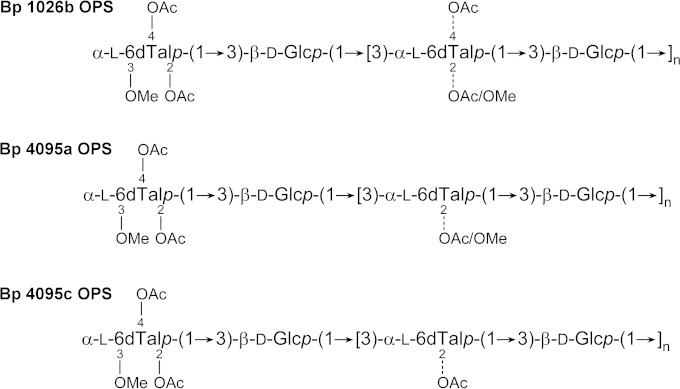Abstract
Burkholderia pseudomallei is a CDC tier 1 select agent that causes melioidosis, a severe disease in humans and animals. Persistent infections are common, and there is currently no vaccine available. Lipopolysaccharide (LPS) is a potential vaccine candidate. B. pseudomallei expresses three serologically distinct LPS types. The predominant O-polysaccharide (OPS) is an unbranched heteropolymer with repeating d-glucose and 6-deoxy-l-talose residues in which the 6-deoxy-l-talose residues are variably replaced with O-acetyl and O-methyl modifications. We observed that primary clinical B. pseudomallei isolates with mucoid and nonmucoid colony morphologies from the same sample expressed different antigenic types distinguishable using an LPS-specific monoclonal antibody (MAb). MAb-reactive (nonmucoid) and nonreactive (mucoid) strains from the same patient exhibited identical LPS banding patterns by silver staining and indistinguishable genotypes. We hypothesized that LPS antigenic variation reflected modification of the OPS moieties. Mutagenesis of three genes involved in LPS synthesis was performed in B. pseudomallei K96243. Loss of MAb reactivity was observed in both wbiA (encoding a 2-O-acetyltransferase) and wbiD (putative methyl transferase) mutants. The structural characteristics of the OPS moieties from isogenic nonmucoid strain 4095a and mucoid strain 4095c were further investigated. Utilizing nuclear magnetic resonance (NMR) spectroscopy, we found that B. pseudomallei 4095a and 4095c OPS antigens exhibited substitution patterns that differed from the prototypic OPS structure. Specifically, 4095a lacked 4-O-acetylation, while 4095c lacked both 4-O-acetylation and 2-O-methylation. Our studies indicate that B. pseudomallei OPS undergoes antigenic variation and suggest that the 9D5 MAb recognizes a conformational epitope that is influenced by both O-acetyl and O-methyl substitution patterns.
INTRODUCTION
Burkholderia pseudomallei is an environmental saprophyte and the cause of melioidosis. This organism is also classified as a CDC tier 1 select agent based on its potential hazard to public health (1). B. pseudomallei infection is acquired by inoculation, inhalation, and ingestion. Once an infection is established, bacteria may disseminate via the bloodstream and affect numerous organs, with common clinical manifestations, including pneumonia and multiple abscesses in the liver and/or spleen. In northeast Thailand, the mortality rate is 40% (2), and clearance of B. pseudomallei is difficult to achieve, leading to the need for prolonged antimicrobial treatment and to relapse in up to 10% of cases. B. pseudomallei is a facultative intracellular organism and can survive in a range of host cell types, including phagocytes (3, 4). Potential mechanisms that contribute to bacterial persistence are poorly understood, although B. pseudomallei has been reported to be highly adaptable and able to survive under extreme conditions (5, 6).
Lipopolysaccharide (LPS) is an important B. pseudomallei virulence factor (7), is a major stimulator of the host immune response (8), and has been considered to be a potential vaccine target (9). Three B. pseudomallei LPS types have been described based on SDS-PAGE pattern (ladder type A, ladder type B, and rough LPS), which have distinct serological reactivities (10). Type A and type B LPSs are composed of three covalently linked domains (lipid A, core-oligosaccharide, and O-polysaccharide [OPS]), while rough LPS lacks OPS (10). Type A is the predominant LPS type expressed by isolates from Thailand (97%) and Australia (80%) (10). The structure of type A OPS is an unbranched polymer consisting of disaccharide repeats having the structure →3)-β-d-glucopyranose-(1→3)-6-deoxy-α-l-talopyranose-(1→, in which the 6-deoxy-α-l-talopyranose (6dTal) residues are variably replaced with O-acetyl and O-methyl modifications (11–14). Antibody to B. pseudomallei LPS is detected in more than 90% of patients with culture-confirmed melioidosis (15).
In several Gram-negative bacteria, LPS variation has been associated with different colony phenotypes (16, 17). This phenomenon generates diversity that confers a survival fitness in different environments. LPS modification can impact antigenicity and also affect serum sensitivity and adhesion to the host cells (18). Our previous study reported that B. pseudomallei has seven colony morphotypes which undergo switching under different laboratory conditions. The type I morphotype represents the predominant type (88% of 241 clinical isolates), but this can switch to other types in vivo and in vitro. The different colonies that were observed on Ashdown selective agar are related to phenotypic variation, including the ability to produce extracellular enzymes, flagella, motility, and biofilm formation, but the relationship between colony morphotype and LPS variation is unclear (19). We have subsequently noted the presence of two distinct colony types in a proportion of clinical samples plated onto nonselective agar, such as Trypticase soy agar (TSA) and blood agar (BA), which are nonselective media commonly used in routine hospital laboratories and in our research laboratory. These two types of colonies are described as mucoid (M) and nonmucoid (NM), although the related phenotypes on these media are unknown.
We hypothesized that different colony types of B. pseudomallei were associated with LPS variation. In the present study, we developed a latex agglutination test based on an LPS-specific monoclonal antibody (MAb) to screen the LPS antigenic variation in B. pseudomallei mucoid and nonmucoid colonies and verified the presence of distinct OPS types by Western blotting. Using a combination of genetic techniques and nuclear magnetic resonance (NMR) spectroscopy, we also demonstrated that O-methyl and O-acetyl modification patterns influenced the ability of the MAb to recognize the OPS antigens.
MATERIALS AND METHODS
Bacterial isolates and culture conditions.
Isolation and cultivation of B. pseudomallei were performed in a class II biosafety cabinet located in a biosafety level 3 (BSL3) containment facility. Three retrospective collections of B. pseudomallei isolates were used, as follows: (i) 200 isolates, one from each of 200 patients presenting to Sappasithiprasong Hospital, Ubon Ratchathani, northeast Thailand, for whom the first episode of melioidosis was between 1986 and 2004 and who did not relapse, as shown by follow-up exams to July 2005 (20); (ii) 166 isolates from 78 melioidosis patients presenting to Sappasithiprasong Hospital with melioidosis who relapsed, as shown by follow-up exams to July 2005 (20); and (iii) 52 isolates from 38 patients, 10 animals, 3 soil samples, and 1 water sample from northern Australia, provided by Bart Currie, Charles Darwin University, in 2002 (10). A fourth prospective collection, in which colonies were picked from primary culture plates (sheep blood agar) of specimens from 40 patients suspected of having melioidosis who presented to Sappasithiprasong Hospital between July 2011 and November 2012, was assembled during this study. The clinical specimens were sputa (n = 13), tracheal secretions (n = 6), pus (n = 11), wound swabs (n = 3), synovial fluid (n = 1), and blood culture (n = 6). Colonies were picked after incubation for 2 days at 37°C in air. These samples were randomly selected from those that grew B. pseudomallei on a primary blood agar plate during routine diagnostic processing. The laboratory strains of B. pseudomallei and Escherichia coli used during the study are described in Table 1. Unless otherwise stated, these were cultured on Trypticase soy agar (TSA) and Luria-Bertani (LB) agar (BD), respectively. All isolates were stored in Trypticase soy broth (TSB) with 15% glycerol at −80°C.
TABLE 1.
Bacterial strains used in this studya
| Strain | Relevant characteristic(s) | Date of isolation (day/mo/yr or yr) | Specimen type | Colony morphology | LPS type | 9D5 reaction | Source and/or reference |
|---|---|---|---|---|---|---|---|
| B. pseudomallei | |||||||
| 4095a (NM) | Clinical strain from a Thai patient | 19/09/2006 | Sputum | NM | A | Positive | Freezer vial |
| 4095c (M) | 11/10/2006 | Pleural fluid | M | A | Negative | Freezer vial | |
| 10457A (NM) | Clinical strain from a Thai patient | 06/07/2011 | Sputum | NM | A | Positive | Primary culture |
| 10457A (M) | M | A | Negative | Primary culture | |||
| 10971B (NM) | Clinical strain from a Thai patient | 10/09/2012 | Tracheal suction | NM | A | Positive | Primary culture |
| 10971B (M) | M | A | Negative | Primary culture | |||
| 11017A (NM) | Clinical strain from a Thai patient | 28/09/2012 | Sputum | NM | A | Positive | Primary culture |
| 11017A (M) | M | A | Negative | Primary culture | |||
| MSHR295 (NM) | Soil in Australia | 2002 | Soil | NM | A | Positive | Freezer vial; 10 |
| MSHR295 (M) | M | A | Negative | Freezer vial; 10 | |||
| K96243 | Wild type | 1996 | NM | A | Positive | 30 | |
| K96243 ΔwbiA | K96243 derivative with ΔwbiA (mutant defective in the acetylation of OPS) | NA | NM | A | Negative | This study | |
| K96243 ΔwbiA complemented strain | K96243 derivative with ΔwbiA complemented with wbiA | NA | NM | A | Positive | This study | |
| K96243 ΔwbiD | K96243 derivative with ΔwbiD (mutant defective in OPS synthesis) | NA | NM | Rough | Negative | This study | |
| K96243 ΔoacA | K96243 derivative with ΔoacA (mutant defective in acetylation of OPS) | NA | NM | A | Positive | This study | |
| 4095a (NM) ΔwcbB | 4095a (NM) derivative with ΔwcbB (mutant defective in CPS) | NA | NM | A | Positive | This study | |
| 4095c (M) ΔwcbB | 4095c (M) derivative with ΔwcbB (mutant defective in CPS synthesis) | NA | M | A | Negative | This study | |
| E. coli | |||||||
| HB101 | ATCC 33694 | NA | NA | NA | ATCC | ||
| DH5α | F′ competent cells | NA | NA | NA | 31 | ||
| RHO3 | Kms SM10 (λpir) Δasd::FRT ΔaphA::FRT | NA | NA | NA | 22 |
FRT, FLP recombination target; NA, not applicable; NM, nonmucoid; M, mucoid; ATCC, American Type Culture Collection.
Latex agglutination assay.
A latex agglutination test was performed as described previously (21). This is based on an MAb termed 9D5, which recognizes B. pseudomallei OPS and is positive for B. pseudomallei and the closely related nonvirulent organism B. thailandensis but negative for B. mallei (21). The MAb was purified using a Hi-trap protein A high-performance (HP) column (GE Healthcare) and sensitized onto latex particles (MAb-latex) according to the manufacturer's instructions (Invitrogen). B. pseudomallei reactivity was assessed by mixing bacterial cells with 10 μl of MAb-latex suspension on a glass slide and observing the cells for agglutination within 2 min.
Isolation and characterization of mucoid and nonmucoid colonies.
B. pseudomallei isolates from the three retrospective collections were streaked from a freezer vial onto TSA and incubated at 37°C in air for 2 days, and the plates were observed for the presence of nonmucoid and mucoid colonies. Nonmucoid colonies were defined as white, opaque, and matte with an umbilicate or umbonate surface, and mucoid colonies were defined as white or yellow and shiny with a smooth surface. Five colonies of each of the two types from the same sample (or all colonies if fewer than 5) were tested with the MAb-latex test. For the prospective study of clinical samples, B. pseudomallei colonies on blood agar were observed for mucoid and nonmucoid colonies after incubation at 37°C in air for 2 days. Ten colonies of each of the two types from the same sample (or all colonies if fewer than 10) were tested with the MAb-latex assay. Separate freezer vials were prepared for all colony picks and stored in TSB with 15% glycerol at −80°C. Each colony was tested for the following: (i) LPS type using 12% SDS-PAGE and silver staining and (ii) expression of the MAb 9D5-specific antigen using Western blotting (10). LPS was extracted using proteinase K (Invitrogen) digestion. LPS types were defined as ladder type A, type B, or rough type (no ladder) (10). In the event that a single sample contained both mucoid and nonmucoid colonies, individual colonies of both types were compared by pulsed-field gel electrophoresis, as previously described (19).
Effects of different culture conditions on B. pseudomallei colony appearance.
The effect of a range of laboratory conditions on colony appearance was tested on the same isogenic pairs. Ten colonies of mucoid or nonmucoid types per strain scraped from TSA were suspended in phosphate-buffered saline (PBS) and adjusted to 1 × 108 CFU/ml using spectrophotometry, with the optical density at 600 nm. Two hundred microliters of suspension of each type was inoculated into 2 ml of distilled water (DW) or TSB and incubated under one of the following conditions: (i) DW at 37°C for 24 h, (ii) TSB (pH 7.4) at 37°C for 24 h, (iii) TSB (pH 4.0) at 37°C for 24 h, (iv) TSB (pH 8.5) at 37°C for 24 h, (v) TSB (pH 7.4) at 42°C for 24 h, (vi) TSB (pH 7.4) plus 350 mM NaCl at 37°C for 24 h, (vii) TSB (pH 7.4) plus 50 mM NaNO2 at 37°C for 24 h, (viii) TSB (pH 7.4) plus 2 mM H2O2 at 37°C for 24 h, (ix) TSB (pH 7.4) at 37°C for 7 days (all incubated in air), and (x) TSB (pH 7.4) at 37°C for 24 h in an anaerobic jar (Oxoid). Thereafter, bacteria were diluted in PBS and spread plated onto TSA and the mucoid and nonmucoid colonies in the sample enumerated. Three separate experiments were performed in duplicate. Five colonies of mucoid and nonmucoid colonies (where present) were tested for each plate using the MAb-latex test.
Bacterial internalization by human monocytes.
Human monocytic cell line THP1 (ATCC TIB-202) was cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal bovine serum albumin (FBS). All cultures were maintained at 37°C in a humidified 5% CO2 incubator. Monocytes (2 × 105 cells) were seeded into each well of a 96-well plate and incubated overnight at 37°C in 5% CO2. Cells were infected with B. pseudomallei at a multiplicity of infection (MOI) of 1:1 at 37°C in 5% CO2 for 2 h. Extracellular bacteria were killed by exposure to medium containing 250 μg/ml kanamycin for a further 2 h. Intracellular bacteria were quantified by lysing infected monocytes with 0.1% Triton X-100 and plating them, followed by colony counting on TSA.
Construction of mutants and complemented strains.
Five B. pseudomallei mutants, including three LPS mutants defective in wbiA (BPSL2680), wbiD (BPSL2677), or oacA (BPSL1936) and two strains of capsule mutants defective in wcbB (BPSL2808), were constructed using a fragment mutagenesis method, as described previously (22, 23). LPS mutants were constructed in B. pseudomallei K96243 (nonmucoid), and capsule mutants were constructed in B. pseudomallei 4095a (nonmucoid) and 4095c (mucoid), which are isogenic pairs from the same patient (Table 1). Gene sequences for wbiA, wbiD, oacA, and wcbB were obtained from GenBank (gene identifiers 3094050, 3093124, 3091646, and 3093047, respectively). PCR primers were designed using Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast) (Table 2). Mutagenesis and complementation was performed using pEXKm5-based allele replacement, as described previously (22, 23). The absence of a 236-bp oriT fragment of the pEXKm5 backbone was examined in all mutants. Phenotypic evaluation of LPS and capsule expression was achieved by SDS-PAGE of proteinase K extracts and Western blot analysis, respectively (21). LPS expression was demonstrated using the 9D5 MAb, while capsule expression was demonstrated using the capsule-specific 4B11 MAb (21).
TABLE 2.
Primer pairs used in this study
| Gene and primer | Sequence (5′–3′) | Positionsa | Product size (bp) |
|---|---|---|---|
| wbiA (BPSL2680) | |||
| wbiA-F1 | CGATTCGATGCCGCCGACGT | 34 to 53 | 297 |
| wbiA-R1 | CAACGCATCCGCTCGCATGC | 311 to 330 | |
| wbiA-F2 | GCATGCGAGCGGATGCGTTGTGCTGCTTGGCTATCCGGCG | 911 to 930 | 305 |
| wbiA-R2 | TCCATCCTTGTCCGGGCCCC | 1176 to 1190 | |
| wbiA-F1 | CGATTCGATGCCGCCGACGT | 34 to 53 | 1,162 |
| wbiA-R2 | TCCATCCTTGTCCGGGCCCC | 1176 to 1190 | |
| wbiA-Fex | GGAATCTGCGTCTCCGGCTT | 256 to 275 | 300 |
| wbiA-Rex | ATAGGGTGTCGTGTCTCGCAG | 535 to 555 | |
| wbiA-seqA | AATGGCTGCACGATGCGGTGT | −104 to −124 | 814 |
| wbiA-seqB | CACGGCAAGCAACACCCTGC | 671 to 690 | |
| wbiA-seqC | TGCTGCGAGACACGACACCC | 533 to 552 | 783 |
| wbiA-seqD | GCGCAGCCGATAAAGCCAGC | +57 to +76 | |
| wbiD (BPSL2677) | |||
| wbiD-F1 | CGGTGTACAGCAATGTCGTT | 41 to 60 | 206 |
| wbiD-R1 | CAGACGGTGCAGGTCGATTC | 227 to 246 | |
| wbiD-F2 | GAATCGACCTGCACCGTCTGTCTACCGAAAGTGGCGTTCC | 1409 to 1428 | 259 |
| wbiD-R2 | CGGATGCCTGACAAAGAACC | 1628 to 1647 | |
| wbiD-F1 | CGGTGTACAGCAATGTCGTT | 41 to 60 | 1,607 |
| wbiD-R2 | CGGATGCCTGACAAAGAACC | 1628 to 1647 | |
| oacA (BPSL1936) | |||
| oacA-F1 | AAAGGCCGACGCATTCCGGG | −162 to −144 | 311 |
| oacA-R1 | AATGCCCGCTGGAGCGTGTC | 130 to 149 | |
| oacA-F2 | GACACGCTCCAGCGGGCATTCCCCAAGCACCATTGGCGGT | 995 to 1014 | 344 |
| oacA-R2 | ATGTGGGCATGGGGAAGCGC | +114 to +133 | |
| oacA-F1 | AAAGGCCGACGCATTCCGGG | −162 to −144 | 1,480 |
| oacA-R2 | ATGTGGGCATGGGGAAGCGC | +114 to +133 | |
| wcbB (BPSL2808) | |||
| wcbB-F1 | CCGGCACTATGGCAGCCGAG | 96 to 115 | 218 |
| wcbB-R1 | CCATTCCGCTGTGGCTTGTATGC | 291 to 313 | |
| wcbB-F2 | GCATACAAGCCACAGCGGAATGGAGCGACCTGGACGTGTTCCG | 940 to 959 | 188 |
| wcbB-R2 | CCACGTCGGTTCGCGGAAGT | 1085 to 1104 | |
| wcbB-F1 | CCGGCACTATGGCAGCCGAG | 96 to 115 | 1,009 |
| wcbB-R2 | CCACGTCGGTTCGCGGAAGT | 1085 to 1104 | |
| 16S rRNA gene | |||
| Univ_16S_F | TGGCTCAGAACGAACGCTGGCGGC | 21 to 44 | 336 |
| Univ_16S_R | CCCACTGCTGCCTCCCGTAGGAGGAGT | 327 to 356 | |
| oriT-F | TCCGCTGCATAACCCTGCTTC | 598 to 578 | 236 |
| oriT-R | CAGCCTCGCAGAGCAGGATTC | 368 to 383 |
Positions correspond to the nucleotide sequences of the wbiA, wbiD, oacA, and wcbB genes of B. pseudomallei K96243 chromosome 1 (NCBI reference sequence gene identifiers 3094050, 3093124, 3091646, and 3093047 for wbiA, wbiD, oacA, and wcbB, respectively). Positions beginning with a minus sign are for genes at the 5′ untranslated region (UTR), and those beginning with a plus sign are genes at the 3′ UTR of the genes.
DNA sequencing and RT-PCR of wbiA.
Genomic DNA was extracted from B. pseudomallei using the QIAamp DNA minikit (Qiagen). B. pseudomallei wbiA was amplified using two overlapping primer pairs (wbiA-seqA and wbiA-seqB, and wbiA-seqC and wbiA-seqD) shown in Table 2. PCR products were cleaned using ExoSAP-IT (Affymetrix) and sequenced by Macrogen (Republic of Korea). One-step reverse transcriptase PCR (RT-PCR) was used to compare wbiA transcription of nonmucoid colony types with that of mucoid colony types. Individual colonies were harvested from TSA, and RNA was extracted using TRIzol reagent (Invitrogen). RT-PCR was performed using 1 μg RNA and a Superscript III one-step RT-PCR system (Invitrogen) with forward primer wbiA-Fex and reverse primer wbiA-Rex (Table 2). The conditions used were cDNA synthesis at 45°C for 30 min; initial denaturation at 95°C for 2 min; 35 cycles of denaturation at 94°C for 15 s, annealing at 58°C for 15 s, and extension at 72°C for 15 s; and a final elongation step at 72°C for 5 min. The transcript reference was RT-PCR for 16S rRNA gene amplification using primers Univ_16S_F and Univ_16S_R (Table 2). The negative control was a wbiA reaction without the reverse transcriptase enzyme.
OPS characterization using NMR spectroscopy.
B. pseudomallei 4095a (NM) ΔwcbB (capsular polysaccharide [CPS]-deficient derivative of 4095a) and 4095c (M) ΔwcbB (CPS-deficient derivative of 4095c) (Table 1) were cultured on TSA, from which colonies were scraped from the plate, and purified LPS and OPS were obtained as previously described (13). LPS was extracted using a modified hot aqueous phenol procedure (12). OPS samples were deuterium exchanged by dissolving them in D2O and lyophilizing, and then the samples were dissolved in 0.27 ml D2O containing 1 μl acetone. One-dimensional (1-D) proton and 2-D gradient-enhanced correlation spectroscopy (gCOSY), total COSY (TOCSY), nuclear Overhauser effect spectroscopy (NOESY), gradient heteronuclear single quantum coherence (gHSQC), and gradient heteronuclear multiple-bond coherence (gHMBC) spectra were obtained on a Varian Inova 600-MHz spectrometer at 50°C using standard Varian pulse sequences. The spectral width was 3.17 kHz in the 1H dimension and 18.1 kHz in the 13C dimension. The numbers of scans and increments were, respectively, 4 and 400 for gCOSY, 8 and 200 for TOCSY and NOESY, 64 and 128 for gHSQC, and 128 and 200 for gHMBC. Acquisition times were 2 s for 1-D 1H, 137 ms for gCOSY, TOCSY, and NOESY, 150 ms for gHSQC, and 128 ms for gHMBC. Mixing times for TOCSY and NOESY experiments were 120 and 300 ms, respectively. Proton chemical shifts were measured relative to internal acetone (δH = 2.218 ppm, δC = 33.0 ppm) (24). Monosaccharide residues in OPS samples were identified using the 2-D HSQC spectra by comparison with the assigned spectra from B. pseudomallei 1026b OPS (14). Integration of the spectra was performed using the Mnova software (Mestrelab Research) after baseplane correction with a third-order Bernstein polynomial.
Susceptibility of mucoid and nonmucoid B. pseudomallei colonies to serum killing.
The susceptibility of B. pseudomallei to killing by normal human serum (NHS) was determined as described previously (7). In brief, B. pseudomallei was harvested from TSA, adjusted to 1 × 106 CFU/ml, and then incubated in 30% NHS for 2 h, after which a viable count (CFU/ml) was determined by plating cells on TSA. E. coli HB101 was used as a susceptible control. Three independent experiments were performed.
Statistical analysis.
Statistical analysis was performed using Stata, version 12 (StataCorp LP, College Station, TX, USA). Fisher's exact test was used to test proportions. Paired t tests were used to assess differences between the isogenic mucoid and nonmucoid strains in the monocyte internalization assays. Quantitative data are presented as means ± standard deviations. Differences were considered statistically significant at a P value of <0.05.
RESULTS
Frequency of mucoid B. pseudomallei colonies.
We previously described 7 different colony types for B. pseudomallei grown on Ashdown agar (19). Using TSA and blood agar in this study, we observed only two colony types, which we describe as mucoid and nonmucoid (Fig. 1). The frequency of these two colony types was determined in two retrospective Thai collections representing isolates from 200 cases of primary infection that was not complicated by relapse and 166 isolates from 78 patients who had at least one episode of relapse. The majority of cultures contained nonmucoid colonies alone, with ≤10% of cultures containing mucoid colonies either alone or mixed with nonmucoid colonies (Table 3). The frequencies of mucoid colonies were not significantly different in the two collections (P = 0.15), and mucoid colonies were observed in both primary (n = 9) and relapse (n = 9) cultures. This indicates a lack of association with clinical relapse. A third Australian collection (n = 52) was also examined, since there is a marked phylogenetic distinction between isolates from Thailand and Australia. As before, the majority of cultures (n = 40, 77%) contained nonmucoid colonies alone, while the remaining cultures originating from humans (n = 10), animals (n = 1), and soil (n = 1) contained a mixture of nonmucoid and mucoid colonies.
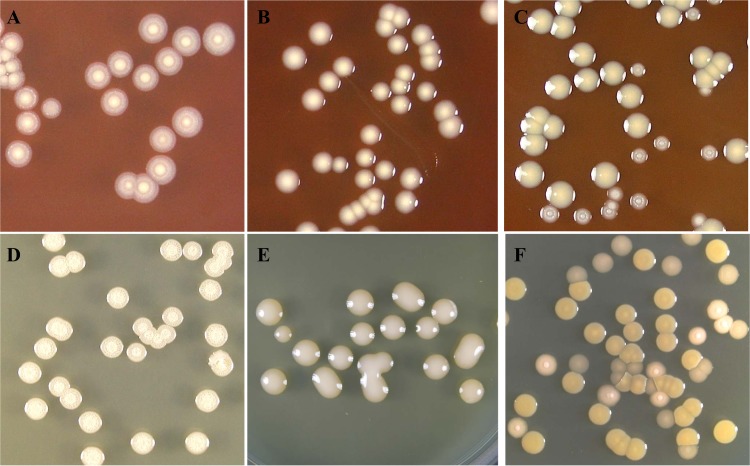
FIG 1.
Colony appearance of B. pseudomallei. Colony morphology was observed after spread plating cells on blood agar and incubation for 4 days at 37°C in air. (A, B, and C, top row) Colonies on blood agar; (D, E, and F, bottom row) colonies on TSA. Nonmucoid colonies (A and D), mucoid colonies (B and E), and mixed colonies (C and F) are shown.
TABLE 3.
Isolate collections used in this study and their colony morphologies, LPS types, and MAb reactivities
| B. pseudomallei isolate source | Total no. of isolates | Colony form observed on solid agar (no. [%] of isolates) | Colony type (no. of isolates) | No. of isolates with indicated LPS type and reaction with MAb 9D5 |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Type A LPS |
Type B LPS |
Rough LPS |
|||||||
| Positive | Negative | Positive | Negative | Positive | Negative | ||||
| Clinical isolates from Thai patients with no relapse (200 patients) | 200 | Single type (191 [95.5]) | M (5) | 0 | 5 | 0 | 0 | 0 | 0 |
| NM (186) | 183 | 0 | 0 | 2 | 0 | 1 | |||
| Mixed types (9 [4.5]) | M (9) | 0 | 9 | 0 | 0 | 0 | 0 | ||
| NM (9) | 9 | 0 | 0 | 0 | 0 | 0 | |||
| Clinical isolates from Thai patients with primary infection followed by at least one episode of relapse (78 patients) | 166 | Single type (153 [92.2]) | M (5) | 0 | 5 | 0 | 0 | 0 | 0 |
| NM (148) | 142 | 0 | 0 | 4 | 0 | 2 | |||
| Mixed types (13 [7.8]) | M (13) | 0 | 13 | 0 | 0 | 0 | 0 | ||
| NM (13) | 13 | 0 | 0 | 0 | 0 | 0 | |||
| Clinical and environmental strains from Australia (38 patients, 10 animals, 4 environmental) | 52 | Single type (40 [76.9]) | M (0) | 0 | 0 | 0 | 0 | 0 | 0 |
| NM (40) | 33 | 0 | 0 | 4 | 0 | 3 | |||
| Mixed types (12 [23.1]) | M (12) | 0 | 9 | 0 | 3 | 0 | 0 | ||
| NM (12) | 9 | 0 | 0 | 3 | 0 | 0 | |||
| Primary culture plate isolates from Thai patients (40 patients) | 40 | Single type (32 [80.0]) | M (0) | 0 | 0 | 0 | 0 | 0 | 0 |
| NM (32) | 32 | 0 | 0 | 0 | 0 | 0 | |||
| Mixed types (8 [20.0]) | M (8) | 0 | 8 | 0 | 0 | 0 | 0 | ||
| NM (8) | 8 | 0 | 0 | 0 | 0 | 0 | |||
Defining the presence and frequency of the two colony types from frozen stocks is potentially flawed in that the number of colonies picked for storage at the time of isolation are usually limited and subject to selection bias. To address this, we undertook a prospective evaluation and determined the frequency of nonmucoid and mucoid colony types in fresh, primary cultures of clinical specimens from 40 patients with melioidosis. The clinical specimens were sputa (n = 13), tracheal secretions (n = 6), pus (n = 11), wound swabs (n = 3), synovial fluid (n = 1), and blood cultures (n = 6). Of these, 32 samples grew only nonmucoid colonies, and the remaining 8 samples contained a mixture of nonmucoid and mucoid colonies, the sample types for which were all sputum (n = 6) and tracheal secretions (n = 2). The mucoid type represented 5% of total B. pseudomallei colonies on the mixed-colony plates.
Taken together, these results indicate that mucoid colonies are in the minority but appear to be widely distributed in the population and are represented in clinical samples. We then considered whether the mixed colony types were isogenic and belonged to the same clone or whether different colony morphologies represented the presence of more than one B. pseudomallei strain. To address this, we selected the 8 prospectively collected clinical samples and genotyped 5 nonmucoid and 5 mucoid colonies from each sample using pulsed-field gel electrophoresis (PFGE). The banding patterns were identical for all of the colonies within a given sample, indicating that the different colony appearances occurred in the same genetic background and could not be explained by mixed infection with two different strains.
Five pairs of mucoid and nonmucoid B. pseudomallei colonies were selected for further detailed phenotypic and genetic analysis in the remainder of the study. These pairs were as follows: 10457A (NM) and 10457A (M), 10971B (NM) and 10971B (M), and 11017A (NM) and 11017A (M), picked from primary clinical cultures containing mixed colony types; MSHR295 (NM) and MSHR295 (M), picked from a culture of B. pseudomallei containing mixed colony types and originating from soil in Australia; and B. pseudomallei 4095a (NM) and 4095c (M), isolated from different samples taken 22 days apart from a Thai patient. PFGE demonstrated that mucoid and nonmucoid colonies from each sample had identical PFGE banding patterns and were considered to be isogenic. Full details of these isolates are shown in Table 1.
Effect of laboratory culture conditions on colony morphology.
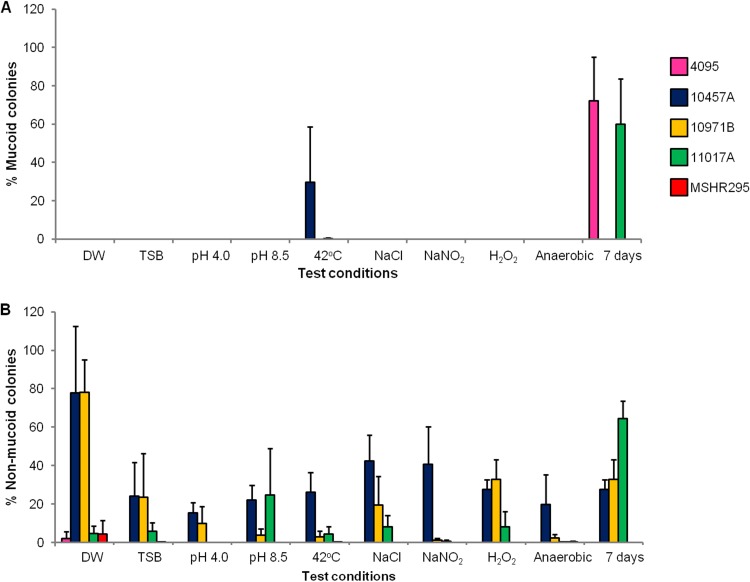
The five nonmucoid/mucoid isolate pairs were used to observe the effect on colony type of growth under 10 different laboratory conditions, as described in Materials and Methods. Starting with nonmucoid isolates, only three isolates (4095a, 10457A, and 11017A) switched to the mucoid type under specific laboratory conditions (Fig. 2A). After the 7-day incubation in TSB, 72% and 60% of the total colony counts for 4095a and 11017A were emergent mucoid colonies, respectively, and after incubation at 42°C, 30% of the colonies of 10457A were emergent mucoid colonies. We did not observe switching to mucoid colonies on the plates for strains 10971B and MSHR295 under any conditions tested.
FIG 2.
Effect of 10 laboratory conditions on colony morphotype switching between nonmucoid and mucoid colonies for 5 paired B. pseudomallei isolates. (A) Colony switching from starting nonmucoid type to mucoid type. (B) Colony switching from starting mucoid type to nonmucoid type. Two hundred microliters of 1 × 108 CFU/ml suspension was inoculated into 2 ml DW or TSB and incubated under one of these following conditions: (i) DW at 37°C for 24 h, (ii) TSB (pH 7.4) at 37°C for 24 h, (iii) TSB (pH 4.0) at 37°C for 24 h, (iv) TSB (pH 8.5) at 37°C for 24 h, (v) TSB (pH 7.4) at 42°C for 24 h, (vi) TSB (pH 7.4) plus 350 mM NaCl at 37°C for 24 h, (vii) TSB (pH 7.4) plus 50 mM NaNO2 at 37°C for 24 h, (viii) TSB (pH 7.4) plus 2 mM H2O2 at 37°C for 24 h, (ix) TSB (pH 7.4) at 37°C for 24 h in an anaerobic jar (Oxoid, United Kingdom), and (x) TSB (pH 7.4) at 37°C for 7 days. Morphotype switching is presented as the proportion (percentage) of the alternate type in relation to the total number of colonies present. Error bars are standard deviations.
Although a variety of conditions were found to influence switching between nonmucoid and mucoid phenotypes, overall patterns of switching varied greatly between the isolates (Fig. 2B). Nonmucoid colonies emerged from parental mucoid colonies under all conditions for strain 10457A and under most conditions for strains 10971B and 11017A. The percentage of nonmucoid colonies per total colony count was highest in DW (78% for both 10457A and 10971B) and after 7 days in TSB (65% for 11017A). Switching from mucoid colonies to nonmucoid colonies was observed for 4095c and MSHR295 in DW only.
Internalization of mucoid and nonmucoid colonies by human monocytes.
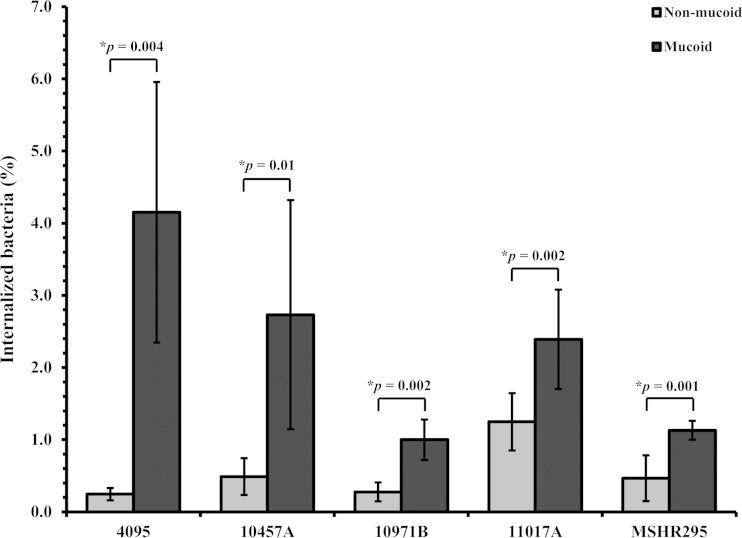
We hypothesized that mucoid and nonmucoid colony types may interact differently with host cells. We evaluated this by comparing rates of internalization of five isogenic mucoid and nonmucoid B. pseudomallei pairs (4095, 10457A, 10971B, 11017A, and MSHR295). Rates of bacterial internalization by human monocyte THP1 cells following 4 h of initial interaction with live bacteria at an MOI of 1:1 varied markedly between bacterial strains (Fig. 3). A consistent finding, however, was a significantly higher number of internalized bacteria for mucoid than for nonmucoid colony types (P < 0.05 for all pairs).
FIG 3.
Internalization of nonmucoid and mucoid colonies by human monocytes. THP1 cells at 2 × 105 cells were incubated for 2 h with B. pseudomallei at an MOI of 1:1, after which nonadherent bacteria were killed by incubating them for 2 h with kanamycin. The bacterial count was enumerated by cell lysis and plating onto TSA. The data represent mean numbers of CFU/ml ± standard deviations for each isogenic pair from 5 B. pseudomallei isolates and are expressed as the percentages of cells internalized at 4 h compared with the number of cells in the inoculum.
Susceptibility of mucoid and nonmucoid colonies to serum killing.
The OPS moiety of LPS is responsible for the resistance of B. pseudomallei to serum (7). We evaluated whether the two different colony types were associated with altered serum resistance using five isogenic mucoid and nonmucoid B. pseudomallei pairs (4095, 10457A, 10971B, 11017A, and MSHR295). The K96243 negative control remained viable in 30% NHS (inoculum, 1.10 × 106 CFU/ml; 1.04 × 106 CFU/ml postexperiment), and the E. coli positive control was killed (inoculum, 1.14 × 106 CFU/ml; 0 CFU/ml postexperiment). Comparison of mucoid and nonmucoid colony pairs for strains 4095, 10971B, 11017A, and MSHR295 indicated that the two colony types had comparable levels of resistance to serum between pairs (P > 0.05 for all pairs, t test) and to the level of resistance of the positive control (data not shown). Unexpectedly, both mucoid and nonmucoid colonies of strain 10457A were susceptible to serum killing (mucoid-colony inoculum, 1.52 × 106 CFU/ml; 1.77 × 102 CFU/ml postexperiment; nonmucoid-colony inoculum, 1.03 × 106 CFU/ml; 4.55 × 102 CFU/ml) postexperiment.
Association between mucoid colonies and LPS antigenic variation.
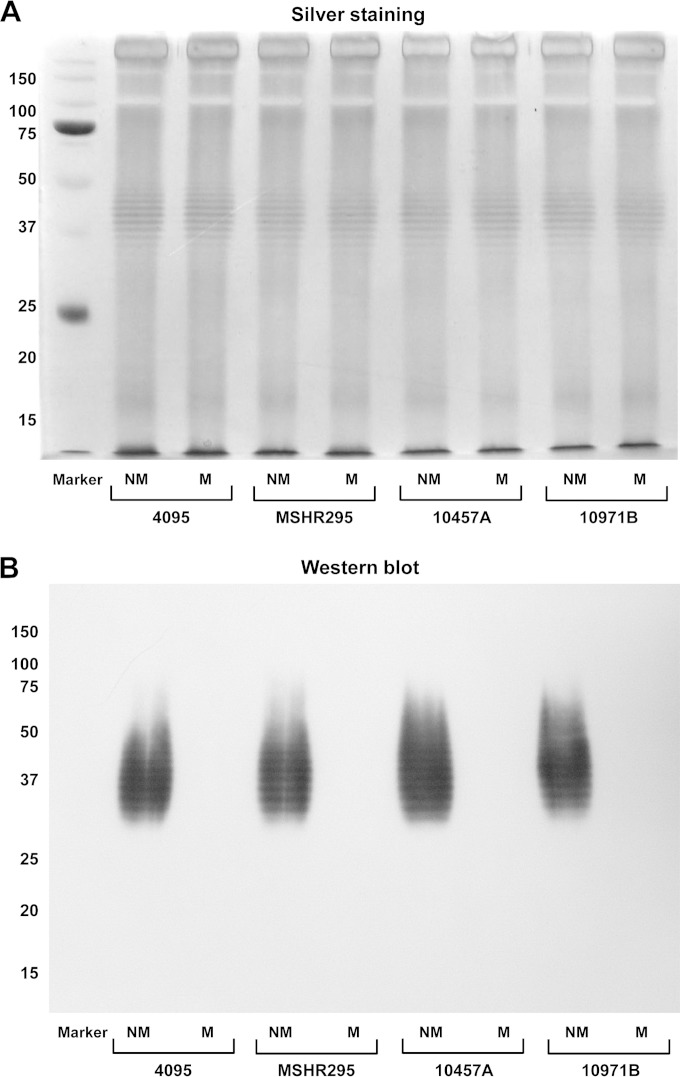
We next considered the phenotypic basis for the difference in colony appearance between nonmucoid and mucoid colonies. LPS is a major surface-expressed determinant of B. pseudomallei, and we hypothesized that variation in LPS structure was responsible for the variable colony appearance. We tested LPS from all isolates from two retrospective Thai collections (200 isolates from cases with no relapse and 166 isolates from patients who relapsed), Australian collections (n = 52), and prospective isolates (n = 40) (Table 3). SDS-PAGE and silver staining of LPS extracts of one nonmucoid and one mucoid colony (when available) for each isolate demonstrated the three LPS types described previously: ladder A, ladder B, and rough LPS (10) (Table 3). Type A and B ladder patterns, which reflect the complete structure of LPS consisting of OPS, core-oligosaccharide, and lipid A, were found in both mucoid and nonmucoid colonies (Fig. 4A). Rough LPS was found in nonmucoid colonies (Table 3). Ten colonies per isolate of all isolates (5 mucoid and 5 nonmucoid) were screened using the MAb-latex agglutination test. This MAb is known to recognize B. pseudomallei OPS type A but not type B or rough LPS by Western blotting (21). The latex agglutination result demonstrated that only nonmucoid colonies of all isolates with LPS type A were positive and that the remainder were negative. The negative result for all mucoid colonies with LPS type A suggests the presence of a modification of OPS which can be distinguished by the MAb. This result was verified using Western blotting, in which the MAb was observed to react with LPS extracted from nonmucoid colonies showing reactions on the type A ladder components but not to react with LPS from mucoid colonies (Fig. 4B). We proposed that the difference in colony morphology variation was due to the antigenic variation of OPS.
FIG 4.
Mucoid colonies are not recognized by OPS-specific monoclonal antibody. (A) SDS-PAGE and silver staining of paired LPS extracts of two colony types of B. pseudomallei from 4 representative paired isolates (4095, MSHR295, 10457A, and 10971B). (B) Western blot of LPS extracts of the same paired isolates probed with 9D5 MAb. Markers represent standard protein markers; numbers at the left are masses (in kilodaltons).
Molecular identification of an OPS molecule that reacts with the 9D5 MAb.
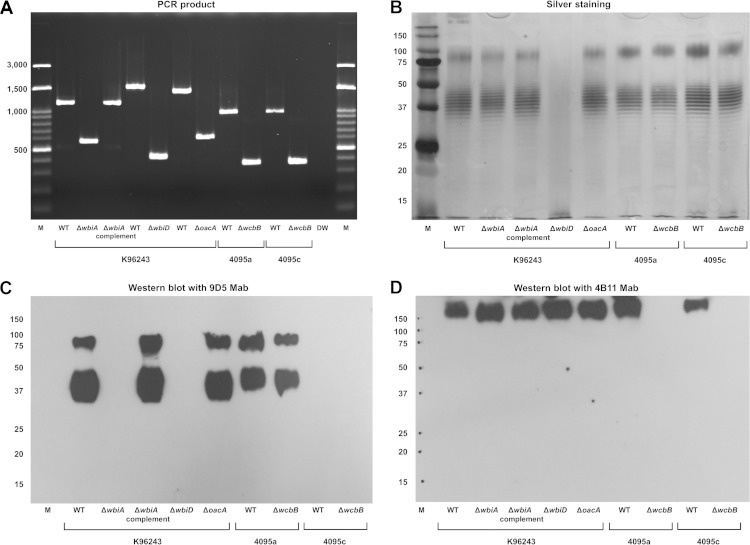
We investigated the nature of the OPS modification(s) associated with varied colony morphologies and latex agglutination reactions. The 6-deoxy-l-talose (6dTal) residues of B. pseudomallei OPS are variably replaced with O-methyl and O-acetyl modifications (11–13). We thus generated a range of allelic-exchange mutants containing unmarked deletions of wbiD, wbiA, and oacA, which are known to be involved in B. pseudomallei LPS synthesis. wbiD is involved in OPS biosynthesis, whereas wbiA and oacA are required for the 2-O- and 4-O-acetylation of OPS, respectively (7, 13). The mutants defective in wbiA (K96243 ΔwbiA), wbiD (K96243 ΔwbiD), and oacA (K96243 ΔoacA) were confirmed to have the desired mutant alleles by PCR (Fig. 5A). SDS-PAGE and silver staining of proteinase K-treated extracts revealed type A ladder patterns for K96243 ΔwbiA and K96243 ΔoacA similar to that of the wild type, but the OPS ladder was absent for K96243 ΔwbiD (Fig. 5B). These findings suggested that while the K96243 ΔwbiA and K96243 ΔoacA mutants expressed OPS moieties, K96243 ΔwbiD did not. Western blot analysis demonstrated that LPS of K96243 ΔoacA and the wild type reacted with the OPS-specific MAb 9D5, but no reaction was observed for the K96243 ΔwbiA and K96243 ΔwbiD mutants (Fig. 5C). These mutants did not change the colony from the nonmucoid to the mucoid type.
FIG 5.
(A) Mutants of the wbiA, wbiD, oacA, and wcbB genes exhibited PCR products of corresponding deletion alleles. Lane M represents a 100-bp ladder marker. Three LPS mutants were constructed with the K96243 wild type (lane WT), the mutant defective in wbiA (BPSL2680) (lane ΔwbiA), the mutant defective in wbiD (BPSL2677) (lane ΔwbiD), and the mutant defective in (BPSL1936) (lane ΔoacA). Lane ΔwbiA complement contains a complemented strain of the wbiA mutant. Two capsule mutants were constructed with the 4095a (NM) wild type and the 4095c (M) wild type. Lanes ΔwcbB contain a mutant defective in ΔwcbB (BPSL2808). (B) SDS-PAGE and a silver stain of LPS extracts of the wbiA, oacA, and wcbB mutants but not the wbiD mutant exhibited an LPS ladder pattern identical to that of the wild types. Lane M contains standard protein markers. Numbers at the left are molecular sizes (in base pairs). (C) Western blotting of LPS extracts of the wbiA, wbiD, oacA, and wcbB mutants probed with OPS-specific MAb 9D5 demonstrated a B. pseudomallei mutant without OPS (ΔwbiD) and the 2-O-acetylation moiety (ΔwbiA) and isolates of 4095c that did not react with MAb. Numbers at the left are molecular sizes (in base pairs). (D) Western blotting of LPS extracts of wbiA, wbiD, oacA, and wcbB mutants probed with capsule-specific MAb 4B11 demonstrated that only wcbB capsule mutants (ΔwcbB) did not react with capsule-specific MAb 4B11. Numbers at the left are molecular sizes (in base pairs).
A complemented strain for the K96243ΔwbiA mutant was constructed and confirmed by PCR (Fig. 5A). Western blot analysis demonstrated that expression of a wild-type wbiA allele in this strain restored the expression of native antigen, as demonstrated by the ladder reaction with MAb 9D5 (Fig. 5C). Taken together, these results suggested that (i) LPS-specific MAb 9D5 reacted with the OPS moiety of LPS and that (ii) 2-O-acetyl modifications (due to wbiA expression) of B. pseudomallei OPS influence the reaction with the MAb.
Analysis of wbiA sequences between mucoid and nonmucoid colonies.
Thirty-four B. pseudomallei cultures containing mucoid and nonmucoid colonies were randomly chosen from the Thai collections, and a colony of each type was tested by PCR for the presence of wbiA. All colonies were positive, and full-length sequencing revealed that all mucoid and nonmucoid pair isolates had identical sequences for the wbiA gene. Using RT-PCR analysis to examine two B. pseudomallei isogenic pair isolates (4095a [NM], 4095c [M], MSHR295 [NM], and MSHR295 [M]), we observed the same levels of wbiA expression in mucoid and nonmucoid isolates. wbiA expression was abolished in the K96243 ΔwbiA mutant negative control. The 16S rRNA positive-control samples all yielded a 336-bp product, and no amplification was detected in the no-RT negative controls (data not shown). These results suggested that wbiA was expressed in mucoid isolates.
Purification of LPS from capsule-deficient strains.
We next investigated the structural differences in OPS between mucoid (4095c) and nonmucoid (4095a) isogenic isolates. To eliminate capsular polysaccharide contamination during LPS purification, B. pseudomallei mutants defective in wcbB were constructed with strains 4095a and 4095c. wcbB encodes a polysaccharide glycosyltransferase biosynthesis protein that is required for capsule production (25). Two mutants, 4095a ΔwcbB and 4095c ΔwcbB, were constructed using a pEXKm5-based allele replacement strategy and then verified by PCR (Fig. 5A). SDS-PAGE and silver staining of proteinase K-treated extracts revealed ladder patterns for 4095a ΔwcbB and 4095c ΔwcbB that were similar to those of the parental strains, suggesting that the mutagenesis of capsule did not alter the synthesis of OPS (Fig. 5B). Western blot analysis was used to examine the reactivity with the LPS-specific MAb 9D5 and the capsule-specific MAb 4B11. As shown in Fig. 5C, LPS MAb 9D5 showed reactivity with 4095a and 4095a ΔwcbB mutant extracts but not with 4095c or 4095c ΔwcbB mutant extracts (Fig. 5C). Capsule was absent in both 4095a ΔwcbB and 4095c ΔwcbB mutants (Fig. 5D). Since the deletion of wcbB did not affect LPS's expression, we used the 4095a ΔwcbB and 4095c ΔwcbB strains to prepare purified LPS for NMR analysis.
NMR analysis of OPS structures.
To investigate the structural characteristics of the OPS antigen expressed by B. pseudomallei 4095a ΔwcbB and 4095c ΔwcbB, OPS was isolated from LPS by mild acid hydrolysis and gel permeation chromatography. A combination of 1-D and 2-D 1H and 13C NMR spectroscopy indicated that 4095a (nonmucoid) and 4095c (mucoid) OPS antigens exhibited 2-O-methyl and 4-O-acetyl substitution patterns that differed from one other and from the predominant OPS serotype expressed by B. pseudomallei 1026b (Fig. 6 and Table 4). Specifically, the 6dTal residues of B. pseudomallei 4095a lacked 4-O-acetyl modifications (except at the nonreducing end residue), while the 6dTal residues of B. pseudomallei 4095c lacked both 4-O-acetyl (except at the nonreducing end residue) and 2-O-methyl modifications. These data indicate that the 9D5 MAb recognizes an epitope that is influenced by the presence/absence of O-acetyl and/or O-methyl substitutions on the 6dTal residues.
FIG 6.
Chemical structures of the O-polysaccharides expressed by B. pseudomallei (Bp) 1026b (prototype strain), 4095a (nonmucoid strain), and 4095c (mucoid strain).
TABLE 4.
Intensity comparisons of the 6dTal anomeric signals in the HSQC spectra of B. pseudomallei OPS as a percentage of the sum of all 6dTal anomericsa
| Residue | % of all 6dTal signals in: |
||
|---|---|---|---|
| 4095a OPS | 4095c OPS | 1026b OPS | |
| →3)-2-O-Me-4-O-Ac-α-l-6dTalp-(1→ | ND | ND | 14.7 |
| →3)-α-l-6dTalp-(1→ | 13.1 | 18.5 | 8.8 |
| →3)-2-O-Ac-α-l-6dTalp-(1→ | 53.4 | 70.4 | 43.0 |
| →3)-2,4-di-O-Ac-α-l-6dTalp-(1→ | ND | ND | 5.4 |
| 3-O-Me-2,4-di-O-Ac-α-l-6dTalp-(1→ | 8.2 | 8.7 | 11.3 |
| →3)-2-O-Me-α-l-6dTalp-(1→ | 23.6b | ND | 14.6b |
| →3)-2-O-Ac-α-l-6dTalp-(1→ | 1.7c | 2.4c | 2.2c |
| Total | 100 | 100 | 100 |
ND, not detected; Me, methyl; Ac, acetyl.
Residues are 1,3-linked to Glc.
Residues are 1,3-linked to GlcNAc.
DISCUSSION
Colony morphology variation in B. pseudomallei occurs during human infections and under challenging environmental conditions (19). We previously defined 7 colony types on Ashdown agar and showed their association with different levels of expression of a range of putative virulence proteins, as well as survival advantages in vivo and under different laboratory conditions (6, 19). Here, we expand the examination of colony variation in nonselective medium and identify mucoid and nonmucoid colony types. On TSA and blood agar, both of these colony types were observed within single samples positive for a pure growth of B. pseudomallei. The colony type can also change between mucoid and nonmucoid colonies under different laboratory conditions. We showed that the mucoid type did not react with a MAb to OPS but had higher internalization by macrophages. The NMR structure analysis of a mucoid variant compared to the nonmucoid type of strain 4095a demonstrated that the variant lacked a 2-O-methyl modification.
B. pseudomallei often persists in the lung as acute or chronic pneumonia, resulting in tissue damage and inflammation (3). In primary cultures of clinical specimens, mixed colony types were observed only in samples collected from respiratory secretions, e.g., sputum and tracheal suction specimens. This finding suggests that some host factor(s), for example, pulmonary deterioration, antimicrobial peptides, or antibodies in respiratory secretions, may enhance the presence of mucoid B. pseudomallei. Indeed, we demonstrated that temperature at 42°C or prolonged incubation (37°C for 7 days) may drive colony alteration of B. pseudomallei toward the mucoid type. It is unclear which factor(s) may trigger these changes in vivo, but the fact that mucoid and nonmucoid colonies were observed in the same specimen suggests that a subpopulation may have a fitness advantage in a particular niche. Mucoid colonies had higher internalization into macrophages than nonmucoid colonies, which could be associated with enhanced survival in the host, since B. pseudomallei is capable of survival and replication in macrophages (4). Once the bacteria were taken up, both mucoid and nonmucoid colonies were able to replicate in THP1 monocytic cell lines (data not shown). The occurrence of mucoid colonies in respiratory secretions has been observed in other bacteria, and it has been associated with persistence. For instance, mixed mucoid and matte colonies were observed in B. cenocepacia after pulmonary challenge in a mouse model with enhanced persistence of mucoid colonies in the lung (26). A large degree of variation in phenotypes of Pseudomonas aeruginosa isolates from individual sputum samples of a patient has been reported (27), and mucoid colonies of P. aeruginosa have been isolated from up to 90% of cystic fibrosis patients, although they are rarely observed from other patient groups (28). This evidence suggests that the lung environment may be a driver for bacterial diversification.
Our results showed that mucoid colonies can switch to nonmucoid colonies after exposure to specific laboratory conditions. The nonmucoid colony was observed as the major type in clinical and environmental B. pseudomallei isolates. Our previous study showed that nonmucoid types of B. pseudomallei, e.g., type I and II on Ashdown agar, produced more biofilm and extracellular enzymes than mucoid types and were more resistant to hydrogen peroxide killing (6, 19), while the mucoid type was associated with flagellum production and motility (19, 23). It is possible that alteration of LPS can occur along with increased expression of other putative virulence factors and may enhance the virulence and persistence of B. pseudomallei in a host. These data support the results of our previous study of a mouse experimental model, in which only nonmucoid colonies were found from cultures of various organs from mice infected intraperitoneally with the mucoid type (19).
DeShazer et al. reported that the OPS moiety of B. pseudomallei LPS is required for serum resistance and virulence in vivo. A B. pseudomallei mutant defective in OPS production was susceptible to killing by 30% NHS (7). In this study, we found that our wbiA mutant defective by lacking a 2-O-acetylation substitution in 6dTal residues of OPS moieties did not change the colony from a nonmucoid to a mucoid type. The mucoid variants did not show greater susceptibility to serum killing. It is possible that the substitutions on OPS moieties did not affect the length of OPS repeating subunits and may still prevent integration of the C5b-C9 complement membrane attack complex (MAC) into the bacterial outer membrane. It is surprising to observe an unusual isolate of B. pseudomallei (10457A) having a full LPS ladder type A but showing susceptibility to serum killing. 10457A also produced capsule, as it demonstrated a reaction with capsule-specific MAb 4B11 (data not shown). This suggests that another unidentified component may be involved in the resistance to serum killing.
We considered that the mechanism of OPS modification was mediated by wbiA expression or gene mutation. However, RT-PCR analysis of mucoid colonies showed that wbiA was still expressed at the same level as in nonmucoid colonies. Moreover, DNA sequencing analysis of wbiA demonstrated no mutations in the mucoid type. It is possible that regulation of colony or OPS variation of B. pseudomallei involves other genes. Several studies have indicated that regulatory genes are essential for colony variation in other bacteria. For instance, a putative transcriptional regulator belonging to the LysR family controls the conversion from rough to shiny colonies of B. cenocepacia (29). Although there might be genetic differences between isolates, PFGE was not able to distinguish all isolate pairs due to lack of resolution. Whole-genome sequencing (WGS) was used to define the difference in 3 pairs of mucoid and nonmucoid colonies in this study, but we did not find consistent results that are relevant to the OPS synthesis genes (data not shown).
We demonstrated that while the 9D5 MAb reacted with the OPS expressed by an oacA (4-O-acetyltransferase) mutant, it was unable to react with the OPS expressed by a wbiA (2-O-acetyltransferase) mutant. NMR analysis of the OPS antigens purified from capsule mutants also revealed that 4095a (a nonmucoid strain) OPS was devoid of internal 4-O-acetyl modifications and that 4095c (a mucoid strain) OPS was devoid of both internal 2-O-methyl and 4-O-acetyl modifications on 6dTal residues. Collectively, these results suggest that the 9D5 MAb recognizes a conformational epitope that is influenced by both O-acetyl and O-methyl substitution patterns. Since B. pseudomallei LPS is a potential vaccine candidate (9), our finding that a mucoid phenotype is associated with antigenic variation of LPS has important implications for vaccine development. In conclusion, we have demonstrated that novel modifications associated with B. pseudomallei OPS antigens can arise during human infection. Such antigenic variation might be a mechanism used by this organism to adapt to changing environments and evade host immune responses.
ACKNOWLEDGMENTS
We are grateful for the support of the staff at Sappasithiprasong Hospital, Ubon Ratchathani, Thailand. We thank the staff of the Mahidol-Oxford Tropical Medicine Research Unit.
This work was supported by the Thailand Research Fund (grant TRG5580004), the Wellcome Trust, United Kingdom, and the Department of Energy-funded Center for Plant and Microbial Complex Carbohydrates (grant DE-FG09-93ER-20097). N.C. was supported by the Wellcome Trust (grant 087769/Z/08/Z).
REFERENCES
- 1.Butler D. 2012. Viral research faces clampdown. Nature 490:456. doi: 10.1038/490456a. [DOI] [PubMed] [Google Scholar]
- 2.Limmathurotsakul D, Wongratanacheewin S, Teerawattanasook N, Wongsuvan G, Chaisuksant S, Chetchotisakd P, Chaowagul W, Day NP, Peacock SJ. 2010. Increasing incidence of human melioidosis in Northeast Thailand. Am J Trop Med Hyg 82:1113–1117. doi: 10.4269/ajtmh.2010.10-0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wiersinga WJ, Currie BJ, Peacock SJ. 2012. Melioidosis. N Engl J Med 367:1035–1044. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 4.Charoensap J, Utaisincharoen P, Engering A, Sirisinha S. 2009. Differential intracellular fate of Burkholderia pseudomallei 844 and Burkholderia thailandensis UE5 in human monocyte-derived dendritic cells and macrophages. BMC Immunol 10:20. doi: 10.1186/1471-2172-10-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pumpuang A, Chantratita N, Wikraiphat C, Saiprom N, Day NP, Peacock SJ, Wuthiekanun V. 2011. Survival of Burkholderia pseudomallei in distilled water for 16 years. Trans R Soc Trop Med Hyg 105:598–600. doi: 10.1016/j.trstmh.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tandhavanant S, Thanwisai A, Limmathurotsakul D, Korbsrisate S, Day NP, Peacock SJ, Chantratita N. 2010. Effect of colony morphology variation of Burkholderia pseudomallei on intracellular survival and resistance to antimicrobial environments in human macrophages in vitro. BMC Microbiol 10:303. doi: 10.1186/1471-2180-10-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.DeShazer D, Brett PJ, Woods DE. 1998. The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol Microbiol 30:1081–1100. doi: 10.1046/j.1365-2958.1998.01139.x. [DOI] [PubMed] [Google Scholar]
- 8.Chantratita N, Tandhavanant S, Myers ND, Seal S, Arayawichanont A, Kliangsa-Ad A, Hittle LE, Ernst RK, Emond MJ, Wurfel MM, Day NP, Peacock SJ, West TE. 2013. Survey of innate immune responses to Burkholderia pseudomallei in human blood identifies a central role for lipopolysaccharide. PLoS One 8:e81617. doi: 10.1371/journal.pone.0081617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burtnick MN, Heiss C, Schuler AM, Azadi P, Brett PJ. 2012. Development of novel O-polysaccharide based glycoconjugates for immunization against glanders. Front Cell Infect Microbiol 2:148. doi: 10.3389/fcimb.2012.00148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Anuntagool N, Wuthiekanun V, White NJ, Currie BJ, Sermswan RW, Wongratanacheewin S, Taweechaisupapong S, Chaiyaroj SC, Sirisinha S. 2006. Lipopolysaccharide heterogeneity among Burkholderia pseudomallei from different geographic and clinical origins. Am J Trop Med Hyg 74:348–352. [PubMed] [Google Scholar]
- 11.Knirel YA, Paramonov NA, Shashkov AS, Kochetkov NK, Yarullin RG, Farber SM, Efremenko VI. 1992. Structure of the polysaccharide chains of Pseudomonas pseudomallei lipopolysaccharides. Carbohydr Res 233:185–193. doi: 10.1016/S0008-6215(00)90930-3. [DOI] [PubMed] [Google Scholar]
- 12.Perry MB, MacLean LL, Schollaardt T, Bryan LE, Ho M. 1995. Structural characterization of the lipopolysaccharide O antigens of Burkholderia pseudomallei. Infect Immun 63:3348–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brett PJ, Burtnick MN, Heiss C, Azadi P, DeShazer D, Woods DE, Gherardini FC. 2011. Burkholderia thailandensis oacA mutants facilitate the expression of Burkholderia mallei-like O polysaccharides. Infect Immun 79:961–969. doi: 10.1128/IAI.01023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heiss C, Burtnick MN, Roberts RA, Black I, Azadi P, Brett PJ. 2013. Revised structures for the predominant O-polysaccharides expressed by Burkholderia pseudomallei and Burkholderia mallei. Carbohydr Res 381:6–11. doi: 10.1016/j.carres.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thepthai C, Smithtikarn S, Suksuwan M, Songsivilai S, Dharakul T. 2005. Serodiagnosis of melioidosis by a competitive enzyme-linked immunosorbent assay using a lipopolysaccharide-specific monoclonal antibody. Asian Pac J Allergy Immunol 23:127–132. [PubMed] [Google Scholar]
- 16.Appelmelk BJ, Shiberu B, Trinks C, Tapsi N, Zheng PY, Verboom T, Maaskant J, Hokke CH, Schiphorst WE, Blanchard D, Simoons-Smit IM, van den Eijnden DH, Vandenbroucke-Grauls CM. 1998. Phase variation in Helicobacter pylori lipopolysaccharide. Infect Immun 66:70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guard-Petter J, Keller LH, Rahman MM, Carlson RW, Silvers S. 1996. A novel relationship between O-antigen variation, matrix formation, and invasiveness of Salmonella enteritidis. Epidemiol Infect 117:219–231. doi: 10.1017/S0950268800001394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van der Woude MW, Baumler AJ. 2004. Phase and antigenic variation in bacteria. Clin Microbiol Rev 17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chantratita N, Wuthiekanun V, Boonbumrung K, Tiyawisutsri R, Vesaratchavest M, Limmathurotsakul D, Chierakul W, Wongratanacheewin S, Pukritiyakamee S, White NJ, Day NP, Peacock SJ. 2007. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J Bacteriol 189:807–817. doi: 10.1128/JB.01258-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Limmathurotsakul D, Chaowagul W, Chierakul W, Stepniewska K, Maharjan B, Wuthiekanun V, White NJ, Day NP, Peacock SJ. 2006. Risk factors for recurrent melioidosis in northeast Thailand. Clin Infect Dis 43:979–986. doi: 10.1086/507632. [DOI] [PubMed] [Google Scholar]
- 21.Anuntagool N, Sirisinha S. 2002. Antigenic relatedness between Burkholderia pseudomallei and Burkholderia mallei. Microbiol Immunol 46:143–150. doi: 10.1111/j.1348-0421.2002.tb02679.x. [DOI] [PubMed] [Google Scholar]
- 22.Lopez CM, Rholl DA, Trunck LA, Schweizer HP. 2009. Versatile dual-technology system for markerless allele replacement in Burkholderia pseudomallei. Appl Environ Microbiol 75:6496–6503. doi: 10.1128/AEM.01669-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chantratita N, Tandhavanant S, Wikraiphat C, Trunck LA, Rholl DA, Thanwisai A, Saiprom N, Limmathurotsakul D, Korbsrisate S, Day NP, Schweizer HP, Peacock SJ. 2012. Proteomic analysis of colony morphology variants of Burkholderia pseudomallei defines a role for the arginine deiminase system in bacterial survival. J Proteomics 75:1031–1042. doi: 10.1016/j.jprot.2011.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wishart DS, Bigam CG, Yao J, Abildgaard F, Dyson HJ, Oldfield E, Markley JL, Sykes BD. 1995. 1H, 13C and 15N chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140. [DOI] [PubMed] [Google Scholar]
- 25.Reckseidler-Zenteno SL, DeVinney R, Woods DE. 2005. The capsular polysaccharide of Burkholderia pseudomallei contributes to survival in serum by reducing complement factor C3b deposition. Infect Immun 73:1106–1115. doi: 10.1128/IAI.73.2.1106-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chu KK, Davidson DJ, Halsey TK, Chung JW, Speert DP. 2002. Differential persistence among genomovars of the Burkholderia cepacia complex in a murine model of pulmonary infection. Infect Immun 70:2715–2720. doi: 10.1128/IAI.70.5.2715-2720.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Workentine ML, Sibley CD, Glezerson B, Purighalla S, Norgaard-Gron JC, Parkins MD, Rabin HR, Surette MG. 2013. Phenotypic heterogeneity of Pseudomonas aeruginosa populations in a cystic fibrosis patient. PLoS One 8:e60225. doi: 10.1371/journal.pone.0060225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Doggett RG. 1969. Incidence of mucoid Pseudomonas aeruginosa from clinical sources. Appl Microbiol 18:936–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernier SP, Nguyen DT, Sokol PA. 2008. A LysR-type transcriptional regulator in Burkholderia cenocepacia influences colony morphology and virulence. Infect Immun 76:38–47. doi: 10.1128/IAI.00874-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Holden MT, Titball RW, Peacock SJ, Cerdeno-Tarraga AM, Atkins T, Crossman LC, Pitt T, Churcher C, Mungall K, Bentley SD, Sebaihia M, Thomson NR, Bason N, Beacham IR, Brooks K, Brown KA, Brown NF, Challis GL, Cherevach I, Chillingworth T, Cronin A, Crossett B, Davis P, DeShazer D, Feltwell T, Fraser A, Hance Z, Hauser H, Holroyd S, Jagels K, Keith KE, Maddison M, Moule S, Price C, Quail MA, Rabbinowitsch E, Rutherford K, Sanders M, Simmonds M, Songsivilai S, Stevens K, Tumapa S, Vesaratchavest M, Whitehead S, Yeats C, Barrell BG, Oyston PC, Parkhill J. 2004. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc Natl Acad Sci U S A 101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liss L. 1987. New M13 host: DH5α F′ competent cells. Focus 9:13. [Google Scholar]