Abstract
Enterotoxigenic Escherichia coli (ETEC), a leading cause of acute diarrhea, colonizes the intestine by means of adhesins. However, 15 to 50% of clinical isolates are negative for known adhesins, making it difficult to identify antigens for broad-coverage vaccines. The ETEC strain 1766a, obtained from a child with watery diarrhea in Chile, harbors the colonization factor CS23 but is negative for other known adhesins. One clone, derived from an ETEC 1766a genomic library (clone G10), did not produce CS23 yet was capable of adhering to Caco-2 cells. The goal of this study was to identify the gene responsible for this capacity. Random transposon-based mutagenesis allowed the identification of a 4,110-bp gene that codes for a homologue of the temperature-sensitive hemagglutinin (Tsh) autotransporter described in avian E. coli strains (97% identity, 90% coverage) and that is called TleA (Tsh-like ETEC autotransporter) herein. An isogenic ETEC 1766a strain with a tleA mutation showed an adhesion level similar to that of the wild-type strain, suggesting that the gene does not direct attachment to Caco-2 cells. However, expression of tleA conferred the capacity for adherence to nonadherent E. coli HB101. This effect coincided with the detection of TleA on the surface of nonpermeabilized bacteria, while, conversely, ETEC 1766a seems to secrete most of the produced autotransporter to the medium. On the other hand, TleA was capable of degrading bovine submaxillary mucin and leukocyte surface glycoproteins CD45 and P-selectin glycoprotein ligand 1 (PSGL-1). These results suggest that TleA promotes colonization of the intestinal epithelium and that it may modulate the host immune response.
INTRODUCTION
Yearly, enterotoxigenic Escherichia coli (ETEC) is responsible for 400 million cases of watery diarrhea and the death of 300,000 children under 5 years of age worldwide (1). ETEC is also the most common etiologic agent of travelers' diarrhea (2, 3). ETEC strains cause watery diarrhea by releasing two toxins, one thermolabile (LT) and the other thermostable (ST), and a given ETEC strain may produce one or both toxins simultaneously (4–7). These toxins recognize specific receptors in the enterocyte membrane, triggering an increase in intracellular cyclic AMP (cAMP) and cGMP and activation of protein kinase A (PKA) and protein kinase GII (PKGII), respectively, in the cell cytoplasm. This leads to the opening of ion channels, such as the cystic fibrosis transmembrane-conductance regulator (CFTR), and finally the release of chloride and loss of water through the intestinal lumen (4, 5).
ETEC strains rely on a diverse adhesin repertoire to colonize the intestine, including 22 classic adhesins or colonization factors (CFs) and 3 nonclassic adhesins (8, 9). In the 1990s, CFs were given the nomenclature “coli surface antigen” (CS) followed by a number according to their order of discovery (10). An ETEC strain may harbor one or more genes that encode CFs in various combinations, with CFA/I, CS1, CS2, CS3, CS4, CS5, CS6, CS7, CS14, CS17, and CS21 being the most common variants worldwide (11). However, international studies have shown that 15 to 50% of clinical ETEC isolates are negative for known CFs (11). Recent studies in Chile reported that 16% of ETEC samples obtained from children with watery diarrhea were negative for classic and nonclassic adhesin genes (9). Therefore, it is likely that these pathogenic strains harbor a number of uncharacterized adhesins.
It should also be noted that nonclassic adhesins are excluded from the ETEC colonization factor nomenclature and are typically overlooked in epidemiological studies (9). Nonclassic adhesins include representatives of various autotransporter families, such as TibA, a member of the adhesin involved in diffuse adherence (AIDA-I) family, and EtpA, part of a two-partner secretion system (12, 13). A lack of knowledge regarding the heterogeneity of ETEC strains, which rely on different repertoires of adhesins, O antigens, and toxins, has been a barrier to developing a vaccine to prevent infection (14, 15). This paper describes the newly identified gene, tleA, which codes for a homologue of the autotransporter Tsh, a member of the SPATE (serine protease autotransporter of Enterobacteriaceae) family described previously in avian pathogenic Escherichia coli (APEC) strains. The discovery of this gene arose from a cosmid clone with adherence capacity (clone G10), selected from a library constructed from the genome of the Chilean strain ETEC 1766a (O4 STh+ LT+ CS23+ [where STh is ST human variant]). TleA is a new autotransporter, closely related to Tsh, that promotes adhesion to Caco-2 epithelial cells and has the capacity to degrade mucin and leukocyte surface glycoproteins.
MATERIALS AND METHODS
Bacterial strains and vectors.
The E. coli strains and DNA vectors used in this study are described in Table 1. ETEC 1766a was isolated from a pediatric patient with acute diarrhea in Santiago, Chile (16). The strains were grown in Luria-Bertani (LB) broth or Dulbecco's modified Eagle medium (DMEM) at 37°C. The antibiotic ampicillin (100 μg/μl), kanamycin (50 μg/μl), or chloramphenicol (50 μg/μl) was added as needed.
TABLE 1.
Vectors and strains
| Vector or strain | Description | Reference or source |
|---|---|---|
| Vectors | ||
| pKD3 | Template plasmid for allelic replacement | 20 |
| pKD46 | Plasmid carrying genes encoding lambda red recombinase system | 20 |
| pHC79-G10 | pHC79::tleA, adherence positive | 16 |
| pHC79-Tn2 | tleA::EZ-Tn5<KAN-2>, adherence negative | This work |
| pTrc99A | Expression vector, Ampr Ptrc lacIq ColE1 ori | 46 |
| Strains | ||
| ETEC 1766a | O4 ETEC LT+ STh+ CS23+ | 16 |
| EHEC E045-00 | EHEC O113:H21 Hek+ isolate from a dysentery case | 47 |
| E. coli HB101 | Nonadherent laboratory strain (supE44 hsdS20(rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 leuB6 thi-1) | 48 |
| ETEC 1766a ΔtleA | ETEC 1766a ΔtleA | This work |
| ETEC 1766a ΔaalB | ETEC 1766a CS23 mutant | 16 |
| ETEC 1766a ΔaalB ΔtleA | CS23-tleA double mutant | This work |
| HB101/pTrc99A-tleA | HB101/Ampr Ptrc lacIq ColE1 ori tleA | This work |
Cell cultures and bacterial adhesion assays.
Caco-2 intestinal epithelial cells derived from human colon carcinomas were cultivated in DMEM supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin at 37°C in a 5% CO2 atmosphere. The cells were seeded for propagation in 75-cm2 vessels and grown to semiconfluence, detached with 0.05% trypsin, and divided in a 1:4 ratio.
For the adhesion assays, cells were seeded on 24-well plates and grown to confluence (approximately 3.5 × 105 cells per well). The bacterial strains were grown overnight (ON) in LB broth at 37°C without agitation and diluted 1:100 with DMEM. The confluent Caco-2 cells were infected with 3.5 × 106 CFU per well (approximate multiplicity of infection [MOI] of 10 bacteria per cell) for 30 min at 37°C in a 5% CO2 atmosphere. The nonadherent (planktonic) bacteria were removed by repeated rinsing with phosphate-buffered saline (PBS), and the adherent bacteria were harvested by lysis with 0.1% Triton X-100. After serial dilutions, adherent bacteria were seeded on LB agar plates and counted after 16 h. The final result was expressed as the percentage of bacteria adhered to the cell layer with respect to total bacteria present after infection (planktonic bacteria plus adherent bacteria). To carry out microscopic analyses, Caco-2 cells were grown in glass coverslips, infected, and, instead of being lysed, fixed with 70% methanol for 7 min at room temperature (RT). To visualize bacteria, coverslips were stained with Giemsa solution in a 1:20 dilution for 40 min at RT. MOIs were increased to 100 bacteria per cell, and infections were extended to 3 h in order to better visualize the bacterial adherence pattern.
Cosmid clone.
The G10 clone was obtained as previously described (16). Briefly, partially digested ETEC 1766a genomic DNA, ranging from 20 to 40 kbp, was ligated to a pHC79 cosmid, packaged in lambda phage heads, and transduced into E. coli HB101. Clones carrying recombinant cosmids were pooled in groups of 50 to perform adhesion assays over Caco-2 cells. Colonies obtained from the groups that displayed a higher adhesion level than that of E. coli HB101 carrying the empty pHC79 cosmid were selected for an additional adhesion assay. After this second round, two clones (G10 and G12) of 1,000 included in the selection process displayed a reproducible (n = 3) adhesion capacity significantly higher than that of E. coli HB101 carrying the empty pHC79. Digestion of the recombinant cosmid carried by G10 (pHC79-G10) with different restriction endonucleases suggests that it contains an insert of about 40 kb.
Sequence identification and analysis.
Sequences determining adhesion capacity within the recombinant G10 cosmid (pHC79-G10) were identified by random insertion of the EZ-Tn5<KAN-2> transposon, according to the manufacturer's instructions (Epicentre, Illumina, Inc., Madison, WI, USA). Briefly, the purified cosmid was incubated with the transposon in the presence of 10 U of transposase for 2 h at RT. After inactivation at 70°C, the mixture was used to electroporate E. coli HB101, and kanamycin/ampicillin-resistant clones were selected (the latter with the goal of discarding insertions in the bla gene) to evaluate their adhesion capacity as described above. The insertion point was determined through sequencing with the primers KAN-2 FP-1 and KAN-2 RP-1, which recognized the transposon, and then with primers Tsh0, Tsh1, Tsh2, Tsh3, Tsh4, and Tsh5 (Table 2) to determine the complete sequence. Identification was then performed by comparison with the blastn database (BLAST).
TABLE 2.
Oligonucleotides used in this study
| Primer | Sequence (5′ to 3′)a | Protocol | Source |
|---|---|---|---|
| Tsh0-F | CCAGCGTACCGACAAGTTCT | Sequencing | This work |
| Tsh1-F | ATTGCCGTATCTGAGTTTGC | Sequencing | This work |
| Tsh2-F | TTATGCCTGATGCCATTGAA | Sequencing | This work |
| Tsh3-R | TTCAATGGCATCAGGCATAA | Sequencing | This work |
| Tsh4-R | TACCAAATGCAGAGCGTTCA | Sequencing | This work |
| Tsh5-F | TGTCAATCCCTGAACAACGA | Sequencing | This work |
| TleA-cmF | AGGCGATAATCTGCATAAAATCGGTGAGGGCACACTGATTGTGTAGGCTGGAGCTGCTTC | Allelic replacement | This work |
| TleA-cmR | TACCAAATGCAGAGCGTTCAACTTCCAGCCCCAGACGCGTCATATGAATATCCTCCTTAG | Allelic replacement | This work |
| TleA-F | TCCGTAAACTGGCAGGTTAA | PCR | This work |
| Cm-R | ATGAAAGACGGTGAGCTGGT | PCR | |
| TleA-NcoI | CGCTCCCATGGACAGAATTTATTCTCT | Cloning | This work |
| TleA-BamHI | GCGGGATCCTCAGAATGAATAACGAATAT | Cloning | This work |
| KAN-2 FP-1 | ACCTACAACAAAGCTCTCATCAACC | Sequencing | Epicentre |
| KAN-2 RP-1 | GCAATGTAACATCAGAGATTTTGAG | Sequencing | Epicentre |
Restriction sites are underlined.
The ORF Finder tool was used to identify possible open reading frames (ORFs) (http://www.ncbi.nlm.nih.gov/projects/gorf/), and ClustalW2 was used for alignment of identified sequences and the APEC Tsh (http://www.ebi.ac.uk/Tools/msa/clustalw2/). The Swiss-Model server was used to obtain the structure of TleA by homology, and Jmol was used for three-dimensional (3D) structure visualization.
Construction of a SPATE phylogenetic tree.
The amino acid sequences of the SPATE passenger domains were obtained from GenBank and aligned using ClustalW2. A distance-based phylogenetic tree was constructed with MEGA, version 5.05, software using the neighbor-joining method and bootstrap confidence intervals (1,000 replications).
Protein extraction by heating.
Surface-exposed proteins were obtained according to the protocol described by Del Canto et al. (16), which has been previously used to obtain autotransporter passenger domains (17, 18). Briefly, bacterial suspensions were centrifuged at 3,000 × g for 10 min, and the supernatant was discarded. Then, the sediment was suspended in saline buffer and heated at 60°C for 30 min. The suspension was then centrifuged at 3,000 × g for 10 min, and the resulting supernatant contained the heat-extracted surface proteins. Protein quantification was performed using the Bradford assay method (19). Approximately 4 μg of protein was mixed with Laemmli sample buffer, boiled for 5 min, separated by SDS-PAGE on a 10% acrylamide gel, and stained with Coomassie blue. The protein bands were cut from the gel for identification and sent for matrix-assisted laser desorption ionization–tandem time of flight (MALDI-TOF/TOF) analysis at the Mass Spectrometry Core, University of Texas Medical Branch (Galveston, TX, USA).
Knockout by allelic replacement.
The tleA gene was inactivated in the ETEC 1766a genome by allelic replacement as previously described (20). Briefly, ETEC 1766a was transformed using the vector pKD46, which codes for the phage lambda-derived Red recombination system, and then transformed with a DNA fragment containing the chloramphenicol resistance (Cmr) gene flanked by 40 nucleotides identical to the tleA sequence in the ETEC 1766a genome (Table 2, primers TleA-cmF and TleA-cmR). The chloramphenicol-resistant clones were analyzed by PCR to verify recombination using a primer that recognizes the Cmr gene and another that flanks tleA (TleA-F) (Table 2). In order to obtain a double mutant, this same procedure was performed starting with ETEC 1766a ΔaalB.
tleA expression in the nonadherent E. coli HB101 strain.
tleA was amplified by PCR using the primers TleA-NcoI and TleA-BamHI (Table 2) with a mixture of Taq and Pfu DNA polymerases (Thermo Scientific High Fidelity) for cloning and expression on the vector pTrc99A. The PCR product and vector were digested with the enzymes NcoI and BamHI. The ligation mixture was used to transform E. coli HB101. tleA expression was induced by adding isopropyl β-d-1-thiogalactopyranoside (IPTG) at a final concentration of 1 mM for 3 h at 37°C.
Hemagglutination assay.
Sheep blood was purchased at the Instituto de Salud Pública (Santiago, Chile). Blood cells were washed three times by centrifugation at 500 × g and suspended in PBS. Bacteria were grown in LB broth at three different temperatures, 26°C, 37°C, and 42°C, and adjusted to an optical density at 600 nm (OD600) of 0.7. Expression of tleA in E. coli HB101 was induced by the addition of IPTG approximately 3 h before the OD600 was adjusted. Undiluted or diluted culture aliquots of 100 μl were dispensed into 96-well round-bottom microtiter plates, mixed with 50 μl of blood cells, and incubated for 1 h at 4°C. A positive result was evidenced by the formation of a red film, and a negative result was denoted by a red point at the bottom, indicating sedimentation of blood cells.
Proteolytic activity.
Bacterial strains were cultured in 40 ml of LB broth for 16 h at 37°C. An additional incubation of 3 h in IPTG (1 mM) was carried out on the E. coli HB101/pTrc99A-tleA strain. The cultures were centrifuged at 3,000 × g for 10 min, and the supernatants were passed through a 0.22-μm-pore-size filter for concentration in a centrifugal filter device with 100-kDa pores (Millipore, Bedford, MA). Proteins were quantified using the Bradford assay method.
To determine the mucinase activity, 100 μg of bovine submaxillary mucin (BSM), 2 μg of CD45 (GenBank accession number P08575) (R&D Systems), and 2 μg of P-selectin glycoprotein ligand 1 (PSGL-1; GenBank accession number NP002997) (R&D Systems) were incubated with 1 μg of protein from the concentrated supernatants. The degradation reaction mixtures were boiled with Laemmli sample buffer, separated using a 4 to 20% gradient SDS-PAGE gel, and stained with Coomassie blue.
Western blotting.
Heat-extracted fractions containing surface proteins or concentrated supernatants were subjected to SDS-PAGE and then transferred to nitrocellulose membranes. TleA was detected using a rabbit anti-vacuolating autotransporter toxin (Vat) serum (1:1,000) directed against the passenger domain. This serum was obtained from Lampire Biological Labs (Pipersville, PA, USA) by immunizing rabbits with the purified recombinant Vat protein (Vat-Ex). Anti-rabbit IgG (1:5,000) conjugated to alkaline phosphatase was used as a secondary antibody, and immunoreactivity was revealed using nitroblue tetrazolium (NBT) and 5-bromo-4-chloro-3-indolylphosphate (BCIP) colorimetric substrates.
Immunogold labeling.
Bacteria were grown ON at 37°C over Formvar-coated nickel grids in 20-μl drops inside a humid chamber. This helped to minimize bacterial loss during washing steps. In the case of E. coli HB101 carrying tleA, expression was induced during the last 2 h by the addition of IPTG to the drop (1 mM final concentration). Grids were blocked with 1% BSA containing 0.01 M glycine for 1 h at RT. TleA was detected by incubating bacteria for 1 h at RT using a 1:10 dilution with rabbit anti-Tsh serum, previously absorbed by ETEC 1766a ΔtleA. Next, after three washes with saline buffer, the sample was incubated for 1 h at RT with anti-rabbit IgG conjugated to 10-nm gold particles in a 1:100 dilution. After three final washes, bacteria were directly analyzed at the Laboratory of Electron Microscopy, Facultad de Ciencias Biológicas, Pontificia Universidad Católica de Chile.
PCR for TleA detection in clinical ETEC isolates.
The presence of tleA was detected by PCR in 102 clinical isolates of ETEC obtained from children under 5 years of age in Santiago, Chile. These isolates were characterized in a previous study (9), and sample collection protocol was approved by the review boards of the Faculty of Medicine, Universidad de Chile (Santiago, Chile) and the University of Maryland (Baltimore, MD), and by the Ethics Committee of the Servicio de Salud Metropolitano Norte, Santiago, Chile. The primers Tsh1-F and Tsh3-R were used (Table 2), allowing amplification of an internal region of the tleA sequence.
Statistical analysis.
Three independent adhesion assays were performed, and the data were analyzed using analysis of variance (ANOVA) and Student's t test, with P values of <0.05 considered statistically significant.
Nucleotide sequence accession number.
The tleA sequence was deposited in GenBank under accession number KF494347.
RESULTS
Identification of adhesion-associated genetic determinants.
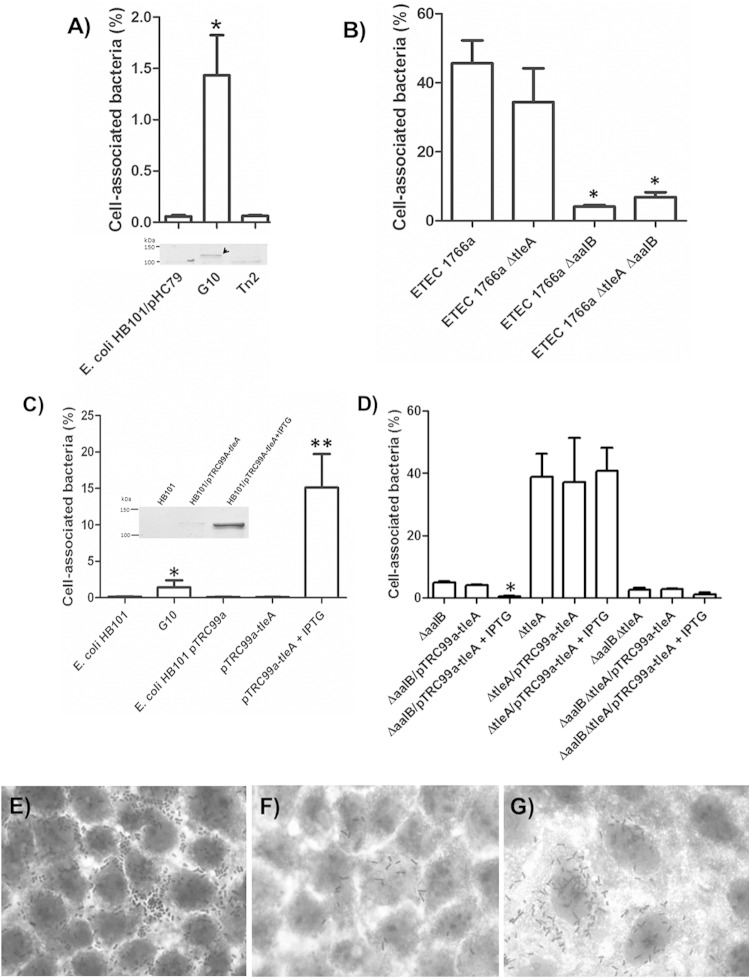
To identify the regions of the G10 clone responsible for conferring Caco-2 cells with adhesion capability, transposon EZ-Tn5<KAN-2> was inserted. From >100 kanamycin-resistant clones obtained, 25 were initially evaluated for their capacity to adhere to Caco-2 cells. Six clones were found to have a significantly lower adhesion capacity than G10 (named Tn2, Tn4, Tn9, Tn10, Tn12, and Tn14) (Fig. 1A).
FIG 1.
Role of TleA in adherence to Caco-2 cells. (A) Adhesion levels of E. coli HB101/pHC79, the cosmid clone carrying the genomic segment containing tleA (G10), and one representative clone which lost the adhesion capacity by insertion of the EZ-Tn5<KAN-2> transposon (Tn2). *, percentage significantly greater than values for E. coli HB101/pHC79 and clones carrying the transposon Ez-Tn5<KAN-2> (n = 3). The lower panel shows extracts enriched with outer membrane proteins (4 μg) obtained from heat preps of E. coli HB101, E. coli HB101/pHC79, G10, and Tn2 strains, separated by SDS-PAGE (10% gel), and stained with Coomassie blue. The arrow indicates the protein identified by mass spectrometry. (B) Adhesion level of mutant ETEC 1766a ΔtleA compared to that of the wild type, the CS23 mutant (ETEC 1766a ΔaalB), and the double mutant (ETEC 1766a ΔaalB ΔtleA) (n = 3). *, percentage significantly lower than values for the wild type and the tleA mutant strain. (C) Adhesion levels of E. coli HB101, G10, E. coli HB101/pTrc99A, and E. coli HB101/pTrc99A-tleA in the presence or absence of 1 mM IPTG. *, percentage significantly greater than that of E. coli HB101; **, percentage significantly greater than values for the remaining four strains (n = 3). The inset shows protein extracts (4 μg,) obtained by heating, from E. coli HB101, E. coli HB101/pTrc99A-tleA, and E. coli HB101/pTrc99A-tleA cultures after induction with 1 mM IPTG for 3 h. Extracts were separated using SDS-PAGE (10% gel) and stained with Coomassie blue. (D) Adhesion levels of single and double mutants of ETEC 1766a transformed with pTrc99A plasmid carrying tleA. *, percentage significantly greater than values for the remaining six strains (n = 3). For panels A to D, results are expressed as percent adhesion with respect to total bacteria present after 30 min of infection at a multiplicity of infection (MOI) of 10 bacteria per cell. (E to G) Giemsa-stained Caco-2 cells infected at an MOI of 100 with ETEC1766a (E), G10 (F), and E. coli HB101/pTrc99A-tleA after induction with 1 mM IPTG for 3 h (G). Magnification, ×1,000.
Next, the outer membrane protein profile was analyzed after extraction by heating. Extracts obtained from G10 showed a band, approximately 110 kDa, that was not observed in the extracts from E. coli HB101/pHC79 or the Tn2 clone (Fig. 1A, bottom panel). MALDI-TOF/TOF analysis of this band identified a homologue of the APEC Tsh protein (O78:K80:H9; GenBank accession number YP_002539316).
Using successive rounds of sequencing from both transposon ends, the clones with lost adhesion capacity were found to possess a sequence containing a 4.11-kb open reading frame that showed 97% identity (90% coverage) with the APEC tsh gene. Considering the high degree of sequence homology, the gene was designated tleA, for tsh-like ETEC autotransporter. Sequences upstream from tleA (743 bp) showed high identity (99%) to six records in the databases. These six records correspond to plasmids carried by the following pathogenic E. coli strains: E. coli EB1, E. coli S286, APEC02, E. coli chi7122, APEC-1, and E. coli ACN001. Sequences downstream (1,246 bp) shared limited identity (56%) with plasmids carried by the following ETEC strains: ETEC H10407, ETEC 1392/75, and ETEC E24377A. Therefore, tleA could also be carried by a plasmid.
Effect of tleA mutation by allelic replacement on ETEC 1766a and tleA expression in E. coli HB101.
The tleA gene was removed from the ETEC 1766a strain by allelic replacement to determine whether it plays a role in adhesion capacity. There was no significant difference in adhesion levels between the mutant and wild-type strains after infection of Caco-2 cells (Fig. 1B) (P > 0.05). Additionally, no loss in adherence capacity was evidenced after knockout of tleA in a mutant ETEC 1766a strain unable to assemble CS23 (ETEC 1766a ΔaalB).
Given that TleA seemed to be directing G10 attachment to Caco-2 cells, we sought to determine whether it was capable of conferring the adhesion capacity on the nonadherent strain E. coli HB101 after being cloned without the context carried by G10 (genomic fragment of ca. 40 kb) and expressed under the control of an inducible promoter. TleA production in E. coli HB101 was confirmed after outer membrane protein extraction by heating (Fig. 1C, inset) and MALDI-TOF/TOF identification. After adhesion assays were performed in Caco-2 cells, a significantly greater adhesion level was found for induced cells than for noninduced controls or for E. coli HB101 cells carrying an empty vector. In fact, the adhesion level was significantly greater than that of G10 (Fig. 1C). These findings coincided with microscopic analysis of infected Caco-2 cells (Fig. 1F and G). However, expression of tleA under the control of the inducible promoter did not have an effect on the adherence level of ETEC 1766a ΔtleA, nor did it increase adhesion of either ETEC 1766a ΔaalB or the double mutant (Fig. 1D). Conversely, expression of tleA seemed to reduce the adhesion level of ETEC 1766a ΔaalB (Fig. 1D) (P < 0.05). These results suggest that TleA is able to increase attachment of E. coli HB101 to Caco-2 cells, but it does not seem to have the same role in the wild-type ETEC 1766a strain.
Bioinformatic analysis of the TleA sequence.
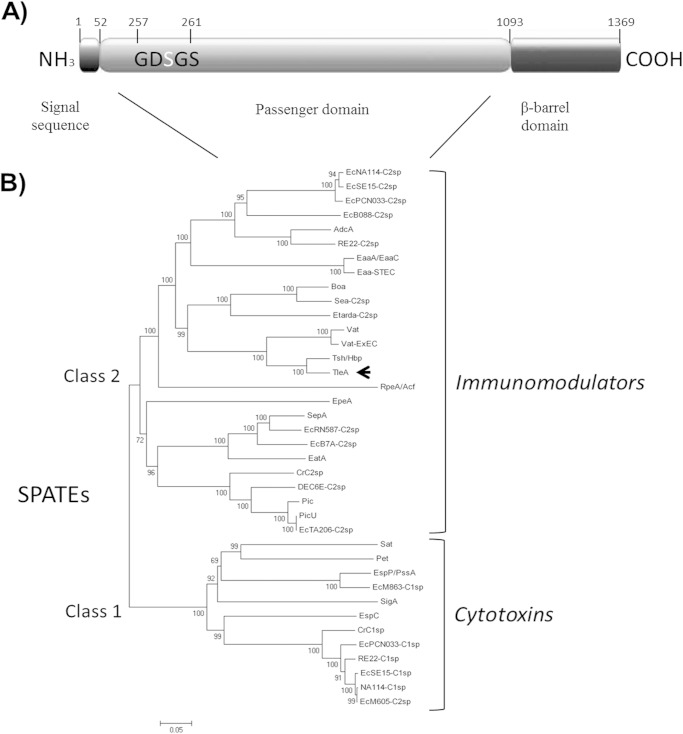
Sequence analyses suggested that TleA is synthesized from a 1,369-amino-acid precursor (147 kDa), with a predicted periplasmic secretion signal sequence from residues 1 to 52, a passenger domain from residues 53 to 1,092, and a β-barrel domain from residues 1,093 to 1,369 (Fig. 2A). TleA shows a 257GDSGS261 serine protease motif in the passenger domain and a cleavage site motif for release of the passenger domain between the asparagine (N) residues (underlined) of 1090EVNNLNKRMGDLRD1103, both of which are conserved among members of the SPATE family. After processing, a 110-kDa free extracellular protein was generated (Fig. 1B), and the β-barrel domain remained anchored to the outer membrane.
FIG 2.
Sequence analysis and phylogenetic relationships of TleA. (A) Schematic representation of TleA precursor domains. Black in the amino terminus represents the signal sequence, and gray represents the passenger domain; black in the carboxyl terminus represents the β-barrel domain. GDSGS, serine protease motif. (B) Phylogenetic relationship among SPATEs. Alignment was performed with MEGA, version 5.05, using ClustalW, based on a previous alignment performed by Ruiz-Perez et al. (21). The arrow indicates the position of TleA.
As mentioned above, the dendrogram of the SPATE passenger domain sequence alignments indicated that TleA clusters with class 2 members (Fig. 2B) and is closely related to APEC Tsh and Vat (vacuolating autotransporter toxin), two SPATE members often found in extraintestinal pathogenic E. coli (21).
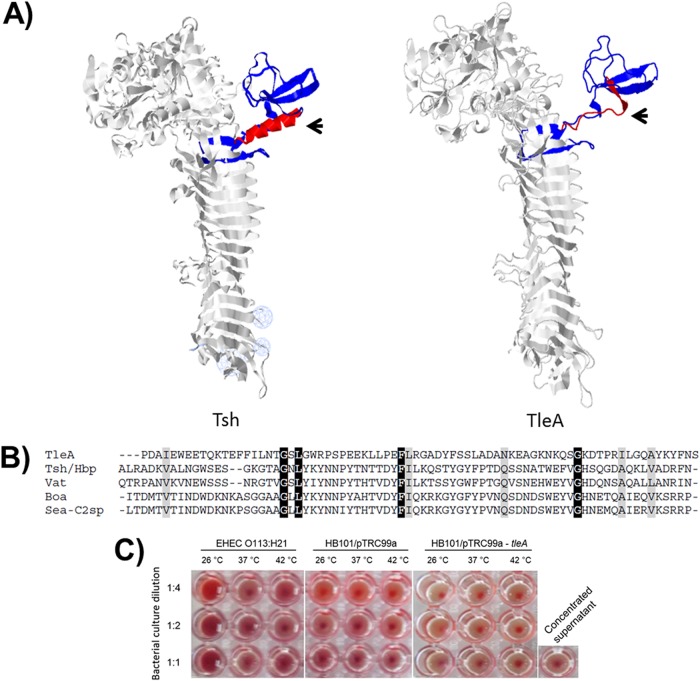
We obtained a hypothetical three-dimensional structure of TleA using the Phyr2 algorithm based on homology with the crystal structure of Hbp (hemoglobin binding protease), a SPATE protein that differs from Tsh by only two amino acids and that was identified in the E. coli EB1 strain isolated from a patient with an infected wound (22). This hypothetical structure revealed that the major discrepancy between the two sequences is in the region known as domain 2, which in the case of Tsh is composed of two β-strands and an α-helix; however, the latter is absent in the case of TleA (residues 481 to 556 in Tsh and Hbp) (Fig. 3A). This 76-amino-acid subdomain, located within a 277-amino-acid residue segment in TleA, shows very little homology with Tsh, Hbp, or other class 2 SPATE proteins (Fig. 3B). Given that it has been suggested that domain 2 plays a role in substrate binding (21), this difference may account for the different functional properties of the two genes. In particular, we found that E. coli HB101 expressing tleA was unable to agglutinate sheep red blood cells (Fig. 3C), a function that has been attributed to Tsh (21). Also, we found that the same strain expressing tleA was unable to bind extracellular matrix proteins (EMPs), laminin, fibronectin, and collagen IV (data not shown).
FIG 3.
Structural analysis of TleA. (A) Hypothetical three-dimensional structure of TleA, constructed based on the crystal structure of Tsh/Hbp (Protein Databank identification number 1WXR) in the Phyre server. Globular subdomain 2 is shown in red α-helical and blue β-strands. Arrows indicate the α-helix of Tsh, which is absent in TleA. (B) Amino acid sequence discrepancy in subdomain 2 among TleA and other SPATEs phylogenetically close to TleA is shown. Identical residues are shown in black, and similar residues are shown in gray. The region shown corresponds to amino acid 533 to amino acid 556 in TleA. (C) Evaluation of the capacity to agglutinate sheep blood cells. Bacteria were grown at 26°C, 37°C, or 42°C and assessed in 1:1, 1:2, and 1:4 dilutions in PBS. Enterohemorrhagic E. coli (EHEC) strain E045-00 (O113:H21) was used as a positive control. A positive result is indicated by formation of a red film, and a negative result is indicated by a red sediment at the bottom of the well.
Localization of TleA.
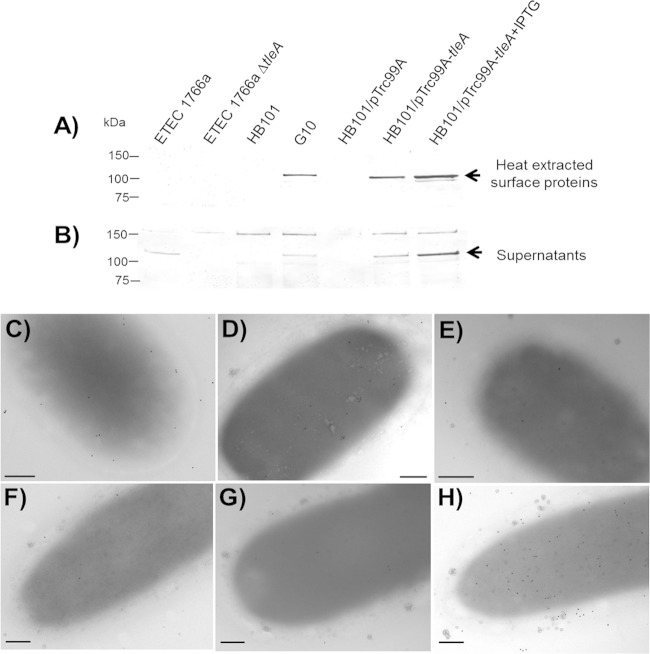
We detected the TleA autotransporter by immunoblotting using an antibody that recognizes the passenger domain of Vat, a highly similar SPATE, in both heat-extracted surface proteins and concentrated culture supernatants (Fig. 4A and B). The size of the protein (approximately 115 kDa) detected in both fractions was consistent with the expected size for the TleA passenger domain and with that described for SPATE proteins such as Tsh, Pet, and others (21). Presence of the immunoreactive band in fractions containing surface proteins seemed to coincide with adherence capacity mediated by TleA. This was the case for the G10 clone and E. coli HB101 carrying the tleA gene under the control of the inducible promoter in pTrc99A (Fig. 4A and B). On the other hand, the presence of the TleA autotransporter was not evidenced by Western blotting on surface fractions of ETEC 1766a; however, TleA was detected in the supernatant, suggesting that the passenger domain is mostly being released into the medium (Fig. 4A and B).
FIG 4.
Detection of TleA. (A and B) Heat-extracted proteins and concentrated supernatants obtained from ETEC 1766a, ETEC 1766aΔtleA, E. coli HB101, G10, E. coli HB101/pTrc99A, and E. coli HB101/pTrc99A-tleA, in presence or absence of 1 mM IPTG, were subjected to SDS-PAGE (10%), and TleA was detected by Western blotting. Lanes are the same for both panels. An antibody against the passenger domain of Vat protein, carried by UPEC strains, which shares 71% identity with TleA was used in both experiments. Arrows indicate the TleA band. (C to H) Detection by immunogold labeling using an antibody directed against the passenger domain of Tsh in nonpermeabilized ETEC 1766a (C), ETEC 1766a ΔaalB (D), ETEC 1766a ΔtleA (E), E. coli HB101 (F), E. coli HB101/pTrc99A (G), and E. coli HB101/pTrc99A-tleA (H). Scale bar, 0.2 μm.
Immunogold labeling using an antibody directed at the Tsh passenger domain allowed detection of the autotransporter decorating the surface of nonpermeabilized ETEC 1766a and the CS23 mutant (Fig. 4C and D). In contrast, only a few gold particles were seen on the ETEC 1766a tleA mutant (Fig. 4E), E. coli HB101, and E. coli HB101 carrying the empty pTrc99A vector (Fig. 4F and G); this may have been a result of residual nonspecific labeling. As expected, labeling was abundant on the surface of E. coli HB101 expressing tleA (Fig. 4H). These results suggest that the TleA passenger domain likely remains associated with the surface of the bacterium after secretion.
Protease activity.
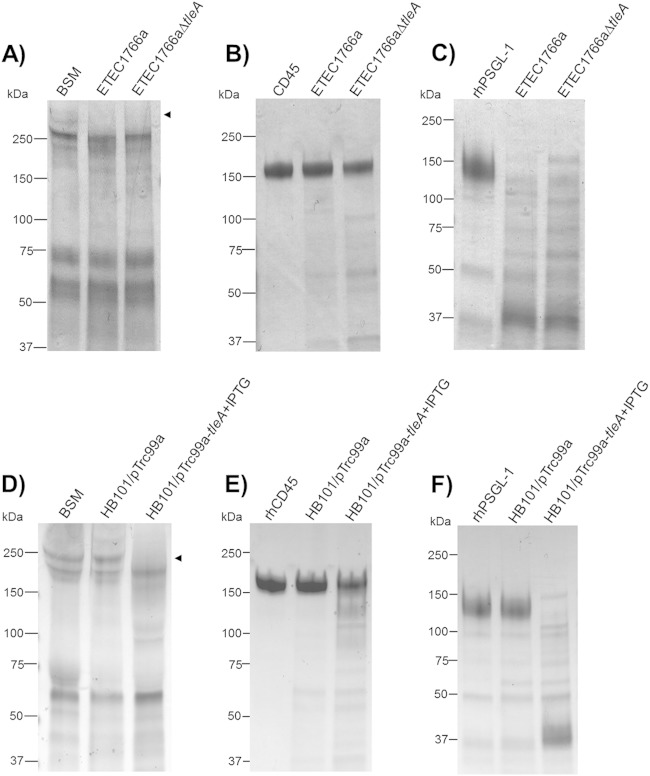
Unlike class 1 SPATEs, some class 2 SPATE proteins possess mucinolytic activity; therefore, mucinase activity assays were performed using concentrated supernatants. After incubation of bovine submaxillary mucin (BSM) with supernatants of ETEC 1766a and the tleA mutant, followed by SDS-PAGE, there was evidence of high-molecular-weight band degradation (Fig. 5A). Degradation was also evident in the tleA mutant, suggesting that the mucinase activity is redundant in this strain (Fig. 5A). Similar results were obtained by incubating concentrated supernatants with the leukocyte glycoproteins CD45 (lymphocyte common antigen) and the P-selectin glycoprotein ligand-1 (PSGL-1), which are normally expressed in all hematopoietic cell lineages and are also cleaved by class 2 SPATEs (Fig. 5B and C) (21). Supernatants of E. coli HB101 expressing tleA also exhibited protease activity over the same three targets, while degradation was not observed in supernatants from E. coli HB101 carrying the empty pTrc99A plasmid (Fig. 5C, D, and E). MALDI-TOF/TOF analysis of the cleaved low-molecular-weight mucin species revealed a resemblance to the bovine Muc19 molecule.
FIG 5.
Mucinolytic activity of TleA on mucin and leukocyte mucin-like proteins. Overnight incubations at 37°C were performed with 100 μg of BSM (A and D), 2 μg of recombinant human CD45 (rhCD45) (B and E), or 2 μg of rhPSGL-1 (C and F) and concentrated supernatant of ETEC 1766a, ETEC 1766a ΔtleA, HB101/pTrc99A, and HB101/pTrc99A-tleA. Samples were analyzed by SDS-PAGE in acrylamide gradient gels (4% to 20%) and stained with Coomassie blue. M, molecular mass marker. Arrows indicate degradation products identified by MALDI-TOF/TOF.
Distribution of TleA in various ETEC isolates.
To determine whether tleA is present in other ETEC strains, PCR was used to detect its presence in 102 strains obtained in Chile. All strains, which included different serogroups and carried different virulence factors, were negative for the presence of tleA. In a set of 86 ETEC strains collected as part of the Global Enterics Multicenter Study (23) from children suffering from diarrhea in developing countries, one strain from Pakistan with a nontypeable O serogroup and carrying genes encoding LT and a human ST variant was positive for detection of tleA.
After BLAST searches were performed, no sequences identical to the tleA sequence were found in the databases, regardless of whether blastn or blastp algorithms were used. Highest identity percentages obtained by blastn, which were similar to those shared between tleA and tsh (97% maximum identity with 90% coverage), corresponded to records of eight APEC genomes and four non-O157 Shiga toxin-producing E. coli (STEC) strains. By using blastp, seven protein records were found in seven different ETEC strains obtained from Bangladesh, and they shared 95 to 99% sequence identity (strains 2785200, MP021561.2, 2780750, 178850, 2865200, P0299917.1 and 2864350; GenBank accession numbers EMW33499.1, EMX29945.1, EMW42412.1, ENG81397.1, EMV83811.1, ENA04112.1 and ENB02748.1, respectively). In all of these cases, sequences of the putative domain 2 (76 amino acids) shared 100% identity with the TleA sequence.
DISCUSSION
Adhesion is a crucial step in the infectious cycle of ETEC and a requisite for efficient release of toxins (7, 24). To date, over 20 adhesins have been identified in ETEC strains, and yet there are clinical strains that do not carry any known adhesin (11). This work attempted to identify a new genetic adherence determinant of ETEC 1766a able to confer the attachment capacity to E. coli HB101. The gene identified, named tleA, encodes a homologue of Tsh, an autotransporter initially described in APEC and uropathogenic E. coli (UPEC) strains (25, 26). Consequently, this is the first study evaluating the activity of a gene homologous to tsh in a diarrheagenic strain of E. coli (ETEC).
Two previously described ETEC non-SPATE proteins exported through type V secretion systems, TibA and EtpA, are known to confer adhesion capacity to intestinal epithelial cells (12, 13). Thus far, the only SPATE identified as an ETEC virulence factor is EatA (ETEC autotransporter A), which does not function directly as an adhesin but, rather, influences the toxigenic effect by degrading EtpA via proteolytic cleavage and colonization by degrading the main component of the intestinal human mucus layer, Muc2 (27, 28). Additionally, another nonautotransporter secreted protein, the metalloprotease YghJ, has recently been described as a mucinase secreted by ETEC strains (29).
SPATEs are secreted and released to the extracellular medium, where they exert their protease activity. There is scarce evidence regarding their adhesin activity, specifically in directing attachment of bacteria to epithelial cells. For example, the protein App, produced by Neisseria meningitidis, is a determinant of adherence capacity to Chang cells (30). On the other hand, RpeA and Hap produced by rabbit enteropathogenic E. coli and Haemophilus influenzae, respectively, are able to direct adherence to epithelial cells when they are expressed in nonadherent strains of their respective species (31, 32). Particularly in the case of Hap, when protein release is stopped by a site-directed mutation affecting the catalytic site, the serine protease remains associated to the membrane, and the adherence capacity of the bacterium is significantly increased (33). TleA was detected when exposed by immunogold labeling at the surface of ETEC 1766a, but knocking out the gene did not significantly alter adhesion compared to that of the wild-type strain. These results may indicate that TleA's effect on the adherence capacity of ETEC 1766a is redundant or masked by other structures present on the bacterial surface. CS23 is known to be largely responsible for this phenotype, given that if the expression of this fimbria is impaired, the capacity to adhere to Caco-2 cells drops dramatically (16). A double mutant ETEC 1766a strain, unable to produce TleA or assemble CS23, showed a capacity for adherence similar to that of the single tleA mutant, suggesting again that this SPATE would not be a key adherence determinant in directing adherence to Caco-2 cells. However, tleA expression confers adherence capacity to E. coli HB101, indicating that TleA may act as an adhesin when it is exposed on the surface. Perhaps the presence of TleA on the bacterial surface of E. coli HB101 is sufficient to confer the adhesive phenotype, something that was not evident in the case of ETEC 1766a. These observations suggest that the main function of TleA in ETEC 1766a is derived from its protease activity in the released form. Further research is needed to establish if the increase observed in E. coli HB101 adherence level derives from direct TleA attachment or from an effect on cell layer integrity. We cannot now rule out a direct role of TleA in bacterial adherence in vivo.
Given the high homology of TleA with Tsh and other SPATE proteins, we considered TleA to be a part of the same family. These molecules are synthesized as precursors for a signal sequence in the N terminus of the bacterium's internal membrane, known as the translocator Sec, and two functional domains: an extracellular passenger domain that contributes to pathogenicity and a β-barrel domain in the C terminus that promotes transit of the passenger domain through the outer membrane (34, 35). SPATEs are categorized phylogenetically into two classes, class 1 and class 2, according to their passenger domain amino acid sequences (21, 36). Class 2 SPATEs, like TleA, have been described as lectin-type immunomodulators because of their role in carbohydrate binding (21). Furthermore, these SPATEs possess proteolytic activity toward different types of mucins and glycoproteins present on the leukocyte surface (21, 37). With respect to protease activity, TleA was observed to degrade mucin, suggesting that it participates in colonization of the intestinal epithelium. Moreover, as with other class 2 SPATEs, TleA degrades O-glycosylated proteins such as CD45 and PSGL-1, suggesting a possible role as an immunomodulator in evading the immune response during infection (37). CD45 is a transmembrane receptor, with tyrosine phosphatase (170 to 220 kDa) expressed in all hematopoietic cells (38). Activation of this type of cell is essential for activities such as adhesion, migration, modulation of cytokine production, and cell signaling (39). In addition, PSGL-1 (220 kDa) is an O-glycosylated protein expressed on the leukocyte surface, which allows recruitment of innate and adaptive immune response factors and induction of cell signaling (40).
It has been reported that APEC Tsh has the capacity to agglutinate red blood cells and also adhere to EMPs, such as fibronectin and collagen IV, forming a protein complex that influences many biological processes, such as adhesion, migration, proliferation, and differentiation of eukaryotic cells (41). Furthermore, the complex serves as a target for colonization by microorganisms (42, 43). TleA differs from Tsh (carried by APEC) by more than 100 amino acids located within the region that includes domain 2, which is conserved in most class 2 SPATEs and has been related to substrate recognition because it shares homology with the chitin-binding domain of chitinase (44). Even though the role of domain 2 in the attachment capacity of class 2 SPATEs has not been directly proved (21), differences between the three-dimensional folding in Tsh and TleA could explain why the latter was not capable of causing agglutination of red blood cells or adhering to EMP-like laminin, collagen IV, or fibronectin under the experimental conditions used in this study. Recently, diverse effects resulting from the incubation of human leukocytes with several purified SPATEs were reported (45). AdcA, a SPATE produced by Citrobacter rodentium, which lacks domain 2, showed a comparable proteolytic activity over mucin and other surface mucin-like receptors. No conclusions could be established regarding the functional consequences of the lack of domain 2 (45).
In conclusion, this work identified the autotransporter TleA, a member of the SPATE family, as a potential new ETEC virulence factor. During the infectious cycle of an ETEC strain, this autotransporter may promote colonization, allowing the bacteria to cross the mucous layer and establish direct contact with the epithelial cells as well as to interfere with the normal immune response.
ACKNOWLEDGMENTS
This research was supported by Fondecyt Grantss 1110260, 1120809, and 3130555, as well as by the Bill and Melinda Gates Foundation grant Global Enterics Multicenter Study (identification number 38874).
We thank Carolina Ponce and Sergio Vargas (Universidad de Chile) for their assistance in microscopic analysis.
REFERENCES
- 1.WHO. 2006. Future directions for research on enterotoxigenic Escherichia coli vaccines for developing countries. Wkly Epidemiol Rec. 81:97–104. [PubMed] [Google Scholar]
- 2.Huilan S, Zhen LG, Mathan MM, Mathew MM, Olarte J, Espejo R, Khin Maung U, Ghafoor MA, Khan MA, Sami Z, Sutton R. 1991. Etiology of acute diarrhoea among children in developing countries: a multicentre study in five countries. Bull World Health Organ 69:549–555. [PMC free article] [PubMed] [Google Scholar]
- 3.Gascón J. 2006. Epidemiology, etiology and pathophysiology of traveler's diarrhea. Digestion 73:102–108. doi: 10.1159/000089785. [DOI] [PubMed] [Google Scholar]
- 4.Nataro JP, Kaper JB. 1998. Diarrheagenic Escherichia coli. Clin Microbiol Rev 11:142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vaandrager AB, Bot AG, De Jonge HR. 1997. Guanosine 3′,5′-cyclic monophosphate-dependent protein kinase II mediates heat-stable enterotoxin-provoked chloride secretion in rat intestine. Gastroenterology 112:437–443. doi: 10.1053/gast.1997.v112.pm9024297. [DOI] [PubMed] [Google Scholar]
- 6.Qadri F, Svennerholm A, Faruque ASG, Sack RB. 2005. Enterotoxigenic Escherichia coli in developing countries: epidemiology, microbiology, clinical features, treatment, and prevention. Clin Microbiol Rev 18:465–483. doi: 10.1128/CMR.18.3.465-483.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fleckenstein JM, Hardwidge PR, Munson GP, Rasko DA, Sommerfelt H, Steinsland H. 2010. Molecular mechanisms of enterotoxigenic Escherichia coli infection. Microbes Infect 12:89–98. doi: 10.1016/j.micinf.2009.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaper JB, Nataro JP, Mobley HL. 2004. Pathogenic Escherichia coli. Nat Rev Microbiol 2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 9.Del Canto F, Valenzuela P, Cantero L, Bronstein J, Blanco JE, Blanco J, Prado V, Levine M, Nataro J, Sommerfelt H, Vidal R. 2011. Distribution of classical and nonclassical virulence genes in enterotoxigenic Escherichia coli isolates from Chilean children and tRNA gene screening for putative insertion sites for genomic islands. J Clin Microbiol 49:3198–3203. doi: 10.1128/JCM.02473-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gaastra W, Svennerholm A. 1996. Colonization factors of human enterotoxigenic Escherichia coli (ETEC). Trends Microbiol 4:444–452. doi: 10.1016/0966-842X(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 11.Isidean SD, Riddle MS, Savarino SJ, Porter CKA. 2011. Systematic review of ETEC epidemiology focusing on colonization factor and toxin expression. Vaccine 29:6167–6178. doi: 10.1016/j.vaccine.2011.06.084. [DOI] [PubMed] [Google Scholar]
- 12.Lindenthal C, Elsinghorst EA. 2001. Enterotoxigenic Escherichia coli TibA glycoprotein adheres to human intestine epithelial cells. Infect Immun 69:52–57. doi: 10.1128/IAI.69.1.52-57.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fleckenstein JM, Roy K, Fischer JF, Burkitt M. 2006. Identification of a two-partner secretion locus of enterotoxigenic Escherichia coli. Infect Immun 74:2245–2258. doi: 10.1128/IAI.74.4.2245-2258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Svennerholm AM, Lundgren A. 2012. Recent progress toward an enterotoxigenic Escherichia coli vaccine. Expert Rev Vaccines 11:495–507. doi: 10.1586/erv.12.12. [DOI] [PubMed] [Google Scholar]
- 15.Fleckenstein JM, Munson GM, Rasko D. 2013. Enterotoxigenic Escherichia coli: orchestrated host engagement. Gut Microbes 4:392–396. doi: 10.4161/gmic.25861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Del Canto F, Botkin DJ, Valenzuela P, Popov V, Ruiz-Perez F, Nataro JP, Levine MM, Stine OC, Pop M, Torres AG, Vidal R. 2012. Identification of coli surface antigen 23, a novel adhesin of enterotoxigenic Escherichia coli. Infect Immun 80:2791–2801. doi: 10.1128/IAI.00263-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Benz I, Schmidt MA. 1992. Isolation and serologic characterization of AIDA-I, the adhesin mediating the diffuse adherence phenotype of the diarrhea-associated Escherichia coli strain 2787 (O126:H27). Infect Immun 60:13–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torres AG, Perna NT, Burland V, Ruknudin A, Blattner FR, Kaper JB. 2002. Characterization of Cah, a calcium-binding and heat-extractable autotransporter protein of enterohaemorrhagic Escherichia coli. Mol Microbiol 45:951–966. doi: 10.1046/j.1365-2958.2002.03094.x. [DOI] [PubMed] [Google Scholar]
- 19.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 20.Datsenko K, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz-Perez F, Nataro JP. 2014. Bacterial serine proteases secreted by the autotransporter pathway: classification, specificity, and role in virulence. Cell Mol Life Sci 71:745–770. doi: 10.1007/s00018-013-1355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Otto BR, van Dooren SJ, Nuijens JH, Luirink J, Oudega B. 1998. Characterization of a hemoglobin protease secreted by the pathogenic Escherichia coli strain EB1. J Exp Med 188:1091–1103. doi: 10.1084/jem.188.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kotloff KL, Nataro JP, Blackwelder WC, Nasrin D, Farag TH, Panchalingam S, Wu Y, Sow SO, Sur D, Breiman RF, Faruque AS, Zaidi AK, Saha D, Alonso PL, Tamboura B, Sanogo D, Onwuchekwa U, Manna B, Ramamurthy T, Kanungo S, Ochieng JB, Omore R, Oundo JO, Hossain A, Das SK, Ahmed S, Qureshi S, Quadri F, Adegbola RA, Antonio M, Hossain MJ, Akinsola A, Mandomando I, Nhampossa T, Acácio S, Biswas K, O'Reilly CE, Mintz ED, Berkeley LY, Muhsen K, Sommerfelt H, Robins-Browne RM, Levine MM. 2013. Burden and aetiology of diarrhoeal disease in infants and young children in developing countries (the Global Enteric Multicenter Study, GEMS): a prospective, case-control study. Lancet 382:209–222. doi: 10.1016/S0140-6736(13)60844-2. [DOI] [PubMed] [Google Scholar]
- 24.Croxen M, Finlay BB. 2010. Molecular mechanisms of Escherichia coli pathogenicity. Nat Rev Microbiol 8:26–38. doi: 10.1038/nrmicro2265. [DOI] [PubMed] [Google Scholar]
- 25.Provence DL, Curtiss R. 1994. Isolation and characterization of a gene involved in hemagglutination by an avian pathogenic Escherichia coli strain. Infect Immun 62:1369–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heimer SR, Rasko DA, Lockatell CV, Johnson DE, Mobley HLT. 2004. Autotransporter genes pic and tsh are associated with Escherichia coli strains that cause acute pyelonephritis and are expressed during urinary tract infection. Infect Immun 72:593–597. doi: 10.1128/IAI.72.1.593-597.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roy K, Kansal R, Bartels SR, Hamilton DJ, Shaaban S, Fleckenstein JM. 2011. Adhesin degradation accelerates delivery of heat-labile toxin by enterotoxigenic Escherichia coli. J Biol Chem 286:29771–29779. doi: 10.1074/jbc.M111.251546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumar P, Luo Q, Vickers TJ, Sheikh A, Lewis WG, Fleckenstein JM. 2014. EatA, an immunogenic protective antigen of enterotoxigenic Escherichia coli degrades intestinal mucin. Infect Immun 82:500–508. doi: 10.1128/IAI.01078-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo Q, Kumar P, Vickers T, Sheikh A, Lewis WG, Rasko DA, Sistrunk J, Fleckenstein JM. 2014. Enterotoxigenic Escherichia coli secrete a highly conserved mucin-degrading metalloprotease to effectively engage intestinal epithelial cells. Infect Immun 82:509–521. doi: 10.1128/IAI.01106-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Serruto D, Adu-Bobie J, Scarselli M, Veggi D, Pizza M, Rappuoli R, Aricò B. 2003. Neisseria meningitidis App, a new adhesin with autocatalytic serine protease activity. Mol Microbiol 48:323–334. doi: 10.1046/j.1365-2958.2003.03420.x. [DOI] [PubMed] [Google Scholar]
- 31.Leyton DL, Adams LM, Kelly M, Sloan J, Tauschek M, Robins-Browne RM, Hartland EL. 2007. Contribution of a novel gene, rpeA, encoding a putative autotransporter adhesin to intestinal colonization by rabbit-specific enteropathogenic Escherichia coli. Infect Immun 75:4664–4669. doi: 10.1128/IAI.00972-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.St Geme JW III, de la Morena ML, Falkow S. 1994. A Haemophilus influenzae IgA protease-like protein promotes intimate interaction with human epithelial cells. Mol Microbiol 14:217–233. doi: 10.1111/j.1365-2958.1994.tb01283.x. [DOI] [PubMed] [Google Scholar]
- 33.Hendrixson DR, St Geme JW III. 1998. The Haemophilus influenzae Hap serine protease promotes adherence and microcolony formation, potentiated by a soluble host protein. Mol Cell 2:841–850. doi: 10.1016/S1097-2765(00)80298-1. [DOI] [PubMed] [Google Scholar]
- 34.Leyton DL, Rossiter AE, Henderson IR. 2012. From self sufficiency to dependence: mechanisms and factors important for autotransporter biogenesis. Nat Rev Microbiol 10:213–225. doi: 10.1038/nrmicro2733. [DOI] [PubMed] [Google Scholar]
- 35.Dautin N, Bernstein HD. 2011. Residues in a conserved α-helical segment are required for cleavage but not secretion of an Escherichia coli serine protease autotransporter passenger domain. J Bacteriol 193:3748–3756. doi: 10.1128/JB.05070-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dutta PR, Cappello R, Navarro-García F, Nataro JP. 2002. Functional comparison of serine protease autotransporters of Enterobacteriaceae. Infect Immun 70:7105–7113. doi: 10.1128/IAI.70.12.7105-7113.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ruiz-Perez F, Wahid R, Faherty CS, Kolappaswamy K, Rodriguez L, Santiago A, Murphy E, Cross A, Sztein MB, Nataro JP. 2011. Serine protease autotransporters from Shigella flexneri and pathogenic Escherichia coli target a broad range of leukocyte glycoproteins. Proc Natl Acad Sci U S A 108:12881–12886. doi: 10.1073/pnas.1101006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hermiston ML, Xu Z, Weiss A. 2003. CD45: a critical regulator of signaling thresholds in immune cells. Annu Rev Immunol 21:107–137. doi: 10.1146/annurev.immunol.21.120601.140946. [DOI] [PubMed] [Google Scholar]
- 39.Lai JCY, Wlodarska M, Liu DJ, Johnson P, Abraham N. 2010. CD45 regulates migration, proliferation, and progression of double negative 1 thymocytes. J Immunol 185:2059–2070. doi: 10.4049/jimmunol.0902693. [DOI] [PubMed] [Google Scholar]
- 40.Carlow DA, Gossens K, Naus S, Veerman KM, Seo W, Ziltener HJ. 2009. PSGL-1 function in immunity and steady state homeostasis. Immunol Rev 230:75–96. doi: 10.1111/j.1600-065X.2009.00797.x. [DOI] [PubMed] [Google Scholar]
- 41.Kostakioti M, Stathopoulos C. 2004. Functional analysis of the Tsh autotransporter from an avian pathogenic Escherichia coli strain. Infect Immun 72:5548–5554. doi: 10.1128/IAI.72.10.5548-5554.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chagnot C, Listrat A, Astruc T, Desvaux M. 2012. Bacterial adhesion to animal tissues: protein determinants for recognition of extracellular matrix components. Cell Microbiol 14:1687–1696. doi: 10.1111/cmi.12002. [DOI] [PubMed] [Google Scholar]
- 43.Farfan MJ, Cantero L, Vidal R, Botkin DJ, Torres AG. 2011. Long polar fimbriae of enterohemorrhagic Escherichia coli O157:H7 bind to extracellular matrix proteins. Infect Immun 79:3744–3750. doi: 10.1128/IAI.05317-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishimura K, Tajima N, Yoon YH, Park SY, Tame JRH. 2010. Autotransporter passenger proteins: virulence factors with common structural themes. J Mol Med (Berl) 88:451–458. doi: 10.1007/s00109-010-0600-y. [DOI] [PubMed] [Google Scholar]
- 45.Ayala-Lujan JL, Vijayakumar V, Gong M, Smith R, Santiago AE, Ruiz-Perez F. 2014. Broad spectrum activity of a lectin-like bacterial serine protease family on human leukocytes. PLoS One 9:e107920. doi: 10.1371/journal.pone.0107920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amann E, Ochs B, Abel KJ. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301–315. doi: 10.1016/0378-1119(88)90440-4. [DOI] [PubMed] [Google Scholar]
- 47.Montero D, Orellana P, Gutiérrez D, Araya D, Salazar JC, Prado V, Oñate A, Del Canto F, Vidal R. 2014. Immunoproteomic analysis to identify Shiga toxin-producing Escherichia coli outer membrane proteins expressed during human infection. Infect Immun 82:4767–4777. doi: 10.1128/IAI.02030-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyer HW, Roulland-Dussoix D. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol 41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]