Abstract
Context:
Delayed puberty (DP) is a common issue and, in the absence of an underlying condition, is typically self limited. Alhough DP seems to be heritable, no specific genetic cause for DP has yet been reported. In contrast, many genetic causes have been found for idiopathic hypogonadotropic hypogonadism (IHH), a rare disorder characterized by absent or stalled pubertal development.
Objective:
The objective of this retrospective study, conducted at academic medical centers, was to determine whether variants in IHH genes contribute to the pathogenesis of DP.
Subjects and Outcome Measures:
Potentially pathogenic variants in IHH genes were identified in two cohorts: 1) DP family members of an IHH proband previously found to have a variant in an IHH gene, with unaffected family members serving as controls, and 2) DP individuals with no family history of IHH, with ethnically matched control subjects drawn from the Exome Aggregation Consortium.
Results:
In pedigrees with an IHH proband, the proband's variant was shared by 53% (10/19) of DP family members vs 12% (4/33) of unaffected family members (P = .003). In DP subjects with no family history of IHH, 14% (8/56) had potentially pathogenic variants in IHH genes vs 5.6% (1 907/33 855) of controls (P = .01). Potentially pathogenic variants were found in multiple DP subjects for the genes IL17RD and TAC3.
Conclusions:
These findings suggest that variants in IHH genes can contribute to the pathogenesis of self-limited DP. Thus, at least in some cases, self-limited DP shares an underlying pathophysiology with IHH.
Delayed puberty offers a unique opportunity to understand the still-elusive factors that initiate puberty. The condition is commonly defined as the lack of sexual maturation at an age greater than 2 SDs above the mean for a given population (1). In conventional clinical practice, this leads to evaluation of boys who have not achieved a testicular volume of 4 mL or greater by 14 years and girls who have not started breast development by 13 years. In the absence of underlying conditions, delayed puberty is typically self limited, with individuals progressing through puberty spontaneously prior to reaching an adult age (commonly defined as 18 years) (2).
The most common cause of delayed puberty is the entity known as constitutional delay of growth and puberty (CDGP), in which children have delayed longitudinal growth patterns in addition to pubertal delay (3). CDGP seems to be highly heritable, as 50–80% of patients with CDGP have a family history of delayed puberty (4, 5). Many CDGP pedigrees show an autosomal-dominant pattern, suggesting that CDGP can be caused by the effects of single-gene mutations (4, 5). Previous investigations have identified a significant association between CDGP and a locus on chromosome 2 (6), but no specific causative gene or pathway has yet been reported for CDGP.
Several observations suggest that there are common pathophysiologic mechanisms that link self-limited delayed puberty and another reproductive endocrine condition called idiopathic hypogonadotrophic hypogonadism (IHH), in which pubertal development is absent or stalled by adult age due to defects in the secretion or action of the master reproductive hormone GnRH (2). Greater than 10% of IHH patients have relatives with a history of delayed puberty, compared with 2.5% of the general population (7, 8). In addition, approximately 20% of IHH individuals undergo “reversal,” with activation of the hypothalamic-pituitary-gonadal axis and normalization of reproductive endocrine function in adulthood (9–15). One difference between reversal of IHH and self-limited delayed puberty is the timing of pubertal initiation: after the age of 18 years in IHH with reversal and before age 18 years in self-limited delayed puberty. Thus, self-limited delayed puberty and IHH may share an underlying pathophysiology, at least in some cases.
Greater than 30 genes have been implicated in the pathogenesis of IHH, and these genes regulate the development, migration, and secretory function of GnRH neurons (2, 16). Of note, IHH with reversal has specifically been associated with rare variants in the genes encoding neurokinin B and its receptor (TAC3 and TACR3, respectively) (13, 15). The phenotypic resemblance between self-limited delayed puberty and IHH with reversal suggests that variants in TAC3 and TACR3 may also contribute to the pathogenesis of self-limited delayed puberty, but prior investigations have not conclusively demonstrated such a link (17, 18).
In this report, we have tested the hypothesis that there is overlap between the genetics of self-limited delayed puberty and that of IHH using two approaches. First, we examined pedigrees with an IHH proband known to carry a potentially pathogenic variant in an IHH gene to determine whether family members with self-limited delayed puberty were more likely than those with normal pubertal timing to share the proband's variant. We also examined probands with delayed puberty (and no family history of IHH) to test whether they were more likely than controls to harbor potentially pathogenic variants in IHH genes, with a specific focus on TAC3 and TACR3 given the association of these genes with IHH with reversal.
Materials and Methods
All studies were approved by the Institutional Review Boards of Massachusetts General Hospital (MGH), Boston Children's Hospital (BCH), and Rainbow Babies and Children's Hospital (RB&C), and written informed consent was obtained for all study participants.
Pedigrees with an IHH proband
Study participants
IHH individuals were either patients at MGH or referred by their physicians to MGH to participate in genetic studies. IHH was defined as low sex steroid levels (T ≤ 100 ng/dL in men; estradiol ≤ 20 pg/mL in women) in the setting of inappropriately normal or low gonadotropin levels at age 18 years or greater in the absence of other identifiable causes of hypogonadotropic hypogonadism.
Family members in IHH pedigrees were evaluated by a staff member of the MGH Reproductive Endocrine Unit (REU) through standardized questionnaires, interviews, and/or review of medical records. Self-limited delayed puberty was defined in this cohort as 1) spontaneous menarche between the ages of 15 and 18 years for girls, initiation of testicular growth between the ages of 14 and 18 years for boys, reported or documented diagnosis of delayed puberty by a physician, and/or self-reported continual height growth past age 16 years, 2) no report of impaired reproductive endocrine function in adulthood, and 3) absence of identifiable underlying causes of delayed puberty. Family members who reported normal pubertal development were used as controls.
Genetic studies
IHH probands had previously been sequenced for variants in 13 IHH genes: FGF8, FGFR1, GNRH1, GNRHR, HS6ST1, KAL1, KISS1, KISS1R, NELF, PROK2, PROKR2, TAC3, and TACR3. Sequence variants were characterized as potentially pathogenic variants by the following criteria: 1) variants with minor allele frequency < 0.1% in the 1000 Genomes Project (19) and National Heart, Lung, and Blood Institute Grand Opportunity Exome Sequencing Project (20); 2) nonsense, frameshift, or splice-site-altering variants, and nonsynonymous missense variants predicted to be deleterious in at least 3/5 in silico prediction programs [PANTHER (21), PolyPhen2 (22), SIFT (23), PMUT (24), and MutationTaster (25)]. Whenever available, previous functional studies of these variants demonstrating loss of function in vitro provided additional evidence of pathogenicity. For each IHH proband with an identified potentially pathogenic variant, family members were screened for the same variant through targeted sequencing as described in Supplemental Materials and Methods.
Delayed puberty probands
Study participants
Delayed puberty participants in the study were either patients at MGH, BCH, or RB&C, or referred to MGH to participate in genetic studies. Some of the delayed puberty subjects from BCH and RB&C were studied in previous reports on the diagnoses and characteristics of adolescents with delayed puberty (3), familial aggregation and inheritance patterns in CDGP pedigrees (4), and the role of sequence variation in IHH genes in pubertal timing (26, 27). Self-limited delayed puberty was defined in this cohort as 1) absence of spontaneous thelarche by age 13 years and spontaneous menarche by age 15 years for girls and testicular length less than 2.5 cm or volume less than 4 mL at age 14 years or older for boys, 2) spontaneous pubertal development prior to age 18 years as evidenced by thelarche and/or spontaneous menarche in girls and testicular length at least 2.5 cm or testicular volume at least 4 mL in volume in boys, and 3) evidence of a normal rate of pubertal progression and/or achievement of normal or near-normal adult sex steroid levels and/or testicular volume in boys, and 4) absence of identifiable underlying causes of delayed puberty. Controls were 33 855 non-Finnish European individuals from the Exome Aggregation Consortium (ExAC) (28).
Genetic studies
Whole-exome sequencing, variant calling, and annotation were performed as described in Supplemental Materials and Methods. Control data were accessed through the ExAC browser. Variants were filtered according to the following parameters: 1) present in one of the following IHH genes: DUSP6, FEZF1, FGF8, FGF17, FGFR1, FLRT3, GNRH1, GNRHR, HS6ST1, IL17RD, KAL1, KISS1, KISS1R, NELF, PROK2, PROKR2, SEMA3A, SPRY4, TAC3, TACR3, and WDR11; 2) quality designation of PASS; 3) variant frequency within the delayed puberty cohort of less than 20% (to exclude sequencing artifacts); and 4) a potentially pathogenic variant as defined above.
Statistical analysis
For analysis of pedigrees with an IHH proband, computer simulations were used to determine P values as detailed in Supplemental Materials and Methods. For analysis of delayed puberty probands, frequencies of potentially pathogenic variants between cases and controls were compared using Fisher's exact test. Comparisons of ages at pubertal milestones were performed using Student's t-test. All tests were two-sided, and a P < .05 was considered statistically significant.
Results
We analyzed two cohorts of subjects with self-limited delayed puberty: family members of an IHH proband who themselves had delayed puberty, and individuals with delayed puberty and no family history of IHH.
Pedigrees with an IHH proband
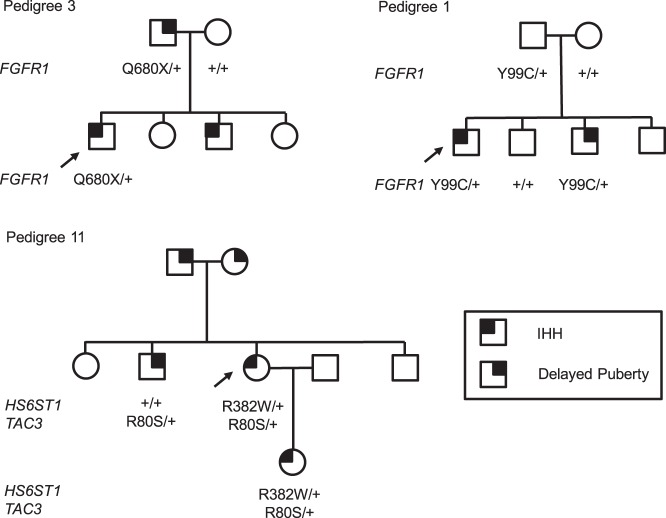
Figure 1 summarizes the process of identifying pedigrees with an IHH proband with a potentially pathogenic variant in an IHH gene and family members with delayed puberty. Of the 15 pedigrees that met criteria for inclusion in this analysis, one had an IHH proband with two potentially pathogenic variants (in TAC3 and HS6ST1) (Figure 2); all other probands had a single potentially pathogenic variant in an IHH gene (Table 1).
Figure 1.
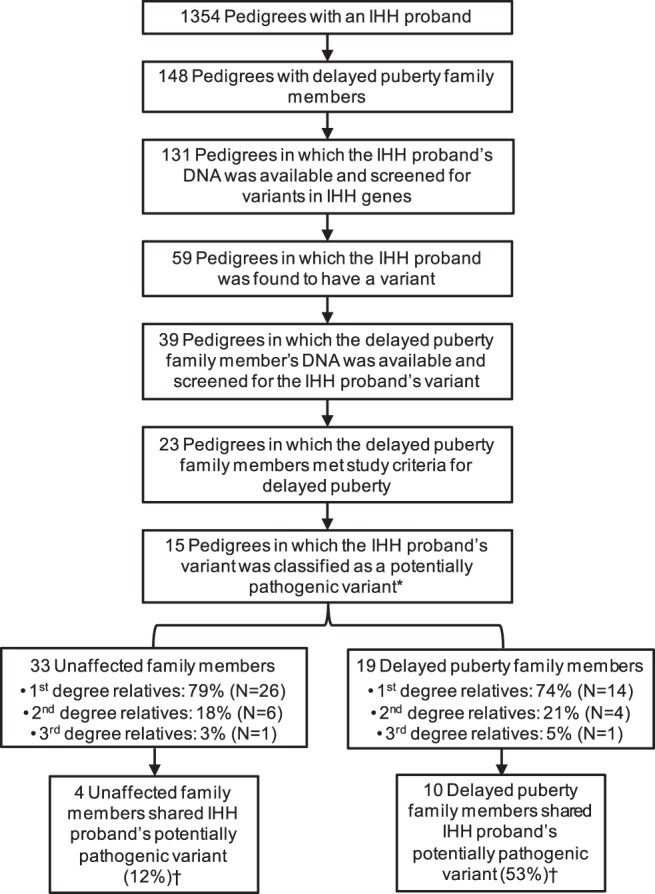
Ascertainment of family members with delayed puberty and normal pubertal timing in pedigrees with IHH probands. N, number of individuals specified in associated rectangle. *, Three pedigrees were excluded: one in which the IHH proband had a potentially pathogenic variant in KAL1 and DNA was available only for male family members, and two in which the IHH probands had biallelic variants in GNRHR and DNA was available for only the proband's parents. †, P = .003.
Figure 2.
Examples of pedigrees with potentially pathogenic variants that segregate with IHH and delayed puberty (Pedigrees 3 and 11 from Table 1) and of a pedigree with a potentially pathogenic variant that does not (Pedigree 1). Arrows identify the IHH proband. Genotypes of subjects for whom DNA was available are listed below each subject's symbol. Plus sign (+) indicates the wild-type allele. Squares represent males; circles represent females.
Table 1.
Potentially Pathogenic Variants in IHH Probands and Family Members
| Pedigree | IHH Proband |
Unaffected Family Members |
Delayed Puberty Family Members |
Reference |
|---|---|---|---|---|
| Potentially Pathogenic Variant | N with a Potentially Pathogenic Variant/Total | N with a Potentially Pathogenic Variant/Total | ||
| 1 | FGFR1 p.Y99C | 1/3 | 1/1 | 38c |
| 2 | FGFR1 p.P366L | 0/3 | 0/1 | — |
| 3 | FGFR1 p.Q680X | 0/1 | 1/1 | 38,39 |
| 4 | FGFR1 p.G237S | 0/3 | 0/2 | 39 |
| 5 | FGFR1 p.E274G | 0/3 | 1/1 | 40 |
| 6 | FGFR1 p.W737R | 0/2 | 0/1 | — |
| 7 | FGFR1 p.R622X | 0/1 | 2/2 | 40 |
| 8 | FGFR1 p.G687R | 0/6 | 0/1 | — |
| 9 | FGFR1 p.I639T | 0/2 | 1/2 | — |
| 10 | FGFR1 p.S346YfsX18 | 0/1 | 1/1 | — |
| 11a | HS6ST p.R382W; TAC3 p.R80S | 0/0 | 1/1 | — |
| 12 | KAL1 p.R457X | 1/2 | 2/2 | 41 |
| 13 | PROKR2 p.S188L | 1/2 | 0/1 | 42c |
| 14 | PROKR2 p.R85C | 0/1 | 0/1 | 42 |
| 15 | TACR3 p.W208X | 1/3 | 0/1 | 13 |
| Total | 4/33 (12%)b | 10/19 (53%)b |
Only the TAC3 p.R80S variant was shared by family members.
P = .003.
Loss-of-function as indicated by significantly decreased performance in in vitro functional assays compared with wild type.
Within these 15 pedigrees, DNA was available from 19 family members with self-limited delayed puberty and 33 family members with normal pubertal timing. To rule out differences in genetic relatedness as a potential confounding factor, the degree of relation of delayed puberty and unaffected family members to the IHH proband was assessed, and no differences were found (Figure 1).
Targeted sequencing was performed to determine whether delayed puberty family members were more likely than unaffected family members to harbor the same variant as the IHH proband. The potentially pathogenic variants found in the IHH probands were shared by 53% (10/19) of delayed puberty family members and in 12% (4/33) of unaffected family members (P = .003, determined by 106 random computer simulations under the null hypothesis as described in Supplemental Materials and Methods). This difference was primarily due to fewer unaffected family members sharing the IHH proband's variant than expected if the variant had no effect on pubertal timing (12% observed vs 44% expected, P < .0001). There was also a suggestion that more delayed puberty family members shared the IHH proband's variant than expected (53% observed vs 44% expected), although this difference was not statistically significant (P = .3).
The genetic variants found to be shared between IHH probands and family members with delayed puberty were found in FGFR1 (seven subjects from six pedigrees), KAL1 (two female subjects from one pedigree), and TAC3 (one subject) (Table 1). In two of these pedigrees, the IHH proband's variant was also found in family members with normal pubertal timing. In addition, in two pedigrees, the IHH proband's variant was shared only with unaffected family members (Table 1).
There were no apparent differences in clinical characteristics (eg, age at menarche, age at testicular growth) between the 10 delayed puberty family members with a potentially pathogenic variant in an IHH gene and the 9 without such a variant (Supplemental Table 1).
Delayed puberty probands
To determine whether variants in IHH genes contribute to self-limited delayed puberty outside the context of pedigrees with an IHH proband, we used whole-exome sequencing to screen for sequence variants in 21 IHH genes in 56 subjects with delayed puberty, no family history of IHH, and self-reported European ancestry; 21 were referred to the MGH REU for clinical care and/or research studies, and 35 were patients at BCH and RB&C. We compared the frequency of potentially pathogenic variants in delayed puberty subjects to that of 33 855 non-Finnish European controls from ExAC (28). Because individual level data was not available in ExAC, each allele with a potentially pathogenic variant was counted as a single individual with such a variant.
Potentially pathogenic variants in IHH genes were identified in 14.3% (8/56) of delayed puberty probands and in 5.6% (1 907/33 855) of controls (P = .01) (Table 2). Heterozygous variants identified in delayed puberty probands were found in GNRHR (n = 1; 1.8% of all delayed puberty subjects), IL17RD (n = 3; 5.4%), SEMA3A (n = 1; 1.8%), TAC3 (n = 2; 3.6%), and TACR3 (n = 1; 1.8%) (Tables 2 and 3). Variants identified in ExAC controls are listed in Supplemental Table 4.
Table 2.
Potentially Pathogenic Variants by Gene in Delayed Puberty Probands
| Gene | Number of Delayed Puberty Subjects (n = 56) with a Potentially Pathogenic Variant | % | Number of Control Subjects (n = 33 855) with a Potentially Pathogenic Variant | % | Pa |
|---|---|---|---|---|---|
| DUSP6 | 0 | 0.0% | 133 | 0.4% | 1 |
| FEZF1 | 0 | 0.0% | 19 | 0.1% | 1 |
| FGF17 | 0 | 0.0% | 13 | 0.0% | 1 |
| FGF8 | 0 | 0.0% | 33 | 0.1% | 1 |
| FGFR1 | 0 | 0.0% | 153 | 0.5% | 1 |
| FLRT3 | 0 | 0.0% | 39 | 0.1% | 1 |
| GNRH1 | 0 | 0.0% | 33 | 0.1% | 1 |
| GNRHR | 1 | 1.8% | 140 | 0.4% | .2 |
| HS6ST1 | 0 | 0.0% | 69 | 0.2% | 1 |
| IL17RD | 3 | 5.4% | 231 | 0.7% | .006 |
| KAL1 | 0 | 0.0% | 145 | 0.4% | 1 |
| KISS1 | 0 | 0.0% | 6 | 0.0% | 1 |
| KISS1R | 0 | 0.0% | 51 | 0.2% | 1 |
| NELF | 0 | 0.0% | 89 | 0.3% | 1 |
| PROK2 | 0 | 0.0% | 72 | 0.2% | 1 |
| PROKR2 | 0 | 0.0% | 146 | 0.4% | 1 |
| SEMA3A | 1 | 1.8% | 257 | 0.8% | .3 |
| SPRY4 | 0 | 0.0% | 62 | 0.2% | 1 |
| TAC3 | 2 | 3.6% | 20 | 0.1% | .0006 |
| TACR3 | 1 | 1.8% | 67 | 0.2% | .1 |
| WDR11 | 0 | 0.0% | 129 | 0.4% | 1 |
| TOTAL | 8 | 14.3% | 1907 | 5.6% | .01 |
| TAC3+TACR3 | 3 | 5.4% | 87 | 0.3% | .0005 |
P values are not corrected for multiple comparisons.
Table 3.
Potentially Pathogenic Variants in Delayed Puberty Probands
| Potentially Pathogenic Variant | Evidence for Pathogenicity |
Identified in IHH Subject(s) (Ref.) | |
|---|---|---|---|
| Severe Variant or LOF (Ref.) | In silico Predictions (Deleterious Predictions/Total Predictions)a | ||
| GNRHR p.L117R | 5/5 | Yes (43) | |
| IL17RD p.K131T | LOF (16) | 3/4 | Yes (16) |
| IL17RD p.P191L | 4/4 | No | |
| IL17RD p.W200X | nonsense | — | No |
| SEMA3A p.T717I | 3/5 | Yes (Unpublished) | |
| TAC3 p.H83R | 5/5 | No | |
| TAC3 g.18595G>T | splice-site | — | No |
| TACR3 p.A171P | 4/5 | No | |
Abbreviations: LOF, loss-of-function as indicated by significantly decreased performance in in vitro functional assays compared with wild-type; Ref., reference; —, not tested.
Not all prediction programs gave predictions for all variants.
Given the association between IHH with reversal and TAC3 and TACR3 (15), we specifically tested whether potentially pathogenic variants in these genes were enriched in patients with self-limited delayed puberty compared with controls. Indeed, 5.4% (3/56) of delayed puberty probands were found to harbor such variants in TAC3 or TACR3 compared with 0.3% (87/33 855) of controls (P = .0005) (Table 2).
The gene with the largest number of potentially pathogenic variants in delayed puberty probands was IL17RD, a modulator of signaling by the fibroblast growth factor receptor, with variants found in three delayed puberty subjects (5.4%) compared with 0.7% (231/33 855) of control subjects (Table 2).
There were no apparent differences in clinical characteristics between subjects with potentially pathogenic variants in IHH genes and subjects without such variants (Supplemental Table 2). There were also no apparent differences in clinical characteristics between subjects recruited from BCH/RB&C and those from the MGH REU (Supplemental Table 3). Of the eight patients with delayed puberty who had potentially pathogenic variants in IHH genes, four patients were from the BCH/RB&C cohort (11%; 4/35), and four patients were from the MGH REU cohort (19%; 4/21, P = .5).
Discussion
We have found that subjects with self-limited delayed puberty are more likely than control subjects to carry potentially pathogenic variants in IHH genes, which are critical to the development and function of the GnRH neuronal network. By implicating these genes in the pathogenesis of delayed puberty, our findings confirm the genetic basis of delayed puberty that had been suggested by previous studies that demonstrated its highly familial nature (4, 5). Furthermore, the genetic link between delayed puberty and IHH suggests that the common, self-limited condition of delayed puberty and the rare, often permanent disorder of IHH share underlying pathophysiologic mechanisms.
Several prior studies, some conducted by members of our group, have tested the hypothesis that either common or rare variants in IHH genes contribute to the pathogenesis of delayed puberty. These studies have examined both rare and common variants in GNRH1 and GNRHR as well as common haplotypes around these genes (26), common variants in a panel of 10 IHH genes (FGFR1, GNRH, GNRHR, GPR54, KAL1, KISS1, LEP, LEPR, PROK2, and PROKR2) (27), variants specifically associated with IHH in KAL1, GNRHR, and FGFR1 (27), and rare variants in FGFR1, GNRHR, TAC3, and TACR3 (17, 18). Although these studies identified occasional rare variants in IHH genes in delayed puberty patients, no study to date has conclusively associated rare variants in IHH genes with self-limited delayed puberty. Given the similarities between IHH and self-limited delayed puberty, the failure to demonstrate such an association was puzzling. The expansion of the IHH gene panel over the years and the use of whole-exome sequencing in this study to screen simultaneously for variants in all genes associated with isolated IHH have now allowed us to demonstrate an enrichment of rare variants in IHH genes in subjects with self-limited delayed puberty.
Our results highlight the variability in phenotypes associated with variants in IHH genes, given that potentially pathogenic variants were found not only in subjects with delayed puberty but also in controls. Because limited phenotypic data was available for the ExAC controls, it is possible that some controls harboring variants in IHH genes in fact had delayed puberty. Furthermore, although these variants were predicted to be deleterious by multiple computer prediction programs, it remains possible that some of these variants have little to no effect on the function of the encoded proteins. Variants in IHH genes may also exhibit variable penetrance and expressivity. Indeed, in two of the IHH pedigrees analyzed in this study, the same heterozygous variant was found in the IHH proband, family members with delayed puberty, and family members with normal pubertal timing. However, in our analysis of pedigrees with an IHH proband, few unaffected family members shared the proband's variant, suggesting that these IHH gene variants tend to produce either delayed puberty or IHH. The phenotypic variability associated with variants in IHH genes limits the clinical utility of genetic testing in IHH and delayed puberty.
Our results provide further evidence for the association between rare variants in the neurokinin B signaling pathway and delayed emergence of reproductive endocrine function, as rare heterozygous variants in TAC3 and TACR3 have now been identified in patients with self-limited delayed puberty as well as in patients with IHH with reversal. Interestingly, a common genetic variant near TACR3 has been shown to be associated with variation in the normal timing of puberty (29). Thus, disruption of the neurokinin B pathway can lead to late-normal pubertal timing, frankly delayed puberty, or even more severely delayed sexual maturation that does not occur until after the age of 18 years, firmly demonstrating a critical role for the neurokinin B pathway in influencing pubertal timing.
We also identified several potential pathogenic variants in IL17RD, a modulator of the FGF8/FGFR1 signaling pathway, which has been suggested to have a role in the fate specification of GnRH neurons (16). A recent study identified a heterozygous variant in IL17RD in both an IHH proband and a parent with delayed puberty and another variant in IL17RD in the homozygous state in an IHH proband and in the heterozygous state in a parent with delayed puberty (16). These findings and our current observations collectively suggest that heterozygous variants in IL17RD may result in reproductive endocrine phenotypes of varying severity.
IHH genes have been broadly classified based on their roles in either the neurodevelopmental ontogeny or neuroendocrine physiology of GnRH neurons (2). It is fairly plausible that TAC3 and TACR3, neuroendocrine genes that seem to regulate GnRH neuronal function, could function as part of the hypothetical “switch” that activates the GnRH neuronal network to initiate puberty. It is less clear, perhaps, how delayed puberty could be caused by deleterious variants in IL17RD, a presumed neurodevelopmental gene involved in the initial fate specification of GnRH neurons (16). One potential mechanism is that IL17RD may also function in other yet-to-be identified pathways critical for activating GnRH neurons at puberty. Another possibility stems from the fact that the hormonal changes underlying puberty can be detected well before the appearance of physical signs of puberty: in boys, nighttime LH secretion has been shown to occur 1–2 years before testicular enlargement can be appreciated (30). Thus, even if the first activation of GnRH neurons occurs at a normal time, impaired activity of the GnRH neuronal network could result in a longer delay between the activation of this diminished GnRH neuronal network and the appearance of the physical changes that are used clinically to mark the start of puberty.
Self-limited delayed puberty has traditionally been described as occurring in the context of CDGP, with associated delays in childhood growth and skeletal maturation. However, in one study, over 50% of individuals with delayed puberty did not exhibit slow prepubertal growth (31). This observation suggests two distinct pathophysiological mechanisms for delayed puberty: one associated with delays in prepubertal growth and the other with isolated pubertal delay. The GnRH neuronal network is relatively quiescent during childhood, so defects in this network would not be expected to cause prepubertal growth delay. Longitudinal growth data were not available for most of our delayed puberty subjects, and future work will determine whether delayed puberty associated with IHH gene variants indeed occurs in the absence of delayed prepubertal growth.
Strengths of our study include the use of exome sequencing to analyze the largest IHH gene panel (21 genes) in a delayed puberty cohort to date, as well as the use of a large, ethnically matched control cohort. Limitations of our study include the relatively small size of the cohorts, which were underpowered to implicate specific genes in the pathogenesis of self-limited delayed puberty, although we have identified candidate genes (TACR3, TAC3, and IL17RD) for future studies. Another limitation is the difference in whole-exome capture techniques used for the delayed puberty cohort and the ExAC controls, which could potentially result in differences in variant detection. However, all potentially pathogenic variants detected in the delayed puberty cases were in genomic regions with adequate coverage in ExAC. A third limitation is selection bias. Because the MGH REU is a referral center for IHH, the referred delayed puberty cases may have been more severe and, thus, not representative of delayed puberty more generally. However, no significant differences were found in clinical characteristics or the frequency of potentially pathogenic variants between the MGH and BCH/RB&C cohorts.
Our findings provide evidence for a genetic link between IHH, a rare disease, and self-limited delayed puberty, a common disorder. Similar genetic links between rare and common disease entities or clinical traits have been previously demonstrated between neonatal diabetes and type 2 diabetes (KCNJ11) (32–34) and between familial dysbetalipoproteinemia and elevated fasting blood lipids (APOE) (35, 36). In reproductive endocrinology, a genetic link has been suggested between IHH and hypothalamic amenorrhea, a type of reproductive endocrine dysfunction that occurs in the setting of excessive exercise, nutritional deprivation, psychological distress, or other stressors (37). Our study serves as an additional example of how genes that underlie a rare genetic disease can also contribute to the genetics of a more common counterpart.
Puberty remains an intriguing scientific mystery, and the factors that initiate puberty and modulate its progression are still largely unknown. IHH has long served as a human disease model to understand the intricacies of the reproductive endocrine system (2). Our report of a significant genetic overlap between self-limited delayed puberty and IHH highlights delayed puberty as an additional human disease model to decipher the intricacies of pubertal timing. Specifically, our study identifies TAC3, TACR3, and IL17RD as candidate delayed puberty genes. Future genetic studies may firmly establish these and other genes in the pathogenesis of delayed puberty, and functional studies will determine how these genes regulate pubertal timing.
Acknowledgments
We thank the following physicians for referring subjects: Melissa Buryk, Ayse Pinar Cemeroglu, Olga Hardy, Susan Kirsch, Abhishek Kulkarni, Veronica Mericq, Paulina M. Merino, Daniel Metzger, J.B. Quintos, Michael Racine, Jose Perez-Rodriguez, Karen Rubin, Menachem Samuel Shapiro, David Schwartz, Phyllis Speiser, Pamela Taxel, and Charles Verge. We thank Matthew Chase for his assistance with Sanger sequencing, Stephanie Hadley for her assistance with ascertainment of pedigrees, and Ines Sedlmeyer for her management of the Boston Children's Hospital cohort. We thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about. We also acknowledge the support of the National Heart, Lung, and Blood Institute (NHLBI) and the contributions of the research institutions, study investigators, field staff, and study participants in creating the Grand Opportunity Exome Sequencing Project (GO ESP) for biomedical research. Funding for GO ESP was provided by NHLBI Grants RC2 HL-103010 (HeartGO), RC2 HL-102923 (LungGO) and RC2 HL-102924 (WHISP), with exome sequencing performed through NHLBI Grants RC2 HL-102925 (BroadGO) and RC2 HL-102926 (SeattleGO).
This work was supported by a Doris Duke Charitable Foundation Clinical Scientist Development Award to Y.-M.C., U54 HD028138 to S.B.S., R01 HD048960 to M.R.P and J.N.H., M01 RR002172 to the Boston Children's Hospital General Clinical Research Center (GCRC), and K23 RR15544 and M01 RR000080 to the William Dahms GCRC at University Hospitals Cleveland. M.H.G. was supported by March of Dimes Grant 6-FY12–393 to J.N.H. Y.-M.C. was supported by a Charles H. Hood Foundation Child Health Research Award and National Institutes of Health/Eunice Kennedy Shriver National Institute of Child Health and Human Development Grant K12 HD052896 (PI Fleisher). S.B.S. was additionally supported by R01 HD043341 and K24 HD 067388.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- BCH
- Boston Children's Hospital
- CDGP
- constitutional delay of growth and puberty
- ExAC
- Exome Aggregation Consortium
- IHH
- idiopathic hypogonadotrophic hypogonadism
- MGH
- Massachusetts General Hospital
- RB&C
- Rainbow Babies and Children's Hospital
- REU
- Reproductive Endocrine Unit.
References
- 1. Palmert MR, Dunkel L. Clinical practice. Delayed puberty. N Engl J Med. 2012;366:443–453. [DOI] [PubMed] [Google Scholar]
- 2. Sykiotis GP, Pitteloud N, Seminara SB, Kaiser UB, Crowley WF., Jr Deciphering genetic disease in the genomic era: The model of GnRH deficiency. Sci Transl Med. 2010;2:32rv32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Sedlmeyer IL, Palmert MR. Delayed puberty: Analysis of a large case series from an academic center. J Clin Endocrinol Metab. 2002;87:1613–1620. [DOI] [PubMed] [Google Scholar]
- 4. Sedlmeyer IL, Hirschhorn JN, Palmert MR. Pedigree analysis of constitutional delay of growth and maturation: Determination of familial aggregation and inheritance patterns. J Clin Endocrinol Metab. 2002;87:5581–5586. [DOI] [PubMed] [Google Scholar]
- 5. Wehkalampi K, Widén E, Laine T, Palotie A, Dunkel L. Patterns of inheritance of constitutional delay of growth and puberty in families of adolescent girls and boys referred to specialist pediatric care. J Clin Endocrinol Metab. 2008;93:723–728. [DOI] [PubMed] [Google Scholar]
- 6. Wehkalampi K, Widén E, Laine T, Palotie A, Dunkel L. Association of the timing of puberty with a chromosome 2 locus. J Clin Endocrinol Metab. 2008;93:4833–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Waldstreicher J, Seminara SB, Jameson JL, et al. The genetic and clinical heterogeneity of gonadotropin-releasing hormone deficiency in the human. J Clin Endocrinol Metab. 1996;81:4388–4395. [DOI] [PubMed] [Google Scholar]
- 8. Schwankhaus JD, Currie J, Jaffe MJ, Rose SR, Sherins RJ. Neurologic findings in men with isolated hypogonadotropic hypogonadism. Neurology. 1989;39:223–226. [DOI] [PubMed] [Google Scholar]
- 9. Bauman A. Markedly delayed puberty or Kallmann's syndrome variant. J Androl. 1986;7:224–227. [DOI] [PubMed] [Google Scholar]
- 10. Finkelstein JS, Spratt DI, O'Dea LS, et al. Pulsatile gonadotropin secretion after discontinuation of long term gonadotropin-releasing hormone (GnRH) administration in a subset of GnRH-deficient men. J Clin Endocrinol Metab. 1989;69:377–385. [DOI] [PubMed] [Google Scholar]
- 11. Quinton R, Cheow HK, Tymms DJ, Bouloux PM, Wu FC, Jacobs HS. Kallmann's syndrome: Is it always for life? Clin Endocrinol (Oxf). 1999;50:481–485. [DOI] [PubMed] [Google Scholar]
- 12. Raivio T, Falardeau J, Dwyer A, et al. Reversal of idiopathic hypogonadotropic hypogonadism. N Engl J Med. 2007;357:863–873. [DOI] [PubMed] [Google Scholar]
- 13. Gianetti E, Tusset C, Noel SD, et al. TAC3/TACR3 mutations reveal preferential activation of gonadotropin-releasing hormone release by neurokinin B in neonatal life followed by reversal in adulthood. J Clin Endocrinol Metab. 2010;95:2857–2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tornberg J, Sykiotis GP, Keefe K, et al. Heparan sulfate 6-O-sulfotransferase 1, a gene involved in extracellular sugar modifications, is mutated in patients with idiopathic hypogonadotrophic hypogonadism. Proc Natl Acad Sci U S A. 2011;108:11524–11529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sidhoum VF, Chan YM, Lippincott MF, et al. Reversal and relapse of hypogonadotropic hypogonadism: Resilience and fragility of the reproductive neuroendocrine system. J Clin Endocrinol Metab. 2014;99:861–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miraoui H, Dwyer AA, Sykiotis GP, et al. Mutations in FGF17, IL17RD, DUSP6, SPRY4, and FLRT3 are identified in individuals with congenital hypogonadotropic hypogonadism. Am J Hum Genet. 2013;92:725–743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vaaralahti K, Wehkalampi K, Tommiska J, Laitinen EM, Dunkel L, Raivio T. The role of gene defects underlying isolated hypogonadotropic hypogonadism in patients with constitutional delay of growth and puberty. Fertil Steril. 2011;95:2756–2758. [DOI] [PubMed] [Google Scholar]
- 18. Tusset C, Noel SD, Trarbach EB, et al. Mutational analysis of TAC3 and TACR3 genes in patients with idiopathic central pubertal disorders. Arq Bras Endocrinol Metabol. 2012;56:646–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abecasis GR, Auton A, Brooks LD, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Exome Variant Server. NHLBI GO Exome Sequencing Project (ESP). http://evs.gs.washington.edu/EVS Accessed October 2014.
- 21. Thomas PD, Kejariwal A, Campbell MJ, et al. PANTHER: A browsable database of gene products organized by biological function, using curated protein family and subfamily classification. Nucleic Acids Res. 2003;31:334–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. [DOI] [PubMed] [Google Scholar]
- 24. Ferrer-Costa C, Gelpí JL, Zamakola L, Parraga I, de la Cruz X, Orozco M. PMUT: A web-based tool for the annotation of pathological mutations on proteins. Bioinformatics. 2005;21:3176–3178. [DOI] [PubMed] [Google Scholar]
- 25. Schwarz JM, Rödelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7:575–576. [DOI] [PubMed] [Google Scholar]
- 26. Sedlmeyer IL, Pearce CL, Trueman JA, et al. Determination of sequence variation and haplotype structure for the gonadotropin-releasing hormone (GnRH) and GnRH receptor genes: Investigation of role in pubertal timing. J Clin Endocrinol Metab. 2005;90:1091–1099. [DOI] [PubMed] [Google Scholar]
- 27. Gajdos ZK, Butler JL, Henderson KD, et al. Association studies of common variants in 10 hypogonadotropic hypogonadism genes with age at menarche. J Clin Endocrinol Metab. 2008;93:4290–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Exome Aggregation Consortium (ExAC). http://exac.broadinstitute.org Accessed November 2014.
- 29. Perry JR, Day F, Elks CE, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wu FC, Butler GE, Kelnar CJ, Sellar RE. Patterns of pulsatile luteinizing hormone secretion before and during the onset of puberty in boys: A study using an immunoradiometric assay. J Clin Endocrinol Metab. 1990;70:629–637. [DOI] [PubMed] [Google Scholar]
- 31. Wehkalampi K, Vangonen K, Laine T, Dunkel L. Progressive reduction of relative height in childhood predicts adult stature below target height in boys with constitutional delay of growth and puberty. Horm Res. 2007;68:99–104. [DOI] [PubMed] [Google Scholar]
- 32. Gloyn AL, Pearson ER, Antcliff JF, et al. Activating mutations in the gene encoding the ATP-sensitive potassium-channel subunit Kir6.2 and permanent neonatal diabetes. N Engl J Med. 2004;350:1838–1849. [DOI] [PubMed] [Google Scholar]
- 33. Gloyn AL, Diatloff-Zito C, Edghill EL, et al. KCNJ11 activating mutations are associated with developmental delay, epilepsy and neonatal diabetes syndrome and other neurological features. Eur J Hum Genet. 2006;14:824–830. [DOI] [PubMed] [Google Scholar]
- 34. Florez JC, Jablonski KA, Kahn SE, et al. Type 2 diabetes-associated missense polymorphisms KCNJ11 E23K and ABCC8 A1369S influence progression to diabetes and response to interventions in the Diabetes Prevention Program. Diabetes. 2007;56:531–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smelt AH, de Beer F. Apolipoprotein E and familial dysbetalipoproteinemia: Clinical, biochemical, and genetic aspects. Semin Vasc Med. 2004;4:249–257. [DOI] [PubMed] [Google Scholar]
- 36. Chang MH, Yesupriya A, Ned RM, Mueller PW, Dowling NF. Genetic variants associated with fasting blood lipids in the U.S. population: Third National Health and Nutrition Examination Survey. BMC Med Genet. 2010;11:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Caronia LM, Martin C, Welt CK, et al. A genetic basis for functional hypothalamic amenorrhea. N Engl J Med. 2011;364:215–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Raivio T, Sidis Y, Plummer L, et al. Impaired fibroblast growth factor receptor 1 signaling as a cause of normosmic idiopathic hypogonadotropic hypogonadism. J Clin Endocrinol Metab. 2009;94:4380–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pitteloud N, Acierno JS, Jr, Meysing A, et al. Mutations in fibroblast growth factor receptor 1 cause both Kallmann syndrome and normosmic idiopathic hypogonadotropic hypogonadism. Proc Natl Acad Sci U S A. 2006;103:6281–6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pitteloud N, Meysing A, Quinton R, et al. Mutations in fibroblast growth factor receptor 1 cause Kallmann syndrome with a wide spectrum of reproductive phenotypes. Mol Cell Endocrinol 2006;254–255:60–69. [DOI] [PubMed] [Google Scholar]
- 41. Oliveira LM, Seminara SB, Beranova M, et al. The importance of autosomal genes in Kallmann syndrome: Genotype-phenotype correlations and neuroendocrine characteristics. J Clin Endocrinol Metab. 2001;86:1532–1538. [DOI] [PubMed] [Google Scholar]
- 42. Cole LW, Sidis Y, Zhang C, et al. Mutations in prokineticin 2 and prokineticin receptor 2 genes in human gonadotrophin-releasing hormone deficiency: Molecular genetics and clinical spectrum. J Clin Endocrinol Metab. 2008;93:3551–3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Gürbüz F, Kotan LD, Mengen E, et al. Distribution of gene mutations associated with familial normosmic idiopathic hypogonadotropic hypogonadism. J Clin Res Pediatr Endocrinol. 2012;4:121–126. [DOI] [PMC free article] [PubMed] [Google Scholar]