Abstract
The interaction of environmental bacteria with unicellular eukaryotes is generally considered a major driving force for the evolution of intracellular pathogens, allowing them to survive and replicate in phagocytic cells of vertebrate hosts. To test this hypothesis on a genome-wide level, we determined for the intracellular pathogen Mycobacterium marinum whether it uses conserved strategies to exploit host cells from both protozoan and vertebrate origin. Using transposon-directed insertion site sequencing (TraDIS), we determined differences in genetic requirements for survival and replication in phagocytic cells of organisms from different kingdoms. In line with the general hypothesis, we identified a number of general virulence mechanisms, including the type VII protein secretion system ESX-1, biosynthesis of polyketide lipids, and utilization of sterols. However, we were also able to show that M. marinum contains an even larger set of host-specific virulence determinants, including proteins involved in the modification of surface glycolipids and, surprisingly, the auxiliary proteins of the ESX-1 system. Several of these factors were in fact counterproductive in other hosts. Therefore, M. marinum contains different sets of virulence factors that are tailored for specific hosts. Our data imply that although amoebae could function as a training ground for intracellular pathogens, they do not fully prepare pathogens for crossing species barriers.
INTRODUCTION
Throughout evolution, pathogens have developed an arsenal of molecular mechanisms to establish a successful infection within the host. Whereas many pathogens are restricted to a single host, some infectious agents have developed ways that allow them to survive in a variety of different hosts (1, 2). The molecular basis of host specificity is poorly understood, and the specific molecular pathways that are used by pathogens in different hosts are largely unexplored. Insight into evolutionary and host adaptation pathways could help to predict emerging pathogens. In addition, knowledge of virulence mechanisms that are essential for survival and pathogenesis across various organisms would be highly valuable for the design of effective drug treatments and vaccination strategies.
One of the diseases for which such knowledge would greatly benefit the design of treatment options is tuberculosis. This deadly infectious disease is caused by Mycobacterium tuberculosis, a facultative intracellular bacterium that has evolved to become one of the most successful human pathogens. As a result of genome downsizing, M. tuberculosis has become restricted to a limited number of mammalian hosts (3). In contrast, Mycobacterium marinum, which is genetically one of the closest relatives of the M. tuberculosis complex (4, 5), has maintained a larger genome and is able to infect a broader range of hosts (5). Both of these mycobacterial species have adapted to an intracellular lifestyle, using primarily macrophages as host cells. However, the ability of M. marinum to adapt to changing environmental conditions makes this pathogen an excellent tool to study mycobacterial adaptation to the host from an evolutionary perspective.
M. marinum causes disease in a large number of poikilothermic animals, including fish, frogs, and reptiles (6–8). The course of infection in these animals is highly similar to that of M. tuberculosis in the human host. In particular, the formation of granuloma-like lesions in the host and the ability to cause both acute and chronic disease are conserved (7, 9, 10). In addition to its natural ectothermic hosts, M. marinum is also able to cause infection in warm-blooded animals. However, due to temperature restrictions, the infection in these species is usually limited to the cooler body parts, such as the skin. M. marinum also has the capacity to infect protozoan hosts, such as the soil amoeba Dictyostelium discoideum and the waterborne amoeba Acanthamoeba castellanii (11, 12). Amoebae have also been suggested to serve as a training ground for intracellular pathogens (13). In fact, the intracellular lifestyle of pathogenic mycobacteria in animals has been hypothesized to be attributable to an evolutionary selection for survival and replication in protozoans (14), although genetic evidence to support this theory is lacking.
In the present work, we have studied on a genome-wide level the molecular mechanisms that are important for M. marinum to survive and replicate in cells derived from a variety of hosts. Since our findings reveal both common and host-specific virulence pathways, we argue that the current perception of amoebae serving merely as a training ground for intramacrophage pathogens (14) might be too simplistic.
MATERIALS AND METHODS
Bacterial strains, cell lines, and culture conditions.
In the present study, we used a transposon library generated in the M. marinum strain E11 (15). This library, consisting of >1.1 × 105 single transposon mutants, was constructed using transposon donor phagemid φMycoMarT7 as described previously (16). Transposon insertions were randomly distributed at TA dinucleotide sites (17). Prior to infection experiments, aliquots of the pooled transposon library were taken from −80°C and grown for 24 h with shaking at 30°C in Middlebrook 7H9 culture medium supplemented with ADC (albumin-dextrose-catalase; BD Biosciences) and 0.05% Tween. The human monocytic cell line THP-1, the mouse macrophage cell line RAW264.7 and the carp leukocytic cell line CLC (18) were cultured in RPMI 1640 with Glutamax-1 (Gibco) supplemented with 10% fetal bovine serum (FBS; Gibco), 100 U of penicillin/ml, and 100 μg of streptomycin/ml at 37°C and 5% CO2. Acanthamoeba castellanii (ATCC 30234) was cultured axenically in proteose-peptone-glucose medium at 30°C. Dictyostelium discoideum (Ax2) was grown axenically in HL5c medium (Formedium) at 25.5°C.
Infection of zebrafish embryos.
In order to visualize bacteria within zebrafish embryos, a pSMT3 plasmid containing a gene encoding the red-fluorescent protein mCherry was introduced into the M. marinum wild-type strain E11 and single transposon mutants of cpsA and eccCb1 by electroporation. Zebrafish embryos were infected with 100 CFU by microinjection in the caudal vein, as described previously (19). After 5 days of infection, zebrafish embryos were analyzed for bacterial load by fluorescence microscopy or bacterial CFU.
Infection of host cells.
For each host cell type, a total of 3 × 107 cells was seeded in T175 flasks (Corning). THP-1 monocytes were differentiated into macrophage-like cells by incubation with 25 ng of phorbol 12-myristate 13-acetate (Sigma)/ml for 24 h. The M. marinum transposon mutant pool was washed in RPMI 1640 (for infection of THP-1, RAW264.7, and CLC cells) or phosphate-buffered saline (PBS; for infection of amoeba species) and added to the cells at a multiplicity of infection of 0.5 for 3 h at 33°C (THP-1 and RAW264.7 cells), 30°C (CLC cells and A. castellanii), and 5% CO2 or 25.5°C (D. discoideum). Infected THP-1, RAW264.7, and CLC cells were washed twice in RPMI 1640 to remove extracellular mycobacteria and incubated at indicated temperatures in RPMI 1640–10% FBS. Amoebae were washed twice by low-speed centrifugation and incubated at the indicated temperatures. After 72 h of infection, cells were collected by scraping with a rubber policeman. After centrifugation at 5,000 rpm, the cell pellets were lysed in 1% Triton X-100 in PBS to release intracellular bacteria. By low-speed centrifugation, cellular debris was removed, and the supernatant was centrifuged at 5,000 rpm to pellet mycobacteria. Mycobacteria were plated on 7H10 agar in 10-fold serial dilutions to determine CFU numbers and isolate DNA. Fluorescence microscopy was performed as described in the supplemental material.
Coinfection.
CLC and THP-1 cells were seeded in 12-well plates at 8 × 105 cells/well. Infection with a 1:1 mixture of the cpsA, eccCb1, espG5, or ppm1a transposon mutant with the mCherry-expressing PE_PGRS15.2 transposon mutant was performed as described above (see the supplemental material). The ratio between selected and PE_PGRS15.2 transposon mutants was determined by performing fluorescence microscopy on at least 300 colonies per condition.
Genome sequencing.
Four micrograms of DNA from M. marinum E11 was used to prepare libraries for next-generation sequencing (NGS) according to the manufacturer's instructions using an Illumina genomic DNA sample preparation kit, a PCR-free sample preparation kit, or a Nextera mate pair sample preparation kit (Illumina Technologies, Inc., San Diego, CA). The DNA libraries were sequenced on Illumina HiSeq or MiSeq sequencers for NGS according to the manufacturer's specifications. A more detailed description of this procedure can be found in the supplemental material. The E11 genome and the plasmid pRAW are assigned in the European Nucleotide Archive under accession numbers HG917972 and HG917973, respectively.
DNA isolation and TraDIS.
For each condition, mycobacterial DNA was isolated from a monolayer of 5 × 106 colonies formed in 7 days on 7H10 plates grown at 30°C. Bacteria were resuspended in a 1:1 mixture of 10 mM Tris, 1 mM EDTA, 100 mM NaCl (pH 8.0), and phenol-chloroform-isoamyl alcohol (50:48:2) and disrupted by bead beating with 0.1-mm zirconia/silica beads (Biospec Products). After centrifugation, the supernatants were extracted with chloroform and centrifuged. DNA was precipitated from the supernatant with isopropanol in the presence of 300 mM sodium acetate. Precipitated DNA was washed with 70% ethanol, air dried, and dissolved in H2O. Transposon-directed insertion site sequencing (TraDIS) was performed on genomic DNA from four or three biological replicates of 5 × 106 colonies of the transposon mutant pool collected prior to and after infection, respectively. Construction of TraDIS libraries and sequencing were carried out essentially as described previously (20). This procedure and the statistical analysis of the data are explained in more detail in the supplemental material.
RESULTS
M. marinum E11 genome sequence.
In order to study pathogen adaptation to different hosts, we used the M. marinum wild-type strain E11. This strain was originally isolated from Dicentrarchus labrax (sea bass) and displays a chronic course of infection with a typical tuberculosis-like signature in zebrafish (5, 7, 10, 15). We first sequenced and annotated the M. marinum E11 genome and, as expected, found that it is highly similar to that of the M. marinum reference strain M (Fig. 1). Its 6.3-Mbp genome contains 5,335 protein coding sequences (CDS). In addition, the genome sequence also includes a 114-kbp plasmid with 98 protein-coding sequences (21). Many chromosomal differences between the two M. marinum strains can be attributed to bacteriophage-related sequences and transposable elements. In addition, there are a number of genes that are clearly recently acquired by horizontal gene transfer as they have no orthologues within closely related mycobacteria, such as a gene encoding the putative ferrous iron transporter FeoB. All unique E11 genes are listed in Data Set S1.1 in the supplemental material. Together, these results show that, in contrast to M. tuberculosis, horizontal gene transfer is probably an ongoing process in M. marinum.
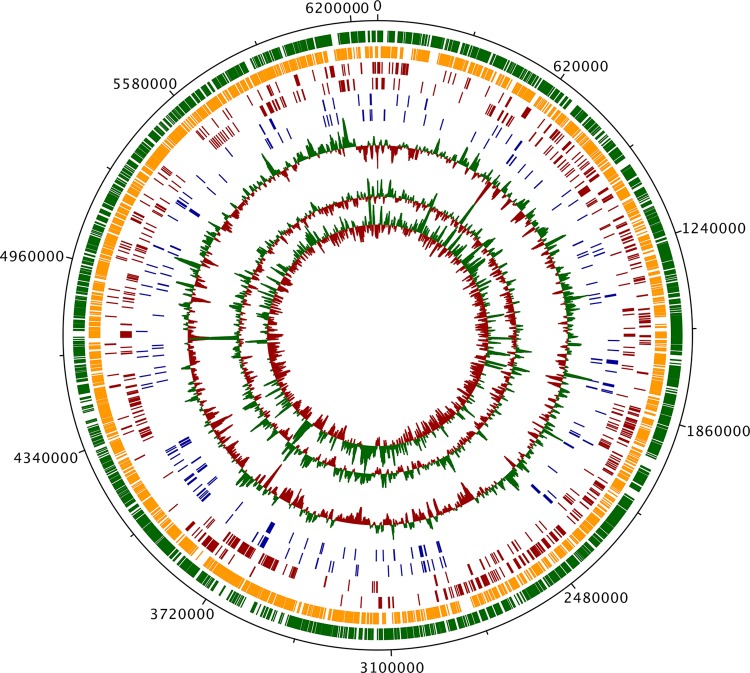
FIG 1.
M. marinum E11 genome. From the outer to inner circles, the tracks signify all non-pseudo-CDS in the forward direction (green), all non-pseudo-CDS in the reverse direction (orange), all CDS orthologous to M. tuberculosis H37Rv essential genes in the forward direction (red), all CDS orthologous to M. tuberculosis H37Rv essential genes in the reverse direction (red), all CDS without orthologues in the M. marinum M strain in the forward direction (blue), all CDS without orthologues in the M. marinum M strain in the reverse direction (blue), the GC plot (red, below average; green, above average), the theoretical distribution of TA sites (red, below average; green, above average), and the actual read coverage from TraDIS inputs (red, below average; green, above average).
M. marinum E11 infection of phagocytic cells.
In order to test whether the M. marinum E11 strain was able to survive in phagocytic cells derived from different hosts, we infected phagocytic cells from three different vertebrates (human, mouse, and fish) and two protozoan species (D. discoideum and A. castellanii) with green fluorescent protein-labeled mycobacteria. By performing fluorescence microscopy and plate count assays, we confirmed the presence and survival of mycobacteria for 3 days (see Fig. S1A in the supplemental material). In order to determine whether infection of protozoan cells showed the same characteristics as infection of vertebrate macrophages, we tested the effect of a mutation in ESX-1 on survival of M. marinum E11 in A. castellanii. M. marinum ESX-1 is essential for the successful infection of both zebrafish and human cell lines (19, 22, 23). We observed that an ESX-1-deficient eccCb1 mutant strain of M. marinum was indeed attenuated during A. castellanii infection (see Fig. S2B in the supplemental material). Together, our data confirm that the E11 strain of M. marinum is able to infect phagocytic cells derived from different hosts and that ESX-1 is important for infection.
Essential genes.
In order to determine the effect of each M. marinum gene on viability and infection, we constructed a pool of 1.1 × 105 single transposon mutants. Given the size of the E11 genome, this number of mutants results in a theoretical average transposon insertion density of one per 57 bp, which means that in principle all 5,433 coding sequences were covered by multiple transposon hits (see Fig. 2 for actual insertion frequencies). The exact locations of all of these transposon insertion sites were determined using TraDIS (20), which resulted in an average of 94% of the sequencing reads mapping to the M. marinum E11 genome (see Fig. S2 in the supplemental material). In order to produce high-confidence data sets, we used four biological replicates of 5 × 106 colonies for these experiments. To identify essential genes, we compared expected and observed maximal lengths of consecutive stretches of TA sites without a hit, using the selection criteria as described by Griffin et al. (24) (see Data Set S1.2 in the supplemental material). This led to the identification of 304 genes that are likely essential for survival of the bacteria when grown in normal culture media. Transposon insertions in these genes were absent or only possible at the terminal ends of these genes (Fig. 3B), which is likely not to affect gene function. Most of these essential genes are shared with M. tuberculosis and are involved in metabolic, respiratory and cell wall-associated processes (Fig. 3A and C) (25). For instance, the esx-3 gene cluster, which encodes an essential type VII secretion system involved in metal uptake in M. tuberculosis (26, 27), is also required for M. marinum viability (see Fig. S3 in the supplemental material). Interestingly, M. marinum contains a relatively large number of essential PE_PGRS genes, of which very few have orthologues in the genome of M. tuberculosis. A comparison of the two species shows that not all of the essential M. tuberculosis genes were found to be equally important for M. marinum (Fig. 3C) (24). This may partly be a result of technical reasons, as we performed the TraDIS with significantly higher sequencing depth, but partially also due to redundancy in the M. marinum genome, which is >2 Mbp larger than that of M. tuberculosis.
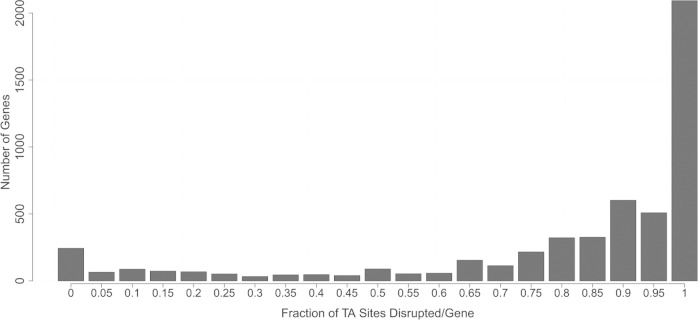
FIG 2.
Frequency distribution of the fraction of disrupted TA sites per gene. For each gene, the fraction of disrupted TA sites was calculated based on the average read coverage from all TraDIS input runs. Displayed are the number of genes that can bear transposon insertions in none to all TA sites, with an interval size of 0.05.
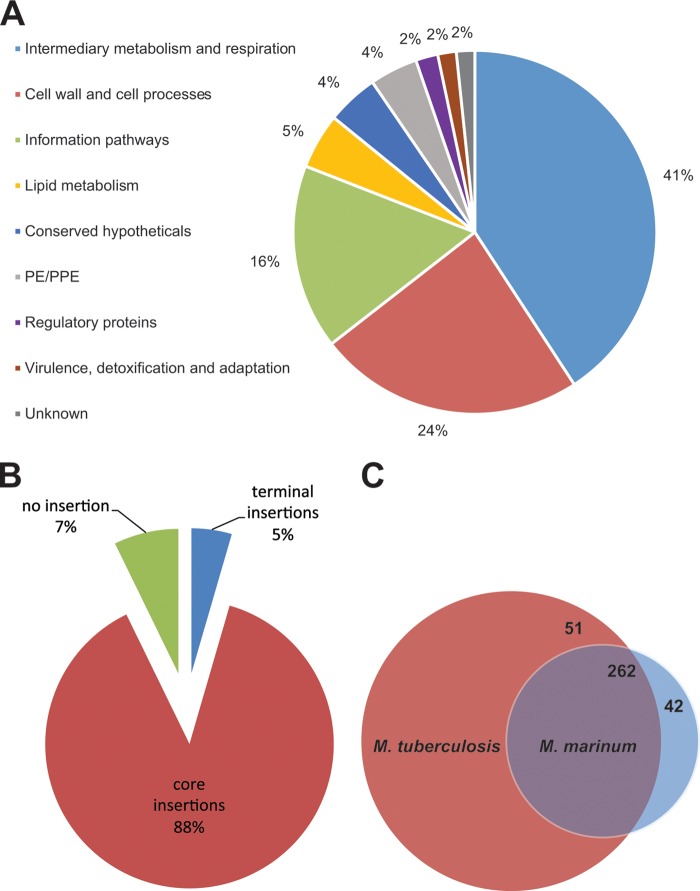
FIG 3.
Essential genes of M. marinum. (A) Pie chart showing a subdivision of all essential M. marinum genes based on functional category. (B) Pie chart showing the percentages of genes that contain core insertions (4,667 genes), terminal insertions (238 genes), and no insertions of transposons (382 genes). A total of 14 genes without any TA sites were excluded. (C) Venn diagram showing the degree of overlap between essential M. tuberculosis genes, as determined by Griffin et al. (24), and M. marinum genes found in the present study. Of the 42 genes that are essential in M. marinum and not in M. tuberculosis, 16 are M. marinum specific, half of which encode PE_PGRS proteins. Of the 511 genes that are specifically required for M. tuberculosis viability, 59 do not have an orthologue in M. marinum.
Hierarchical clustering of host cells based on infection-associated M. marinum genes.
Given the broad host range of M. marinum, the virulence mechanisms used may differ between hosts. To study these differences in detail, we infected phagocytic cells derived from human (THP-1), mouse (RAW264.7), and fish (CLC) origin and two amoeba species (A. castellanii and D. discoideum) with the complete pool of M. marinum transposon mutants. Bacterial growth rate, and therefore the putative number of generations within 3 days, increases with temperature and was highest in the mammalian cell lines, which were infected at 33°C (see Fig. S4 in the supplemental material). The experiments were performed with high bacterial numbers (1.5 × 107), but with a low multiplicity of infection (0.5) to ensure maximal statistical significance and minimal interference caused by different bacterial mutants infecting the same host cell (i.e., avoiding compensation of the defect by other bacteria, also known as the wild-type helper effect [28]). By performing TraDIS and comparing the transposon mutants that were present before and after 3 days of infection (see Fig. S5 in the supplemental material), we could identify the bacterial factors required, advantageous or disadvantageous in specific hosts. Importantly, the analysis of three individual biological replicates gave highly reproducible results (see Fig. S6 in the supplemental material), which means that the effect of different genes on intracellular survival and outgrowth can be predicted with high confidence. Based on the behavioral patterns of the M. marinum transposon mutant pool, we could cluster the results by the phagocytic cells from the five different hosts (Fig. 4). This revealed a high degree of similarity between the two mammalian cell lines, which seems logical since they are evolutionarily the most closely related. Furthermore, the two protozoan species also cluster, indicating functional similarities in these host cells. The phagocytic cells derived from fish show an intermediate behavior.
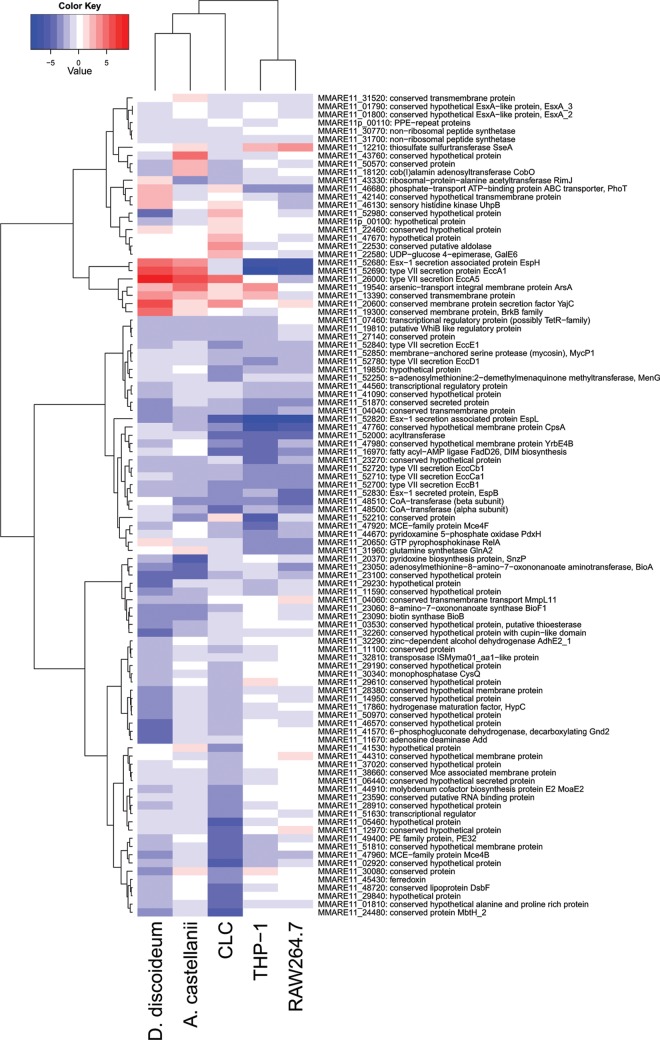
FIG 4.
Clustering of differentially affected genes and the host phagocytes. Heat map showing the clustering of the first 100 genes that display the most significant effects in the different phagocytic host cells. The degree of similarity between phagocytic cells is based on the (log2-transformed) effects of the transposon mutants in these cells. The effects were calculated per gene as the median effect over all TA sites covering the gene. Blue bars indicate attenuation of transposon mutants of the respective gene, whereas red bars point to an infection advantage.
Validation and identification of a novel virulence factor.
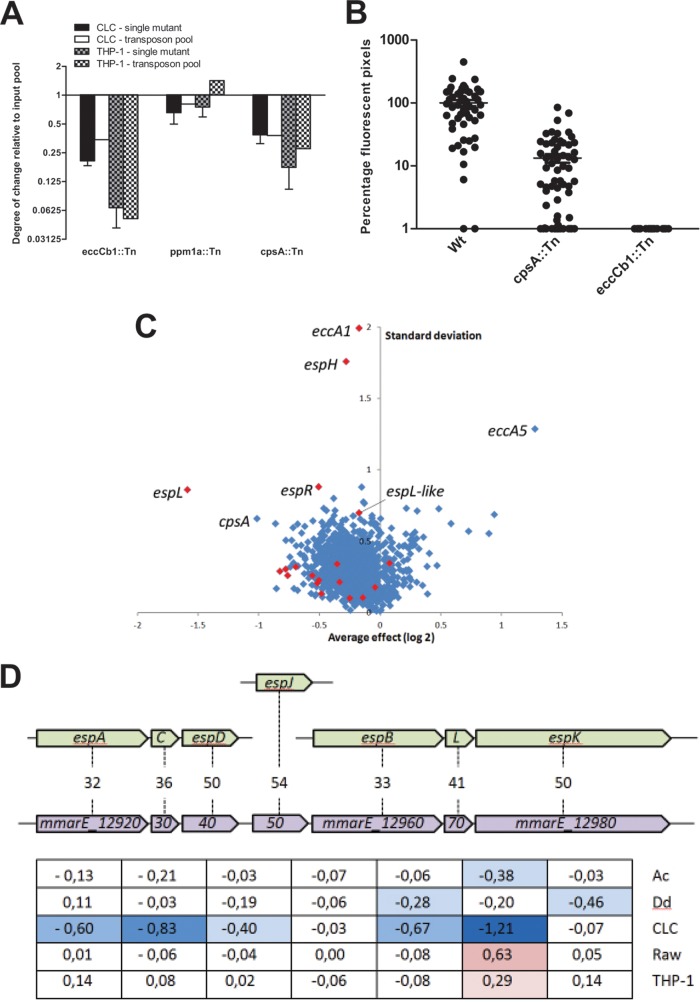
Although our TraDIS analysis was highly reproducible, we set out to validate the reliability of the data by testing three single M. marinum transposon mutants of genes that showed a clear phenotype in specific host cells. In a coinfection experiment, we infected human and fish cells with a 1:1 mixture of selected mutants and a transposon mutant showing no apparent phenotype and which we had available in our lab (PE_PGRS15.2). As selected mutants, we used single transposon mutants of the eccCb1 gene of the esx-1 locus, ppm1a (encoding a polyprenol-monophosphomannose synthase), and cpsA. The cpsA gene encodes a protein that belongs to the LytR family of cell envelope-bound transcriptional regulators, although the exact function is unknown. Specific characteristics of the transposon insertion mutants of these genes and the TraDIS effect on host cell infection are shown in Table S1 in the supplemental material. After 3 days of coinfection, the same time point as used for TraDIS, we determined relative bacterial growth. The mutants with a transposon insertion in cpsA and eccCb1 showed an attenuation pattern that was largely in agreement with the TraDIS data (Fig. 5A). Only for the third mutant, with a transposon insertion in ppm1a, we could not confirm the putative growth advantage in the THP-1 cell line. This may be due to the relatively small size of the effect and/or because the statistical significance of the TraDIS data for this gene was not below 0.05 (see Table S1 in the supplemental material). The P values of cpsA and eccCb1 were a factor 1,000 lower (see Table S1 in the supplemental material). Because CpsA was a novel virulence factor candidate for M. marinum, we also performed zebrafish embryo infection studies with the cpsA mutant and found that it was significantly attenuated in vivo (Fig. 5B). Together, these data show that the TraDIS results for M. marinum E11 are reliable and can be used to identify the specific contribution of known and novel virulence genes on a genome-wide scale.
FIG 5.
Specific effects of M. marinum transposon mutants. (A) Four single transposon mutants that showed a specific phenotype in the TraDIS analysis were tested for attenuation during 3 days of infection of human and mouse phagocytic cells. (B) A single cpsA transposon mutant was tested for attenuation in zebrafish embryos. The degree of infection after 5 days was determined by fluorescent pixel counts. The fluorescent pixel counts of the M. marinum wild-type (Wt) strain were set to 100%, and an eccCb1 transposon mutant was taken along as a negative control. Shown are the means ± standard errors of the mean of three biological replicates. (C) Average effects and variations of mutants that were significantly attenuated or enriched in one or more host cells in our TraDIS analysis. The average effect size and standard deviation of the log2-transformed data of all five host cells were taken for the 1,459 genes that had a P value below 0.05 in at least one condition. Genes of interesting outliers discussed in the text are indicated, and esx-1 genes are shown in red. (D) M. marinum-specific gene cluster (in purple) of which transposon mutants show a specific underrepresentation after infection of the CLC cells compared to the other cell lines. The percent identity with Esp homologues (in brown) is indicated, and the effect on different host cells (log2-transformed) is also shown. The blue represents growth disadvantage; red indicates growth advantage. Ac, A. castellanii; Dd, D. discoideum.
General and conserved virulence factors.
The disruption of several M. marinum genes and pathways was found to have similar effects in multiple hosts, indicating shared virulence mechanisms. These conserved virulence mechanisms range from secreted effector molecules to cell envelope biogenesis and adaptation to cell physiology. For example, interruption of transport and biosynthesis of the cell wall-associated lipid phthiocerol dimycocerosates (PDIM) results in attenuation of M. marinum in mammalian, fish, and protozoan cells (see Data Set S1.3 in the supplemental material), demonstrating that this virulence factor is essential for the pathogen. PDIM mutants are known to have a leaky outer membrane, which could affect intracellular survival. In M. tuberculosis, PDIM has been found to be important for infection as well (29, 30), indicating that PDIM function is conserved across mycobacterial species. A similar conclusion can be drawn for the ESAT-6 secretion system 1 (ESX-1). Transposon mutants of genes encoding the membrane components and some of the substrates of this type VII secretion system are highly attenuated in virulence in all tested phagocytic cells (see Data Set S1.3 in the supplemental material). Numerous studies have previously shown that ESX-1 is crucial for virulence of both M. marinum and M. tuberculosis (reviewed in reference 31). In addition to PDIM and ESX-1, the mammalian cell entry 4 (mce4) and mce1 gene clusters could also be identified as general virulence factors of both M. marinum and M. tuberculosis (29, 32–34). mce4 encodes a (chole)sterol import system that can provide energy to mycobacteria during infection (35). We found that transposon insertions in several genes of the M. marinum mce4 gene cluster led to attenuation in phagocytic cells of human, mouse and fish origin (see Data Set S1.3 in the supplemental material). In fact, a large number of genes required for M. tuberculosis growth on cholesterol (24) showed the same phenotype (see Data Set S1.4 in the supplemental material). Since we observed a strong selection of these genes within the 3-day time frame of the experiment, the dietary switch to cholesterol or other sterols (in amoeba) not only is a crucial step for intracellular survival and outgrowth but also occurs shortly after infection. Similarly to Mce4, Mce1 may act as a lipid import system as well and has been implicated in virulence of M. tuberculosis (32, 33, 36, 37). In M. marinum, we found a general attenuation pattern of mce1 transposon mutants (see Data Set S1.5 in the supplemental material). Together, our data indicate that a successful M. marinum infection in any of the tested hosts is dependent upon several general virulence mechanisms that are conserved in M. tuberculosis.
Host-specific virulence factors.
Although several general virulence genes and mechanisms could be discerned, most virulence genes were found to have a very strong but specific effect in a limited number of host cells and may therefore be markers for bacterial host adaptation (Fig. 5C). A list of the most variable infection-associated genes in the different host species can be found in Data Set S1.6 in the supplemental material. Most of the host-specific effects were observed in the CLC cell line. Of all tested phagocytes, the CLC cell line probably most closely resembles a natural host cell because it originates from carp, a natural host for M. marinum. Therefore, it is not surprising that a large number of the specific adaptations of M. marinum were found using these cells. One of the CLC-specific effects was observed in the lipooligosaccharide (LOS) biosynthesis pathway, which remarkably appears to be disadvantageous for infection. LOS is a cell wall-localized glycolipid that has been suggested to mask other surface-associated factors (38). Transposon insertions in several genes of the LOS region resulted in a marked increased fitness of the pathogen (MMARE11_22530-MMARE11_22600 [see Data Set S1.5 in the supplemental material]). Interestingly, not all LOS genes show this phenotype, only a specific set of genes involved in the biosynthesis and addition of caryophyllose, of which the abrogation results in a shorter version of LOS (39).
Another interesting cluster that was specifically affected in CLC cells was an M. marinum-specific set of genes that are all homologues of ESX-1 substrates (Fig. 5D). Because of their similarity, these genes probably also encode ESX-1 substrates. However, unlike the known ESX-1 substrates, these proteins seem to play a very specific role in host adaptation. Orthologues of their coding genes were present only in M. marinum or closely related species with a similar host range, such as Mycobacterium liflandii.
In protozoa, M. marinum is highly dependent on biotin synthesis for intracellular survival. On the other hand, disruption of the enzymes of this pathway had only a minor effect in phagocytes derived from the other hosts. This differential dependency on de novo biotin synthesis points to various levels of exogenous biotin in the diverse host cells, which may have to be considered when this pathway is explored as a potential drug target (40).
In contrast to biotin biosynthesis, inactivation of the vitamin B12 (cobalamin) biosynthesis pathway seems to provide M. marinum with a specific growth advantage in Acanthamoeba (see Fig. S7 in the supplemental material). In this host species, transposon insertions in multiple genes encoding enzymes of the vitamin B12 biosynthesis pathway, such as CobO, CobB, and CobH, give rise to highly increased fitness. Interestingly, this effect is very host-specific and is not, or to a very limited degree, observed in the other host species. Further analysis of mutants with a growth advantage in Acanthamoeba, combined with data on genomic localization and gene similarity, enabled us to detect additional factors that are probably involved in the cobalamin biosynthesis pathway. The newly identified CobF (MMARE11_43760) and a conserved transmembrane protein (MMARE11_3152; Rv2206) could be linked to this pathway as a direct result of TraDIS, providing a more complete picture of vitamin B12 biosynthesis in mycobacteria. Although the effect of inactivation of de novo vitamin B12 synthesis on infection is highly significant, the basis for this response is less clear. Interestingly, mycobacteria can also utilize vitamin B12 supplied by the host.
Unexpectedly, genes associated with the mycobacterial type VII secretion system ESX-1 were found among the genes with the most significant species-specific effect on virulence. Many studies, including this one, have firmly established a role for ESX-1 in virulence, both in M. marinum and in M. tuberculosis (29, 31, 41–43). This system, which is encoded by the esx-1 gene cluster, is responsible for the secretion of several virulence factors. Since we have made use of multiple M. marinum transposon mutants of each gene of the esx-1 locus, TraDIS allowed us to compare the contributions of individual ESX-1 system components during infection. Transposon insertions in genes encoding the conserved components that form the actual secretion machinery (44) result in strong attenuation of M. marinum in all hosts (see Data Set S1.3 and S1.5 in the supplemental material). Furthermore, disruption of genes encoding individual substrates of ESX-1 also have a general impact on virulence, albeit the effect is somewhat lower. Of the known substrates, EspB seems to be the most crucial for virulence, but it has to be noted that M. marinum has, in contrast to M. tuberculosis, two nearly identical copies of the best-known ESX-1 substrates esxAB, which could imply redundancy. The extra set of esxAB homologues (MMARE11_01760-MMARE11_01800) appears to be specifically important for infection of the fish cell line CLC, suggesting a host-specific ESX-1 function. However, the most dramatic effects are observed for esx-1 genes that are neither known substrates nor structural components of the transmembrane machinery (see Fig. S8 and Data Set S1.6 in the supplemental material). For instance, while disruption of espH and eccA1 leads to severe attenuation in both mammalian cell lines, it provides M. marinum with a strong growth advantage in the two protozoan species. In fact, these mutations cause the most disparate effects in the different hosts. Another striking observation is that the ESX-1-associated regulator EspR (encoded by MMARE11_52210) seems to be disadvantageous for infection of fish cells, whereas it is required for virulence in human cells and in A. castellanii. Possibly, espR regulation may go beyond ESX-1 and involve other host-specific processes (45). Together, these data show that although ESX-1-mediated virulence of M. marinum is crucial in phagocytic cells, fine-tuning of this system has major effects on virulence. Therefore, the ESX-1 system is probably subjected to strong selection during transmission to different hosts in order to adapt to these species.
DISCUSSION
The rapid development of deep-sequencing techniques has opened new possibilities for functional genomics studies. The recently developed TraDIS technique enables the high-throughput analysis of bacterial gene requirements using a large pool of transposon mutants (20, 24, 46, 47). Here, we used this technique to determine essentiality of gene function in M. marinum. Moreover, we used it to identify the factors that are important for infection of five different host species of this broad-host-range pathogen. A comparison of the genetic requirements of M. marinum in phagocytic cells derived from several organisms reveals important information that can help to elucidate the process of adaptation to new host species. Since our experiments gave highly reproducible results, TraDIS is an excellent tool to predict pathogen-host adaptation and to study virulence mechanisms in bacterial pathogens.
Our data showed that the M. marinum genome contains 304 genes that are essential for survival of the organism in vitro, which represents 6% of the total coding sequences. This number is significantly lower than the 19% for essential genes in M. tuberculosis (24), which may in part be attributable to higher sequence coverage used in the present study. However, since the genome of M. marinum is considerably larger than that of M. tuberculosis, it may contain alternatives for otherwise essential genes.
In order to identify the factors involved in intracellular survival and replication of M. marinum, we infected macrophages derived from three different host species and two protozoan species with the pool of transposon mutants, followed by TraDIS. Although we kept the infection conditions as similar as possible, the culture medium and infection temperature of the different host cells may influence the fitness of specific mutants and the number of replication rounds. We observed that the balance between bacterial replication and killing in Dictyostelium discoideum led to an equal number of bacteria before and after 3 days of infection (see Fig. S4 in the supplemental material). Although the infection temperature of 25.5°C is quite low for M. marinum to grow optimally, the M strain of this species has been described to replicate efficiently in Dictyostelium at this temperature (12). Our observation that the E11 strain seems to be less virulent in Dictyostelium demonstrates that strain-specific differences can have a large impact on the virulence of bacteria.
Overall, we found that the virulence mechanisms used by M. marinum are quite similar to those used by M. tuberculosis. Our data clearly demonstrate that ESX-1, PDIM, and cholesterol utilization are crucial for M. marinum during infection. We also identified cpsA as a novel virulence factor. In a large transposon site hybridization study, the cpsA gene of M. tuberculosis was previously found to be important for growth in the mouse spleen (29), indicating that CpsA is a shared virulence factor. CpsA is a putative transcriptional regulator localized in the membrane and has been suggested to be involved in capsule biosynthesis (48).
Being a broad-host-range pathogen, M. marinum has developed several host species-specific virulence mechanisms. Genes that were found to be disadvantageous for the infection of fish-derived cells include those that are involved in LOS biosynthesis. Interestingly, LOS is absent in M. tuberculosis, whereas it is present in the closely related M. canetti (49). The latter species is an early branching lineage of the M. tuberculosis complex and, as such, probably still has a broad environmental adaptability (50). Therefore, the loss of LOS may reflect pathogen specialization to a specific host. A recent study showed that variations in LOS resulted in increased phagocytosis of M. marinum by host cells (51), which could explain the observed phenotypes.
Inactivation of vitamin B12 biosynthesis was found to increase fitness of M. marinum in amoebae. In M. tuberculosis, vitamin B12 has been shown to serve as a cofactor for activity of the methionine synthase MetH, the methylmalonyl coenzyme A mutase MutA/B, and the class II RNR NrdZ (52, 53). MutA/B are part of the methylmalonyl pathway, which is important for relieving metabolic stress imposed by a dietary switch to cholesterol (54). The exact reason for the counterselection of vitamin B12 biosynthesis is not known, but considering the size of the observed effect and a similar effect on the recently identified vitamin B12 transporter (55), it is probably more than just energy conservation.
By using TraDIS, we could determine the relative contribution of each component of the virulence-associated protein secretion system ESX-1. Our data confirmed previous reports that the secretion machinery itself is essential for virulence, in all host species tested (19, 29, 41–43). Surprisingly, we found that not the classical ESX-1-secreted proteins but the secretion-associated proteins are responsible for the most disparate effects in the different models. This suggests that these secretion-associated proteins, such as EspH, EccA1, and EspL and the newly identified EspL-like homologue (Fig. 5D and see Fig. S8 in the supplemental material), play an auxiliary role in the secretion process and that it is the fine-tuning of secretion by these proteins that primarily determines virulence. Consequently, the most important effector proteins and mechanisms of ESX-1 may differ between host species, which should be taken into account when using mutants with a complete defect of the ESX-1 machinery. In addition, we identified a set of putative new ESX-1 substrates that do seem to play a role in host adaptation.
In the present study, we determined whether intracellular survival of the broad-host-range pathogen M. marinum involves general or host-specific virulence mechanisms and found that both apply to this pathogen. The observation that M. marinum uses host-specific mechanisms indicates that it adapts to different intracellular environments. Therefore, it may not be correct to consider amoebae as a training ground for intracellular pathogens, where observed virulence strategies can be translated one on one to a natural and clinically more relevant environment. In M. marinum, we found specific genetic requirements for infection of mammalian cells that were not applicable to protozoa and vice versa. These genetic requirements can be used to study host adaptation and may be useful for the identification of new drug targets. The use of TraDIS in infection studies can be highly instrumental in this process.
Supplementary Material
ACKNOWLEDGMENTS
We thank Janneke Maaskant for technical assistance. We also gratefully acknowledge Nicole van der Wel for her valuable help with the confocal fluorescence microscopy.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.03050-14.
REFERENCES
- 1.Relman DA, Hamburg MA, Choffnes ER, Mack A. 2009. Pathogen evolution, p 121–157. In Microbial evolution and co-adaptation: a tribute to the life and scientific legacies of Joshua Lederberg. National Academies Press, Washington, DC. [PubMed] [Google Scholar]
- 2.Bowden SE, Drake JM. 2013. Ecology of multi-host pathogens of animals. Nat Education Knowledge 4:5 http://www.nature.com/scitable/knowledge/library/ecology-of-multi-host-pathogens-of-animals-105288915 [Google Scholar]
- 3.Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST. 2002. A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci U S A 99:3684–3689. doi: 10.1073/pnas.052548299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stahl DA, Urbance JW. 1990. The division between fast- and slow-growing species corresponds to natural relationships among the mycobacteria. J Bacteriol 172:116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stinear TP, Seemann T, Harrison PF, Jenkin GA, Davies JK, Johnson PD, Abdellah Z, Arrowsmith C, Chillingworth T, Churcher C, Clarke K, Cronin A, Davis P, Goodhead I, Holroyd N, Jagels K, Lord A, Moule S, Mungall K, Norbertczak H, Quail MA, Rabbinowitsch E, Walker D, White B, Whitehead S, Small PL, Brosch R, Ramakrishnan L, Fischbach MA, Parkhill J, Cole ST. 2008. Insights from the complete genome sequence of Mycobacterium marinum on the evolution of Mycobacterium tuberculosis. Genome Res 18:729–741. doi: 10.1101/gr.075069.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clark H, Shepard C. 1963. Effect of environmental temperatures on infection with Mycobacterium marinum (Balnei) of mice and a number of poikilothermic species. J Bacteriol 86:1057–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Sar AM, Abdallah AM, Sparrius M, Reinders E, Vandenbroucke-Grauls CMJE, Bitter W. 2004. Mycobacterium marinum strains can be divided into two distinct types based on genetic diversity and virulence. Infect Immun 72:6306–6312. doi: 10.1128/IAI.72.11.6306-6312.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cosma CL, Swaim LE, Volkman H, Ramakrishnan L, Davis JM. 2006. Zebrafish and frog models of Mycobacterium marinum infection. Curr Protoc Microbiol Chapter 10:Unit 10B.2. doi: 10.1002/0471729256.mc10b02s3. [DOI] [PubMed] [Google Scholar]
- 9.Swaim LE, Connolly LE, Volkman HE, Humbert O, Born DE, Ramakrishnan L. 2006. Mycobacterium marinum infection of adult zebrafish causes caseating granulomatous tuberculosis and is moderated by adaptive immunity. Infect Immun 74:6108–6117. doi: 10.1128/IAI.00887-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weerdenburg EM, Abdallah AM, Mitra S, de Punder K, van der Wel NN, Bird S, Appelmelk BJ, Bitter W, van der Sar AM. 2012. ESX-5-deficient Mycobacterium marinum is hypervirulent in adult zebrafish. Cell Microbiol 14:728–739. doi: 10.1111/j.1462-5822.2012.01755.x. [DOI] [PubMed] [Google Scholar]
- 11.Kennedy GM, Morisaki JH, Champion PA. 2012. Conserved mechanisms of Mycobacterium marinum pathogenesis within the environmental amoeba Acanthamoeba castellanii. Appl Environ Microbiol 78:2049–2052. doi: 10.1128/AEM.06965-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Solomon JM, Leung GS, Isberg RR. 2003. Intracellular replication of Mycobacterium marinum within Dictyostelium discoideum: efficient replication in the absence of host coronin. Infect Immun 71:3578–3586. doi: 10.1128/IAI.71.6.3578-3586.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molmeret M, Horn M, Wagner M, Santic M, Abu Kwaik Y. 2005. Amoebae as training grounds for intracellular bacterial pathogens. Appl Environ Microbiol 71:20–28. doi: 10.1128/AEM.71.1.20-28.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Primm TP, Lucero CA, Falkinham JO III. 2004. Health impacts of environmental mycobacteria. Clin Microbiol Rev 17:98–106. doi: 10.1128/CMR.17.1.98-106.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puttinaowarat S, Thompson K, Lilley J, Adams A. 1999. Characterization of Mycobacterium spp. isolated from fish by pyrolysis mass spectrometry (PyMS) analysis. AAHRI Newslett 8:4–8. [Google Scholar]
- 16.Sassetti CM, Boyd DH, Rubin EJ. 2001. Comprehensive identification of conditionally essential genes in mycobacteria. Proc Natl Acad Sci U S A 98:12712–12717. doi: 10.1073/pnas.231275498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rubin EJ, Akerley BJ, Novik VN, Lampe DJ, Husson RN, Mekalanos JJ. 1999. In vivo transposition of mariner-based elements in enteric bacteria and mycobacteria. Proc Natl Acad Sci U S A 96:1645–1650. doi: 10.1073/pnas.96.4.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faisal M, Ahne W. 1990. A cell line (CLC) of adherent peripheral blood mononuclear leucocytes of normal common carp Cyprinus carpio. Dev Comp Immunol 14:255–260. doi: 10.1016/0145-305X(90)90097-X. [DOI] [PubMed] [Google Scholar]
- 19.Stoop EJM, Schipper T, Rosendahl Huber SK, Nezhinsky AE, Verbeek FJ, Gurcha SS, Besra GS, Vandenbroucke-Grauls CMJE, Bitter W, van der Sar AM. 2011. Zebrafish embryo screen for mycobacterial genes involved in granuloma formation reveals a novel ESX-1 component. Dis Models Mech 4:526–536. doi: 10.1242/dmm.006676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Langridge GC, Phan MD, Turner DJ, Perkins TT, Parts L, Haase J, Charles I, Maskell DJ, Peters SE, Dougan G, Wain J, Parkhill J, Turner AK. 2009. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res 19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ummels R, Abdallah AM, Kuiper V, Aajoud A, Sparrius M, Naeem R, Spaink HP, van Soolingen D, Pain A, Bitter W. 2014. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio 5:e01744-14. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao LY, Guo S, McLaughlin B, Morisaki H, Engel JN, Brown EJ. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol Microbiol 53:1677–1693. doi: 10.1111/j.1365-2958.2004.04261.x. [DOI] [PubMed] [Google Scholar]
- 23.Volkman HE, Clay H, Beery D, Chang JCW, Sherman DR, Ramakrishnan L. 2004. Tuberculous granuloma formation is enhanced by a Mycobacterium virulence determinant. PLoS Biol 2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Griffin JE, Gawronski JD, Dejesus MA, Ioerger TR, Akerley BJ, Sassetti CM. 2011. High-resolution phenotypic profiling defines genes essential for mycobacterial growth and cholesterol catabolism. PLoS Pathog 7:e1002251. doi: 10.1371/journal.ppat.1002251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sassetti CM, Boyd DH, Rubin EJ. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol Microbiol 48:77–84. doi: 10.1046/j.1365-2958.2003.03425.x. [DOI] [PubMed] [Google Scholar]
- 26.Serafini A, Boldrin F, Palu G, Manganelli R. 2009. Characterization of a Mycobacterium tuberculosis ESX-3 conditional mutant: essentiality and rescue by iron and zinc. J Bacteriol 191:6340–6344. doi: 10.1128/JB.00756-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Siegrist MS, Unnikrishnan M, McConnell MJ, Borowsky M, Cheng TY, Siddiqi N, Fortune SM, Moody DB, Rubin EJ. 2009. Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc Natl Acad Sci U S A 106:18792–18797. doi: 10.1073/pnas.0900589106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Autret N, Charbit A. 2005. Lessons from signature-tagged mutagenesis on the infectious mechanisms of pathogenic bacteria. FEMS Microbiol Rev 29:703–717. doi: 10.1016/j.femsre.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Sassetti CM, Rubin EJ. 2003. Genetic requirements for mycobacterial survival during infection. Proc Natl Acad Sci U S A 100:12989–12994. doi: 10.1073/pnas.2134250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cox JS, Chen B, McNeil M, Jacobs WR Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 31.Stoop EJ, Bitter W, van der Sar AM. 2012. Tubercle bacilli rely on a type VII army for pathogenicity. Trends Microbiol 20:477–484. doi: 10.1016/j.tim.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 32.Shimono N, Morici L, Casali N, Cantrell S, Sidders B, Ehrt S, Riley LW. 2003. Hypervirulent mutant of Mycobacterium tuberculosis resulting from disruption of the mce1 operon. Proc Natl Acad Sci U S A 100:15918–15923. doi: 10.1073/pnas.2433882100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchida Y, Casali N, White A, Morici L, Kendall LV, Riley LW. 2007. Accelerated immunopathological response of mice infected with Mycobacterium tuberculosis disrupted in the mce1 operon negative transcriptional regulator. Cell Microbiol 9:1275–1283. doi: 10.1111/j.1462-5822.2006.00870.x. [DOI] [PubMed] [Google Scholar]
- 34.Senaratne RH, Sidders B, Sequeira P, Saunders G, Dunphy K, Marjanovic O, Reader JR, Lima P, Chan S, Kendall S, McFadden J, Riley LW. 2008. Mycobacterium tuberculosis strains disrupted in mce3 and mce4 operons are attenuated in mice. J Med Microbiol 57:164–170. doi: 10.1099/jmm.0.47454-0. [DOI] [PubMed] [Google Scholar]
- 35.Pandey AK, Sassetti CM. 2008. Mycobacterial persistence requires the utilization of host cholesterol. Proc Natl Acad Sci U S A 105:4376–4380. doi: 10.1073/pnas.0711159105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Joshi SM, Pandey AK, Capite N, Fortune SM, Rubin EJ, Sassetti CM. 2006. Characterization of mycobacterial virulence genes through genetic interaction mapping. Proc Natl Acad Sci U S A 103:11760–11765. doi: 10.1073/pnas.0603179103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casali N, White AM, Riley LW. 2006. Regulation of the Mycobacterium tuberculosis mce1 operon. J Bacteriol 188:441–449. doi: 10.1128/JB.188.2.441-449.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Belisle JT, Brennan PJ. 1989. Chemical basis of rough and smooth variation in mycobacteria. J Bacteriol 171:3465–3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van der Woude AD, Sarkar D, Bhatt A, Sparrius M, Raadsen SA, Boon L, Geurtsen J, van der Sar AM, Luirink J, Houben ENG, Besra GS, Bitter W. 2012. Unexpected link between lipooligosaccharide biosynthesis and surface protein release in Mycobacterium marinum. J Biol Chem 287:20417–20429. doi: 10.1074/jbc.M111.336461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woong PS, Klotzsche M, Wilson DJ, Boshoff HI, Eoh H, Manjunatha U, Blumenthal A, Rhee K, Barry CE, Aldrich CC III, Ehrt S, Schnappinger D. 2011. Evaluating the sensitivity of Mycobacterium tuberculosis to biotin deprivation using regulated gene expression. PLoS Pathog 7:e1002264. doi: 10.1371/journal.ppat.1002264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brodin P, Majlessi L, Marsollier L, de Jonge MI, Bottai D, Demangel C, Hinds J, Neyrolles O, Butcher PD, Leclerc C, Cole ST, Brosch R. 2006. Dissection of ESAT-6 system 1 of Mycobacterium tuberculosis and impact on immunogenicity and virulence. Infect Immun 74:88–98. doi: 10.1128/IAI.74.1.88-98.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Guinn KM, Hickey MJ, Mathur SK, Zakel KL, Grotzke JE, Lewinsohn DM, Smith S, Sherman DR. 2004. Individual RD1-region genes are required for export of ESAT-6/CFP-10 and for virulence of Mycobacterium tuberculosis. Mol Microbiol 51:359–370. doi: 10.1046/j.1365-2958.2003.03844.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohol YM, Goetz DH, Chan K, Shiloh MU, Craik CS, Cox JS. 2010. Mycobacterium tuberculosis MycP1 protease plays a dual role in regulation of ESX-1 secretion and virulence. Cell Host Microbe 7:210–220. doi: 10.1016/j.chom.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Houben EN, Bestebroer J, Ummels R, Wilson L, Piersma SR, Jimenez CR, Ottenhoff TH, Luirink J, Bitter W. 2012. Composition of the type VII secretion system membrane complex. Mol Microbiol 86:472–484. doi: 10.1111/j.1365-2958.2012.08206.x. [DOI] [PubMed] [Google Scholar]
- 45.Blasco B, Chen JM, Hartkoorn R, Sala C, Uplekar S, Rougemont J, Pojer F, Cole ST. 2012. Virulence regulator EspR of Mycobacterium tuberculosis is a nucleoid-associated protein. PLoS Pathog 8:e1002621. doi: 10.1371/journal.ppat.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luan SL, Chaudhuri RR, Peters SE, Mayho M, Weinert LA, Crowther SA, Wang J, Langford PR, Rycroft A, Wren BW, Tucker AW, Maskell DJ. 2013. Generation of a Tn5 transposon library in Haemophilus parasuis and analysis by transposon-directed insertion-site sequencing (TraDIS). Vet Microbiol 166:558–566. doi: 10.1016/j.vetmic.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 47.van Opijnen T, Camilli A. 2013. Transposon insertion sequencing: a new tool for systems-level analysis of microorganisms. Nat Rev Microbiol 11:435–442. doi: 10.1038/nrmicro3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cieslewicz MJ, Kasper DL, Wang Y, Wessels MR. 2001. Functional analysis in type Ia group B Streptococcus of a cluster of genes involved in extracellular polysaccharide production by diverse species of streptococci. J Biol Chem 276:139–146. doi: 10.1074/jbc.M005702200. [DOI] [PubMed] [Google Scholar]
- 49.Daffe M, McNeil M, Brennan PJ. 1991. Novel type-specific lipooligosaccharides from Mycobacterium tuberculosis. Biochemistry 30:378–388. doi: 10.1021/bi00216a011. [DOI] [PubMed] [Google Scholar]
- 50.Gutierrez MC, Brisse S, Brosch R, Fabre M, Omais B, Marmiesse M, Supply P, Vincent V. 2005. Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathog 1:e5. doi: 10.1371/journal.ppat.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Alibaud L, Pawelczyk J, Gannoun-Zaki L, Singh VK, Rombouts Y, Drancourt M, Dziadek J, Guerardel Y, Kremer L. 2014. Increased phagocytosis of Mycobacterium marinum mutants defective in lipooligosaccharide production: a structure-activity relationship study. J Biol Chem 289:215–228. doi: 10.1074/jbc.M113.525550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Warner DF, Savvi S, Mizrahi V, Dawes SS. 2007. A riboswitch regulates expression of the coenzyme B12-independent methionine synthase in Mycobacterium tuberculosis: implications for differential methionine synthase function in strains H37Rv and CDC1551. J Bacteriol 189:3655–3659. doi: 10.1128/JB.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Savvi S, Warner DF, Kana BD, McKinney JD, Mizrahi V, Dawes SS. 2008. Functional characterization of a vitamin B12-dependent methylmalonyl pathway in Mycobacterium tuberculosis: implications for propionate metabolism during growth on fatty acids. J Bacteriol 190:3886–3895. doi: 10.1128/JB.01767-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee W, Vanderven BC, Fahey RJ, Russell DG. 2013. Intracellular Mycobacterium tuberculosis exploits host-derived fatty acids to limit metabolic stress. J Biol Chem 288:6788–6800. doi: 10.1074/jbc.M112.445056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gopinath K, Venclovas C, Ioerger TR, Sacchettini JC, McKinney JD, Mizrahi V, Warner DF. 2013. A vitamin B12 transporter in Mycobacterium tuberculosis. Open Biol 3:120175. doi: 10.1098/rsob.120175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.