Abstract
Urinary tract infections (UTIs) are among the most common infectious diseases of humans, with Escherichia coli responsible for >80% of all cases. One extreme of UTI is asymptomatic bacteriuria (ABU), which occurs as an asymptomatic carrier state that resembles commensalism. To understand the evolution and molecular mechanisms that underpin ABU, the genome of the ABU E. coli strain VR50 was sequenced. Analysis of the complete genome indicated that it most resembles E. coli K-12, with the addition of a 94-kb genomic island (GI-VR50-pheV), eight prophages, and multiple plasmids. GI-VR50-pheV has a mosaic structure and contains genes encoding a number of UTI-associated virulence factors, namely, Afa (afimbrial adhesin), two autotransporter proteins (Ag43 and Sat), and aerobactin. We demonstrated that the presence of this island in VR50 confers its ability to colonize the murine bladder, as a VR50 mutant with GI-VR50-pheV deleted was attenuated in a mouse model of UTI in vivo. We established that Afa is the island-encoded factor responsible for this phenotype using two independent deletion (Afa operon and AfaE adhesin) mutants. E. coli VR50afa and VR50afaE displayed significantly decreased ability to adhere to human bladder epithelial cells. In the mouse model of UTI, VR50afa and VR50afaE displayed reduced bladder colonization compared to wild-type VR50, similar to the colonization level of the GI-VR50-pheV mutant. Our study suggests that E. coli VR50 is a commensal-like strain that has acquired fitness factors that facilitate colonization of the human bladder.
INTRODUCTION
Urinary tract infections (UTIs) are among the most common infectious diseases of humans and a major cause of morbidity. It is estimated that 40 to 50% of all adult women experience at least one UTI episode in their lifetime (1). In addition to well-documented symptomatic infections, many UTIs are asymptomatic. These infections, referred to as asymptomatic bacteriuria (ABU), represent a carrier state that resembles commensalism and occur in a percentage of the population, depending on age and gender. ABU patients may carry >105 CFU of a single bacterial strain/ml of urine for months or years without significant symptoms.
Escherichia coli causes more than 80% of all symptomatic and asymptomatic UTIs. In general, strains that cause symptomatic UTI are collectively described as uropathogenic E. coli (UPEC), while strains that cause asymptomatic UTI are referred to as ABU E. coli. Both UPEC and ABU E. coli strains exhibit a high degree of genetic diversity that is largely attributed to the presence of virulence/fitness genes on mobile genetic elements referred to as pathogenicity islands (PAIs) or genomic islands (GIs) (2, 3). While no single virulence factor is uniquely definitive for UPEC or ABU E. coli, the ability to colonize the urinary tract is enhanced by a number of factors, including fimbriae (e.g., type 1, P, F1C, and Afa), autotransporter proteins (e.g., Ag43, UpaB, and UpaH), cell surface polysaccharides (e.g., O antigen), and siderophores (e.g., enterobactin, salmochelin, aerobactin, and yersiniabactin) (4–8).
The clinically benign nature of ABU was initially explained by a lack of virulence, since phenotypically, many ABU E. coli strains lack adhesins and toxins commonly associated with virulence. However, there are inconsistencies in these observations with respect to strain genotype, and it is now apparent that many ABU E. coli strains have arisen from virulent UPEC strains that have become attenuated through gene loss or deletion (9, 10). This is supported by the observation that ABU strains often possess virulence genes, such as those encoding type 1 fimbriae, P fimbriae, or hemolysin, but do not express the associated phenotype (9–12). Molecular and genomic analyses of the ABU prototype E. coli strain 83972 also support this observation. E. coli 83972 is a clinical isolate capable of long-term bladder colonization, as shown in human inoculation studies (13–15). The virulence-associated adhesin genes of E. coli 83972 have become attenuated through a series of independent mutational events. The type 1 fimbrial system has been inactivated by a major deletion encompassing 4.5 kb of the fim gene cluster affecting all genes except those encoding the minor components, namely, fimF, fimG, and fimH (16). The F1C system has been inactivated by point mutations in the fimbrial transport system affecting the focD gene encoding the usher protein (17). Finally, the P fimbrial system has been inactivated by point mutations in the papG gene that have rendered the PapG adhesin nonfunctional (16). Thus, the ancestor of E. coli 83972, which belongs to the B2 clonal group, was most likely a virulent UPEC strain that has become attenuated (18). Recently, E. coli 83972 adaptation to the human urinary tract has also been linked to its ability to suppress RNA polymerase II-dependent host gene expression following human bladder colonization, thereby preventing the activation of host proinflammatory responses during infection (19).
A second group of ABU E. coli strains comprises commensal-like strains that have acquired fitness factors that contribute to colonization of the bladder (10). Little is understood about the genetic makeup of these strains or the factors that contribute to their prolonged survival without inducing a proinflammatory host response. In this study, we obtained a complete genome sequence of the ABU E. coli strain VR50. We show that E. coli VR50 is most closely related to commensal E. coli strains, particularly the K-12 strain MG1655, with the major difference being the presence of mobile genetic elements, including a large 94-kb genomic island (GI-VR50-pheV), prophage regions, and plasmids. GI-VR50-pheV has a mosaic composition of genes, and we show that the genes encoding the Afa adhesin (which are located within GI-VR50-pheV) contribute significantly to VR50 adherence to bladder epithelial cells and colonization of the mouse bladder.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
E. coli VR50 (serotype OR:K1:H−) was isolated from a 30-year-old otherwise healthy woman who had carried it for at least 1 year without any symptoms (20). E. coli 83972 and UPEC CFT073 have been described previously (21, 22). Bacteria were routinely grown at 37°C on solid or in liquid Luria-Bertani (LB) medium supplemented with appropriate antibiotics unless otherwise stated (23).
DNA manipulations and genetic techniques.
Chromosomal DNA was purified using the Ultraclean Microbial DNA isolation kit (Mo Bio Laboratories). PCR was performed using Taq polymerase according to the manufacturer's instructions (Roche, Australia). Sequencing of PCR products was performed by the Australian Genome Research Facility. The E. coli VR50 phylogenetic group of origin was determined using a three-locus PCR-based method (24).
Construction of mutants.
E. coli VR50 deletion mutants (VR50pheV-GI, VR50afa, VR50afaE, VR50fim, and VR50fimH) were constructed using the λ-Red recombinase gene replacement system (25). Briefly, the kanamycin gene from plasmid pKD4 was amplified using primers containing 50-nucleotide (nt) homology extensions to the beginnings and ends of the gene clusters to be deleted. The following primers were used: VR50pheV-GI, 1772 (5′-CTGTGCAACATACTCACCATTATGGTAAGCGTGCAGCAAGAACCGTATTGGGCCTGGTGATGATGGCGGGATCG) and 1773 (5′-CCATCAGCCAGCTTATCATTCAGTAGAAGTTGATAAGCGGGTGTTGCCAGTCAGAAGAACTCGTCAAGAAGGCG); VR50afa, 490 (5′-ATGAGGGAGCGATATCTGTATCTTGCTGACACCCCTCAGGGGATACTGATGTGTAGGCTGGAGCTGCTTC) and 491 (5′-TCAATTTGTCCAGTAACCGCCAGTCAGTGTTAAAGTGTAATTACCAGTCGCATATGAATATCCTCCTTAG); VR50afaE, 827 (5′-AAAAACGCAGCGCCGGTATGAATGAATTACGTCATCCGGGAAGCACACAGGTGTAGGCTGGAGCTGCTTC) and 828 (5′-GCTCCTTTGTTGATCTATTTTTTATTATCGGCCAGTGATTGATTGGTTCCCATATGAATATCCTCCTTAG); VR50fim, 244 (5′-GTCGATTGAGGATTTCGGATATTGATCTTAAGGCAAAGTGGTGTAGGCTGGAGCTGCTTC) and 245 (5′-GCTCCTAACGATACCGTGTTATTCGCTGGAATAATCGTACCATATGAATATCCTCCTTAG); and VR50fimH, 787 (5′-AGTGATTAGCATCACCTATACCTACAGCTGAACCCGAAGAGATGATTGTAGTGTAGGCTGGAGCTGCTTC) and 788 (5′-TAGCTTCAGGTAATATTGCGTACCAGCATTAGCAATGTCCTGTGATTTCTCATATGAATATCCTCCTTAG). The primers were used to amplify a 1.6-kb PCR product from plasmid pKD4, representing the kanamycin resistance cassette from pKD4 and additional 50-bp overhang regions at the 5′ and 3′ ends of the PCR product complementary to the target genes in VR50. The mutants were constructed by transforming VR50(pKD46) with the PCR product containing the homology arms and selection of kanamycin-resistant colonies. The kanamycin cassette was then removed using plasmid pCP20 (25). All deletions were confirmed by PCR and sequencing.
Agglutination assays.
Afa-mediated mannose-resistant hemagglutination (MRHA) was assessed as previously described (26). Briefly, a 5% suspension (10 μl) of human type A red blood cells (RBCs) washed in phosphate-buffered saline (PBS) was mixed with a 10-μl bacterial suspension on glass slides in the presence and absence of d-mannose. The bacterial suspension was prepared by transferring cells from a freshly grown LB agar colony into 50 μl PBS.
Adhesion assays.
E. coli binding to human HeLa and T24 bladder epithelial cells was assessed by a quantitative adhesion assay using confluent monolayers of epithelial cells (12). Epithelial cells were seeded into 24-well cell culture plates (Corning Inc., NY) at a concentration of 3 × 105 cells per well and incubated overnight at 37°C in 5% CO2 prior to infection. The monolayers were washed with PBS and inoculated with bacteria at a multiplicity of infection of 50 bacteria per epithelial cell for HeLa cell assays and 10 bacteria per epithelial cell for T24 cell assays. The bacteria were prepared by washing an overnight culture grown in LB once in PBS (3,500 × g; 15 min; 4°C) and resuspended to a concentration of 1.5 × 107 CFU/ml in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 2 mM glutamine (HeLa cell assays) or McCoy's 5A modified medium (Life Technologies) supplemented with 10% heat-inactivated FBS (T24 cell assays). The infected monolayers were incubated at 37°C for 2 h (HeLa cells) or 1 h (T24 cells) in 5% CO2 and then washed four times with PBS to remove nonadherent bacteria. The monolayers (n = 4) were lysed with 0.01% Triton X-100 in distilled water, and the lysates were diluted in PBS to obtain quantitative colony counts of bacteria on agar. Data pooled from a minimum of three independent experiments are presented as the mean number of CFU/ml plus the standard error of the mean (SEM).
Genome sequencing and assembly.
The genome sequence of E. coli VR50 was determined using a combination of 454 (Roche) pyrosequencing (∼24× coverage) and traditional Sanger sequencing (∼1× coverage) (Australian Genome Research Facility). The following reads were included in the final assembly. (i) 1.2 million 454 GS20 shotgun reads (average read length, 101 bp); (ii) 85,155 454 GS-FLX 20-bp mate pair reads (average distance between reads, 2 kb); (iii) 7,680 Sanger mate pair reads from a pUC clone library with an insert size range of 5 to 8 kb; and (iv) 384 Sanger end sequences from 192 gap-spanning PCR products. A hybrid assembly of 25 scaffolds (230 contigs) using GS-20, GS-FLX, and Sanger reads was produced using the GS De Novo Assembler (version 1.1.02.15) (Roche). The physical map of the chromosome was also determined by OpGen Technologies, Inc. (Madison, WI), using the restriction enzyme NcoI and the optical-mapping technique. The order and orientations of the scaffolds were confirmed by aligning the scaffolds on the optical map using Opgen Mapviewer. Gaps between linked contigs were closed either by walking on gap-spanning clones or with PCR products generated from genomic DNA. While the chromosome was finished to high quality, all attempts to unambiguously resolve the plasmid-related contigs were unsuccessful due to their highly repetitive nature. Therefore, two PacBio single-molecule real-time (SMRT) cells were carried out using the Pacific Biosciences RSII platform to close the plasmids and complete the genome.
PacBio sequencing was carried out by shearing two aliquots of ∼4 μg genomic E. coli VR50 DNA, using g-Tube (Covaris), into fragments size targeted at 10 kb. The sheared samples were then purified and concentrated using washed Agencourt AMPure XP magnetic beads (Beckman Coulter Inc.) with size selection at 0.45-fold volume. Subsequently, SMRTbell template libraries were prepared using the commercial template preparation kit from Pacific Biosciences Inc. In brief, the DNA was end repaired, and adapters were ligated, followed by exonuclease digestion of incompletely ligated products. Using the provided P4-C2 DNA/polymerase binding kit from Pacific Biosciences, 0.83 nM libraries was then annealed with sequencing primers and bound to 50 nM P4 DNA polymerase. For enhanced loading efficiency into the sequencing zero-mode waveguides (ZMWs), 15 pM bound complexes was immobilized into Magbeads (Pacific Biosciences Inc.) according to the accompanying protocols. During sequencing, the duration of the sequence collection was set at 120 min with the stage start option. Upon acquisition of the sequencing data, short reads that were less than 50 bp were filtered off, and the minimum polymerase read quality was set at 0.75.
De novo genome assemblies of PacBio sequence reads were produced using SMRT Portal (v2.0.0) and the hierarchical genome assembly process (HGAP) (27), with default settings and a seed read cutoff length of 5,000 bp to ensure accurate assembly across E. coli rRNA operons. The hybrid 454/Sanger and PacBio assemblies were compared using MUMmer3 (28), Artemis Comparison Tool (29), and mauve (30), and discrepancies between the assemblies were investigated at the read level using BAMview (31).
Detection of DNA modifications was carried out using the RS_Modification_and_Motif_Analysis.1 tool from the SMRT analysis package version 2.2.0. In brief, PacBio reads were mapped to the complete VR50 genome. The polymerase kinetics, interpulse durations (IPDs) (32), were measured for each base, and methylated bases were identified by comparison of IPD ratios against an in silico kinetic reference model (details are available from Pacific Biosciences) and against the expected IPD signatures of the three bacterial methylation types: m6A, m4C, and m5C (33). Sequence motifs were identified using PacBio Motif finder v1 with a quality value (QV) cutoff of 30.
Genome annotation and comparative analysis.
Annotation of the VR50 genome was performed by transferring the high-quality annotation of E. coli K-12 MG1655, used as a main reference, and E. coli EDL933, UTI89, CFT073, and 536 when relevant, using the Rapid Annotation Transfer Tool (RATT) (34). Complementary annotation data were provided by the Rapid Annotations Using Subsystems Technology (RAST) server (35). Manual curation was also performed to ensure the accuracy of the annotations transferred, with particular attention to regions of differences, prophage-related regions, and pseudogenes. BRIG (36), Artemis Comparison Tool (29), and mauve (30) were used to visually compare the VR50 assembly with various other genomes in order to identify regions of similarity and difference. High-quality single-nucleotide polymorphisms (SNPs) (bounded by 20 exact base pair matches on both sides) in pairwise genome comparisons were calculated using MUMmer3 (28).
An in silico multilocus sequence type (MLST) comparison of seven housekeeping genes from E. coli VR50 and representative complete E. coli genomes (Table 1) was carried out by retrieving the sequence fragments for adk, fumC, gyrB, icd, mdh, purA, and recA (http://mlst.warwick.ac.uk/mlst/). To more accurately determine the phylogenetic relatedness of E. coli VR50 to other E. coli strains, a SNP-based tree was constructed. In silico-simulated reads for five fully sequenced group A strains (HS, BW2952, DH10, W3110, and MG1655) and 12 additional E. coli strains (IAI1, EDL933, Sakai, UMN026, IAI39, SE15, S88, APEC O1, UTI89, ED1a, CFT073, and S38) were mapped against the complete genome of VR50 using SHRIMP 2.0 (37). Simulated 150-bp paired reads with an insert length of 250 bp were generated at 150-fold coverage using a custom program (available on request). SNP calling and indel prediction were performed using the Nesoni package (Victorian Bioinformatics Consortium), and all conserved polymorphic positions in the data set were identified using the Nesoni n-way pairwise comparative-analysis tool. Polymorphic substitution sites were extracted and concatenated, and the resulting alignment was used for phylogenetic-tree construction. An unrooted maximum-likelihood (ML) phylogenetic tree based on the SNP alignment was constructed with RAxML 7.2.8 (38) using the GTR substitution model and 1,000 bootstrap replicates. The tree was plotted using FigTree 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/).
TABLE 1.
Summary of genome features of publicly available E. coli strains used in this study
| Organism | Pathotypea (host) | Size of chromosome (bp)b | GC content (%)b | No. of genesb | No. of pseudogenesb | No. of rRNAsb | No. of tRNAsb | No. of prophage regionsb | No. of plasmidsb | GenBank accession no. |
|---|---|---|---|---|---|---|---|---|---|---|
| E. coli OR:K1:H− strain VR50 | ABU (human) | 5,000,386 | 50.8 | 4,711 | 125 | 22 | 85 | 8 | 9 | CP011134 |
| E. coli O18:K1:H7 strain UTI89 | UPEC (human) | 5,065,741 | 50.6 | 5,066 | NAc | 22 | 88 | NA | 1 | CP000243.1 |
| E. coli O6:K15:H31 strain 536 | UPEC (human) | 4,938,920 | 50.5 | 4,685 | NA | 22 | 81 | 1 | 0 | CP000247.1 |
| E. coli OR:K5:H− strain ABU 83972 | ABU (human) | 5,131,397 | 50.6 | 4,793 | NA | 22 | 88 | 6 | 1 | CP001671 |
| E. coli O6:H1:K2 strain CFT073 | UPEC (human) | 5,231,428 | 50.5 | 5,379 | 94 | 21 | 89 | 5 | 0 | AE014075.1 |
| E. coli O7:K1 strain IAI39 | UPEC (human) | 5,132,068 | 50.6 | 5,132 | 80 | 22 | 88 | NA | 0 | CU928164.2 |
| E. coli O17:K52:H18 strain UMN026 | UPEC (human) | 5,202,090 | 50.7 | 4,918 | 45 | 22 | 88 | NA | 2 | CU928163.2 |
| E. coli O25b:H4-ST131 strain EC958 | UPEC (human) | 5,109,767 | 50.7 | 4,981 | 25 | 22 | 89 | 7 | 2 | HG941718.1 |
| E. coli O18:K1:H7 strain IHE3034 | NMEC (human) | 5,108,383 | 50.7 | 4,757 | NA | 21 | 97 | NA | 0 | CP001969 |
| E. coli O45:K1 strain S88 | NMEC (human) | 5,032,268 | 50.7 | 5,049 | 90 | 22 | 91 | NA | 1 | CU928161.2 |
| E. coli OR:H48 strain K-12 substrain W3110 | K-12 (human) | 4,646,332 | 50.8 | 4,227 | NA | 22 | 86 | NA | 0 | AP009048.1 |
| E. coli OR:H48 strain K-12 substrain MG1655 | K-12 (human) | 4,639,675 | 50.8 | 4,294 | 81 | 22 | 89 | 10 | 0 | U00096.2 |
| E. coli OR:H48 strain K-12 substrain DH10B | K-12 (human) | 4,686,137 | 50.8 | 4,200 | NA | 22 | 86 | 10 | 0 | CP000948.1 |
| E. coli OR:H48 strain K-12 substrain BW2952 | K-12 (human) | 4,578,159 | 50.8 | 4,098 | NA | 22 | 88 | NA | 0 | CP001396.1 |
| E. coli BL21(DE3) | Commensal (human) | 4,570,938 | 50.8 | 4,336 | 70 | 22 | 85 | NA | 0 | CP001509 |
| E. coli B strain REL606 | Commensal (human) | 4,629,812 | 50.8 | 4,383 | 67 | 22 | 85 | 10 | 2 | CP000819.1 |
| E. coli O152:H28 strain SE11 | Commensal (human) | 4,887,515 | 50.7 | 4,679 | NA | 22 | 86 | 7 | 6 | AP009240.1 |
| E. coli O8 strain IAI1 | Commensal (human) | 4,700,560 | 50.8 | 4,491 | 51 | 22 | 86 | NA | 0 | CU928160.2 |
| E. coli O9:H4 strain HS | Commensal (human) | 4,643,538 | 50.8 | 4,478 | 94 | 22 | 88 | NA | 0 | CP000802.1 |
| E. coli O81 strain ED1a | Commensal (human) | 5,209,548 | 50.7 | 4,915 | 95 | 22 | 91 | NA | 1 | CU928162.2 |
| E. coli O150:H5 strain SE15 | Commensal (human) | 4,717,338 | 50.7 | 4,338 | NA | 22 | 84 | 2 | 1 | AP009378 |
| E. coli W | W (human) | 4,900,968 | 50.8 | 4,764 | 91 | 22 | 87 | 7 | 2 | CP002185 |
| E. coli ATCC 8739 | C (human) | 4,746,218 | 50.9 | 4,409 | 82 | 22 | 87 | 8 | 0 | CP000946.1 |
| E. coli O19:H34 strain SMS-3-5 | Environmental | 5,068,389 | 50.5 | 4,743 | 78 | 22 | 90 | 4 | 4 | CP000970.1 |
| E. coli O178:H11 strain H10407 | ETEC (human) | 5,153,435 | 51.2 | 4,746 | NA | 22 | 87 | 9 | 4 | FN649414 |
| E. coli O139:H28 strain E24377A | ETEC (human) | 4,979,619 | 50.6 | 4,873 | 118 | 21 | 91 | NA | 6 | CP000800.1 |
| E. coli O55:H7 strain CB9615 | EPEC (human) | 5,386,352 | 50.5 | 5,028 | 69 | 22 | 101 | 19 | 1 | CP001846.1 |
| E. coli O127:H6 strain E2348/69 | EPEC (human) | 4,965,553 | 50.8 | 4,703 | 145 | 22 | 92 | 13 | 2 | FM180568.1 |
| E. coli O157:H7 strain EC4115 | EHEC (human) | 5,572,075 | 50.4 | 5,315 | NA | 22 | 110 | NA | 2 | CP001164.1 |
| E. coli O157:H7 strain TW14359 | EHEC (human) | 5,528,136 | 50.5 | 5,263 | NA | 22 | 106 | NA | 1 | CP001368.1 |
| E. coli O157:H7 strain Sakai | EHEC (human) | 5,498,450 | 50.5 | 5,361 | 247 | 22 | 105 | 18 | 1 | BA000007.2 |
| E. coli O157:H7 strain EDL933 | EHEC (human) | 5,528,445 | 50.4 | 5,349 | NA | 22 | 98 | 24 | 2 | AE005174.2 |
| E. coli O103:H2 strain 12009 | EHEC (human) | 5,449,314 | 50.6 | 5,054 | 209 | 22 | 98 | 15 | 1 | AP010958.1 |
| E. coli O26:H11 strain 11368 | EHEC (human) | 5,697,240 | 50.6 | 5,364 | 254 | 22 | 101 | 21 | 4 | AP010953.1 |
| E. coli O111:H− strain 11128 | EHEC (human) | 5,371,077 | 50.4 | 4,972 | 291 | 22 | 106 | 17 | 5 | AP010960.1 |
| E. coli O44:H18 strain 042 | EAEC (human) | 5,241,977 | 50.5 | 4,810 | 110 | 22 | 93 | 9 | 1 | FN554766 |
| E. coli 55989 | EAEC (human) | 5,154,862 | 50.7 | 4,969 | 79 | 22 | 94 | NA | 1 | CU928145.2 |
| E. coli O1:K1:H7 strain APECO1 | APEC (chicken) | 5,082,025 | 50.6 | 4,467 | NA | 22 | 93 | 10 | 4 | CP000468.1 |
| E. coli UM146 | AIEC (human) | 4,993,013 | 50.6 | 4,650 | NA | 22 | 86 | NA | 1 | CP002167 |
| E. coli O83:H1 strain NRG 857C | AIEC (human) | 4,747,819 | 50.7 | 4,431 | NA | 22 | 84 | 5 | 1 | CP001855 |
| E. coli O83:H1 strain LF82 | AIEC (human) | 4,773,108 | 49.9 | 4,376 | NA | 22 | 84 | 4 | 1 | CU651637 |
AIEC, adherent-invasive E. coli; APEC, avian pathogenic E. coli; EAEC, enteroaggregative E. coli; EHEC, enterohemorrhagic E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli; NMEC, neonatal meningitis E. coli; UPEC, uropathogenic E. coli.
Values are given according to information retrieved from the sequence annotation combined with data from the literature, when available.
NA, not available.
Quantification of secreted IL-6.
The level of interleukin 6 (IL-6) secreted by HeLa epithelial cells was quantified by enzyme-linked immunosorbent assay (ELISA) as described by the manufacturer (R&D Systems). Culture supernatants from E. coli-stimulated epithelial cells were collected 2 h following infection.
Mouse model of UTI.
The mouse model of UTI was used as described previously (39). Female C57BL/6 mice (8 to 10 weeks old) were purchased from the Animal Resources Center, Western Australia, and housed in sterile cages with ad libitum access to sterile water. An inoculum of 25 μl, containing 5 × 108 CFU of bacteria in PBS, was instilled directly into the bladder using a 1-ml tuberculin syringe attached to the catheter. Urine was collected from each mouse 18 h after inoculation for quantitative colony counts. Groups of mice were euthanized by cervical dislocation 18 h after challenge; the bladders were then excised aseptically, weighed, and homogenized in PBS. The bladder homogenates were serially diluted in PBS and plated onto LB agar for colony counts. Data are expressed as the total number of CFU per gram of bladder tissue for each mouse. Experiments were performed with a minimum group size of eight.
Nucleotide sequence accession numbers.
The complete chromosome (GenBank accession no. CP011134) and plasmid (GenBank accession no. CP011135 to CP011143) sequences have been deposited at DDBJ/EMBL/GenBank (Bioproject PRJEA61445). The version described in this paper is version 1.
RESULTS
Phenotypic characteristics of E. coli VR50.
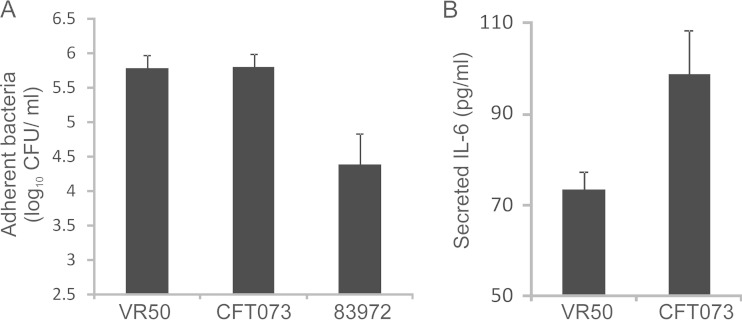
E. coli VR50 (OR:K1:H−) is an ABU isolate that belongs to phylogroup A. The strain was isolated from a 30-year-old otherwise healthy woman who had carried it for at least 1 year without any adverse effects. E. coli VR50 causes mannose-sensitive (MS) agglutination of yeast cells (indicative of type 1 fimbria expression) and mannose-resistant (MR) hemagglutination of human type A red blood cells (20). E. coli VR50 is also nonmotile and adheres strongly to HeLa epithelial cells, with the level of adhesion equal to that of the well-characterized pyelonephritis strain CFT073 and significantly greater than that of the ABU strain E. coli 83972 (Fig. 1A). Since E. coli VR50 is an ABU strain that was responsible for a prolonged infection in the absence of symptoms, we hypothesized that it might fail to trigger a strong proinflammatory response in comparison to other virulent UPEC strains. To test this, we compared the abilities of E. coli VR50 and CFT073 to stimulate IL-6 production following adhesion to epithelial cells. The level of secreted IL-6, which is an important proinflammatory cytokine associated with UTI, was quantified by ELISA. Despite equivalent adherence, the level of IL-6 secreted following adhesion of E. coli VR50 was significantly lower than the level of IL-6 secreted following adhesion of CFT073 (P < 0.001) (Fig. 1B). Taken together, these characteristics prompted us to perform an in-depth analysis of E. coli VR50, as described below.
FIG 1.
(A) Adhesion of E. coli VR50 and 83972 (ABU strains) and CFT073 (UPEC strain) to HeLa human epithelial cells. Epithelial cells were inoculated with bacteria and incubated for 1 h, with adherent bacteria enumerated by direct plating and colony counts. The results are the averages of two independent experiments plus standard errors of the mean. E. coli VR50 adhered to the cells in equal numbers to the UPEC strain CFT073. (B) ELISA demonstrating levels of IL-6 secreted by HeLa epithelial cells infected with E. coli VR50 and CFT073. The results are the averages of four replicates plus standard errors of the mean. E. coli VR50 stimulated a significantly reduced IL-6 response compared to CFT073, despite a similar level of adherence (P < 0.001).
Genome features of E. coli VR50.
The complete genome of E. coli VR50 was determined and includes a 5,000,366-bp circular chromosomal sequence and 9 plasmid sequences. The VR50 chromosome consists of 4,711 annotated protein-coding regions, including 125 predicted pseudogenes, 22 rRNAs, and 85 tRNAs, with a G+C content of 50.78%. In addition to the chromosome, we also identified several plasmid-associated sequences. Besides 7 small plasmids, with sizes ranging from ∼1.7 kb up to ∼7 kb, we identified two large plasmids, pVR50A and pVR50B, with estimated sizes of 70.5 kb and 97.9 kb, respectively. The detailed results of genome analysis of VR50 and 40 other sequenced E. coli strains can be found in Table 1.
In addition to determining the VR50 genome sequence, we took advantage of PacBio SMRT sequencing to detect DNA base modifications and to produce a genome-wide methylation profile. The genome of VR50 had adenine methylated exclusively, with the majority (96.3%) of m6A modifications occurring at 5′-Gm6ATC-3′ deoxyadenosine methylase (Dam) motifs (Table 2). A total of 39,361 (98%) of 40,144 Dam sites contained m6A modifications. This frequency of Dam methylation in VR50 is consistent with previously characterized E. coli methylomes (40–42). In addition to Dam, VR50 encodes a single active type I restriction-modification (R-M) system in a typical type I operon (hsdR-hsdM-hsdS) (Table 2). The coding sequences in the operon share 100% amino acid sequence identity with the previously characterized type I R-M system EcoB from E. coli B; this methylase is predicted to recognize the motif 5′-TGm6AN8TGCT-3′ (methylated base on opposite strand underlined) (Table 2). It should be noted that no ten-eleven translocation (TET)-converted sample DNA was sequenced, and thus, m5C methylated bases could not be discriminated from unmodified bases.
TABLE 2.
Active methyltransferase enzymes identified in VR50
| Locus tag | Coordinates | Type | R-M/orphan | Target sequencea | No. in genome | No. detectedb | % Methylatedb | Comments |
|---|---|---|---|---|---|---|---|---|
| ECVR50_3796 | 3831641–3832477 | II | Orphan | Gm6ATC | 40,144 | 39,361 | 98 | 100% amino acid sequence identity to M.EcoKDam from E .coli K-12 MG1655 |
| ECVR50_4827 to ECVR50_4829 | 4942889–4946401 | II | R-M | TGm6AN8TGCT | 775 | 755 (751) | 97.4 (96.9) | 100% amino acid sequence identity to M.EcoBI and S.EcoBI from E. coli B |
Underlined nucleotides reflect methylated bases on opposite DNA strand.
Figures for the complementary strand are in parentheses and suggest hemimethylation at 4 of 755 TGm6AN8TGCT sites.
The E. coli VR50 genome is most similar to that of the K-12 strain MG1655.
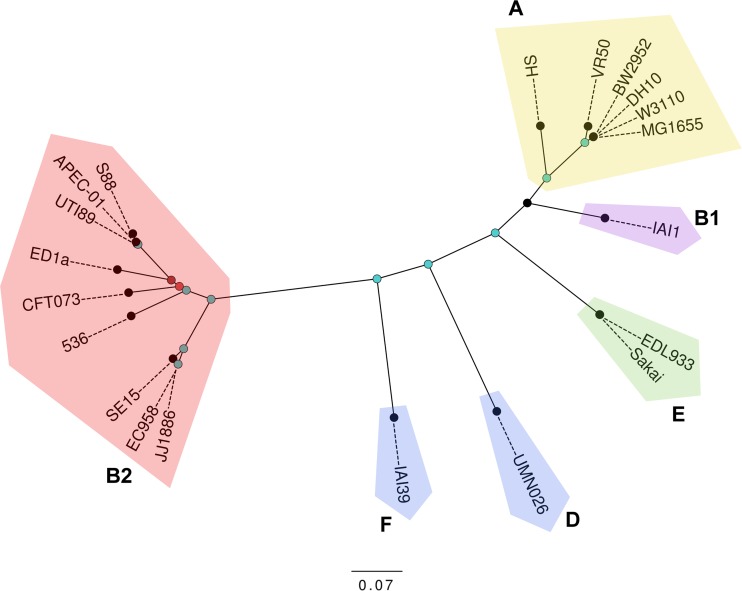
To further investigate the phylogenetic relationship of E. coli VR50 to other sequenced E. coli strains, an in silico MLST comparison was carried out. VR50 was found to belong to the sequence type (ST) 10 complex and was indistinguishable from the K-12 strains MG1655 and W3110. Additional housekeeping genes (i.e., arcA, aroA, mtlD, pgi, and rpoS) and the fimH adhesin gene (43) were also found to be identical between the K-12 and VR50 genomes. Whole-genome nucleotide comparisons confirmed that VR50 belongs to phylogroup A and is most closely related to K-12 strains (Fig. 2). Pairwise comparisons between complete genomes of phylogroup A E. coli strains indicated that the genomes of VR50 and K-12 MG1655 differ by 3,953 high-quality SNPs, with both strains differing from E. coli HS, the most divergent phylogroup A strain, by approximately 14,500 SNPs (Fig. 2). Although the genomes of VR50 and K-12 MG1655 are largely syntenic, VR50 exhibits two large inversions (12.4 kb and 54.2 kb) flanked by IS629 elements. The IS629 element has expanded to 19 chromosomal copies after acquisition by VR50, contributing to the larger number of pseudogenes observed in the genome (Table 1).
FIG 2.
Whole-genome nucleotide comparison of VR50 with selected sequenced E. coli genomes. Shown is a maximum-likelihood phylogenetic comparison of VR50 with 5 phylogroup A and 14 other representative E. coli strains. Phylogenetic relationships were inferred from 212,716 polymorphic SNPs and 1,000 bootstrap replicates. Major E. coli phylogroups are labeled. Nodes are colored according to bootstrap support: 1,000 (blue), 960 (dark red), and 630 (light red). The scale bar corresponds to the mean number of nucleotide substitutions per site.
E. coli VR50 has acquired multiple mobile genetic elements.
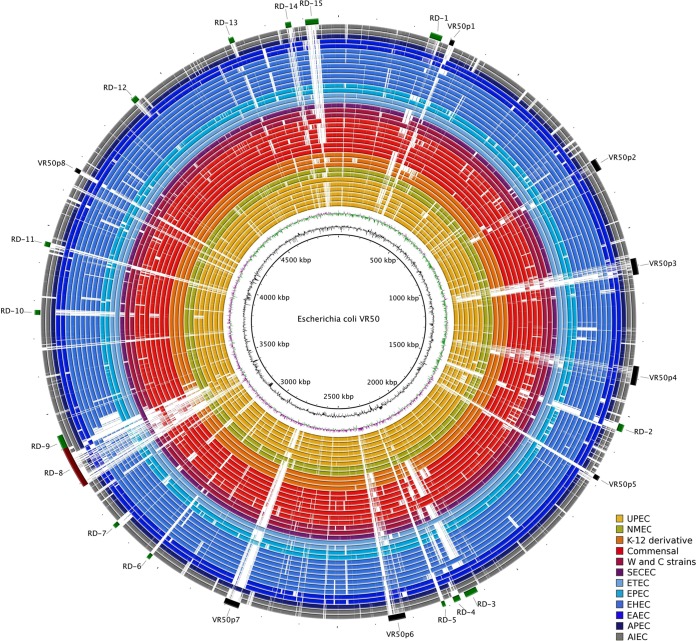
To identify genomic regions that may be associated with the ability to persist in the urinary tract, we compared the VR50 chromosome to other representative complete E. coli genomes using BLAST. We defined 15 regions of differences (RDs) (Table 3) and 8 prophages (Table 4) that were differentially distributed among 39 complete representative E. coli genomes (Fig. 3). RD-8 is the largest of these regions and carries a 94-kb phe tRNA-associated integrative island that we refer to as GI-VR50-pheV. VR50 also carries a 32-kb asn tRNA-associated island (RD-3) that shares 99% identity with the high-pathogenicity island from Yersinia pestis (44). While RD-3 is present in all UPEC strains and absent from most K-12 derivative strains and commensals, it is found in the laboratory strain BL21(DE3) and the B strain REL606.
TABLE 3.
Major regions of differences identified in VR50 compared to other E. coli strains
| Identifier | Start nucleotide | End nucleotide | Size (bp) | Insertion site | Virulence-related or other notable gene(s) | Description |
|---|---|---|---|---|---|---|
| RD-1 | 244012 | 277704 | 33,692 | aspV tRNA | yafT | Lipoprotein |
| RD-2 | 1527896 | 1547892 | 19,996 | ydbL_ynbG | feaRB, paaABCDEFGHIJKXY | Phenylacetic acid degradation |
| RD-3 | 2125253 | 2160033 | 34,780 | asnT tRNA | Yersiniabactin-biosynthetic cluster | 99% similar to high-pathogenicity island (HPI) from Yersinia pestis |
| RD-4 | 2172863 | 2192851 | 19,988 | yeeHa | flu1 | Antigen 43 phase-variable biofilm formation autotransporter |
| RD-5 | 2216480 | 2225524 | 9,044 | gnd_galF | O-antigen region | |
| RD-6 | 3037592 | 3047232 | 9,640 | iap_cysH | CRISPR locus | CRISPR 2.1 |
| RD-7 | 3157781 | 3169951 | 12,170 | glyU tRNA | ETT2 cluster remnant | T3SSb-related genes; similar to ETT2 sepsis (eprKa, epaS1a, eivJ2a, and eivICAEGF missing) |
| RD-8 | 3295104 | 3407870 | 112,766 | pheV tRNA | pap cluster remnant; iuc, iha, flu2, sat, and afaABCDE kps cluster | GI-VR50 pheV capsular biosynthesis gene cluster |
| RD-9 | 3407872 | 3442920 | 35,048 | kpsM_yghG | gsp1 glcABGFEDC | T2SSc-related genes (general secretory pathway); glycolate metabolism genes |
| RD-10 | 3770072 | 3782784 | 12,712 | rpsJ_bfr | gsp2 cryptic cluster | T2SS-related genes (general secretory pathway; cryptic) |
| RD-11 | 3952087 | 3965106 | 13,019 | yhiM_yhiN | Tn7-related genes | |
| RD-12 | 4403223 | 4419794 | 16,571 | typA_dtd | yihLMN, ompL, and yihOPQRSTUVWXY | Sugar catabolism |
| RD-13 | 4704726 | 4718639 | 13,913 | pheU tRNA | fecIRABCDE regulon | Iron dicitrate transport |
| RD-14 | 4859525 | 4874849 | 15,324 | leuX tRNA | Putative oxidoreductase putative transporters | |
| RD-15 | 4912259 | 4947503 | 35,244 | nanS_yjiA | hsdRMS, mrr, and mcrBC sgcXBCQAER | Restriction-modification enzymes pentose and pentitol sugar breakdown |
Pseudogene.
T3SS, type III secretion system.
T2SS, type II secretion system.
TABLE 4.
Prophage-related regions identified in VR50
| Identifier | Start nucleotide | End nucleotide | Size (bp) | Insertion site | Direct repeat (bp) | Type | Virulence-related or other notable gene(s) |
|---|---|---|---|---|---|---|---|
| VR50p1 | 300479 | 312346 | 11,868 | thrW tRNA | 16 | P4-like | |
| VR50p2 | 800678 | 832594 | 31,917 | ybhJb_ybhB | NDc | Lambdoid, remnant | Outer-membrane porin proteins; Lom-like outer membrane proteins |
| VR50p3a | 1082316 | 1125788 | 43,473 | torS_torT | ND | P22-like | |
| VR50p4a | 1369562 | 1420434 | 50,873 | ompW | 39 | Lambdoid | Lom-like outer membrane proteins |
| VR50p5 | 1678552 | 1691947 | 13,396 | yneLb_hipAb | ND | Untypeable; remnant | |
| VR50p6a | 2324694 | 2371668 | 46,705 | mrlAb | 18 | Lambdoid | Putative DinI-like damage-inducible proteins; outer-membrane porin proteins; Lom-like outer-membrane proteins |
| VR50p7a | 2770918 | 2811852 | 40,935 | yfgI_guaA | ND | Epsilon-like | DNA adenine methylase |
| VR50p8 | 4163270 | 4174321 | 11,052 | selC tRNA | ND | P4-like |
Prophage genomes appear to have all the genes necessary to produce fully functional phages.
Pseudogene.
ND, not determined.
FIG 3.
BRIG visualization of the E. coli VR50 genome compared with selected complete E. coli genomes colored according to the strain group or pathotype. The innermost circles represent the GC content (black) and GC skew (purple/green) of the E. coli VR50 chromosome. BRIG shows the results of unfiltered BLASTn searches of E. coli strains against VR50, arranged from inner to outer colored circles as follows: UPEC strains UTI89, 536, ABU83972, CFT073, IAI39, and UMN026 (yellow); NMEC strains IHE3034 and S88 (green); K-12 derivative strains W3110, MG1655, and DH10B (orange); commensal strains BL21(DE3), REL606, SE11, IAI1, HS, ED1a, and SE15 (red); clone W and ATCC 8739 (clone C) strains (maroon); strain SMS-3-5 (purple); ETEC (light blue); EPEC (aqua); enterohemorrhagic E. coli (EHEC) (blue); EAEC (dark blue); APEC (navy); and adherent-invasive E. coli (AIEC) (gray). The color intensity is proportional to the BLASTn identity, with dark regions having high nucleotide identity and light regions having little or no nucleotide identity. E. coli VR50 genomic features are annotated around the outermost circle: prophage regions, VR50p1 to -8, in black and regions of differences, RD-1 to -15 in green, with RD-8 depicted in brown to allow differentiation from the contiguous RD-9.
Most of the prophage-related regions identified in the VR50 genome (VR50p1 to VR50p8) (Table 4) were not found in K-12 or other commensal strains. Of these, 3 are lambdoid prophages (VR50p2, VR50p4, and VR50p6) and have similarity to each other and to the lambdoid prophages in other E. coli genomes, with the exception of most K-12 derivative strains, from which they are absent. Of note, VR50p6 is inserted in the mrlA gene, which encodes a regulator required for curli production and extracellular matrix formation (45). VR50p3 is a P22-like phage, sharing similarities with the only P22-like phage found in UTI89, whereas prophages VR50p1 and VR50p8 are similar to the satellite phage P4 (Fig. 3). VR50p7 is related to the serotype-converting Salmonella phage epsilon 15 and to Eco39I-1 in UPEC strain IAI39 but does not carry any of the genes involved in serotype conversion.
The plasmid pVR50A is an F-like conjugative plasmid, sharing nearly 90% of its backbone with pUTI89, with a nucleotide similarity of ∼99%. A significant portion of pUTI89 is missing in pVR50A and is replaced by a multidrug resistance cluster that includes several antibiotic resistance genes, including strAB (which confers resistance to streptomycin) and folP (which confers resistance to sulfonamide). Of note, a copy of the blaTEM-1 gene, associated with ampicillin resistance, was also identified, but a truncation of its N terminus suggests its inactivation, as confirmed by antibiotic sensitivity phenotypic tests (data not shown). The other large plasmid found in VR50, pVR50B, possesses a complex mosaic structure comprising multiple insertion sequence (IS) elements, with regions of similarity to numerous UPEC-, neonatal meningitis E. coli (NMEC)-, and commensal-associated plasmids, including pCE10A (strain CE10; NMEC), pECOED (strain ED1a; commensal), pSE11-2 (SE11; commensal), and pUTI89 (UTI89; UPEC). Plasmid pVR50B carries a tra region that is almost identical to the corresponding region in pVR50A; the sequences of these two plasmids could only be properly resolved using PacBio sequencing. While pVR50B does not contain any antibiotic resistance genes, several putative virulence-associated cargo genes were identified, including senB (which encodes a putative enterotoxin), imm (which encodes a colicin Ib immunity protein), and the toxin-antitoxin phd-doc genes. Of note, VR50 also harbors several small plasmids that are very similar to cryptic plasmids often associated with other commensal strains; for example, pVR50C and pVR50D are similar to pColE1-like plasmids, and pVR50F is similar to pSE11-6.
Taken together, our data suggest E. coli VR50 is a commensal strain, remarkably similar to E. coli K-12, which has gained the ability to persist in the urinary tract through gene acquisition. This is in stark contrast to the prototypical ABU E. coli strain 83972, which appears to have evolved through mutation and gene loss from a virulent UPEC strain (9, 10, 15–17, 46).
The pheV genomic island in E. coli VR50 includes uropathogenicity genes.
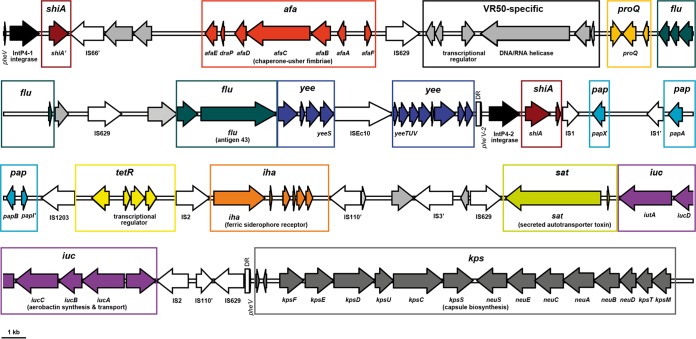
The most notable chromosomal variation between E. coli VR50 and MG1655 is a contiguous gene cluster in VR50 comprising the 94-kb genomic island GI-VR50-pheV (RD-8), a kps capsular biosynthesis gene cluster, and a gsp type II secretion system gene cluster (RD-9). GI-VR50-pheV is located between the pheV tRNA gene and the ksp gene cluster (Fig. 4). It is delimited by direct repeats (DR) that encompass the last 19 bp of the pheV tRNA gene and a truncated tRNA gene (phe′V), equivalent to attL and attR, left and right junction sites generated by phage insertion (47). Consistent with the features of many other integrative islands, a P4-like integrase gene is located immediately adjacent to the 5′ end of GI-VR50-pheV. The GC content of GI-VR50-pheV is 48.11% compared with the E. coli VR50 genomic average of 50.78%. GI-VR50-pheV contains several genes associated with UPEC virulence, including (i) remnants of the pap cluster; (ii) the iuc aerobactin biosynthesis locus and the iha siderophore receptor; (iii) two autotransporter (AT)-encoding genes, flu (encoding antigen 43 [Ag43]) and sat (encoding a secreted AT toxin); and (iv) the afaABCDE genes, the Afa/Dr adhesin family chaperone-usher fimbrial genes.
FIG 4.
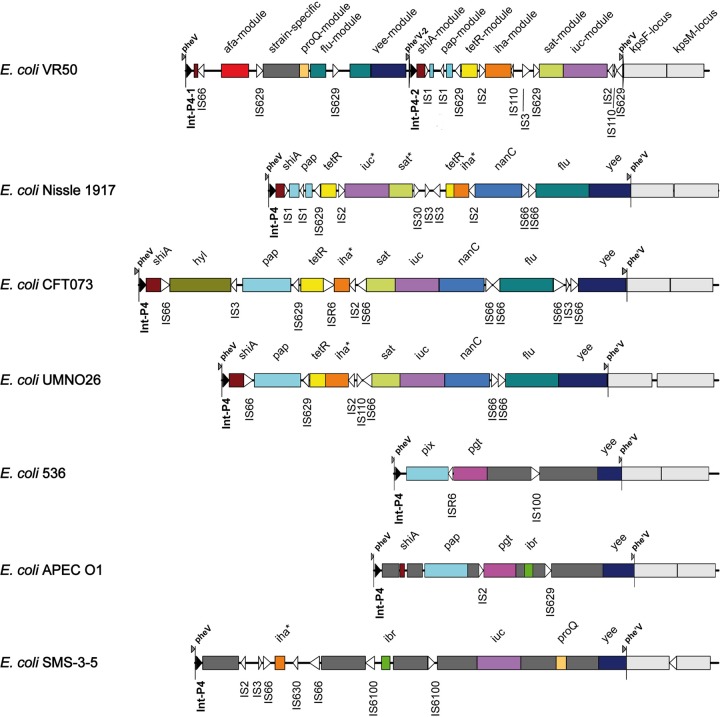
Genetic locus map of the 94-kb GI-VR50-pheV island and adjacent kps capsular biosynthesis gene cluster from E. coli VR50 (drawn to scale). The arrows showing genes are colored according to the modules shown in Fig. 5. IntP-like integrase genes, the pheV tRNA gene, and pheV repeats (phe′V and phe′V -2) are colored black. Insertion sequences are shown as white arrows and may contain more than one coding sequence (CDS). Direct repeats (172 nt) are shown as boxes adjacent to phe′V and phe′V -2. Degenerate or truncated genes or insertion sequences are labeled with an apostrophe. GI-VR50-pheV is a composite genomic island formed from two independent integrase-mediated insertions.
The gene content of GI-VR50-pheV is most similar to those of pheV-associated genomic islands in the probiotic strain Nissle 1917, as well as UPEC strains CFT073, UMN026, 536, and IAI39 (Table 5 and Fig. 5). Like other pheV islands, GI-VR50-pheV has a mosaic structure of defined sequence modules separated by a variety of insertion sequences (Fig. 5). Modules encoding virulence factors are highly conserved at the nucleotide level (i.e., typically >95% nucleotide identity), although not necessarily in the same order or orientation or at the same level of completeness. Intervening regions typically contain strain-specific sequences or insertion sequence elements that presumably facilitate the many rearrangements observed between islands. For example, the 28,718-nt region containing the sat, iuc, and iha modules in VR50 is 99% identical to equivalent modules in the Nissle 1917, CFT073, and UMN026 pheV islands; however, sat and iuc are inverted in Nissle 1917 and iha is inverted in VR50 (Fig. 5) (48). Although in Nissle 1917 this region is also flanked by IS2-like sequences, the insertion sequences found between these modules are different in all four strains, reflecting the dynamic evolution of genomic islands (Fig. 5).
TABLE 5.
Characteristics of pheV genomic islands from different E. coli strains
| Genomic islanda | Alternate name | Size (nt)b | Genome position (nt) | Accession no.c |
|---|---|---|---|---|
| GI-VR50-pheV | NAd | 93,983 | 3295095–3389077 | This study |
| GI-Nissle1917-pheV | GEI-IINISSLE1917 | 77,685 | NA | (AJ586888) |
| GI-CFT073-pheV | PAI-ICFT073 | 104,676 | 3406225–3510900 | AE014075 (AF081285) |
| GI-UMN026-pheV | NA | 82,784 | 3445924–3528707 | CU928163 |
| GI-536-pheV | PAI-V536 | 48,805 | 3128084–3176888 | CP000247 (AJ617685) |
| GI-APECO1-pheV | NA | 55,888 | 3305017–3360904 | CP000468 (DQ095216) |
| GI-SMS-3-5-pheV | NA | 91,940 | 3197159–3289098 | CP000970 |
| GI-S88-pheV | NA | 55,896 | 3219801–3275696 | CU928161 |
| GI-ED1a-pheV | NA | 135,753 | 3374807–3510560 | CU928162 |
| GI-55989-pheV | NA | 110,593 | 3339841–3450433 | CU928145 |
| GI-IAI39-pheV | NA | 38,534 | 3545466–3583999 | CU928164 |
| GI-E24377A-pheV | NA | 101,766 | 3325510–3427276 | CP000800 |
The APECO1 and S88 pheV islands are virtually identical (99% at the nucleotide level); the transposition of a 2,581-nt insertion sequence is the only substantial difference (data not shown).
The size was calculated from the first nucleotide of pheV to the last nucleotide of the phe′V direct repeat. No repeat was identified in GI-ED1a-pheV or GI-IAI39-pheV, and the right-hand junction was taken as the breakpoint adjacent to the kpsF locus. E24377A contains a composite arrangement of two islands, similar to VR50 (data not shown).
The GenBank accession number for the complete genome sequence, where available. Accession numbers in parentheses refer to GenBank accession numbers for genomic islands, if published separately prior to the genome sequence. AF081285 covers only 13,710 bp of the CFT073 genomic island.
NA, not applicable.
FIG 5.
Comparison of the GI-VR50-pheV island with pheV genomic islands from other E. coli strains. The schematic diagram shows pheV genomic islands from E. coli strains VR50, Nissle 1917, CFT073, UMN026, 536, APECO1, and SMS-3-5. Conserved modules are colored the same and named for the best-known locus or gene. Strain-specific modules are colored dark gray. Insertion sequences (often comprising more than one CDS) are represented as unfilled arrowheads. The P4-like integrase sequences are represented as black arrowheads. The pheV and phe′V direct repeats denote the boundaries of each island. The kpsF and kpsM modules, which are not considered part of the island, are colored light gray; variation in the gene content of the kpsM module is not shown. Inversions of individual modules relative to the VR50 genome are shown with an asterisk adjacent to the module name. The diagram is drawn to scale.
The archetypal pheV island is from CFT073 (PAI-ICFT073). PAI-ICFT073 contains the sat, iha, iut, and flu gene modules with a high degree of identity to islands from Nissle 1917 and VR50; in addition, it includes the pap operon and a hylCABD operon. The mosaic structure of the pheV island is further illustrated by the fact that, although it is not present in the well-characterized UPEC strain UTI89, several modules similar to those on GI-VR50-pheV are found on other UTI89 islands (e.g., leuX). Although the K-12 and HS commensal strains do not carry a pheV island, it is found in the sequenced ED1a commensal strain (49). GI-ED1a-pheV contains fragments of modules typically associated with UPEC (i.e., iha and sat), with full-length iha and iuc loci on the nearby pheU island. The genomes of the Shiga toxin-producing E. coli (STEC) strains Sakai, EDL933, and EC4115 carry a large deletion encompassing pheV and the adjacent kps capsular and gsp type II secretion loci. However, the pheV tRNA gene is included in a nonsyntenic locus and is known to be a hot spot for insertion of the locus for enterocyte effacement (LEE) (50, 51). The afa-8 locus on a pheV genomic island has been described previously (52), although the associated int gene is like intP4-2, not intP4-1, and the VR50 afa locus is quite divergent from afa-8 (see below).
GI-VR50-pheV is formed from two independent integration events.
GI-VR50-pheV is a composite structure with evidence of two independent pheV island insertions mediated by different IntP4-like integrase genes (Fig. 5). As observed in the well-characterized GI-536-pheV pathogenicity island (PAI V536), GI-VR50-pheV is bound by direct repeats corresponding to the 3′ end of the pheV tRNA gene (Table 6). Like pheV islands in CFT073, Nissle 1917, and UMN026, the right-hand direct repeat (phe′V) is 19 nt and contains a substitution (Table 6). Strains 536 and APECO1 share a different phe′V sequence, corresponding to a duplication of the 23 nt at the 3′ end of pheV and a single-nucleotide deletion (Table 6) (4, 53), whereas, GI-SMS-3-5-pheV is bounded by a set of perfect 19-nt direct repeats. Intriguingly, within GI-VR50-pheV there is a perfect 19-nt direct repeat of the pheV 3′ region (phe′V -2) immediately upstream of a different P4-like integrase gene (intP4-2) (Fig. 5 and Table 6). IntP4-1 and IntP4-2 share only 43% amino acid identity, making it certain that they are not the result of an internal duplication. A comparison of integrase sequences associated with pheV islands indicated that VR50 intP4-1 shares 94 to 95% nucleotide identity with the pheV-associated integrase genes of SMS-3-5 and E24377A. VR50 intP4-2 shares at least 94% nucleotide identity with the integrase genes of most other pheV islands from E. coli genomes (e.g., Int PAI V536) and 98 to 99% identity with int genes from Nissle 1917, CFT073, and UMN026 pheV islands.
TABLE 6.
Alignment of pheV and phe′V DR at genomic-island boundaries
| pheV island | Direct-repeat sequencea | Descriptor |
|---|---|---|
| VR50 pheV | TTCGATTCCGAGTCCGGGCACCA | 54–76 |
| VR50 phe′V | TTTCATTCCGATTCCGGGCACCA | 19/19-nt DR |
| VR50 phe′V2 | TTTCATTCCGAGTCCGGGCACCA | 19/19-nt DR |
| Nissle pheV | TTCGATTCCGAGTCCGGGCACCA | 54–76 |
| Nissle phe′V | TTTCATTCCGATTCCGGGCACCA | 19/19-nt DR |
| CFT073 pheV | TTCGATTCCGAGTCCGGGCACCA | 54–76 |
| CFT073 phe′V | TTTCATTCCGATTCCGGGCACCA | 19/19-nt DR |
| UMN026 pheV | TTCGATTCCGAGTCCGGGCACCA | 54–76 |
| UMN026 phe′V | TTTCATTCCGATTCCGGGCACCA | 19/19-nt DR |
| 536 pheV | TTCGATTCCGAGTCCGGGCACCA | 54–76 |
| 536 phe′V | TTCGATTCCGAGTCCGG-CACCA | 23/22-nt DR |
| APECO1 pheV | TTCGATTCCGAGTCCGGGCACCA | 54–76 |
| APECO1 phe′V | TTCGATTCCGAGTCCGG-CACCA | 23/22-nt DR |
| SMS_3_5 pheV | TTCGATTCCGAGTCCGGGCACCA | 54–76 |
| SMS_3_5 phe′V | TTTTATTCCGAGTCCGGGCACCA | 19/19-nt DR |
The columns in boldface contain differences (underlined) in phe′V (the direct repeat at the 3′ end of island) relative to the pheV tRNA gene. The nucleotides in italics are not considered part of the phe′V repeat. The complete 76-nt pheV tRNA gene is 100% identical in all the E. coli strains shown here.
The PAI-536-pheV (PAI V536) island has been extensively studied and found to excise at lower frequencies than other E. coli 536 pathogenicity islands, possibly due to a single-nucleotide deletion in the right-hand DR (54). Interestingly, the left-hand DR of pheV islands from VR50, Nissle 1917, CFT073, and UMN026 contain a single-nucleotide substitution that may also increase the stability of the islands in these strains and suggests recent inheritance from a common ancestor (Table 6). The close relationship between these islands is borne out by the high nucleotide identity of the shared modules. The composite structure of GI-VR50-pheV suggests that there have been 2 independent insertions of unrelated pheV islands, although the actual polarity of this process is unclear. The existence of composite pheV islands in E. coli highlights the capacity of these elements to generate variability.
GI-VR50-pheV contributes to colonization of the mouse urinary tract.
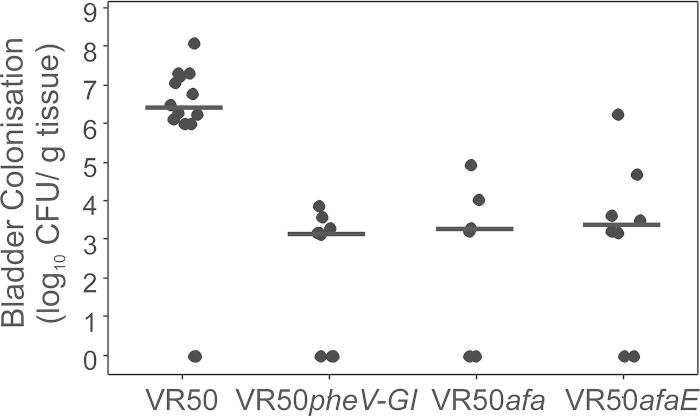
To assess the contribution of GI-VR50-pheV to the ability of E. coli VR50 to colonize the mouse urinary tract, we deleted the island using λ-Red-mediated homologous recombination. This resulted in the replacement of the entire 94-kb island with a kanamycin resistance gene cassette (referred to as VR50pheV-GI). E. coli VR50 colonized the mouse bladder in high numbers, demonstrating its suitability in this infection model. However, we observed significantly reduced colonization of the bladder by VR50pheV-GI compared to the wild-type VR50 strain (P = 0.002) (Fig. 6). Thus, GI-VR50-pheV contributes to the ability of E. coli VR50 to colonize the mouse bladder. Based on our bioinformatic analysis of GI-VR50-pheV, we predicted that this phenotype was directly associated with production of the Afa adhesin and tested this hypothesis (see below).
FIG 6.
Persistence of E. coli VR50, VR50pheV-GI, VR50afa, and VR50afaE in the bladders of C57BL/6 mice 18 h after intraurethral challenge. The results represent log10 CFU/0.1 g bladder tissue of individual mice, and the horizontal bars mark group medians. A minimum of 8 mice were assessed per group. E. coli VR50 was recovered from the bladders of infected mice in significantly higher numbers than each of the mutant strains (P = 0.002; Kruskal-Wallis test).
Phylogenetic analysis of the AfaE adhesin.
We performed a phylogenetic analysis of the E. coli VR50 adhesin-encoding afaE gene. The closest homologues of AfaE were the group 254 Dr family adhesins, which were sequenced as part of a larger study investigating the diversity of Dr family adhesin-encoding genes from 100 E. coli isolates (55). The authors divided the Dr family into 8 distinct groups (afaE-1, afaE-2, afaE-5, nfaE-111, daaE, drbE, draE, and 254). A multiple-sequence alignment of VR50 afaE with the group 254 adhesin gene sequences showed only between five and nine nucleotide substitutions relative to other group members (>98% nucleotide sequence identity). Four of these substitutions are unique to VR50 and are found in the 3′ region; two VR50 substitutions cause nonsynonymous replacements of the terminal residues (GGYWAK to GGYWTN). Interestingly, the same substitutions are found in another subgroup (DaaE). Although no binding function has been described for the C-terminal residues of the Dr adhesin, the observation that positive selection of point mutations drives changes for adhesin specificity in the DraE subfamily (55) leads us to speculate that this region may be important for binding of VR50 in the urinary tract. As yet, no information has been provided about the origins of the isolates carrying group 254 adhesins; however, the closest functionally characterized homologue is AfaE-3 (GenBank accession no. CAA54121).
The Afa adhesin is functionally expressed on the surface of E. coli VR50.
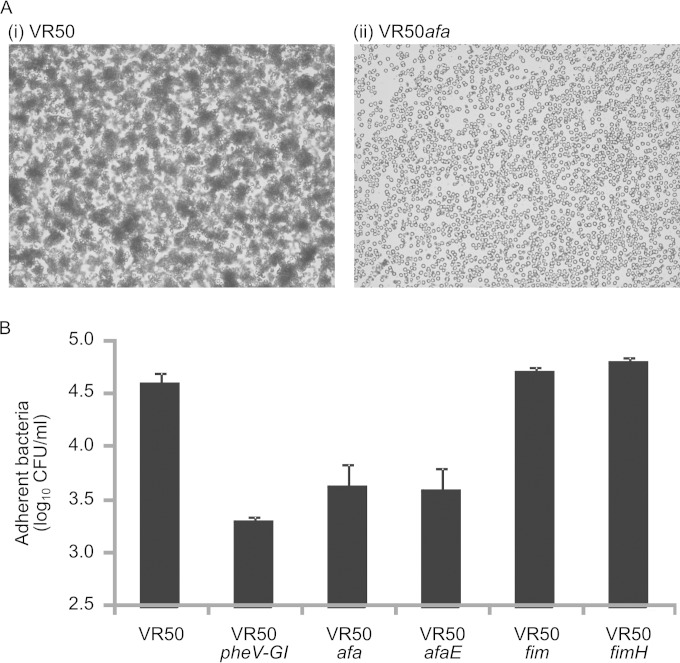
Phenotypic assays previously demonstrated that E. coli VR50 causes weak MR hemagglutination of human type A RBCs (20). This property is consistent with Afa adhesin surface expression. To confirm this, we constructed mutants with afaABCDE deleted (VR50afa; Afa gene cluster mutant) and with the afaE gene deleted (VR50afaE) using λ-Red-mediated homologous recombination. In contrast to E. coli VR50, the VR50afa and VR50afaE mutants did not cause MR hemagglutination of human type A RBCs (Fig. 7A), thus confirming the functional expression of Afa by E. coli VR50.
FIG 7.
(A) VR50 expresses functional Afa. Shown is hemagglutination of human RBCs by E. coli VR50 (i) in contrast to E. coli VR50afa (ii), which was unable to mediate hemagglutination of human RBCs under the same conditions. The images were viewed under ×63 magnification. (B) Adhesion of E. coli VR50, VR50pheV-GI, VR50afa, VR50afaE, VR50fim, and VR50fimH to T24 human bladder epithelial cells. The epithelial cells were inoculated with bacteria and incubated for 1 h, with adherent bacteria enumerated by direct plating and colony counts. The results are the averages of two independent experiments plus standard errors of the mean. Mutant strains VR50pheV-GI, VR50afa, and VR50afaE adhered significantly less to T24 cells than wild-type VR50 and the two type 1 fimbria-deficient mutants, VR50fim and VR50fimH (P < 0.001; analysis of variance [ANOVA]).
Afa mediates adherence of E. coli VR50 to human bladder epithelial cells.
To examine the adhesion phenotype of E. coli VR50 with respect to the urinary tract, we tested its ability to adhere to T24 bladder epithelial cells. E. coli VR50 adhered strongly to these cells (Fig. 7B). Next, we examined the adhesion phenotype of the VR50pheV-GI, VR50afa, and VR50afaE mutants. All three mutants displayed a significant reduction in adherence to T24 bladder epithelial cells (P < 0.001) (Fig. 7B). Interestingly, mutants of VR50 with the entire type 1 fimbrial gene cluster (VR50fim) or the fimH gene (VR50fimH) deleted did not show any significant difference in adherence to T24 bladder epithelial cells compared to the parent VR50 strain. Taken together, the data support a role for the AfaE adhesin of E. coli VR50 in adhesion to human bladder epithelial cells.
Afa contributes to colonization of the mouse bladder by E. coli VR50.
In light of the above in vitro adherence data and the in vivo mouse colonization phenotype of VR50pheV-GI, we tested E. coli VR50afa and VR50afaE for the ability to colonize and survive in the mouse urinary tract following transurethral infection (Fig. 6). E. coli VR50afa and VR50afaE were both significantly attenuated for colonization of the mouse bladder. The degree of attenuation was identical to that observed for E. coli VR50pheV-GI, demonstrating a direct role for the Afa adhesin in colonization. No significant colonization of the kidneys was observed for VR50 or any of the mutants; this is consistent with previous data from our laboratory using C57BL/6 mice (56, 57).
DISCUSSION
ABU E. coli strains, which colonize the human urinary tract without causing symptoms, comprise strains that have become attenuated through gene loss or deletion (e.g., E. coli 83972), as well as commensal-like strains that have acquired fitness factors that facilitate colonization of the human bladder. Here, we demonstrate that E. coli VR50 is an example of a commensal E. coli strain that has gained important genes to enable it to survive in the urinary bladder.
The complete genome sequence of VR50 revealed multiple accessory genes that may contribute to its survival in the human urinary tract. In general, these genes were associated with a range of mobile genetic elements, including plasmids, prophages, and genomic islands. We identified 8 prophage regions in the VR50 chromosome, 4 of which are likely to be functional, with another 2 encoding satellite phage. Several putative cargo gene products are encoded within the prophage regions, most of which have unknown functions, but some have similarity to known proteins, e.g., DinI-like damage-inducible proteins (VR50p6), outer-membrane porin proteins (VR50p2 and VR50p6), Lom-like outer-membrane proteins (VR50p2, VR50p4, and VR50p6), and a DNA adenine methylase (VR50p7). E. coli VR50 also carries two large plasmids, one of which contains several antibiotic resistance genes. Prophages, genomic islands, and plasmids are themselves comprised of smaller mobile genetic elements, some of which appear to have undergone expansion in VR50 since its divergence from other group A E. coli strains. For example, the IS629 element is not found in E. coli K-12 MG1655, but there are 19 copies distributed throughout the E. coli VR50 chromosome, including disruptive insertions within genes encoding putative outer-membrane proteins and chaperone-usher fimbriae (see Data Set S1 in the supplemental material).
We focused a major part of our analysis on the GI-VR50-pheV island, which contains multiple virulence-associated genes, including those encoding Afa, Ag43, Sat, and aerobactin. Lloyd et al. recommended that GIs are differentiated from PAIs by the fact that the latter contain established or putative virulence genes whereas the former contain genes with unknown functions (58). The original criteria for classifying pathogenicity islands suggested that the term should be reserved for islands preferentially associated with pathogenic strains (59). It is clear from a simple comparison of pheV-associated islands in commensal, pathogenic, and asymptomatic strains that there are no apparent qualitative differences between GIs and PAIs, so use of the more neutral GI terminology may be more appropriate to describe integrated islands in the VR50 genome. Direct repeats can often be used to distinguish between integrative islands that appear to play such a dynamic role in the adaptation to new environments (e.g., PAI-536-pheV and PAI-CFT073-pheV) and lineage-specific indels. Delineating these boundaries is not always trivial, as the GI-pheV case demonstrates. For example, the kps capsule biosynthesis locus is included within the island boundaries in the original report of the Nissle 1917 pheV island (48). Similarly, a comparison of several pheV islands does not distinguish between the integrative island at the pheV gene and downstream variability in the kps and gsp loci, even though much of the gsp secretion locus is present in a syntenic location in the Escherichia fergusonii genome (49). Recent studies of the E. coli ST131 lineage by our group have begun to reveal the extent of recombination associated with GI-pheV and other mobile genetic elements (60).
The GI-VR50-pheV island is very similar to pheV-associated islands from a wide variety of sequenced strains, e.g., the probiotic E. coli Nissle 1917, extraintestinal pathogenic E. coli (ExPEC)/UPEC (CFT073, UMN026, IA39, S88, 536, and APECO1), enterotoxigenic E. coli (ETEC) (E24377A), enteroaggregative E. coli (EAEC) (55989), and commensal (ED1a) and environmental (SMS-3-5) strains. Remarkably, the LEE has been associated with pheV islands in rabbit enteropathogenic (EPEC) (51) and STEC (61). It appears that these integrative islands are modular, and this mosaicism could be the key factor in their success, allowing rapid insertion and deletion of new elements that provide a selective advantage in different environments. In the case of E. coli VR50, our molecular analyses suggest that a selective advantage is provided by the acquisition of the Afa/Dr adhesin locus. Another interesting consequence of this modularity is that there is extensive exchange between islands via homologous recombination. For example, although E. coli UTI89 lacks the pheV island, many of the core modules found on GI-VR50-pheV (e.g., sat, iut, and flu) are found on other UTI89 islands. As the number of complete genomes increases, it should be possible to obtain a clearer picture of the role that integrative islands play in pathogen evolution.
It is an open question as to what role the other pheV island-encoded “pathogenicity factors,” such as the sat gene product and aerobactin, play in the life styles of VR50 and other intestinal commensal organisms. Given the ease with which elements or the entire island are lost from these loci in some strains (54), it appears that these elements are under strong selective pressure to be retained. In the case of VR50, the presence of a complete aerobactin gene locus and the gene encoding the Iha siderophore receptor within GI-VR50-pheV suggests these genes might contribute to its overall fitness in the bladder. Iron is a limiting nutrient in the human urinary tract, and UTI E. coli strains often produce several siderophores to acquire ferric iron (Fe3+) from the host. Four siderophore systems have been described from UTI E. coli, i.e., enterobactin (which is common to all strains), salmochelin, yersiniabactin, and aerobactin (62, 63). Indeed, UPEC mutants with genes encoding the production of siderophores deleted are less virulent in the mouse urinary tract (64). An enhanced ability of ABU E. coli strains to acquire iron has also been proposed as a mechanism by which they outcompete UPEC during growth in human urine (39, 63, 65). We did not observe any significant difference in the overall level of siderophores produced by E. coli strains MG1655 and VR50 as measured by the chrome azurol S (CAS) assay (reference 66 and data not shown); however, this is most likely due to the common production of enterobactin by both strains.
GI-VR50-pheV was shown to contribute to the ability of E. coli VR50 to colonize the mouse urinary tract, and we further defined Afa as a GI-encoded factor that mediates adherence of VR50 to T24 bladder epithelial cells. The Afa (and Dr) adhesins comprise a family of surface-located factors encoded by the afa (67–71), dra (72, 73), and daa (74, 75) operons. These operons possess similar genetic structures and have been associated with E. coli strains of diarrheagenic and UTI origin (76, 77). Many Afa adhesins recognize as a receptor the decay-accelerating factor (DAF), a complement-regulatory protein present on the surfaces of a range of human epithelial cells (including epithelial cells of the urinary tract) (78, 79). Some Afa variants also mediate binding to type IV collagen and carcinoembryonic antigen-related cell adhesion molecules (80, 81). We demonstrated a role for Afa (but not type 1 fimbriae) in the adhesion of E. coli VR50 to T24 bladder epithelial cells. Afa has been shown to mediate adherence and invasion of E. coli in HeLa and Chinese hamster ovary cells (82, 83). In the case of VR50, we previously reported that the strain does not readily invade T24 cells, with low numbers observed intracellularly (84). However, it is possible that the E. coli VR50 cells recovered from HeLa cells in our assay may represent both adhered and intracellular bacteria. In the mouse UTI model, E. coli VR50 colonized the bladder with high efficiency, and the overall cell numbers were very similar to the level of colonization we routinely observe for CFT073 (56, 57, 85). In contrast, VR50pheV-GI, VR50afa, and VR50afaE all displayed significantly attenuated colonization of the mouse bladder, strongly suggesting that the afa genes within GI-VR50-pheV enhance the fitness of VR50 in the urinary tract.
The innate immune response is essential for defense against UTI, and the associated tissue inflammation is a major cause of symptoms, leading to tissue damage and contributing to the severity of disease. The innate response is affected by the virulence of the infecting strain, and many well-characterized virulence factors act by triggering inflammation. UPEC triggers the secretion of epithelial cytokines and chemokines that contribute to inflammation and pathology. For example, IL-6 levels in the urine correlate with disease severity, and IL-8 results in recruitment of neutrophils to the site of infection (86–88). Despite equivalent adherence to epithelial cells, E. coli CFT073 triggered a significantly stronger IL-6 cytokine response than E. coli VR50. This property may be associated with the ability of E. coli VR50 to colonize the bladder in high numbers without provoking a host inflammatory response and is consistent with the low IL-6 response observed in individuals deliberately colonized with E. coli 83972 (89). It remains to be determined if VR50 is able to suppress RNA polymerase II-dependent host gene expression, as has been reported recently for E. coli 83972 (19). Furthermore, host genetic variation that leads to suppression of the innate immune response to UTI (e.g., via Toll-like receptor 4 [TLR4] promoter polymorphisms) also contributes to protection against symptomatic disease in some ABU patients (90–93).
ABU occurs in up to 6% of healthy individuals, 18% of diabetics (mostly women), and 20% of elderly individuals (women more often than men). E. coli is also the most frequent cause of ABU. An understanding of the molecular mechanisms that underpin ABU would have a major impact on current approaches to treating and preventing symptomatic UTI for several reasons. First, ABU strains provide a unique model to identify factors that enable UPEC to colonize the urinary tract. Second, the outcomes of ABU vary; in some patients, ABU can predispose to kidney infection, while in other patients, ABU can prevent colonization by more virulent strains. A greater understanding of the risk factors associated with ABU is therefore required to guide treatment decisions. Third, the unnecessary treatment of ABU in some patients may be associated with the rapid spread of genes coding for antibiotic resistance, particularly in health care settings. In this respect, it is interesting that E. coli VR50 contains a truncated copy of the blaTEM-1 gene. Although not functional, a significant association between ampicillin resistance and the presence of the afa and dra genes has been reported previously (94). Finally, ABU E. coli represents a viable prophylactic approach for the prevention and treatment of chronic and recurrent UTIs. The use of ABU E. coli as a prophylactic agent could address important concerns about antibiotic resistance in patients with UTI recalcitrant to treatment with currently available antibiotics. In the case of VR50, its capacity to express Afa, which contributes to chronic pyelonephritis (77, 95) and recurrent cystitis (96), suggests that it would not constitute a suitable prophylactic ABU strain for human use.
In summary, our study has determined the complete genome sequence of E. coli VR50 and demonstrated that it is a commensal-like strain that has acquired a number of UPEC-associated virulence factors. One of these factors, Afa, contributes specifically to VR50 colonization of the mouse bladder.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from the National Health and Medical Research Council (NHMRC) of Australia (511224, 569676, 631654, and APP1033799). M.A.S. is supported by an Australian Research Council (ARC) Future Fellowship (FT100100662), S.A.B. is supported by an NHMRC Career Development Fellowship (APP1090456), and M.T. is supported by an ARC Discovery Early Career Researcher Award (DE130101169). PacBio sequencing was carried out with the support of UM-MOHE HIR grant no. H-50001-A000027.
We thank John Davis and Annette McGrath from the Australian Genome Research Facility for assistance in library preparation and 454/Sanger hybrid assembly. We are indebted to the contribution of Per Klemm, who sadly passed away during the course of this study.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02810-14.
REFERENCES
- 1.Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am J Med 113(Suppl 1A):5S–13S. doi: 10.1016/S0002-9343(02)01054-9. [DOI] [PubMed] [Google Scholar]
- 2.Dobrindt U, Agerer F, Michaelis K, Janka A, Buchrieser C, Samuelson M, Svanborg C, Gottschalk G, Karch H, Hacker J. 2003. Analysis of genome plasticity in pathogenic and commensal Escherichia coli isolates by use of DNA arrays. J Bacteriol 185:1831–1840. doi: 10.1128/JB.185.6.1831-1840.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dobrindt U, Hochhut B, Hentschel U, Hacker J. 2004. Genomic islands in pathogenic and environmental microorganisms. Nat Rev Microbiol 2:414–424. doi: 10.1038/nrmicro884. [DOI] [PubMed] [Google Scholar]
- 4.Brzuszkiewicz E, Bruggemann H, Liesegang H, Emmerth M, Olschlager T, Nagy G, Albermann K, Wagner C, Buchrieser C, Emody L, Gottschalk G, Hacker J, Dobrindt U. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc Natl Acad Sci U S A 103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klemm P, Schembri MA. 2000. Bacterial adhesins: function and structure. Int J Med Microbiol 290:27–35. doi: 10.1016/S1438-4221(00)80102-2. [DOI] [PubMed] [Google Scholar]
- 6.Totsika M, Moriel DG, Idris A, Rogers BA, Wurpel DJ, Phan MD, Paterson DL, Schembri MA. 2012. Uropathogenic Escherichia coli mediated urinary tract infection. Curr Drug Targets 13:1386–1399. doi: 10.2174/138945012803530206. [DOI] [PubMed] [Google Scholar]
- 7.Hannan TJ, Totsika M, Mansfield KJ, Moore KH, Schembri MA, Hultgren SJ. 2012. Host-pathogen checkpoints and population bottlenecks in persistent and intracellular uropathogenic Escherichia coli bladder infection. FEMS Microbiol Rev 36:616–648. doi: 10.1111/j.1574-6976.2012.00339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ulett GC, Totsika M, Schaale K, Carey AJ, Sweet MJ, Schembri MA. 2013. Uropathogenic Escherichia coli virulence and innate immune responses during urinary tract infection. Curr Opin Microbiol 16:100–107. doi: 10.1016/j.mib.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Salvador E, Wagenlehner F, Kohler CD, Mellmann A, Hacker J, Svanborg C, Dobrindt U. 2012. Comparison of asymptomatic bacteriuria Escherichia coli isolates from healthy individuals versus those from hospital patients shows that long-term bladder colonization selects for attenuated virulence phenotypes. Infect Immun 80:668–678. doi: 10.1128/IAI.06191-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zdziarski J, Svanborg C, Wullt B, Hacker J, Dobrindt U. 2008. Molecular basis of commensalism in the urinary tract: low virulence or virulence attenuation? Infect Immun 76:695–703. doi: 10.1128/IAI.01215-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Plos K, Carter T, Hull S, Hull R, Svanborg Eden C. 1990. Frequency and organization of pap homologous DNA in relation to clinical origin of uropathogenic Escherichia coli. J Infect Dis 161:518–524. doi: 10.1093/infdis/161.3.518. [DOI] [PubMed] [Google Scholar]
- 12.Mabbett AN, Ulett GC, Watts RE, Tree JJ, Totsika M, Ong CL, Wood JM, Monaghan W, Looke DF, Nimmo GR, Svanborg C, Schembri MA. 2009. Virulence properties of asymptomatic bacteriuria Escherichia coli. Int J Med Microbiol 299:53–63. doi: 10.1016/j.ijmm.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Hull R, Rudy D, Donovan W, Svanborg C, Wieser I, Stewart C, Darouiche R. 2000. Urinary tract infection prophylaxis using Escherichia coli 83972 in spinal cord injured patients. J Urol 163:872–877. doi: 10.1016/S0022-5347(05)67823-8. [DOI] [PubMed] [Google Scholar]
- 14.Sunden F, Hakansson L, Ljunggren E, Wullt B. 2006. Bacterial interference—is deliberate colonization with Escherichia coli 83972 an alternative treatment for patients with recurrent urinary tract infection? Int J Antimicrob Agents 28(Suppl 1):S26–S29. doi: 10.1016/j.ijantimicag.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 15.Zdziarski J, Brzuszkiewicz E, Wullt B, Liesegang H, Biran D, Voigt B, Gronberg-Hernandez J, Ragnarsdottir B, Hecker M, Ron EZ, Daniel R, Gottschalk G, Hacker J, Svanborg C, Dobrindt U. 2010. Host imprints on bacterial genomes—rapid, divergent evolution in individual patients. PLoS Pathog 6:e1001078. doi: 10.1371/journal.ppat.1001078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klemm P, Roos V, Ulett GC, Svanborg C, Schembri MA. 2006. Molecular characterization of the Escherichia coli asymptomatic bacteriuria strain 83972: the taming of a pathogen. Infect Immun 74:781–785. doi: 10.1128/IAI.74.1.781-785.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roos V, Schembri MA, Ulett GC, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152:1799–1806. doi: 10.1099/mic.0.28711-0. [DOI] [PubMed] [Google Scholar]
- 18.Klemm P, Hancock V, Schembri MA. 2007. Mellowing out: adaptation to commensalism by Escherichia coli asymptomatic bacteriuria strain 83972. Infect Immun 75:3688–3695. doi: 10.1128/IAI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lutay N, Ambite I, Gronberg Hernandez J, Rydstrom G, Ragnarsdottir B, Puthia M, Nadeem A, Zhang J, Storm P, Dobrindt U, Wullt B, Svanborg C. 2013. Bacterial control of host gene expression through RNA polymerase II. J Clin Invest 123:2366–2379. doi: 10.1172/JCI66451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roos V, Nielsen EM, Klemm P. 2006. Asymptomatic bacteriuria Escherichia coli strains: adhesins, growth and competition. FEMS Microbiol Lett 262:22–30. doi: 10.1111/j.1574-6968.2006.00355.x. [DOI] [PubMed] [Google Scholar]
- 21.Mobley HL, Green DM, Trifillis AL, Johnson DE, Chippendale GR, Lockatell CV, Jones BD, Warren JW. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect Immun 58:1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andersson P, Engberg I, Lidin-Janson G, Lincoln K, Hull R, Hull S, Svanborg C. 1991. Persistence of Escherichia coli bacteriuria is not determined by bacterial adherence. Infect Immun 59:2915–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertani G. 1951. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol 62:293–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clermont O, Bonacorsi S, Bingen E. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl Environ Microbiol 66:4555–4558. doi: 10.1128/AEM.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagberg L, Jodal U, Korhonen TK, Lidin-Janson G, Lindberg U, Svanborg Eden C. 1981. Adhesion, hemagglutination, and virulence of Escherichia coli causing urinary tract infections. Infect Immun 31:564–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chin CS, Alexander DH, Marks P, Klammer AA, Drake J, Heiner C, Clum A, Copeland A, Huddleston J, Eichler EE, Turner SW, Korlach J. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 28.Delcher AL, Salzberg SL, Phillippy AM. 2003. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics Chapter 10:Unit 10.3. doi: 10.1002/0471250953.bi1003s00. [DOI] [PubMed] [Google Scholar]
- 29.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis Comparison Tool. Bioinformatics 21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 30.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS One 5:e11147. doi: 10.1371/journal.pone.0011147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carver T, Harris SR, Otto TD, Berriman M, Parkhill J, McQuillan JA. 2013. BamView: visualizing and interpretation of next-generation sequencing read alignments. Brief Bioinform 14:203–212. doi: 10.1093/bib/bbr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Flusberg BA, Webster DR, Lee JH, Travers KJ, Olivares EC, Clark TA, Korlach J, Turner SW. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7:461–465. doi: 10.1038/nmeth.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clark TA, Murray IA, Morgan RD, Kislyuk AO, Spittle KE, Boitano M, Fomenkov A, Roberts RJ, Korlach J. 2012. Characterization of DNA methyltransferase specificities using single-molecule, real-time DNA sequencing. Nucleic Acids Res 40:e29. doi: 10.1093/nar/gkr1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Otto TD, Dillon GP, Degrave WS, Berriman M. 2011. RATT: Rapid Annotation Transfer Tool. Nucleic Acids Res 39:e57. doi: 10.1093/nar/gkq1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.David M, Dzamba M, Lister D, Ilie L, Brudno M. 2011. SHRiMP2: sensitive yet practical SHort Read Mapping. Bioinformatics 27:1011–1012. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- 38.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 39.Roos V, Ulett GC, Schembri MA, Klemm P. 2006. The asymptomatic bacteriuria Escherichia coli strain 83972 outcompetes uropathogenic E. coli strains in human urine. Infect Immun 74:615–624. doi: 10.1128/IAI.74.1.615-624.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ, Deikus G, Clark TA, Luong K, Murray IA, Davis BM, Keren-Paz A, Chess A, Roberts RJ, Korlach J, Turner SW, Kumar V, Waldor MK, Schadt EE. 2012. Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol 30:1232–1239. doi: 10.1038/nbt.2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cooper KK, Mandrell RE, Louie JW, Korlach J, Clark TA, Parker CT, Huynh S, Chain PS, Ahmed S, Carter MQ. 2014. Comparative genomics of enterohemorrhagic Escherichia coli O145:H28 demonstrates a common evolutionary lineage with Escherichia coli O157:H7. BMC Genomics 15:17. doi: 10.1186/1471-2164-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Powers JG, Weigman VJ, Shu J, Pufky JM, Cox D, Hurban P. 2013. Efficient and accurate whole genome assembly and methylome profiling of E. coli. BMC Genomics 14:675. doi: 10.1186/1471-2164-14-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sokurenko EV, Chesnokova V, Dykhuizen DE, Ofek I, Wu XR, Krogfelt KA, Struve C, Schembri MA, Hasty DL. 1998. Pathogenic adaptation of Escherichia coli by natural variation of the FimH adhesin. Proc Natl Acad Sci U S A 95:8922–8926. doi: 10.1073/pnas.95.15.8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gehring AM, DeMoll E, Fetherston JD, Mori I, Mayhew GF, Blattner FR, Walsh CT, Perry RD. 1998. Iron acquisition in plague: modular logic in enzymatic biogenesis of yersiniabactin by Yersinia pestis. Chem Biol 5:573–586. doi: 10.1016/S1074-5521(98)90115-6. [DOI] [PubMed] [Google Scholar]
- 45.Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol 41:349–363. doi: 10.1046/j.1365-2958.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- 46.Vejborg RM, Friis C, Hancock V, Schembri MA, Klemm P. 2010. A virulent parent with probiotic progeny: comparative genomics of Escherichia coli strains CFT073, Nissle 1917 and ABU 83972. Mol Genet Genomics 283:469–484. doi: 10.1007/s00438-010-0532-9. [DOI] [PubMed] [Google Scholar]
- 47.Hochhut B, Wilde C, Balling G, Middendorf B, Dobrindt U, Brzuszkiewicz E, Gottschalk G, Carniel E, Hacker J. 2006. Role of pathogenicity island-associated integrases in the genome plasticity of uropathogenic Escherichia coli strain 536. Mol Microbiol 61:584–595. doi: 10.1111/j.1365-2958.2006.05255.x. [DOI] [PubMed] [Google Scholar]
- 48.Grozdanov L, Raasch C, Schulze J, Sonnenborn U, Gottschalk G, Hacker J, Dobrindt U. 2004. Analysis of the genome structure of the nonpathogenic probiotic Escherichia coli strain Nissle 1917. J Bacteriol 186:5432–5441. doi: 10.1128/JB.186.16.5432-5441.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Touchon M, Hoede C, Tenaillon O, Barbe V, Baeriswyl S, Bidet P, Bingen E, Bonacorsi S, Bouchier C, Bouvet O, Calteau A, Chiapello H, Clermont O, Cruveiller S, Danchin A, Diard M, Dossat C, Karoui ME, Frapy E, Garry L, Ghigo JM, Gilles AM, Johnson J, Le Bouguenec C, Lescat M, Mangenot S, Martinez-Jehanne V, Matic I, Nassif X, Oztas S, Petit MA, Pichon C, Rouy Z, Ruf CS, Schneider D, Tourret J, Vacherie B, Vallenet D, Medigue C, Rocha EP, Denamur E. 2009. Organised genome dynamics in the Escherichia coli species results in highly diverse adaptive paths. PLoS Genet 5:e1000344. doi: 10.1371/journal.pgen.1000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rumer L, Jores J, Kirsch P, Cavignac Y, Zehmke K, Wieler LH. 2003. Dissemination of pheU- and pheV-located genomic islands among enteropathogenic (EPEC) and enterohemorrhagic (EHEC) E. coli and their possible role in the horizontal transfer of the locus of enterocyte effacement (LEE). Int J Med Microbiol 292:463–475. doi: 10.1078/1438-4221-00229. [DOI] [PubMed] [Google Scholar]
- 51.Tauschek M, Strugnell RA, Robins-Browne RM. 2002. Characterization and evidence of mobilization of the LEE pathogenicity island of rabbit-specific strains of enteropathogenic Escherichia coli. Mol Microbiol 44:1533–1550. doi: 10.1046/j.1365-2958.2002.02968.x. [DOI] [PubMed] [Google Scholar]
- 52.Lalioui L, Le Bouguenec C. 2001. afa-8 Gene cluster is carried by a pathogenicity island inserted into the tRNA(Phe) of human and bovine pathogenic Escherichia coli isolates. Infect Immun 69:937–948. doi: 10.1128/IAI.69.2.937-948.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson TJ, Kariyawasam S, Wannemuehler Y, Mangiamele P, Johnson SJ, Doetkott C, Skyberg JA, Lynne AM, Johnson JR, Nolan LK. 2007. The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes. J Bacteriol 189:3228–3236. doi: 10.1128/JB.01726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Middendorf B, Hochhut B, Leipold K, Dobrindt U, Blum-Oehler G, Hacker J. 2004. Instability of pathogenicity islands in uropathogenic Escherichia coli 536. J Bacteriol 186:3086–3096. doi: 10.1128/JB.186.10.3086-3096.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Korotkova N, Chattopadhyay S, Tabata TA, Beskhlebnaya V, Vigdorovich V, Kaiser BK, Strong RK, Dykhuizen DE, Sokurenko EV, Moseley SL. 2007. Selection for functional diversity drives accumulation of point mutations in Dr adhesins of Escherichia coli. Mol Microbiol 64:180–194. doi: 10.1111/j.1365-2958.2007.05648.x. [DOI] [PubMed] [Google Scholar]
- 56.Valle J, Mabbett AN, Ulett GC, Toledo-Arana A, Wecker K, Totsika M, Schembri MA, Ghigo JM, Beloin C. 2008. UpaG, a new member of the trimeric autotransporter family of adhesins in uropathogenic Escherichia coli. J Bacteriol 190:4147–4161. doi: 10.1128/JB.00122-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ulett GC, Valle J, Beloin C, Sherlock O, Ghigo JM, Schembri MA. 2007. Functional analysis of antigen 43 in uropathogenic Escherichia coli reveals a role in long-term persistence in the urinary tract. Infect Immun 75:3233–3244. doi: 10.1128/IAI.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lloyd AL, Rasko DA, Mobley HL. 2007. Defining genomic islands and uropathogen-specific genes in uropathogenic Escherichia coli. J Bacteriol 189:3532–3546. doi: 10.1128/JB.01744-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hacker J, Kaper JB. 2000. Pathogenicity islands and the evolution of microbes. Annu Rev Microbiol 54:641–679. doi: 10.1146/annurev.micro.54.1.641. [DOI] [PubMed] [Google Scholar]
- 60.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan MD, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Bano J, Pascual A, Pitout JD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jores J, Rumer L, Kiessling S, Kaper JB, Wieler LH. 2001. A novel locus of enterocyte effacement (LEE) pathogenicity island inserted at pheV in bovine Shiga toxin-producing Escherichia coli strain O103:H2. FEMS Microbiol Lett 204:75–79. doi: 10.1111/j.1574-6968.2001.tb10866.x. [DOI] [PubMed] [Google Scholar]
- 62.Braun V. 2003. Iron uptake by Escherichia coli. Front Biosci 8:s1409–s1421. doi: 10.2741/1232. [DOI] [PubMed] [Google Scholar]
- 63.Watts RE, Totsika M, Challinor VL, Mabbett AN, Ulett GC, De Voss JJ, Schembri MA. 2012. Contribution of siderophore systems to growth and urinary tract colonization of asymptomatic bacteriuria Escherichia coli. Infect Immun 80:333–344. doi: 10.1128/IAI.05594-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Torres AG, Redford P, Welch RA, Payne SM. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect Immun 69:6179–6185. doi: 10.1128/IAI.69.10.6179-6185.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roos V, Klemm P. 2006. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect Immun 74:3565–3575. doi: 10.1128/IAI.01959-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwyn B, Neilands JB. 1987. Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- 67.Garcia MI, Gounon P, Courcoux P, Labigne A, Le Bouguenec C. 1996. The afimbrial adhesive sheath encoded by the afa-3 gene cluster of pathogenic Escherichia coli is composed of two adhesins. Mol Microbiol 19:683–693. doi: 10.1046/j.1365-2958.1996.394935.x. [DOI] [PubMed] [Google Scholar]
- 68.Labigne-Roussel A, Schmidt MA, Walz W, Falkow S. 1985. Genetic organization of the afimbrial adhesin operon and nucleotide sequence from a uropathogenic Escherichia coli gene encoding an afimbrial adhesin. J Bacteriol 162:1285–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Labigne-Roussel AF, Lark D, Schoolnik G, Falkow S. 1984. Cloning and expression of an afimbrial adhesin (AFA-I) responsible for P blood group-independent, mannose-resistant hemagglutination from a pyelonephritic Escherichia coli strain. Infect Immun 46:251–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lalioui L, Jouve M, Gounon P, Le Bouguenec C. 1999. Molecular cloning and characterization of the afa-7 and afa-8 gene clusters encoding afimbrial adhesins in Escherichia coli strains associated with diarrhea or septicemia in calves. Infect Immun 67:5048–5059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Le Bouguenec C, Garcia MI, Ouin V, Desperrier JM, Gounon P, Labigne A. 1993. Characterization of plasmid-borne afa-3 gene clusters encoding afimbrial adhesins expressed by Escherichia coli strains associated with intestinal or urinary tract infections. Infect Immun 61:5106–5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nowicki B, Barrish JP, Korhonen T, Hull RA, Hull SI. 1987. Molecular cloning of the Escherichia coli O75X adhesin. Infect Immun 55:3168–3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pham TQ, Goluszko P, Popov V, Nowicki S, Nowicki BJ. 1997. Molecular cloning and characterization of Dr-II, a nonfimbrial adhesin-I-like adhesin isolated from gestational pyelonephritis-associated Escherichia coli that binds to decay-accelerating factor. Infect Immun 65:4309–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bilge SS, Apostol JM Jr, Fullner KJ, Moseley SL. 1993. Transcriptional organization of the F1845 fimbrial adhesin determinant of Escherichia coli. Mol Microbiol 7:993–1006. doi: 10.1111/j.1365-2958.1993.tb01191.x. [DOI] [PubMed] [Google Scholar]
- 75.Bilge SS, Clausen CR, Lau W, Moseley SL. 1989. Molecular characterization of a fimbrial adhesin, F1845, mediating diffuse adherence of diarrhea-associated Escherichia coli to HEp-2 cells. J Bacteriol 171:4281–4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Servin AL. 2005. Pathogenesis of Afa/Dr diffusely adhering Escherichia coli. Clin Microbiol Rev 18:264–292. doi: 10.1128/CMR.18.2.264-292.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Servin AL. 2014. Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dr adhesins (Afa/Dr DAEC): current insights and future challenges. Clin Microbiol Rev 27:823–869. doi: 10.1128/CMR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nowicki B, Hart A, Coyne KE, Lublin DM, Nowicki S. 1993. Short consensus repeat-3 domain of recombinant decay-accelerating factor is recognized by Escherichia coli recombinant Dr adhesin in a model of a cell-cell interaction. J Exp Med 178:2115–2121. doi: 10.1084/jem.178.6.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Medof ME, Walter EI, Rutgers JL, Knowles DM, Nussenzweig V. 1987. Identification of the complement decay-accelerating factor (DAF) on epithelium and glandular cells and in body fluids. J Exp Med 165:848–864. doi: 10.1084/jem.165.3.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Korotkova N, Cota E, Lebedin Y, Monpouet S, Guignot J, Servin AL, Matthews S, Moseley SL. 2006. A subfamily of Dr adhesins of Escherichia coli bind independently to decay-accelerating factor and the N-domain of carcinoembryonic antigen. J Biol Chem 281:29120–29130. doi: 10.1074/jbc.M605681200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Berger CN, Billker O, Meyer TF, Servin AL, Kansau I. 2004. Differential recognition of members of the carcinoembryonic antigen family by Afa/Dr adhesins of diffusely adhering Escherichia coli (Afa/Dr DAEC). Mol Microbiol 52:963–983. doi: 10.1111/j.1365-2958.2004.04033.x. [DOI] [PubMed] [Google Scholar]
- 82.Rana T, Hasan RJ, Nowicki S, Venkatarajan MS, Singh R, Urvil PT, Popov V, Braun WA, Popik W, Goodwin JS, Nowicki BJ. 2014. Complement protective epitopes and CD55-microtubule complexes facilitate the invasion and intracellular persistence of uropathogenic Escherichia coli. J Infect Dis 209:1066–1076. doi: 10.1093/infdis/jit619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goluszko P, Popov V, Selvarangan R, Nowicki S, Pham T, Nowicki BJ. 1997. Dr fimbriae operon of uropathogenic Escherichia coli mediate microtubule-dependent invasion to the HeLa epithelial cell line. J Infect Dis 176:158–167. doi: 10.1086/514018. [DOI] [PubMed] [Google Scholar]
- 84.Bokil NJ, Totsika M, Carey AJ, Stacey KJ, Hancock V, Saunders BM, Ravasi T, Ulett GC, Schembri MA, Sweet MJ. 2011. Intramacrophage survival of uropathogenic Escherichia coli: differences between diverse clinical isolates and between mouse and human macrophages. Immunobiology 216:1164–1171. doi: 10.1016/j.imbio.2011.05.011. [DOI] [PubMed] [Google Scholar]
- 85.Tree JJ, Ulett GC, Ong CL, Trott DJ, McEwan AG, Schembri MA. 2008. Trade-off between iron uptake and protection against oxidative stress: deletion of cueO promotes uropathogenic Escherichia coli virulence in a mouse model of urinary tract infection. J Bacteriol 190:6909–6912. doi: 10.1128/JB.00451-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wullt B, Bergsten G, Connell H, Rollano P, Gebratsedik N, Hang L, Svanborg C. 2001. P-fimbriae trigger mucosal responses to Escherichia coli in the human urinary tract. Cell Microbiol 3:255–264. doi: 10.1046/j.1462-5822.2001.00111.x. [DOI] [PubMed] [Google Scholar]
- 87.Svanborg C, Frendeus B, Godaly G, Hang L, Hedlund M, Wachtler C. 2001. Toll-like receptor signaling and chemokine receptor expression influence the severity of urinary tract infection. J Infect Dis 183(Suppl 1):S61–S65. doi: 10.1086/318858. [DOI] [PubMed] [Google Scholar]
- 88.Bergsten G, Wullt B, Schembri MA, Leijonhufvud I, Svanborg C. 2007. Do type 1 fimbriae promote inflammation in the human urinary tract? Cell Microbiol 9:1766–1781. doi: 10.1111/j.1462-5822.2007.00912.x. [DOI] [PubMed] [Google Scholar]
- 89.Gronberg-Hernandez J, Sunden F, Connolly J, Svanborg C, Wullt B. 2011. Genetic control of the variable innate immune response to asymptomatic bacteriuria. PLoS One 6:e28289. doi: 10.1371/journal.pone.0028289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ragnarsdottir B, Fischer H, Godaly G, Gronberg-Hernandez J, Gustafsson M, Karpman D, Lundstedt AC, Lutay N, Ramisch S, Svensson ML, Wullt B, Yadav M, Svanborg C. 2008. TLR- and CXCR1-dependent innate immunity: insights into the genetics of urinary tract infections. Eur J Clin Invest 38(Suppl 2):S12–S20. doi: 10.1111/j.1365-2362.2008.02004.x. [DOI] [PubMed] [Google Scholar]
- 91.Ragnarsdottir B, Jonsson K, Urbano A, Gronberg-Hernandez J, Lutay N, Tammi M, Gustafsson M, Lundstedt AC, Leijonhufvud I, Karpman D, Wullt B, Truedsson L, Jodal U, Andersson B, Svanborg C. 2010. Toll-like receptor 4 promoter polymorphisms: common TLR4 variants may protect against severe urinary tract infection. PLoS One 5:e10734. doi: 10.1371/journal.pone.0010734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ragnarsdottir B, Lutay N, Gronberg-Hernandez J, Koves B, Svanborg C. 2011. Genetics of innate immunity and UTI susceptibility. Nat Rev Urol 8:449–468. doi: 10.1038/nrurol.2011.100. [DOI] [PubMed] [Google Scholar]
- 93.Ragnarsdottir B, Samuelsson M, Gustafsson MC, Leijonhufvud I, Karpman D, Svanborg C. 2007. Reduced Toll-like receptor 4 expression in children with asymptomatic bacteriuria. J Infect Dis 196:475–484. doi: 10.1086/518893. [DOI] [PubMed] [Google Scholar]
- 94.Hart A, Nowicki BJ, Reisner B, Pawelczyk E, Goluszko P, Urvil P, Anderson G, Nowicki S. 2001. Ampicillin-resistant Escherichia coli in gestational pyelonephritis: increased occurrence and association with the colonization factor Dr adhesin. J Infect Dis 183:1526–1529. doi: 10.1086/320196. [DOI] [PubMed] [Google Scholar]
- 95.Goluszko P, Moseley SL, Truong LD, Kaul A, Williford JR, Selvarangan R, Nowicki S, Nowicki B. 1997. Development of experimental model of chronic pyelonephritis with Escherichia coli O75:K5:H-bearing Dr fimbriae: mutation in the dra region prevented tubulointerstitial nephritis. J Clin Invest 99:1662–1672. doi: 10.1172/JCI119329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Foxman B, Zhang L, Tallman P, Palin K, Rode C, Bloch C, Gillespie B, Marrs CF. 1995. Virulence characteristics of Escherichia coli causing first urinary tract infection predict risk of second infection. J Infect Dis 172:1536–1541. doi: 10.1093/infdis/172.6.1536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.