Abstract
Acute ethanol intoxication suppresses the host immune responses against Streptococcus pneumoniae. As interleukin 17 (IL-17) is a critical cytokine in host defense against extracellular pathogens, including S. pneumoniae, we hypothesized that ethanol impairs mucosal immunity against this pathogen by disrupting IL-17 production or IL-17 receptor (IL-17R) signaling. A chronic ethanol feeding model in simian immunodeficiency virus (SIV)-infected rhesus macaques and acute ethanol intoxication in a murine model were used. Transcriptome analysis of bronchial brushes in the nonhuman primate model showed downregulation of the expression of IL-17-regulated chemokines in ethanol-fed animals, a finding also replicated in the murine model. Surprisingly, recombinant CXCL1 and CXCL5 but not IL-17 or IL-23 plus IL-1β rescued bacterial burden in the ethanol group to control levels. Taken together, the results of this study suggest that ethanol impairs IL-17-mediated chemokine production in the lung. Thus, exogenous luminal restoration of IL-17-related chemokines, CXCL1 and CXCL5, improves host defenses against S. pneumoniae.
INTRODUCTION
Bacterial pneumonia is a complication of ethanol abuse, and this susceptibility was documented both in animal models and in human studies more than a century ago (1, 2). Excessive ethanol consumption remains a public health problem responsible for approximately 79,000 deaths in the United States each year and for an estimated economic cost of $223.5 billion in 2006 (3).
Streptococcus pneumoniae is a Gram-positive bacterium and the leading etiology of community-acquired pneumonia, especially in older adults and alcoholic patients. Innate immune responses are important to control infections by S. pneumoniae, as evidenced by the roles of Toll-like receptors, MyD88, and IRAK4 in both humans and animal models (4, 5). More recently, evidence has emerged to support the fact that interleukin 17 (IL-17) plays a critical role in the regulation of S. pneumoniae colonization and infection. IL-17 and CD4+ T cells, and specifically the Th17 subset, are important for clearance of nasopharyngeal colonization with S. pneumoniae (a prerequisite of pneumococcal pneumonia) during primary and secondary infection (6, 7).
IL-17 is a cytokine produced by activated CD4+ T cells and innate lymphocytes (γδ+ T cells, NK cells, and innate lymphoid cells) and drives inflammation and autoimmunity (8). In the lung, the airway epithelium plays an important role in orchestrating the production of C-X-C chemokines, and these cells express both IL-17RA and IL-17RC and can produce both C-X-C chemokines and granulocyte colony-stimulating factor (G-CSF) in response to IL-17, leading to neutrophil recruitment in vivo (9–11).
As the lung epithelium is a major site of chemokine production during bacterial pneumonia (12), we examined the effect of ethanol on gene expression in the airway epithelium during experimental S. pneumoniae infection in a nonhuman primate model of ethanol consumption and pneumococcal infection (13, 14). We analyzed bronchial brush samples for gene expression using RNA sequencing and determined that ethanol consumption was associated with downregulation of IL-17 inducible genes in the bronchial epithelium 24 h postinfection. Based on these data, we hypothesized that ethanol impairs IL-17 immunity upon pneumococcal challenge by altering chemokine production from the lung epithelium.
To model the findings in our nonhuman primate model, we used a mouse model of ethanol exposure followed by intrapulmonary S. pneumoniae inoculation. The ethanol group demonstrated higher bacterial burdens and decreased IL-17 related pulmonary chemokine responses after bacterial challenge. Surprisingly, neither recombinant IL-17 nor IL-23 plus IL1B, which can activate γδ T cells to produce IL-17 (15), improved bacterial clearance in ethanol-treated mice. However, recombinant CXCL1 and CXCL5 but not CXCL2 decreased S. pneumoniae burden in the ethanol group to close to control levels.
Collectively, the data from this study not only demonstrate that ethanol impairs mucosal immunity against S. pneumoniae infection by disrupting IL-17 gene expression and signaling but also suggest that IL-17-mediated chemokine release by lung epithelium appears to be critical for mucosal defense against S. pneumoniae. Interestingly, only certain CXCR2 ligands (9) were capable of restoring mucosal immunity in the setting of ethanol intoxication.
MATERIALS AND METHODS
Experimental S. pneumoniae in rhesus macaques.
Bronchial brush samples were obtained from a cohort of male rhesus macaques between 3 and 6 years of age which were part of a larger study (13). All experiments were approved by the Institutional Animal Care and Use Committee at both the Tulane National Primate Research Center (TNPRC) in Covington, LA, and the Louisiana State University Health Sciences Center in New Orleans, LA, and adhered to the NIH guidelines for the use of experimental animals. Three months after initiation of the chronic binge ethanol (a half-hour daily gastric infusions to achieve plasma alcohol concentrations of 50 to 60 mM) or sucrose administration protocols, animals were intrarectally inoculated with 100 TCID50 (50% tissue culture infectious doses) of SIVmac251. Approximately two and a half months after SIV inoculation, antiretroviral therapy (ART) was initiated in animals used in this study. ART consisted of tenofovir (9-R-2-phosphonomenthoxypropyl adenine [PMPA]) and emtricitabine (beta-2, 3-dideoxy-3-fluoro-5-thiacytidine [FTC]). ART was continued throughout the study period. At four and a half months of SIV infection, animals were inoculated with 2 × 106 CFU of S. pneumoniae, serotype 19F (ATCC 6319), into a subsegment of the lower lobe of the right lung using a pediatric bronchoscope as previously described (14). Monkeys underwent bronchoscopies and bronchial brushings prepneumonia and day 1 postpneumonia. RNA was extracted from bronchial brushes, and a cDNA library was obtained. RNA samples from this project at different time points were archived.
RNA sequencing of rhesus macaques bronchial brushes.
Ten RNA samples from bronchial brushes of the following groups were processed: (i) sucrose before infection with S. pneumoniae (S/Pre) (n = 2), (ii) sucrose on day 1 after infection with S. pneumoniae (S/Post) (n = 2), (iii) ethanol before infection with S. pneumoniae (EtOH/Pre) (n = 3), and (iv) ethanol on day 1 after infection with S. pneumoniae (EtOH/Post) (n = 3). The RNA yield ranged from 700 ng to 200 ng with ratios of optical density at 260 nm (OD260) to OD280 above 1.8. Agilent analysis revealed that the RNA integrity numbers were between 3.1 and 6.7. mRNA was poly(A) selected. Following this, RNA was fragmented to an average length of 260 nucleotides (nt) by magnesium-catalyzed hydrolysis at 94°C and then converted to cDNA by random priming. The cDNA was adapted into a molecular library for Illumina sequencing on a Illumina GA IIx. The resulting single-read 50-bp reads were mapped to the rhesus macaque reference genome using a Burrows-Wheeler aligner (whole-transcriptome sequencing [WTS] BWA GATK v3).
RNA sequencing (RNA-seq) data analysis.
After FASTQ files were aligned to the reference genome, transcript abundance was quantified using Genesifter (Geospiza, Seattle, WA) software. Gene expression in samples was analyzed by measuring the frequency of reads mapping to the specific gene similar to SAGE tag quantification. First, the S/Pre group was compared to their respective paired S/Post counterparts using a paired t test (P < 0.1), having at least 20 reads per gene in one group with an induction of 2-fold. This analysis gave 432 differentially expressed genes. These genes were analyzed using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, which showed 23 (out of 432) genes involved in cytokine-cytokine receptor interaction and which were extrapolated to the ethanol group (EtOH/Pre and EtOH/Post) for comparison, and a paired t test (P < 0.05) was run for each group. To assess pathway enrichment, paired preinfection samples from sucrose- and ethanol-fed animals were compared to the respective postinfection samples, with parameters as mentioned above, and the sets of genes were exported to Ingenuity Pathway Analysis (IPA). Canonical analysis identified pathways from the IPA library that were most significant to the data set. The significance of the association between the data set and the canonical pathway was measured by a ratio of the number of genes from the data set that map to the pathway, divided by the total number of genes that map to the canonical pathway. Furthermore, Fischer's exact test followed by Benjamini-Hochberg (BH) multiple testing correction was used to calculate a P value.
Mice.
Eight- to 12-week-old female wild-type C57BL/6 mice obtained from Jackson Laboratories were housed in specific-pathogen-free rooms within animal facilities at the Rangos Research Building at Children's Hospital of Pittsburgh. To model acute ethanol intoxication, wild-type mice received 5 g of 20% ethanol (vol/vol) per kg of body weight in phosphate-buffered saline (PBS) administered intraperitoneally. Mice were monitored for 30 min and then inoculated with 1 × 107 to 2 × 107 CFU of live S. pneumoniae 19F (ATCC 6319) by intranasal aspiration. All protocols were approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Pittsburgh.
Experimental S. pneumoniae mouse infection.
One fresh colony of S. pneumoniae 19F was grown in 100 ml of Todd-Hewitt broth for 18 h at 37oC and 5% CO2. The bacterial culture was centrifuged at 1,000 × g for 15 min and washed twice with PBS. The bacterial pellet was resuspended in PBS, and concentration was determined by measuring the absorbance at 600 nm. An absorbance of 0.1 was about 2 × 107 CFU/ml. A dose of 1 × 107 to 2 × 107 CFU/mouse was given by intranasal aspiration (in 50 μl of PBS). The working stock was serially diluted and plated on Trypticase soy agar with 5% sheep blood plates (TSAII) to determine the exact concentration of the inoculum. For IL-17RC blockade, anti-murine IL-17RC (anti-mIL-17RC) antibody (20 μg/mouse; R&D) or goat IgG control (20 μg/mouse) was given by oropharyngeal aspiration 10 h after bacterial infection. For rescue experiments, recombinant IL-23 (1 μg/mouse in 50 μl of PBS) plus IL-1β (25 ng/mouse in 50 μl PBS) or IL-17 (25 ng/mouse in 50 μl PBS), CXCL1, CXCL5, or CXCL2 (all chemokines, 1 μg/mouse in 50 μl PBS; R&D) or PBS (vehicle) was given by oropharyngeal aspiration 2 h after S. pneumoniae infection.
Lung bacterial burden, mRNA, and protein analysis.
Bacterial lung burden, measured as number of CFU, was determined at 5 and 24 h after infection. Lungs were harvested in PBS, homogenized with a glass homogenizer, and plated on TSAII plates in three serial dilutions. Colonies were counted after overnight incubation at 37°C. Gene expression was determined by RNA extraction with TRIzol (Invitrogen), reverse transcription by iScript (Bio-Rad), and quantitative PCR (qPCR) with TaqMan probes (Applied Biosystems; Tnfa, Mm00443258_m1; IL17a, Mm00439618_m1; IL17f, Mm00521423_m1; Cxcl1, Mm04207460_m1; Cxcl2, Mm00436450_m1;Csf3, Mm00438334_m1; Il1b, Mm00434228_m1; Hprt, Mm01545399_m1; Ifng, Mm01168134_m1; Il12a, Mm00434165_m1; Il12b, Mm01288993_m1; Il6, Mm00446190_m1; Il23a, Mm01160011_g1; CpsA, 6-carboxyfluorescein [FAM]-[5′-AATGTTACGCAACTGACGAG-MGBNFQ1-3′]) on an iCycler thermocycler (Bio-Rad). Threshold cycle (CT) values were normalized to Hprt or GAPDH. Lung homogenates were analyzed using enzyme-linked immunosorbent assay (ELISA) for protein levels of IL-17.
Statistical analyses.
Unpaired, one-tailed Student's t tests (P < 0.05) were used to assess whether the means of two normally distributed groups differed significantly. Analysis of variance (ANOVA) or a Kruskal-Wallis test was used to compare multiple means. Significance is indicated by a P value of <0.05; in the figures, all error bars represent standard errors (SE).
RESULTS
Ethanol downregulates IL-17-related chemokine gene expression in rhesus macaques after experimental pneumococcal infection.
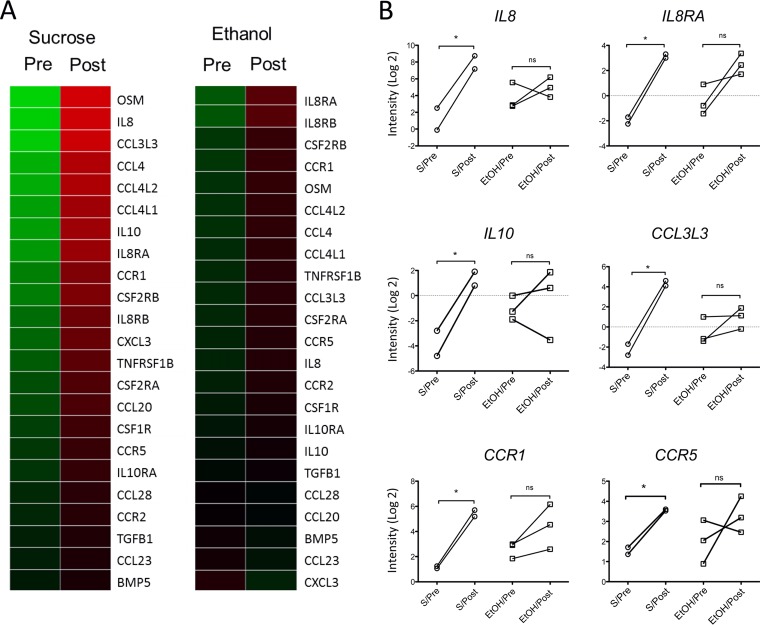
Airway transcriptome analysis from sucrose-fed rhesus macaques before and after pneumococcal infection suggests that S. pneumoniae drives the upregulation of genes in the chemokine and cytokine pathway and that this expression was suppressed in the group assigned to chronic ethanol intake. First, to assess the effect of S. pneumoniae inoculation, macaque samples from the sucrose group prior to (S/Pre) and 1 day after (S/Post) pneumococcal infection were compared. This comparison revealed 432 differentially expressed genes, which likely were driven by pneumococcal infection. The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway showed that 23 (out of 432) genes were involved in the cytokine-cytokine receptor interaction pathway (Fig. 1A) and that some of these genes encoded IL-17-related cytokines and chemokines, such as IL8, CXCL3, CCL3, and CCL4. Prior to S. pneumoniae challenge, the expression of most of these 23 genes did not differ between sucrose and ethanol treatment groups (Fig. 1A), which supports the fact that the difference observed in the sucrose group is likely due to pneumococcal infection. Expression of six of these 23 genes (Fig. 1A) did not increase in response to S. pneumoniae infection in the ethanol postinfection group, as they did in the control group (Fig. 1B); among these genes were IL8, IL8RA, CCL3L3, and IL10. Other chemokine genes such as CXCL1 and CXCL2 from the sucrose group had a trend toward upregulation compared to the ethanol group, but this trend did not reach statistical significance. Furthermore, Ingenuity Pathway Analysis (IPA) showed enrichment for the role of pattern recognition receptors in recognition of bacteria and viruses and IL-8 signaling pathways, among others (see Table S2 in the supplemental material).
FIG 1.
Gene expression of macaques bronchial brushes. (A) Heat map of genes involved in cytokine-cytokine receptor interaction differentially expressed in S/Pre and S/Post groups (n = 2) compared to those of the EtOH/Pre and EtOH/Post groups (n = 3); P < 0.1 (paired t test). (B) Gene expression data from panel A that reached significance (*, P < 0.05 [paired t test]). S/Pre, sucrose group before S. pneumoniae infection; S/Post, sucrose group 1 day after S. pneumoniae infection; ETOH/Pre, ethanol group before S. pneumoniae infection; EtOH/Post, ethanol group 1 day after S. pneumoniae infection; ns, not significant.
Based on this information, we generated the hypothesis that ethanol may impair lung mucosal immunity by downregulating IL-17-related chemokines. For this purpose, we utilized a mouse model of acute ethanol intoxication followed by S. pneumoniae 19F lung infection that allowed access to lung parenchymal cells as well as the ability to perform immune reconstitution experiments that were not possible in the macaque cohort.
IL-17RC blockade impairs S. pneumoniae 19F clearance.
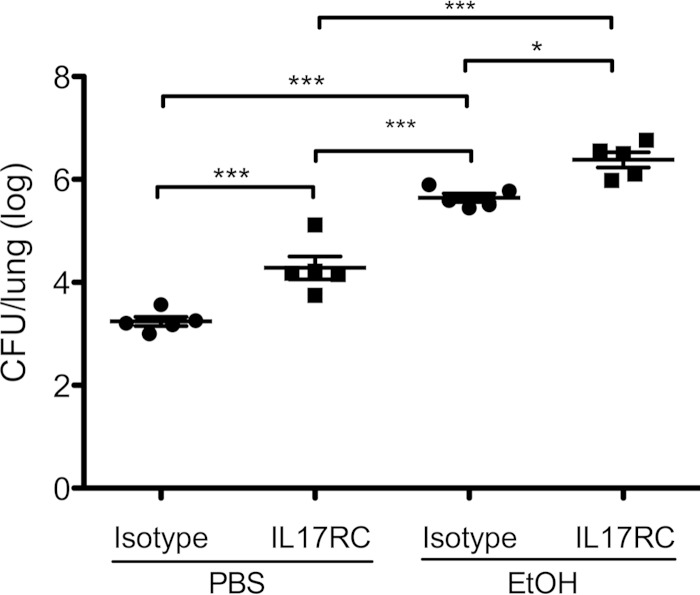
Wild-type mice were given anti-IL-17RC antibody or an isotype control 10 h after S. pneumoniae infection. Both groups were given either intraperitoneal PBS or ethanol (5 g/kg), and all received S. pneumoniae 19F by the intranasal route. Mice were sacrificed 24 h postinfection. Wild-type mice that were not ethanol intoxicated but had an IL-17RC blockade displayed higher bacterial burdens than the isotype control group. The administration of ethanol lessened even further the capacity to clear bacteria. The group with ethanol intoxication and the IL-17RC blockade showed the highest bacterial burden (Fig. 2), suggesting that ethanol exacerbates S. pneumoniae infection in an IL-17RC-dependent and -independent fashion.
FIG 2.
IL-17RC blockade impairs S. pneumoniae 19F clearance. Mice were given IL-17RC antibody or an isotype control 10 h after infection. Mice were randomized to control (PBS) or ethanol (EtOH) groups. All mice received 1 × 107 to 2 × 107 CFU of S. pneumoniae 19F. Each symbol represents one mouse. *, P < 0.5; ***, P < 0.0001 (one-way ANOVA). Experiments were repeated at least twice.
Ethanol impairs S. pneumoniae 19F clearance and IL-17-related genes in lung tissue.
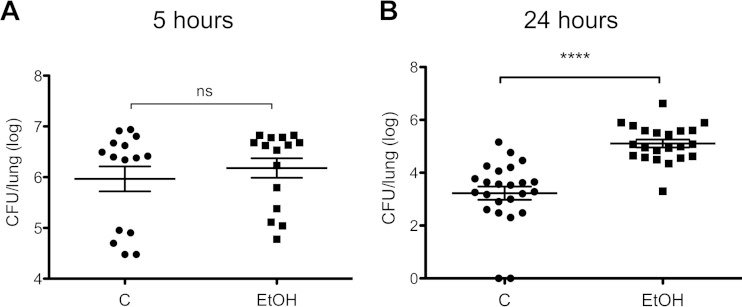
Mice with acute ethanol intoxication displayed higher bacterial burdens at 24 h postinfection than control mice. We randomized wild-type B6 mice to control groups that received either PBS or ethanol intraperitoneally, and then both groups received intranasal S. pneumoniae 19F. Mice were sacrificed at 5 or 24 h postinfection. At the 5-h time point, there was no difference in bacterial burden between the groups (Fig. 3A). However, at 24 h, the ethanol group had significantly higher numbers of CFU postinfection (Fig. 3B) than the control group. We used the 5-h time point to analyze gene expression by qPCR because the difference we would see would not be driven by changes in bacterial burden.
FIG 3.
Ethanol impairs S. pneumoniae 19F clearance. (A) Mice randomized to control or EtOH groups were sacrificed at 5 h postinfection. ns, not significant. (B) Mice randomized to control or EtOH groups were sacrificed at 24 h postinfection. Mice received 1 × 107 to 2 × 107 CFU of S. pneumoniae 19F each. Each symbol represents one mouse. C, control group; EtOH, ethanol group. ****, P < 0.0001 (unpaired t test, one tailed).
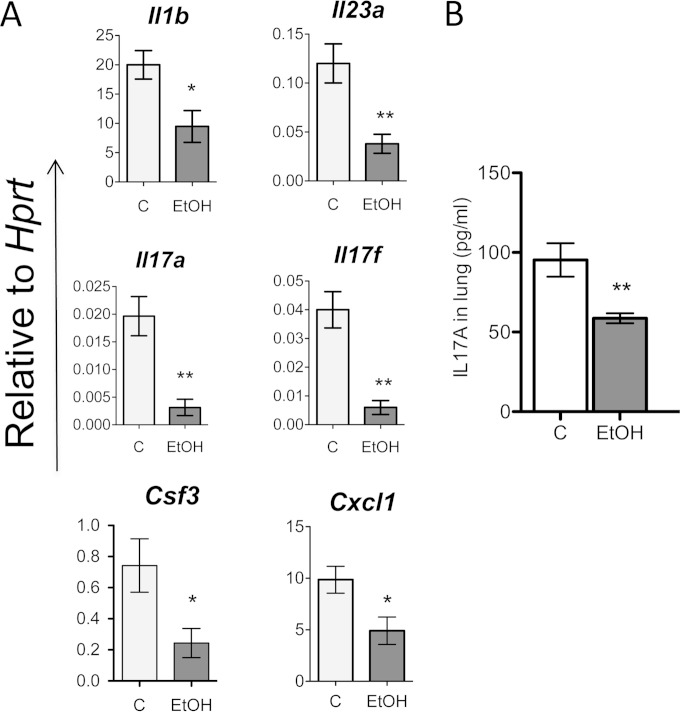
We detected that IL-17-related genes were significantly downregulated in the lung tissues of the ethanol group compared to the control group. This group of genes included Il17a and Il17f, upstream genes like Il23a and Il1b, and downstream genes such as Csf3 and Cxcl1 (Fig. 4A). IL-17A protein expression in lung tissue was also decreased in the ethanol group compared to the control group (Fig. 4B).
FIG 4.
Ethanol suppresses type 17 responses at 5 h after pneumococcal infection. (A) Gene expression relative to Hprt as determined by qPCR data from lung tissue of the control group (C) or the ethanol group (EtOH) after S. pneumoniae infection. (B) Protein expression, as determined by ELISA, from lung homogenates from control and EtOH groups infected with S. pneumoniae. *, P < 0.05; **, P < 0.01 (unpaired t test). Experiments were repeated at least 3 times.
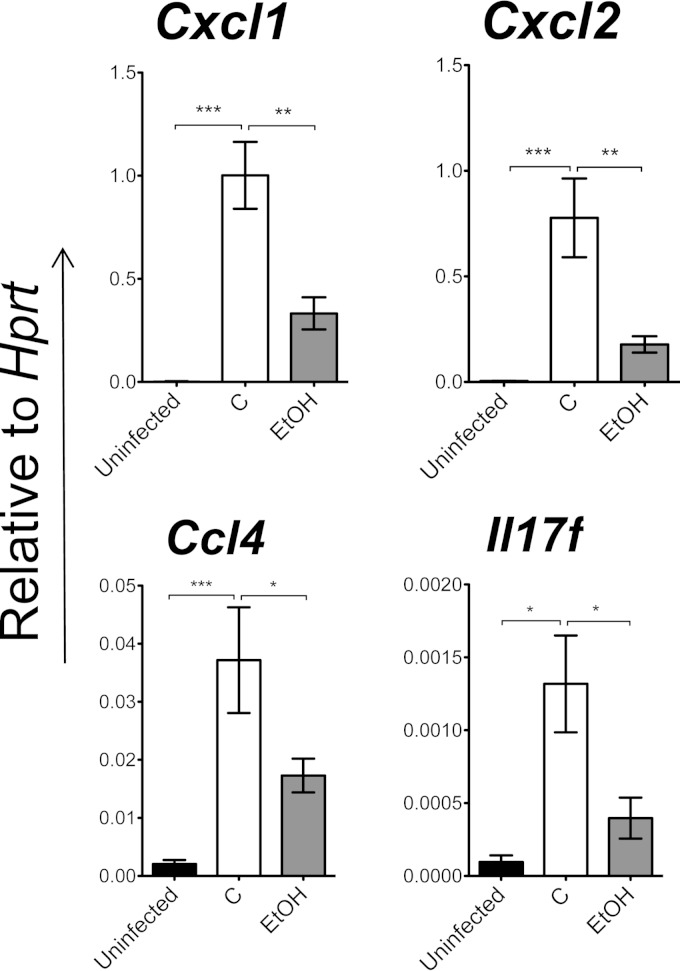
Ethanol suppresses chemokine expression in the lung epithelium.
Based on the data we obtained from the rhesus macaque bronchial brushes, in which there was downregulation of chemokine genes, we analyzed chemokine expression in the airway epithelium in addition to lung tissue in the murine model. Uninfected, control, and ethanol-treated mice underwent bronchial brushings 5 h postinfection, and the ethanol-treated mouse group showed significant downregulation of transcripts encoding mucosal chemokines (Cxcl1, Cxcl2, and Ccl4) as well as IL17f (Fig. 5). This trend correlated with the RNA-seq data obtained from rhesus macaques, which supports the importance of these chemokines during acute pneumococcal infection.
FIG 5.
Ethanol suppresses chemokine expression in the lung epithelium 5 h postinfection. Gene expression relative to Hprt by qPCR data from bronchial brushes of the uninfected group (n = 5), the control group (C) (n = 5), and the ethanol group (EtOH) (n = 5) after S. pneumoniae lung infection is shown. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (one-way ANOVA). Experiments were repeated at least twice.
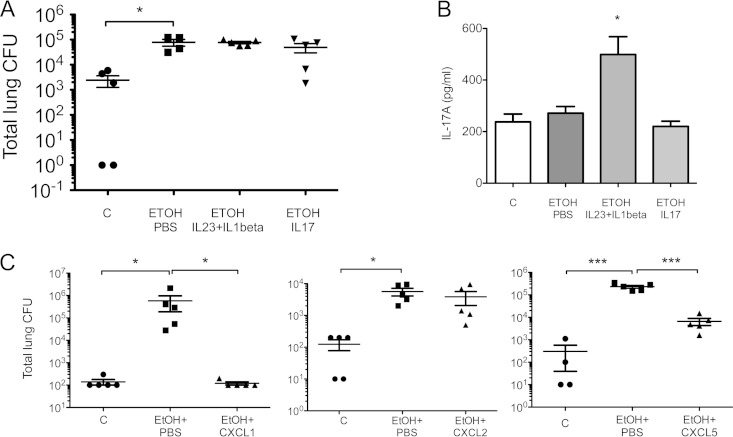
Local chemokine administration but not type 17 cytokines rescued bacterial burden phenotype 24 h postinfection.
Since IL-17 and related genes were downregulated in the lung tissue of ethanol-treated mice, we examined if rescuing with recombinant IL-17 in the lungs would decrease bacterial burden in these animals. To restore IL-17 to a physiologically relevant concentration, we gave the ethanol-treated mice recombinant IL-23 and IL-1B, which are known to induce IL-17 production mostly by γδ T cells in the lung (15), or recombinant IL-17. There was no change in bacterial clearance (Fig. 6A), despite the fact that IL-17 protein levels were equally increased in the animals that were rescued with recombinant IL-17 and in controls. We even observed much higher IL-17 levels in mice treated with IL-23 plus IL-1B (Fig. 6B), yet this combination of cytokines failed to rescue bacterial clearance. These data suggested that host defense pathways, other than IL-17 production per se, were also affected by ethanol intoxication. As IL-17 is a key regulator of chemokine gene expression in lung epithelium and the chemokine pathway was inhibited by ethanol, we next assessed whether restoring chemokine levels would correct the host defense defect. As CXCR2 ligands were particularly downregulated in airway epithelium, we investigated whether recombinant CXCL1, CXCL2, or CXCL5 could rescue impaired bacterial clearance in the ethanol group. Local delivery of recombinant CXCL1 and CXCL5 (doses used in previous studies [16–18]) but not CXCL2 significantly decreased bacterial burden compared to the ethanol group that received vehicle control (Fig. 6C).
FIG 6.
Recombinant CXCL1 chemokine and not IL-17 cytokines rescue bacterial phenotype. (A) Mice were randomized to four groups: controls (C), ethanol treatment rescued with PBS (EtOH PBS), ethanol treatment rescued with IL-23+IL1B (EtOH IL-23+IL1B), and ethanol treatment rescued with IL-17 (EtOH IL-17). (B) IL-17 protein expression in lung tissue from mice sacrificed 24 h postinfection in the four groups described above. (C) Mice were randomized to the control group (C), the ethanol group rescued with PBS (EtOH+PBS), the ethanol group rescued with CXCL1 (EtOH+CXCL1), the ethanol group rescued with CXCL2 (EtOH+CXCL2), or the ethanol group rescued with CXCL5 (EtOH+CXCL5). Cytokines and chemokines used for rescue were given 2 h postinfection. *, P < 0.05; ***, P < 0.001 (Kruskal-Wallis test or one-way ANOVA). Experiments were repeated at least twice.
Taken together, these experiments demonstrated that ethanol impairs host defense against S. pneumoniae by damping IL-17 cytokine production as well as suppressing the epithelial responses regulated by IL-17. So far, improvement of host defense against S. pneumoniae bacterial burden seems to be a response of CXCL1 and CXCL5 but not CXCL2.
DISCUSSION
Our study suggests that ethanol attenuates an early type 17 immune response in the lung epithelium, which may result in a decreased luminal chemokine gradient for neutrophil effector function into the lung. Our data support the concept that ethanol disrupts the mucosal immune response in the lung at least at two levels. One, ethanol decreases the expression of upstream regulators of type 17 immunity, Il23a and Il1b, by lung antigen-presenting cells, resulting in less IL-17 induction in vivo. However, since IL-17 was unable to rescue ethanol-treated animals, our data suggest that ethanol may also impair IL-17 signaling in epithelial cells, which in turn decreases the production of mucosal chemokines. We have evidence that this may be one mechanism, as HBE-1 cells (immortalized human bronchial epithelial cell line) incubated with 100 mM ethanol and stimulated with IL-17, TNF-α, or a combination had decreased CXCL1 protein expression at 24 h after cytokine stimulation (see Fig. S6 in the supplemental material).
Previous studies have shown that ethanol impairs lung chemokine protein secretion (CXCL1 and CXCL2) in rodent models of ethanol consumption and bacterial pneumonia, to which the authors ascribed decreased neutrophil recruitment into the lung probably due to impaired upregulation of adhesion molecule expression on the surface of neutrophils, resulting in poorer bacterial clearance and outcomes (19–21). Also, evidence obtained with a murine model showed that ethanol inhibits bone marrow cells through the enhancement of STAT3 activation following S. pneumoniae lung infection, thus decreasing leukocyte recruitment to the site of infection and worsening outcomes (22). In our mouse study, we obtained further evidence that ethanol decreases the expression not only of Cxcl1 and Cxcl2 but also of other genes encoding mucosal chemokines (Ccl3 and Ccl4) and cytokines (Il1b and Il6) which are related to IL-17. The alteration in gene expression pattern was seen in the primate model mostly for CXCL8 and CCL4. Preliminary observations from our experiments suggest that neutrophil recruitment is similar at 5 h in control and ethanol groups (our unpublished data) in the mouse model, which indicates that neutrophil function and not recruitment may also be affected in our model. Chemokines not only recruit leukocytes but also induce the secretion of leukocyte granular contents (23, 24), and these properties are concentration dependent (23).
At 24 h, when the ethanol group had significantly higher numbers of CFU/lung, we also observed equal or higher levels of IL-17 in the ethanol group compared to the control group. This is most likely due to the higher bacterial burden in the ethanol group. Based on this, we assessed IL-17 and chemokine expression at 5 h postinfection, because at this time point, bacterial burdens were similar in both groups, so the differences observed would not be biased by pneumococcal burden. At this time point, ethanol suppressed IL-17 levels (Fig. 4B).
Of interest, both CXCL1 and CXCL5 were able to rescue bacterial burden to control levels; however, a similar dose of CXCL2 did not have this effect. This effect may be related to the fact that CXCL1 has a noncompartmentalized effect outside the lung; thus, there may be further recruitment of innate immune cells (25, 26). In contrast, CXCL2 has a compartmentalized effect upon LPS challenge, as plasma CXCL2 levels are not detected upon intratracheal CXCL2 delivery (26). Thus, our data show that specific chemokines and not all CXCR2 ligands play critical roles in lung immunity against S. pneumoniae.
The advantage of analyzing samples from a nonhuman primate model of S. pneumoniae infection and ethanol consumption is that the rhesus macaque genome is closer to the human genome than that of other animal models. Therefore, we can speculate that the findings are more reliable and applicable to human disease. It is important to note that our primate cohort was SIV infected; this adds one more variable to the nonhuman primate data, and although it is possible that SIV infection may have had an effect on chemokine expression, we tried to overcome this problem by analyzing just the macaques that received ART. Also, the primate cohort underwent chronic ethanol consumption before pneumococcal infection, while our mouse model was of acute ethanol intoxication; nevertheless, our animal models receiving ethanol treatment showed similar effects on the bronchial epithelium transcriptome. We intend to investigate further mechanisms involved in chemokine regulation upon pneumococcal infection in a mouse model of chronic ethanol consumption, since the latter may represent a more advantageous model to study further the mechanisms involving ethanol and the immune system (27–29). Another important factor to consider is the small nonhuman primate sample size analyzed, which does not give us enough power to reach a definite conclusion, but we tried to overcome this problem by using a mouse model, which allowed us to perform further experiments.
In summary, ethanol impairs IL-17 cross talk to the lung epithelium, which translates into defective luminal chemokine expression and gradient. Restoration of luminal chemokines in a mouse model enhanced host defenses against S. pneumoniae. This paper highlights the important role of type 17 immune responses and specific CXCR2 ligands in orchestrating mucosal lung immunity during acute S. pneumoniae infection.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by grants from NIAAA (P60 AA009803) and NHLBI (R37HL079142).
We thank Gilead Sciences, Inc., for providing tenofovir and emtricitabine for these studies. We thank Larissa Devlin, Wayne A. Cyprian, and Nancy Dillman from the TNPRC (Covington, LA) for excellent care of study animals, including the delivery of alcohol and sucrose. We are thankful for the technical support of Jean Carnal, Jane Schexnayder, Amy B. Weinberg, and Rhonda R. Martinez from LSUHSC-NO.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/IAI.02869-14.
REFERENCES
- 1.Abbott AC. 1896. The influence of acute alcoholism on the normal vital resistance of rabbits to infection. J Exp Med 1:447–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rubin G. 1904. The influence of alcohol, ether, and chloroform on natural immunity in its relation to leucocytosis and phagocytosis. J Infect Dis 1:425–444. doi: 10.1093/infdis/1.3.425. [DOI] [Google Scholar]
- 3.Bouchery EE, Harwood HJ, Sacks JJ, Simon CJ, Brewer RD. 2011. Economic costs of excessive alcohol consumption in the U.S., 2006. Am J Prev Med 41:516–524. doi: 10.1016/j.amepre.2011.06.045. [DOI] [PubMed] [Google Scholar]
- 4.Picard C, Puel A, Bonnet M, Ku CL, Bustamante J, Yang K, Soudais C, Dupuis S, Feinberg J, Fieschi C, Elbim C, Hitchcock R, Lammas D, Davies G, Al-Ghonaium A, Al-Rayes H, Al-Jumaah S, Al-Hajjar S, Al-Mohsen IZ, Frayha HH, Rucker R, Hawn TR, Aderem A, Tufenkeji H, Haraguchi S, Day NK, Good RA, Gougerot-Pocidalo MA, Ozinsky A, Casanova JL. 2003. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science 299:2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 5.von Bernuth H, Picard C, Jin Z, Pankla R, Xiao H, Ku CL, Chrabieh M, Mustapha IB, Ghandil P, Camcioglu Y, Vasconcelos J, Sirvent N, Guedes M, Vitor AB, Herrero-Mata MJ, Arostegui JI, Rodrigo C, Alsina L, Ruiz-Ortiz E, Juan M, Fortuny C, Yague J, Anton J, Pascal M, Chang HH, Janniere L, Rose Y, Garty BZ, Chapel H, Issekutz A, Marodi L, Rodriguez-Gallego C, Banchereau J, Abel L, Li X, Chaussabel D, Puel A, Casanova JL. 2008. Pyogenic bacterial infections in humans with MyD88 deficiency. Science 321:691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z, Clarke TB, Weiser JN. 2009. Cellular effectors mediating Th17-dependent clearance of pneumococcal colonization in mice. J Clin Invest 119:1899–1909. doi: 10.1172/JCI36731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu YJ, Gross J, Bogaert D, Finn A, Bagrade L, Zhang Q, Kolls JK, Srivastava A, Lundgren A, Forte S, Thompson CM, Harney KF, Anderson PW, Lipsitch M, Malley R. 2008. Interleukin-17A mediates acquired immunity to pneumococcal colonization. PLoS Pathog 4:e1000159. doi: 10.1371/journal.ppat.1000159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gu C, Wu L, Li X. 2013. IL-17 family: cytokines, receptors and signaling. Cytokine 64:477–485. doi: 10.1016/j.cyto.2013.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laan M, Cui ZH, Hoshino H, Lotvall J, Sjostrand M, Gruenert DC, Skoogh BE, Linden A. 1999. Neutrophil recruitment by human IL-17 via C-X-C chemokine release in the airways. J Immunol 162:2347–2352. [PubMed] [Google Scholar]
- 10.Ye P, Rodriguez FH, Kanaly S, Stocking KL, Schurr J, Schwarzenberger P, Oliver P, Huang W, Zhang P, Zhang J, Shellito JE, Bagby GJ, Nelson S, Charrier K, Peschon JJ, Kolls JK. 2001. Requirement of interleukin 17 receptor signaling for lung CXC chemokine and granulocyte colony-stimulating factor expression, neutrophil recruitment, and host defense. J Exp Med 194:519–527. doi: 10.1084/jem.194.4.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McAllister F, Henry A, Kreindler JL, Dubin PJ, Ulrich L, Steele C, Finder JD, Pilewski JM, Carreno BM, Goldman SJ, Pirhonen J, Kolls JK. 2005. Role of IL-17A, IL-17F, and the IL-17 receptor in regulating growth-related oncogene-alpha and granulocyte colony-stimulating factor in bronchial epithelium: implications for airway inflammation in cystic fibrosis. J Immunol 175:404–412. doi: 10.4049/jimmunol.175.1.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aujla SJ, Chan YR, Zheng M, Fei M, Askew DJ, Pociask DA, Reinhart TA, McAllister F, Edeal J, Gaus K, Husain S, Kreindler JL, Dubin PJ, Pilewski JM, Myerburg MM, Mason CA, Iwakura Y, Kolls JK. 2008. IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat Med 14:275–281. doi: 10.1038/nm1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Molina PE, Amedee AM, Veazey R, Dufour J, Volaufova J, Bagby GJ, Nelson S. 2014. Chronic binge alcohol consumption does not diminish effectiveness of continuous antiretroviral suppression of viral load in SIV-infected macaques. Alcohol Clin Exp Res 9:2335–2344. doi: 10.1111/acer.12507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson S, Happel KI, Zhang P, Myers L, Dufour JP, Bagby GJ. 2013. Effect of bacterial pneumonia on lung simian immunodeficiency virus (SIV) replication in alcohol consuming SIV-infected rhesus macaques. Alcohol Clin Exp Res 37:969–977. doi: 10.1111/acer.12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubin PJ, Martz A, Eisenstatt JR, Fox MD, Logar A, Kolls JK. 2012. Interleukin-23-mediated inflammation in Pseudomonas aeruginosa pulmonary infection. Infect Immun 80:398–409. doi: 10.1128/IAI.05821-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finsterbusch M, Voisin MB, Beyrau M, Williams TJ, Nourshargh S. 2014. Neutrophils recruited by chemoattractants in vivo induce microvascular plasma protein leakage through secretion of TNF. J Exp Med 211:1307–1314. doi: 10.1084/jem.20132413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yao L, Yago T, Shao B, Liu Z, Silasi-Mansat R, Setiadi H, Lupu F, McEver RP. 2013. Elevated CXCL1 expression in gp130-deficient endothelial cells impairs neutrophil migration in mice. Blood 122:3832–3842. doi: 10.1182/blood-2012-12-473835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boespflug ND, Kumar S, McAlees JW, Phelan JD, Grimes HL, Hoebe K, Hai T, Filippi MD, Karp CL. 2014. ATF3 is a novel regulator of mouse neutrophil migration. Blood 123:2084–2093. doi: 10.1182/blood-2013-06-510909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boe DM, Nelson S, Zhang P, Bagby GJ. 2001. Acute ethanol intoxication suppresses lung chemokine production following infection with Streptococcus pneumoniae. J Infect Dis 184:1134–1142. doi: 10.1086/323661. [DOI] [PubMed] [Google Scholar]
- 20.Boe DM, Nelson S, Zhang P, Quinton L, Bagby GJ. 2003. Alcohol-induced suppression of lung chemokine production and the host defense response to Streptococcus pneumoniae. Alcohol Clin Exp Res 27:1838–1845. doi: 10.1097/01.ALC.0000095634.82310.53. [DOI] [PubMed] [Google Scholar]
- 21.Quinton LJ, Nelson S, Zhang P, Happel KI, Gamble L, Bagby GJ. 2005. Effects of systemic and local CXC chemokine administration on the ethanol-induced suppression of pulmonary neutrophil recruitment. Alcohol Clin Exp Res 29:1198–1205. doi: 10.1097/01.ALC.0000171927.66130.AA. [DOI] [PubMed] [Google Scholar]
- 22.Siggins RW, Melvan JN, Welsh DA, Bagby GJ, Nelson S, Zhang P. 2011. Alcohol suppresses the granulopoietic response to pulmonary Streptococcus pneumoniae infection with enhancement of STAT3 signaling. J Immunol 186:4306–4313. doi: 10.4049/jimmunol.1002885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smith RJ, Sam LM, Leach KL, Justen JM. 1992. Postreceptor events associated with human neutrophil activation by interleukin-8. J Leukoc Biol 52:17–26. [DOI] [PubMed] [Google Scholar]
- 24.Barlic J, Andrews JD, Kelvin AA, Bosinger SE, DeVries ME, Xu L, Dobransky T, Feldman RD, Ferguson SS, Kelvin DJ. 2000. Regulation of tyrosine kinase activation and granule release through beta-arrestin by CXCRI. Nat Immunol 1:227–233. doi: 10.1038/79767. [DOI] [PubMed] [Google Scholar]
- 25.Zhang P, Nelson S, Holmes MC, Summer WR, Bagby GJ. 2002. Compartmentalization of macrophage inflammatory protein-2, but not cytokine-induced neutrophil chemoattractant, in rats challenged with intratracheal endotoxin. Shock 17:104–108. doi: 10.1097/00024382-200202000-00004. [DOI] [PubMed] [Google Scholar]
- 26.Quinton LJ, Nelson S, Zhang P, Boe DM, Happel KI, Pan W, Bagby GJ. 2004. Selective transport of cytokine-induced neutrophil chemoattractant from the lung to the blood facilitates pulmonary neutrophil recruitment. Am J Physiol Lung Cell Mol Physiol 286:L465–L472. doi: 10.1152/ajplung.00153.2003. [DOI] [PubMed] [Google Scholar]
- 27.Cook RT. 1998. Alcohol abuse, alcoholism, and damage to the immune system—a review. Alcohol Clin Exp Res 22:1927–1942. doi: 10.1111/j.1530-0277.1998.tb05900.x. [DOI] [PubMed] [Google Scholar]
- 28.Bertola A, Mathews S, Ki SH, Wang H, Gao B. 2013. Mouse model of chronic and binge ethanol feeding (the NIAAA model). Nat Protoc 8:627–637. doi: 10.1038/nprot.2013.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mathews S, Xu M, Wang H, Bertola A, Gao B. 2014. Animals models of gastrointestinal and liver diseases. Animal models of alcohol-induced liver disease: pathophysiology, translational relevance, and challenges. Am J Physiol Gastrointest Liver Physiol 306:G819–G823. doi: 10.1152/ajpgi.00041.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.