Abstract
Streptococcus pneumoniae is a major bacterial pathogen in humans. Its polysaccharide capsule is a key virulence factor that promotes bacterial evasion of human phagocytic killing. While S. pneumoniae produces at least 94 antigenically different types of capsule, the genes for biosynthesis of almost all capsular types are arranged in the same locus. The transcription of the capsular polysaccharide (cps) locus is not well understood. This study determined the transcriptional features of the cps locus in the type 2 virulent strain D39. The initial analysis revealed that the cps genes are cotranscribed from a major transcription start site at the −25 nucleotide (G) upstream of cps2A, the first gene in the locus. Using unmarked chromosomal truncations and a luciferase-based transcriptional reporter, we showed that the full transcription of the cps genes not only depends on the core promoter immediately upstream of cps2A, but also requires additional elements upstream of the core promoter, particularly a 59-bp sequence immediately upstream of the core promoter. Unmarked deletions of these promoter elements in the D39 genome also led to significant reduction in CPS production and virulence in mice. Lastly, common cps gene (cps2ABCD) mutants did not show significant abnormality in cps transcription, although they produced significantly less CPS, indicating that the CpsABCD proteins are involved in the encapsulation of S. pneumoniae in a posttranscriptional manner. This study has yielded important information on the transcriptional characteristics of the cps locus in S. pneumoniae.
INTRODUCTION
Streptococcus pneumoniae (pneumococcus) is a major cause of bacterial infections in humans, including otitis media, pneumonia, bacteremia, and meningitis (1). As the outermost structure, the capsule is the major virulence factor of S. pneumoniae and protects the bacterium by interfering with host phagocytic killing (2). Virtually all of the clinical isolates are encapsulated, and mutations in the cps locus result in the loss of virulence (3). The capsule of S. pneumoniae is composed of capsular polysaccharides (CPSs). The CPSs are immunogenic and thus serve as the target antigens for the current pneumococcal vaccines (4). Due to host immune selection that targets the capsule, the CPSs have undergone extensive chemical and antigenic variation via genetic diversity in the CPS biosynthesis genes. At least 94 antigenically distinct capsular serotypes have been identified in S. pneumoniae (5, 6). While the CPSs of types 3 and 37 are produced as relatively simple repeats of polysaccharides by the synthase pathway (7, 8), all other capsule types are much more complex and are synthesized by WZY-dependent polymerization, a mechanism of capsule production that is widely found in other bacteria (9–11).
Except for type 37, all of the CPS types in S. pneumoniae are determined by a set of capsule biosynthesis genes, which are historically designated cps or cap (12). Bentley et al. recently renamed the pneumococcal capsule genes on the basis of their overall functions in prokaryotic polysaccharide synthesis (5). We use the cps nomenclature in this study for convenience in discussion. All encapsulated pneumococcal strains carry the cps genes as a gene cluster in the same locus of the genome between the dexB and aliA genes (5, 6), referred to as the cps locus here. In all type 37 strains, the genes in the dexB-aliA locus are degenerated; the type 37 synthase gene (tts) is located elsewhere on the chromosome (8). The cps genes of S. pneumoniae are highly variable, ranging from a single gene in type 37 to a locus of many genes in the vast majority of other serotypes. The sizes of the cps loci range from approximately 2.2 kb (type 37) to 30 kb (type 38), with an average of 21 kb (5). The cps locus of the type 2 strain D39, an extensively studied virulent S. pneumoniae strain, contains at least 17 genes (20.316 kb in size) (13).
In all WZY-dependent CPS types, the cps locus consists of the common genes (cpsA, cpsB, cpsC, and cpsD) and downstream type-specific genes that are involved in CPS synthesis and export (11). Previous studies showed that mutations in each of the S. pneumoniae cpsABCD genes lead to significant reduction in capsule synthesis, as well as impaired nasopharyngeal colonization and virulence in animal models (14–16). Homologs of the cpsABCD genes are also present in the capsule loci of other encapsulated streptococci (17–19), as well as the loci for exopolysaccharide synthesis of oral streptococci (20, 21). CpsA is homologous to the transcriptional regulator LytR of Bacillus subtilis (3). The cpsA mutations in Streptococcus agalactiae lead to significant reductions in transcription of the cps genes and capsule production (22). Recent studies have shown specific binding of recombinant CpsA to the promoter sequences of the cps locus in Streptococcus iniae and S. agalactiae (23, 24), suggesting that CpsA is a transcriptional regulator of the cps loci. CpsB (phosphotyrosine phosphatase), CpsC (tyrosine kinase), and CpsD constitute a phosphorelay system, which regulates the phosphorylation status of CpsD and thereby CPS production (16, 25–28). The level of CpsD phosphorylation is positively correlated with S. pneumoniae encapsulation in clinical isolates, and CpsD phosphorylation is suppressed by atmospheric oxygen conditions (29). The oxygen concentration appears to affect CpsD phosphorylation by modulation of Cps2B-mediated phosphatase activity (28). The precise functions of CpsABCD and their homologs in encapsulation or exopolysaccharide synthesis remain undefined.
Previous studies have demonstrated that the amount of CPS produced by S. pneumoniae differs under various conditions (30–32). There is a reduction in capsule formation in the airways and whenever the bacteria are in close contact with host cells (31–34). In contrast, dissemination of S. pneumoniae to the blood leads to a significant increase in capsule production (35, 36). The well-characterized phase variation in pneumococcal colony opacity is associated with the changes in the amount of CPS (30). Compared with their isogenic transparent counterparts, the opaque variants produce significantly more CPS under standard culture conditions (37) and even more CPS under anaerobic growth conditions (29). As a result, the opaque variants are less susceptible to opsonophagocytic killing (36) and more virulent in mouse infection models (37). These phenotypic changes have indicated that CPS production is subject to regulation by environmental conditions. Ogunniyi et al. reported that the cps locus of strain D39 is transcriptionally upregulated during pneumococcal infection in the bloodstream (38). However, the transcription and regulation of the cps genes in S. pneumoniae are poorly understood.
The transcription start sites of the S. pneumoniae cps locus in the type 1 strain 13868 (39) and the type 4 strain TIGR4 (40) have been mapped to the 23rd and 24th nucleotides upstream of the cpsA coding sequence, respectively. Shainheit et al. showed that the sequence between the −35 promoter motif and the transcription start site of the cps locus in the type 4 strain TIGR4 is essential for the transcription of the cps gene cluster (40). There is extensive sequence diversity upstream of the transcription start sites. Moscoso and Garcia divide the sequences between dexB and cpsA from 115 S. pneumoniae strains into 26 sequence organizations (SOs) (41). The 1st SO is the most frequently reported among all 26 of the SOs, which include the type 2 strain D39 and 21 other strains. The majority of the SOs consists of one or more insertion elements (IE), a repeat unit of pneumococcus (RUP), and a conserved sequence immediately upstream of cpsA that contains the predicted −10 and −35 promoter motifs. Except for the core promoter recently defined in TIGR4 (40), the precise constituents of the cps promoter in S. pneumoniae remain undefined.
In this context, we sought to characterize the transcription of the cps locus in the type 2 strain D39, a virulent strain widely used in studying pneumococcal biology and pathogenesis (42). Intergenic reverse transcription (RT)-PCR, Northern blotting, and primer extension analyses revealed that the cps gene cluster is cotranscribed as an operon from a major transcription start site at the −25th nucleotide (G) upstream of cps2A. Unmarked truncations in the intergenic region between the dexB and cps2A coding sequences showed that the full transcription of the cps genes and encapsulation in D39 not only depend on the core promoter immediately upstream of cps2A, but also require additional sequence elements upstream of the core promoter, particularly a 59-bp spacing sequence. Lastly, in-frame deletion of cps2A in D39 did not significantly affect the transcription of the cps locus, although the mutant produced significantly less CPS, indicating that CpsA is not a transcriptional regulator. This study has yielded important information on the transcriptional characteristics of the cps locus in S. pneumoniae.
MATERIALS AND METHODS
Bacterial strains, cultivation, and chemical reagents.
S. pneumoniae was cultured at 37°C under aerobic conditions with 5% CO2 or under anaerobic conditions as described previously (29, 43). The growth kinetics of D39 derivatives were determined in Todd-Hewitt broth supplemented with yeast extract (THY broth) as follows. Fresh THY broth (4 ml) was inoculated with the frozen stock (1 ml) of each strain, and the bacteria were allowed to grow at 37°C to an optical density at 620 nm (OD620) of 0.4, pelleted by centrifugation at 3,000 × g for 5 min, and resuspended in 1 ml fresh THY broth as seeding cultures. Subsequent cultures were initiated by mixing THY broth with predetermined dilutions of the seeding cultures to an OD620 of 0.002 to 0.006. Growth was monitored hourly by measuring the OD620 value of each culture. Escherichia coli DH5α was used to construct plasmids. All the bacterial strains and plasmids used in this study are described in Table 1. All the chemicals used in this work were obtained from Sigma-Aldrich (Shanghai, China) unless otherwise stated. All the restriction enzymes were purchased from New England BioLabs (NEB) (Beijing, China).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Antibiotic resistancea | Reference or source |
|---|---|---|---|
| D39 | TH4533 S. pneumoniae strain D39, serotype 2, encapsulated | NCTC | |
| D39s | D39 derivative; rpsL1 | Smr | This study |
| ST196 | S. pneumoniae, serotype 6A, encapsulated | 73 | |
| ST395 | S. pneumoniae, strain TIGR4, serotype 4, encapsulated | ATCC | |
| ST556 | S. pneumoniae, serotype19F, encapsulated | 43, 74 | |
| ST588 | D39s derivative; ΔcbpA::JCb rpsL1 | Kanr | 44 |
| ST787 | TH4533 derivative; the RUP region is replaced with JC ΔRUP::JC, a T-C mutation in the −10 promoter motif of the cps locus; unencapsulated | Kanr | This study |
| TH4317 | D39s derivative; the entire cps2ABCD region is replaced with JC; Δcps2ABCD::JC | Kanr | This study |
| TH4319 | TH4317 derivative, the cps2A region is removed; Δcps2A | Smr | This study |
| TH4323 | TH4317 derivative; the cps2C region is removed; Δcps2C | Smr | This study |
| TH4325 | TH4317 derivative; the cps2D region is removed; Δcps2D | Smr | This study |
| TH4455 | TH4317 derivative; the cps2B region is removed; Δcps2B | Smr | This study |
| TH4458 | TH4317 derivative; the cps2ABCD region is removed; Δcps2ABCD | Smr | This study |
| TH4702 | D39s derivative; the dexB-cps2A region is replaced with JC; ΔdexB-cps2A::JC | Kanr | This study |
| TH4525 | TH4702 derivative; the entire dexB-cps2A region is removed; ΔdexB-cps2A | Smr | This study |
| TH5048 | TH4702 derivative; the IE region is removed; ΔIE | Smr | This study |
| TH5049 | TH4702 derivative; the IE-RUP region is removed; ΔIE-RUP | Smr | This study |
| TH5050 | TH4702 derivative; the IE-RUP-SS region is removed; ΔIE-RUP-SS | Smr | This study |
| TH5615 | TH4702 derivative; the RUP region is removed; ΔRUP | Smr | This study |
| TH5618 | TH4702 derivative; the core promoter is removed; Δcore | Smr | This study |
| TH5621 | TH4702 derivative; the RUP-SS region is removed; ΔRUP-SS | Smr | This study |
| TH5648 | TH4702 derivative; the SS region is removed; ΔSS | Smr | This study |
| TH4766 | TH4702 derivative; the dexB-cps2A region is replaced with the corresponding region, dexB-cps4A, of TIGR4 | Smr | This study |
| TH5966 | TH4702 derivative; the SS region of TIGR4 is removed; ΔSS | Smr | This study |
| pVA838 | E. coli-S. pneumoniae shuttle vector | Cmr | 54 |
| pST1111 | pVA838 derivative containing a 265-bp insertion sequence of the D39 cps locus | Cmr | This study |
| pR424 | ColE1 derivative carrying an S. pneumoniae 5′-ssbB-targeting fragment driving luc | Cmr | 47 |
| pIB166 | E. coli-S. pneumoniae shuttle vector | Cmr | 46 |
| pTH3932 | pIB166 derivative containing the sequence between the stop codon of dexB and the putative ribosomal binding site of cps2A and luc | Cmr | This study |
| pTH3937 | pTH3932 derivative; the original promoter is removed; WT | Cmr | This study |
| pTH6460 | pTH3937 derivative; the 3′ 20-bp SS segment is replaced | Cmr | This study |
| pTH6461 | pTH3937 derivative; the middle 20-bp SS segment is replaced | Cmr | This study |
| pTH6462 | pTH3937 derivative; the 5′19-bp SS segment is replaced | Cmr | This study |
| pTH6463 | pTH3937 derivative; the 10-bp SS segment adjacent to the −35 promoter motif is replaced | Cmr | This study |
| pTH6491 | pTH3937 derivative; the entire SS element is removed; ΔSS | Cmr | This study |
| pTH6494 | pTH3937 derivative; the 10-bp SS and 10-bp segment distal to the −35 promoter motif are replaced | Cmr | This study |
Antibiotic resistance markers: Kanr, kanamycin; Smr, streptomycin; Cmr, chloramphenicol.
JC, Janus cassette.
Primers and DNA sequence analysis.
All of the primers used in this study were synthesized by Sangon Biotech (Beijing, China) and are described in Table S1 in the supplemental material. To minimize sequence errors during PCR amplification, a high-fidelity PrimeStar DNA polymerase (TaKaRa, Dalian, China) was used for all PCRs in this study. All DNA mutations were verified by PCR amplification and DNA sequencing analysis. Rho-independent transcription terminators were identified by the Internet-based ARNold program (http://rna.igmors.u-psud.fr/toolbox/arnold/index.php#Results).
Construction of pneumococcal mutants.
All mutagenesis work in the cps locus was carried out in D39s, a streptomycin-resistant derivative of D39, essentially as described previously (44). All unmarked deletions in the dexB-cps2A intergenic region were constructed in strain TH4702, a promoter-null D39s derivative in which the dexB-cps2A region was replaced by a Janus cassette (JC) (45). TH4702 was established as follows. The up- and downstream sequences of the dexB-cps2A region were amplified with primer pairs Pr7339/Pr7345 and Pr7348/Pr7344, respectively, from genomic DNA of D39. The amplicon was ligated to the 5′ and 3′ ends of the JC by fusion PCR with primers Pr7339 and Pr7344, as described previously (44), and transformed into strain D39s as described previously (44). The JC was amplified with primers Pr7346 and Pr7347 from genomic DNA of strain ST588 (44).
To generate unmarked deletions in the dexB-cps2A region, the up- and downstream sequences of the target sequences were amplified with the following primer pairs: IS630-Spn1 (IE), Pr7339/Pr7177 and Pr7178/Pr7344; RUP, Pr7339/Pr8584 and Pr8585/Pr7344; spacing sequence (SS), Pr7339/Pr8586 and Pr8587/Pr7344; core promoter, Pr7339/Pr8588 and Pr8589/Pr7344; the entire dexB-cps2A region, Pr7339/Pr7183 and Pr7184/Pr7344; IE-RUP, Pr7339/Pr7179 and Pr7180/Pr7344; RUP-SS, Pr7339/Pr8590 and Pr8591/Pr7344; and IE-RUP-SS, Pr7339/Pr7181 and Pr7182/Pr7344. The up- and downstream amplicons were ligated by fusion PCR with primers Pr7339 and Pr7344 before being transformed in strain TH4702 (44). A similar process was used to construct the RUP-null strain ST787. The up- and downstream sequences were amplified with primer pairs Pr646/Pr652 and Pr653/Pr649. The two amplicons were ligated by fusion PCR with primers Pr646 and Pr649. Sequencing analysis showed that ST787 carried the expected RUP deletion, as well as an accidental point mutation in the −10 promoter motif of the cps locus (data not shown). In the same manner, the entire dexB-cps2A region in D39 was replaced with the dexB-cps4A sequence of TIGR4 in its entirety (TH4766) or a truncated form lacking the 61-bp SS element (TH5966). The full dexB-cps4A sequence was amplified with primers Pr7341 and Pr7342 from genomic DNA of TIGR4. The partial dexB-cps4A construct in strain TH5966 was sequentially generated by amplifying the flanking sequences of the 61-bp SS element from genomic DNA of strain TH4766 with Pr7339/Pr8586 and Pr8587/Pr7344, ligating the two amplicons by fusion PCR with primers Pr7339 and Pr7344, transforming the final amplicon in strain TH4702, and counterselection for streptomycin resistance.
Unmarked deletions in the cps2ABCD region were generated in D39s in a similar fashion. First, strain TH4317 lacking the cps2ABCD genes was constructed in D39s by allelic replacement with the JC as follows. The up- and downstream sequences of cps2ABCD were amplified from genomic DNA of D39 with primer pairs Pr6854/Pr6852 and Pr6853/Pr1276 and fused to the 5′ and 3′ ends of the JC amplicon by fusion PCR using primers Pr6854 and Pr1276. The JC was amplified from genomic DNA of ST588 (44) with primers Pr6850 and Pr6851. The final amplicon was transformed in D39s to obtain the kanamycin-resistant and streptomycin-sensitive strain TH4317, in which the cps2ABCD coding region was replaced by the JC. TH4317 was used to reconstitute various segments of the cps2ABCD region and thereby establish desirable in-frame deletions in each of the cps2ABCD genes by homologous recombination and streptomycin counterselection. For in-frame deletion of cps2A, the up- and downstream sequences were amplified with primer pairs Pr6854/Pr6754 and Pr6755/Pr1276, respectively, and linked by fusion PCR with primers Pr6854 and Pr1276. The final PCR product was used to transform strain TH4317, resulting in strain TH4319. The D39s derivatives lacking cps2B (TH4455), cps2C (TH4323), cps2D (TH4325), or cps2ABCD (TH4458) were constructed in strain TH4317 in a similar manner. The 5′ and 3′ sequences of the other target genes were amplified with the following primer sets: cps2B, Pr6854/Pr6756 and Pr6767/Pr1276; cps2C, Pr6854/Pr6758 and Pr6769/Pr1276; cps2D, Pr6854/Pr6760 and Pr6761/Pr1276; and cps2ABCD, Pr6854/Pr7022 and Pr7023/Pr1276. The flanking amplicons of each target gene were covalently linked by fusion PCR with primers Pr6854 and Pr1276 before being used to transform TH4317.
Transcriptional reporter constructs and luciferase assay.
Luciferase reporter constructs of the cps promoter region were prepared in the shuttle vector pIB166 (46). The promoterless firefly luciferase gene (luc) and the sequence between the stop codon of dexB and the putative ribosomal binding site of cps2A were individually amplified from plasmid pR424 (47) and genomic DNA of strain D39 using primer pairs Pr6214/Pr5791 and Pr6212/Pr6213, respectively. The cps promoter and luc sequences were digested with BamHI/XmaI and XmaI/SacI, respectively, before being coligated to BamHI/SacI-digested pIB166, resulting in plasmid pTH3932 (Table 1). The original promoter upstream of the multiple cloning site in pTH3932 was removed by digestion with ApaI/BamHI, treatment with Klenow DNA polymerase, and religation, resulting in plasmid pTH3937, carrying luc driven by the wild-type (WT) dexB-cps2A sequence. The mutations in the SS element were introduced by PCR amplification of pTH3937 with primer pairs Pr9638/Pr9639, Pr9641/Pr9642, Pr9643/Pr9644, Pr9645/Pr9646, Pr9647/Pr9648, and Pr9649/Pr9650, resulting in plasmids pTH6460, pTH6461, pTH6462, pTH6463, pTH6494, and pTH6491, respectively, after the PCR products were circularized in E. coli as described previously (48). The irrelevant DNA sequences were from a previous study of the upstream (UP) promoter element (49). The promoterless luc construct pTH3936 (vector control) was generated by digestion with ApaI/XmaI, treatment with Klenow DNA polymerase, and religation. The luciferase assay was performed as described previously (47). All assays were repeated at least three separate times. The results of representative experiments are presented as means of three replicates plus standard deviations.
RNA extraction and Northern blotting.
Total pneumococcal RNA was purified using the TRIzol reagent (Invitrogen, Beijing, China) according to the supplier's instructions. Northern blotting was carried out as described previously (50). The following DNA probe fragments were amplified with the indicated primers: cps2A (Pr894/Pr895), cps2B (Pr1168/Pr1169), cps2C (Pr1170/Pr1171), cps2D (Pr1172/Pr1173), cps2O (Pr972/Pr973), and cbpA (Pr74/Pr758). Each of the PCR products was used to generate 32P-labeled DNA probes by random priming using the NEBlot kit (NEB) according to the manufacturer's instructions.
RT-PCR.
The cotranscription of the cps genes in D39 was determined by intergenic RT-PCR essentially as described previously (51). The cDNA molecules (cDNAs) of D39 were prepared using total RNA preparation (described above) with an iScript cDNA synthesis kit (Bio-Rad, Beijing, China) as instructed by the supplier. This cDNA pool was used to amplify the intergenic regions of the D39 cps genes with the following primer pairs: Pr8067/Pr8068, cps2A-cps2B; Pr8069/Pr8070, cps2B-cps2C; Pr8071/Pr8072, cps2C-cps2D; Pr8073/Pr8074, cps2D-cps2E; Pr8075/Pr8076, cps2E-cps2T; Pr8077/Pr8078, cps2T-cps2F; Pr8079/Pr8080, cps2F-cps2G; Pr8081/Pr8082, cps2G-cps2H; Pr8083/Pr8084, cps2H-cps2I; Pr8085/Pr8086, cps2I-cps2J; Pr8087/Pr8088, cps2J-cps2K; Pr8089/Pr8090, cps2K-cps2P; Pr8091/Pr8092, cps2P-cps2L; Pr8093/Pr8094, cps2L-cps2M; Pr8095/Pr8096, cps2M-cps2N; and Pr8097/Pr8098, cps2N-cps2O. As a positive control for cotranscription, the intergenic region of the previously characterized SPD2018-cbpA operon (52) was also amplified with primers Pr8105 and Pr8106. The mRNA level of the cps locus was quantified by quantitative RT-PCR (qRT-PCR) as described previously (40). Primer pairs Pr7705/Pr7706 and Pr7707/Pr7708 were used to amplify cps2A and cps2E, respectively. rplI, encoding a ribosomal protein, was amplified with primers Pr7709 and Pr7710 as an internal control for the constitutively expressed gene. The relative expression level was calculated using the average mean cycle threshold (CT) value for the genes of interest for each sample and rplI. The following equation was used to obtain the normalized values of gene transcription: 1.8e (CT rplI − CT gene of interest). The results of representative experiments are presented as means of three replicates plus standard deviations.
Immunoblotting.
Pneumococcal CPS was detected by immunoblotting essentially as described previously (40). For aerobic cultures, all D39 derivatives were grown to the same phase (OD620, ∼0.4) before being processed for CPS detection. Because the anaerobic culture device was not equipped for frequent monitoring of bacterial density by spectrophotometry, all anaerobic cultures were pelleted by centrifugation and resuspended to an OD620 of 0.4 in phosphate-buffered saline (PBS) before being used for CPS immunoblotting. The type 2 CPS was detected with a rabbit anti-serotype 2 serum (Statens Serum Institut, Copenhagen, Denmark) and peroxidase-conjugated goat anti-rabbit IgG(H+L) (Bio-Rad) at dilutions of 1:20,000 and 1:10,000, respectively. Binding of the CPS and antibody was visualized with a Clarity Western ECL reagent (ChemiDOC XRS+) from Bio-Rad according to the supplier's instructions. The CPS on each spot was digitized with ImageJ software (ImageJ 1.47v; National Institutes of Health) on the basis of its chemiluminescence intensity level. The results of representative experiments are presented as means of at least three replicates plus standard deviations.
Primer extension.
Primer extension was carried out with primer Pr1025 (located in the complementary strand of the cps2A 5′ coding region) as described previously (53). For a DNA ladder, plasmid pST1111 was incubated with the same radiolabeled Pr1025 and sequenced using a sequencing kit (USB, Cleveland, OH) according to the supplier's instructions. pST1111 was a pVA838 (54) derivative containing a 265-bp insertion sequence of the D39 cps locus, which consisted of a 217-bp dexB-cps2A sequence and a 48-bp cps2A 5′ coding sequence. The cps insertion sequence was amplified from genomic DNA of D39 with primers Pr1025 and Pr1026. The gel was dried and analyzed with a Molecular Dynamics Storm 820 PhosphorImager.
Host cell adhesion.
Pneumococcal adhesion to A549, a human lung alveolar carcinoma epithelial cell line, was assessed in 24-well plates as described previously (55). Each monolayer of A549 cells was infected with 107 CFU of pneumococci. The results of representative experiments are presented as means of four replicates plus standard deviations.
Mouse infection.
Pneumococcal virulence was determined in a coinfection mouse bacteremia model as described previously (56). All animal infection procedures were in compliance with the guidelines of the Institutional Animal Care and Use Committee of Tsinghua University. Groups of 10 female CD1 mice (6 to 8 weeks old; Vital River, Beijing, China) were infected with approximately 500 CFU of pneumococci (∼250 CFU each for the mutant and parent strains) by intraperitoneal (i.p.) inoculation. The 50% lethal dose (LD50) of D39 by the i.p. infection route was approximately 250 CFU (data not shown). Blood samples (∼50 μl) were collected by cardiac puncture and mixed with 3 μl of 500 mM EDTA to enumerate CFU. The level of attenuation was expressed as a competitive index (CI). The CI was defined as the output CFU ratio (mutant/wild type) divided by the input CFU ratio (mutant/wild type), as described previously (56). To differentiate parent and mutant strains in bacterial mixtures in animal experiments, we performed all coinfections for the D39s derivatives by mixing each of them (streptomycin resistant) with the wild-type D39 (streptomycin sensitive). Potential attenuation of D39s (streptomycin resistant) was predetermined by performing coinfection with the mixtures of the wild-type D39 and D39s in a 1:1 ratio. The final CI values of the D39s derivatives were obtained after normalization with the mean CI of D39s (mutant CI/D39s mean CI). When only the wild-type pneumococci (no mutants) were recovered from a mouse in the coinfection experiments, a value of 1 was given to the output mutant for convenience of data presentation in common logarithms.
Statistical analysis.
The statistical significance of the data from qRT-PCR, the luciferase data, and the immunoblotting data was determined by two-tailed unpaired Student's t tests. The data from animal infection experiments and cell adhesion were analyzed using Wilcoxon signed-rank tests and one-way analysis of variance (ANOVA), respectively. Significant differences in statistics were defined by P values of <0.05, <0.01, and <0.001.
RESULTS
Transcription of the cps gene cluster as an operon.
Based on the short intergenic sequences and functional relationship of the genes in the cps locus, it is predicted that the genes in the locus are transcribed as a single transcript from a common promoter upstream of cpsA (3, 13). However, the cotranscription of the cps genes has not been experimentally demonstrated. In this study, we tested the cotranscription of the cps gene cluster in the type 2 strain D39 by RT-PCR with the primers spanning the intergenic regions of the adjacent cps genes. The locus contains 19,984 nucleotides, which comprise 19 open reading frames (ORFs) between the flanking dexB and aliA genes, as annotated in the genome sequence of D39 (42) (illustrated in Fig. 1A). The first 17 genes are predicted to be involved in CPS production; the last two ORFs (SPD332 and SPD333) are homologous to the IS66 family insertion element and may not be related to pneumococcal encapsulation.
FIG 1.
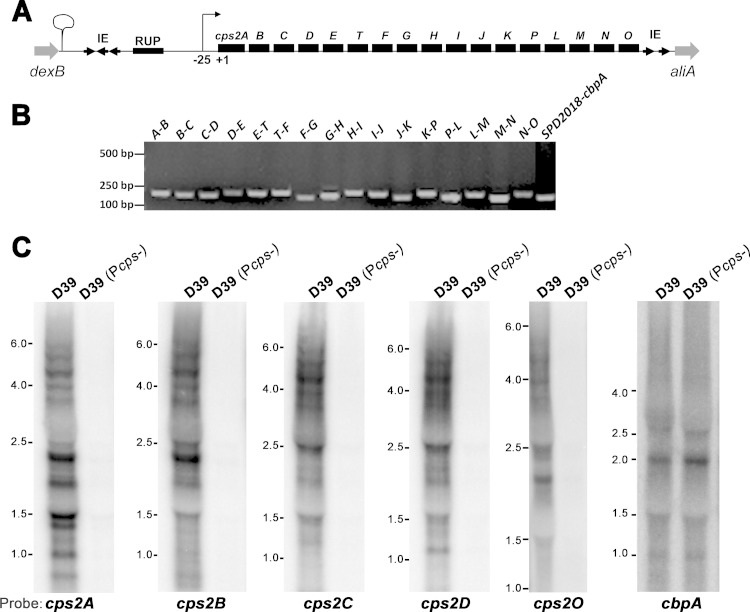
Detection of cotranscription between adjacent cps genes. (A) Schematic map of the type 2 cps gene locus in D39. The orientations of the demarcating genes (dexB and ailA) and the ORFs in the IE are indicated with arrows. The transcription start site of cps2A (Fig. 2) is marked according to its relative distance (in base pairs) from the ATG start codon of cps2A. A predicted transcription terminator downstream of dexB is indicated with a hairpin. (B) Detection of cotranscription between the adjacent cps genes in D39 by RT-PCR. Potentially cotranscribed mRNAs from adjacent genes in the total RNA of D39 were sequentially converted to cDNAs and PCR products by reverse transcription and PCR, respectively. The intergenic PCR products were separated by DNA gel electrophoresis and are identified by the abbreviations of the adjacent cps genes. Molecular markers are indicated on the left. (C) Detection of cps transcription by Northern blotting. Total RNA preparations of D39 and its unencapsulated derivative ST787, D39 (Pcps-), were separated by RNA gel electrophoresis, blotted to nylon membranes, and hybridized with 32P-labeled probes of the cps genes or cbpA (an RNA loading control) of D39. The sizes of RNA standards are marked in kilonucleotides on the left of each gel.
The RT-PCR experiment yielded a PCR product with the predicted molecular size for each pair of adjacent genes in the cps locus (Fig. 1B). As negative controls, no PCR products were detected for all of the primer pairs in similar reactions with the reverse transcription mixture lacking reverse transcriptase (data not shown). Because the capsule gene operons in other bacteria can be driven by a major promoter and additional internal promoters, as exemplified in Staphylococcus aureus (57–59), we also tested the transcription of the cps genes from potential internal promoters in the operon of D39 by performing the same set of RT-PCR with an RNA preparation from strain ST787, a D39 derivative with undetectable mRNAs of the cps genes (see Fig. 1C). None of these reactions yielded detectable PCR products (data not shown), strongly suggesting that there are no strong internal promoters in the cps locus. Lastly, the positive control of the SPD2018-cbpA operon produced a predicted RT-PCR product (Fig. 1B, far-right lane).
We attempted to detect a single large mRNA species from D39 by Northern blotting without success. Instead, the experiment identified multiple bands ranging from approximately 1,000 to 6,000 nucleotides, which were completely undetectable in the unencapsulated D39 derivative ST787 due to a point mutation in the −10 promoter sequence (Fig. 1C). As an RNA loading control, the major cbpA transcript was detected in both D39 and ST787. It is worth noting that the probes representing five different cps genes (cps2A, cps2B, cps2C, cps2D, and cps2O) revealed similar but not identical banding patterns. This may be caused by degradation of the large cps transcript. These results demonstrated that the 17 genes in the cps locus of D39 are transcribed as an operon from the common promoter upstream of cps2A.
Identification of the transcription start site in the cps locus.
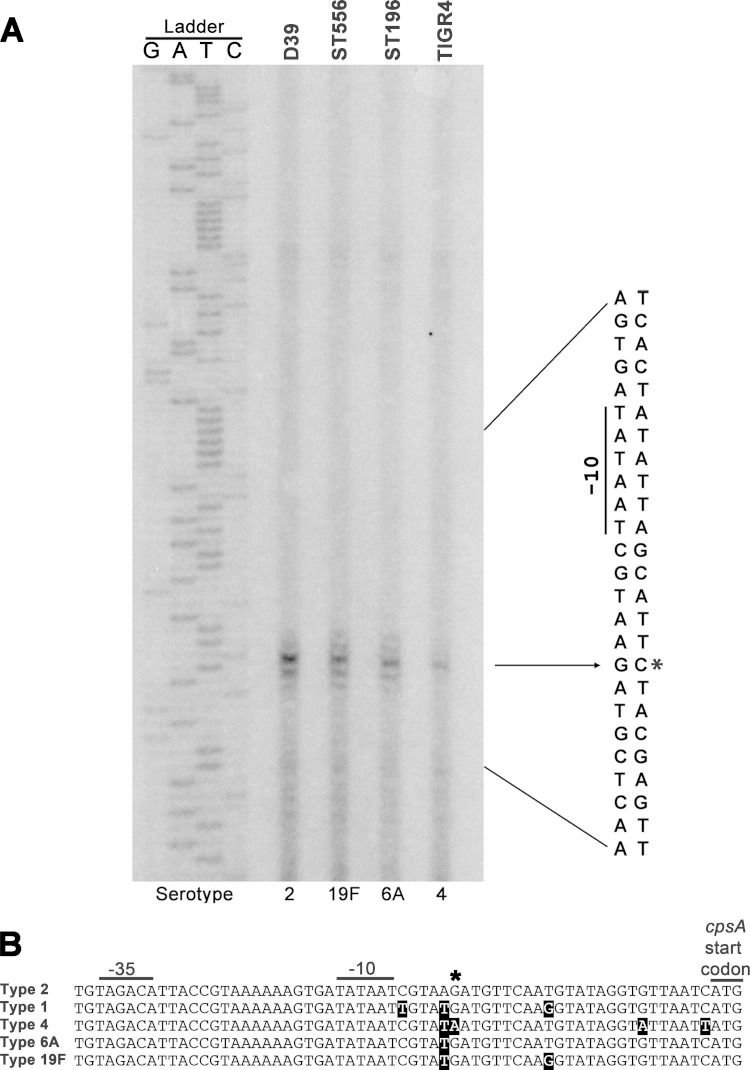
Previous studies have determined the transcription start sites for the type 1 and 4 cps loci of S. pneumoniae (39, 40). The major transcript is initiated at the 23rd (T) or 24th (A) nucleotide upstream of cpsA in the cps locus of the type 1 strain 13868 (39) or the type 4 strain TIGR4 (40), respectively. Because of extensive sequence variability in the cps locus (5, 41), we sought to determine the transcription start sites of the cps loci in strains D39 (type 2), TIGR4 (type 4), ST196 (type 6A), and ST556 (type 19F) by primer extension with primer Pr1025, which is located in the 5′ complementary strand of cps2A in D39 (SPD0315). The result showed a major band and multiple minor bands around the major band from the primer extension reaction mixtures with the RNA preparations of the four strains (Fig. 2A). The major band for D39 represented a transcript starting at the −25th nucleotide (G) upstream of the cps2A start codon. As shown in Fig. 2B, there is an identical sequence in the region of the RT primer Pr1205 and the same number of nucleotides between the primer Pr1205 and the predicted −35 motif. It is reasonable to conclude that, in strains TIGR4, ST196, and ST556, the transcription of the cps locus starts in the same position upstream of cpsA as in D39. Based on this result, the major cps mRNA in D39 is 1 or 2 nucleotides longer than that reported for the type 4 strain TIGR4 (40) or the type 1 strain 13868 (39), respectively. These discrepancies may be explained by the observation of minor transcription start sites around the major start position for all S. pneumoniae strains tested in this study (Fig. 2A). There are 6 nucleotides that separate the major transcription start site of cpsA from the previously predicted −10 promoter motif of the type 2 cps locus (13).
FIG 2.
Mapping the transcription start sites in the cps locus of D39 by primer extension. (A) Detection of the transcription start sites for the cpsA gene by primer extension in strains D39 (type 2), TIGR4 (type 4), ST196 (type 6A), and ST556 (type 19F). The major transcription start site of cps2A in strain D39 is indicated by an arrow and an asterisk. The predicted −10 and −35 promoter motifs of the cps promoter are marked by lines. (B) Alignment of the upstream noncoding sequences of the cpsA genes among five capsular serotypes (1, 2, 4, 6A, and 19F) of S. pneumoniae. The transcription start site of the cps locus identified for strain D39 in panel A is indicated by an asterisk. The predicted promoter motifs (−10 and −35) and the translation start codon of cpsA are marked with lines. The nucleotides that are different from those in D39 are shaded.
Mapping the promoter activity in the dexB-cps2A region of strain D39.
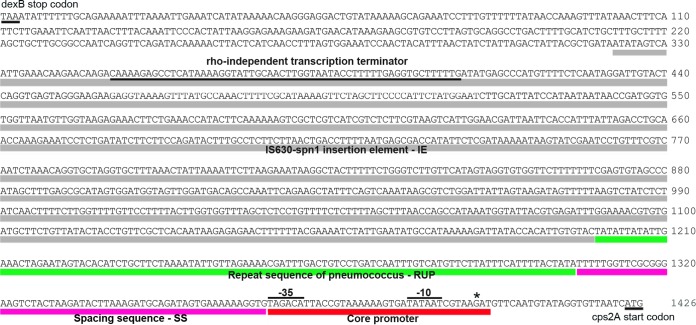
There are 1,420 bp between the stop codon of dexB and the start codon of cpsA, the first gene in the cps locus in D39 (Fig. 3), referred to as the dexB-cps2A region here. This region contains a 5′ IS630-Spn1 insertion element (881 bp), a RUP (107 bp), a 59-bp SS element, and a 3′ 59-bp segment proximal to cps2A that contains the predicted −10 and −35 promoter motifs and the transcription start site. Shainheit et al. have recently shown that the counterpart of the last sequence in strain TIGR4, referred to as the core promoter, is able to drive the transcription of the cps locus (40). Therefore, we refer to this region as the core promoter region here. Bioinformatic analysis identified a putative Rho-independent transcription terminator in the 5′ region of IS630-Spn1 (Fig. 3), which is likely to terminate transcription from the upstream dexB gene.
FIG 3.
Sequence annotation of the dexB-cps2A region in strain D39. The four sequence elements (IS630-Spn1, RUP, SS element, and core promoter) are differently colored. The sequences of the −10 and −35 promoter motifs, predicted Rho-independent transcription terminator, dexB stop codon, and cps2A start codon are indicated with lines. The transcription start site is marked with an asterisk.
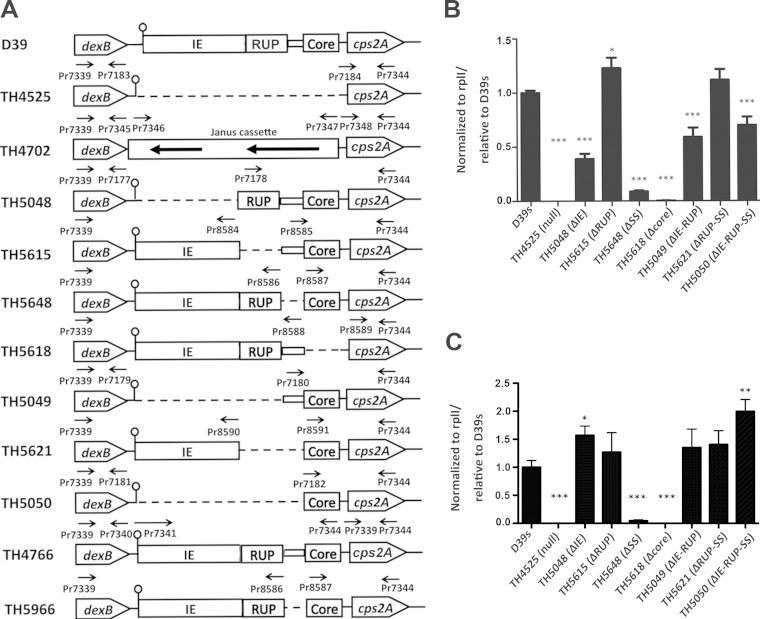
To define the impacts of these sequence elements on the transcription of the cps operon, we generated a series of unmarked deletions in the dexB-cps2A region of the D39 genome by sequence replacement and counterselection (Fig. 4A) and determined the transcription level of cps2A in the resulting D39 derivatives by qRT-PCR. Since the locus is transcribed as an operon, we used the mRNA level of the cps2A gene as a marker for the transcription of the locus in this study. To preserve the Rho-independent transcription terminator in the 5′ region of IS630-Spn1, we deleted only the 3′ IE segment immediately after the terminator (Fig. 3).
FIG 4.
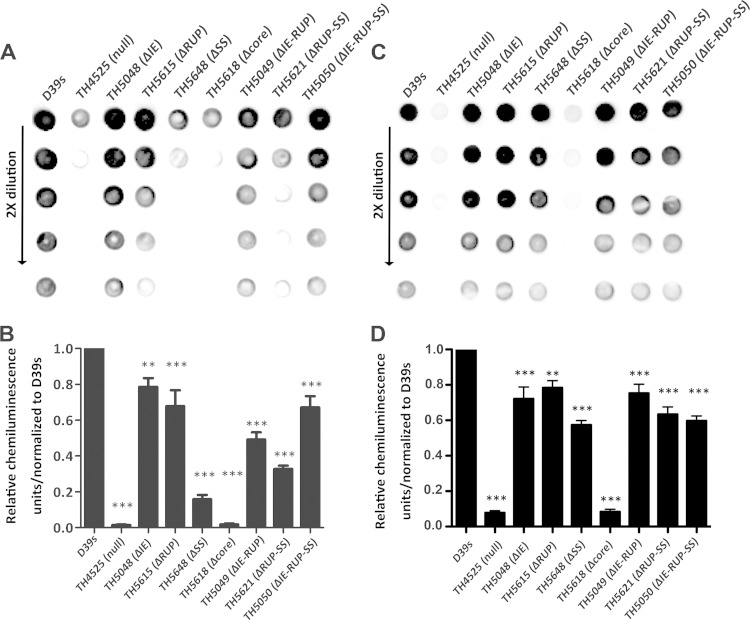
Detection of cps2A mRNA in D39s and its mutants with unmarked deletions in the dexB-cps2A region. (A) Schematic illustration of deletion constructs in the dexB-cps2A region. The deletions in the IS630-Spn1 (IE) RUP, SS element (rectangle), and core promoter (Core) elements upstream of cps2A are indicated with dashed lines. The matchsticks represent the predicted terminator downstream of dexB. The primers used to amplify individual regions are shown above the target sequences. (B) Quantification of cps2A mRNA by qRT-PCR in the cps promoter mutants of D39s cultured under aerobic conditions. A segment of the cps2A mRNA in total RNA preparations of D39s and its mutants was sequentially converted to a cDNA and a PCR product by reverse transcription and PCR, respectively. The amount of the cps2A PCR product in each sample was quantified by its average mean cycle threshold value and normalized with that of the housekeeping gene rplI in the same reaction. (C) Same as panel B, except that pneumococci cultured under anaerobic conditions were used. *, P <0.05; **, P < 0.01; ***, P < 0.001.
We first determined the impact of deletion mutations in the dexB-cps2A region on cps2A transcription in THY broth under aerobic growth conditions with 5% CO2. As expected, removing the entire dexB-cps2A region (strain TH4525) abrogated the transcription of cps2A compared with the parent strain, D39s (Fig. 4B). We next individually deleted each of the sequence elements in this region to define their contributions to cps transcription. While deleting IS630-Spn1 (ΔIE; TH5048) reduced the level of the cps2A mRNA by 60%, the strain lacking a RUP (ΔRUP; TH5615) showed a small but statistically significant increase (22%) in cps2A transcription. The mutant TH5648, lacking the 59-bp SS element (ΔSS), showed a much more significant reduction (10.1-fold; P < 0.001) in cps2A transcription. The greatest impact on cps transcription was observed with the mutant TH5618, which did not show detectable transcription of cps2A. TH5618 lost a 41-bp sequence in the core promoter region, which contains the putative −10 and −35 promoter motifs, and the transcription start site of the cps locus. This result is consistent with the recent study by Shainheit et al. (40), in which the corresponding sequence deleted in TH5618 is essential for the transcription of the cps locus in the type 4 strain TIGR4. As exemplified in Fig. 2 and Fig. S1 in the supplemental material, the core promoter region is highly conserved among the WZY-dependent CPS types of S. pneumoniae, including the 12 CPS types that are most frequently associated with pneumococcal disease and covered by the latest 13-valent childhood vaccine (60). According to the boundary of the cps core promoter described by Shainheit et al. (40), there are only two nucleotide polymorphisms between D39 and TIGR4 in this region. Interestingly, one of the two variable nucleotides coincides with the transcription start site of the cps locus in TIGR4 (40). These results indicated that the core promoter is essential for the cps promoter activity in D39, as reported for strain TIGR4 (40), but its upstream sequence elements, particularly the SS element, can influence cps transcription.
To further define the contributions of the sequence elements upstream of the core promoter, we performed additional unmarked deletions in the region. Removing the IE-RUP (TH5049) or IE-RUP-SS (TH5050) segment led to small but significant reductions in cps2A transcription (Fig. 4B). Surprisingly, the RUP-SS mutant (TH5621) showed a level of cps2A transcription similar to that of the parent strain, namely, the transcription activity for the fusion construct of the IE and core promoter sequences was as strong as that for the native dexB-cps2A region. These data suggest that the SS element enhances the strength of cps transcription by improving the sequence and/or structural context of the core promoter for the optimal activity of the RNA polymerase (RNAP) in the cps locus.
Because the pneumococci produce more CPS under anaerobic growth conditions (29), we wondered whether this effect occurs at the transcription level. We thus repeated the above-described experiments with the same D39 mutants cultured under anaerobic conditions. Similar to the results obtained with the pneumococci grown under aerobic conditions (Fig. 4B), deleting the core promoter or SS element from the dexB-cps2A region virtually abolished cps2A transcription in the bacteria exposed to anaerobic conditions (Fig. 4C). This result thus confirmed that the core promoter and SS elements are essential for cps transcription under both aerobic and anaerobic conditions. The only major difference between the two growth conditions was in the ΔIE mutant (TH5048), in which cps2A transcription was significantly reduced under aerobic conditions but increased under anaerobic conditions (Fig. 4C). This observation suggests that the IE sequence plays different roles in cps transcription under different environmental conditions. Lastly, comparative analysis of D39 cell growth under aerobic or anaerobic conditions did not reveal a significant condition-dependent difference in the cps2A mRNA levels (data not shown), indicating that the transcription in the cps locus does not significantly contribute to the increased CPS production under anaerobic conditions.
Confirming the importance of the SS element in the promoter activity of the cps locus.
Based on the intriguing results of the mutagenesis experiments (Fig. 4), we sought to verify the essential role of the SS element in the transcription of the cps locus with the dexB-cps4A sequence in the serotype 4 strain TIGR4. TIGR4 and D39 share similar sequence features in the dexB-cps4A region, except for an extra IS1380-Spn1 sequence (1,708 bp) upstream of the RUP in TIGR4. We determined whether the SS element of strain TIGR4 is required for the transcription of the cps locus. When the dexB-cps4A region of TIGR4 was placed in the corresponding position in the cps locus of strain D39, the resulting strain (TH4766) showed cps2A transcription comparable to that of its D39 counterpart (data not shown). However, the isogenic SS mutant (TH5966) had a 9.4-fold (P < 0.001) reduction in transcription of cps2A (see Fig. S2 in the supplemental material). This result further confirmed that the SS element is important for the full activity of the cps promoter in other serotypes of S. pneumoniae.
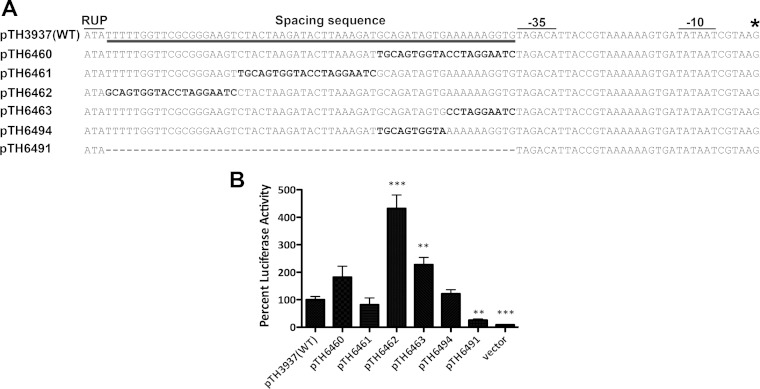
To test the contributions of various SS segments to cps transcription, we generated luciferase-based transcriptional reporter constructs by replacing the SS segments of D39 with the same number of irrelevant nucleotides (Fig. 5A). Consistent with the chromosomal deletion of the SS element (Fig. 4), the reporter construct lacking the entire SS element showed only 25.9% luciferase activity compared with the wild type (Fig. 5B) (P < 0.01). While replacing the 5′ (19 bp; pTH6462) and 3′ (20 bp; pTH6460) ends of the SS element resulted in 3.3-fold (P < 0.001) and 0.82-fold (P > 0.05) increases in luciferase activity, respectively, sequence replacement in the middle segment (20 bp; pTH6461) had no significant impact (relative luciferase activity, 81.8%). Since the 3′ segment is relatively conserved in the cps locus among different serotypes of S. pneumoniae (see Fig. S1 in the supplemental material), we subdivided the region into two 10-bp segments in the reporter system. Replacement of the segment proximal to the core promoter (pTH6463) enhanced the luciferase activity by 1.27-fold (P < 0.01); a similar construct for the distal segment (pTH6494) showed a level of luciferase activity similar to that of the wild type (relative luciferase activity, 121.7%) (Fig. 5B). In summary, sequence substitution of individual SS segments by the luciferase reporter did not identify a specific sequence that explains the essential role of the entire 59-bp SS element in cps transcription. The reporter experiment further supports the notion that the SS element promotes cps transcription by improving the sequence and/or structural context of the core promoter for optimal activity of the RNA polymerase.
FIG 5.
Impact of sequence deletion and substitutions in the SS element on transcription activity of the cps promoter as assessed by a luciferase reporter. (A) WT sequence of the SS and core promoter region and its derivatives with sequence deletion (TH6491) or substitutions in the SS element. The substituted nucleotides are highlighted in boldface. The transcription start site is marked with an asterisk. (B) Luciferase activities of D39 carrying the pID166 plasmid containing the luciferase gene driven by the WT or mutant forms of the SS element and the cps core promoter. All values were converted to percent luciferase activity relative to that of the WT. **, P < 0.01; ***, P < 0.001.
Characterization of CPS production in the unmarked dexB-cps2A region deletion mutants.
We determined the impacts of the sequence elements in the dexB-cps2A region on the production of the capsular polysaccharide by immunoblotting with an antibody against type 2 pneumococcal CPS. All of the dexB-cps2A sequence mutants showed significant reductions in the CPS level under either aerobic or anaerobic growth conditions, but the extent of the impairment varied among different strains and/or growth conditions (Fig. 6). In agreement with the qRT-PCR results (Fig. 4), the D39 derivatives lacking the entire region (TH4525) or the core promoter (TH5618) showed undetectable CPS under both growth conditions. Similarly, the SS mutant (TH5648) was significantly impaired in CPS production under aerobic (18%) or anaerobic (66%) conditions (Fig. 6). The RUP-SS (TH5621) and IE-RUP-SS (TH5050) mutants also displayed significant reductions in the CPS level under both growth conditions. To a lesser extent, the CPS levels in the IS630-Spn1 (TH5048) and RUP (TH5615) mutants were also reduced. Lastly, the strain carrying the dexB-cps4A sequence from the type 4 strain TIGR4 produced an amount of type 2 CPS comparable to that of D39 (data not shown). In contrast, deleting the SS element from the dexB-cps4A region led to a 78% decrease in the CPS levels (see Fig. S2 in the supplemental material), demonstrating the importance of the SS element in CPS production in multiple S. pneumoniae serotypes.
FIG 6.
Detection of CPS production in D39s and its mutants by immunoblotting. (A) Image of a representative immunoblot. (B) Quantitative chemiluminescence intensity levels from the immunoblotting results represented in panel A. The CPS on each spot was quantified with ImageJ software on the basis of its chemiluminescence intensity level and normalized to the values of D39s in the corresponding spots. (C) Same as panel A, except that pneumococci cultured under anaerobic conditions were used. (D) Same as panel B, except that the data from panel C were used. **, P < 0.01; ***, P < 0.001.
Impacts of the common cps genes (cpsABCD) on cps transcription and capsule production.
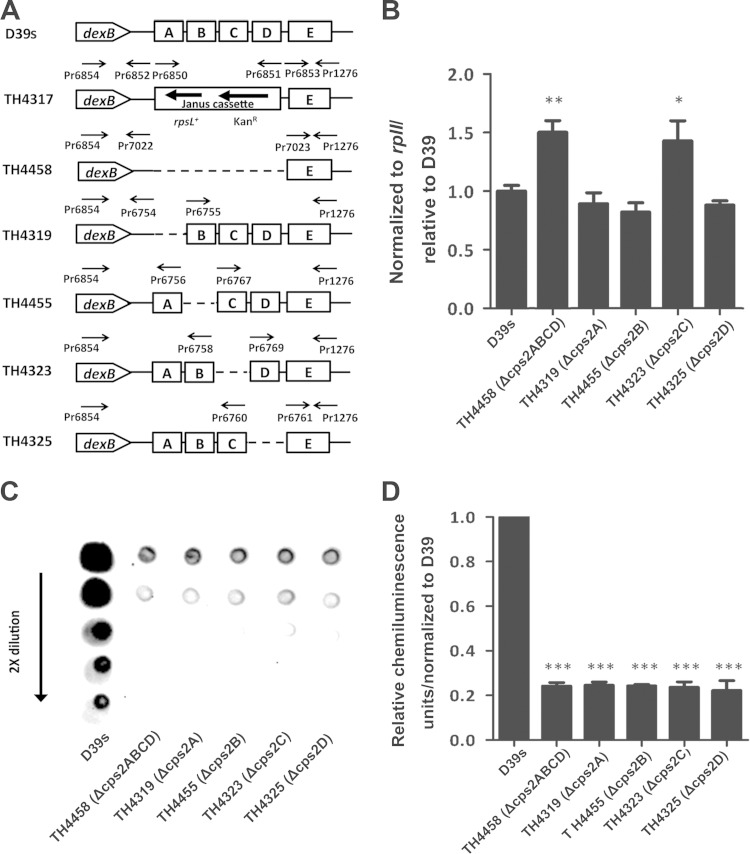
Previous studies have demonstrated that the common S. pneumoniae cpsABCD gene mutants have significantly impaired CPS production in the strain backgrounds of multiple serotypes (14–16). However, it is unclear precisely how the cpsABCD genes contribute to the encapsulation of S. pneumoniae. To determine whether the impairment in CPS production of the cpsABCD mutants is due to their roles in the transcription of the cps operon, we constructed individual in-frame cps2ABCD deletions in D39 (Fig. 7A). This procedure deleted the 1,362-bp, 648-bp, 609-bp, 597-bp, and 3,486-bp sequences in the coding regions of cps2A (total, 1,446 bp; TH4319), cps2B (732 bp; TH4455), cps2C (693 bp; TH4323), cps2D (681 bp; TH4323), and cps2ABCD (3,570 bp; TH4458), respectively. The impact of each deletion on cps transcription was assessed by qRT-PCR. Compared with the parent strain, removing cps2A (TH4319), cps2B (TH4455), or cps2D (TH4325) from the cps locus did not significantly affect the transcription of the operon (Fig. 7B). The mutants lacking cps2C (TH4323) or cps2ABCD (TH4458) showed marginal increases in cps transcription over the parent strain. This result indicated that Cps2A and other proteins encoded by the common cps genes are not involved in the transcription of the cps locus in D39 and, perhaps, other WZY-dependent serotypes of S. pneumoniae.
FIG 7.
Quantitative detection of the cps2E mRNA and CPS levels in D39s and its mutants with in-frame deletions in cps2ABCD. (A) Schematic illustration of the in-frame deletion constructs in cps2ABCD. Deleted regions are indicated with dashed lines. The primers used to amplify individual regions are shown above the target sequences. (B) Quantification of cps2E mRNA in cps2ABCD region deletion mutants by qRT-PCR. Similar to Fig. 4B, except that total RNA preparations from different strains were used. (C) Image of a representative immunoblot. Similar to Fig.6A, except that different strains were used. (D) Quantitative presentation of the results in panel C. Similar to Fig. 6B, except that different strains were used. *, P <0.05; **, P < 0.01; ***, P < 0.001.
We further assessed CPS production in the cps2ABCD deletion mutants of D39 by immunoblotting. In sharp contrast to the qRT-PCR results (Fig. 7B), the D39 derivatives lacking cps2A (TH4319), cps2B (TH4455), cps2C (TH4323), cps2D (TH4325), or cpsABCD (TH4458) produced only approximately 30% as much CSP as the parent strain and thus had severe impairment in CPS production (Fig. 7C and D). The immunoblotting result confirmed the previous observations that the common cps2ABCD genes are necessary for the full encapsulation of S. pneumoniae (14–16). The impaired CPS production in the cps2ABCD mutants is also reflected in the increased levels of adhesion to host cells (Fig. 8B). Because deletions in the cps2ABCD coding sequences showed no or marginal effects on the transcription of the cps locus (Fig. 7B), we conclude that CpsA, CpsB, CpsC, and CpsD are involved in CPS production in a posttranscriptional manner.
FIG 8.
Adhesion of D39s and the isogenic mutants to host cells. (A) Adhesion to A549 cells of D39s or the isogenic dexB-cps2A region mutants. Bacteria were incubated with A549 cells for 1 h, followed by extensive washing and lysis of the cells. The adherent bacteria were quantified by plating the cell lysates on tryptic soy agar blood plates and counting the CFU. (B) Adhesion to A549 cells of D39s or the isogenic cps2ABCD mutants. Same as panel A, except that different D39s mutants were used. **, P < 0.01; ***, P < 0.001.
Impact of the dexB-cps2A sequence elements and cps2ABCD on pneumococcal adhesion to respiratory epithelial cells.
It is well documented that the polysaccharide capsule reduces pneumococcal attachment to host cells (61). This property facilitates bacterial evasion of killing by host phagocytes, but also hinders pneumococcal adhesion to mucosal epithelial cells. In this study, we used epithelial adhesion as a functional marker to determine the impacts of the sequence deletions in the cps locus on the degree of pneumococcal encapsulation. We first tested the impacts of the dexB-cps2A deletions on pneumococcal adhesion to A549 human alveolar lung epithelial cells. As expected, the parent strain, D39s, displayed a low level of adhesion to A549 cells (Fig. 8A). However, all five of the dexB-cps2A mutants showed significantly higher levels of adhesion. The mutants lacking the core promoter (TH5618) or the entire dexB-cps2A sequence (TH4525) showed 268.2-fold (P < 0.001) and 152.9-fold (P < 0.001) increases in adhesion, respectively. Significant increases in adhesion were also observed with the the IS630-Spn1 (TH5048; 2.0-fold increase; P < 0.001), RUP (TH5615; 6.7-fold increase; P < 0.001), or SS (TH5648; 9.5-fold increase; P < 0.001) element mutants. In full agreement with the qRT-PCR (Fig. 4) and immunoblotting (Fig. 6) results, the deletion of the SS element in combination with the RUP (ΔRUP-SS; TH5621) or IE-RUP (TH5050) led to significantly increased adhesion to A549 cells, but the levels of increase in these mutants were substantially lower than that in the mutant lacking only the SS element (TH5648) (Fig. 8A). This result demonstrated that all four of the sequence elements in the dexB-cps2A region are necessary for pneumococcal evasion of attachment to host cells and thus indicated the importance of these elements in the full encapsulation of D39.
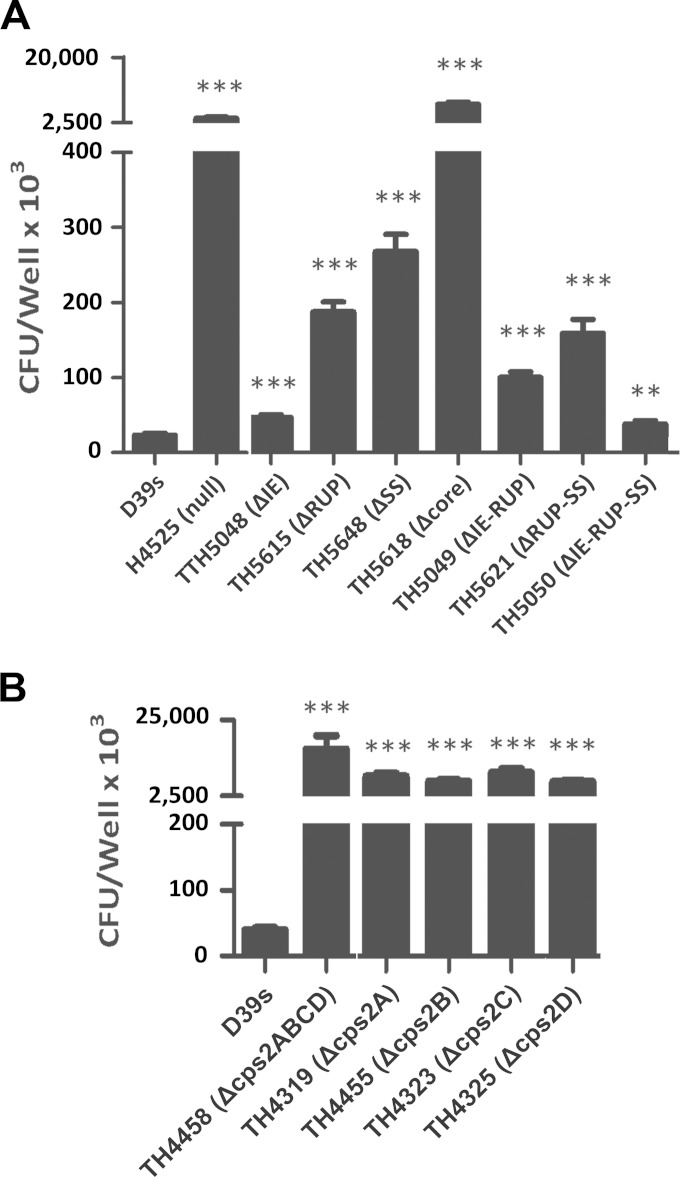
We also determined the epithelial adhesion of the cps2ABCD mutants in a similar manner. As expected from the CPS immunoblotting test (Fig. 7C and D), epithelial adhesion was dramatically increased in the unmarked cps2A (TH4319; 170-fold; P < 0.001), cps2B (TH4455; 211-fold; P < 0.001), cps2C (TH4323; 175-fold; P < 0.001), cps2D (TH4325; 238-fold; P < 0.001), and cps2ABCD (TH4458; 416-fold; P < 0.001) deletion mutants (Fig. 8B). The significantly increased adhesion of the cps2ABCD mutants to host cells is fully explained by the severe impairment of these strains in encapsulation (Fig. 7C and D). The adhesion experiment thus confirmed the importance of the common cpsABCD genes in pneumococcal evasion of attachment to host cells by facilitating the encapsulation of the pathogen.
Impacts of the dexB-cps2A sequence elements on pneumococcal virulence.
We finally assessed the impacts of the dexB-cps2A elements on pneumococcal virulence. We initially assessed the growth properties of the eight D39 derivatives with different truncations in the cps promoter region in THY broth. None of these strains showed major growth defects, although the unencapsulated strains lacking the core promoter (TH5618) or the entire dexB-cps2A region (TH4525) displayed slower growth and earlier autolysis (Fig. 9A). We next determined the relative virulence levels of these strains by coinfection with the parent strain, D39. Each of the deletion mutants (streptomycin resistant) in this region was mixed with strain D39 (streptomycin sensitive) in a 1:1 ratio before being used to infect CD1 mice by intraperitoneal inoculation. The wild-type and mutant pneumococci were differentiated by their sensitivity/resistance to streptomycin. Because the deletion mutants were constructed in D39s, a streptomycin-resistant D39 derivative carrying the rpsL1 allele (see Materials and Methods), we performed a similar experiment with D39s to assess the impact of the rpsL1 allele. The experiment showed that D39s was slightly attenuated compared with the wild-type D39 (mean CI = 0.5) (data not shown). We thus calculated the competitive-index values of the D39s derivatives in consideration of this observation (Fig. 9B).
FIG 9.
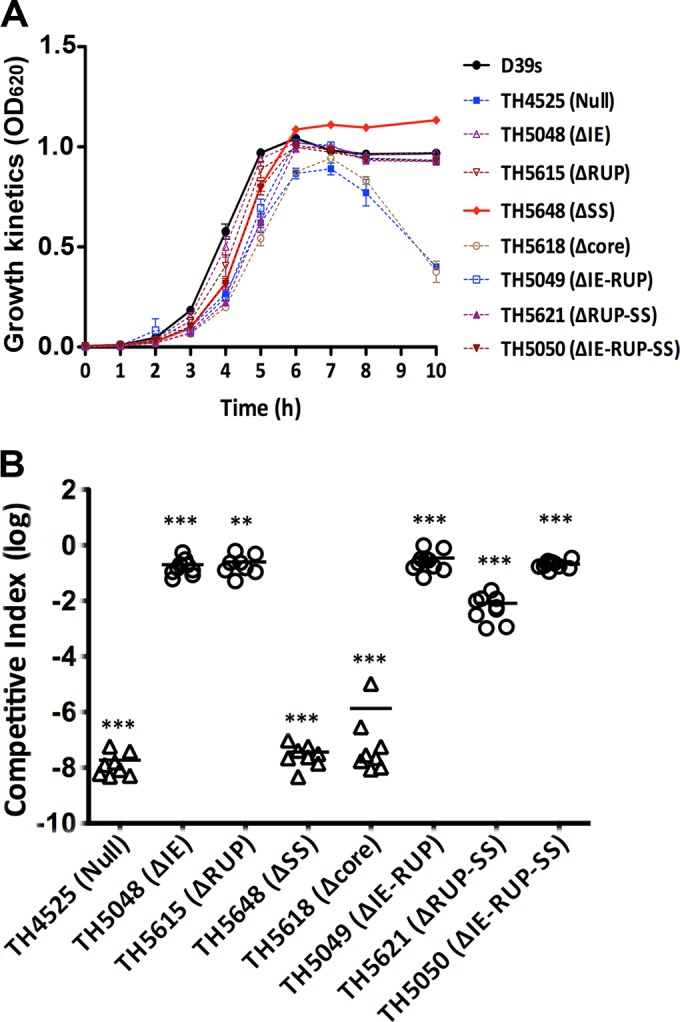
Quantification of bacteremia in mice infected with mixtures of wild-type D39 and unmarked dexB-cps2A region mutants. (A) Growth kinetics of the D39s derivatives under aerobic conditions. The ticks indicate standard deviations. (B) Bacteremia levels of the D39s derivatives. CD1 mice were intraperitoneally infected with a mixture of D39 and the D39s mutant in a 1:1 ratio. The pneumococci in the bloodstreams of the mice were enumerated 21 h postinfection as for Fig. 6A. Each symbol represents a relative bacteremia level or CI value of a single mouse. Each triangle represents a mouse from which no mutant pneumococci were recovered. The horizontal bars indicate the medians in individual groups of mice. **, P < 0.01; ***, P < 0.001.
The coinfection experiment did not recover any streptomycin-resistant pneumococci from the mice infected with the mixture of D39 and the dexB-cps2A mutant (TH4525) or D39 and the core promoter mutant (TH5618), although the wild-type D39 pneumococci were consistently recovered at levels of 105 to 109 CFU/ml from the blood samples from these mice (Fig. 9B and data not shown), indicating that these mutants lost the capability to survive and replicate in the bloodstream of mice and thus became completely avirulent. The result observed with the core promoter mutant of D39 (TH5618) confirmed the previous study in TIGR4 by Shainheit et al. showing that the core promoter is essential for virulence of S. pneumoniae (40). To our surprise, deleting the SS element (TH5648) also led to complete loss of virulence, because only the wild-type D39 pneumococci were recovered from the mice infected with the mixture of D39 and TH5648. Given the fact that TH5648 produced approximately 18% of the parental level of CPS, the avirulent phenotype of the mutant indicated that this level of encapsulation is not sufficient to fulfill the immune evasion function of the capsule.
To much lesser extents, all of the mutants lacking single or multiple sequence elements of the dexB-cps2A region also showed significantly reduced bacteremia levels compared with the parent strain, D39s (Fig. 9B). A comparable level of attenuation in bacteremia was observed with the mutant lacking the IE (TH5048; mean CI = 0.18), ΔRUP (TH5615; mean CI = 0.26), IE-RUP (TH5049; mean CI = 0.35), or IE-RUP-SS (TH5050; mean CI = 0.21). The bacteremia levels of these strains were generally 6- to 10-fold lower than those of the coinfected D39 in the same mice. Furthermore, the mean number of ΔRUP-SS mutants (TH5621) recovered from the mice was approximately 120-fold lower than that of D39s (mean CI = 8.27 × 10−3). In summary, for all of the dexB-cps2A mutants, the relative levels of impairment in the bacteremia model (Fig. 9B) are generally consistent with those in encapsulation (Fig. 6). These data, for the first time, demonstrate that the cis-acting elements upstream of the core promoter (IE, RUP, and SS) significantly contribute to the full pneumococcal virulence capacity.
DISCUSSION
Despite the importance of the polysaccharide capsule in pneumococcal immune evasion and virulence, the transcription of the cps locus remains largely undefined. This study has demonstrated that the 17 genes in the cps locus of the type 2 strain D39 are cotranscribed as an operon. The major transcript of the cps locus in D39 started at the −25th nucleotide (G) upstream of the cpsA start codon. We constructed an unmarked truncation in each of the four sequence elements in the intergenic region between dexB and cps2A: IS630-Spn1, RUP, SS, and core promoter. Comprehensive transcription and functional analysis of these mutants showed that the transcription of the cps locus is predominantly driven by the SS element and core promoter region, but the IE and RUP elements are necessary for adequate transcription of the cps locus and thereby full encapsulation of D39. Furthermore, we have demonstrated that cps transcription does not depend on cps2ABCD, the common genes in the cps loci of all WZY-dependent CPS serotypes in S. pneumoniae. Our results have provided insight and information leading to a complete understanding the basic transcription and regulation of the cps locus. To our knowledge, this is the first comprehensive experimental analysis of the cps promoter activity in S. pneumoniae.
This study shows that the core promoter region of the type 2 strain D39 is important for the transcription of the cps locus and CPS production. This region contains the previously predicted −35 and −10 promoter motifs (3, 13) and the transcription start site identified in this study. This result is consistent with the recent study by Shainheit et al. (40), which demonstrated that a similar core promoter of the cps locus in the type 4 strain TIGR4 is necessary for the transcription of the locus. Because the core promoter is highly conserved among the WZY-dependent CPS types, the region should play an important role in the transcription of the cps genes in many other serotypes of S. pneumoniae.
Our results indicate that the sequence elements upstream of the core promoter are necessary for the full transcription of the cps operon in D39 and, perhaps, other WZY-dependent CPS types of S. pneumoniae. The mutant TH5648, lacking the SS element immediately upstream of the core promoter, showed the most striking phenotypes. The absence of the SS element in TH5648 resulted in dramatic reduction in the transcription of cps2A. Consistently, placing a similar deletion construct of TIGR4 in the dexB-cps2A region of D39 also led to a similar reduction in the transcription of the cps locus. These results obtained with chromosomal deletion mutants of D39 were fully verified with the transcriptional reporter constructs of the dexB-cps2A sequence. The importance of the SS element in cps transcription is further reflected by the other results obtained with the ΔSS mutant, including >80% decrease in CPS production, significant enhancement of adhesion to host cells, and complete loss of virulence in the mouse bacteremia model. These lines of evidence indicate that the SS element represents a previously unrecognized region that is important for the adequate expression of the cps operon and full encapsulation of S. pneumoniae.
The SS element may enhance cps transcription by improving the sequence and/or structural context of the core promoter for the optimal activity of the RNA polymerase. This can be achieved by serving as a new recognition motif(s) for S. pneumoniae RNAP and/or a potential regulator(s). Certain bacterial and phage promoters consist of three RNAP recognition sequences: the −10 motif, the −35 motif, and the UP element (62). For these promoters, the −10 and −35 motifs interact with the RNAP σ subunit (63), whereas the UP element, located upstream of the −35 motif, is recognized by the RNAP α subunit (64, 65). The UP elements contain a consensus sequence: 5′-nnAAA(A/T)(A/T)T(A/T)TTTTnnAAAAnnn-3′ (n = A, T, G, or C) (66). Interestingly, the SS element partially matches the 3′ segment of the consensus sequences of the UP sites. However, replacing this segment of the SS element with irrelevant sequences did not reproduce the effect of the deletions on cps transcription in the transcriptional reporter system. This observation thus casts doubt on the possibility that the SS element contains an authentic UP sequence or encodes a regulator. It remains to be determined, through biochemical analysis of binding interactions, whether this region directly interacts with S. pneumoniae RNAP. It is puzzling that the double (ΔRUP-SS; TH5621) and triple (ΔIE-RUP-SS; TH5050) SS element deletion mutants consistently showed milder phenotypes than the mutant lacking only the SS element (ΔSS; TH5648) in cps transcription, CPS production, host cell adhesion, and virulence. It is possible that these deletions resulted in direct fusion of different sequences to the 5′ end of the core promoter and created various sequence contexts upstream of the core promoter, thereby influencing the interactions between the cps promoter elements and the subunits of S. pneumoniae RNAP.
IS630-Spn1 and a RUP in the dexB-cpsA region are necessary for adequate transcription of the cps locus and full encapsulation in D39. Although deleting the RUP or IS630-Spn1 yielded small impacts on cps transcription, the ΔIE and ΔRUP strains showed reproducible impairment in CPS production, increased adhesion to host cells, and significantly attenuated virulence in the mouse bacteremia model. Despite the extensive sequence variation in the dexB-cpsA region (41), IS630-Spn1 and the RUP are found in the cps loci of many pneumococcal strains (41). IS630-Spn1 is a putative insertion element of S. pneumoniae, ranging from 880 to 899 bp in size (67). There are 5 nearly identical copies of IS630-Spn1 in the genome of D39 (42), including the one in the cps locus. The RUP is a 107-bp repeat sequence that is present in the cps locus and many other loci of S. pneumoniae (39, 68, 69). There are 84 and 127 copies of the RUP in the genomes of D39/R6 and TIGR4, respectively (42, 68, 69). Oggioni and Claverys identified significant homology between the RUP and IS630-Spn1 (see below) (67), supporting the proposal that the RUP serves as an insertion site for IS630-Spn1 and thus promotes DNA rearrangements (67). Other investigators postulate that these repetitive sequences may have regulatory roles for their neighboring genes, since virtually all the RUP elements are located in the intergenic spaces (8, 68, 70). In particular, a RUP sequence overlaps the transcription start sites of the tts gene (8). tts encodes a β-glucosyltransferase that synthesizes the type 37 CPS, because the “conventional” type 37 cps locus is inactive (61). Llull et al. showed that three of the five transcription start sites for the tts gene are located in an upstream RUP element (7). This study presents the first experimental evidence to support the hypothesis that IS630-Spn1 and the RUP influence gene expression and contribute to the biology of S. pneumoniae. It is possible that IS630-Spn1 and the RUP fine tune the transcription of the cps locus and CPS production by recruiting a regulatory factor(s), encoding regulatory RNA species, or forming a certain DNA configuration(s) that influences the cps promoter activity. Many noncoding small RNAs have recently been discovered to regulate bacterial genes, including virulence factors, as exemplified by the RNA III molecule of S. aureus (71). It should be noted that the sequence of the RUP is predicted to form a stable stem-loop structure (67). Taken together, the sequence diversity in the dexB-cpsA region among pneumococcal strains/serotypes may be a mechanism for phenotypic diversification of S. pneumoniae in encapsulation and thereby for evolutionary fitness. Future studies are warranted to determine how the sequence elements contribute to pneumococcal encapsulation.
Our intergenic PCR result indicates that the 17 genes in the cps locus of D39 are transcribed as an operon. Based on the short intergenic gaps between the cps genes and their functional relationships, it has long been predicted that the genes in the cps locus of S. pneumoniae are cotranscribed (3), but we have provided the first experimental evidence to prove this prediction. We made numerous attempts to detect the complete mRNA species of this operon by Northern blotting without success, which might be due to the potential instability or the fragile nature of the long mRNA molecule during RNA isolation, separation, and/or transfer.
The data from this study show that cps2ABCD, the common genes of all WZY-dependent CPS types, are not involved in the transcription of the cps genes in D39. As one of the four common cps genes among the >90 WZY-dependent CPS types (5, 6), cps2A encodes a highly conserved protein in the genetic loci for capsule polysaccharide or exopolysaccharide synthesis in many other streptococcal species, such as S. agalactiae (17), Streptococcus suis (18), S. iniae (19), and many oral streptococci (20, 21). CpsA may function as a transcriptional regulator due to its homology with the transcriptional regulator LytR of B. subtilis (3), the negative impact of the cpsA mutations on the transcription of the cps genes in S. agalactiae (22) and on CPS production in S. pneumoniae and S. agalactiae (16, 22, 27), and binding of recombinant CpsA to the capsule promoter sequences of S. iniae (23) and S. agalactiae (24). However, deleting 94.3% of the cps2A coding sequence in D39 did not significantly alter transcription in the cps locus in this study (Fig. 7). In contrast, the same mutant displayed approximately 30% of the CPS produced by the parent strain and greatly enhanced adhesion to host cells (another sign of the loss of CPS production). This is the first study to demonstrate experimentally that the impaired CPS production in the cpsA mutant of S. pneumoniae is not caused by the previously perceived contribution of CpsA to the transcription of the cps genes. Instead, these lines of evidence support an alternative hypothesis that CpsA and the proteins encoded by the other common cps genes are involved in transferring the capsular polysaccharides from the lipid-linked precursor to cell wall peptidoglycan in S. pneumoniae, as described for the CpsA homologs in the synthesis of other cell wall-associated bacterial polymers (72).
Supplementary Material
ACKNOWLEDGMENTS
We thank Marc Prudhomme and Jean-Pierre Claverys for providing plasmid pR424.
This work was supported by grants from the Ministry of Science and Technology of China (no. 2012CB518702), the Tsinghua University Collaborative Research Program (no. 2011Z23153), the Center for Marine Medicine and Rescue of Tsinghua University (no. 20124812029), and the Grand Challenges Exploration of the Bill and Melinda Gates Foundation (no. OPP1021992).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02944-14.
REFERENCES
- 1.Alonso de Velasco E, Verheul AF, Verhoef J, Snippe H. 1995. Streptococcus pneumoniae: virulence factors, pathogenesis, and vaccines. Microbiol Rev 59:591–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hyams C, Camberlein E, Cohen JM, Bax K, Brown JS. 2010. The Streptococcus pneumoniae capsule inhibits complement activity and neutrophil phagocytosis by multiple mechanisms. Infect Immun 78:704–715. doi: 10.1128/IAI.00881-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guidolin A, Morona JK, Morona R, Hansman D, Paton JC. 1994. Nucleotide sequence analysis of genes essential for capsular polysaccharide biosynthesis in Streptococcus pneumoniae type 19F. Infect Immun 62:5384–5396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Feldman C, Anderson R. 2014. Review: current and new generation pneumococcal vaccines. J Infect 69:309–325. doi: 10.1016/j.jinf.2014.06.006. [DOI] [PubMed] [Google Scholar]
- 5.Bentley SD, Aanensen DM, Mavroidi A, Saunders D, Rabbinowitsch E, Collins M, Donohoe K, Harris D, Murphy L, Quail MA, Samuel G, Skovsted IC, Kaltoft MS, Barrell B, Reeves PR, Parkhill J, Spratt BG. 2006. Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2:e31. doi: 10.1371/journal.pgen.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Park IH, Park S, Hollingshead SK, Nahm MH. 2007. Genetic basis for the new pneumococcal serotype, 6C. Infect Immun 75:4482–4489. doi: 10.1128/IAI.00510-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Llull D, Garcia E, Lopez R. 2001. Tts, a processive beta-glucosyltransferase of Streptococcus pneumoniae, directs the synthesis of the branched type 37 capsular polysaccharide in Pneumococcus and other gram-positive species. J Biol Chem 276:21053–21061. doi: 10.1074/jbc.M010287200. [DOI] [PubMed] [Google Scholar]
- 8.Llull D, Munoz R, Lopez R, Garcia E. 1999. A single gene (tts) located outside the cap locus directs the formation of Streptococcus pneumoniae type 37 capsular polysaccharide. Type 37 pneumococci are natural, genetically binary strains. J Exp Med 190:241–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wen Z, Zhang J-R. 2015. Bacterial capsules, p 33–52. In Tang Y-W, Sussman M, Liu D, Poxton I, Schwartzman J (ed), Molecular medical microbiology 2nd ed, vol 1 Academic Press, Amsterdam, The Netherlands. [Google Scholar]
- 10.Whitfield C. 2006. Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu Rev Biochem 75:39–68. doi: 10.1146/annurev.biochem.75.103004.142545. [DOI] [PubMed] [Google Scholar]
- 11.Yother J. 2011. Capsules of Streptococcus pneumoniae and other bacteria: paradigms for polysaccharide biosynthesis and regulation. Annu Rev Microbiol 65:563–581. doi: 10.1146/annurev.micro.62.081307.162944. [DOI] [PubMed] [Google Scholar]
- 12.Arrecubieta C, Lopez R, Garcia E. 1994. Molecular characterization of cap3A, a gene from the operon required for the synthesis of the capsule of Streptococcus pneumoniae type 3: sequencing of mutations responsible for the unencapsulated phenotype and localization of the capsular cluster on the pneumococcal chromosome. J Bacteriol 176:6375–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iannelli F, Pearce BJ, Pozzi G. 1999. The type 2 capsule locus of Streptococcus pneumoniae. J Bacteriol 181:2652–2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morona JK, Miller DC, Morona R, Paton JC. 2004. The effect that mutations in the conserved capsular polysaccharide biosynthesis genes cpsA, cpsB, and cpsD have on virulence of Streptococcus pneumoniae. J Infect Dis 189:1905–1913. doi: 10.1086/383352. [DOI] [PubMed] [Google Scholar]
- 15.Morona JK, Morona R, Paton JC. 2006. Attachment of capsular polysaccharide to the cell wall of Streptococcus pneumoniae type 2 is required for invasive disease. Proc Natl Acad Sci U S A 103:8505–8510. doi: 10.1073/pnas.0602148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bender MH, Cartee RT, Yother J. 2003. Positive correlation between tyrosine phosphorylation of CpsD and capsular polysaccharide production in Streptococcus pneumoniae. J Bacteriol 185:6057–6066. doi: 10.1128/JB.185.20.6057-6066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Madoff LC, Paoletti LC, Kasper DL. 2006. Surface structures of group B Streptococci important in human immunology, p 169–185. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-positive pathogens, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 18.Okura M, Takamatsu D, Maruyama F, Nozawa T, Nakagawa I, Osaki M, Sekizaki T, Gottschalk M, Kumagai Y, Hamada S. 2013. Genetic analysis of capsular polysaccharide synthesis gene clusters from all serotypes of Streptococcus suis: potential mechanisms for generation of capsular variation. Appl Environ Microbiol 79:2796–2806. doi: 10.1128/AEM.03742-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe BA, Miller JD, Neely MN. 2007. Analysis of the polysaccharide capsule of the systemic pathogen Streptococcus iniae and its implications in virulence. Infect Immun 75:1255–1264. doi: 10.1128/IAI.01484-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kilian M, Poulsen K, Blomqvist T, Havarstein LS, Bek-Thomsen M, Tettelin H, Sorensen UB. 2008. Evolution of Streptococcus pneumoniae and its close commensal relatives. PLoS One 3:e2683. doi: 10.1371/journal.pone.0002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin AM, Morris VJ, Gasson MJ. 1996. The cpsABCDE genes involved in polysaccharide production in Streptococcus salivarius ssp. thermophilus strain NCBF 2393. Gene 183:23–27. doi: 10.1016/S0378-1119(96)00405-2. [DOI] [PubMed] [Google Scholar]
- 22.Cieslewicz MJ, Kasper DL, Wang Y, Wessels MR. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J Biol Chem 276:139–146. doi: 10.1074/jbc.M005702200. [DOI] [PubMed] [Google Scholar]
- 23.Hanson BR, Lowe BA, Neely MN. 2011. Membrane topology and DNA-binding ability of the streptococcal CpsA protein. J Bacteriol 193:411–420. doi: 10.1128/JB.01098-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanson BR, Runft DL, Streeter C, Kumar A, Carion TW, Neely MN. 2012. Functional analysis of the CpsA protein of Streptococcus agalactiae. J Bacteriol 194:1668–1678. doi: 10.1128/JB.06373-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bender MH, Yother J. 2001. CpsB Is a modulator of capsule-associated tyrosine kinase activity in Streptococcus pneumoniae. J Biol Chem 276:47966–47974. doi: 10.1074/jbc.M105448200. [DOI] [PubMed] [Google Scholar]
- 26.Morona JK, Morona R, Miller DC, Paton JC. 2002. Streptococcus pneumoniae capsule biosynthesis protein CpsB is a novel manganese-dependent phosphotyrosine-protein phosphatase. J Bacteriol 184:577–583. doi: 10.1128/JB.184.2.577-583.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morona JK, Paton JC, Miller DC, Morona R. 2000. Tyrosine phosphorylation of CpsD negatively regulates capsular polysaccharide biosynthesis in Streptococcus pneumoniae. Mol Microbiol 35:1431–1442. doi: 10.1046/j.1365-2958.2000.01808.x. [DOI] [PubMed] [Google Scholar]
- 28.Geno KA, Hauser JR, Gupta K, Yother J. 2014. Streptococcus pneumoniae phosphotyrosine phosphatase CpsB and alterations in capsule production resulting from changes in oxygen availability. J Bacteriol 196:1992–2003. doi: 10.1128/JB.01545-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiser JN, Bae D, Epino H, Gordon SB, Kapoor M, Zenewicz LA, Shchepetov M. 2001. Changes in availability of oxygen accentuate differences in capsular polysaccharide expression by phenotypic variants and clinical isolates of Streptococcus pneumoniae. Infect Immun 69:5430–5439. doi: 10.1128/IAI.69.9.5430-5439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiser JN. 2006. Phase variation of Streptococcus pneumoniae, p 268–274. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-positive pathogens. ASM Press, Washington, DC. [Google Scholar]
- 31.Briles DE, Novak L, Hotomi M, van Ginkel FW, King J. 2005. Nasal colonization with Streptococcus pneumoniae includes subpopulations of surface and invasive pneumococci. Infect Immun 73:6945–6951. doi: 10.1128/IAI.73.10.6945-6951.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hammerschmidt S, Wolff S, Hocke A, Rosseau S, Muller E, Rohde M. 2005. Illustration of pneumococcal polysaccharide capsule during adherence and invasion of epithelial cells. Infect Immun 73:4653–4667. doi: 10.1128/IAI.73.8.4653-4667.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cundell DR, Gerard NP, Gerard C, Idanpaan-Heikkila I, Tuomanen EI. 1995. Streptococcus pneumoniae anchor to activated human cells by the receptor for platelet-activating factor. Nature 377:435–438. doi: 10.1038/377435a0. [DOI] [PubMed] [Google Scholar]
- 34.Weiser JN, Austrian R, Sreenivasan PK, Masure HR. 1994. Phase variation in pneumococcal opacity: relationship between colonial morphology and nasopharyngeal colonization. Infect Immun 62:2582–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weiser JN, Markiewicz Z, Tuomanen EI, Wani JH. 1996. Relationship between phase variation in colony morphology, intrastrain variation in cell wall physiology, and nasopharyngeal colonization by Streptococcus pneumoniae. Infect Immun 64:2240–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim JO, Romero-Steiner S, Sorensen UB, Blom J, Carvalho M, Barnard S, Carlone G, Weiser JN. 1999. Relationship between cell surface carbohydrates and intrastrain variation on opsonophagocytosis of Streptococcus pneumoniae. Infect Immun 67:2327–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim JO, Weiser JN. 1998. Association of intrastrain phase variation in quantity of capsular polysaccharide and teichoic acid with the virulence of Streptococcus pneumoniae. J Infect Dis 177:368–377. doi: 10.1086/514205. [DOI] [PubMed] [Google Scholar]
- 38.Ogunniyi AD, Giammarinaro P, Paton JC. 2002. The genes encoding virulence-associated proteins and the capsule of Streptococcus pneumoniae are upregulated and differentially expressed in vivo. Microbiology 148:2045–2053. [DOI] [PubMed] [Google Scholar]
- 39.Munoz R, Mollerach M, Lopez R, Garcia E. 1997. Molecular organization of the genes required for the synthesis of type 1 capsular polysaccharide of Streptococcus pneumoniae: formation of binary encapsulated pneumococci and identification of cryptic dTDP-rhamnose biosynthesis genes. Mol Microbiol 25:79–92. doi: 10.1046/j.1365-2958.1997.4341801.x. [DOI] [PubMed] [Google Scholar]
- 40.Shainheit MG, Mule M, Camilli A. 2014. The core promoter of the capsule operon of Streptococcus pneumoniae is necessary for colonization and invasive disease. Infect Immun 82:694–705. doi: 10.1128/IAI.01289-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moscoso M, Garcia E. 2009. Transcriptional regulation of the capsular polysaccharide biosynthesis locus of Streptococcus pneumoniae: a bioinformatic analysis. DNA Res 16:177–186. doi: 10.1093/dnares/dsp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lanie JA, Ng WL, Kazmierczak KM, Andrzejewski TM, Davidsen TM, Wayne KJ, Tettelin H, Glass JI, Winkler ME. 2007. Genome sequence of Avery's virulent serotype 2 strain D39 of Streptococcus pneumoniae and comparison with that of unencapsulated laboratory strain R6. J Bacteriol 189:38–51. doi: 10.1128/JB.01148-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen H, Ma Y, Yang J, O'Brien CJ, Lee SL, Mazurkiewicz JE, Haataja S, Yan JH, Gao GF, Zhang JR. 2008. Genetic requirement for pneumococcal ear infection. PLoS One 3:e2950. doi: 10.1371/journal.pone.0002950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lu L, Ma Y, Zhang JR. 2006. Streptococcus pneumoniae recruits complement factor H through the amino terminus of CbpA. J Biol Chem 281:15464–15474. doi: 10.1074/jbc.M602404200. [DOI] [PubMed] [Google Scholar]
- 45.Sung CK, Li H, Claverys JP, Morrison DA. 2001. An rpsL cassette, janus, for gene replacement through negative selection in Streptococcus pneumoniae. Appl Environ Microbiol 67:5190–5196. doi: 10.1128/AEM.67.11.5190-5196.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biswas I, Jha JK, Fromm N. 2008. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 154:2275–2282. doi: 10.1099/mic.0.2008/019265-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Prudhomme M, Attaiech L, Sanchez G, Martin B, Claverys JP. 2006. Antibiotic stress induces genetic transformability in the human pathogen Streptococcus pneumoniae. Science 313:89–92. doi: 10.1126/science.1127912. [DOI] [PubMed] [Google Scholar]
- 48.Davies DH, Liang X, Hernandez JE, Randall A, Hirst S, Mu Y, Romero KM, Nguyen TT, Kalantari-Dehaghi M, Crotty S, Baldi P, Villarreal LP, Felgner PL. 2005. Profiling the humoral immune response to infection by using proteome microarrays: high-throughput vaccine and diagnostic antigen discovery. Proc Natl Acad Sci U S A 102:547–552. doi: 10.1073/pnas.0408782102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ross W, Gourse RL. 2005. Sequence-independent upstream DNA-alphaCTD interactions strongly stimulate Escherichia coli RNA polymerase-lacUV5 promoter association. Proc Natl Acad Sci U S A 102:291–296. doi: 10.1073/pnas.0405814102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sertil O, Vemula A, Salmon SL, Morse RH, Lowry CV. 2007. Direct role for the Rpd3 complex in transcriptional induction of the anaerobic DAN/TIR genes in yeast. Mol Cell Biol 27:2037–2047. doi: 10.1128/MCB.02297-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Charaniya S, Mehra S, Lian W, Jayapal KP, Karypis G, Hu WS. 2007. Transcriptome dynamics-based operon prediction and verification in Streptomyces coelicolor. Nucleic Acids Res 35:7222–7236. doi: 10.1093/nar/gkm501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ma Z, Zhang JR. 2007. RR06 activates transcription of spr1996 and cbpA in Streptococcus pneumoniae. J Bacteriol 189:2497–2509. doi: 10.1128/JB.01429-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Arrecubieta C, Garcia E, Lopez R. 1995. Sequence and transcriptional analysis of a DNA region involved in the production of capsular polysaccharide in Streptococcus pneumoniae type 3. Gene 167:1–7. doi: 10.1016/0378-1119(95)00657-5. [DOI] [PubMed] [Google Scholar]
- 54.Macrina FL, Tobian JA, Jones KR, Evans RP, Clewell DB. 1982. A cloning vector able to replicate in Escherichia coli and Streptococcus sanguis. Gene 19:345–353. doi: 10.1016/0378-1119(82)90025-7. [DOI] [PubMed] [Google Scholar]
- 55.Zhang JR, Mostov KE, Lamm ME, Nanno M, Shimida S, Ohwaki M, Tuomanen E. 2000. The polymeric immunoglobulin receptor translocates pneumococci across human nasopharyngeal epithelial cells. Cell 102:827–837. doi: 10.1016/S0092-8674(00)00071-4. [DOI] [PubMed] [Google Scholar]
- 56.Su J, Yang J, Zhao D, Kawula TH, Banas JA, Zhang JR. 2007. Genome-wide identification of Francisella tularensis virulence determinants. Infect Immun 75:3089–3101. doi: 10.1128/IAI.01865-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ouyang S, Lee CY. 1997. Transcriptional analysis of type 1 capsule genes in Staphylococcus aureus. Mol Microbiol 23:473–482. doi: 10.1046/j.1365-2958.1997.d01-1865.x. [DOI] [PubMed] [Google Scholar]
- 58.Ouyang S, Sau S, Lee CY. 1999. Promoter analysis of the cap8 operon, involved in type 8 capsular polysaccharide production in Staphylococcus aureus. J Bacteriol 181:2492–2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sau S, Sun J, Lee CY. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J Bacteriol 179:1614–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Plosker GL. 2013. 13-valent pneumococcal conjugate vaccine: a review of its use in infants, children, and adolescents. Paediatr Drugs 15:403–423. doi: 10.1007/s40272-013-0047-z. [DOI] [PubMed] [Google Scholar]
- 61.Paton JC, Morona JK. 2006. Streptococcus pneumoniae capsular polysaccharide, p 241–252. In Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI (ed), Gram-positive pathogens, 2nd ed ASM Press, Washington, DC. [Google Scholar]
- 62.Gourse RL, Ross W, Gaal T. 2000. UPs and downs in bacterial transcription initiation: the role of the alpha subunit of RNA polymerase in promoter recognition. Mol Microbiol 37:687–695. doi: 10.1046/j.1365-2958.2000.01972.x. [DOI] [PubMed] [Google Scholar]
- 63.Dombroski AJ, Walter WA, Record MT Jr, Siegele DA, Gross CA. 1992. Polypeptides containing highly conserved regions of transcription initiation factor sigma 70 exhibit specificity of binding to promoter DNA. Cell 70:501–512. doi: 10.1016/0092-8674(92)90174-B. [DOI] [PubMed] [Google Scholar]
- 64.Ross W, Gosink KK, Salomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse RL. 1993. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science 262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 65.Blatter EE, Ross W, Tang H, Gourse RL, Ebright RH. 1994. Domain organization of RNA polymerase alpha subunit: C-terminal 85 amino acids constitute a domain capable of dimerization and DNA binding. Cell 78:889–896. doi: 10.1016/S0092-8674(94)90682-3. [DOI] [PubMed] [Google Scholar]
- 66.Estrem ST, Gaal T, Ross W, Gourse RL. 1998. Identification of an UP element consensus sequence for bacterial promoters. Proc Natl Acad Sci U S A 95:9761–9766. doi: 10.1073/pnas.95.17.9761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Oggioni MR, Claverys JP. 1999. Repeated extragenic sequences in prokaryotic genomes: a proposal for the origin and dynamics of the RUP element in Streptococcus pneumoniae. Microbiology 145:2647–2653. [DOI] [PubMed] [Google Scholar]
- 68.Hoskins J, Alborn WE Jr, Arnold J, Blaszczak LC, Burgett S, DeHoff BS, Estrem ST, Fritz L, Fu DJ, Fuller W, Geringer C, Gilmour R, Glass JS, Khoja H, Kraft AR, Lagace RE, LeBlanc DJ, Lee LN, Lefkowitz EJ, Lu J, Matsushima P, McAhren SM, McHenney M, McLeaster K, Mundy CW, Nicas TI, Norris FH, O'Gara M, Peery RB, Robertson GT, Rockey P, Sun PM, Winkler ME, Yang Y, Young-Bellido M, Zhao G, Zook CA, Baltz RH, Jaskunas SR, Rosteck PR Jr, Skatrud PL, Glass JI. 2001. Genome of the bacterium Streptococcus pneumoniae strain R6. J Bacteriol 183:5709–5717. doi: 10.1128/JB.183.19.5709-5717.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tettelin H, Nelson KE, Paulsen IT, Eisen JA, Read TD, Peterson S, Heidelberg J, DeBoy RT, Haft DH, Dodson RJ, Durkin AS, Gwinn M, Kolonay JF, Nelson WC, Peterson JD, Umayam LA, White O, Salzberg SL, Lewis MR, Radune D, Holtzapple E, Khouri H, Wolf AM, Utterback TR, Hansen CL, McDonald LA, Feldblyum TV, Angiuoli S, Dickinson T, Hickey EK, Holt IE, Loftus BJ, Yang F, Smith HO, Venter JC, Dougherty BA, Morrison DA, Hollingshead SK, Fraser CM. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498–506. doi: 10.1126/science.1061217. [DOI] [PubMed] [Google Scholar]
- 70.Croucher NJ, Vernikos GS, Parkhill J, Bentley SD. 2011. Identification, variation and transcription of pneumococcal repeat sequences. BMC Genomics 12:120. doi: 10.1186/1471-2164-12-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A, Gaspin C, Vandenesch F, Romby P. 2007. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev 21:1353–1366. doi: 10.1101/gad.423507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kawai Y, Marles-Wright J, Cleverley RM, Emmins R, Ishikawa S, Kuwano M, Heinz N, Bui NK, Hoyland CN, Ogasawara N, Lewis RJ, Vollmer W, Daniel RA, Errington J. 2011. A widespread family of bacterial cell wall assembly proteins. EMBO J 30:4931–4941. doi: 10.1038/emboj.2011.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.King SJ, Hippe KR, Gould JM, Bae D, Peterson S, Cline RT, Fasching C, Janoff EN, Weiser JN. 2004. Phase variable desialylation of host proteins that bind to Streptococcus pneumoniae in vivo and protect the airway. Mol Microbiol 54:159–171. doi: 10.1111/j.1365-2958.2004.04252.x. [DOI] [PubMed] [Google Scholar]
- 74.Li G, Hu FZ, Yang X, Cui Y, Yang J, Qu F, Gao GF, Zhang JR. 2012. Complete genome sequence of Streptococcus pneumoniae strain ST556, a multidrug-resistant isolate from an otitis media patient. J Bacteriol 194:3294–3295. doi: 10.1128/JB.00363-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.