Abstract
Tryptophan-rich proteins play important biological functions for the Plasmodium parasite. Plasmodium vivax contains remarkably large numbers of such proteins belonging to the “Pv-fam-a” family that need to be characterized. Earlier, we reported the presence of memory T cells and naturally acquired antibodies against 15 of these proteins in P. vivax malaria-exposed individuals (M. Zeeshan, H. Bora, and Y. D. Sharma, J Infect Dis 207:175–185, 2013, http://dx.doi.org/10.1093/infdis/jis650). Here, we sought to characterize and ascertain the cross talk between effector responses of T and B cells in malarial patients against all Pv-fam-a family proteins. Therefore, we expressed the remaining 21 of these proteins in Escherichia coli and studied the humoral and cellular immune responses based on the same parameters used in our previous study. Naturally acquired IgG antibodies were detected against all 21 antigens in P. vivax patient sera (37.7 to 94.4% seropositivity). These antigens were able to activate the lymphocytes of P. vivax-exposed individuals, and the activated CD4+ T lymphocytes produced higher levels of Th1 (interleukin-2 [IL-2] and gamma interferon [IFN-γ]) and Th2 (IL-4 and IL-10) cytokines than the healthy controls, but the response was Th2 biased. The combined results of present and previous studies seem to suggest a striking link between induction of the CD4+ T cell response and naturally acquired antibodies against all 36 proteins of the Pv-fam-a family, the majority of them having conserved sequences in the parasite population. Further work is required to utilize this information to develop immunotherapeutic treatments for this disease.
INTRODUCTION
Despite continued efforts to control malaria, it remains a major health problem in affected tropical countries. There is an urgent need to develop an effective malaria vaccine and newer antimalarial drugs to control this disease. This requires the identification and characterization of newer parasite molecules that play significant biological function for the survival of the parasite and could generate cellular and humoral immune responses in humans to provide protection against the disease. Previous studies have shown that humoral and T-cell-mediated immune responses are induced against malaria parasite, and these responses were found to be antigen and stage specific (1). Antibodies come to play their role as soon as the parasite enters the human body. These antibodies not only clear the parasite by opsonization but also block the invasion of host cells by the parasite. Therefore, the humoral immune response plays a critical role in controlling the parasite.
A role for dendritic cells, monocyte/macrophages, B cells, and several groups of T cells has been proposed in the cellular immune response against malaria (1–4). Some specific cytokines are produced from peripheral blood mononuclear cells in response to parasite infection, which further activates the host's macrophages, neutrophils, T cells, and natural killer cells to react against the parasites (5). Monocytes/macrophages play an important role in the clearance of parasites by phagocytosing whole infected erythrocytes via opsonic or nonopsonic phagocytosis (6). Nitric oxide produced by macrophages in response to parasitic components and T cell gamma interferon (IFN-γ) have antiparasitic effects (1, 7, 8).
Being multistagic, the parasite encounters different components of the immune system, and thus selection of a vaccine candidate may be stage specific or multistagic, targeted to one or several antigens. We need a suitable vaccine candidate(s) that can elicit strong and protective immune response with conserved sequences. Although a number of vaccine candidates of Plasmodium falciparum have made it into clinical trials, few of them have shown promising immunogenicity and protection. In the absence of efficient and continuous in vitro culture of Plasmodium vivax, the development of a vaccine against this most widespread human malaria parasite has been more difficult and challenging. As a result, there are fewer P. vivax than P. falciparum vaccine candidate antigens being tested in clinical trials. In fact, very few P. vivax antigens have been immunologically characterized to determine their potential as vaccine candidates.
Tryptophan-rich proteins have been identified in murine and human malaria parasites (9–19). Across the Plasmodium species, these proteins have positionally conserved tryptophan residues. They are highly immunogenic in nature, and some of them are able to provide protection against Plasmodium yoelii in a murine malaria model (11, 20). For this reason, these antigens have been proposed as potential malaria vaccine candidates (9, 11, 12, 14, 18, 20, 21). Remarkably, a large number (n = 36) of tryptophan-rich protein coding genes have been identified in the P. vivax genome. These P. vivax tryptophan-rich antigens (PvTRAgs) are classified as “Pv-fam-a” family proteins (www.plasmodb.org). Several of these proteins have shown the stage-specific expression, and some of them are expressed at merozoite stage (22). Earlier, we showed that 10 of these 36 PvTRAgs, including those expressed by the merozoites, bind to uninfected human erythrocytes that can be inhibited by the patients' sera, thereby indicating their biological role in the parasite life cycle (17, 23). We also immunologically characterized 15 of these proteins and showed that P. vivax-exposed individuals produce antibodies and generate the memory T-cell responses against them (9, 14, 24). This still leaves the majority of PvTRAgs (21 of 36) immunologically uncharacterized.
Although antibody levels in patient sera and cytokine levels against some P. vivax antigens have been studied (16, 25, 26), there is a lack of information about the induction of CD4+ T cells and their cross talk with B cells in malaria patients which could generate stronger immune response to minimize P. vivax infection. Here, we analyzed antibody responses and the effector immune responses of CD4+ T cells among P. vivax-exposed individuals against the remaining 21 PvTRAgs, in addition to the genetic polymorphism. We deliberately used the same parameters as in our previous study (24) so as to draw a conclusion on immune responses generated against all 36 proteins of the Pv-fam-a family. The results, combined with previous data on 15 PvTRAgs (24), indicate the possibility of cross talk between effector responses of T and B cells in P. vivax-exposed individuals against all antigens of the Pv-fam-a family.
MATERIALS AND METHODS
Subjects and sample collection.
Individuals living in Northern India were screened for the clinical symptoms of malaria and the presence of malarial parasites by light microscopy. For genetic polymorphism and serological studies, 200 to 500 μl of heparinized blood was collected from microscopically confirmed P. vivax malaria patients prior to the start of antimalarial drugs. Patients were treated with antimalarial drugs according to the national drug policy (http://nvbdcp.gov.in/malaria-new.html). The same amount of heparinized blood was also collected from uninfected healthy individuals from the same area. For cellular immune response studies, 3 ml of heparinized blood was collected from malaria-naive volunteers and individuals who had recovered from their last P. vivax malaria episode about 8 to 12 weeks prior to sample collection. Samples were stored on ice and transported to the lab within 8 h.
Preparation of recombinant PvTRAgs and endotoxin determination.
PCR cloning, expression, and purification of histidine-tagged recombinant proteins of 21 PvTRAgs have been described elsewhere (23). These purified recombinant proteins were quantified by BCA kit according to the manufacturer's instructions (Thermo Scientific, Rockford, IL) and used for Western blot analysis, enzyme-linked immunosorbent assay (ELISA), and peripheral blood mononuclear cell stimulation. The endotoxin levels in the purified protein preparation were measured with a Limulus amebocyte lysate chromogenic endotoxin quantification kit according to the manufacturer's protocol (Pierce Biotechnology, Rockford, IL).
Western blot analysis.
The purified recombinant histidine-tagged PvTRAg (5 μg) was loaded into each well for SDS–12% PAGE. The protein bands were transferred from the gel to nitrocellulose filter paper using a semidry Western blot apparatus (Bio-Rad, Inc., Hercules, CA). The filter paper was developed as described earlier using pooled patients' sera at a 1:200 dilution (13).
ELISA.
ELISA was performed according to a standardized method (24). Briefly, a 96-well microtiter plate (BD Biosciences, San Diego, CA) was coated in triplicate with purified recombinant PvTRAgs (100 ng/well). Serum samples from P. vivax malaria patients and malaria-naive individuals were used as a primary antibody at a dilution of 1:200. Horseradish peroxidase-conjugated rabbit anti-human immunoglobulin G (IgG) was used as a secondary antibody (Thermo Fisher Scientific, Inc., Rockford, IL) at a dilution of 1:2,000, and o-phenylenediamine dihydrochloride was used as a substrate. The absorbance was measured at 595 nm in a microplate ELISA reader (Bio-Rad Laboratories).
Flow cytometry.
A whole-blood assay was performed according to a previously described protocol (24). Briefly, whole blood (100 μl) cells diluted (1:1) in RPMI 1640 (total volume, 200 μl) were stimulated with purified and filter-sterilized recombinant PvTRAgs (10 μg/ml) or phytohemagglutinin 1 (5 μg/ml) or with medium alone in the 96-well culture plates at 37°C in a humidified chamber with 5% CO2 for 60 h. The secretion inhibitor brefeldin A (eBioscience, Inc., San Diego, CA) was added at a concentration of 10 μg/ml, and the plates were incubated further for 4 h at 37°C. At 64 h, the erythrocytes were lysed, and the remaining cells were immunolabeled with surface markers using CD3 PE-Cy5, CD4 APC-Cy7, fluorescein isothiocyanate (FITC) CD69, and intracellular cytokines with anti-IL-4 FITC, anti-IL-10 PE, anti-IFN-γ PE-Cy7, and anti-IL-2 APC monoclonal antibodies. After labeling, the cells were washed, fixed with 1% paraformaldehyde, and kept in the dark at 4°C until acquisition. The data acquisition was carried out on a BDLSRII flow cytometer (Becton Dickinson Immunocytometry Systems, Palo Alto, CA) using FACSDiva software. Twenty-thousand events were acquired for all tests. For data analysis, CD3+ CD4+ events were gated, and the percentages of the cytokines were determined.
Genetic polymorphism.
The parasite DNA from clinical isolates was used to amplify the same region described above for cloning each of the 21 pvtrag genes. These PCR products were gel purified and sequenced as described elsewhere (24). The products were sequenced from both strands using nested primers described earlier (23) and an ABI BigDye terminator ready reaction kit version 3.1 on an ABI Prism 3130xl genetic analyzer (PE Applied Biosystems, Foster City, CA). The BioEdit sequence alignment editor and GeneDoc version 2.6.002 were used to analyze the sequencing electropherograms and generate sequence alignment, respectively.
Statistical analysis.
Statistical analysis was performed with SPSS software (v13). Descriptive data were expressed as the means ± the standard errors of the mean for nonparametric distributions. Differences in the levels of antibody and cytokines between groups were compared by using an unpaired Student t test. A P value of <0.05 was considered significant.
Nucleotide sequence accession numbers.
All of the sequences generated here have been submitted to the NCBI GenBank database under the accession numbers indicated in Table 1.
TABLE 1.
Genetic polymorphism in Plasmodium vivax tryptophan-rich antigens among clinical isolatesa
| Antigen (n) | Region analyzed (aa position) | Haplotypes (% prevalence) | Positions of amino acid substitutions | Accession no. (source or reference) |
|---|---|---|---|---|
| PvTRAg38.7 (31) | 60–315 | HI (70.9) | Conserved as Salvador-1 strain | KF178648 to KF178649 (this study) |
| HII (29.1) | E293K | |||
| PvTRAg35.7 (30) | 24–287 | HI (100) | Conserved as Salvador-1 strain | KF032070 (this study) |
| PvTRAg40.8 (33) | 60–330 | HI (100) | K312E | KF178645 (this study) |
| PvTRAg37.4 (37) | 1–294 | HI (59.4) | Conserved as Salvador-1 strain | KF178650 to KF178651 (this study) |
| HII (40.6) | T10N | |||
| PvTRAg34.9 (30) | 18–279 | HI (100) | Conserved as Salvador-1 strain | KF178652 to KF178653 (this study) |
| PvTRAg42.9a (30) | 38–358 | HI (30) | Conserved as Salvador-1 strain | KF233580 to KF233586 (this study) |
| HII (10) | N178T | |||
| HIII (3.4) | N178T, N216Q | |||
| HIV (10) | G208E, N214G, N216Q | |||
| HV (23.4) | N178T, G208E, N214G, N216Q | |||
| HVI (16.6) | N178T, C204F, G208E, N214G, N216Q | |||
| HVII (6.6) | C204F, G208E, N214G, N216Q | |||
| PvTRAg26.3 (34) | 60–209 | HI (55.9) | Conserved as Salvador-1 strain | KF032068 to KF032069 (this study) |
| HII (44.1) | D187G | |||
| PvTRAg36.6 (32) | 30–313 | HI (100) | Conserved as Salvador-1 strain | KF178653 (this study) |
| PvTRAg36 (33) | 30–312 | HI (30.3) | Conserved as Salvador-1 strain | KF178646 to KF178647 (this study) |
| HII (69.7) | D65E, 68(K) deletion, D70G, N81S, G82T, K85N, K93N, K95N, E98A, L106P | |||
| PvTRAg34 (30) | 30–302 | HI (6.7) | Conserved as Salvador-1 strain | KF207910 to KF207915 (this study) |
| HII (40) | G46A | |||
| HIII (6.7) | G46A and D205E | |||
| IV (10) | G46A, E68D, insertion at codon 75 (DKTTEKTTDKTTEKTT), P90A, and D205E | |||
| HV (23.3) | G46A, insertion at codon 75 (EKTADKATDKATDKPTDKTT) and D205E | |||
| HVI (13.3) | G46A, E68D, insertion at codon 75 (EKTTDKTTEKTTDKTTEKTT), P90A, and D205E | |||
| PvTRAg39.8a (30) | 29–327 | H1 (100) | Conserved as Salvador-1 strain | KF268447 (this study) |
| PvTRAg35.2a (33) | 30–288 | HI (30.3) | Conserved as Salvador-1 strain | KF178657 to KF178658 (this study) |
| HII (69.7) | T194K | |||
| PvTRAg56.2 (32) | 61–457 | HI (100) | Conserved as Salvador-1 strain | KF178654 (this study) |
| PvTRAg157 (31) | 1176–1414 | HI (54.8) | Conserved as Salvador-1 strain | KF178655 to KF178656 (this study) |
| HII (45.2) | H1378Q | |||
| PvTRAg36.7 (32) | 30–313 | HI (46.8) | Conserved as Salvador-1 strain | KF178659 to KF178660 (this study) |
| HII (45.2) | A244E | |||
| PvTRAg38.8 (42) | 24–339 | HI (28.6) | Conserved as Salvador-1 strain | KF233575 to KF233576 (this study) |
| HII (71.4) | S71P | |||
| PvTRAg38.5 (32) | 31–335 | H1 (100) | Conserved as Salvador-1 strain | KF233587 (this study) |
| PvTRAg309 (30) | 2388–2662 | HI (70) | Conserved as Salvador-1 strain | KF178661, KF207909 (this study) |
| HII (30) | K2466E | |||
| PvTRAg73.4 (31) | 64–612 | HI (77.4) | Conserved as Salvador-1 strain | KF233577 to KF233578 (this study) |
| HII (22.6) | E539K | |||
| PvTRAg99.6 (33) | 556–869 | HI (100) | Insertion at 764 IKW | KF233574 (this study) |
| PvTRAg33.6 (31) | 70–828 | HI (100) | Conserved as Salvador-1 strain | KF233579 (this study) |
| PvTRAg (33) | 65–324 | HI (3.3) | N180L, A293T | Y18842, AY576437, AY575008 to Y575013, AY570515 to Y570519, AY753149 to AY753168 (13) |
| HII (9.09) | A293T | |||
| HIII (81.8) | N186K, A293T | |||
| HIV (6.6) | Same as Salvador-1 strain type | |||
| PvTRAg69.4 (32) | 322–588 | HI (56.2) | Same as Salvador-1 strain type | JQ321368 to JQ321369 (24) |
| HII (43.7) | N340S | |||
| PvTRAg40 (35) | 85–321 | HI (100) | Same as Salvador-1 strain type | EF472686 to EF472720 (16) |
| PvTRAg38 (31) | 58–316 | HI (70.9) | Q200P | JQ321361 to JQ321364 (24) |
| HII (29.0) | Q200P, V270E | |||
| PvTARAg55 (31) | 94–478 | HI (83.3) | Same as Salvador-1 strain type | EF547891 to EF547921 (21) |
| HII (9.6) | Insertion of heptapeptide (GVAAAPG) at aa position 331 | |||
| P411S, A413T, deletion of A at aa position 414, deletion of heptapeptide (EETAASS) at 420–426 aa, T428A, T430P | ||||
| HIII (3.2) | Deletion of heptapeptide (TVNPEAT) at 429–435 aa | |||
| HIV (3.2) | ||||
| PvTRAg80.6 (31) | 59–678 | HI (90.3) | Same as Salvador-1 strain type | GU229675–GU229706 (14) |
| HII (9.6) | Insertion of K at 173 aa | |||
| PvTRAg53.7 (32) | 59–444 | HI (100) | Same as Salvador-1 strain type | JQ321360 (24) |
| PvTRAg39.8 (31) | 1–322 | HI (96.7) | Same as Salvador-1 strain type | EU446027 to EU446057 (12) |
| HII (32.2) | S32R | |||
| PvATRAg74 (32) | 187–635 | HI (6.2) | E339V | EU274314 to EU274328 (9) |
| HII (9.3) | Same as Salvador-1 strain type | |||
| HIII (21.8) | S293R | |||
| HIV (15.6) | E219G, S234G, S293R, K337E, T348A, K395E | |||
| HV (3.1) | S194R, S293R | |||
| HVI (6.2) | S293R, R338H | |||
| HVII (6.2) | S293R, G342E | |||
| HVIII (9.3) | S293R, I375 M, S387P | |||
| HIX (3.1) | S194R, S293R, T348A, K395E | |||
| HX (6.2) | S293R, K337E, T348A, K395E | |||
| HXI (3.1) | E219G, S234G, S293R, I386V | |||
| HXII (3.1) | G237R, S293R, T348A, I375 M ,K395E | |||
| HXIII (3.1) | E339V, deletion of 2 octapeptides (KSDASGVA and KSDASAVA) at 305–320 aa | |||
| HXIV (3.1) | L360I, deletion of octapeptide (KSDASAVA) at 305–312 aa | |||
| PvTRAg43.1 (30) | 1–356 | HI (3.3) | Deletion of octapeptide (AKAIQQAD) at 95–102 aa | JQ321351 to JQ321359 (24) |
| HII (16.6) | D35E, D43E, deletion of 3 octapeptide repeats (TKVAE/DKTG) at 47–70 aa | |||
| A121V, D43E, E51D, insertion of 2 octapeptide repeats (TKVAE/DKTG) at codon 70 | ||||
| HIII (13.3) | D35E, D59E, deletion of 1 octapeptide repeat (TKVAEKTG) at 63–70 aa | |||
| D27E, D43E, deletion of 2 octapeptide repeats (TKVAE/DKTG) at 55–70 aa | ||||
| HIV (33.3) | D35E, deletion of 2 octapeptide repeats (TKVAE/DKTG) at 55–70 aa and AKAIQQAD at 95–102 aa, deletion of 2 octapeptide repeats (TKVAE/DKTG) at 55–70 aa and AKAIQQAD at 95–102 aa | |||
| HV (6.6) | ||||
| HVI (10) | ||||
| HVII (6.6) | ||||
| HVIII (3.3) | D43E, E51D, D59E, deletion of 1 octapeptide repeat (TKVAEKTG) at 63–70 aa and AKAIQQAD at 95–102 aa | |||
| A26S, D35E, D59E, deletion of 1 octapeptide repeat (TKVAEKTG) at 63–70 aa | ||||
| HIX (3.3) | ||||
| PvTRAg42.9 (32) | 61–346 | HI (100) | Same as Salvador-1 strain type | JQ321365 (24) |
| PvTRAg39.9 (37) | 60–326 | HI (21.6) | Same as Salvador-1 strain type | JQ321370, JQ321348 to JQ321350 (24) |
| HII (27) | V153F | |||
| HIII (35.1) | N311K | |||
| HIV (16.2) | V153F & N311K | |||
| PvTRAg32.4 (31) | 1–262 | HI (100) | Same as Salvador-1 strain type | EF547859 to EF547889 (24) |
| PvTRAg35.2 (33) | 24–274 | HI (100) | Same as Salvador-1 strain type | GU229707 to GU229738 (14) |
| PvTRAg33.5 (31) | 24–268 | HI (100) | Same as Salvador-1 strain type | EU529797 to EU529826 (24) |
The amino acid positions shown here are for full-length proteins. aa, amino acid(s); n, number of isolates.
RESULTS
Effector B cell response against PvTRAgs.
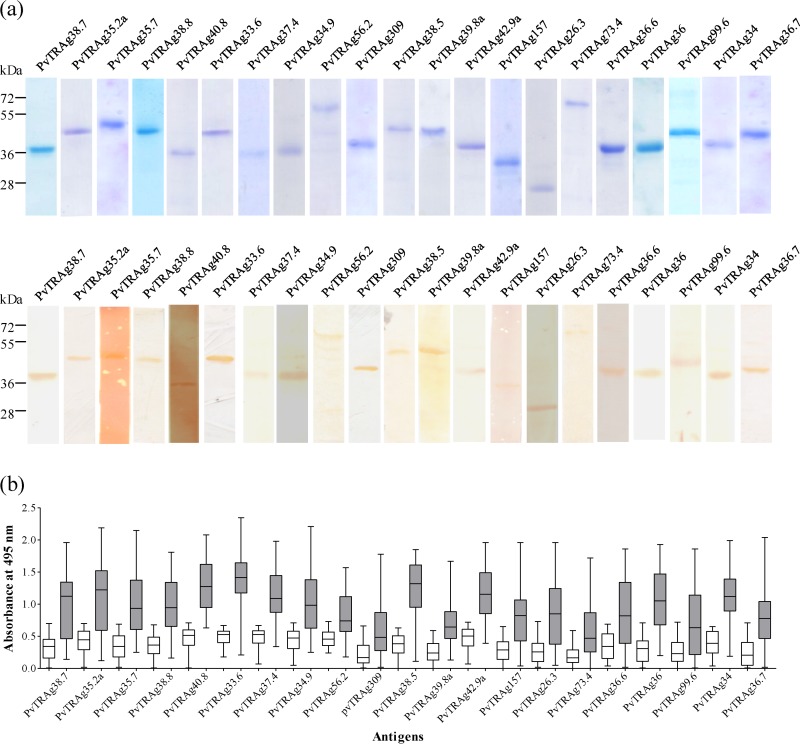
The current genomic database of P. vivax shows 36 PvTRAgs belonging to the Pv-fam-a family of proteins (www.plasmodb.org). The schematic diagram showing genomic organization of all 36 PvTRAgs is given in Fig. 1. The recombinant proteins derived from the exon 2 or tryptophan-rich domain encoded regions of 21 PvTRAgs were produced in Escherichia coli and affinity-purified proteins (Fig. 2A, upper panel) were used for immunological studies here (exon 1 is very small and encodes signal peptide in the majority of them). The endotoxin contents in these purified recombinant PvTRAgs were determined, and these values ranged from 1.8 to 7.3 endotoxin units (EU)/100 μg of protein (see Table S1 in the supplemental material). Therefore, the endotoxin levels of 0.036 to 0.146 EU/2 μg of purified recombinant PvTRAgs used here were in the permissive range (http://www.protocol-online.org/biology-forums/posts/1544.html). All of these 21 purified recombinant PvTRAgs were found to react with pooled P. vivax patient sera in Western blot analysis (Fig. 2A, lower panel). Naturally acquired anti-PvTRAgs antibody (IgG) levels were found to be elevated in patients (n = 54) in comparison to healthy controls (n = 40) (Fig. 2B). Seropositivity rates varied from 37.7% (for PvTRAg309) to 94.4% (for PvTRAg33.6) among the antigens. The level of the antibodies, in terms of the mean absorbance at 495 nm, also varied from 0.58 (PvTRAg73.4) to 1.38 (PvTRAg33.6). Taking together the results from this and a previous study (24), we found that all 36 PvTRAgs generated potent humoral immune responses during the course of a natural P. vivax infection (see Table S2 in the supplemental material).
FIG 1.
Gene organization of P. vivax tryptophan-rich antigens (PvTRAgs). The sequences of these antigens are retrieved from the PlasmoDB database (www.plasmodb.org). PvTRAgs are divided here into three groups based on the number of exons present. (a) PvTRAgs with one exon; (b) PvTRAgs with two exons; (c) PvTRAgs with three exons. Exons and introns are indicated by boxes and lines, respectively. Exon and intron sizes in base pairs are indicated at the top, while the encoded amino acid (aa) length is indicated at the bottom of the boxes. The predicted signal sequence in exon 1 and the tryptophan-rich domain in exon 2 or 3 are indicated by hatched boxes. “n” is the number of amino acids present in PvTRAgs. PvTRAg nomenclature is assigned according to molecular weight prefix.
FIG 2.
Naturally acquired antibodies against different PvTRAgs. (a) SDS-PAGE and Western blot analysis. Each of the purified histidine-tagged PvTRAg (5 μg/well) was separated by using SDS–12% PAGE (upper panel). Protein bands were transferred to nitrocellulose membrane, and blots were developed with 1:200 dilutions of pooled sera from five P. vivax patients. Molecular mass markers are indicated on the left in kilodaltons. (b) ELISA results. ELISA was performed using serum samples at a 1:200 dilution from P. vivax-exposed (n = 54) and malaria-naive individuals (n = 40) against different PvTRAgs. The box represents the interquartile range or the middle 50% of observations. The horizontal line represents the median. Whiskers represent the minimum and maximum observations.
PvTRAg-specific cell proliferative responses.
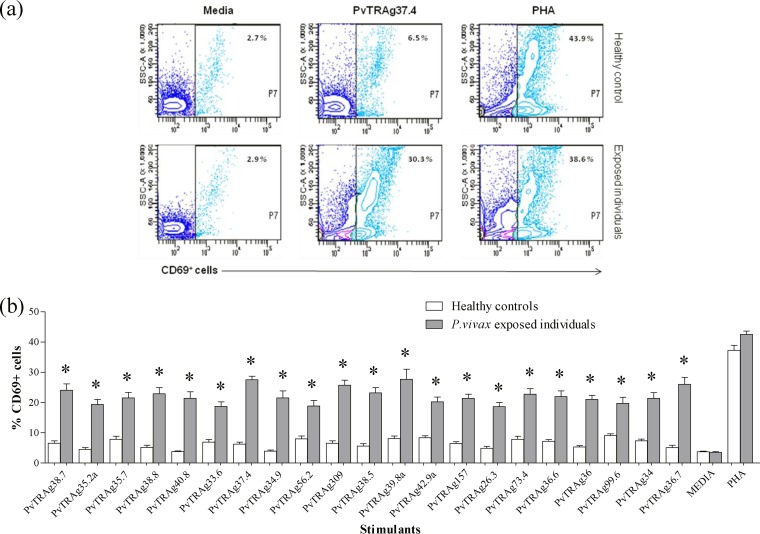
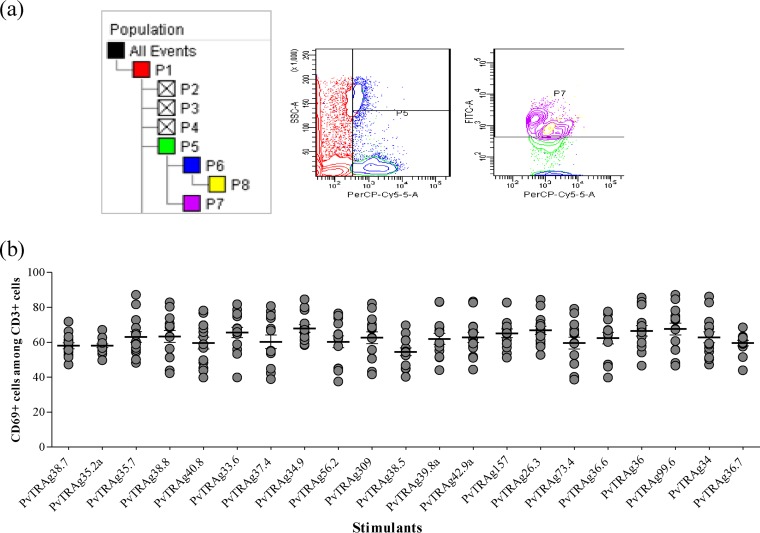
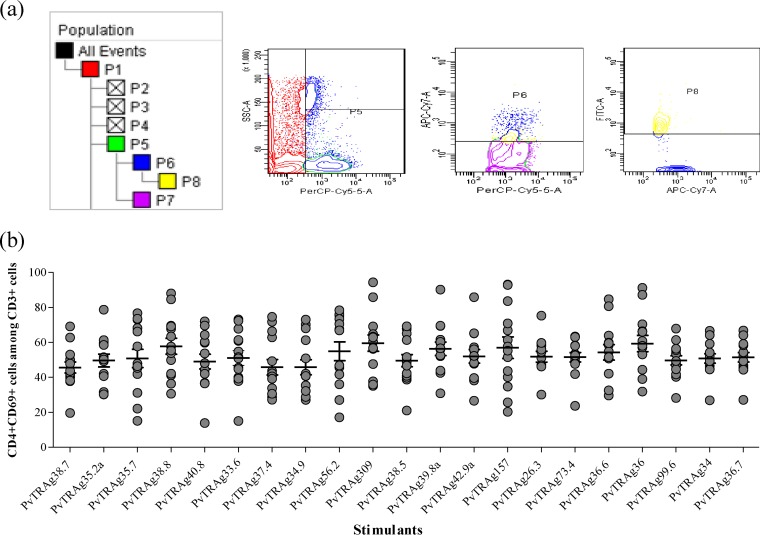
Antigen-specific cell proliferation was studied in terms of expression of CD69 in response to different stimuli (PvTRAgs). The frequency of CD69+ cells was significantly greater (P < 0.05) against all 21 PvTRAgs among P. vivax-exposed individuals (n = 10) than among the malaria-naive individuals (n = 14) (Fig. 3). The pattern of CD69 expression in patients against different PvTRAgs (including the previously characterized 15 PvTRAgs) was more or less similar. The relative levels of CD69 expression in patient blood were measured in terms of the mean fluorescence index (MFI). The MFI was generated by measuring the mean fluorescence of all CD69+ cells in stimulated blood samples of P. vivax individuals and dividing this value by the mean fluorescence of all CD69+ cells from nonstimulated blood samples of the same patient. These MFI values for P. vivax exposed individuals increased several times after stimulation with PvTRAgs compared to healthy controls (see Table S2 in the supplemental material). The CD69 expression level was ca. 60% (range, 54 to 68%) of CD3+ cells (Fig. 4) and 50% (range, 45 to 60%) of CD3+ CD4+ cells in the patient group (Fig. 5).
FIG 3.
Expression level of CD69 in leukocytes. (a) Representative contour plots showing the frequency of CD69+ cells (P7) for different stimulants (media, specific antigen, and phytohemagglutinin [PHA]) in blood samples of healthy controls and P. vivax-exposed individuals. (b) Comparative analysis of CD69 expression on leukocytes in response to different stimulants and media only, in cultures of whole blood of vivax malaria recovered patients and healthy controls. Each bar indicates the mean percentages ± the standard errors of the mean. Values that are statistically different between healthy controls and P. vivax exposed individuals are indicated by an asterisk (P < 0.05).
FIG 4.
Expression level of CD69 among CD3+ cells. (a) Representative contour plots showing the frequency of CD69 expression among CD3+ cells. The percentage of cells expressing CD69 (P7) was analyzed by flow cytometry on gated CD3+ T cells (P5). (b) Expression pattern of activation antigen (CD69) among CD3+ cells of P. vivax-exposed individuals in response to different PvTRAgs. Each bar indicates the mean percentage ± the standard error of the mean.
FIG 5.
Expression level of CD69 among CD3+ CD4+ cells. (a) Representative contour plots showing the frequency of CD69 expression among CD3+ CD4+ cells. The percentage of cells expressing CD69 (P8) was analyzed by flow cytometry on gated CD3+ CD4+ T cells (P6). (b) Pattern of the expression of CD69 among CD3+ CD4+cells of P. vivax-exposed individuals in response to different PvTRAgs. Each bar indicates the mean percentage ± the standard error of the mean.
Effector responses of PvTRAg-specific T cells.
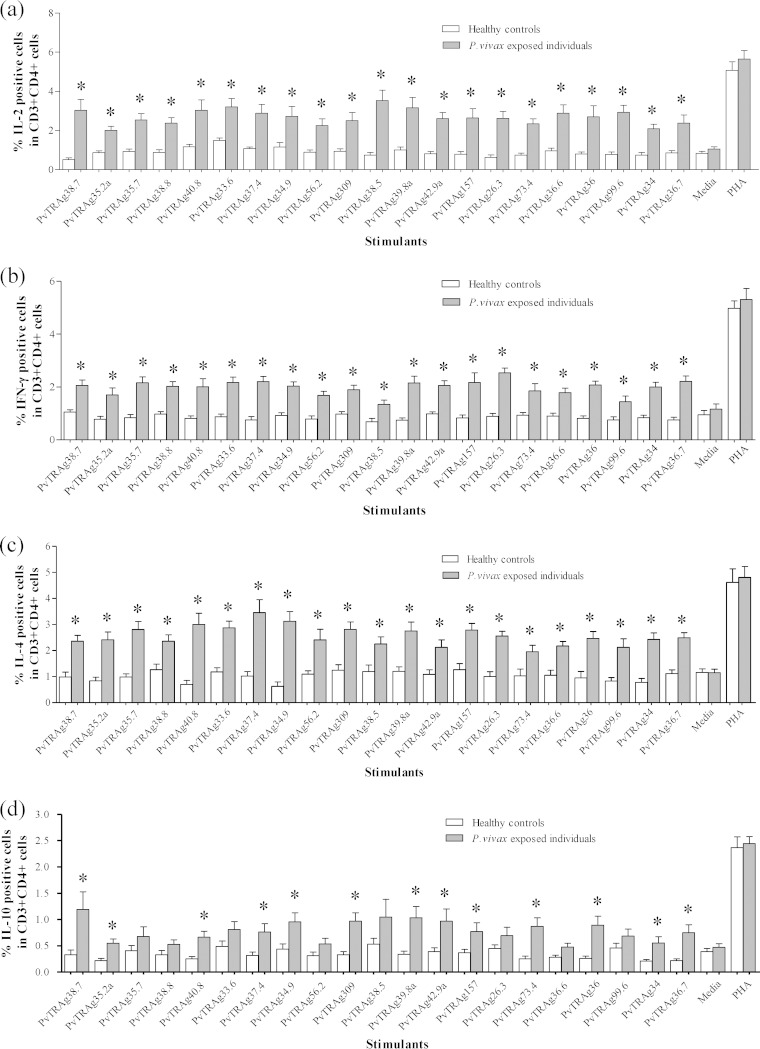
The frequency of cytokine-producing CD3+CD4+ cells was determined after stimulation with different PvTRAgs. We studied both Th1 (interleukin-2 [IL-2] and IFN-γ) and Th2 (IL-4 and IL-10) cytokines. The percentage of IL-2-producing cells was significantly higher (P < 0.05) against all 21 PvTRAgs in P. vivax-exposed individuals than in healthy controls. The level of intracellular IL-2 varies from antigen to antigen (2.01 to 3.53%) and was highest against PvTRAg38.5 (Fig. 6a). The mean percentage of IFN-γ-producing CD3+ CD4+ cells was also significantly higher (P < 0.05) among P. vivax-exposed individuals than the healthy controls against all 21 PvTRAgs (Fig. 6b) that ranged from 1.34% (PvTRAg38.5) to 2.54% (PvTRAg26.3), but these levels were lower than those for IL-2. Similarly, the number of cells that produced IL-4 was significantly higher (P < 0.05) among P. vivax-exposed individuals than among healthy controls against all 21 PvTRAgs (Fig. 6c). IL-10-positive cells were significantly higher in P. vivax-exposed individuals than in healthy controls against only 13 PvTRAgs (P < 0.05) (Fig. 6d). The mean percentage of IL-10-positive cells was much lower than IL-4-, IL-2-,and IFN-γ-positive cells in response to PvTRAgs. Taking together these data and those from a previous study (24), the CD3+ CD4+ cells of P. vivax-exposed individuals produced higher levels of IL-4, IL-2, and IFN-γ, whereas the level of IL-10 was lowest against most of these PvTRAgs (see Table S2 in the supplemental material).
FIG 6.
Expression level of cytokines in CD3+ CD4+ T cells. (a to d) Frequency of IL-2 (a)-, IFN-γ (b)-, IL-4 (c)-, and IL-10 (d)-expressing CD3+ CD4+ T cells obtained after stimulation with different stimulants (specific antigen, media, and PHA) in blood samples of P. vivax-recovered patients and healthy controls. Each bar indicates the mean percentage ± the standard error of the mean. Values that are statistically different between patients and controls are indicated by an asterisk (P < 0.05).
Genetic variations in PvTRAgs.
In the present study, the genetic variation of 21 PvTRAg genes was studied among Indian P. vivax clinical isolates. However, to give a complete picture of all 36 PvTRAgs, we included previously described data on remaining PvTRAgs (referenced in Table 1) for comparison purposes. The tryptophan residues of all the PvTRAgs were conserved among the field isolates. The genes encoding 15 different PvTRAgs were completely conserved in the parasite population; 13 of them had the same haplotype as the reference strain, whereas the sequences of PvTRAg40.8 and PvTRAg99.6 were different. PvTRAg40.8 also had one synonymous mutation (cytosine to adenine) at nucleotide position 528 (Table 1). Two haplotypes were observed for 14 different PvTRAgs, where one haplotype was the same as that of the reference strain, while the other haplotype was different. The second haplotype showed a single nucleotide polymorphism (SNP) for these PvTRAgs except PvTRAg38 (two SNPs) and PvTRAg36 (nine SNPs). The deletion or insertion of one lysine residue was seen in PvTRAg36 and PvTRAg80.6, respectively (Table 1). In PvTRAg38.7, one synonymous mutation (cytosine to guanine) was also found at nucleotide position 541. More than two haplotypes were observed in the parasite population for seven PvTRAgs (Table 1). These PvTRAgs were also showing substitutions, deletions, and insertions (Table 1).
DISCUSSION
The pathogenesis of malaria is complex because this parasite undergoes both extracellular and intracellular phases during its life cycle in the host. Thus, both humoral and cellular arms of the host immune system have to be mobilized to fight this parasitic infection. The serologic data of the present study demonstrate that distinct anti-PvTRAg antibody responses occur during the course of natural P. vivax infection since there were clear differences in anti-PvTRAg antibody levels between infected and uninfected group of individuals (Fig. 2). Similar results were obtained earlier against 15 PvTRAgs (24), indicating that all 36 PvTRAgs of Pv-fam-a family induce similar types of humoral immune responses in humans during P. vivax infection. Malaria-exposed individuals mount an antibody response to several antigens present on sporozoites, on merozoites, and on infected erythrocytes. This antibody-dependent mechanism plays an important role in reduction of parasitemia and can diminish clinical symptoms (27, 28). During asexual parasite development, proposed vaccines may induce specific antibodies that block merozoite invasion of erythrocytes (29), facilitate erythrophagocytosis (30), induce complement-mediated erythrocyte lysis, or inhibit intraerythrocytic parasite development through soluble immune mediators, such as oxygen radicals that are released in response to antibody-dependent cell inhibition (31).
The expression of significantly higher levels of CD69 on the leukocytes of P. vivax-exposed individuals than on healthy malaria-naive controls observed in the present and in previous (24) studies suggests that all 36 PvTRAgs have the ability to activate T lymphocytes (Fig. 3). However, the expression pattern of CD69 among CD3+ and CD4+ cells indicates that, in addition to T cells, some other cells, such as NK cells and B cells, may also express this activation marker but to a lesser extent (32). Activated T cells undergo proliferation and further differentiation to give effectors function. Naive T cells take time to start proliferation, whereas memory T lymphocytes, on secondary infection, can initiate a faster and more specific proliferation under the same condition that prevents or reduces the renewed occurrence of disease symptoms. Our results showed strong recall response, suggesting the presence of memory T cells against all the PvTRAgs in P. vivax-exposed individuals. In addition to antibodies, T-cell-mediated immune responses also play an important role in the control of malaria. Cell-mediated immunity, in which T cells play a major role, targets hepatic stages of the parasite and may be involved in providing the blood-stage immunity. Thus, some peripheral blood cells (neutrophils, macrophages, natural killers, etc.) (6) and lymphocyte subsets are thought to play a major role in malarial parasite control (1, 33).
Several studies, both in humans and in animals, have shown that CD4+ T cells play a major role in protection against Plasmodium infection (34, 35). Phenotype of CD4+ T cells can be determined by the cytokines expressed by them viz. IFN-γ and IL-2 for Th1 response and IL-4 and IL-10 for Th2 response (36, 37). The pattern of cytokine expression by CD4+ T cells of malaria-exposed individuals after stimulation with PvTRAgs observed here was strongly dominated by IL-4, followed by IL-2 and IFN-γ (see Table S2 in the supplemental material). Several studies indicate that IL-4 helps to sustain the growth and prolongs the survival of CD4+ T and B cells (38). It has also been reported that IL-4-secreting CD4+ T cells are crucial to the development of CD8+ T-cell responses against liver stages of the parasite (39). IL-2 levels were also found to be higher in patients' T cells for these PvTRAgs. IL-2 is a major T cell growth factor and is essential for the homeostasis, proliferation, and differentiation of CD4+ and CD8+ T cells (40). IL-2 secreted from CD4+ T cells is required for Foxp3 induction and contributes in maintaining the balance between CD4+ Foxp3+ regulatory T cells and effector CD4+ T cells required for immune control of blood-stage malaria infection (37, 41). PvTRAgs were producing a mixed—i.e., a Th1 and Th2 type—response. However, a geometric mean ratio of IFN-γ to IL-4 of <1 is an indication of the predominance of Th2 response (see Table S2 in the supplemental material).
Activation of CD4+ T cells has been linked to induction of a humoral immune response by promoting antibody class switch, affinity maturation, and induction of memory B cells which play an important role in the induction of high titers of antigen-specific IgG upon secondary exposure (42, 43). We observed that most of the PvTRAgs were inducing higher levels of IL-4-producing CD4+ T cells and naturally acquired IgG. This is similar to reports that suggest that a relationship exists between the activation of IL-4-producing T-cell subsets and antibody production in a human system in which the immune response is induced by natural infection (44). Similarly, the number of IL-2-producing CD4+ T cells was significantly higher against most of these PvTRAgs, a finding which is supported by a report that IL-2 produced by antigen-specific CD4+ T cells promotes the growth and differentiation of antigen-specific B cells (45). These findings indicate a link between the activation of CD4+ T cells and the generation of antibodies against PvTRAgs. Further, it has been reported that B cells mediate optimal proliferation of CD4+ T cells and can enforce or stabilize the differentiation of T cells into polarized effector cell subsets (34, 46–48). B cells also influence the development of CD4+ T cell memory (48–50). The presence of naturally acquired antibodies shows the induction of B cell response against PvTRAgs. It is tempting to speculate that this cross talk between the effector responses of CD4+ T cells and B cells may generate strong immune responses to minimize P. vivax infection.
Antigenic variation in malaria plays an important role in immune evasion, and most of the malarial vaccine candidates have shown such variation in the parasite population. This has hampered the development of a universal malaria vaccine. Such studies provide information about the frequency and dynamics of vaccine antigen polymorphisms that can be used to make informed decisions about which parasite allele(s) should be included in vaccine formulations (24, 51). We observed that sequences of the majority of PvTRAgs were mostly conserved in the parasite population (Table 1). The variations shown by PvTRAgs in the present study were mainly SNPs, except certain deletions/insertions observed among some of them (9, 12, 13, 24).
In conclusion, Pv-fam-a family proteins are antigenic and immunogenic, and their epitopes are conserved, having very limited sequence variations among P. vivax isolates. Effector responses of B cell and CD4+ T cells in response to PvTRAgs may generate a strong immune response to minimize this parasitic infection. Further studies analyzing the relationship between antigen-specific antibody levels and CD4+ T cell subsets, including follicular T helper cells, are required to demonstrate these correlations to establish their relevance for immunity to P. vivax infections.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by an Indian Council of Medical Research grant (61/4/2012/-BMS to Y.D.S.), a Department of Biotechnology grant (BT/PR9800/MED/29/44/2007 to Y.D.S.), a Senior Research Fellowship grant (M.Z.), and a Council for Scientific and Industrial Research Senior Research Fellowship grant (K.T.).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We are grateful to all patients who donated their blood samples for this study. We also acknowledge M. K. Das for helpful discussions, Poonia Ram for blood collection, and Shalini Narang for helping in the preparation of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03095-14.
REFERENCES
- 1.Troye-Blomberg M, Riley EM, Kabilan L, Holmberg M, Perlmann H, Andersson U, Heusser CH, Perlmann P. 1990. Production by activated human T cells of interleukin 4 but not gamma interferon is associated with elevated levels of serum antibodies to activating malaria antigens. Proc Natl Acad Sci U S A 87:5484–5488. doi: 10.1073/pnas.87.14.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casares S, Richie TL. 2009. Immune evasion by malaria parasites: a challenge for vaccine development. Curr Opin Immunol 21:321–330. doi: 10.1016/j.coi.2009.05.015. [DOI] [PubMed] [Google Scholar]
- 3.Douradinha B, Doolan DL. 2011. Harnessing immune responses against Plasmodium for rational vaccine design. Trends Parasitol 27:274–283. doi: 10.1016/j.pt.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Haque A, Best SE, Amante FH, Mustafah S, Desbarrieres L, de Labastida F, Sparwasser T, Hill GR, Engwerda CR. 2010. CD4+ natural regulatory T cells prevent experimental cerebral malaria via CTLA-4 when expanded in vivo. PLoS Pathog 6:e1001221. doi: 10.1371/journal.ppat.1001221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malaguarnera L, Musumeci S. 2002. The immune response to Plasmodium falciparum malaria. Lancet Infect Dis 2:472–478. doi: 10.1016/S1473-3099(02)00344-4. [DOI] [PubMed] [Google Scholar]
- 6.Chua CL, Brown G, Hamilton JA, Rogerson S, Boeuf P. 2013. Monocytes and macrophages in malaria: protection or pathology? Trends Parasitol 29:26–34. doi: 10.1016/j.pt.2012.10.002. [DOI] [PubMed] [Google Scholar]
- 7.Boutlis CS, Lagog M, Chaisavaneeyakorn S, Misukonis MA, Bockarie MJ, Mgone CS, Wang Z, Morahan G, Weinberg JB, Udhayakumar V, Anstey NM. 2003. Plasma interleukin-12 in malaria-tolerant Papua New Guineans: inverse correlation with Plasmodium falciparum parasitemia and peripheral blood mononuclear cell nitric oxide synthase activity. Infect Immun 71:6354–6357. doi: 10.1128/IAI.71.11.6354-6357.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor-Robinson AW. 1998. Immunoregulation of malarial infection: balancing the vices and virtues. Int J Parasitol 28:135–148. doi: 10.1016/S0020-7519(97)00173-2. [DOI] [PubMed] [Google Scholar]
- 9.Alam MT, Bora H, Singh N, Sharma YD. 2008. High immunogenicity and erythrocyte-binding activity in the tryptophan-rich domain (TRD) of the 74-kDa Plasmodium vivax alanine-tryptophan-rich antigen (PvATRAg74). Vaccine 26:3787–3794. doi: 10.1016/j.vaccine.2008.05.059. [DOI] [PubMed] [Google Scholar]
- 10.Burns JM, Adeeku EK, Belk CC, Dunn PD. 2000. An unusual tryptophan-rich domain characterizes two secreted antigens of Plasmodium yoelii-infected erythrocytes. Mol Biochem Parasitol 110:11–21. doi: 10.1016/S0166-6851(00)00252-8. [DOI] [PubMed] [Google Scholar]
- 11.Burns JM Jr, Dunn PD, Russo DM. 1997. Protective immunity against Plasmodium yoelii malaria induced by immunization with particulate blood-stage antigens. Infect Immun 65:3138–3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garg S, Chauhan SS, Singh N, Sharma YD. 2008. Immunological responses to a 39.8-kDa Plasmodium vivax tryptophan-rich antigen (PvTRAg39.8) among humans. Microbes Infect 10:1097–1105. doi: 10.1016/j.micinf.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 13.Jalah R, Sarin R, Sud N, Alam MT, Parikh N, Das TK, Sharma YD. 2005. Identification, expression, localization, and serological characterization of a tryptophan-rich antigen from the human malaria parasite Plasmodium vivax. Mol Biochem Parasitol 142:158–169. doi: 10.1016/j.molbiopara.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 14.Mittra P, Singh N, Sharma YD. 2010. Plasmodium vivax: immunological properties of tryptophan-rich antigens PvTRAg 35.2 and PvTRAg 80.6. Microbes Infect 12:1019–1026. doi: 10.1016/j.micinf.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Ntumngia FB, Bouyou-Akotet MK, Uhlemann AC, Mordmuller B, Kremsner PG, Kun JF. 2004. Characterization of a tryptophan-rich Plasmodium falciparum antigen associated with merozoites. Mol Biochem Parasitol 137:349–353. doi: 10.1016/j.molbiopara.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 16.Siddiqui AA, Bora H, Singh N, Dash AP, Sharma YD. 2008. Expression, purification, and characterization of the immunological response to a 40-kilodalton Plasmodium vivax tryptophan-rich antigen. Infect Immun 76:2576–2586. doi: 10.1128/IAI.00677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tyagi RK, Sharma YD. 2012. Erythrocyte binding activity displayed by a selective group of Plasmodium vivax tryptophan-rich antigens is inhibited by patients' antibodies. PLoS One 7:e50754. doi: 10.1371/journal.pone.0050754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uhlemann AC, Oguariri RM, McColl DJ, Coppel RL, Kremsner PG, Anders RF, Kun JF. 2001. Properties of the Plasmodium falciparum homologue of a protective vaccine candidate of Plasmodium yoelii. Mol Biochem Parasitol 118:41–48. doi: 10.1016/S0166-6851(01)00370-X. [DOI] [PubMed] [Google Scholar]
- 19.Siddiqui AA, Khan F, Sharma YD. 2014. Humoral immune responses to a recombinant Plasmodium vivax tryptophan-rich antigen among Plasmodium vivax-infected patients and its localization in the parasite. Appl Biochem Biotechnol 175:2166–2177. doi: 10.1007/s12010-014-1428-7. [DOI] [PubMed] [Google Scholar]
- 20.Burns JM Jr, Adeeku EK, Dunn PD. 1999. Protective immunization with a novel membrane protein of Plasmodium yoelii-infected erythrocytes. Infect Immun 67:675–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Siddiqui AA, Singh N, Sharma YD. 2007. Expression and purification of a Plasmodium vivax antigen: PvTARAg55 tryptophan- and alanine-rich antigen and its immunological responses in human subjects. Vaccine 26:96–107. doi: 10.1016/j.vaccine.2007.10.042. [DOI] [PubMed] [Google Scholar]
- 22.Bozdech Z, Mok S, Hu G, Imwong M, Jaidee A, Russell B, Ginsburg H, Nosten F, Day NP, White NJ, Carlton JM, Preiser PR. 2008. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci U S A 105:16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zeeshan M, Tyagi RK, Tyagi K, Alam MS, Sharma YD. 2015. Host-parasite interaction: selective Pv-fam-a family proteins of Plasmodium vivax bind to a restricted number of human erythrocyte receptors. J Infect Dis 211:1111–1120. doi: 10.1093/infdis/jiu558. [DOI] [PubMed] [Google Scholar]
- 24.Zeeshan M, Bora H, Sharma YD. 2013. Presence of memory T cells and naturally acquired antibodies in Plasmodium vivax malaria-exposed individuals against a group of tryptophan-rich antigens with conserved sequences. J Infect Dis 207:175–185. doi: 10.1093/infdis/jis650. [DOI] [PubMed] [Google Scholar]
- 25.Bitencourt AR, Vicentin EC, Jimenez MC, Ricci R, Leite JA, Costa FT, Ferreira LC, Russell B, Nosten F, Renia L, Galinski MR, Barnwell JW, Rodrigues MM, Soares IS. 2013. Antigenicity and immunogenicity of Plasmodium vivax merozoite surface protein-3. PLoS One 8:e56061. doi: 10.1371/journal.pone.0056061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen JH, Wang Y, Ha KS, Lu F, Suh IB, Lim CS, Park JH, Takeo S, Tsuboi T, Han ET. 2011. Measurement of naturally acquired humoral immune responses against the C-terminal region of the Plasmodium vivax MSP1 protein using protein arrays. Parasitol Res 109:1259–1266. doi: 10.1007/s00436-011-2370-z. [DOI] [PubMed] [Google Scholar]
- 27.Leoratti FM, Durlacher RR, Lacerda MV, Alecrim MG, Ferreira AW, Sanchez MC, Moraes SL. 2008. Pattern of humoral immune response to Plasmodium falciparum blood stages in individuals presenting different clinical expressions of malaria. Malar J 7:186. doi: 10.1186/1475-2875-7-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mackintosh CL, Christodoulou Z, Mwangi TW, Kortok M, Pinches R, Williams TN, Marsh K, Newbold CI. 2008. Acquisition of naturally occurring antibody responses to recombinant protein domains of Plasmodium falciparum erythrocyte membrane protein 1. Malar J 7:155. doi: 10.1186/1475-2875-7-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnot DE, Cavanagh DR, Remarque EJ, Creasey AM, Sowa MP, Morgan WD, Holder AA, Longacre S, Thomas AW. 2008. Comparative testing of six antigen-based malaria vaccine candidates directed toward merozoite-stage Plasmodium falciparum. Clin Vaccine Immunol 15:1345–1355. doi: 10.1128/CVI.00172-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.La Raja M. 2002. Erythrophagocytosis by peripheral monocytes in Plasmodium falciparum malaria. Haematologicaae 87:EIM14. [PubMed] [Google Scholar]
- 31.Legorreta-Herrera M, Rivas-Contreras S, Ventura-Gallegos J, Zentella-Dehesa A. 2011. Nitric oxide is involved in the upregulation of IFN-gamma and IL-10 mRNA expression by CD8+ T cells during the blood stages of P. chabaudi AS infection in CBA/Ca mice. Int J Biol Sci 7:1401–1411. doi: 10.7150/ijbs.7.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ziegler SF, Ramsdell F, Alderson MR. 1994. The activation antigen CD69. Stem Cells 12:456–465. doi: 10.1002/stem.5530120502. [DOI] [PubMed] [Google Scholar]
- 33.Tran TM, Samal B, Kirkness E, Crompton PD. 2012. Systems immunology of human malaria. Trends Parasitol 28:248–257. doi: 10.1016/j.pt.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fonseca L, Seixas E, Butcher G, Langhorne J. 2007. Cytokine responses of CD4+ T cells during a Plasmodium chabaudi chabaudi (ER) blood-stage infection in mice initiated by the natural route of infection. Malar J 6:77. doi: 10.1186/1475-2875-6-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oliveira GA, Kumar KA, Calvo-Calle JM, Othoro C, Altszuler D, Nussenzweig V, Nardin EH. 2008. Class II-restricted protective immunity induced by malaria sporozoites. Infect Immun 76:1200–1206. doi: 10.1128/IAI.00566-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acacia de Sa Pinheiro A, Morrot A, Chakravarty S, Overstreet M, Bream JH, Irusta PM, Zavala F. 2007. IL-4 induces a wide-spectrum intracellular signaling cascade in CD8+ T cells. J Leukoc Biol 81:1102–1110. doi: 10.1189/jlb.0906583. [DOI] [PubMed] [Google Scholar]
- 37.Scholzen A, Mittag D, Rogerson SJ, Cooke BM, Plebanski M. 2009. Plasmodium falciparum-mediated induction of human CD25Foxp3 CD4 T cells is independent of direct TCR stimulation and requires IL-2, IL-10, and TGFβ. PLoS Pathog 5:e1000543. doi: 10.1371/journal.ppat.1000543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nelms K, Keegan AD, Zamorano J, Ryan JJ, Paul WE. 1999. The IL-4 receptor: signaling mechanisms and biologic functions. Annu Rev Immunol 17:701–738. doi: 10.1146/annurev.immunol.17.1.701. [DOI] [PubMed] [Google Scholar]
- 39.Carvalho LH, Sano G, Hafalla JC, Morrot A, Curotto de Lafaille MA, Zavala F. 2002. IL-4-secreting CD4+ T cells are crucial to the development of CD8+ T-cell responses against malaria liver stages. Nat Med 8:166–170. doi: 10.1038/nm0202-166. [DOI] [PubMed] [Google Scholar]
- 40.Bachmann MF, Oxenius A. 2007. Interleukin 2: from immunostimulation to immunoregulation and back again. EMBO Rep 8:1142–1148. doi: 10.1038/sj.embor.7401099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berretta F, St-Pierre J, Piccirillo CA, Stevenson MM. 2011. IL-2 contributes to maintaining a balance between CD4+ Foxp3+ regulatory T cells and effector CD4+ T cells required for immune control of blood-stage malaria infection. J Immunol 186:4862–4871. doi: 10.4049/jimmunol.1003777. [DOI] [PubMed] [Google Scholar]
- 42.Kalia V, Sarkar S, Gourley TS, Rouse BT, Ahmed R. 2006. Differentiation of memory B and T cells. Curr Opin Immunol 18:255–264. doi: 10.1016/j.coi.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 43.McAdam AJ, Greenwald RJ, Levin MA, Chernova T, Malenkovich N, Ling V, Freeman GJ, Sharpe AH. 2001. ICOS is critical for CD40-mediated antibody class switching. Nature 409:102–105. doi: 10.1038/35051107. [DOI] [PubMed] [Google Scholar]
- 44.Gascan H, Gauchat JF, Roncarolo MG, Yssel H, Spits H, de Vries JE. 1991. Human B cell clones can be induced to proliferate and to switch to IgE and IgG4 synthesis by interleukin 4 and a signal provided by activated CD4+ T cell clones. J Exp Med 173:747–750. doi: 10.1084/jem.173.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Litjens NH, Huisman M, Hijdra D, Lambrecht BM, Stittelaar KJ, Betjes MG. 2008. IL-2 producing memory CD4+ T lymphocytes are closely associated with the generation of IgG-secreting plasma cells. J Immunol 181:3665–3673. doi: 10.4049/jimmunol.181.5.3665. [DOI] [PubMed] [Google Scholar]
- 46.Crawford A, Macleod M, Schumacher T, Corlett L, Gray D. 2006. Primary T cell expansion and differentiation in vivo requires antigen presentation by B cells. J Immunol 176:3498–3506. doi: 10.4049/jimmunol.176.6.3498. [DOI] [PubMed] [Google Scholar]
- 47.Lagrelius M, Jones P, Franck K, Gaines H. 2006. Cytokine detection by multiplex technology useful for assessing antigen specific cytokine profiles and kinetics in whole blood cultured up to 7 days. Cytokine 33:156–165. doi: 10.1016/j.cyto.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 48.Lund FE, Randall TD. 2010. Effector and regulatory B cells: modulators of CD4+ T cell immunity. Nat Rev Immunol 10:236–247. doi: 10.1038/nri2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Iijima N, Linehan MM, Zamora M, Butkus D, Dunn R, Kehry MR, Laufer TM, Iwasaki A. 2008. Dendritic cells and B cells maximize mucosal Th1 memory response to herpes simplex virus. J Exp Med 205:3041–3052. doi: 10.1084/jem.20082039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Linton PJ, Harbertson J, Bradley LM. 2000. A critical role for B cells in the development of memory CD4 cells. J Immunol 165:5558–5565. doi: 10.4049/jimmunol.165.10.5558. [DOI] [PubMed] [Google Scholar]
- 51.Bastos MS, da Silva-Nunes M, Malafronte RS, Hoffmann EH, Wunderlich G, Moraes SL, Ferreira MU. 2007. Antigenic polymorphism and naturally acquired antibodies to Plasmodium vivax merozoite surface protein 1 in rural Amazonians. Clin Vaccine Immunol 14:1249–1259. doi: 10.1128/CVI.00243-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.