Abstract
Infection with HBV is common worldwide, with over 350 million chronic carriers. Chronic HBV infection is associated with cirrhosis and hepatocellular carcinoma. All currently available oral antivirals are directed against the HBV polymerase enzyme, a reverse transcriptase. HBV polymerase contains several important domains and motifs which define its functions and reveal ways to further target it. This enzyme executes many functions required for the HBV replication cycle, including viral RNA binding, RNA packaging, protein priming, template switching, DNA synthesis and RNA degradation. In addition, HBV polymerase must interact with host proteins for its functions. Future therapeutics may inhibit not only the DNA synthesis steps which are carried out by the reverse transcriptase domain (as all current antivirals do) but other domains, functions and interactions which are essential to the HBV replication cycle.
Keywords: antiviral targets, drug resistance, HBV, replication cycle, reverse transcriptase
Despite an effective vaccine which was introduced in 1986, HBV remains an important cause of morbidity and mortality in the world. Many millions of people become infected every year, and HBV is a major cause of cirrhosis and hepatocellular carcinoma. Chronic hepatitis B is the resultant condition for over 350 million people worldwide. Chronic hepatitis B may entail lifelong monitoring and treatment in order to impact disease outcomes [1]. Several treatment options exist, including antivirals which directly target HBV.
Members of Hepadnaviridae such as HBV are pararetroviruses – DNA viruses with an RNA intermediate [2]. They infect several bird and mammal species. HBV is small in both size and genome. This enveloped virus produces spherical infectious particles of 42 nm, as well as non-infectious particles of varying size and morphology [3]. The HBV genome is 3.2 kb and contains four open reading frames which encode the polymerase (HP), core, surface and X genes. The core gene encodes the core protein, or c antigen (HBcAg), and also encodes the secreted nonstructural e antigen (HBeAg). The surface gene can also produce different products – the small, middle and large surface antigens (HBsAg). Other than the enigmatic X protein, all of HBV's genes have been successfully targeted for diagnostic, vaccine or treatment aims.
The environmental reservoir for HBV is infected individuals; fomites are also a potential issue since HBV can survive outside the body for at least a week [4]. The most common transmission routes are vertical transmission during delivery and horizontal transmission by blood contact. When an infection is established, individuals may become chronic carriers [5]. Although this infection may be subclinical, liver damage can occur at each stage of infection. There are four progressive stages of chronic HBV infection. The stages are, first, an immune tolerance phase, then immune clearance of HBeAg as infected liver cells are attacked. Next comes a nonreplicative stage with low levels of HBV DNA and the presence of HBeAg antibodies; finally a reactivation phase may occur when core mutants arise that prevent HBeAg production while allowing HBV replication [6]. Although these four stages of chronic hepatitis B are usually linear in their progression, individuals may move forward or backward on this scale. The real danger of HBV infection is that between 25 and 40% of chronically infected patients are predisposed to developing liver cirrhosis and hepatocellular carcinoma [7]. Fortunately, there are treatments which combat HBV, ameliorating the symptoms associated with HBV infection.
Current treatments for chronic hepatitis B
Treatment is indicated for HBV infection in many cases [8]. Several treatments are currently in use or are being developed. These drugs may modulate the immune system or target HBV more directly. The cytokine interferon was one of the earliest treatment options for viral hepatitis such as HBV. Today use of interferon (and PEGylated interferon, which has reduced renal clearance [9]) is still common. In fact, PEGylated interferon is indicated as a first line treatment. It may work best for those infected with HBV genotype A, and treatment outcomes may be better in those with certain IL28B genotypes [10]. Other immunomodulatory treatments are in clinical trials including thymosin α1, IL-7 and a Toll-like receptor 7 (TLR7) agonist [11,12]. These immunomodulatory drugs do not target a specific function of HBV, but instead augment the body's natural immune defense. As such they are not associated with specific HBV mutations.
Other therapeutics have been designed to inhibit a specific HBV protein or defined steps in its replication cycle. Currently approved drugs in this category are limited to one class – nucleos(t) ide analog reverse transcriptase inhibitors (NRTIs). NRTIs are inhibitors of the polymer-ase enzyme, a reverse transcriptase with both RNA-directed and DNA-directed DNA polymerase activity. The effect is at the initial polymerization step, protein priming or the subsequent polymerization step, DNA strand elongation. Although NRTIs may suppress viral replication and thus affect disease outcomes, it is important to note that they do not represent a true cure of the virus infection.
The nucleoside and nucleotide analogs currently used against HBV in the clinic are clevu-dine, emtricitabine, entecavir, lamivudine and telbivudine (nucleosides); and adefovir dipivoxil and tenofovir disoproxil fumarate (nucleotides). The two NRTIs suggested as first-line treatment (in addition to interferon) have very low cumulative resistance rates, entecavir less than 2% at 5 years [8] and tenofovir at 0% at 8 years [13]. When considering their ability to suppress serum levels of HBV DNA, entecavir and tenofovir are over 90% effective [14,15]. Tenofovir monotherapy has not been directly associated with tenofovir resistance mutations at the time of this report. Mutations have been found in patients being treated with tenofovir; however, the mutant genotypes may [16–19] or may not [20,21] be associated with a tenofovir-resistant phenotype when later tested in cell culture experiments. Although the cure rate is low, these inhibitors are effective at relieving disease symptoms such as normalization of alanine aminotransferase levels and improved liver histology. Additionally, they are important in delaying or preventing liver cirrhosis and hepatocellular carcinoma [8,22].
Adefovir, emtricitabine, lamivudine and telbivudine all have higher 5-year resistance rates, ranging between 30 and 70% [8]. Adefovir and lamivudine were the first generation of HBV drugs, US FDA approved in 2002 and 1998, respectively. They are associated with resistance at a greater frequency than later generation drugs. Telbivudine is similar to lamivudine, but more potent and associated with less resistance [23]. Clevudine is approved for use in South Korea and the Philippines, however due to resistance and myopathy in some individuals [24,25], it is being tested at lower doses and in combination therapies for continued use. In combination with adefovir, it did not yield any resistance after 96 weeks [26]. Emtricitabine is only approved for treatment against HIV, however it is known to inhibit HBV due to studies involving co-infected patients. The list of anti-HBV NRTIs will likely expand as more drugs in this chemical class are developed.
Before they can be incorporated into the DNA strand, nucleos(t)ide analogs must first be converted into their active triphosphate forms by kinases which are provided by the cell. They then compete with natural dNTPs for incorporation into the DNA strand being synthesized. However, NRTIs are usually chain terminators due to lack of a 3′ hydroxyl group. The first bases incorporated during protein-primed initiation of HBV reverse transcription (protein priming, see below) are dGMP followed by dAMPs, forming the initiating GAA trinucleotide. Therefore guanosine and adenosine analogs may be more inhibitory to HBV, due to their effect on both the initial priming reaction as well as downstream DNA synthesis. This may be due to the fact that priming and subsequent DNA synthesis require different reverse transcriptase (RT) conformations, potentially raising the bar to resistance. Indeed, the two NRTIs which are most effective, entecavir and tenofovir (respectively, G and A analogs), inhibit both priming and DNA synthesis [27]. Entecavir was shown to terminate synthesis in a delayed fashion – DNA polymerization is interrupted only after adding entecavir and several nucleotides more [27–29]. The mechanism of clevudine is less well understood. It is a thymidine analog; however, it inhibits GAA synthesis during priming without being incorporated into the DNA. Therefore it may function as a non-nucleoside inhibitor although its chemical structure is a nucleoside analog [27].
NRTIs can lose effectiveness due to viral mutations. HBV polymerase has a high mutation rate due to lack of proofreading ability. For every replication cycle, there is a mutation in approximately every 2 × 105 DNA bases [30]. Thus, each possible mutation occurs each day in an active infection. If the mutations are favorable for resistance, a patient will experience increases in clinical markers of infection, such as viremia, as the mutant HBV strain expands. Drug-resistance associated mutations will continue to be an issue as more treatments become available against chronic HBV. Fortunately, once resistance occurs with one drug, another may still be effective. This may suggest the usage of combination therapy from the outset of treatment; however, combination therapy has not been shown to prevent resistance in treatment-naive patients [1]. This may not be true for all cases [26], and combination therapy merits continued testing.
Multiple roles of HBV polymerase represent both realized & potential drug targets
To determine how to inhibit the polymer-ase enzyme, it is important to appreciate its many functions during HBV replication. These functions are viral RNA binding, RNA packaging (or encapsidation), protein priming, template switching, DNA synthesis and RNA degradation. The only HBV-specific drugs currently approved are NRTIs which inhibit the DNA synthesis steps – priming and strand elongation of DNA. This narrow scope of the functions that are inhibited is in contrast to the many additional functions of the polymerase enzyme. Furthermore, HBV polymerase (HP) must associate with host-cell proteins for proper function, and these interactions could be disrupted. The next generation of anti-HBV drugs could target not only DNA synthesis steps, but any of polymerase's functions or interactions.
HBV polymerase is the only enzymatically active protein and participates in many steps of the viral replication cycle (Figure 1). In an established infection, covalently-closed circular DNA (cccDNA) is maintained in infected cells’ nuclei. This cccDNA is transcribed by host factors into all viral mRNAs, including the pregenomic RNA (pgRNA). pgRNA is both the template for translation of the viral polymerase and core protein, as well as the template RNA which will be reverse transcribed into viral DNA.
Figure 1. Polymerase-dependent events in the HBV replication cycle.
Beginning at the cccDNA which is found in the nucleus of infected cells, HBV pgRNA is transcribed. It is exported to the cytoplasm and then translated. The translated polymerase binds to pgRNA at the 5′ ε stem-loop structure. This interaction facilitates the encapsidation, or packaging, of both polymerase and pgRNA. Priming may occur before, concurrent with or after nucleocapsid assembly. Using a free hydroxyl group on Y63 of the polymerase as primer, DNA is attached to HP using ε as a template. After the initiating GAA nucleotides are laid down, HP and these bases switch templates to the 3′ end and continue minus strand DNA synthesis. As the template pgRNA is copied, it is concurrently degraded by the RNase H activity of the polymerase. The resultant ssDNA-containing nucleocapsid is then further matured as the polymerase copies the second strand of DNA, yielding an rcDNA-containing nucleocapsid. This maturation of the nucleocapsid leads to its destabilization, which may facilitate disassembly (uncoating) to liberate the rcDNA for conversion to more cccDNA in the nucleus. The polymerase must be removed in order to allow cccDNA formation. cccDNA: Covalently-closed circular DNA; ε: Epsilon RNA secondary structure motif; HP: HBV polymerase; pgRNA: Pregenomic RNA; rcDNA: Relaxed circular DNA.
Once HP is translated, its binding to HBV RNA is a critical early step in virus assembly and DNA replication. When associated with host cell chaperone proteins such as Hsp90, HP binds to pgRNA at an RNA structural motif, called ε [31–34]. This ε RNA is a region of secondary structure which forms a stem-loop that is recognized by the HBV polymerase. Next the interaction of both HP and ε RNA is necessary for their packaging into assembling nucleocapsids. HBV ε RNA contains the initial template for reverse transcription, where its UUC template is copied as a GAA trinucleotide. The primer for DNA synthesis is a free hydroxyl on the polymerase enzyme itself, and the template DNA is thus covalently linked to the HP enzyme – a process called protein priming [35].
After the initial priming reaction yields the first DNA bases, HP translocates with these bases to the 3′ end of the pgRNA, where it continues to synthesize minus-strand DNA. This template-switching reaction is dependent on the activity of the polymerase in studies with duck HBV [36]. As HP reverse transcribes the minus strand of DNA, it concurrently degrades the template pgRNA owing to its RNase H activity. Then plus strand DNA is made using full-length minus strand DNA as template. These various roles of the HBV polymerase all must function in order to complete a successful replication cycle. HP also exhibits transferase activity in vitro, which is nontemplate-directed synthesis, but its significance is unknown in vivo [37]. Distinct regions of the polymerase protein are important for each of these step.
Importance of different domains, motifs & residues
The HBV polymerase is composed of 4 domains (Figures 2B & 3A). They are a terminal protein (TP) at the amino end which is important in initial DNA synthesis, a spacer domain, the RT domain which houses the polymerase activity and an RNase H domain at the carboxy-terminus which is critical for removing template RNA. Although much of the spacer domain is not critical for polymerase function, the TP, RT and RNase H domains are essential. Each is a potential therapeutic target.
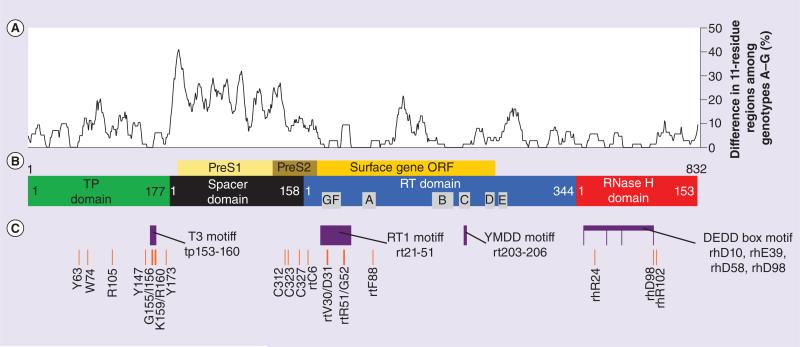
Figure 2. Sequence conservation and important domains, motifs and residues of HBV polymerase.
(A) Genotype homology mapping. Amino acid residues of seven reference strains of HBV representing genotypes A–G were mapped according to differences among 11 amino acid regions (considering a single residue with five amino acids upstream and five downstream). Each point along the graph represents the percentage of amino acid differences within the corresponding 11 amino acid region. Higher graph values thus represent less homology in that region among genotypes. GenBank IDs are: X02763.1, D00329.1, X01587.1, V01460.1, X75657.1, X75658.1 and AF160501.1 for genotypes A–G, respectively. (B) The domains and conserved RT boxes A–G of HP are shown. Numbering is according to genotype D for the TP domain and spacer, and follows standardized numbering for the RT and RNase H domains [38]. The position of the surface gene, which is in a different reading frame, is also shown. (C) Functionally important motifs and residues revealed through in vitro mutant screens. Several regions are important for the functional roles of HP, including the T3 and RT1 motifs which facilitate ε RNA binding. See text for specific details. Drawn to scale. ORF: Open reading frame; RT: Reverse transcriptase; TP: Terminal protein. Reference genotype list is taken from Stuyver et al. [38].
Figure 3. Structure of the HBV polymerase and treatment-associated mutations in its reverse transcriptase domain.
(A) Illustration of the four domains, including the three RT subdomains. Minus-strand DNA is attached at the TP, and DNA or RNA passes through the ‘hand’ which synthesizes DNA. Most nucleotide interactions are at the palm subdomain. (B) Subdomain organization of the RT domain of HBV polymerase. Boxes A–G are part of the enzyme's catalytic core, and they are homologous to other RT enzymes. Each nucleos(t)ide analog is shown with its treatment-associated resistance mutations. Drawn to scale.
†Tenofovir mutations have not been shown to confer in vivo resistance (see text for details).
AA: Amino acid; RT: Reverse transcriptase; TP: Terminal protein.
There are many ways to determine the important residues, motifs and domains of HBV polymerase. One method is to compare sequences among isolates of this highly mutable virus (Figure 2A). Another way to determine important regions is through in vitro mutational experiments. Truncations or specific amino acid changes can be made and evaluated using functional assays (Figure 2C). During the course of treatments which inhibit the polymerase, escape mutants can arise that are rendered less susceptible to the drug. These mutations can be mapped to identify important areas of the protein. This type of information can help decide which secondary treatment option is best (Figure 3B). These differences among isolates or mutants can reveal regions important for HBV polymerase function.
TP domain
Attached to the main RT portion of the enzyme is a functional domain simply called the terminal protein. It shows no homology to RT enzymes outside Hepadnaviridae. If named according to its function, it might be called the priming domain. Template-directed polymerization begins when the first DNA nucleotide, dGTP, attaches to the free hydroxyl group on tyrosine 63 (numbering will follow genotype D for TP and spacer [GenBank ID V01460.1], and following standard nomenclature from Stuyver et al. for RT and RNase H [38]). Attachment onto HP is referred to as protein priming since Y63 on the protein itself serves as the primer. Priming encompasses ε RNA-directed synthesis, of both the first nucleotide of viral DNA (initiation) and the subsequent two nucleotides (polymerization) (Figure 1 inset) [39]. Despite the enzyme being attached to the DNA, it continues to synthesize both strands of DNA. The minus strand DNA continues its attachment to TP until cccDNA synthesis, at which point HP must be removed from the DNA before circularization. As the nucleocapsid undergoes this process of maturation, it becomes less stable [40], leading to the release of rcDNA for conversion to cccDNA in the nucleus (Figure 1).
TP is essential for ε RNA binding, RNA packaging and protein priming. It has several important residues and motifs. The attachment to DNA is made through the Y63 residue, and there is a T3 motif which is important in ε binding, RNA packaging and protein priming. This T3 motif is thought to interact with the RT1 motif in the RT domain, thus aiding ε binding [41,42]. Adjacent to the T3 domain are several key residues: R105 and Y173 are required for pgRNA packaging, and W74 and Y147 are required for DNA synthesis (Figure 2C) [43,44].
There have been no treatment-associated mutations reported in the TP region, but this region is not usually screened by sequencing when HBV overcomes drug susceptibility. In conclusion, the TP region has a high potential for new drug targets, however little homology to other proteins places TP in an uncharted area of drug screening. New functional assays, such as the in vitro RNA binding and protein priming assay developed by Jones et al. would help in screening compounds that affect the critical TP domain [31]. Carbonyl J compounds and porphyrin compounds affect polymerase binding to ε RNA, however they are in the preclinical evaluation stage [45,46].
Spacer domain
The spacer domain may be little more than a bridge between the TP and RT domains. A greater amount of sequence variation exists among different HBV genotypes in this domain compared with the other three domains (Figure 2A). Although this may suggest this region is unimportant and genetic drift has not affected function, there is additional selection pressure in the spacer due to overlapping reading frames with the surface gene (Figure 2B) [47]. Therefore, although mutations within the spacer or its partial removal may not render an inactive protein when considering polymerase only [48], mutations in the spacer may affect the function of HBsAg in the context of an infection. Indeed, the PreS1 and PreS2 regions which overlap the spacer are used to form the large (L) and middle surface antigens, and L is the main surface protein involved in attachment to target cells [49,50]. In short, the spacer may not be critical for polymerase function, but it cannot be easily altered without affecting HBsAg function.
There are several cysteine residues which are technically in the spacer region according to the current numbering system [38]. However, using modeling with the RT of HIV as a template, these cysteine residues have been mapped to the fingers subdomain of the RT domain (Figures 2C & 3A) [51]. These cysteines are thought to maintain an active conformation, and have been shown to be essential for ε binding, RNA packaging and priming (Figure 2B) [42].
The spacer domain is not commonly screened for mutations when drug resistance appears, and thus no treatment-associated mutations have been found here. Other than the cysteines mentioned above, this domain may not hold significance to polymerase–drug interactions. Therefore the potential for drug targeting is likely low for the spacer domain.
RT domain
The RT domain is the main catalytic center of the enzyme. Three subdomains are present, common to all DNA or RNA polymerases – the fingers, palm and thumb subdomains (Figure 3). These subdomains in HBV have a high degree of homology with HIV's RT enzyme. Also using HIV as a model, conserved boxes A through G have been characterized as the catalytic core of the RT enzyme [52–56]. These boxes process the incoming template strand and incorporate natural nucleotide substrates or nucleos(t)ide analog inhibitors into the elongating strand (Figure 3B).
The importance of these conserved boxes is demonstrated by their functions. Box A was shown to interact with box C and possibly box G, forming the dNTP binding pocket [57]. Box A contains a phenylalanine residue at rt88 which discriminates between dNTP and NTP in studies using the duck HBV polymerase [58]. This is important since HBV polymerase only synthesizes DNA, not RNA. Box B is involved with base pairing the primer and template strand in HIV. The next box, C, contains the conserved tyrosine, methionine, two aspartate (YMDD) motif. This motif coordinates the binding of two magnesium ions and is the enzyme's active site. Although box D does not directly affect dNTP binding, it likely contributes to overall structure. Box E contains the primer grip which aids polymerase-ε binding and DNA synthesis [59]. Box F was reported to facilitate interactions between the incoming dNTP and its template nucleotide [55].
The RT1 motif contains the conserved boxes F and G and is thought to interact with the T3 motif in TP to facilitate ε binding and protein priming [41]. Mutations within RT1 at rtV30/rtD31 disrupted protein priming, and at rtR51/rtG52 disrupted priming and RNA packaging [42]. Together, the above motifs and boxes govern the catalytic function of the reverse transcriptase.
Although there is no crystal structure available for any domain of the polymerase, several groups have modeled the RT domain using the homologous HIV RT, whose crystal structure is solved [51,60–62]. These models are useful for mapping key regions that interact with nucleos(t)ide analog inhibitors. Almost all of the drug treatment-associated mutations of HBV polymerase are in the palm subdomain (Figure 3A), which is also the site of nucleotide recognition and strand synthesis.
The most common single mutations are in the catalytic core of the RT domain, at rtM204, rtL180 and rtA181. The most common combination mutations include amino acid changes at rt180 + rt204, rt181 + rt236 and rt173 + rt180 + rt204 [63]. The methionine at rt204 is part of the YMDD active site in box C; the rt173, rt180 and rt181 positions are in box B; and rt236 is in box D. In addition to these most common mutations, several mutations have been associated with NRTI treatment [60,64–68]. Earlier generation drugs, such as adefovir and lamivudine are associated with a larger number of mutations, as well as a higher rate of mutation. A summary of currently known mutations is mapped graphically in Figure 3B to demonstrate the regions affected by drug resistance, which align to a great degree with the catalytic core boxes.
The causative mutations are not always known when virological breakthrough occurs. This may be due to the lack of sufficient sequencing coverage for these clinical samples. Perhaps mutations in regions outside the RT domain are leading to viral resistance as well. Despite the important issue of drug resistance, the RT domain remains an excellent target for drug development, as evidenced by the success of current RT inhibitors.
RNase H domain
Also key to the replication cycle of HBV is the RNase H domain of the polymerase. The RNase H domain removes the pgRNA template but leaves intact the 5′ end of the pgRNA (Figure 1). In this manner it both clears the way for DNA synthesis of the plus strand and creates the plus strand primer from the 5′ end of pgRNA. This region is homologous to the HIV RNase H domain, which performs a similar function. Indeed, HBV RNase H has been found to be sensitive to certain compounds first designed for HIV [69].
The RNase H domain contains a conserved DEDD box motif, which is important for metal ion co-factor binding [70]. The RNase H domain is also needed for RNA packaging and priming [42,71–73] and residues rhR24, rhR98 and rhR102 are critical for DNA synthesis (Figure 2B) [74]. Owing to the amount of knowledge we have due to its homology with other RNase enzymes, and its essential role in the HBV replication cycle, it is an attractive target for the next generation of anti-HBV drugs. In fact, several compounds have shown activity against HBV's RNase H domain, including β-thujaplicinol and HID compounds [69,75,76]. Testing these RNase H-directed compounds is facilitated by functional assays using purified RNase H from HBV. These compounds and the screening methods used represent a newly paved path for drug development against this essential HP domain.
Interactions with host factors as a potential target
Many host factors have been shown to interact with HBV polymerase. Since many interactions with host proteins are involved in virus replication, these interactions may be altered to affect HBV infection therapeutically. In addition to the direct protein–protein interactions mentioned below, the HP enzyme must be enzymatically cleaved from the minus strand DNA in order to form cccDNA. The host enzyme TDP2 has been implicated in this deproteination step (Figure 1) [31,77,78].
Hsp90 is a cellular protein complex which associates with HP and is required to keep the polymerase in a conformation which allows for ε interaction [79,80]. This Hsp90 complex is packaged with HP into the viral nucleocapsid [32,33]. Another host protein packaged into nucleocapsids is eIF4E, but the functional consequence is unknown [81]. DDX3 is an immunoregula-tory protein, and it is required for both hepatitis C virus and HIV replication [82,83]. DDX3 directly promotes IRF3 phosphorylation by TBK1, leading to interferon production [84]. DDX also interacts with HP independently of pgRNA and is incorporated into nucleocapsids [85]. However, its immune-signaling function may be disrupted by HP, leading to decreased interferon β promoter activity [86]. APOBEC3G may also be packaged into nucleocapsids due to its association with HP. APOBEC3G is a cyti-dine deaminase which induces DNA hypermutation. It binds to HP in an RNA-independent manner and is packaged into nucleocapsids in an HP- and ε-dependent manner [87]. Although the deaminase effect of APOBEC3G is potently antiviral, it was shown to block HBV replication independent of its deaminase activity, instead inhibiting early stage minus-strand DNA synthesis [88,89]. Thus it should be noted that host factors may have inhibitory or proviral effects. In the future, additional proteins which are provided by the host cell will likely be found to interact with HP. Some of these interactions may be a potential target for viral therapies.
Conclusion
Infection with HBV leads to a chronic state of infection in millions of people worldwide. It is controlled but not cured by current antiviral therapies. All HBV-directed medicines target the HBV polymerase enzyme. By using comparative genomics, mutational assays and sequencing of clinical isolates, many important regions of HBV polymerase have been found which may reveal novel ways to inhibit it. Understanding the many functions of HP also assists in finding ways to target it. These functional roles include viral RNA binding, RNA packaging, protein priming, template switching, DNA synthesis and RNA degradation. Also, HP interacts with host proteins while performing these functions. Since current drugs only target the DNA synthesis function of the RT domain, future HP inhibitors may target the RNase H domain or TP domains as well, exploiting their essential functional roles.
Future perspective
Understanding the domains, motifs and amino acid residues which are important for the distinct roles of the HBV polymerase inform future studies and drug development. Continued use of mutant screens, comparative genotyping and treatment-associated mutation monitoring will prove useful in future HBV research.
As suggested by clevudine's function as a non-NRTI, other non-NRTI compounds could be found in the future to inhibit HBV. Several non-NRTIs are already in use against HIV and are in development for hepatitis C virus [90,91]. NRTIs continue to be a fertile ground for anti-HBV drug development. Many new agents are in clinical trials, including new tenofovir prodrugs and novel inhibitors besifovir, elvucitabine, lagociclovir valactate, pradefovir mesylate and valtorcitabine [65]. In addition to these NTRIs, which target the RT domain of the HP enzyme, several compounds have been found to inhibit the function of the RNase H domain or RNA binding through the TP domain; however, these compounds are in early preclinical stages of development. Beyond antipolymerase drugs, treatment options may be forthcoming such as RNAi which targets HBV RNA, cccDNA inhibitors or capsid inhibitors [92]. Interactions with host proteins could also be inhibited, however toxicity may be an issue when targeting interactions between host proteins and viral components.
Although vaccination strategies and effective antivirals exist, infections are unlikely to be cured by current therapies. Current HBV-specific treatments, which are directed against the polymerase enzyme, help control the devastating effects of chronic HBV and reduce spread by lowering serum viral load. However, continued development should also target the RNase H domain as well as the TP domain. It is important to note that polymerase inhibitors alone are unlikely to eradicate HBV infection. Finding ways to directly deplete the cccDNA pool [93] will be critical since cccDNA is the source of all viral RNA and may survive in hepatocytes during drug treatment. A combination of prevention and increasingly effective treatment can control and may eventually eliminate HBV infection.
EXECUTIVE SUMMARY.
Current treatments for chronic hepatitis B
Immune-modulator drugs such as interferon do not cause any viral resistance, however their efficacies are more dependent on the host and can be highly variable.
For now, nucleos(t)ide analog reverse transcriptase inhibitors are the only approved class of drugs which target HBV. They include adefovir, clevudine (approved in South Korea and The Philippines), emtricitabine (approved for HIV), entecavir, lamivudine, telbivudine and tenofovir.
The first line drugs are entecavir and tenofovir. They are over 90% effective and have resistance rates less than 2%.
Multiple roles of HBV polymerase represent potential drug targets
Current HBV-specific antivirals target only one domain of the polymerase (the reverse transcriptase domain) and inhibit only one of its activities (DNA polymerization).
The functional roles of HBV polymerase are viral ε RNA binding, pgRNA packaging, protein priming, template switching, DNA synthesis and RNA degradation.
Importance of HBV polymerase's domains, motifs & residues
The terminal protein (TP) domain is unique to Hepadnaviridae and contains critical residues for priming, ε binding, RNA packaging and DNA synthesis. TP could prove to be a novel therapeutic target. Carbonyl J and porphyrin compounds affect TP's RNA binding activity.
The spacer domain contains cysteine residues near the RT domain which may form disulfide bonds in the RT domain; they aid ε binding, RNA packaging and priming. The spacer region contains the most genetic variation among clinical isolates, and is the least important region functionally. It is not a likely drug target.
The reverse transcriptase (RT) domain is the target of all currently approved HBV-specific drugs. It forms the fingers, palm and thumb subdomains and contains several important conserved boxes. Several mutations commonly occur which are associated with drug treatment. Sequencing beyond the RT domain may reveal mutations in other important regions of the polymerase that may also be relevant for drug resistance.
The RNase H domain is also critical for the HBV life cycle. It degrades the template viral RNA and is needed for RNA packaging, priming and DNA synthesis. This domain will likely be a fruitful region for future drug development. Several compounds have already been reported to have anti-RNase H activity.
Interactions with host factors as a potential target
The viral polymerase must be removed (deproteination) to allow formation of cccDNA. This may occur through host phosphodiesterases, such as TDP2.
Hsp90, eIF4E, DDX3 and APOBEC3G associate with the polymerase and may affect its function. Knowledge concerning these host-virus interactions may be useful for future therapeutic development.
Conclusion
The RT domain remains an excellent target for anti-HBV inhibitors. The RNase H domain and the TP domain are viable future targets.
Acknowledgments
This work was supported by Public Health Service grant R01 AI074982 to J Hu from the NIH. DN Clark was supported by the ‘Viruses and Cancer’ training grant 5 T32 CA60395 from the National Cancer Institute. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Footnotes
Financial & competing interests disclosure
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Paul N, Han SH. Combination therapy for chronic hepatitis B: current indications. Curr. Hepat. Rep. 2011;10(2):98–105. doi: 10.1007/s11901-011-0095-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Summers J, Mason WS. Replication of the genome of a hepatitis B – like virus by reverse transcription of an RNA intermediate. Cell. 1982;29(2):403–415. doi: 10.1016/0092-8674(82)90157-x. [DOI] [PubMed] [Google Scholar]
- 3.Seeger C, Mason WS. Hepatitis B virus biology. Microbiol. Mol. Biol. Rev. 2000;64(1):51–68. doi: 10.1128/mmbr.64.1.51-68.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bond W, Favero M, Petersen N, Gravelle C, Ebert J, Maynard J. Survival of hepatitis B virus after drying and storage for one week. Lancet. 1981;317(8219):550–551. doi: 10.1016/s0140-6736(81)92877-4. [DOI] [PubMed] [Google Scholar]
- 5.McMahon BJ, Alward WLM, Hall DB, et al. Acute hepatitis B virus infection: relation of age to the clinical expression of disease and subsequent development of the carrier state. J. Infect. Dis. 1985;151(4):599–603. doi: 10.1093/infdis/151.4.599. [DOI] [PubMed] [Google Scholar]
- 6.Hadziyannis SJ, Vassilopoulos D, Hadziyannis E. Chapter seven – The natural course of chronic hepatitis B virus infection and its management. In: Erik De C, editor. Advances in Pharmacology. Academic Press; MA, USA: 2013. pp. 247–291. [DOI] [PubMed] [Google Scholar]
- 7.Kim BK, Han KH, Ahn SH. Prevention of hepatocellular carcinoma in patients with chronic hepatitis B virus infection. Oncology. 2011;81(Suppl. 1):41–49. doi: 10.1159/000333258. [DOI] [PubMed] [Google Scholar]
- 8.Lok AS, McMahon BJ. Chronic hepatitis B. update 2009. Hepatology. 2009;50(3):661–662. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- 9.Veronese FM, Harris JM. Introduction and overview of peptide and protein PEGylation. Adv. Drug Deliv. Rev. 2002;54(4):453–456. doi: 10.1016/s0169-409x(02)00020-0. [DOI] [PubMed] [Google Scholar]
- 10.Sonneveld MJ, Wong VWS, Woltman AM, et al. Polymorphisms near IL28B and serologic response to PEGinterferon in HBeAg-positive patients with chronic hepatitis B. Gastroenterology. 2012;142(3):513–520. e511. doi: 10.1053/j.gastro.2011.11.025. [DOI] [PubMed] [Google Scholar]
- 11.Lanford RE, Guerra B, Chavez D, et al. GS-9620, an oral agonist of Toll-like receptor-7, induces prolonged suppression of hepatitis B virus in chronically infected chimpanzees. Gastroenterology. 2013;144(7):1508–1517. e1501–1510. doi: 10.1053/j.gastro.2013.02.003. 1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang YF, Ma ZH, Zhao PW, et al. Effect of thymosin-alpha(1) on T-helper 1 cell and T-helper 2 cell cytokine synthesis in patients with hepatitis B virus e antigen-positive chronic hepatitis B. J. Int. Med. Res. 2010;38(6):2053–2062. doi: 10.1177/147323001003800620. [DOI] [PubMed] [Google Scholar]
- 13.Corsa AC, Liu Y, Flaherty JF, Marcellin P, Miller MD, Kitrinos KM. No detectable resistance to tenofovir disoproxil fumarate in HBeAg+ and HBeAg− patients with chronic hepatitis B after eight years of treatment. American Association for the Study of Liver Diseases.. Presented at: The Liver Meeting 2014; Boston, MA, USA. 2014 (Poster 1707). [Google Scholar]
- 14.Tang CM, Yau TO, Yu J. Management of chronic hepatitis B infection. current treatment guidelines, challenges, and new developments. World J. Gastroenterol. 2014;20(20):6262–6278. doi: 10.3748/wjg.v20.i20.6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahon BJ. Chronic hepatitis B virus infection. Med. Clin. North Am. 2014;98(1):39–54. doi: 10.1016/j.mcna.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 16.Qin B, Budeus B, Cao L, et al. The amino acid substitutions rtP177G and rtF249A in the reverse transcriptase domain of hepatitis B virus polymerase reduce the susceptibility to tenofovir. Antiviral Res. 2013;97(2):93–100. doi: 10.1016/j.antiviral.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 17.Karatayli E, Karayalcin S, Karaaslan H, et al. A novel mutation pattern emerging during lamivudine treatment shows cross-resistance to adefovir dipivoxil treatment. Antivir. Ther. 2007;12(5):761–768. [PubMed] [Google Scholar]
- 18.Villet S, Pichoud C, Billioud G, et al. Impact of hepatitis B virus rtA181V/T mutants on hepatitis B treatment failure. J. Hepatol. 2008;48(5):747–755. doi: 10.1016/j.jhep.2008.01.027. [DOI] [PubMed] [Google Scholar]
- 19.Sheldon J, Camino N, Rodes B, et al. Selection of hepatitis B virus polymerase mutations in HIV-coinfected patients treated with tenofovir. Antivir. Ther. 2005;10(6):727–734. [PubMed] [Google Scholar]
- 20•.Corsa AC, Liu Y, Flaherty JF, et al. No resistance to tenofovir disoproxil fumarate through 96 weeks of treatment in patients with lamivudine-resistant chronic hepatitis B. Clin. Gastroenterol. Hepatol. 2014;12(12):2106–2112. doi: 10.1016/j.cgh.2014.05.024. [This and other papers demonstrate the effectiveness of tenofovir, a milestone in drug development, and an HBV success story.] [DOI] [PubMed] [Google Scholar]
- 21.Zhu Y, Curtis M, Borroto-Esoda K. The YMDD and rtA194T mutations result in decreased replication capacity in wild-type HBV as well as in HBV with precore and basal core promoter mutations. Antivir. Chem. Chemother. 2011;22(1):13–22. doi: 10.3851/IMP1791. [DOI] [PubMed] [Google Scholar]
- 22.Halegoua-De Marzio D, Hann HW. Prevention of hepatocellular carcinoma and its recurrence with anti-hepatitis B viral therapy. Minerva Gastroenterol. Dietol. 2014;60(3):191–200. [PubMed] [Google Scholar]
- 23.Lai CL, Leung N, Teo EK, et al. A 1-year trial of telbivudine, lamivudine, and the combination in patients with hepatitis B e antigen-positive chronic hepatitis B. Gastroenterology. 2005;129(2):528–536. doi: 10.1016/j.gastro.2005.05.053. [DOI] [PubMed] [Google Scholar]
- 24.Kim SB, Song IH, Kim YM, et al. Long-term treatment outcomes of clevudine in antiviral-naive patients with chronic hepatitis B. World J. Gastroenterol. 2012;18(47):6943–6950. doi: 10.3748/wjg.v18.i47.6943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seok JI, Lee DK, Lee CH, et al. Long-term therapy with clevudine for chronic hepatitis B can be associated with myopathy characterized by depletion of mitochondrial DNA. Hepatology. 2009;49(6):2080–2086. doi: 10.1002/hep.22959. [DOI] [PubMed] [Google Scholar]
- 26.Tak WY, Yang JM, Kim BI, et al. A randomized, open-label study comparing low-dose clevudine plus adefovir combination therapy with clevudine monotherapy in naive chronic hepatitis B patients. Hepatol. Int. 2014;8(3):375–381. doi: 10.1007/s12072-014-9537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones SA, Murakami E, Delaney W, Furman P, Hu J. Non-competitive Inhibition of hepatitis B virus reverse transcriptase protein priming and DNA synthesis by the nucleoside analog clevudine. Antimicrob. Agents Chemother. 2013;57(9):4181–4189. doi: 10.1128/AAC.00599-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langley DR, Walsh AW, Baldick CJ, et al. Inhibition of hepatitis B virus polymerase by entecavir. J. Virol. 2007;81(8):3992–4001. doi: 10.1128/JVI.02395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchesnokov EP, Obikhod A, Schinazi RF, Gotte M. Delayed chain termination protects the anti-hepatitis B virus drug entecavir from excision by HIV-1 reverse transcriptase. J. Biol. Chem. 2008;283(49):34218–34228. doi: 10.1074/jbc.M806797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nowak MA, Bonhoeffer S, Hill AM, Boehme R, Thomas HC, Mcdade H. Viral dynamics in hepatitis B virus infection. Proc. Natl Acad. Sci. USA. 1996;93(9):4398–4402. doi: 10.1073/pnas.93.9.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31••.Jones SA, Boregowda R, Spratt TE, Hu J. In vitro epsilon RNA-dependent protein priming activity of human hepatitis B virus polymerase. J. Virol. 2012;86(9):5134–5150. doi: 10.1128/JVI.07137-11. [The authors develop an assay to test HBV polymerase function in vitro which, similar to in vivo requirements, is ε dependent and includes host chaperones.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu J, Anselmo D. In vitro reconstitution of a functional duck hepatitis B virus reverse transcriptase: posttranslational activation by Hsp90. J. Virol. 2000;74(24):11447–11455. doi: 10.1128/jvi.74.24.11447-11455.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu J, Toft DO, Seeger C. Hepadnavirus assembly and reverse transcription require a multi-component chaperone complex which is incorporated into nucleocapsids. EMBO J. 1997;16(1):59–68. doi: 10.1093/emboj/16.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hu J, Flores D, Toft D, Wang X, Nguyen D. Requirement of heat shock protein 90 for human hepatitis B virus reverse transcriptase function. J. Virol. 2004;78(23):13122–13131. doi: 10.1128/JVI.78.23.13122-13131.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang GH, Seeger C. The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell. 1992;71(4):663–670. doi: 10.1016/0092-8674(92)90599-8. [DOI] [PubMed] [Google Scholar]
- 36.Gong Y, Yao E, Stevens M, Tavis JE. Evidence that the first strand-transfer reaction of duck hepatitis B virus reverse transcription requires the polymerase and that strand transfer is not needed for the switch of the polymerase to the elongation mode of DNA synthesis. J. Gen. Virol. 2000;81(8):2059–2065. doi: 10.1099/0022-1317-81-8-2059. [DOI] [PubMed] [Google Scholar]
- 37.Jones SA, Hu J. Protein-primed terminal transferase activity of hepatitis B virus polymerase. J. Virol. 2013;87(5):2563–2576. doi: 10.1128/JVI.02786-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stuyver LJ, Locarnini SA, Lok A, et al. Nomenclature for antiviral-resistant human hepatitis B virus mutations in the polymerase region. Hepatology. 2001;33(3):751–757. doi: 10.1053/jhep.2001.22166. [DOI] [PubMed] [Google Scholar]
- 39••.Jones SA, Hu J. Hepatitis B virus reverse transcriptase. diverse functions as classical and emerging targets for antiviral intervention. Emerg. Microbes Infect. 2013;2:e56. doi: 10.1038/emi.2013.56. [Excellent review of the domains, interactions and functions of HBV polymerase.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui X, Ludgate L, Ning X, Hu J. Maturation-associated destabilization of hepatitis B virus nucleocapsid. J. Virol. 2013;87(21):11494–11503. doi: 10.1128/JVI.01912-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Badtke MP, Khan I, Cao F, Hu J, Tavis JE. An interdomain RNA binding site on the hepadnaviral polymerase that is essential for reverse transcription. Virology. 2009;390(1):130–138. doi: 10.1016/j.virol.2009.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones SA, Clark DN, Cao F, Tavis JE, Hu J. Comparative analysis of hepatitis B virus polymerase sequences required for viral RNA binding, RNA packaging, and protein priming. J. Virol. 2014;88(3):1564–1572. doi: 10.1128/JVI.02852-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shin YC, Ko C, Ryu WS. Hydrophobic residues of terminal protein domain of hepatitis B virus polymerase contribute to distinct steps in viral genome replication. FEBS Lett. 2011;585(24):3964–3968. doi: 10.1016/j.febslet.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Shin YC, Park S, Ryu WS. A conserved arginine residue in the terminal protein domain of hepatitis B virus polymerase is critical for RNA pre-genome encapsidation. J. Gen. Virol. 2011;92(Pt 8):1809–1816. doi: 10.1099/vir.0.031914-0. [DOI] [PubMed] [Google Scholar]
- 45.Lin L, Hu J. Inhibition of hepadnavirus reverse transcriptase-epsilon RNA interaction by porphyrin compounds. J. Virol. 2008;82(5):2305–2312. doi: 10.1128/JVI.02147-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang YX, Wen YM, Nassal M. Carbonyl J acid derivatives block protein priming of hepadnaviral P protein and DNA-dependent DNA synthesis activity of hepadnaviral nucleocapsids. J. Virol. 2012;86(18):10079–10092. doi: 10.1128/JVI.00816-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen P, Gan Y, Han N, et al. Computational evolutionary analysis of the overlapped surface (S) and polymerase (P) region in hepatitis B virus indicates the spacer domain in P is crucial for survival. PLoS ONE. 2013;8(4):e60098. doi: 10.1371/journal.pone.0060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Radziwill G, Tucker W, Schaller H. Mutational analysis of the hepatitis B virus P gene product. domain structure and RNase H activity. J. Virol. 1990;64(2):613–620. doi: 10.1128/jvi.64.2.613-620.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meier A, Mehrle S, Weiss TS, Mier W, Urban S. Myristoylated PreS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology. 2013;58(1):31–42. doi: 10.1002/hep.26181. [DOI] [PubMed] [Google Scholar]
- 50.Glebe D, Urban S, Knoop EV, et al. Mapping of the hepatitis B virus attachment site by use of infection-inhibiting preS1 lipopeptides and tupaia hepatocytes. Gastroenterology. 2005;129(1):234–245. doi: 10.1053/j.gastro.2005.03.090. [DOI] [PubMed] [Google Scholar]
- 51.Das K, Xiong X, Yang H, et al. Molecular modeling and biochemical characterization reveal the mechanism of hepatitis B virus polymerase resistance to lamivudine (3TC) and emtricitabine (FTC). J. Virol. 2001;75(10):4771–4779. doi: 10.1128/JVI.75.10.4771-4779.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA. Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science. 1992;256(5065):1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 53.Jacobo-Molina A, Ding J, Nanni RG, et al. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc. Natl Acad. Sci. USA. 1993;90(13):6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tantillo C, Ding J, Jacobo-Molina A, et al. Locations of anti-AIDS drug binding sites and resistance mutations in the three-dimensional structure of HIV-1 reverse transcriptase. Implications for mechanisms of drug inhibition and resistance. J. Mol. Biol. 1994;243(3):369–387. doi: 10.1006/jmbi.1994.1665. [DOI] [PubMed] [Google Scholar]
- 55.Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase. implications for drug resistance. Science. 1998;282(5394):1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 56.Xiong Y, Eickbush TH. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 1990;9(10):3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bartholomeusz A, Tehan BG, Chalmers DK. Comparisons of the HBV and HIV polymerase, and antiviral resistance mutations. Antivir. Ther. 2004;9(2):149–160. [PubMed] [Google Scholar]
- 58.Beck J, Vogel M, Nassal M. dNTP versus NTP discrimination by phenylalanine 451 in duck hepatitis B virus P protein indicates a common structure of the dNTP-binding pocket with other reverse transcriptases. Nucleic Acids Res. 2002;30(7):1679–1687. doi: 10.1093/nar/30.7.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang YX, Luo C, Zhao D, Beck J, Nassal M. Extensive mutagenesis of the conserved box E motif in duck hepatitis B virus P protein reveals multiple functions in replication and a common structure with the primer grip in HIV-1 reverse transcriptase. J. Virol. 2012;86(12):6394–6407. doi: 10.1128/JVI.00011-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghany M, Liang TJ. Drug targets and molecular mechanisms of drug resistance in chronic hepatitis B. Gastroenterology. 2007;132(4):1574–1585. doi: 10.1053/j.gastro.2007.02.039. [DOI] [PubMed] [Google Scholar]
- 61.Walsh AW, Langley DR, Colonno RJ, Tenney DJ. Mechanistic characterization and molecular modeling of hepatitis B virus polymerase resistance to entecavir. PLoS ONE. 2010;5(2):e9195. doi: 10.1371/journal.pone.0009195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Van Hemert FJ, Berkhout B, Zaaijer HL. Differential binding of tenofovir and adefovir to reverse transcriptase of hepatitis B virus. PLoS ONE. 2014;9(9):e106324. doi: 10.1371/journal.pone.0106324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tan YW, Ge GH, Zhao W, et al. YMDD motif mutations in chronic hepatitis B antiviral treatment naive patients. a multi-center study. Brazilian J. Infect. Dis. 2012;16(3):250–255. [PubMed] [Google Scholar]
- 64.Mukaide M, Tanaka Y, Shin IT, et al. Mechanism of entecavir resistance of hepatitis B virus with viral breakthrough as determined by long-term clinical assessment and molecular docking simulation. Antimicrob. Agents Chemother. 2010;54(2):882–889. doi: 10.1128/AAC.01061-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65••.Menendez-Arias L, Alvarez M, Pacheco B. Nucleoside/nucleotide analog inhibitors of hepatitis B virus polymerase. mechanism of action and resistance. Current opinion in virology. 2014;8C:1–9. doi: 10.1016/j.coviro.2014.04.005. [Excellent review of the current state of drug development and treatment-associated mutations.] [DOI] [PubMed] [Google Scholar]
- 66.Shaw T, Bartholomeusz A, Locarnini S. HBV drug resistance. Mechanisms, detection and interpretation. J. Hepatol. 2006;44(3):593–606. doi: 10.1016/j.jhep.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 67.Li X, Liu Y, Zhao P, et al. Investigation into drug-resistant mutations of HBV from 845 nucleos(t)ide analogue-naive Chinese patients with chronic HBV infection. Antivir. Ther. 2014 doi: 10.3851/IMP2813. doi:10.3851/IMP2813 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 68.Liu B-M, Li T, Xu J, et al. Characterization of potential antiviral resistance mutations in hepatitis B virus reverse transcriptase sequences in treatment-naïve Chinese patients. Antiviral Res. 2010;85(3):512–519. doi: 10.1016/j.antiviral.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 69••.Tavis JE, Cheng X, Hu Y, et al. The hepatitis B virus ribonuclease H is sensitive to inhibitors of the human immunodeficiency virus ribonuclease H and integrase enzymes. PLoS Pathog. 2013;9(1):e1003125. doi: 10.1371/journal.ppat.1003125. [Development of functional assay using purified RNase H; the authors found inhibitors using this low-throughput screening assay.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nowotny M, Gaidamakov SA, Crouch RJ, Yang W. Crystal structures of RNase H bound to an RNA/DNA hybrid. substrate specificity and metal-dependent catalysis. Cell. 2005;121(7):1005–1016. doi: 10.1016/j.cell.2005.04.024. [DOI] [PubMed] [Google Scholar]
- 71.Hirsch RC, Lavine JE, Chang LJ, Varmus HE, Ganem D. Polymerase gene products of hepatitis B viruses are required for genomic RNA packaging as well as for reverse transcription. Nature. 1990;344(6266):552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 72.Chen Y, Robinson WS, Marion PL. Selected mutations of the duck hepatitis B virus P gene RNase H domain affect both RNA packaging and priming of minus-strand DNA synthesis. J. Virol. 1994;68(8):5232–5238. doi: 10.1128/jvi.68.8.5232-5238.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seeger C, Leber EH, Wiens LK, Hu J. Mutagenesis of a hepatitis B virus reverse transcriptase yields temperature-sensitive virus. Virology. 1996;222(2):430–439. doi: 10.1006/viro.1996.0440. [DOI] [PubMed] [Google Scholar]
- 74.Ko C, Shin YC, Park WJ, Kim S, Kim J, Ryu WS. Residues Arg703, Asp777, and Arg781 of the RNase H domain of hepatitis B virus polymerase are critical for viral DNA synthesis. J. Virol. 2014;88(1):154–163. doi: 10.1128/JVI.01916-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cai CW, Lomonosova E, Moran EA, et al. Hepatitis B virus replication is blocked by a 2-hydroxyisoquinoline-1,3(2H,4H)-dione (HID) inhibitor of the viral ribonuclease H activity. Antiviral Res. 2014;108:48–55. doi: 10.1016/j.antiviral.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hu Y, Cheng X, Cao F, Huang A, Tavis JE. beta-Thujaplicinol inhibits hepatitis B virus replication by blocking the viral ribonuclease H activity. Antiviral Res. 2013;99(3):221–229. doi: 10.1016/j.antiviral.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 77.Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5’-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461(7264):674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- 78.Königer C, Wingert I, Marsmann M, Rösler C, Beck J, Nassal M. Involvement of the host DNA-repair enzyme TDP2 in formation of the covalently closed circular DNA persistence reservoir of hepatitis B viruses. Proc. Natl Acad. Sci. USA. 2014;111(40):E4244–E4253. doi: 10.1073/pnas.1409986111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Stahl M, Beck J, Nassal M. Chaperones activate hepadnavirus reverse transcriptase by transiently exposing a C-proximal region in the terminal protein domain that contributes to epsilon RNA binding. J. Virol. 2007;81(24):13354–13364. doi: 10.1128/JVI.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hu J, Lin L. RNA-protein interactions in hepadnavirus reverse transcription. Front. Biosci. 2009;14:1606–1618. doi: 10.2741/3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim S, Wang H, Ryu WS. Incorporation of eukaryotic translation initiation factor eIF4E into viral nucleocapsids via interaction with hepatitis B virus polymerase. J. Virol. 2010;84(1):52–58. doi: 10.1128/JVI.01232-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ariumi Y, Kuroki M, Abe K, et al. DDX3 DEAD-box RNA helicase is required for hepatitis C virus RNA replication. J. Virol. 2007;81(24):13922–13926. doi: 10.1128/JVI.01517-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yedavalli VS, Neuveut C, Chi YH, Kleiman L, Jeang KT. Requirement of DDX3 DEAD box RNA helicase for HIV-1 Rev-RRE export function. Cell. 2004;119(3):381–392. doi: 10.1016/j.cell.2004.09.029. [DOI] [PubMed] [Google Scholar]
- 84.Gu L, Fullam A, Brennan R, Schroder M. Human DEAD box helicase 3 couples IkappaB kinase epsilon to interferon regulatory factor 3 activation. Mol. Cell. Biol. 2013;33(10):2004–2015. doi: 10.1128/MCB.01603-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang H, Kim S, Ryu WS. DDX3 DEAD-Box RNA helicase inhibits hepatitis B virus reverse transcription by incorporation into nucleocapsids. J. Virol. 2009;83(11):5815–5824. doi: 10.1128/JVI.00011-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang H, Ryu WS. Hepatitis B virus polymerase blocks pattern recognition receptor signaling via interaction with DDX3. implications for immune evasion. PLoS Pathog. 2010;6(7):e1000986. doi: 10.1371/journal.ppat.1000986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nguyen DH, Hu J. Reverse transcriptase- and RNA packaging signal-dependent incorporation of APOBEC3G into hepatitis B virus nucleocapsids. J. Virol. 2008;82(14):6852–6861. doi: 10.1128/JVI.00465-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nguyen DH, Gummuluru S, Hu J. Deamination-independent inhibition of hepatitis B virus reverse transcription by APOBEC3G. J. Virol. 2007;81(9):4465–4472. doi: 10.1128/JVI.02510-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Turelli P, Mangeat B, Jost S, Vianin S, Trono D. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303(5665):1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- 90.Gentile I, Buonomo AR, Borgia G. Dasabuvir. A non-nucleoside inhibitor of NS5B for the treatment of hepatitis C virus infection. Rev. Recent Clin. Trials. 2014 doi: 10.2174/1574887109666140529222602. doi:10.2174/1574887109666140529222602 (Epub ahead of print) [DOI] [PubMed] [Google Scholar]
- 91.De Béthune M-P. Non-nucleoside reverse transcriptase inhibitors (NNRTIs), their discovery, development, and use in the treatment of HIV-1 infection. A review of the last 20 years (1989–2009). Antiviral Res. 2010;85(1):75–90. doi: 10.1016/j.antiviral.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 92.Wooddell CI, Rozema DB, Hossbach M, et al. Hepatocyte-targeted RNAi therapeutics for the treatment of chronic hepatitis B virus infection. Mol. Ther. 2013;21(5):973–985. doi: 10.1038/mt.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93•.Lucifora J, Xia Y, Reisinger F, et al. Specific and nonhepatotoxic degradation of nuclear hepatitis B virus cccDNA. Science. 2014;343(6176):1221–1228. doi: 10.1126/science.1243462. [Important study suggesting how covalently-closed circular DNA may be targeted.] [DOI] [PMC free article] [PubMed] [Google Scholar]