Abstract
Unlike mammals, the CNS of the medicinal leech can regenerate damaged neurites, thus restoring neural functions after lesion. We previously demonstrated that the injured leech nerve cord is able to mount an immune response promoting the regenerative processes. Indeed neurons and microglia express sensing receptors like Hm-TLR1, a leech TLR ortholog, associated with chemokine release in response to a septic challenge or lesion. To gain insights into the TLR signaling pathways involved during these neuroimmune responses, members of the MyD88 family were investigated. In the present study, we report the characterization of Hm-MyD88 and Hm-SARM. The expression of their encoding gene was strongly regulated in leech CNS not only upon immune challenge but also during CNS repair, suggesting their involvement in both processes. This work also showed for the first time that differentiated neurons of the CNS could respond to LPS through a MyD88-dependent signalling pathway, while in mammals, studies describing the direct effect of LPS on neurons and the outcomes of such treatment are scarce and controversial. In the present study, we established that this PAMP induced the relocalization of Hm-MyD88 in isolated neurons.
Innate immunity corresponds to the first line of defense common to all metazoans. To sense invading pathogens, animals align a panel of germline-encoded receptors called pattern recognition receptors (PRRs)1. Among them, the family of TLRs is the best characterized. TLRs are transmembrane proteins containing a Leucine-Rich Repeat (LRR) domain and a cytoplasmic Toll/Il1 receptor (TIR) domain. They detect and distinguish pathogen-associated molecular patterns (PAMPs) derived from various microbial pathogens including viruses, bacteria, protozoa and fungi2. The recognition of these PAMPs triggers the associated signaling pathways to activate downstream immune responses and eliminate invading pathogens3. Among the molecules recruited by activated TLRs are the members of the MyD88 family4. In mammals, it includes 5 adaptor proteins containing a TIR domain: myeloid differentiation factor 88 (MyD88), MyD88-adapter-like (Mal), TIR-domain-containing adaptor protein-inducing IFN beta (TRIF), TRIF-related adaptor molecule (TRAM) and sterile alpha and amardillo-motif-containing protein (SARM). All TLRs, except TLR3, recruit MyD88 to mediate innate immune signaling. MyD88 exhibits an N-terminal death domain (DD) and a C-terminal TIR domain. Upon stimulation of TLRs, MyD88 interacts with the cytosolic part of TLR through a homophilic interaction of the TIR domains. Its DD, in turn, associates with the DD of interleukin-1 receptor associated kinase (IRAK) to trigger downstream signaling cascades that lead to the activation of NF-κB5,6,7. The structure of MyD88 is extremely well conserved across evolution and its key role in immunity has been demonstrated in both bilaterian and non bilaterian species8,9,10,11,12,13,14. The second member of the MyD88 family particularly well conserved throughout the animal kingdom is SARM. Indeed, it is the only TIR domain-containing adaptor conserved from C. elegans to mammals15,16. SARM consists of two sterile-alpha motif (SAM) domains that are flanked by an N-terminal Armadillo motif (ARM) and a C-terminal TIR domain17. With this unique combination of three protein-protein interaction domains, SARM can bind a greater variety of interactants. Thus, SARM associates with mitochondria and/or microtubules in neurons, T-cells, Hela and kidney cells18,19,20,21,22. Moreover, in mammals18,23,24; protochordates19,25 and arthropods16,26, SARM also interacts with MyD88 and/or TRIF and/or TRAF6 to down-regulate TLR signaling. Noteworthy, SARM is the only negative regulator of the five TLR adaptor proteins. In contrast, SARM protects C. elegans against bacterial and fungal infection in a Tol-1-independent manner15,27 and restricts viral infections in mice brain28.
In the present study, we investigate such TLR adaptors in leech at the level of the CNS after infection or lesion. In fact, we previously established that the leech nerve cord is able to establish a specific neuroimmune response by discriminating microbial components29,30 or after lesion29,31,32. Indeed leech neurons and microglia express PRRs, and secrete antimicrobial peptides as well as chemokines in response to a septic challenge or injury29,31,33. A survey of the medicinal leech transcriptome and genome databases also reveals the presence of the main signaling molecules involved in the canonical TLR pathways33. We report here the characterization of two members of the MyD88 family: Hm-MyD88 and Hm-SARM. We demonstrated that their respective genes are tightly regulated during an immune challenge and CNS repair. A stimulation of leech isolated neurons with lipopolysaccharide (LPS) also triggered a redistribution of Hm-MyD88 at the cell surface. To the best of our knowledge, this clearly showed for the first time that differentiated neurons of the CNS could respond to LPS through a MyD88-dependent signaling pathway. Previously, we have also observed that stimulation of leech CNS with Muramyl dipeptide (MDP) induced the expression of the gene encoding Hm-TLR130. We report here that this PAMP induced a relocalization of Hm-TLR1 and Hm-MyD88 in isolated neurons. A putative priming effect of MDP on TLR pathways and the possible existence of a cross talk between NLR sensor and TLRs in leech are discussed.
Methods
Microorganisms
The Gram-positive and Gram-negative bacteria, Micrococcus nishinomiyaensis and Aeromonas hydrophila respectively, were isolated from the natural environment of Hirudo medicinalis as previously described34. These bacterial colonies, which live in freshwater, were selected from agar plates under aerobic conditions at room temperature using a random isolation grid.
Animals and treatments
Adult H. medicinalis were purchased from Ricarimpex (Bordeaux, France) and maintained in autoclaved pond water changed daily, for 1 week before starting any experimental procedure.
Microbial challenges ex vivo
Ten dissected nerve cords were collected per condition. Connectives between ganglia were injured in a standard manner using a pair of sterilized fine iridectomy scissors. Axotomized nerve cords were separately incubated in L-15 media containing different microbial components: 3 × 107 CFU/ml of heat-killed Gram positive (M. nishinomiyaensis) or negative (A. hydrophila) bacteria, 100 ng/ml of E. coli LPS (0111:B4 strain, Invivogen), 100 μg/ml of zymosan (Invivogen), 10 μg/ml of Muramyl dipeptide (MDP, Invivogen), 2 μg/ml of lipoteichoic acid (LTA) (Invivogen) or 10 μg/ml of Poly(I:C) (Invivogen) for different time (T0, t = 6 h) at room temperature. The supernatant of culture of cells infected with Vesicular stomatitis virus (VSV) for several days was used to stimulate the axotomized nerve cord ex vivo. Incubations without microbial components were performed in the same conditions as controls (H2O).
Regeneration process
Collected nerve cords were axotomized between each ganglion and cultured up to 8 days under sterile conditions. Ganglia were collected for RNA preparation or protein extraction at point 0 (T0), 6 h, 1day, 3days, 4 days and 8 days. Total RNA from 6 nerve cords were used in the RT-qPCR reactions. For western blot analysis, proteins were extracted from 3 isolated nerve cords.
cDNA cloning
Partial sequences encoding Hm-MyD88 and Hm-SARM have been retrieved from the leech H. medicinalis nervous system EST database33. Full length cDNAs were generated by 5′-RACE using GeneRacer Core Kit (Invitrogen). Double stranded cDNAs from leech nervous systems were ligated to adaptors and these templates were used to PCR amplify 5′-RACE fragments using adaptor specific primers and gene-specific primers deduced from the initial fragment sequences. 5′-RACE-PCR were performed using 2.5 units of Platinum Taq polymerase (Invitrogen) in 1.5 mM of MgCl2. The cycling parameters were: 96°C/30sec; 5 cycles at 96°C/10sec and 72°C/4 min; 5 cycles at 96°C/10sec and 70°C/4 min; 25 cycles at 96°C/10sec and 68°C/4 min. All PCR products were cloned using pGemT-easy vector (according to the protocol provided by the manufacturer) and transformed into competent Escherichia coli JM 109 cells (Promega). Plasmids DNA were sequenced with a FM13/RM13 sequencing kit (Applied Biosystems) according to the manufacturer's instructions.
Sequence and phylogenetic analyses
Translated sequences of Hm-MyD88 and Hm-SARM were used to search for conserved domains using the Simple Molecular Architecture Research Tool SMART™ 35 web server. Orthologous sequences from various vertebrates and invertebrates were retrieved from GenBank and used for the construction of phylogenetic trees using PhyML 3.0, www.phylogeny.fr36.
Gene expression analysis
Gene expression in purified cells
The ganglia from 9 isolated nerve cords were carefully decapsulated by removing the collagen layer that envelops the nerve cord with microscissors. Neurons (>10 μm) and microglial cells (5 μm) were mechanically dissociated and resuspended in 200 μl of complete L-15 medium. The cells were then filtered through a 7 μm nylon mesh, as described previously37. According to their small size, purified microglial cells were collected with the eluate, while purified neurons were retained on the mesh. The latter were collected by gently scraping the inner face of the mesh in clean culture medium. RNA from purified cells were then extracted (Qiazol, Qiagen) and used for cDNA synthesis (Superscript II, Invitrogen) and PCR amplification (GoTaq, Promega). cDNA amplification was performed for 40 cycles at 95°C/30 s, 54°C (Hm-MyD88 and Hm-SARM) or 60°C (18 s reference gene)/2 min and 72°C/45 s (Hm-MyD88 and Hm-SARM) or 30 s (18S reference gene). These RT-PCR were performed with the following primers: Hm-MyD88 (forward primer: 5′-CTTCAAGATCCAAATGATGG-3′; reverse primer: 5′-AGTCTTCAGAGTAACAATCG-3′); Hm-SARM (forward primer: 5′-ACAACTTGCAAGTCTTCT-3′; reverse primer: 5′-GTAGTCATGTATCCATCTGATT-3′) the 18S reference gene (forward primer: 5′-TGCGGTTATTTCGATTGTCA-3′, reverse primer: 5′-AGACAAATCGCTCCACCAAC-3′).
Real time PCR quantification
20 axotomized nerve cords cultured at different times in the presence or absence of microbial components were used per condition. RNA extraction, cDNA synthesis, and real time PCR procedures were realized as already described34. The primers used for Hm-MyD88 (forward primer: 5′-GGAACTGGAAGACAAACAA-3′; reverse primer: 5′-GGGCTTAGGACGACAATGA-3′) and Hm-SARM (forward primer: 5′-TGCAACATCATTCCAGTCAT-3′; reverse primer: 5′-AATTGTTCCAGCTTGTCCAC-3′) quantification were designed with the Primer3 Input software38,39. The 18S was used as the reference gene (18S forward: 5′-TGCGGTTATTTCGATTGTCA-3′, 18S reverse: 5′-AGACAAATCGCTCCACCAAC-3′). Real Time reactions were conducted on a CFX96 qPCR system (BioRad) using a hot start, then 40 cycles at 94°C, 15 s; 56°C, 30 s; 72°C, 30 s, and a final extension step at 72°C for 3 min. Analysis of relative gene expression data was performed using the ΔΔCt method.
Immunodetection of Hm-MyD88
Antisera
Hm-TLR1 protein was detected using the mouse antiserum as previously characterized by Ref. 40. The Hm-MyD88 antiserum was produced by Agrobio. The chemically synthesized immunogenic sequence (CIPDRDFLPGPPKYEAIT) was coupled to BSA and used for the immunization procedure of two rabbits.
Western blot analysis
For Hm-MyD88 characterization, 3 isolated nerve cords were disrupted in lysis buffer (6 M Urea, 2 M Thiourea, 40 mM Tris base) containing a cocktail of protease inhibitors (Sigma Fast Protease Inhibitor cocktail tablets EDTA free, Sigma-Aldrich). 20 μg of proteins were subjected to 14% SDS-PAGE and transferred to a nitrocellulose membrane (Protran, Whatman) by electroblotting. After transfer, the membrane was blocked for 1 h in PBS containing 0.1% Tween 20 and 5% skimmed milk and then probed either with the anti-Hm-MyD88 Ab or the anti-Hm-MyD88 Ab preincubated with the blocking peptide or the preimmune serum at a dilution of 1/10000 in blocking solution overnight at 4°C. After intensive washes with PBS/0.1% Tween20, the immunolabeled bands were detected using a peroxidase-conjugated anti-rabbit secondary Ab (1/50000 – Jackson ImmunoResearch; 60 min at room temperature). An ECL Western blotting kit (Amersham Biosciences) was used for chemoluminescence visualization with Kodak X-Omat AR film.
For protein expression analysis during CNS repair, 3 nerve cords were collected at each time point. CNS were then disrupted in PBS containing a cocktail of protease inhibitors (Sigma Fast Protease Inhibitor cocktail tablets EDTA free, Sigma-Aldrich). Proteins were precipitated with Acetone-10% TCA and resuspended in a solution containing 7 M Urea, 2 M Thiourea and 4% CHAPS. 20 μg (Hm-MyD88) or 30 μg (Hm-SARM) of proteins were subjected to western blot analysis as described above. The antibodies used were rabbit anti-Hm-MyD88 (1/10000) and rabbit anti-SARM (1 μg/mL, Novus Biologicals). To assess that an equal amount of proteins was loaded on gels, membranes were stripped with 0,2 M citric acid and reprobed with mouse anti-Actin (1/400, Thermo-Fisher).
Immunofluorescence
Neurons mechanically dissociated from nerve cords were fixed and centrifuged on slides. They were subjected to immunofluorescence procedure as follows. Cells were first blocked during 1 h with a blocking solution (PBS 1X, 1% Normal Donkey Serum, 0.01% Triton, 1% BSA) and then incubated overnight at 4°C either with rabbit anti-Hm-MyD88 or preimmune serum diluted 1/1200 in blocking solution. After intensive washes with PBS 1X, a biotin-conjugated donkey anti-rabbit (Jackson ImmunoResearch) diluted at 1/250 in blocking solution was applied for 1 h. After intensive washes with PBS 1X, the immune complex was revealed with a Rhodamine Red-X-Streptavidin (Jackson ImmunoResearch) at a concentration of 0.9 μg/mL. Slices were then mounted in glycergel (Dako) and examined using a confocal microscope (ZeissLSM510).
For immune challenge, isolated neurons were stimulated with 100 ng/mL lipopolysaccharide (LPS) or 10 μg/mL muramyl dipeptide (MDP) for 1 or 5 minutes respectively and double labeled with anti-Hm-MyD88 and anti-Hm TLR1 (1/100).
Results
Characterization of two MyD88 family members, Hm-MyD88 and Hm-SARM, in leech CNS
Analysis of the cDNA library of the leech CNS coupled to 5′ RACE-PCR allowed us to obtain the complete sequence of a molecule exhibiting the hallmarks of MyD88 (Supplementary Material 1): A death domain at the N-terminus and a C-terminal TIR domain. Thus, the leech molecule was named Hm-MyD88. This bioinformatics analysis revealed that Hm-MyD88 does not exhibit a signal peptide and is predicted as cytoplasmic. Interestingly, Hm-MyD88 also possesses the three highly conserved regions (Box 1, Box 2 and Box 3) present in most TIR domains. Moreover, the R-D-x-L-P-G motif, where x represents any amino acid, can be found in the Box 2. Indeed, in mammals, this motif is essential for the interaction of TLRs with MyD8841 and for MyD88 dimerization7. Hm-MyD88 also displays an additional C-terminal extension (CTE) as was described in the mussel Mytilus galloprovincialis42, the fruit fly Drosophila melanogaster43 and the mud crab Scylla paramamosain44. In the two arthropods, the CTE anchors MyD88 at the plasma membrane. Then, MyD88 recruits its activated Toll receptor and downstream cytosolic adaptors to trigger Toll signaling in response to an immune challenge45.
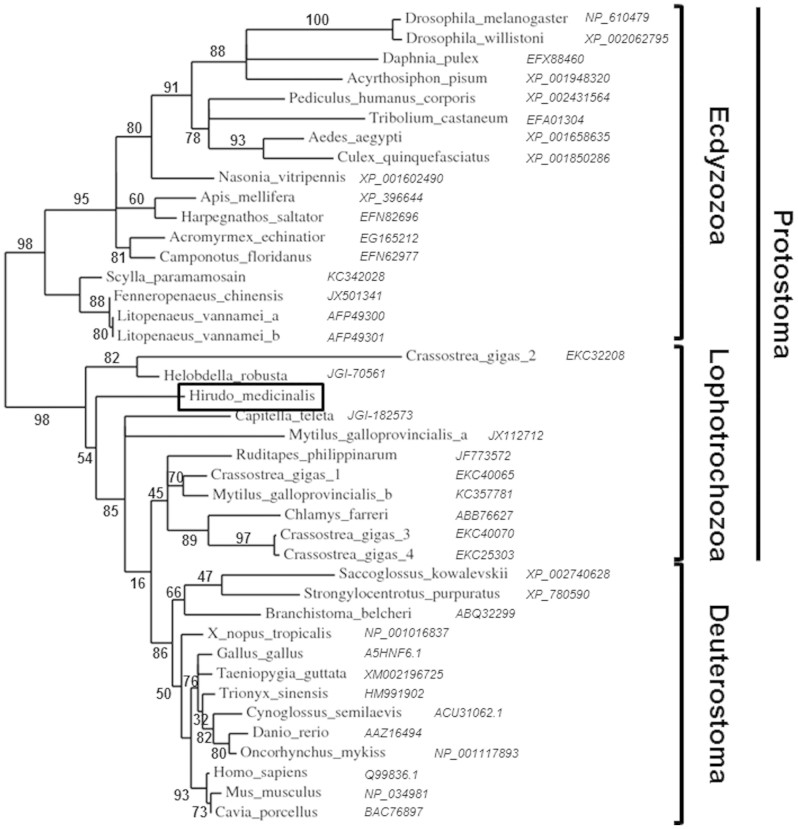
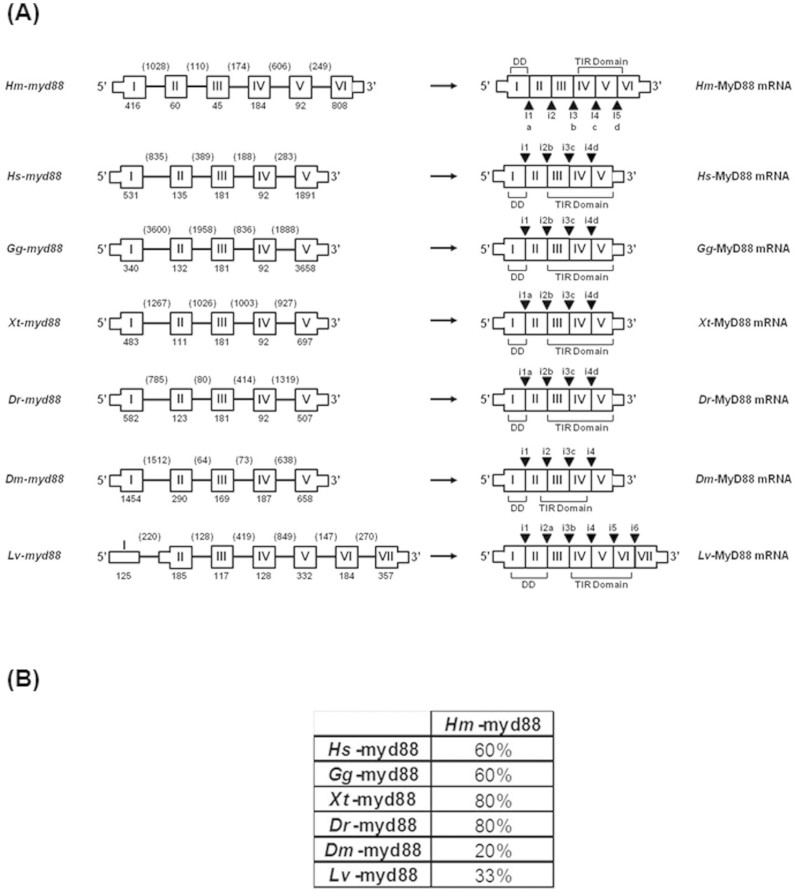
A search in the EMBL/Genbank databanks revealed 36%–18% identity and 53–37% similarity of Hm-MyD88 with vertebrate, protochordate and invertebrate MyD88s (Supplementary Material 1). Surprisingly, Hm-MyD88 shares higher sequence identities and similarities with deuterostomian orthologs than with protostomian MyD88s. Phylogenetic analysis was carried out to investigate the relationship between vertebrate and invertebrate MyD88s (Figure 1). The resulting tree exhibits three distinct groups: the deuterostomian group, the lophotrocozoan group and the ecdyzozoan group. Note that the lophotrocozoan group clusters closer to that of the deuterostomian group. In addition, this analysis showed that annelid MyD88s including Hm-MyD88 emerged early during evolution, before the divergence between Lophotrochozoa and Deuterostoma. To deal with this phylogenetic analysis in depth, the structural organization of the Hm-MyD88 gene was compared to those of the vertebrate and ecdyzozoa genes (Figure 2A). This revealed that Hm-myd88 exhibits a different organization. It displays 6 exons and 5 introns while the vertebrate and Drosophila genes have 5 exons and 4 introns and the shrimp gene possesses 7 exons and 6 introns. However, 60 to 80% of exon-intron boundaries in Hm-myd88 occur at the same position as those of vertebrate genes versus only 20 to 33% for the ecdyzozoa genes (Figures 2A, 2B and Supplementary data 3). This again proves the higher phylogenetic proximity of Hm-MyD88 with the deuterostomian orthologs.
Figure 1. Phylogenetic representation of the relationship between Hirudo medicinalis MyD88 (Hm-MyD88), vertebrate MyD88 and invertebrate MyD88.
The tree was built using the phylogeny.fr web server. Numbers correspond to the percentages of bootstrap values over 100 replicates.
Figure 2. Comparison of the MyD88 gene structures in leech, vertebrates and ecdyzozoa.
(A) The structural organization of Hm-myd88 was deduced after the analysis of the leech genomic library. The exons are boxes marked I-VI; I-V or I-VII. Thick boxes represent the coding regions and thin boxes represent the non coding regions. The introns are given as solid lines. The numbers specify the length (bp) of the exons and introns. On the transcripts, black Δ marked i1-i5; i1-i4 or i1-6 indicate the position of the intron-exon junctions. Letters a, b, c and d point out which leech intron-exon junction is conserved in vertebrate or ecdyzozoa genes. For example, i2b in the human gene indicates that the position of the second intron occurs at the same position as the third intron in leech gene. (B) Percentage of conserved intron-exon junctions between leech gene and vertebrate or ecdyzozoa genes.
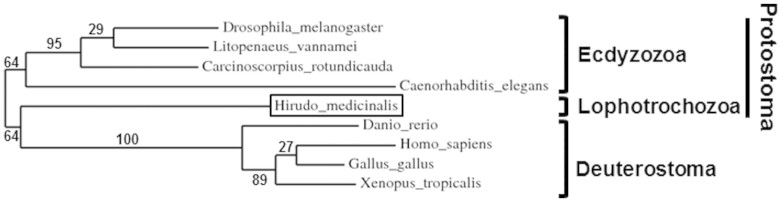
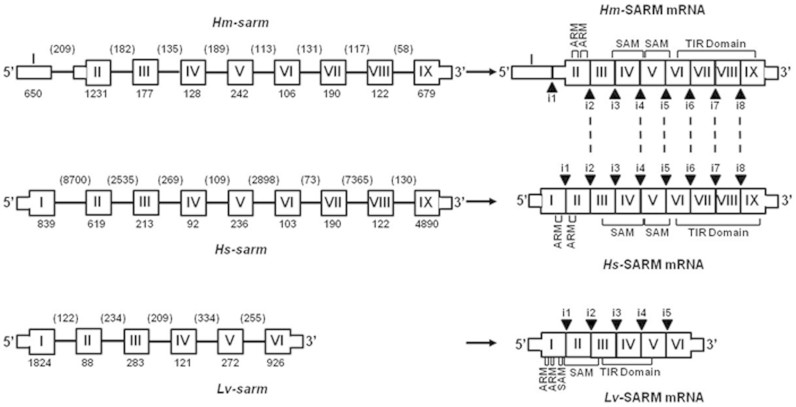
In parallel to the characterization of Hm-MyD88, a partial sequence orthologous to vertebrate and invertebrate SARM was retrieved from the cDNA library of the leech CNS. From this cDNA fragment, a complete sequence encoding Hm-SARM was cloned using 5′ RACE-PCR (Supplementary Material 2). It exhibits the hallmarks of SARM, that is, the two sterile-alpha motif (SAM) domains that are flanked by an N-terminal Armadillo motif (ARM) and a C-terminal TIR domain17. A search in the EMBL/Genbank databanks revealed 40%–23% identity and 55–34% similarity of Hm-Sarm with vertebrate and invertebrate SARM (Supplementary Material 2). Phylogenetic analysis was carried out to investigate the relationship between vertebrate and invertebrate SARM (Figure 3). The resulting tree exhibits three distinct groups as was described for Hm-MyD88: the deuterostomian group, the lophotrocozoan group and the ecdyzozoan group. As in Hm-MyD88, the lophotrocozoan group clusters closer to that of the deuterostomian group. Again, the hypothesis of a higher phylogenetic proximity between these two groups is supported by the study of the gene structure organizations (Figure 4). Indeed, Hm-sarm exhibits the same organization as in the human gene, i.e. 9 exons and 8 introns, while the shrimp gene displays 6 exons and 5 introns. Moroever, 6 of the 8 exon-intron boundaries in Hm-sarm and the human gene occur at the same position whereas none occurs in the shrimp gene (Figures 4 and Supplementary data 4).
Figure 3. Phylogenetic representation of the relationship between Hirudo medicinalis SARM (Hm-SARM), vertebrate SARM and invertebrate SARM.
The tree was built using the phylogeny.fr web server. Numbers correspond to the percentages of bootstrap values over 100 replicates. Accession numbers in EMBL/Genbank: HsSARM: Homo sapiens (Q6SZW1); GgSARM: Gallus gallus (A5HNF6.1); XtSARM: Xenopus tropicalis (XM_002937143.2); DrSARM: Danio rerio (B3DK97); DmSARM: Drosophila melanogaster (Q6IDD9); LvSARM: Litopenaeus vannamei (G8GV23); CrSARM: Carcinoscorpius rotundicauda (A9X3T4); CeTIR-1: Caenorhabditis elegans (Q86DA5).
Figure 4. Comparison of the sarm gene structures in leech, human and shrimp.
The structural organization of Hm-sarm was deduced after analysis of the leech genomic library. The exons are boxes marked I-IX or I-VI. Thick boxes represent the coding regions and thin boxes represent the non coding regions. The introns are given as solid lines. The numbers specify the length (bp) of the exons and introns. On the transcripts, black Δ marked i1-i9 or i1-i5, indicate the position of the intron-exon junctions. Dashed lines point out that the position of an intron-exon junction is the same in leech and human genes.
Expression of Hm-MyD88 and Hm-SARM molecules in leech CNS
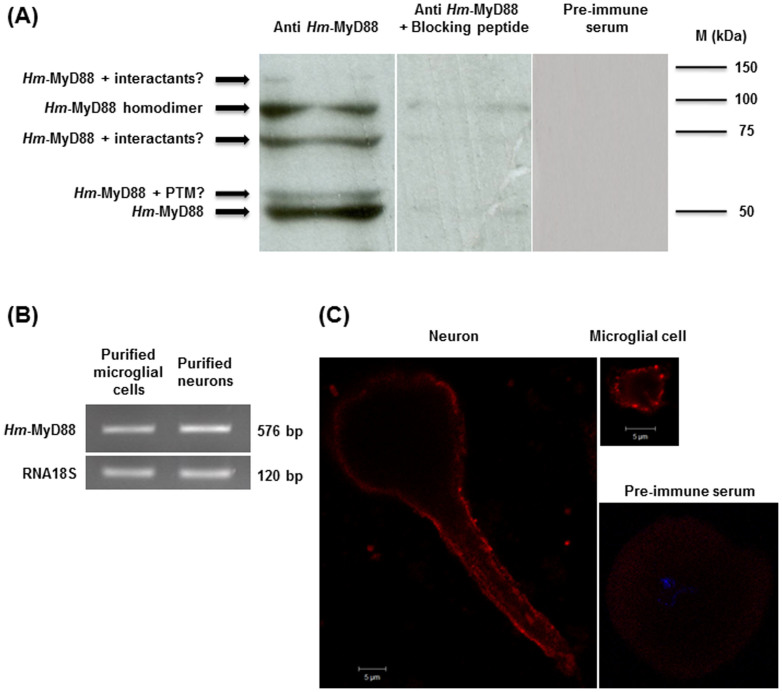
To continue the characterization of Hm-MyD88, a Western blot analysis of leech CNS extracts was performed using an anti-Hm-MyD88 (Figure 5A). To assess the specificity of Hm-MyD88 recognition, two controls were used (i) A reaction with the Hm-MyD88 antibody pre-incubated with the blocking peptide and (ii) a reaction with the pre-immune serum. Five specific bands were detected. The most intense band at 45,4 kDa is consistent with the predicted mass deduced from Hm-MyD88 cDNA. The upper band at 50 kDa may correspond to Hm-MyD88 bearing post translational modifications such as phosphorylations. Indeed, bioinformatics analysis revealed several potential sites of phosphorylation of serine, threonine and tyrosine residues. In mammals, MyD88 phosphorylation is crucial to allow its recruitment and activation4. Besides, mammalian MyD88 function as homodimers7. The presence of the R-D-x-L-P-G motif (Supplementary Material 1) and a band around 90 kDa suggest that Hm-MyD88 seems also to function as a dimer interface. The two other bands at 75 kDa and 150 kDa could correspond to Hm-MyD88 still bound to its interactants.
Figure 5. Hm-MyD88 is expressed in the central nervous system of the leech.
(A) Western blot analysis of leech CNS extracts with anti-Hm-MyD88. To assess the specificity of anti-Hm-MyD88, two controls were performed: An incubation with Hm-MyD88 antibody pre-incubated with the blocking peptide and a reaction with the pre-immune serum. PTM: Post translational modifications. (B) Detection of Hm-MyD88 transcript by RT-PCR in leech purified neurons and microglial cells. (C) Cellular localization of Hm-MyD88 in neurons and microglia was determined by immunofluorescence and confocal microscopy analyses. No immunolabeling was observed when incubated isolated neurons with the pre-immune serum.
As shown by RT-PCR, Hm-MyD88 transcript was detected in purified neurons and microglial cells in basal conditions (Figure 5B). Cellular localization of Hm-MyD88 in the absence of infection was further investigated in neurons and microglia by immunofluorescence and confocal microscopy analyses (Figure 5C). In microglia, Hm-MyD88 was localized adjacent to the plasma membrane. In neurons, the protein was detected in the cell bodies and along the axons as aggregates in the vicinity of the cell surface. This is consistent with the presence of a CTE that could anchor Hm-MyD88 at the plasma membrane and it corroborates the results of the Western blot. In Hela cells, MyD88 associated with beta-actin46. Interestingly, some Hm-MyD88 positive fibers were also revealed along the axons Thus, it may also interact with beta-actin. This suggests the antero-grade transport of this adapter at the cut-ends of the axons or its internalization.
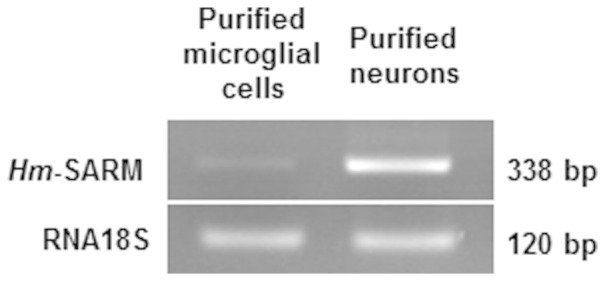
The pattern of expression of Hm-SARM in leech CNS was also carried out by RT-PCR (Figure 6). Interestingly, SARM transcripts were strongly detected in neurons and were only slightly expressed in microglial cells. This is reminiscent of the situation found in mouse brain20.
Figure 6. Detection of Hm-SARM transcript by RT-PCR in leech purified neurons and microglial cells.
Modulation of Hm-myd88 and Hm-sarm gene expression upon microbial challenge of the leech CNS
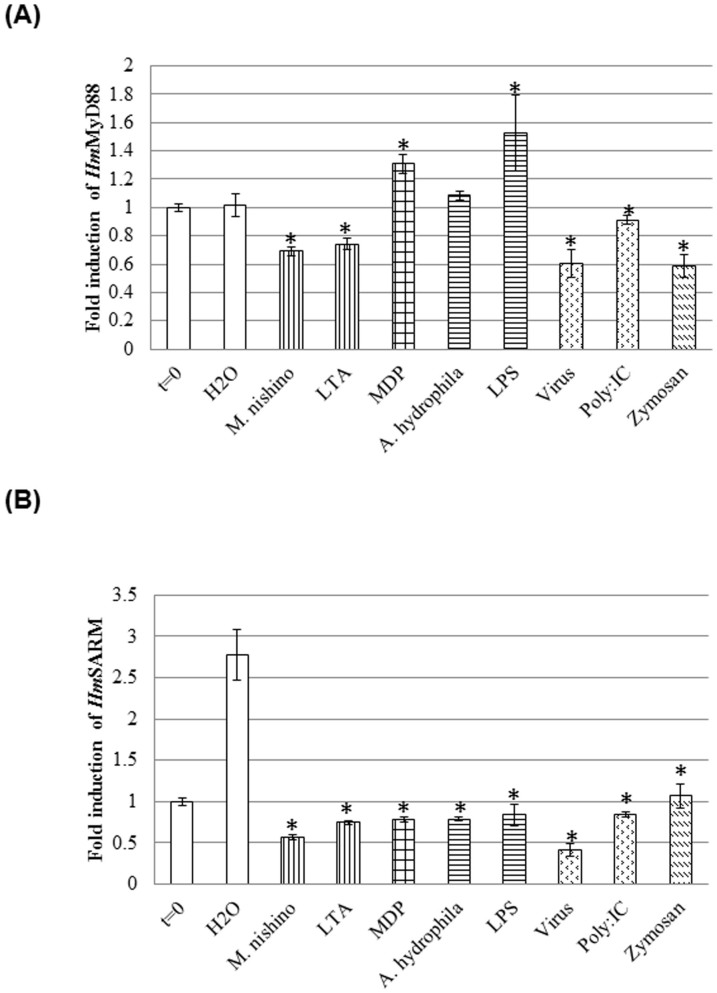
We previously showed that the leech nerve cord is able to discriminate microbial components present in its environment29. Indeed, the presence of various components that derive from or mimic the presence of different microorganisms e.g. bacteria, fungi or viruses, modulated the expression of the genes encoding various PRRs, AMPs and chemokines29,30,31. To assess if such experimental infections also modulate the gene expression patterns of Hm-MyD88 and Hm-SARM, quantitative RT-PCR experiments were carried out (Figure 7A). As described earlier30,34,40, incubation for 6 h was chosen to distinguish the effects due to the presence of microorganisms from those related to the trauma itself. In agreement with that observation, no induction of Hm-myd88 expression was measured 6 h following dissection under sterile conditions. These experiments also revealed that Hm-myd88 expression was strongly and differently modulated according to the immune challenge applied. Indeed, Hm-MyD88 gene expression was induced by LPS and MDP. On the contrary, it was repressed during stimulation with Gram positive bacteria, viruses and yeast. Unlike Hm-myd88, Hm-sarm was only and strongly expressed 6 h following dissection under sterile conditions (Figure 7B). Moreover, when H. medicinalis nerve cords were experimentally infected by various pathogens and microbial components, the up-regulation of Hm-sarm was abolished and quite contrarily its expression was repressed.
Figure 7. Modulation of expression upon experimental microbial challenges.
The modulation of (A) Hm-MyD88 and (B) Hm-SARM gene expression were investigated in nerve cords incubated for 6 h with various microbial components (LTA: lipoteichoïc acid; MDP: muramyl dipeptide; LPS: lipopolysaccharides; zymosan), killed microorganisms (M.nishino: Micrococcus nishinomiyaensis; A. hydrophila: Aeromonas hydrophila; VSV: Vesicular stomatitis virus) or viral mimetic poly(I:C). Graphics represent the best results of two independent experiments that displayed similar variations; p-values from Student's T-tests were calculated versus the control treatment (H2O), based on experimental measurements performed in triplicate (*p < 0.05).
Leech neuronal response to lipopolysaccharide (LPS) may involve an Hm-MyD88-dependent signaling pathway
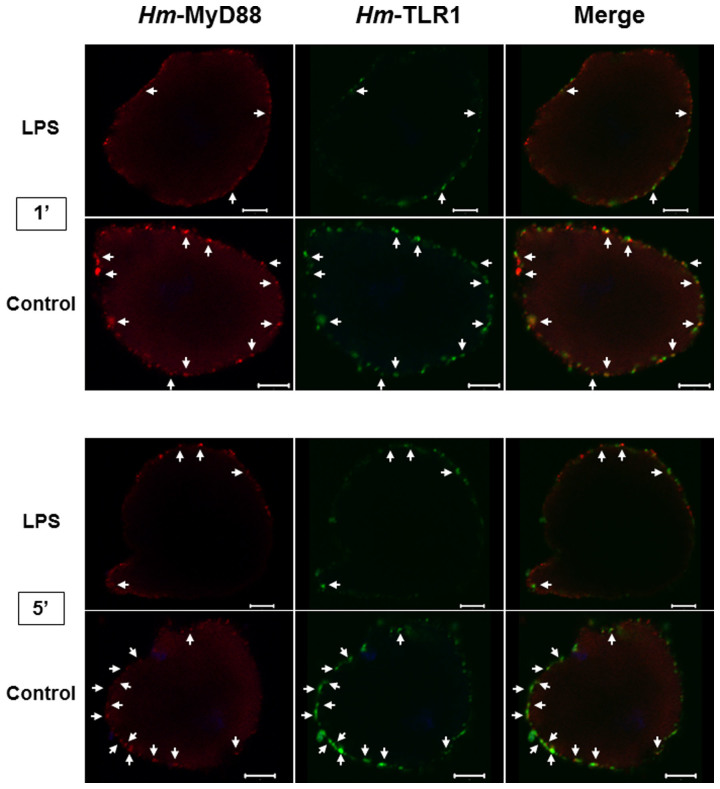
Quantitative RT-PCR results suggested that the MyD88 pathway may be conserved in leech CNS challenged with LPS. Therefore, to determine if leech neurons could indeed respond to LPS through Hm-MyD88, isolated neurons were challenged with LPS for 1 and 5 minutes and labelled with an anti-Hm-MyD88 (Figure 8). A detection of the endosomal Hm-TLR130 was also carried out to assess the specificity of the reaction. Hm-TLR1 is intracellular and thus unlikely to respond directly to LPS and to launch a subsequent Hm-MyD88-dependent signalling pathway.
Figure 8. Neurons could respond to lipopolysaccharide (LPS) through a Hm-MyD88 dependent signaling pathway.
Dissociated neurons were incubated with 100 ng/mL LPS for 1 and 5 minutes and double-labeled with anti-Hm-MyD88 (red) and anti-Hm-TLR1 (green). Untreated neurons served as controls. Arrows indicate regions where Hm-MyD88 and Hm-TLR1 show partial co-localization. Scale bar = 5 μm.
In untreated neurons, Hm-MyD88 were detected as aggregates in the vicinity of the cell surface and in close localization with Hm-TLR1 (arrows). In contrast, Hm-MyD88 labelling was more diffuse in neurons treated with LPS for 1 and 5 minutes. As expected, co-localization between Hm-MyD88 and Hm-TLR1 disappeared. The movement of this adaptor in such a short period of time strongly suggests its recruitment by an uncharacterized receptor involved in LPS recognition. It also suggests that the response of leech neurons to LPS may involve an Hm-MyD88-dependent signaling pathway.
Muramyl dipeptide (MDP) induces a redistribution of Hm-TLR1 and Hm-MyD88 in leech isolated neurons
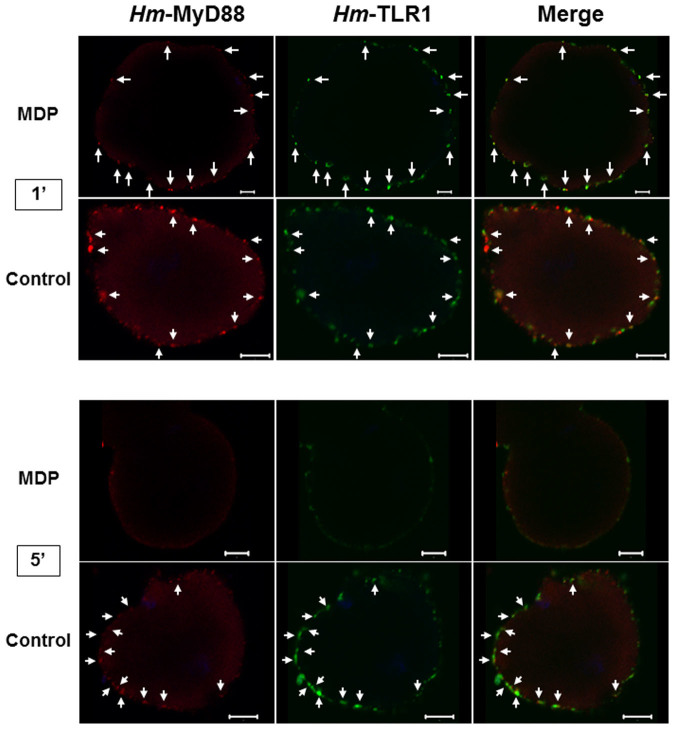
Several studies support the role of muramyl dipeptide (MDP) in the process of neuro-immune modulation47,48,49. In mammals, MDP is a ligand for the intracellular sensor NOD2, a member of the NLR family. The cooperation between NOD2 and TLR is now clearly demonstrated in various cell types including microglia50,51,52. Interestingly, stimulation of leech CNS with MDP induced the expression of the genes encoding Hm-TLR1, the cytokine Hm-EMAPII40 and Hm-MyD88 (Figure 7A). It is to be noted that, during a bacterial challenge, Hm-emapii is under the control of Hm-TLR1 signaling pathway. This raises the likehood that leech CNS is primed by MDP and exhibits a NLR signaling pathway. In addition, these results suggest that cooperation between NLR and TLR also exists. Thus, to test a putative MDP priming effect on leech TLR signaling pathways, isolated neurons were challenged with MDP for 1 and 5 minutes and double-labeled with anti-Hm-MyD88 and anti-Hm-TLR1 (Figure 9). The controls used were the same as those presented for the LPS challenge.
Figure 9. Muramyl dipeptide (MDP) induces a redistribution of Hm-MyD88 and Hm-TLR1 in neurons.
Dissociated neurons were incubated with 10 μg/mL MDP for 1 and 5 minutes and double-labeled with anti-Hm-MyD88 (red) and anti-Hm-TLR1 (green). Untreated neurons served as controls. Arrows indicate regions where Hm-MyD88 and Hm-TLR1 show partial co-localization. Scale bar = 5 μm.
Compared with untreated neurons, Hm-MyD88 and Hm-TLR1 underwent a potent redistribution when the cells were treated with MDP. At 1′, some aggregates were still apparent for both proteins. Interestingly, significant co-localization between the receptor and the adaptor were still observed (arrows). From 1′ to 5′, the movement of Hm-MyD88 and Hm-TLR1 continued and at 5′ the two proteins were hardly detectable. This reveals that MDP priming on leech CNS affected not only delayed physiological processes (e.g. genes expression) but also early responses such as the movement of key proteins involved in TLR signalling pathways. This also supports the idea that the cooperation between leech NLR sensor and leech TLR may be a general characteristic of leech CNS immune response.
Leech CNS repair may involve activation of Hm-MyD88 and Hm-SARM pathways
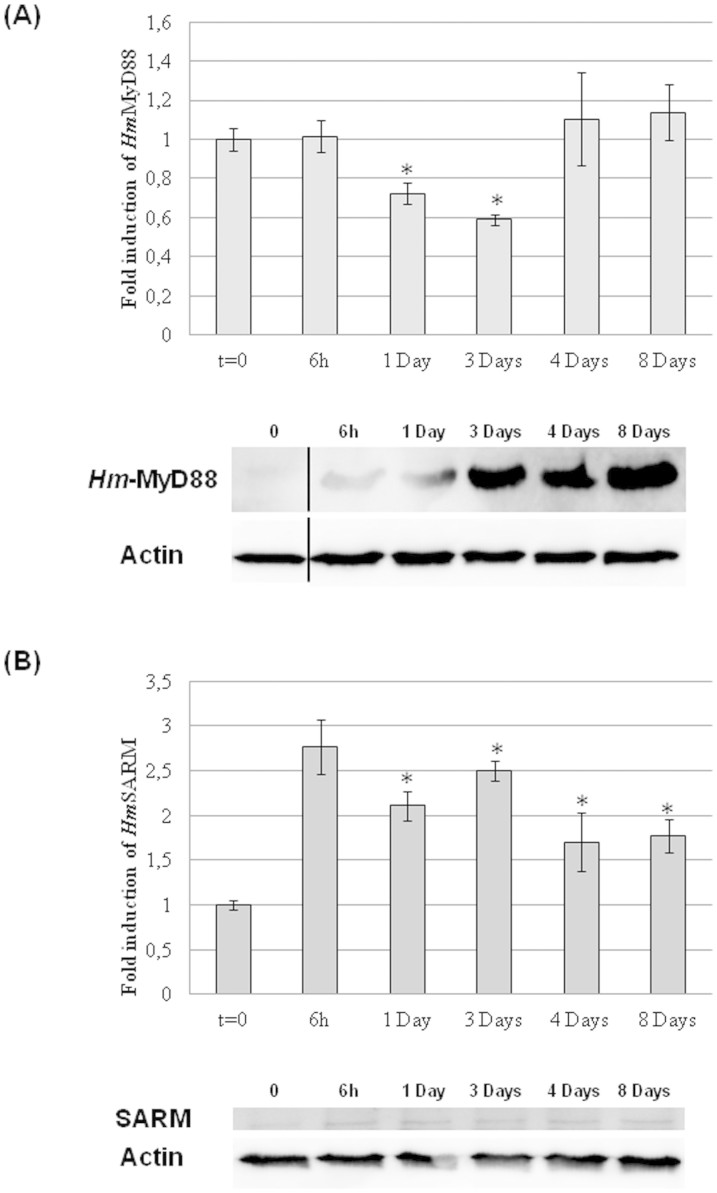
The medicinal leech is able to regenerate its CNS after a lesion. Our previous studies have shown that this regenerative process was enhanced when the nerve cord was also facing a microbial challenge34. This was linked to the release of antimicrobial peptides exhibiting neurotrophic properties. The dual role of these molecules in leech CNS immunity and repair raises the likelihood that common signaling pathways are triggered after an infection or lesion. This was confirmed by our quantitative RT-PCR and western blot experiments performed in conditions of neural regeneration (Figure 10). For this purpose, nerve cords maintained in culture for up to eight days under sterile conditions were lesioned at T0 by cutting completely through half of a connective nerve that links two adjacent ganglia. The time intervals were chosen in reference to the observations reported by Müller, who demonstrated that synaptic connections and normal functions of axotomized leech neurons were restored eight days after injury53. Cultures were stopped at different time post axotomy. Our real-time quantitative RT-PCR experiment showed that the level of Hm-MyD88 mRNA decreased progressively during the first three days of repair and returned to the basal level on the fourth day and until repair is achieved (Figure 10A). This decrease reflects the synthesis of the protein from pre-existing messengers as revealed by western blot analysis (Figure 10A). Indeed, Hm-MyD88 protein level increased progressively in course of regeneration and peaked when regeneration is achieved. Concerning Hm-SARM, we observed that both mRNA and protein levels increased as soon as six hours and remained elevated all along the repair process. Thus, Hm-sarm gene expression may be induced to maintain constant the amount of Hm-SARM protein and control CNS repair (Figure 10B).
Figure 10. Kinetics of expression in the course of neural regeneration.
(A) Hm-MyD88 and (B) Hm-SARM gene and protein expressions were assessed by qRT-PCR and western blot in isolated nerve cords cultured ex vivo for up to 8 days under sterile conditions. Samples were collected at 0 h (T0), 6 h, 1 day, 3 days and 8 days. qRT-PCR results are presented as graphics. Graphics represent the best results of two independent experiments that displayed similar variations; p-values from Student's T-tests were calculated between the different conditions, based on experimental measurements performed in triplicate (*p < 0.05). For western blot experiments, membranes were incubated with anti-Hm-MyD88 and anti-SARM. To assess that an equal amount of proteins was loaded on gels, membranes were stripped and reprobed with anti-Actin.
Discussion
In the present study, we present the identification of the first lophotrocozoan SARM. We also report for the first time the characterization of MyD88 in the CNS of an invertebrate. To the best of our knowledge, we clearly showed for the first time that differentiated neurons of the CNS could respond to LPS through a MyD88-dependent signaling pathway. Such a recruitment of a MyD88 ortholog after LPS challenge was also never demonstrated in any protostomian or non-mammalian deuterostome. In vertebrates and invertebrates, MyD88 plays a central role in the innate immune system. It is the adaptor molecule in majority of the TLRs and it transmits the signals necessary to mount an efficient immune response. Consequently, stimulation of TLR signaling by various microbial derivatives strongly modulates MyD88 gene expression10,11,13,42,54,55. Our quantitative RT-PCR experiment revealed that the expression of Hm-MyD88 encoding gene was also tightly regulated in leech CNS experimentally infected by various pathogens and microbial components (Figure 7A). Undeniably, its expression was induced during challenges with LPS and MDP, while it was repressed during stimulation with Gram-positive bacteria, viruses and yeast. A survey of leech genome and transcriptome databases reveals the presence of at least five TLRs and several splice variants33. This and the differential pattern of expression of Hm-Myd88 gene suggest that different PRRs are involved to detect these various microbial compounds and to launch an accurate immune response against bacteria, viruses and fungi. This immune reaction will in turn induce the differential secretion of antimicrobial peptides (AMPs) such as neuromacin and Hm-lumbricin34 as well as the release of the chemokine Hm-EMAPII40.
The observed induction of Hm-myd88 after LPS and MDP challenge prompted us to examine the role of this adaptor at the functional level. To achieve this study, isolated neurons were stimulated with LPS or MDP for 1 and 5 minutes and then labeled with an anti-Hm-MyD88 (Figure 8 and 9). We report that stimulation of leech neurons with LPS triggered a redistribution of Hm-MyD88 at the cell surface. This suggests its recruitment by a receptor involved in LPS recognition. It shows for the first time that a non-mammalian CNS could respond to LPS through a MyD88-dependent signaling pathway. In mammals, the effect of LPS challenge on CNS has been tested extensively. However, studies describing the direct effect of LPS on neurons and the outcomes of such treatment are scarce and controversial. Indeed, some works demonstrated that neurons were unresponsive to this PAMP56,57, while other studies depicted their possible involvement as key sensors of LPS to launch CNS inflammation58,59,60. Besides, whereas LPS treatment on cortical neurons caused their fragility61, survival of P60 DRG neurons was not affected58. Moreover, signals mediated by TLR4 in neurons are not well understood and were depicted as non-canonical for a long time62. Nevertheless, it was shown that LPS activated NF-κB in chemosensory neurons from NPJgc63 and P60 DRG neurons58, suggesting the involvement of MyD88-dependent mechanisms. To the best of our knowledge, our study is the first to demonstrate directly that differentiated neurons of the CNS could respond to LPS through a MyD88-dependent signaling pathway. It gives therefore an important clue regarding molecular mechanisms underlying LPS response in differentiated neurons of the CNS. Keeping in mind that the medicinal leech is able to regenerate its CNS and that chemokines favour this repair31, LPS treatment on leech neurons may therefore contribute to a better understanding of the effect of this PAMP on axon growth, neuron survival and inflammatory response.
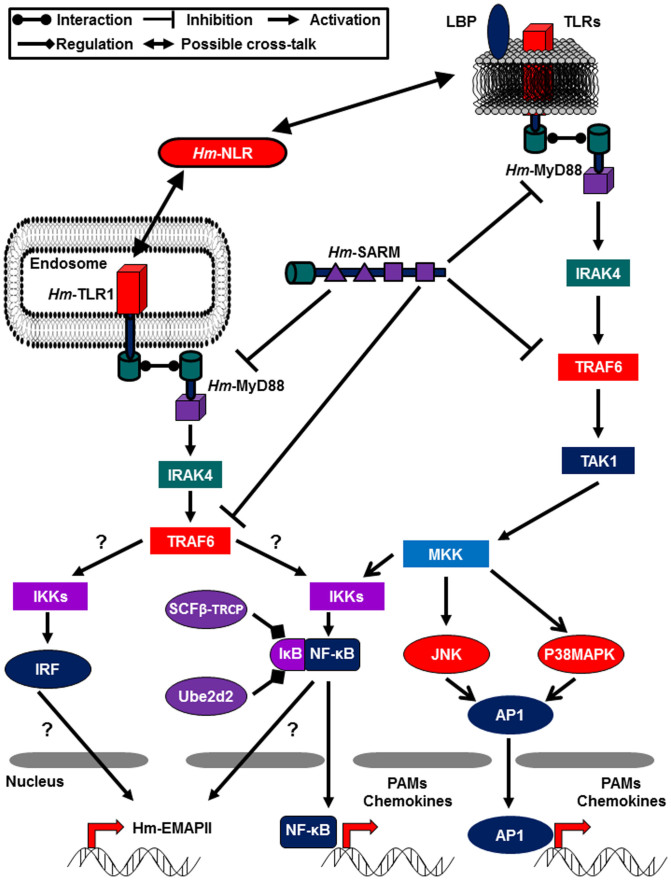
In mammalian CNS, the response to LPS is mediated by TLR4 expressed at the cell surface of astrocytes64, microglia65, and neurons60. Analysis of leech genome and transcriptome databases33 reveals the presence of key molecules implicated in LPS recognition and signalling in mammals, i.e. a TLR closely related to mammalian TLR4, several LBP/BPI, Hm-MyD88, Traf6, IκBα, and two IκBα regulators: the E3 ubiquitin ligase SCFβ-TrCP and the E2 ubiquitin conjugating enzyme Ube2d23,66,67. This suggests that a true TLR4-like pathway could exist in leech (Figure 11). Indeed, apart from mammals and birds68, where the LPS receptor is clearly a TLR, it signals through a TLR-independent pathway in other species9,10,69,70,71. For example, in sponge, LPS is bound by the specific poriferan molecule SLIP to trigger a MyD88-dependent signaling pathway10. The existence of alternative receptors has also been evocated in the horseshoe crab72 and fish73 i.e. factor C and beta-2 integrins, respectively.
Figure 11. The putative TLR4-like and Hm-TLR1 pathways in leech neurons.
The survey of H. medicinalis databases and our experiments30,33 pointed out that leech possesses (i) the main components of the canonical TLR4/LPS pathway and (ii) the molecules recruited by endosomal TLRs. “?” indicates that stimulation of Hm-TLR1 may activate an IRF or a NF-κB signaling.
Data obtained with Muramyl dipeptide (MDP) further suggests the similarity of the leech PRR pathways to its mammalian counterpart. In vertebrate CNS, MDP, the ligand of NOD2, is a potent immune modulator47,48,49 and exerts priming and synergistic effects for TLR ligand activities50,51,52,74. In this line, in human monocytic cells in culture, MDP also up-regulated MyD88 mRNA expression74. Similarly, we showed that stimulation of leech CNS with MDP induced the expression of the genes encoding Hm-TLR130, the cytokine Hm-EMAPII40 and Hm-MyD88 (Figure 7A). Our immunofluorescence experiment also revealed that a stimulation of leech neurons with MDP triggered a potent redistribution of Hm-MyD88 and Hm-TLR1 in the cells (Figure 9). It is the first time that such movements of a TLR and MyD88 are demonstrated in neurons. This raises the likehood that leech CNS is primed by MDP, exhibits a NLR signaling pathway and that cooperation between NLR and TLR exists (Figure 11). Despite the fact that the leech receptor for MDP is still unknown, the recent characterization of Hm-NLR, a NLR-like sensor ortholog, and its common pattern of expression with Hm-TLR1 in neurons support this hypothesis30. This also supports the idea that cooperation between leech NLR sensor and leech TLR may be a general characteristic of leech CNS immune response. However, in leech injured CNS, MDP blocked microglia recruitment to the lesion site29. Thus, MDP may exert a dual effect on leech CNS: (i) prime neurons to ensure an efficient immune response and (ii) control the amount of microglia attracted to avoid an over inflammation deleterious for the nervous system.
Another key mechanism to balance the inflammatory response and avoid an immune signalling storm is to shut down TLR pathways75. Among negative regulators of TLR signalling, SARM is the most conserved across evolution. Indeed, in mammals18,23,24; protochordates19,25 and arthropods16,26, SARM inhibits the TLR signaling pathways specifically via MyD88 and/or TRIF and/or TRAF6. To control this precise switch between pathway activation and repression, the time-course of SARM expression is tightly regulated in immune cells during an immune challenge16,18,19,23,24,26. Our quantitative RT-PCR experiment revealed that the levels of Hm-SARM messengers significantly diminished in leech nerve cords experimentally infected during 6 hours by various pathogens and microbial components (Figure 7B). At this time point, we have previously demonstrated that H. medicinalis CNS released beneficial immune factors to mount an immune response and restore homeostasis29. Therefore, we may expect Hm-SARM to be synthesized from a pre-existing pool of messengers to slow down TLR pathways by its interaction with Hm-MyD88 and/or Hm-TRAF633. This may adjust the amount of AMPs and chemokines secreted. However, to prevent Hm-SARM from abolishing TLRs signalling, its encoding gene is repressed to avoid a subsequent synthesis of the protein. On the contrary, we can't exclude that Hm-SARM is required to initiate an innate immune response in leech CNS infected with various pathogens as it was observed in C. elegans challenged with bacteria and fungi15,27 and during a viral infection of mammalian brain28,76. Thus, the down-regulation of leech Hm-SARM gene may suppress excessive innate immunity and prevent tissue damage. Indeed, in brains of mice experimentally infected with vesicular stomatitis virus, an excessive inflammatory response involving SARM signaling led to neuronal injury76.
Moreover, the present study demonstrated that a tight link between CNS repair and neuroimmunity exists in this lophotrochozoan model. We showed that the amount of Hm-MyD88 mRNA decreased progressively in course of regeneration while the protein level increased and peaked when regeneration is achieved (Figure 10A). Hm-MyD88 pathway may favour the regeneration by stimulating the synthesis of antimicrobial peptides as well as chemokines like Hm-EMAPII known to be critical for regeneration processes29,31,32,34,40,77. Concerning Hm-SARM, we observed that its gene is induced all along the repair process in order to sustain the protein synthesis (Figure 10B). In mammals, SARM regulates the morphology of hippocampal neurons78 and controls their death during metabolic stress20. It also activates an injury-induced axon death pathway known as Wallerian degeneration in mice and drosophila79. In leech, isolated neurons such as Retzius neurons and anterior pagoda neurons start sprouting a few minutes after being plated80 and neurite growth is not random81. In accordance with the literature mentioned above, we may expect Hm-SARM to control the complex dynamics of neurite elongation and retraction as well as the elimination of misdirected sprouts observed during axon growth80,82.
Taken together, our data revealed that Hm-MyD88 and Hm-SARM pathways may fully contribute to set a balance between stimulatory and inhibitory signals in leech to ensure CNS repair and defense.
Accession codes:
Hm-MyD88 mRNA, Hm-SARM mRNA, Hm-myd88 gene and Hm-sarm gene sequences were deposited in the NCBI GenBank under accession no. KM233119; KM233120; KM233121 and KM233122 respectively.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by grants from Le Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche (MENESR, France), the Centre National de la Recherche Scientifique (CNRS), l'Université de Lille 1 and from L'Agence Nationale de la Recherche (ANR-11-JSV4-005). The authors are indebted to Dr. J. Quanico for carefully reading the manuscript and to Elodie Richard of the CCMIC–Université de Lille 1 (BiCel) for acces to confocal microscopy facilities.
Footnotes
The authors declare no competing financial interests.
Author Contributions F.R. has done experiments and has written the paper. A.T., C.B.W., C.V.C., C.V. and C.S. have done experiments. M.S. has got financial support to the project and corrected the manuscript. All authors have reviewed the manuscript.
References
- Medzhitov R. & Janeway C. Innate immune recognition: mechanisms and pathways. Immunol. Rev. 173, 89–97 (2000). [DOI] [PubMed] [Google Scholar]
- Kawai T. & Akira S. Signaling to NF-kappaB by Toll-like receptors. Trends Mol. Med. 13, 460–9 (2007). [DOI] [PubMed] [Google Scholar]
- Kawai T. & Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat. Immunol. 11, 373–84 (2010). [DOI] [PubMed] [Google Scholar]
- O'Neill L. A. J. & Bowie A. G. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat. Rev. Immunol. 7, 353–64 (2007). [DOI] [PubMed] [Google Scholar]
- Burns K. et al. Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J. Exp. Med. 197, 263–8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiarro M. et al. Identification of critical residues of the MyD88 death domain involved in the recruitment of downstream kinases. J. Biol. Chem. 284, 28093–103 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiarro M. et al. Peptide-mediated interference of TIR domain dimerization in MyD88 inhibits interleukin-1-dependent activation of NF-{kappa}B. J. Biol. Chem. 280, 15809–14 (2005). [DOI] [PubMed] [Google Scholar]
- Medzhitov R. et al. MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2, 253–8 (1998). [DOI] [PubMed] [Google Scholar]
- Tauszig-Delamasure S., Bilak H., Capovilla M., Hoffmann J. A. & Imler J.-L. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat. Immunol. 3, 91–7 (2002). [DOI] [PubMed] [Google Scholar]
- Wiens M. et al. Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway. Induction of a perforin-like molecule. J. Biol. Chem. 280, 27949–59 (2005). [DOI] [PubMed] [Google Scholar]
- Qiu L. et al. Identification and characterization of a myeloid differentiation factor 88 (MyD88) cDNA from Zhikong scallop Chlamys farreri. Fish Shellfish Immunol. 23, 614–23 (2007). [DOI] [PubMed] [Google Scholar]
- Yuan S. et al. An amphioxus TLR with dynamic embryonic expression pattern responses to pathogens and activates NF-kappaB pathway via MyD88. Mol. Immunol. 46, 2348–56 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang S. et al. Identification and function of myeloid differentiation factor 88 (MyD88) in Litopenaeus vannamei. PLoS One 7, e47038 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzenburg S. et al. MyD88-deficient Hydra reveal an ancient function of TLR signaling in sensing bacterial colonizers. Proc. Natl. Acad. Sci. U. S. A. 109, 19374–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couillault C. et al. TLR-independent control of innate immunity in Caenorhabditis elegans by the TIR domain adaptor protein TIR-1, an ortholog of human SARM. Nat. Immunol. 5, 488–94 (2004). [DOI] [PubMed] [Google Scholar]
- Belinda L. W.-C. et al. SARM: a novel Toll-like receptor adaptor, is functionally conserved from arthropod to human. Mol. Immunol. 45, 1732–42 (2008). [DOI] [PubMed] [Google Scholar]
- Jenkins K. A. & Mansell A. TIR-containing adaptors in Toll-like receptor signalling. Cytokine 49, 237–44 (2010). [DOI] [PubMed] [Google Scholar]
- Zhou X. et al. Molecular characterization of porcine SARM1 and its role in regulating TLRs signaling during highly pathogenic porcine reproductive and respiratory syndrome virus infection in vivo. Dev. Comp. Immunol. 39, 117–26 (2013). [DOI] [PubMed] [Google Scholar]
- Yuan S. et al. Amphioxus SARM involved in neural development may function as a suppressor of TLR signaling. J. Immunol. 184, 6874–81 (2010). [DOI] [PubMed] [Google Scholar]
- Kim Y. et al. MyD88-5 links mitochondria, microtubules, and JNK3 in neurons and regulates neuronal survival. J. Exp. Med. 204, 2063–74 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C., Hsieh Y.-W., Lesch B. J., Bargmann C. I. & Chuang C.-F. Microtubule-based localization of a synaptic calcium-signaling complex is required for left-right neuronal asymmetry in C. elegans. Development 138, 3509–18 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panneerselvam P. et al. T-cell death following immune activation is mediated by mitochondria-localized SARM. Cell Death Differ. 20, 478–89 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carty M. et al. The human adaptor SARM negatively regulates adaptor protein TRIF-dependent Toll-like receptor signaling. Nat. Immunol. 7, 1074–81 (2006). [DOI] [PubMed] [Google Scholar]
- Peng J. et al. SARM inhibits both TRIF- and MyD88-mediated AP-1 activation. Eur. J. Immunol. 40, 1738–47 (2010). [DOI] [PubMed] [Google Scholar]
- Yang M. et al. Characterization of bbtTICAM from amphioxus suggests the emergence of a MyD88-independent pathway in basal chordates. Cell Res. 21, 1410–23 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.-H. et al. Litopenaeus vannamei sterile-alpha and armadillo motif containing protein (LvSARM) is involved in regulation of Penaeidins and antilipopolysaccharide factors. PLoS One 8, e52088 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberati N. T. et al. Requirement for a conserved Toll/interleukin-1 resistance domain protein in the Caenorhabditis elegans immune response. Proc. Natl. Acad. Sci. U. S. A. 101, 6593–8 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szretter K. J. et al. The immune adaptor molecule SARM modulates tumor necrosis factor alpha production and microglia activation in the brainstem and restricts West Nile Virus pathogenesis. J. Virol. 83, 9329–38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasiemski A. & Salzet M. Leech immunity: from brain to peripheral responses. Adv. Exp. Med. Biol. 708, 80–104 (2010). [DOI] [PubMed] [Google Scholar]
- Cuvillier-Hot V., Boidin-Wichlacz C., Slomianny C., Salzet M. & Tasiemski A. Characterization and immune function of two intracellular sensors, HmTLR1 and HmNLR, in the injured CNS of an invertebrate. Dev. Comp. Immunol. 35, 214–26 (2011). [DOI] [PubMed] [Google Scholar]
- Le Marrec-Croq F., Drago F., Vizioli J., Sautière P.-E. & Lefebvre C. The leech nervous system: a valuable model to study the microglia involvement in regenerative processes. Clin. Dev. Immunol. 2013, 274019 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boidin-Wichlacz C. et al. Morphological and functional characterization of leech circulating blood cells: role in immunity and neural repair. Cell. Mol. Life Sci. 69, 1717–31 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macagno E. R. et al. Construction of a medicinal leech transcriptome database and its application to the identification of leech homologs of neural and innate immune genes. BMC Genomics 11, 407 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikorski D. et al. Microbial challenge promotes the regenerative process of the injured central nervous system of the medicinal leech by inducing the synthesis of antimicrobial peptides in neurons and microglia. J. Immunol. 181, 1083–95 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz J., Milpetz F., Bork P. & Ponting C. P. SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U. S. A. 95, 5857–64 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A. et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36, W465–9 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahtouh M. et al. Evidence for a novel chemotactic C1q domain-containing factor in the leech nerve cord. Mol. Immunol. 46, 523–31 (2009). [DOI] [PubMed] [Google Scholar]
- Koressaar T. & Remm M. Enhancements and modifications of primer design program Primer3. Bioinformatics 23, 1289–91 (2007). [DOI] [PubMed] [Google Scholar]
- Untergasser A. et al. Primer3--new capabilities and interfaces. Nucleic Acids Res. 40, e115 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schikorski D. et al. Deciphering the immune function and regulation by a TLR of the cytokine EMAPII in the lesioned central nervous system using a leech model. J. Immunol. 183, 7119–28 (2009). [DOI] [PubMed] [Google Scholar]
- Xu Y. et al. Structural basis for signal transduction by the Toll/interleukin-1 receptor domains. Nature 408, 111–5 (2000). [DOI] [PubMed] [Google Scholar]
- Toubiana M. et al. Toll-like receptors and MyD88 adaptors in Mytilus: complete cds and gene expression levels. Dev. Comp. Immunol. 40, 158–66 (2013). [DOI] [PubMed] [Google Scholar]
- Horng T. & Medzhitov R. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc. Natl. Acad. Sci. U. S. A. 98, 12654–8 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X.-C. et al. A novel myeloid differentiation factor 88 homolog, SpMyD88, exhibiting SpToll-binding activity in the mud crab Scylla paramamosain. Dev. Comp. Immunol. 39, 313–22 (2013). [DOI] [PubMed] [Google Scholar]
- Marek L. R. & Kagan J. C. Phosphoinositide binding by the Toll adaptor dMyD88 controls antibacterial responses in Drosophila. Immunity 36, 612–22 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaunin F., Burns K., Tschopp J., Martin T. E. & Fakan S. Ultrastructural distribution of the death-domain-containing MyD88 protein in HeLa cells. Exp. Cell Res. 243, 67–75 (1998). [DOI] [PubMed] [Google Scholar]
- Dougherty P. M. & Dafny N. Microiontophoretic application of muramyl-dipeptide upon single cortical, hippocampal and hypothalamic neurons in rats. Neuropharmacology 29, 973–81 (1990). [DOI] [PubMed] [Google Scholar]
- Dougherty P. M. & Dafny N. Muramyl-dipeptide, a macrophage-derived cytokine, alters neuronal activity in hypothalamus and hippocampus but not in the dorsal raphe/periaqueductal gray of rats. J. Neuroimmunol. 28, 201–8 (1990). [DOI] [PubMed] [Google Scholar]
- Plata-Salamán C. R., Ilyin S. E., Gayle D. & Flynn M. C. Gram-negative and gram-positive bacterial products induce differential cytokine profiles in the brain: analysis using an integrative molecular-behavioral in vivo model. Int. J. Mol. Med. 1, 387–97 (1998). [DOI] [PubMed] [Google Scholar]
- Sterka D. & Marriott I. Characterization of nucleotide-binding oligomerization domain (NOD) protein expression in primary murine microglia. J. Neuroimmunol. 179, 65–75 (2006). [DOI] [PubMed] [Google Scholar]
- Chauhan V. S., Sterka D. G., Furr S. R., Young A. B. & Marriott I. NOD2 plays an important role in the inflammatory responses of microglia and astrocytes to bacterial CNS pathogens. Glia 57, 414–23 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K. et al. Cooperation between NOD2 and Toll-like receptor 2 ligands in the up-regulation of mouse mFPR2, a G-protein-coupled A 42 peptide receptor, in microglial cells. J. Leukoc. Biol. 83, 1467–1475 (2008). [DOI] [PubMed] [Google Scholar]
- Muller K. J. & Carbonetto S. The morphological and physiological properties of a regenerating synapse in the C.N.S. of the leech. J. Comp. Neurol. 185, 485–516 (1979). [DOI] [PubMed] [Google Scholar]
- Liu Y. et al. A unique feature of Toll/IL-1 receptor domain-containing adaptor protein is partially responsible for lipopolysaccharide insensitivity in zebrafish with a highly conserved function of MyD88. J. Immunol. 185, 3391–400 (2010). [DOI] [PubMed] [Google Scholar]
- Ding X. et al. Orange-spotted grouper (Epinephelus coioides) toll-like receptor 22: molecular characterization, expression pattern and pertinent signaling pathways. Fish Shellfish Immunol. 33, 494–503 (2012). [DOI] [PubMed] [Google Scholar]
- Tang S.-C. et al. Pivotal role for neuronal Toll-like receptors in ischemic brain injury and functional deficits. Proc. Natl. Acad. Sci. U. S. A. 104, 13798–803 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun E. et al. Toll-like receptors in neurodegeneration. Brain Res. Rev. 59, 278–92 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta C. & Davies A. Bacterial lipopolysaccharide regulates nociceptin expression in sensory neurons. J. Neurosci. Res. 86, 1077–86 (2008). [DOI] [PubMed] [Google Scholar]
- Diogenes A., Ferraz C. C. R., Akopian A. N., Henry M. A. & Hargreaves K. M. LPS sensitizes TRPV1 via activation of TLR4 in trigeminal sensory neurons. J. Dent. Res. 90, 759–64 (2011). [DOI] [PubMed] [Google Scholar]
- Leow-Dyke S. et al. Neuronal Toll-like receptor 4 signaling induces brain endothelial activation and neutrophil transmigration in vitro. J. Neuroinflammation 9, 230 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen K. et al. TLR-4-dependent and -independent mechanisms of fetal brain injury in the setting of preterm birth. Reprod. Sci. 19, 839–50 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okun E., Griffioen K. J. & Mattson M. P. Toll-like receptor signaling in neural plasticity and disease. Trends Neurosci. 34, 269–81 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández R. et al. Lipopolysaccharide signaling in the carotid chemoreceptor pathway of rats with sepsis syndrome. Respir. Physiol. Neurobiol. 175, 336–48 (2011). [DOI] [PubMed] [Google Scholar]
- Gorina R., Font-Nieves M., Márquez-Kisinousky L., Santalucia T. & Planas A. M. Astrocyte TLR4 activation induces a proinflammatory environment through the interplay between MyD88-dependent NFκB signaling, MAPK, and Jak1/Stat1 pathways. Glia 59, 242–55 (2011). [DOI] [PubMed] [Google Scholar]
- Esen N. & Kielian T. Central role for MyD88 in the responses of microglia to pathogen-associated molecular patterns. J. Immunol. 176, 6802–11 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O. & Akira S. Pattern recognition receptors and inflammation. Cell 140, 805–20 (2010). [DOI] [PubMed] [Google Scholar]
- De Arras L. et al. An evolutionarily conserved innate immunity protein interaction network. J. Biol. Chem. 288, 1967–78 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukui A. et al. Molecular cloning and functional characterization of chicken toll-like receptors. A single chicken toll covers multiple molecular patterns. J. Biol. Chem. 276, 47143–9 (2001). [DOI] [PubMed] [Google Scholar]
- Sun H., Bristow B. N., Qu G. & Wasserman S. A. A heterotrimeric death domain complex in Toll signaling. Proc. Natl. Acad. Sci. U. S. A. 99, 12871–6 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepulcre M. P. et al. Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-kappaB activation. J. Immunol. 182, 1836–45 (2009). [DOI] [PubMed] [Google Scholar]
- Aballay A., Drenkard E., Hilbun L. R. & Ausubel F. M. Caenorhabditis elegans innate immune response triggered by Salmonella enterica requires intact LPS and is mediated by a MAPK signaling pathway. Curr. Biol. 13, 47–52 (2003). [DOI] [PubMed] [Google Scholar]
- Inamori K., Ariki S. & Kawabata S. A Toll-like receptor in horseshoe crabs. Immunol. Rev. 198, 106–15 (2004). [DOI] [PubMed] [Google Scholar]
- Iliev D. B., Roach J. C., Mackenzie S., Planas J. V. & Goetz, F. W. Endotoxin recognition: in fish or not in fish? FEBS Lett. 579, 6519–28 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. et al. Synergistic effect of muramyldipeptide with lipopolysaccharide or lipoteichoic acid to induce inflammatory cytokines in human monocytic cells in culture. Infect. Immun. 69, 2045–53 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liew F. Y., Xu D., Brint E. K. & O'Neill L. A. J. Negative regulation of toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5, 446–58 (2005). [DOI] [PubMed] [Google Scholar]
- Hou Y.-J. et al. SARM is required for neuronal injury and cytokine production in response to central nervous system viral infection. J. Immunol. 191, 875–83 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arafah K. et al. Involvement of nitric oxide through endocannabinoids release in microglia activation during the course of CNS regeneration in the medicinal leech. Glia 61, 636–649 (2013). [DOI] [PubMed] [Google Scholar]
- Chen C.-Y., Lin C.-W., Chang C.-Y., Jiang S.-T. & Hsueh Y.-P. Sarm1, a negative regulator of innate immunity, interacts with syndecan-2 and regulates neuronal morphology. J. Cell Biol. 193, 769–84 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterloh J. M. et al. dSarm/Sarm1 is required for activation of an injury-induced axon death pathway. Science 337, 481–4 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- De-Miguel F. F. Steps in the formation of neurites and synapses studied in cultured leech neurons. Braz. J. Med. Biol. Res. 33, 487–97 (2000). [DOI] [PubMed] [Google Scholar]
- Chiquet M. & Nicholls J. G. Neurite outgrowth and synapse formation by identified leech neurones in culture. J. Exp. Biol. 132, 191–206 (1987). [DOI] [PubMed] [Google Scholar]
- Muller K. J., McGlade-McCulloh E. & Mason A. Tinkering with successful synapse regeneration in the leech: adding insult to injury. J. Exp. Biol. 132, 207–21 (1987). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information