SNP content and an additional cocaine addiction sample (Levran et al., 2009; Levran et al., 2014b) and follows our association studies in subjects with European ancestry (Levran et al., 2008; Levran et al., 2014a).
MATERIALS AND METHODS
Subjects
The study sample (n = 801) was divided into three main samples based on addiction status and drug of addiction (heroin or cocaine) (Table 1). The control sample included 207 subjects. All subjects (n = 315) in the heroin addiction sample (#1, “OD±CD”) were current addicts in methadone maintenance treatment programs and reported heroin as their drug of choice. This sample is composed of two subsamples: (#1a) “OD only”; including subjects with no cocaine addiction (CD) and current or past alcohol addiction (AD) (5%) (n = 138, 44%), and (#1b) “OD+CD”; including subjects with current or past CD, with or without AD (n = 177, 56%). All subjects (n = 279) in the cocaine addiction sample (#2, “CD only”) had current or past CD and reported cocaine as their drug of choice. Some of them (29%) also had current or past AD and none of them had OD. The sex ratios were 0.52, 0.37, and 0.37 females in the controls, “OD±CD” and “CD only” samples, respectively. The mean ages (± SD) when blood was obtained were 33 ±12, 49 ±10, and 43 ± 7 years in the control, “OD±CD” and “CD only” samples, respectively. The age range was 18-76 years. Cannabis use/abuse was assessed but was not taken into account in the present analysis.
Table 1.
Groups description
| Sample |
Diagnosis |
|||||
|---|---|---|---|---|---|---|
| # | Drug “symbol” |
Symbol | 1 | 2 | n | f |
| 1 | Heroin “OD±CD” |
315 | ||||
| 1a | “OD only” | 138 | 0.44 | |||
| OD# | - | 123 | 0.39 | |||
| OD# | AD* | 15 | 0.05 | |||
| 1b | “OD+CD” | 177 | 0.56 | |||
| OD# | CD* | 76 | 0.24 | |||
| OD# | CD*/AD* | 101 | 0.32 | |||
| 2 | Cocaine “CD only” |
279 | ||||
| CD* | - | 198 | 0.71 | |||
| CD* | AD* | 81 | 0.29 | |||
| 3 | Control | - | - | 207 | ||
Abbreviation: OD, heroin addiction; CD, cocaine addiction; AD, Alcohol addiction; n, number of subjects; f,
frequency; current,
past and current
This study is an expansion of our previous study of OD in AA (Levran et al., 2009) for which we added 481 new subjects. Most subjects were self-identified as African Americans (AA) and showed > 50% African ancestry contribution by STRUCTURE analysis (see below). A total of 46 subjects who self-identified as AA were excluded because they showed < 50% African ancestry contribution. Seventy subjects whose self-identified ancestry was different from AA (ambiguous, Caribbean African non-Hispanic, European, or Native American) were included based on > 50% African ancestry contribution. Hispanic subjects were not included.
Ascertainment was made by extensive personal interview, using the following instruments: the Addiction Severity Index (McLellan et al., 1992), the Kreek-McHugh-Schluger-Kellogg Scale (KMSK) (Kellogg et al., 2003) and the Diagnostic and Statistical Manual of Mental Disorders, 4th Edition (DSM-IV).
Subjects were excluded from the control category with (1) >1 instance of drinking to intoxication or any illicit drug use in the last month; (2) a history of alcohol drinking to intoxication or illicit drug use more than twice a week for more than 6 consecutive months and (3) cannabis use for more than 12 days in the last month or past cannabis use for > 2 days/week for > 4 years. The former heroin addicts had a history of > 1 year of daily multiple uses of heroin and were treated at a methadone maintenance treatment program (MMTP).
Subjects were recruited at the Rockefeller University Hospital, the Manhattan Campus of the VA New York Harbor Health Care System and the Dr. Miriam and Sheldon G. Adelson Clinic for Drug Abuse Treatment and Research in Las Vegas (LV), NV. The Institutional Review Boards of the Rockefeller University Hospital and the VA New York Harbor Healthcare System approved the study. The Rockefeller University IRB also reviews the Adelson Clinic, LV. All subjects signed informed consent for genetic studies.
Genes/SNPs and Genotyping
A new custom array (GS0013101-OPA) was designed based on the “addiction” array (GS0007064-OPA; Illumina, San Diego, CA, USA) (Hodgkinson et al., 2008) that was used in our previous studies (Levran et al., 2008; Levran et al., 2009). The original design included tagging SNPs with a minor allele frequency (MAF) > 0.005 that aimed to capture the full haplotype information of the HapMap Yoruban population (Hodgkinson et al., 2008).
The 13 genes associated with the dopaminergic pathway in this array were included in this analysis (Table 2). X chromosome genes (e.g., MAOA) were not included. Based on current HapMap data for the African YRI sample, the mean gene coverage for 10 of the genes is ~70%. For 3 genes (DRD4, DRD5 and PPP1R1B) there is not enough data to calculate % coverage.
Table 2.
Dopaminergic pathway genes analyzed in the study
| Symbol | Name |
|---|---|
| ANKK1 | ankyrin repeat and kinase domain containing 1 |
| COMT | catechol-O-methyltransferase |
| CSNK1E | casein kinase 1, epsilon |
| DBH | dopamine beta-hydroxylase |
| DDC | DOPA decarboxylase |
| DRD1 | dopamine receptor D1 |
| DRD2 | dopamine receptor D2 |
| DRD3 | dopamine receptor D3 |
| DRD4 | dopamine receptor D4 |
| DRD5 | dopamine receptor D5 |
| PPP1R1B | protein phosphatase 1, regulatory (inhibitor) subunit 1B (DARPP-32) |
| SLC6A3 | solute carrier family 6 member 3 (dopamine transporter, DAT) |
| TH | tyrosine hydroxylase |
Fourteen SNPs from these genes were excluded from the new array based on low quality in the original array, and 19 SNPs were excluded based on low MAF in the population of this study. Nine SNPs were added, based on functionality or reports of association with related phenotypes, including three ANKK1 SNPs, with a total of 118 SNPs (Supplement Table 1).
Genotyping was performed using an Illumina GoldenGate Custom Panel of 1536 SNPs at the Rockefeller University Genomics Resource Center. Random samples were genotyped in duplicate. Analysis was performed with BeadStudio software v2.3.43 (Illumina) and all cluster plots were manually inspected.
Assessment of percentage of African Ancestry Using Ancestry Informative Markers (AIMs)
A set of 155 AIMs was used for STRUCTURE 2.2 analysis (Pritchard et al., 2000). Biographic Ancestry Scores (e.g., fractions of genetic affiliation of the individual in each cluster) were estimated under the assumption of seven clusters (k). Each subject was anchored against genotypes of 1051 samples from the Human Genome Diversity Cell Line Panel (http://www.cephb.fr/HGDP-CEPH-Panel), as described (Ducci et al., 2009). To be included in the study, an individual had to show > 50% African ancestry contribution.
Statistical analysis
The statistical power was estimated for the different case samples with the standard test for difference between proportions, under additive, dominant or recessive logistic regression model, assuming odds ratios (OR) between 1.2 and 2.2 and MAF within the range 0.05-0.45, at a fixed significance level of 0.05 and power = 0.80 (Sham & Purcell 2014). The same control sample size (n = 207) was used in each calculation. All samples have sufficient power to detect ORs as low as 1.2 for a SNP with MAF as high as 0.45. For SNPs with MAF as low as 0.05, the combined sample (“OD±CD” & “CD only”, n = 594) can detect ORs as low as 2.0, the “OD±CD” sample (n = 315) can detect OR as low as 2.2, and the “CD only” sample (n = 279) can detect OR as low as 1.8. The “OD only” sample of (n = 138) can detect OR as low as 2.1 for a SNP with MAF as low as 0.1.
Deviation from Hardy–Weinberg equilibrium (HWE) was examined in the control sample by the exact test using the PLINK program (Purcell et al., 2007). Pairwise LD and LD blocks were estimated using Haploview 4.2 (Barrett et al., 2005) based on ‘Solid Spine of LD’ algorithm. Association analyses were performed by two methods:
ANOVA with the data as a categorical variable: (a) “OD only” #1a, (b) “OD+CD” #1b, (c) “CD only” #2, and (d) control (see Table 1), followed by Post hoc pair comparisons for the significant associations; The group is used as a fixed factor in a logistic regression model and the genotype as the binary response variable according to the following codification. We fitted logistic regression models for all SNPs and used the chi-square to test the significance of the model (the alternative hypothesis that the genotypes are different across the 4 groups). P-values were corrected according to the Benjamin-Hochberg methodology (FDR).
Independent logistic regression analyses for the “OD±CD”, “OD only”, “CD only”, and “OD±CD” + “CD only” samples. Logistic regression was performed for each SNP separately using the PLINK program, and as two groups reflecting dominant or recessive inheritance model. The direction of the regression coefficient represents the minor allele. Sex was included as a covariate in the initial model but had no significant effect on the results so it was not included in the final analysis. Adjustment for multiple testing was also performed by permutation test (n = 100,000) for each model of inheritance, using PLINK.
RESULTS
Case control association analysis was conducted in 801 subjects (315 subjects with heroin addiction, with or without cocaine or alcohol addiction, 279 subjects with cocaine addiction, with or without alcohol addiction, and 207 controls)(Table 1). The heroin sample (#1) is comprised of two subsamples: #1a, “OD only”; including subjects with no past or current CD (n = 138), and #1b, “OD+CD”; including subjects with current or past CD (n = 177).” One hundred and eighteen SNPs from 13 genes related to the dopaminergic pathway were genotyped (Table 2, Supplement Table 1). Twenty SNPs were excluded based on low quality. The remaining 98 SNPs were analyzed separately for association with heroin or cocaine addiction. One SNP (ANKK1 rs7118900) showed significant deviation from HWE in controls (P = 0.007) and the rest of the SNPs showed no significant deviation (P > 0.01). There was no evidence for population substructure among the three samples (heroin, cocaine and controls) based on Structure analysis of 155 AIMs. There was an average of 81±10% African ancestry and similar admixture pattern among the samples, as was shown in our previous study of this cohort (Levran et al., 2014b). Analysis of LD indicated 26 haplotype blocks, including 17 highly correlated (r2 > 0.7) SNPs (7 pairs and one triplet) (Supplement Table 2 & Supplement Figure 1).
ANOVA assuming four categories: (#1a) “OD only”, (#1b) “OD+CD”, (2) “CD only”, and controls, revealed 16 SNPs with nominally significant differences among the groups under at least one model of inheritance (Supplement Table 3). None remain significant after correction for multiple testing. Post hoc pairwise comparisons of these SNPs revealed nominally significant differences of 11 SNPs, including five SNPs (ANKK1 rs7118900, DRD2 rs1079596 and rs2587548 DRD3 rs167771, and COMT rs4818) between the control sample and the “OD only” sample, one SNP (CSNK1E rs5757037) between “CD only” and “OD only”, and two SNPs (COMT rs2239393 and rs4818) between the “OD only” sample and “OD+CD” sample (Supplement Table 3). None remained significant after correction for multiple testing.
Logistic regression under two models of inheritance (dominant/recessive) for “OD only” (1a), “OD±CD” (1a &1b), the “CD only” (2), and “OD±CD” & “CD only” (1 & 2) samples revealed nominally significant differences in genotype frequencies for 24 SNPs in nine genes (DRD1, ANKK1, DRD2, DRD3, DRD5, DBH, DDC, COMT and CSNK1E)(Table 3). No signal survived correction for multiple testing. Of those, four SNPs (in ANKK1, DBH and COMT) are coding SNPs (two non-synonymous and two synonymous). All the SNPs indicated by ANOVA were also indicated by the logistic regression analyses.
Table 3.
Nominally significant associations using logistic regression
| Gene | SNP | Location |
P
1
|
Model | OR2 | L953 | U954 | |||
|---|---|---|---|---|---|---|---|---|---|---|
| “CD only” |
“OD only” |
“OD±CD” | “OD±CD” & “CD only” |
|||||||
| 2 | 1a | 1a & 1b | 1 & 2 | |||||||
| DRD1 |

|
3′ UTR | 0.014 | 0.031 | D | 1.61 | 1.10 | 2.35 | ||
| 5′ UTR | 0.006 | D | 1.87 | 1.16 | 3.02 | |||||
| ANKK1 | rs71189005 | Ala239Thr | 0.008 | 0.006 | 0.010 | R | 0.49 | 0.29 | 0.81 | |
| DRD2 |

|
intron | 0.038 | 0.047 | D | 1.78 | 1.03 | 3.08 | ||
| intron | 0.038 | 0.047 | D | 1.78 | 1.03 | 3.08 | ||||
| intron | 0.022 | 0.005 | 0.018 | D | 0.51 | 0.32 | 0.81 | |||
| intron | 0.032 | 0.011 | 0.043 | D | 0.55 | 0.35 | 0.87 | |||
| intron | 0.007 | 0.006 | 0.019 | 0.005 | D | 1.63 | 1.16 | 2.29 | ||
| intron | 0.011 | 0.010 | 0.028 | 0.009 | D | 1.58 | 1.12 | 2.21 | ||
| intron | 0.045 | 0.029 | 0.029 | D | 1.48 | 1.04 | 2.10 | |||
| rs4648318 | intron | 0.039 | R | 1.58 | 1.02 | 2.45 | ||||
| rs4274224 | intron | 0.025 | R | 2.14 | 1.10 | 4.16 | ||||
| rs1799978 | intron | 0.017 | 0.029 | D | 1.64 | 1.09 | 2.46 | |||
| DRD3 | rs3773678 | intron | 0.016 | R | 2.17 | 1.16 | 4.07 | |||
| rs167771 | intron | 0.007 | 0.012 | 0.048 | R | 4.83 | 1.53 | 15.31 | ||
| DRD5 | rs2867383 | downstream | 0.031 | 0.013 | 0.010 | D | 0.64 | 0.46 | 0.90 | |
| DBH | rs1108580 | Glu162= | 0.042 | 0.046 | R | 0.58 | 0.34 | 0.98 | ||
| DDC | rs2122822 | intron | 0.046 | D | 0.67 | 0.45 | 0.99 | |||
| rs2329341 | intron | 0.031 | D | 0.68 | 0.48 | 0.97 | ||||
| COMT | rs933271 | intron | 0.039 | R | 1.68 | 1.03 | 2.75 | |||

|
intron | 0.047 | D | 1.65 | 1.01 | 2.71 | ||||
| Leu126= | 0.002 | D | 1.98 | 1.27 | 3.07 | |||||
| rs4680 | Val158Met | 0.043 | R | 0.38 | 0.15 | 0.97 | ||||
| CSNK1E | rs5757037 | upstream | 0.012 | D | 1.78 | 1.14 | 2.77 | |||
P-Value in bold are the lowest obtained for the specific SNP; Dash lines box represents haplotype block
The lowest nominal P value obtained from testing two models of inheritance: dominant and recessive. Empty cells indicate P ≥ 0.05.
OR for the lowest P value for this SNP. OR < 1 represent protective effect of the minor allele, OR > 1 represent risk effect of the minor allele
95% confidence interval lower value
95% confidence interval upper value
This SNP showed significant deviation from HWE in controls (P = 0.007)
These SNPs were associated with heroin addiction in subjects with European ancestry (Levran et al. 2014c)
Abbreviations: D, dominant; R, recessive; Chr, chromosome; OR, odds ratio
LD analysis showed that some of these SNPs are part of three haplotype blocks, including seven DRD2 SNPs, DRD1 SNPs rs686 and rs5326, as well as COMT SNPs rs2239393 and rs4818, respectively. Among the DRD2 haplotype block there are two SNP pairs and one triplet in strong correlation (r2 > 0.72) (Table 3, Figure 1, Supplement Table 2, & Supplement Figure 1).
Figure 1. DRD2 pairwise Linkage Disequilibrium.
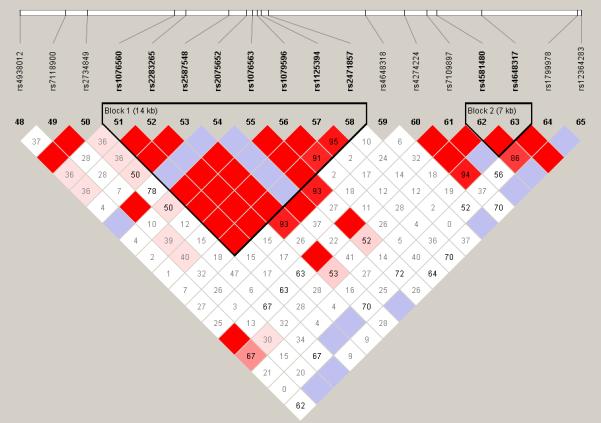
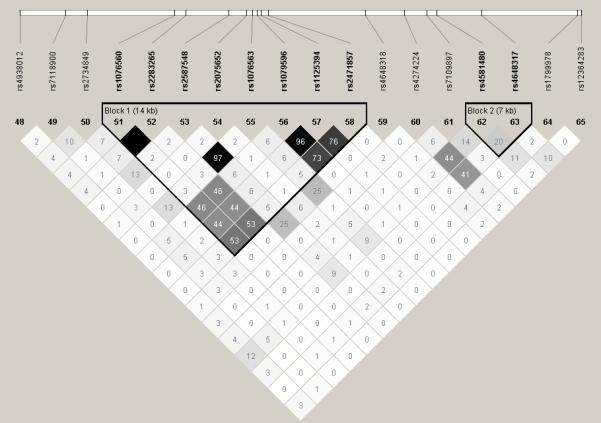
The pairwise correlation between SNPs was measured as D’ (a) or r2 (b). The values are shown (x100) in each box. The color scheme indicates the magnitude of the value. When the value is equal to 1.0 no number is given.
The comparison of results of the different analyses indicated several SNPs that were only detected in a specific sample (e.g., “OD only”), and SNPs for which the P-values were lower in the “OD only” or “CD only” compared with other samples, despite the smaller sample size (Table 3). Other SNPs showed lower signals in the combined addiction group including DRD2 and DRD5 SNPs.
The association signal, under recessive model, of the functional COMT SNP rs4680 (Val158Met) was driven by lower frequency (0.04) of the AA genotype (encoding Met/Met) in the “OD only” group than in the control group (0.11) indicating protective effect (OR = 0.38; 95% CI 0.15-0.97) (Supplement Table 4).
DISCUSSION
This study provides evidence for nominally significant associations of several SNPs in genes involved in the dopaminergic pathway with heroin and/or cocaine addiction in African Americans. This study can be viewed as a major expansion of our previous study of OD in African Americans (Levran et al., 2009). The genes included in the current study, except for ANKK1, were included in the previous study, although the SNP content was modified to exclude redundant and non-informative markers and to include additional SNPs. We have not previously studied cocaine addiction in this cohort.
The two DRD1 SNPs identified in association with heroin and/or cocaine addictions are located in the 5′ and the 3′ UTR of the gene, respectively. They are in the same haplotype block, but they are not strongly correlated (r2 < 0.2) in both African and European populations. SNP rs5326 has a comparable allele frequency in different populations (0.14- 0.2 in HapMap CEU, YRI and CHB). DRD1 rs686 is located in a sequence complementary to the seed sequence of miR-504 and showed differential expressions of the two variants (Huang & Li, 2009). The study results are in accordance with our previous finding of association of DRD1 SNP rs5326 with OD in AA (Levran et al., 2009). It is also in line with a study showing association of this SNP with time to develop to heroin dependence in Han Chinese (Peng et al., 2013). DRD1 SNP rs686 was associated with nicotine dependence in AA (Huang et al., 2008) and alcohol dependence in Europeans (Batel et al., 2008). It was also associated with duration of transition from the first use to opioid dependence in Han Chinese (Zhu et al., 2013), and with opioid abuse in AA (Jacobs et al., 2013b).
One ANKK1 SNP and 10 DRD2 SNPs showed association with heroin and/or cocaine addiction, including seven SNPs that are part of one haplotype block, suggesting a related signal. The non-synonymous ANKK1 SNP rs7118900 (Ala239Thr) identified in this study is in perfect LD with SNP rs1800497 (TaqIA) in the HapMap European population (CEU), but in low LD (r2 < 0.2) with this SNP in the HapMap African population (YRI) suggesting a potentially different effect of this SNP on DRD2 expression. SNP rs1800497 was excluded from this analysis based on low quality. The association of ANKK1 SNP rs7118900 should be interpreted with caution since the SNP showed significant deviation from HWE in the control sample, although there was no indication of low genotyping quality and this SNP was in HWE in another control sample of European ancestry that was genotyped with the same array (Levran et al. 2014c). An in vitro study showed differential gene expression of the two variants of SNP rs7118900 at baseline and in response to stimulation with apomorphine. The variant 239Thr is predicted to create a new phosphorylation site (Garrido et al., 2011). DRD2 has two major splice isoforms with distinct functions: D2 long (D2L) and D2 short (D2S). The DRD2 SNPs rs1076560 and rs2283265 that are in perfect LD, were shown to shift splicing from D2S to D2L changing their expression ratio. These variants were associated with differences in brain functions and psychiatric disorders (Zhang et al. 2007, Moyer et al., 2011).
Previous association studies of DRD2 with heroin and cocaine addictions produced mixed results that may be partly explained by small sample size, population substructure and suboptimal ascertainment (for additional references and discussion see Moyer et al., 2011). The results of this study are compatible with several studies. DRD2 SNPs rs2283265 and rs10765560 were associated with OD in a Jordanian sample (Al-Eitan et al., 2012), and in Europeans (Jacobs et al., 2013a). They were also associated with cocaine abuse and dependence in European Americans (Moyer et al., 2011). SNP rs10765560 has been recently associated with opioid dependence, but not with cocaine dependence, in European and African Americans (Clarke et al., 2014). Of note, the OD sample used by Clarke et al. includes a substantive number of DNA samples from our laboratory that are part of the NIDA Center for Genetic Studies DNA Repository, so it cannot be considered an independent sample. DRD2 SNP rs1079597 (also called “TaqIB”) has been previously associated with heroin dependence in a Hungarian sample (Vereczkei et al., 2013). Although this SNP was not included in the study, it is in strong LD, in the HapMap African sample (YRI), with SNP rs1076560. DRD3 SNP rs167771 indicated in this study was previously associated with nicotine dependence (Wei et al., 2012), smoking behavior (Yu et al., 2006), and autism (Toma et al., 2013).
The functional rs4680 (Val158Met) identified in this study in association with OD, is one of the most a well-studied SNP (Montag et al., 2012)(Tammimaki & Mannisto 2010). The frequency of the A allele (encoding Met) and the AA genotype was lower in subjects with OD (without CD), compared with controls or subjects with CD or OD+CD. This allele is associated with lower enzymatic activity, higher dopamine levels, enhanced vulnerability to stress, as well as increased susceptibility to white matter structural alterations in the context of addiction (Zhang et al., 2013). It was associated with CD in AA (Lohoff et al., 2008), and cocaine-induced paranoia in EA and AA (Ittiwut et al., 2011). It has been suggested that environmental factors impact the detection of small effects of this variant on alcohol addiction vulnerability (Schellekens et al., 2012). COMT SNPs rs4680 and rs933271, indicated in this study, were predicted to have potential functionality based on a signature of positive selection in EA and AA (Ittiwut et al., 2011). SNP rs737866 that was associated with earlier age of opiate onset in Chinese (Li et al., 2012) is in strong LD (D’ = .85) with SNP rs933271 in this sample.
The suggested association of a SNP upstream of CSNK1E with OD is intriguing since we have found association of another CSNK1E SNP and haplotype with OD, in EA (Levran et al., 2014c) and yet another SNP was associated with OD in Han Chinese (Wang et al., 2014). There is no LD between these SNPs in this sample and no information on the SNP identified. CSNK1E interacts with circadian rhythms and DARPP-32 and has been implicated in negative regulation of sensitivity to opioids in rodents (Bryant et al., 2012; Cheong & Virshup 2011).
DDC SNP rs2329341 that was indicated in this study was recently shown to be associated with cocaine dependence subtype, by interacting with GABRG3 SNP (Bi et al., 2014). A recent report suggested an association of DBH SNP rs1611115 with progressive OD in Han Chinese (Xie et al., 2013). This SNP was included in this study but did not show significant association.
Cocaine vs. heroin addiction
The model of common (non-drug specific) liability to addiction hypothesizes the presence of a non-specific liability to all drug addictions based on shared biological mechanisms and high genetic correlations between liabilities to specific drug addictions (Vanyukov et al., 2012). Decrements in DRD2 receptors in the striatum were observed in addicts to several drugs of abuse (Koob & Volkow, 2010). Although cocaine and heroin addiction share psychological processes and neurobiological substrates, they differ in neurobiological mechanisms and environmental influence (Badiani et al., 2011; Peters et al., 2013). For example, chronic heroin or cocaine abusers showed unique gene expression profiles in the post-mortem nucleus accumbens (Albertson et al., 2006). In this study, we have analyzed different combinations of heroin and cocaine addiction, in order to identify SNPs with either drug-specific or non-drug-specific effect. Comparison of the association signals in the different samples suggests several shared signals (e.g. DRD2 SNPs rs1079596 and rs1125394) that might reflect an association with a general phenotype of substance addiction, and also signals that might be unique to the CD or OD (e.g. COMT rs4680 Val158Met).
European vs. African Americans (heroin addiction)
Although heroin and/or cocaine addiction have similar prevalence in different ancestral populations, these populations may have distinct as well as shared genetic risk factors. The causes for the distinct factors may include different allele frequencies, LD patterns, modifier genes and/or gene-environment interactions.
We have reported association analyses of OD using similar marker set in subjects of predominantly European ancestry (Levran et al., 2008, Levran et al. 2014c). Comparison of the findings in the two ancestral populations reveals mostly distinct polymorphisms. We have reported an association of the casein kinase 1ε gene CSNK1E SNP rs1534891, and related haplotype, with OD in subjects with European ancestry. This SNP was not indicated in this study, although that may be partly explained by the low MAF of this SNP in African populations. This study indicated the common SNP rs5757037 in association with OD, but this SNP was not identified in EA. There is no LD between these SNPs.
Nevertheless, in both populations there were common association signals in DRD2 (rs1076563, rs2587548). Although the two DRD2 SNPs are in strong LD in both populations, their allele frequency is different and their minor alleles in EA are the major alleles in AA. Another potential common association is the DRD2 SNP rs1076560. This SNP (or SNPs in strong LD with this SNP) were associated with OD in subjects with European ancestry by other groups (Jacobs et al., 2013a, Vereczkei et al., 2013, Clarke et al., 2014).
The general LD pattern of the ANKK1/DRD2 region is different between the two populations. Thus, the ANKK1 SNP rs7118900, which was identified in this study, is in strong LD with the two DRD2 SNPs mentioned above and the ANKK1 SNP rs1800497 (TaqIA) only in population of European ancestry, suggesting a potentially different effect of this SNP on gene expression. There is no LD between the DRD3 SNPs indicated in EA and AA.
Some limitations should be considered when interpreting the results of this study. Although nominally significant, the associations may be due to chance and further testing in independent populations and/or meta-analysis are needed for confirmation. The study is underpowered to detect variants with small effects and it is possible that significant associations were not detected because of limited power due to the sample size and partial gene coverage. Although there was no indication of population substructure between cases and controls, the admixed nature of the African American populations could be a source of error for SNPs that differ in their allele frequencies between the two ancestral populations. In addition, because of the high substance-related comorbidity and the study design, the labeling of shared or common liability may not be accurate. Despite these limitations, the study has important strengths, including rigorous ascertainment, reliable phenotype data and stringent inclusion/exclusion criteria.
In summary, this study provides evidence for nominally significant associations of specific variants in DA-related genes with heroin and cocaine addictions that are common and/or specific to the African American population. African Americans are significantly under-represented in association studies and this study helps to address this gap. The findings suggest the presence of both shared and distinct genetic liability for heroin and cocaine addictions as well as for African and European Americans. Several SNPs of potential importance were indicated and further studies are warranted to confirm the results.
Supplementary Material
ACKNOWLEDGEMENTS
We thank all the clinical staff including S Linzy, RN, E Ducat, NP and B Ray, NP. We are grateful to P-H Shen, PhD, and D Goldman, PhD from the NIH/NIAAA for STRUCTURE analysis. We thank C Zhao, PhD, and B Zhang, from the Rockefeller Genomic Resource Center, for their excellent assistance in genotyping. This work was supported by the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation, the Clinical and Translational Science Award UL1RR024143 from the National Center for Advancing Translational Sciences of the National Institutes of Health (JC) and by NSFC grant 31470070 from the Chinese Government (JO).
Footnotes
Conflict of interest The authors declare no conflict of interest
Supporting Information Additional supporting information may be found at the publisher’s web-site:
Supplement Table 1. SNP list
Supplement Table 2. LD data
Supplement Table 3. ANOVA
Supplement Table 4. COMT Val158Met genotype frequencies
Supplement Fig 1. Pairwise Linkage Disequilibrium of all the genes analyzed
All authors declare no conflict of interest.
References
- Al-Eitan LN, Jaradat SA, Hulse GK, Tay GK. Custom genotyping for substance addiction susceptibility genes in Jordanians of Arab descent. BMC Res Notes. 2012;5:497. doi: 10.1186/1756-0500-5-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. 2006;31:2304–2312. doi: 10.1038/sj.npp.1301089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badiani A, Belin D, Epstein D, Calu D, Shaham Y. Opiate versus psychostimulant addiction: the differences do matter. Nat Rev Neurosci. 2011;12:685–700. doi: 10.1038/nrn3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Batel P, Houchi H, Daoust M, Ramoz N, Naassila M, Gorwood P. A haplotype of the DRD1 gene is associated with alcohol dependence. Alcohol Clin Exp Res. 2008;32:567–572. doi: 10.1111/j.1530-0277.2008.00618.x. [DOI] [PubMed] [Google Scholar]
- Bi J, Gelernter J, Sun J, Kranzler HR. Comparing the utility of homogeneous subtypes of cocaine use and related behaviors with DSM-IV cocaine dependence as traits for genetic association analysis. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:148–156. doi: 10.1002/ajmg.b.32216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant CD, Parker CC, Zhou L, Olker C, Chandrasekaran RY, Wager TT, Bolivar VJ, Loudon AS, Vitaterna MH, Turek FW, Palmer AA. Csnk1e is a genetic regulator of sensitivity to psychostimulants and opioids. Neuropsychopharmacology. 2012;37:1026–1035. doi: 10.1038/npp.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong JK, Virshup DM. Casein kinase 1: Complexity in the family. Int J Biochem Cell Biol. 2011;43:465–469. doi: 10.1016/j.biocel.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Clarke TK, Weiss AR, Ferarro TN, Kampman KM, Dackis CA, Pettinati HM, O’Brien CP, Oslin DW, Lohoff FW, Berrettini WH. The dopamine receptor D2 (DRD2) SNP rs1076560 is associated with opioid addiction. Ann Hum Genet. 2014;78:33–39. doi: 10.1111/ahg.12046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Chiara G, Bassareo V, Fenu S, De Luca MA, Spina L, Cadoni C, Acquas E, Carboni E, Valentini V, Lecca D. Dopamine and drug addiction: the nucleus accumbens shell connection. Neuropharmacology. 2004;47(Suppl 1):227–241. doi: 10.1016/j.neuropharm.2004.06.032. [DOI] [PubMed] [Google Scholar]
- Ducci F, Roy A, Shen PH, Yuan Q, Yuan NP, Hodgkinson CA, Goldman LR, Goldman D. Association of substance use disorders with childhood trauma but not African genetic heritage in an African American cohort. Am J Psychiatry. 2009;166:1031–1040. doi: 10.1176/appi.ajp.2009.08071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Castillo N, Ribases M, Roncero C, Casas M, Gonzalvo B, Cormand B. Association study between the DAT1, DBH and DRD2 genes and cocaine dependence in a Spanish sample. Psychiatr Genet. 2010;20:317–320. doi: 10.1097/YPG.0b013e32833b6320. [DOI] [PubMed] [Google Scholar]
- Fernandez-Castillo N, Roncero C, Grau-Lopez L, Barral C, Prat G, Rodriguez-Cintas L, Sanchez-Mora C, Gratacos M, Ramos-Quiroga JA, Casas M, Ribases M, Cormand B. Association study of 37 genes related to serotonin and dopamine neurotransmission and neurotrophic factors in cocaine dependence. Genes Brain Behav. 2013;12:39–46. doi: 10.1111/gbb.12013. [DOI] [PubMed] [Google Scholar]
- Garrido E, Palomo T, Ponce G, Garcia-Consuegra I, Jimenez-Arriero MA, Hoenicka J. The ANKK1 protein associated with addictions has nuclear and cytoplasmic localization and shows a differential response of Ala239Thr to apomorphine. Neurotox Res. 2011;20:32–39. doi: 10.1007/s12640-010-9219-6. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Sherva R, Koesterer R, Almasy L, Zhao H, Kranzler HR, Farrer L. Genome-wide association study of cocaine dependence and related traits: FAM53B identified as a risk gene. Mol Psychiatry. 2013;6:717–723. doi: 10.1038/mp.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkinson CA, Yuan Q, Xu K, Shen PH, Heinz E, Lobos EA, Binder EB, Cubells J, Ehlers CL, Gelernter J, Mann J, Riley B, Roy A, Tabakoff B, Todd RD, Zhou Z, Goldman D. Addictions biology: haplotype-based analysis for 130 candidate genes on a single array. Alcohol Alcohol. 2008;43:505–515. doi: 10.1093/alcalc/agn032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenicka J, Quinones-Lombrana A, Espana-Serrano L, Alvira-Botero X, Kremer L, Perez-Gonzalez R, Rodriguez-Jimenez R, Jimenez-Arriero MA, Ponce G, Palomo T. The ANKK1 gene associated with addictions is expressed in astroglial cells and upregulated by apomorphine. Biol Psychiatry. 2010;67:3–11. doi: 10.1016/j.biopsych.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Huang W, Ma JZ, Payne TJ, Beuten J, Dupont RT, Li MD. Significant association of DRD1 with nicotine dependence. Hum Genet. 2008;123:133–140. doi: 10.1007/s00439-007-0453-9. [DOI] [PubMed] [Google Scholar]
- Huang W, Li MD. Differential allelic expression of dopamine D1 receptor gene (DRD1) is modulated by microRNA miR-504. Biol Psychiatry. 2009;65:702–705. doi: 10.1016/j.biopsych.2008.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ittiwut R, Listman JB, Ittiwut C, Cubells JF, Weiss RD, Brady K, Oslin D, Farrer LA, Kranzler HR, Gelernter J. Association between polymorphisms in catechol-O-methyltransferase (COMT) and cocaine-induced paranoia in European-American and African-American populations. Am J Med Genet B Neuropsychiatr Genet. 2011;156:651–660. doi: 10.1002/ajmg.b.31205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MM, Murray J, Byrd DA, Hurd YL, Morgello S. HIV-related cognitive impairment shows bi-directional association with dopamine receptor DRD1 and DRD2 polymorphisms in substance-dependent and substance-independent populations. J Neurovirol. 2013a;19:495–504. doi: 10.1007/s13365-013-0204-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs MM, Okvist A, Horvath M, Keller E, Bannon MJ, Morgello S, Hurd YL. Dopamine receptor D1 and postsynaptic density gene variants associate with opiate abuse and striatal expression levels. Mol Psychiatry. 2013b;18:1205–1210. doi: 10.1038/mp.2012.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg SH, McHugh PF, Bell K, Schluger JH, Schluger RP, LaForge KS, Ho A, Kreek MJ. The Kreek-McHugh-Schluger-Kellogg scale: a new, rapid method for quantifying substance abuse and its possible applications. Drug Alcohol Depend. 2003;69:137–150. doi: 10.1016/s0376-8716(02)00308-3. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Karkowski LM, Neale MC, Prescott CA. Illicit psychoactive substance use, heavy use, abuse, and dependence in a US population-based sample of male twins. Arch Gen Psychiatry. 2000;57:261–269. doi: 10.1001/archpsyc.57.3.261. [DOI] [PubMed] [Google Scholar]
- Koob GF, Volkow ND. Neurocircuitry of addiction. Neuropsychopharmacology. 2010;35:217–238. doi: 10.1038/npp.2009.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreek MJ, Levran O, Reed B, Schlussman SD, Zhou Y, Butelman ER. Opiate addiction and cocaine addiction: underlying molecular neurobiology and genetics. J Clin Invest. 2012;122:3387–3393. doi: 10.1172/JCI60390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawford BR, Young RM, Noble EP, Sargent J, Rowell J, Shadforth S, Zhang X, Ritchie T. The D(2) dopamine receptor A(1) allele and opioid dependence: association with heroin use and response to methadone treatment. Am J Med Genet. 2000;96:592–598. doi: 10.1002/1096-8628(20001009)96:5<592::aid-ajmg3>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- Levran O, Londono D, O’Hara K, Nielsen DA, Peles E, Rotrosen J, Casadonte P, Linzy S, Randesi M, Ott J, Adelson M, Kreek MJ. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes Brain Behav. 2008;7:720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Londono D, O’Hara K, Randesi M, Rotrosen J, Casadonte P, Linzy S, Ott J, Adelson M, Kreek MJ. Heroin addiction in African Americans: a hypothesis-driven association study. Genes Brain Behav. 2009;8:531–540. doi: 10.1111/j.1601-183X.2009.00501.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Peles E, Randesi M, Li Y, Rotrosen J, Ott J, Adelson M, Kreek MJ. Stress-related genes and heroin addiction: a role for a functional FKBP5 haplotype. Psychoneuroendocrinology. 2014a;45:67–76. doi: 10.1016/j.psyneuen.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Randesi M, Li Y, Rotrosen J, Ott J, Adelson M, Kreek MJ. Drug addiction and stress-response genetic variability: association study in African Americans. Ann Hum Genet. 2014b;78:290–298. doi: 10.1111/ahg.12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levran O, Peles E, Randesi, Correa da Rosa J, Ott J, Rotrosen J, Adelson M, Kreek MJ. Dopaminergic pathway polymorphisms and heroin addiction: further support for association of Casein Kinase 1ε variants. Pharmacogenomics. 2014c;15:2001–2009. doi: 10.2217/pgs.14.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Du J, Yu S, Jiang H, Fu Y, Wang D, Sun H, Chen H, Zhao M. Pathways to age of onset of heroin use: a structural model approach exploring the relationship of the COMT gene, impulsivity and childhood trauma. PLoS One. 2012;7:e48735. doi: 10.1371/journal.pone.0048735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Shao C, Zhang D, Zhao M, Lin L, Yan P, Xie Y, Jiang K, Jin L. The effect of dopamine D2, D5 receptor and transporter (SLC6A3) polymorphisms on the cue-elicited heroin craving in Chinese. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:269–273. doi: 10.1002/ajmg.b.30264. [DOI] [PubMed] [Google Scholar]
- Lohoff FW, Weller AE, Bloch PJ, Nall AH, Ferraro TN, Kampman KM, Pettinati HM, Oslin DW, Dackis CA, O’Brien CP, Berrettini WH. Association between the catechol-O-methyltransferase Val158Met polymorphism and cocaine dependence. Neuropsychopharmacology. 2008;33:3078–3084. doi: 10.1038/npp.2008.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLellan AT, Kushner H, Metzger D, Peters R, Smith I, Grissom G, Pettinati H, Argeriou M. The Fifth Edition of the Addiction Severity Index. J Subst Abuse Treat. 1992;9:199–213. doi: 10.1016/0740-5472(92)90062-s. [DOI] [PubMed] [Google Scholar]
- Montag C, Jurkiewicz M, Reuter M. The role of the catechol-O-methyltransferase (COMT) gene in personality and related psychopathological disorders. CNS Neurol Disord Drug Targets. 2012;11:236–250. doi: 10.2174/187152712800672382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer RA, Wang D, Papp AC, Smith RM, Duque L, Mash DC, Sadee W. Intronic polymorphisms affecting alternative splicing of human dopamine D2 receptor are associated with cocaine abuse. Neuropsychopharmacology. 2011;36:753–762. doi: 10.1038/npp.2010.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EC, Lynskey MT, Heath AC, Wray N, Agrawal A, Shand FL, Henders AK, Wallace L, Todorov AA, Schrage AJ, Saccone NL, Madden PA, Degenhardt L, Martin NG, Montgomery GW. ANKK1, TTC12, and NCAM1 polymorphisms and heroin dependence: importance of considering drug exposure. JAMA Psychiatry. 2013;70:325–333. doi: 10.1001/jamapsychiatry.2013.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng S, Du J, Jiang H, Fu Y, Chen H, Sun H, Wang D, Yu S, Zhao M. The dopamine receptor D1 gene is associated with the length of interval between first heroin use and onset of dependence in Chinese Han heroin addicts. J Neural Transm. 2013;120:1591–1598. doi: 10.1007/s00702-013-1029-6. [DOI] [PubMed] [Google Scholar]
- Perez de los Cobos J, Baiget M, Trujols J, Sinol N, Volpini V, Banuls E, Calafell F, Luquero E, del Rio E, Alvarez E. Allelic and genotypic associations of DRD2 TaqI A polymorphism with heroin dependence in Spanish subjects: a case control study. Behav Brain Funct. 2007;3:25. doi: 10.1186/1744-9081-3-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J, Pattij T, De Vries TJ. Targeting cocaine versus heroin memories: divergent roles within ventromedial prefrontal cortex. Trends Pharmacol Sci. 2013;34:689–695. doi: 10.1016/j.tips.2013.10.004. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellekens AF, Franke B, Ellenbroek B, Cools A, de Jong CA, Buitelaar JK, Verkes RJ. Reduced dopamine receptor sensitivity as an intermediate phenotype in alcohol dependence and the role of the COMT Val158Met and DRD2 Taq1A genotypes. Arch Gen Psychiatry. 2012;69:339–348. doi: 10.1001/archgenpsychiatry.2011.1335. [DOI] [PubMed] [Google Scholar]
- Shahmoradgoli Najafabadi M, Ohadi M, Joghataie MT, Valaie F, Riazalhosseini Y, Mostafavi H, Mohammadbeigi F, Najmabadi H. Association between the DRD2 A1 allele and opium addiction in the Iranian population. Am J Med Genet B Neuropsychiatr Genet. 2005;134B:39–41. doi: 10.1002/ajmg.b.30117. [DOI] [PubMed] [Google Scholar]
- Sham PC, Purcell SM. Statistical power and significance testing in large-scale genetic studies. Nat Rev Genet. 2014;15:335–346. doi: 10.1038/nrg3706. [DOI] [PubMed] [Google Scholar]
- Tammimaki AE, Mannisto PT. Are genetic variants of COMT associated with addiction? Pharmacogenet Genomics. 2010;20:717–741. doi: 10.1097/FPC.0b013e328340bdf2. [DOI] [PubMed] [Google Scholar]
- Toma C, Hervas A, Balmana N, Salgado M, Maristany M, Vilella E, Aguilera F, Orejuela C, Cusco I, Gallastegui F, Perez-Jurado LA, Caballero-Andaluz R, Diego-Otero Y, Guzman-Alvarez G, Ramos-Quiroga JA, Ribases M, Bayes M, Cormand B. Neurotransmitter systems and neurotrophic factors in autism: association study of 37 genes suggests involvement of DDC. World J Biol Psychiatry. 2013;14:516–527. doi: 10.3109/15622975.2011.602719. [DOI] [PubMed] [Google Scholar]
- Vanyukov MM, Tarter RE, Kirillova GP, Kirisci L, Reynolds MD, Kreek MJ, Conway KP, Maher BS, Iacono WG, Bierut L, Neale MC, Clark DB, Ridenour TA. Common liability to addiction and "gateway hypothesis": theoretical, empirical and evolutionary perspective. Drug Alcohol Depend. 2012;123(Suppl 1):S3–17. doi: 10.1016/j.drugalcdep.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vereczkei A, Demetrovics Z, Szekely A, Sarkozy P, Antal P, Szilagyi A, Sasvari-Szekely M, Barta C. Multivariate analysis of dopaminergic gene variants as risk factors of heroin dependence. PLoS One. 2013;8:e66592. doi: 10.1371/journal.pone.0066592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhu Y, Wang W, Wu F, Cui H, Xun X, Lai J. A population-based association study of casein kinase 1 epsilon loci with heroin dependence in Han Chinese. J Mol Neurosci. 2014;53:143–149. doi: 10.1007/s12031-013-0186-2. [DOI] [PubMed] [Google Scholar]
- Wei J, Chu C, Wang Y, Yang Y, Wang Q, Li T, Zhang L, Ma X. Association study of 45 candidate genes in nicotine dependence in Han Chinese. Addict Behav. 2012;37:622–626. doi: 10.1016/j.addbeh.2012.01.009. [DOI] [PubMed] [Google Scholar]
- Xie X, Xu L, Liu H, Chen W, Zhuang D, Zhang J, Duan S, Zhou W. Positive association between--1021TT genotype of dopamine beta hydroxylase gene and progressive behavior of injection heroin users. Neurosci Lett. 2013;541:258–262. doi: 10.1016/j.neulet.2013.02.049. [DOI] [PubMed] [Google Scholar]
- Xu K, Lichtermann D, Lipsky RH, Franke P, Liu X, Hu Y, Cao L, Schwab SG, Wildenauer DB, Bau CH, Ferro E, Astor W, Finch T, Terry J, Taubman J, Maier W, Goldman D. Association of specific haplotypes of D2 dopamine receptor gene with vulnerability to heroin dependence in 2 distinct populations. Arch Gen Psychiatry. 2004;61:597–606. doi: 10.1001/archpsyc.61.6.597. [DOI] [PubMed] [Google Scholar]
- Yu Y, Panhuysen C, Kranzler HR, Hesselbrock V, Rounsaville B, Weiss R, Brady K, Farrer LA, Gelernter J. Intronic variants in the dopa decarboxylase (DDC) gene are associated with smoking behavior in European-Americans and African-Americans. Hum Mol Genet. 2006;15:2192–2199. doi: 10.1093/hmg/ddl144. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lee MR, Salmeron BJ, Stein DJ, Hong LE, Geng X, Ross TJ, Li N, Hodgkinson C, Shen PH, Yang Y, Goldman D, Stein EA. Prefrontal white matter impairment in substance users depends upon the catechol-o-methyl transferase (COMT) val158met polymorphism. Neuroimage. 2013;69:62–69. doi: 10.1016/j.neuroimage.2012.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bertolino A, Fazio L, Blasi G, Rampino A, Romano R, Lee ML, Xiao T, Papp A, Wang D, Sadee W. Polymorphisms in human dopamine D2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proc Natl Acad Sci U S A. 2007;104:20552–20557. doi: 10.1073/pnas.0707106104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F, Yan CX, Wen YC, Wang J, Bi J, Zhao YL, Wei L, Gao CG, Jia W, Li SB. Dopamine D1 receptor gene variation modulates opioid dependence risk by affecting transition to addiction. PLoS One. 2013;8:e70805. doi: 10.1371/journal.pone.0070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


