Abstract
A variety of techniques for strain engineering in Saccharomyces cerevisiae have recently been developed. However, especially when multiple genetic manipulations are required, strain construction is still a time-consuming process. This study describes new CRISPR/Cas9-based approaches for easy, fast strain construction in yeast and explores their potential for simultaneous introduction of multiple genetic modifications. An open-source tool (http://yeastriction.tnw.tudelft.nl) is presented for identification of suitable Cas9 target sites in S. cerevisiae strains. A transformation strategy, using in vivo assembly of a guideRNA plasmid and subsequent genetic modification, was successfully implemented with high accuracies. An alternative strategy, using in vitro assembled plasmids containing two gRNAs, was used to simultaneously introduce up to six genetic modifications in a single transformation step with high efficiencies. Where previous studies mainly focused on the use of CRISPR/Cas9 for gene inactivation, we demonstrate the versatility of CRISPR/Cas9-based engineering of yeast by achieving simultaneous integration of a multigene construct combined with gene deletion and the simultaneous introduction of two single-nucleotide mutations at different loci. Sets of standardized plasmids, as well as the web-based Yeastriction target-sequence identifier and primer-design tool, are made available to the yeast research community to facilitate fast, standardized and efficient application of the CRISPR/Cas9 system.
Keywords: CRISPR/Cas9; S. cerevisiae, gRNA; genetic modification; webtool; plasmid
CRISPR/Cas9 like a Swiss army knife enables molecular biologists to quickly introduce simultaneous multiple and diverse genetic modifications in baker's yeast Saccharomyces cerevisiae.
Graphical Abstract Figure.
CRISPR/Cas9 like a Swiss army knife enables molecular biologists to quickly introduce simultaneous multiple and diverse genetic modifications in baker's yeast Saccharomyces cerevisiae.
INTRODUCTION
For decades, Saccharomyces cerevisiae has been successfully used as a model organism to decipher biological processes in higher eukaryotes (Botstein and Fink 2011) and as a popular metabolic engineering platform (Nielsen et al. 2013). Expression and optimization of heterologous product pathways in S. cerevisiae (see e.g. Paddon et al. 2013; Beekwilder et al. 2014) requires introduction of multiple (successive) genetic modifications, including integration of product pathway genes at multiple genetic loci and rewiring central metabolism by modifying properties of specific metabolic reactions (e.g. via gene deletion, changing regulatory properties or replacement of native genes by heterologous counterparts) (Wieczorke et al. 1999; Ro et al. 2006). Introduction of the required genetic modifications has so far remained a time-consuming and labour-intensive process, as each individual alteration requires a cycle of transformation, selection and confirmation. Furthermore, since each modification is accompanied by the integration of a selection marker gene, the maximum number of sequential modifications may be limited by selection marker availability. This limitation stimulated extensive research into the identification of novel genetic markers for S. cerevisiae (Chee and Haase 2012; Solis-Escalante et al. 2013; Siewers 2014). Additionally, multiple strategies for the recycling of genetic markers have been developed, such as homologous recombination (HR)-mediated counter selection (gene ‘loop-out’) and use of recombinases such as the Cre/loxP or 2 μm-plasmid-based Flp/FRT methods (Güldener et al. 1996; Storici, Coglievina and Bruschi 1999; Hegemann and Heick 2011). These recombinase-based methods leave a copy of a repeat sequence (e.g. loxP or FRT site) in the genome, which leads to genome instability after multiple, repeated rounds of marker recovery (Delneri et al. 2000; Solis-Escalante et al. 2014b). ‘Scarless’ removal of counter-selectable markers has been made possible via the delitto perfetto method (Storici, Lewis and Resnick 2001), while a recently reported marker-recovery method based on generation of I-SceI-induced double-stranded breaks even allows simultaneous, seamless removal of multiple markers (Solis-Escalante et al. 2014a). While these methods largely eliminate limitations by marker-gene availability, substitution of target genes by marker cassettes remained a time-consuming process, due to the absence of robust methods for simultaneous introduction of multiple genetic modifications in a single transformation step. Alternative methods such as meganucleases, zinc finger nucleases (ZFNs) (Urnov et al. 2010; Carroll 2011) and transcription activator-like effector nucleases (TALENs) (Christian et al. 2010; Miller et al. 2011; Mussolino et al. 2011) utilize double-stranded DNA breaks (DSBs) for site-directed genome editing. Due to the lethal nature of DSBs in yeast, these methods could theoretically be used for marker-free modifications. However, for each genetic modification, a new ZFN or TALEN protein has to be designed and generated.
Bacteria have developed several systems to degrade foreign DNA. Very quickly after their discovery, restriction enzymes became the ‘workhorses of molecular biology’ (reviewed by Roberts 2005). Another prokaryotic immune mechanism, consisting of Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated (Cas) systems, was discovered in 2007 (Barrangou et al. 2007; Brouns et al. 2008; Marraffini and Sontheimer 2008). To function in vivo, the type-II bacterial CRISPR system of Streptococcus pyogenes requires the Cas9 nuclease and the RNA complex that guides it to a specific sequence of the (foreign) DNA. This RNA complex generally consists of two RNA molecules: the CRISPR RNA (crRNA) and the trans-activating CRISPR RNA (tracrRNA). The crRNA contains the 20–30 base pairs (bp) target sequence and a sequence that binds to the tracrRNA, resulting in a duplex RNA complex, recognized by the Cas9 nuclease. When directly after the target sequence a proper protospacer adjacent motif (PAM) is present (in case of the S. pyogenes Cas9 this sequence is NGG), Cas9 will bind to and restrict the target sequence of the (invading) DNA (Deltcheva et al. 2011). A unique feature of this system is the RNA dependence for targeting of the nuclease Cas9, which makes selective targeting of any locus for the introduction of DSBs possible. Since its discovery, Cas9-based systems have been used for the construction of (multiplexed) genetic modifications in a variety of organisms, including human pluripotent stem cells (González et al. 2014), zebrafish (Hwang et al. 2013), plants (Feng et al. 2013), flies (Gratz et al. 2013) and mice (Wang et al. 2013; for a more extensive list, see Hsu, Lander and Zhang 2014).
In 2013, DiCarlo et al. (2013b) employed the CRISPR/Cas9 system for the introduction of DSBs in S. cerevisiae. In a strain expressing a plasmid-borne cas9 gene from S. pyogenes, a second plasmid was introduced, containing the SNR52 promoter followed by a sequence encoding for a chimeric crRNA-tracrRNA, or guide-RNA (gRNA) with a 20 bp targeting sequence for CAN1. The gRNA was recognized by the Cas9 protein, resulting in a double-strand break at the CAN1 locus. Subsequently, this otherwise lethal break was repaired by the yeast HR machinery, using a co-transformed repair fragment that bridged the flanking regions of the break. Since the repair fragment was designed to introduce a premature stop codon, introduction and repair of the DSB resulted in colonies that were resistant to canavanine. It has recently been shown that the CRISPR/Cas9 system can be used for making up to three simultaneous gene deletions in yeast (Bao et al. 2014).
The goal of the present study is to explore the use of CRISPR/Cas9 for standardized (multiplexed) construction of gene deletions, multipathway integrations and site-directed single-nucleotide mutagenesis. To this end, we present a web-based CRISPR tool to facilitate selection of suitable targets and to design the primers necessary for construction of plasmids that express specific gRNAs. Furthermore, we report on the construction of standardized plasmids for expression of one or two gRNAs and explore their use for multiplexed gene deletions, both alone and in combination with multigene chromosomal integrations and/or with the introduction of single-nucleotide changes.
MATERIALS AND METHODS
Strains, growth conditions and storage
The S. cerevisiae strains used in this study (Table 1) share the CEN.PK genetic background (Entian and Kötter 2007; Nijkamp et al. 2012). Shake flask cultures were grown at 30 °C in 500 mL flasks containing 100 mL synthetic medium (SM) (Verduyn et al. 1992) with 20 g·L−1 glucose in an Innova incubator shaker (New Brunswick Scientific, Edison, NJ, USA) set at 200 rpm. When required, auxotrophic requirements were complemented via addition of 150 mg·L−1 uracil, 100 mg·L−1 histidine, 500 mg·L−1 leucine, 75 mg·L−1 tryptophan (Pronk, 2002) or by growth in YP medium (demineralized water, 10 g·L−1 Bacto yeast extract, 20 g·L−1 Bacto peptone). As a carbon source, 20 g·L−1 glucose was used. Frozen stocks were prepared by addition of glycerol (30% v/v) to exponentially growing shake-flask cultures of S. cerevisiae and overnight cultures of Escherichia coli and stored aseptically in 1 mL aliquots at –80 °C.
Table 1.
Saccharomyces cerevisiae strains used in this study.
| Name (Accession no.) | Relevant genotype | Parental strain | Origin |
|---|---|---|---|
| CEN.PK113-7D | MATa URA3 TRP1 LEU2 HIS3 | P. Kötter | |
| CEN.PK113-5D | MATa ura3-52 TRP1 LEU2 HIS3 | P. Kötter | |
| CEN.PK122 | MATa/MATα URA3/URA3 TRP1/TRP1 LEU2/LEU2 HIS3/HIS3 | P. Kötter | |
| CEN.PK2-1C | MATa ura3-52 trp1-289 leu2-3,112 his3Δ | P. Kötter | |
| CEN.PK115 | MATa/MATα ura3-52/ura3-52 TRP1/TRP1 LEU2/LEU2 HIS3/HIS3 | P. Kötter | |
| IMX585 (Y40592) | MATa can1Δ::cas9-natNT2 URA3 TRP1 LEU2 HIS3 | CEN.PK113-7D | This study |
| IMX581 (Y40593) | MATa ura3-52 can1Δ::cas9-natNT2 TRP1 LEU2 HIS3 | CEN.PK113-5D | This study |
| IMX664 (Y40594) | MATa/MATα CAN1/can1Δ::cas9-natNT2 URA3/URA3 TRP1/TRP1 LEU2/LEU2 HIS3/HIS3 | CEN.PK122 | This study |
| IMX672 (Y40595) | MATa ura3-52 trp1-289 leu2-3,112 his3Δ can1Δ::cas9-natNT2 | CEN.PK2-1C | This study |
| IMX673 (Y40596) | MATa/MATα ura3-52/ ura3-52 CAN1/can1Δ::cas9-natNT2 TRP1/TRP1 LEU2/LEU2 HIS3/HIS3 | CEN.PK115 | This study |
| IMX711 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ can1Δ::cas9-natNT2 mch1Δ pMEL10-gRNA-MCH1 | IMX672 | This study |
| IMX712 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ can1Δ::cas9-natNT2 mch2Δ pMEL10-gRNA-MCH2 | IMX672 | This study |
| IMX713 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ can1Δ::cas9-natNT2 mch5Δ pMEL10-gRNA-MCH5 | IMX672 | This study |
| IMX714 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ can1Δ::cas9-natNT2 mch1Δ mch5Δ pMEL10-gRNA-MCH1 pMEL10-gRNA-MCH5 | IMX672 | This study |
| IMX715 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ can1Δ::cas9-natNT2 itr1Δ pdr12Δ pUDR005 | IMX672 | This study |
| IMX716 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ can1Δ::cas9-natNT2 mch1Δ mch2Δ itr1Δ pdr12Δ pUDR002 pUDR005 | IMX672 | This study |
| IMX717 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ can1Δ::cas9-natNT2 mch1Δ mch2Δ mch5Δ aqy1Δ itr1Δ pdr12Δ pUDR002 pUDR004 pUDR005 | IMX672 | This study |
| IMX718 | MATa ura3-52 trp1-289 leu2-3,112 his3Δ can1Δ::cas9-natNT2 GET4G315C NAT1C1139G pUDR020 | IMX672 | This study |
| IMX719 | MATa can1Δ::cas9-natNT2 URA3 TRP1 LEU2 HIS3 acs1Δ acs2Δ::(pADH1-aceF-tPGI1 pPGI1-lplA2-tPYK1 pPGK1-lplA-tPMA1 pTDH3-pdhB-tCYC1 pTEF1-lpd-tADH1 pTPI1-pdhA-tTEF1) | IMX585 | This study |
Strains with an accession number have been deposited at Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/).
Plasmid construction
Construction of the single gRNA plasmid series (pMEL10–pMEL17)
The single gRNA plasmids (pMEL10–pMEL17) were constructed via Gibson assembly (New England Biolabs, Beverly, MA, USA) of a marker cassette with one fragment containing both the gRNA CAN1.Y (DiCarlo et al. 2013b) and the 2 μm replication sequence. This fragment was obtained by PCR from plasmid p426-SNR52p-gRNA.CAN1.Y-SUP4t, using primers 6845 & 6846 (Table S1, Supplementary data). The various marker cassettes were PCR amplified from plasmid templates pUG72, pUG-amdSYM, pUG-hphNT1, pUG6, pUG73 and pUG-natNT2 with primers 3093 & 3096 resulting in the KlURA3, amdSYM, hphNT1, kanMX, KlLEU2 and natNT2 cassettes, respectively. The HIS3 and TRP1 cassettes were obtained by PCR with primers 6847 & 6848 on plasmid templates pRS423 and pRS424. Assembly of the single gRNA plasmids was done by combining the appropriate marker cassette with the backbone containing the gRNA CAN1.Y and 2 μm sequences in a Gibson assembly reaction, following the manufacturer's recommendations. For each single gRNA plasmid (pMEL10–pMEL17), an E. coli clone containing the correctly assembled plasmid (confirmed by restriction analysis) was selected, stocked and deposited at EUROSCARF (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/).
Construction of the double gRNA plasmid series (pROS10 – pROS17)
To construct the double gRNA plasmids (pROS10–pROS17), an intermediate plasmid was first constructed, carrying two gRNA cassettes that both targeted CAN1.Y (DiCarlo et al. 2013b). This intermediate plasmid was assembled out of four different overlapping fragments: the two gRNA cassettes overlapping with each other in the 2 μm replicon, one URA3 marker cassette and a cassette containing all sequences for amplification in E. coli. The gRNA cassettes were obtained in a two-step PCR approach. First, a 2 μm fragment was obtained from pUD194 (Table 2) with primers 3289 & 4692 and two different gRNA cassettes were PCR amplified from p426-SNR52p-gRNA.CAN1.Y-SUP4t with primers 5972 & 5976 for the first cassette and 5977 & 5973 for the second cassette. Each gRNA cassette was separately pooled with the 2 μm fragment and in a second PCR reaction, the gRNA cassettes were extended with either the 5′ or 3′ halve of the 2 μm fragment, resulting in two different gRNA cassettes, overlapping in the 2 μm sequence, by using primer pair 5975 & 4068 for the first and 5974 & 3841 for the second fragment. The marker fragment containing URA3 was obtained from pUD192 with primers 3847 & 3276 (Table 2) and the fragment containing all sequences for amplification in E. coli was PCR amplified from pUD195 (Table 2) with primers 3274 & 3275. Using Gibson assembly, the four overlapping fragments were assembled into the intermediate plasmid pUDE330. To obtain pROS10, pUDE330 was linearized by PCR amplification of the backbone, excluding the gRNA fragments, and co-transformed with two gRNA cassettes for in vivo assembly by HR in yeast (Kuijpers et al. 2013b). For linearizing the backbone, a single primer was used (5793) and the gRNA fragments were obtained by PCR from pUDE330 with primers 6008 & 5975 and 6007 & 5974. The plasmid was extracted from yeast and transformed into E. coli for storage and plasmid propagation. The other double gRNA plasmids were assembled by the Gibson assembly method with a marker cassette and the pROS10 plasmid backbone fragment. This backbone fragment was obtained by linearization of pROS10 with restriction enzymes PvuII and NotI. The various marker cassettes were PCR amplified from plasmid templates pUG-amdSYM, pUG-hphNT1, pUG6, pUG73 and pUG-natNT2 with primers 3093 & 3096 resulting in the amdSYM, hphNT1, kanMX, KlLEU2 and natNT2 cassettes, respectively. The HIS3 and TRP1 cassettes were obtained by PCR with primers 6847 & 6848 on plasmid templates pRS423 and pRS424. Combining the appropriate marker cassette with the pROS10 backbone fragment in a Gibson assembly reaction, following the manufacturer's recommendations, resulted in pROS11–pROS17. For each of these double gRNA plasmids, an E. coli clone containing the correctly assembled plasmid was selected, stocked and deposited at EUROSCARF (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/).
Table 2.
Plasmids used in this study.
| Name (Accession no.) | Relevant characteristics | Origin |
|---|---|---|
| pUG6 | Template for A-kanMX-B† cassette | (Gueldener et al. 2002) |
| pUG72 | Template for A-KlURA3-B cassette | (Gueldener et al. 2002) |
| pUG73 | Template for A-KlLEU2-B cassette | (Gueldener et al. 2002) |
| pUG-hphNT1 | Template for A-hphNT1-B cassette | (de Kok et al. 2011) |
| pUG-natNT2 | Template for A-natNT2-B cassette | (de Kok et al. 2012) |
| pUG-amdSYM | Template for A-amdSYM-B cassette | (Solis-Escalante et al. 2013) |
| pRS423 | Template for A-HIS3-B cassette | (Christianson et al. 1992) |
| pRS424 | Template for A-TRP1-B cassette | (Christianson et al. 1992) |
| p414-TEF1p-Cas9-CYC1t | CEN6/ARS4 ampR TRP1 pTEF1-cas9-tCYC1 | (DiCarlo et al. 2013a) |
| p426-SNR52p-gRNA.CAN1.Y-SUP4t | 2μm ampR URA3 gRNA-CAN1.Y | (DiCarlo et al. 2013a) |
| pUD192 | pUC57 + URA3 | (Kozak et al. 2014a) |
| pUD194 | pUC57 + 2μm | (Kozak et al. 2014a) |
| pUD195 | pUC57 + pMB1 + ampR | (Kozak et al. 2014a) |
| pUD301 | pUC57 + pTPI1-pdhA E. faecalis-tTEF1 | (Kozak et al. 2014a) |
| pUD302 | pUC57 + pTDH3-pdhB E. faecalis -tCYC1 | (Kozak et al. 2014a) |
| pUD303 | pUC57 + pADH1-aceF E. faecalis -tPGI1 | (Kozak et al. 2014a) |
| pUD304 | pUC57 + pTEF1-lpd E. faecalis -tADH1 | (Kozak et al. 2014a) |
| pUD305 | pUC57 + pPGK1-lplA E. faecalis -tPMA1 | (Kozak et al. 2014a) |
| pUD306 | pUC57 + pPGI1-lplA2 E. faecalis -tPYK1 | (Kozak et al. 2014a) |
| pUDE330 | 2μm ampR URA3 gRNA-CAN1.Y [2x] | This study |
| pMEL10 (P30779) | 2μm ampR KlURA3 gRNA-CAN1.Y | This study |
| pMEL11 (P30780) | 2μm ampR amdSYM gRNA-CAN1.Y | This study |
| pMEL12 (P30781) | 2μm ampR hphNT1 gRNA-CAN1.Y | This study |
| pMEL13 (P30782) | 2μm ampR kanMX gRNA-CAN1.Y | This study |
| pMEL14 (P30783) | 2μm ampR KlLEU2 gRNA-CAN1.Y | This study |
| pMEL15 (P30784) | 2μm ampR natNT2 gRNA-CAN1.Y | This study |
| pMEL16 (P30785) | 2μm ampR HIS3 gRNA-CAN1.Y | This study |
| pMEL17 (P30786) | 2μm ampR TRP1 gRNA-CAN1.Y | This study |
| pROS10 (P30787) | 2μm ampR URA3 gRNA-CAN1.Y gRNA-ADE2.Y | This study |
| pROS11 (P30788) | 2μm ampR amdSYM gRNA-CAN1.Y gRNA-ADE2.Y | This study |
| pROS12 (P30789) | 2μm ampR hphNT1 gRNA-CAN1.Y gRNA-ADE2.Y | This study |
| pROS13 (P30790) | 2μm ampR kanMX gRNA-CAN1.Y gRNA-ADE2.Y | This study |
| pROS14 (P30791) | 2μm ampR KlLEU2 gRNA-CAN1.Y gRNA-ADE2.Y | This study |
| pROS15 (P30792) | 2μm ampR natNT2 gRNA-CAN1.Y gRNA-ADE2.Y | This study |
| pROS16 (P30793) | 2μm ampR HIS3 gRNA-CAN1.Y gRNA-ADE2.Y | This study |
| pROS17 (P30794) | 2μm ampR TRP1 gRNA-CAN1.Y gRNA-ADE2.Y | This study |
| pUDR002 | 2μm ampR TRP1 gRNA-MCH1 gRNA-MCH2 | This study |
| pUDR004 | 2μm ampR HIS3 gRNA-MCH5 gRNA-AQY1 | This study |
| pUDR005 | 2μm ampR URA3 gRNA-ITR1 gRNA-PDR12 | This study |
| pUDR020 | 2μm ampR URA3 gRNA-NAT1 gRNA-GET4 | This study |
| pUDR022 | 2μm ampR kanMX gRNA-ACS1 gRNA-ACS2 | This study |
†A and B refer to 60 bp tags that are incorporated via PCR, enabling homologous recombination.
Plasmid with an accession number have been deposited at Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/).
Strain construction
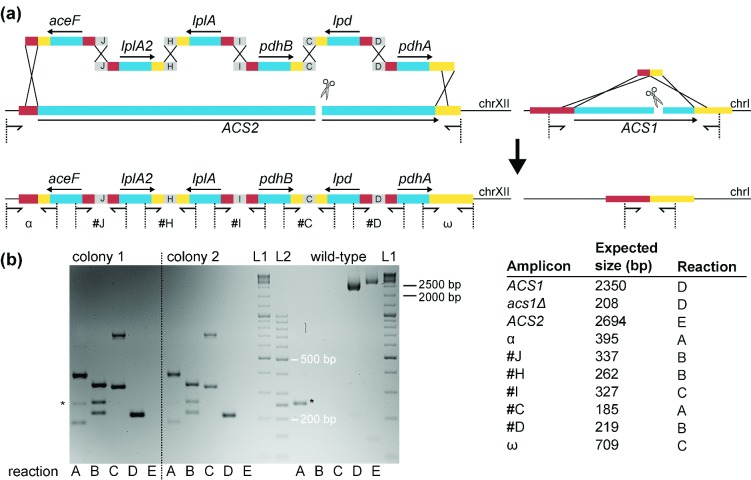
Saccharomyces cerevisiae strains were transformed according to Gietz and Woods (2002). Mutants were selected on solid YP medium (demineralized water, 10 g·L−1 Bacto yeast extract, 20 g·L−1 Bacto peptone, 2% (w/v) agar), supplemented with 200 mg·L−1 G418, 200 mg·L−1 hygromycin B or 100 mg·L−1 nourseothricin (for dominant markers) or on SM supplemented with appropriate auxotrophic requirements (Verduyn et al. 1992). In all cases, gene deletions and integrations were confirmed by colony PCR on randomly picked colonies, using the diagnostic primers listed in Table S1 (Supplementary data). Integration of cas9 into the genome was achieved via assembly and integration of two cassettes containing cas9 and the natNT2 marker into the CAN1 locus. The cas9 cassette was obtained by PCR from p414-TEF1p-cas9-CYC1t (DiCarlo et al. 2013b), using primers 2873 & 4653. The natNT2 cassette was PCR amplified from pUG-natNT2 with primers 3093 & 5542. 2.5 μg cas9 and 800 ng natNT2 cassette were pooled and used for each transformation. Correct integration was verified by colony PCR (Supplementary data) using the primers given in Table S1 (Supplementary data), the resulting strains have been deposited at EUROSCARF. IMX719 was constructed by co-transformation of pUDR022 (see below) with genes required for functional Enterococcus faecalis PDH expression (Kozak et al. 2014b). The gene cassettes were obtained by PCR using plasmids pUD301–pUD306 as template (Table 2) with the primers indicated in Table S1 (Supplementary data) and the ACS1 dsDNA repair fragment, obtained by annealing two complementary single-stranded oligos (6422 & 6423). After confirmation of the relevant genotype (Fig. 4B), the pUDR022 plasmid was removed as explained in Supplementary data.
Figure 4.
Multiplexing CRISPR/Cas9 in S. cerevisiae. (a) Chromosomal integration of the six genes required for expression of a functional E. faecalis pyruvate dehydrogenase complex in the yeast cytosol. All six genes are flanked by 60 bp sequences enabling HR (indicated with black crosses). The first and the last fragments are homologous to 60 bp just up- and downstream of the ACS2 ORF, respectively, thus enabling repair of the Cas9-induced double strand break by HR (left panel). Deletion of ACS1 using a 120 bp dsDNA repair fragment is shown in the right panel. (b) Multiplex colony PCR was performed on 10 transformants to check their genotypes. Results are shown for two representative colonies, confirming the intended genotype. The PCR on the wild-type strain (CEN.PK113-7D) shows the predicted bands for the presence of the wild-type ACS1 and ACS2 alleles. Two DNA ladders were used; L1 refers to the GeneRuler DNA ladder (Thermo Scientific) and L2 to the GeneRuler 50 bp DNA ladder (Thermo Scientific). (The bands indicated with an asterisk reflect aspecific PCR products).
Single gRNA method (cloning-free)
The yeast HR machinery was used to assemble plasmids with specific gRNA sequences out of two different fragments: a plasmid backbone and a gRNA target sequence. Depending on the preferred selectable marker, the linearized plasmid backbone was obtained via PCR using the appropriate single gRNA plasmid (pMEL10–pMEL17) as a template, with primers 6005 & 6006. To obtain the double-stranded gRNA cassettes (the target sequences are listed in Table 3), two complementary single-stranded oligos (Table S1, Supplementary data) were mixed in a 1:1 molar ratio, heated to 95°C and then cooled down to room temperature. The resulting gRNA fragments contained the 20 bp gRNA recognition sequences, flanked by 50 bp overlaps with the linearized plasmid backbone. The 120 bp repair fragments were obtained by following the same procedure and were identical to the up- and downstream regions of the DSB break, allowing for effective repair by the HR-machinery. For each transformation, a linearized plasmid backbone, a double-stranded cassette containing the gRNA sequence of choice and the double-stranded DNA cassette for repair of the DSB were pooled and co-transformed to the appropriate strain.
Table 3.
Target sequences used in this study.
| Locus | Sequence (including PAM) | Restriction site | AT score | RNA score |
|---|---|---|---|---|
| MCH1 | TATTGGCAATAAACATCTCGAGG | 0.65 | 0.50 | |
| MCH2 | ATCTCGATCGAGGTGCCTGATGG | PvuI | 0.45 | 0.30 |
| MCH5 | ACTCTTCCGTTTTAGATATCTGG | EcoRV | 0.65 | 0.50 |
| AQY1 | ACCATCGCTTTAAAATCTCTAGG | DraI | 0.65 | 0.50 |
| ITR1 | ATACATCAACGAATTCCAACCGG | EcoRI | 0.65 | 0.60 |
| PDR12 | GCATTTTCGGTACCTAACTCCGG | KpnI | 0.55 | 0.65 |
| NAT1 | AAAGGAATTGGATCCTGCGTAGG | BamHI | 0.55 | 0.60 |
| GET4 | GGGCTCGCTAGGATCCAATTCGG | BamHI | 0.45 | 0.50 |
| ACS1 | TTCTTCACAGCTGGAGACATTGG | PvuII | 0.55 | 0.45 |
| ACS2 | TCCTTGCCGTTAAATCACCATGG | 0.55 | 0.75 |
Double gRNA method
Plasmids with two gRNAs were assembled in vitro, using Gibson assembly of a 2 μm fragment containing the two gRNA sequences and a double gRNA (pROS10–pROS17) plasmid backbone. The 2 μm fragment was obtained via PCR, using pROS10 as a template with two primers containing the 20 bp gRNA recognition sequences and a 50 bp sequence, homologous to the linearized plasmid backbone (Table S1, Supplementary data). The linearized plasmid backbone was obtained via PCR using one of the double gRNA plasmids (pROS10–pROS17, depending on the preferred selective marker) as a template with a single primer (6005), binding at each of the two SNR52 promoters (Supplementary data). The two fragments were combined using Gibson assembly, followed by transformation to E. coli for storage and plasmid propagation. Since both of the gRNA containing primers could bind on either side of the 2 μm fragment, it was important to check that the final plasmid contained one copy of each gRNA (theoretically this would be the case in 50% of the E. coli transformants). To simplify this confirmation step, the gRNA target sequences were selected for the presence of a restriction site. Alternatively, diagnostic primers specific for the introduced 20 bp recognition sequences were used for the identification of correctly assembled gRNA plasmids using PCR (see Protocol in Supplementary data).
To construct the plasmid pUDR002 (targeting MCH1&MCH2, TRP1 marker), the 2 μm fragment was amplified using DreamTaq (Fisher Scientific) from pROS10 using primers 6835 & 6837 (Supplementary data). The backbone of pROS17 was amplified using the Phusion polymerase (Fisher Scientific) with a single primer 6005 (Supplementary data). The two fragments were assembled using Gibson assembly and confirmed via restriction analysis. Similarly, the following plasmids were constructed: pUDR004 (targeting MCH5&AQY1, HIS3 marker), pUDR005 (targeting ITR1&PDR12, KlURA3 marker), pUDR020 (targeting NAT1&GET4, URA3 marker) and pUDR022 (targeting ACS1&ACS2, kanMX marker). Transformations using the double gRNA method required co-transformation of 2 μg of (each) pUDR plasmid together with 1 μg of (each) corresponding double-stranded DNA cassette for DSB repair.
Molecular biology techniques
PCR amplification with Phusion® Hot Start II High Fidelity Polymerase (Thermo Fisher Scientific) was performed according to the manufacturer's instructions using PAGE-purified oligonucleotide primers (Sigma-Aldrich, St. Louis, MO, USA). Diagnostic PCR was done via colony PCR on randomly picked yeast colonies, using DreamTaq (Thermo Fisher Scientific) and desalted primers (Sigma-Aldrich). The primers used to confirm successful deletions by one of the two described methods can be found in Table S1 (Supplementary data). DNA fragments obtained by PCR were separated by gel electrophoresis on 1% (w/v) agarose gels (Thermo Fisher Scientific) in TAE buffer (Thermo Fisher Scientific) at 100 V for 30 minutes. Fragments were excised from gel and purified by gel purification (Zymoclean™, D2004, Zymo Research, Irvine, CA, USA). Plasmids were isolated from E. coli with Sigma GenElute Plasmid kit (Sigma-Aldrich) according to the supplier's manual. Yeast plasmids were isolated with Zymoprep Yeast Plamid Miniprep II Kit (Zymo Research). Escherichia coli DH5α (18258–012, Life Technologies) was used for chemical transformation (T3001, Zymo Research) or for electroporation. Chemical transformation was done according to the supplier's instructions. Electrocompetent DH5α cells were prepared according to Bio-Rad's protocol, with the exception that the cells were grown in LB medium without NaCl. Electroporation was done in a 2 mm cuvette (165–2086, Bio-Rad, Hercules, CA, USA) using a Gene Pulser Xcell Electroporation System (Bio-Rad), following the manufacturer's protocol.
Yeastriction webtool
The tool is written in Javascript and based on the MEAN.io stack (MongoDB, Express, AngularJS and Node.js). The source code is available at https://github.com/hillstub/Yeastriction. Genome and ORF sequences were downloaded from SGD (http://www.yeastgenome.org) in GFF and FASTA file format, respectively. ORFs, including their 1 kb up- and downstream sequences, were extracted and imported into Yeastriction, with the aid of an in-house script.
Yeastriction extracts all possible Cas9 target sequences (20 bp followed by NGG) from a specified ORF and from its complementary strand. Subsequently, sequences containing six or more Ts are discarded as this can terminate transcription (Braglia, Percudani and Dieci 2005; Wang and Wang 2008). Target sequences are then tested for off-targets (an off-target is defined as a sequence with either the NGG or NAG PAM sequence and 17 or more nucleotides identical to the original 20 bp target sequence; Hsu et al. 2013) by matching the sequences against the reference genome using Bowtie (version 1) (Langmead et al. 2009). If any off-target is found, the original target sequence is discarded. In a next step, the AT content is calculated for the target sequence. Using the RNAfold library (essentially with the parameters –MEA –noLP –temp = 30.) (Lorenz et al. 2011), the maximum expected accuracy structure of each RNA molecule is calculated. The target sequence is also searched for the presence of restriction sites based on a default list or a user-defined list. The targets can be ranked based on the presence of restriction sites (1 for containing and 0 for lacking a restriction site), AT content (1 having the highest AT content and 0 for the lowest AT content) and secondary structure (1 having the lowest amount of pairing nucleotides and 0 for the highest number of nucleotides involved in secondary structures, indicated by brackets). The range for every parameter is determined per locus and used to normalize the values. Subsequently, the target sequences are ranked by summation of the score for each parameter. These ranking scores should only be used to order the targets from a single locus and not to compare targets for different loci. The application can be accessed at the following URL: http://yeastriction.tnw.tudelft.nl/.
RESULTS AND DISCUSSION
Yeastriction: a CRISPR design tool
To streamline design and construction of CRISPR/Cas9 gRNA plasmids for introduction of (multiple) genetic modifications, the Yeastriction webtool (http://yeastriction.tnw.tudelft.nl) was developed. This webtool was designed to be compatible with the single and double gRNA plasmid series (pMEL10–pMEL17 and pROS10–pROS17, respectively), as described below. Because the CRISPR/Cas9 system can be highly sequence specific, it is crucially important to select target sequences based on correct reference genome sequence information. A difference of a single nucleotide in the gRNA, as compared to the genomic target sequence, can already completely abolish Cas9 nuclease activity (Hsu et al. 2013). To make Yeastriction useful for the entire yeast community, a set of 33 S. cerevisiae genomes (www.yeastgenome.org) was implemented. In the first step, the user can select the desired reference genome and enter (multiple) systematic names (e.g. YDL054C, YKL221W) or gene names (e.g. MCH1, MCH2). For each entered gene, the tool matches every potential target sequence to the selected reference genome. If there are potential off-targets (other sequences present in the same genome with either NAG or NGG as PAM sequence and with a 0–3 nucleotide difference in the 20 bp target sequence, Hsu et al. 2013), the target sequence is discarded. Sequences that contain six or more consecutive Ts are also discarded as they may cause transcript termination (Braglia, Percudani and Dieci 2005; Wang and Wang 2008). A recent report indicates that the AT content of Cas9 target sequences should preferably be above 65% (Lin et al. 2014). Furthermore, there are indications that target sequences without obvious nucleotide interactions in secondary structures are more efficient (Zhang 2014). The presence of unique restriction sites within the target sequence simplifies verification of correct plasmid assembly. Yeastriction therefore ranks potential Cas9 target sequences according to AT content, secondary structures and the presence of restriction sites. To increase flexibility, the user can also choose to leave out one of the parameters in the final ranking. For the top-ranked target sequence, the tool automatically designs the oligonucleotide primers required for plasmid construction and the oligonucleotides (which can be ordered as primers) needed to form the repair fragment when a gene deletion is desired. To increase flexibility, the user can also choose another target site than the top-ranked sequence (Supplementary data). In comparison with existing tools such as ChopChop (Montague et al. 2014), Yeastriction combines several features to improve gRNA sequence selection for S. cerevisiae: (i) yeast strain specificity, (ii) elimination of spacer sequences with potential off-targets, (iii) ranking of the remaining gRNA sequences based on RNA structure and AT content and (iv) direct generation of primer sequences, compatible with the transformation methods described below.
Construction of a set of plasmids for transformation with one or two gRNAs
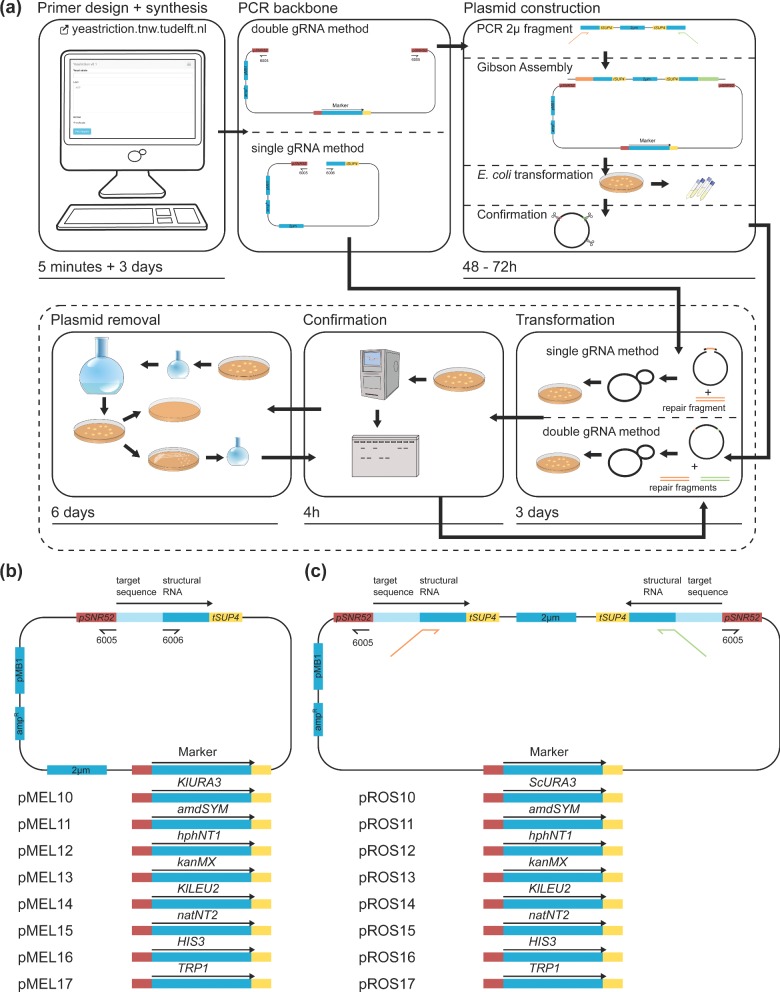
DiCarlo et al. (2013b) described construction of plasmid p426-SNR52p-gRNA.CAN1.Y-SUP4t (Addgene www.addgene.org/43803/) for expression of a single gRNA in yeast, using the SNR52 promoter and SUP4 terminator. We hypothesized that the gRNA could be easily changed by in vivo HR via co-transformation of the linearized p426-SNR52p-gRNA.CAN1.Y-SUP4t backbone (from which the 20 bp recognition sequence ‘CAN1.Y’ was omitted) together with a new 20 bp gRNA fragment, flanked by 50 bp overlaps with the plasmid backbone. In order to further increase the flexibility of this system and to allow its use with genetic markers other than URA3, a standardized set of plasmids was constructed (Fig. 1B). To this end, a linearized plasmid backbone of the p426-SNR52p-gRNA.CAN1.Y-SUP4t plasmid that excluded the URA3 marker gene was obtained by PCR. Subsequently, eight different genetic markers (KlURA3, amdSYM, hphNT1, kanMX, KlLEU2, natNT2, HIS3 and TRP1) were PCR amplified and (re-)introduced in this backbone using Gibson assembly. The resulting plasmids were named pMEL10 to pMEL17 (Fig. 1B, Table 2)
Figure 1.
Workflow for CRISPR/Cas9 modification of S. cerevisiae genome using single and double gRNA plasmid series (pMEL10–pMEL17 and pROS10–pROS17, respectively). (a) All oligonucleotides, required for targeting the gene(s) of interest (GOI), can be automatically designed with the Yeastriction webtool (http://yeastriction.tnw.tudelft.nl). For the single gRNA method, the tool designs complementary oligonucleotides that can be annealed to form (i) a double-stranded repair fragment and (ii) a double-stranded insert which contains the target sequence for the GOI. For expression of the gRNA, a plasmid backbone containing the genetic marker of choice is amplified from a single gRNA plasmid (pMEL10–pMEL17). Gene deletion is achieved via co-transformation of the plasmid backbone, the dsDNA insert (containing the gRNA target sequence, flanked by sequences identical to both sides of the linearized plasmid backbone) and the repair fragment. For the double gRNA method, Yeastriction designs two sets of oligonucleotides, the first oligonucleotide binds to the 2 μm fragment and has a tail containing the desired target sequence for the GOI and a sequence identical to both sides of a linearized double gRNA plasmid backbone (pROS10–pROS17), the second set of oligonucleotides can be annealed to form the dsDNA repair fragment(s). To construct a gRNA plasmid with two target sequences, first a double gRNA plasmid backbone with the appropriate marker (pROS10–pROS17) is amplified by PCR, excluding the 2 μm fragment. Then, the 2 μm fragment is PCR amplified using the primers harbouring the targets for the GOIs and sequences identical to the plasmid backbone. The final plasmid is then constructed in vitro using the plasmid backbone and the 2 μm fragment, e.g. with the Gibson assembly method. After confirmation of correct plasmid assembly using restriction analysis or PCR, the resulting plasmid is transformed to yeast, together with the appropriate 120 bp dsDNA repair fragment(s). After transformation, the desired genetic modification(s) are checked by PCR, Southern blot analysis or sequencing. Subsequently, the strain can be modified again in a new round of transformation (preferably using plasmids with other markers). Before physiological analysis, the gRNA plasmid(s) are preferably removed. This can be done by growing the strains in liquid media without selection pressure or, if possible, by counter-selection pressure (with 5-fluoroorotic acid, fluoroacetamide or 5-fluoroanthranilic acid for pMEL10+pROS10 plasmids, pMEL11+pROS11 plasmids and pMEL17+pROS17 plasmids, respectively). After confirming plasmid removal by restreaking the same colony on selective and non-selective medium and/or PCR analysis, the resulting strain is re-grown in liquid medium and stored at –80 °C. (b) Architecture of the single gRNA plasmid series (pMEL10–pMEL17). The primers used for PCR amplification of the plasmid backbone are indicated by black arrows. (c) Architecture of the double gRNA plasmid series (pROS10–pROS17) with two gRNA cassettes. The plasmid backbone can be PCR amplified with a single primer (indicated with a black arrow). The 2 μm fragment is amplified with primers designed using Yeastriction (indicated in orange and light green coloured arrows).
The use of in vivo recombination enabled the introduction of one gRNA sequence per single gRNA plasmid backbone (pMEL10–pMEL17), without the need of prior cloning. When multiple genetic modifications are required, marker availability might become a limitation. In this case, plasmids containing multiple gRNA sequences would be preferred as this allows introduction of more genetic modifications before plasmid removal is required. For this purpose, a second set of plasmids was constructed that contained two gRNA sequences, between separate promoters and terminators.
First, pROS10 was constructed (Fig. 1C), containing the URA3 marker, ampR for selection in E. coli and a 2 μm fragment flanked by two pSNR52-gRNA-tSUP4 cassettes in opposite directions, with one of these cassettes containing the target sequence CAN1.Y and the other ADE2.Y (DiCarlo et al. 2013b). This design enables construction of plasmids that target other loci, by first PCR amplifying the 2 μm fragment, using primers that incorporate the new target sites, followed by assembly into a linearized backbone that contains the desired marker gene via an in vitro method such as Gibson assembly. To generate plasmids with marker genes other than URA3, the backbone of pROS10 was first digested with restriction enzymes to remove the URA3 marker. Subsequently, other markers were PCR amplified and inserted using Gibson assembly. This yielded plasmids pROS11 to pROS17 (Fig. 1C, Table 2), which contained the marker genes amdSYM, hphNT1, kanMX, KlLEU2, natNT2, HIS3 and TRP1, respectively. Both plasmid series (pMEL10–pMEL17 and pROS10–pROS17) have been made available through EUROSCARF.
Construction of various cas9-expressing CEN.PK strains
In the cas9-bearing strains described by DiCarlo et al. (2013b), a centromeric plasmid was used to express cas9, either from a variant of the inducible GAL1 promoter or from the constitutive TEF1 promoter. When multiple rounds of transformation and gRNA plasmid removal are desired, a stable integrated copy of cas9 is preferred, since this allows growth of strains on complex medium, which enables faster growth and efficient plasmid recycling. To this end, a cas9 gene under the control of the TEF1 promoter was integrated into the CAN1 locus together with the natNT2 marker. In order to increase the flexibility of the CRISPR/Cas9 system, cas9 was integrated in the haploid strains CEN.PK113–7D and CEN.PK113–5D (Ura−) and the diploid strains CEN.PK122 and CEN.PK115 (Ura−). Additionally, cas9 was integrated in the quadruple auxotrophic strain CEN.PK2–1C (Ura−, Leu−, Trp−, His−), resulting in strain IMX672, which was used to test the efficiencies of the gRNA plasmids developed in this study. All these strains are deposited at EUROSCARF. Constitutive expression of cas9 in CEN.PK113–7D (IMX585) did not affect the maximum specific growth rate. IMX585 grew at 0.37 ± 0.003 h−1 (value and mean deviation are based on two independent experiments) on glucose SM in shake-flask cultures while the growth rate of the reference CEN.PK113–7D was 0. 39 ± 0.01 h−1.
Seamless and markerless gene deletion using in vivo assembled plasmids containing single gRNAs
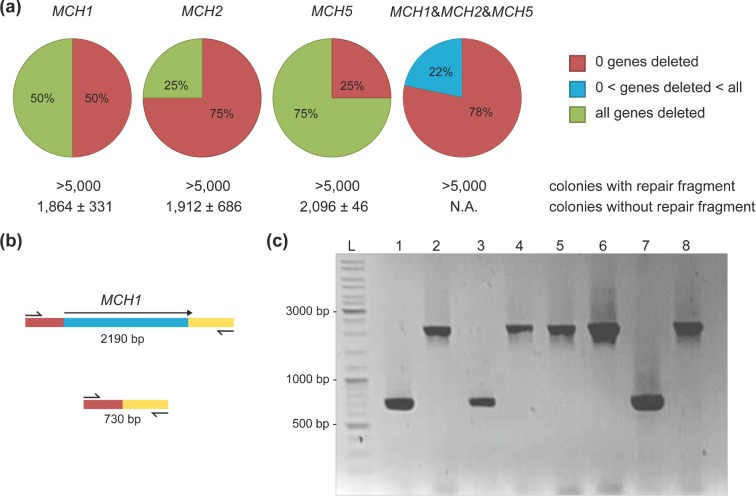
The first plasmid series, pMEL10–pMEL17, was designed to perform gene deletions from plasmids assembled in vivo, harnessing the high frequency and fidelity of HR in S. cerevisiae (Orr-Weaver and Szostak 1983; Orr-Weaver, Szostak and Rothstein 1983; Kunes, Botstein and Fox 1985; Kuijpers et al. 2013b) and obviating the need for prior cloning of the gRNA sequence. To test whether in vivo assembly of a plasmid carrying the gRNA sequence could be combined with CRISPR-mediated genetic modification, a plasmid backbone and insert containing the gRNA of choice were co-transformed with a 120 bp ‘repair fragment’ for markerless and scarless gene deletion. The gRNA sequence targeting the gene MCH1 (Table 3) was selected based on a high score (AT content 0.65, RNA score 0.50) in Yeastriction and contained 50 bp overlaps to each side of the linearized plasmid backbone with the KlURA3 marker, facilitating HR. Transformation of IMX672 (ura3–52 trp1–289 leu2–3,112 his3Δ, can1Δ::cas9) resulted in >5000 colonies per plate and colony PCR was performed to confirm successful gene deletion. Out of 24 randomly tested colonies, 12 (50%) showed the intended single gene deletion of MCH1 (Fig. 2). The colonies that did not show the intended deletion could be caused by misassembly of the plasmid, as omitting either the insert or the repair fragment from the transformation mixture yielded ∼2000 colonies. To test whether these results could be reproduced when different loci were targeted, the same strategy was employed to target MCH2 and MCH5. A gRNA recognition sequence was selected for each gene using Yeastriction, based on a low score for MCH2 (AT content 0.45, RNA score 0.30) and a high score for MCH5 (AT content 0.65, RNA score 0.50) targeting sequence, respectively. Transformation with gRNA inserts targeting either of these loci and the corresponding repair fragments resulted in similar colony counts as observed for MCH1. Furthermore, colony PCR determined successful deletions for both loci at varying efficiencies, 25% and 75% for MCH2 and MCH5, respectively (Fig. 2). This observed difference in deletion efficiency was in line with the quality scores predicted by Yeastriction for MCH2- and MCH5-selected gRNAs. This cloning-free approach might be extended to enable co-transformation of multiple gRNA inserts and their corresponding repair fragments. By assembly of multiple plasmids bearing different gRNA sequences in a single cell, this might facilitate the simultaneous introduction of multiple gene deletions. To test this possibility, the same plasmid backbone, containing the KlURA3 marker, was co-transformed with three different gRNA fragments (MCH1, MCH2 & MCH5) and their corresponding repair fragments. Over 5000 colonies were obtained after transformation and 24 randomly picked colonies were tested via colony PCR. Transformation with three inserts led to the identification of four mutants containing a single and one mutant containing a double deletion (24 colonies tested, Fig. 2A) but none with three deletions. Only one of the six identified deletions was found in the MCH1 locus (targeted by a gRNA with a low Yeastriction score, Table 3), while three and two deletions corresponded to ORFs of MCH1 and MCH5 respectively. These results indicated that mutants containing multiple gene deletions could be obtained via combined in vivo recombination and gene disruption, although at low (∼4%) efficiencies. This low efficiency of multiple gene disruption might have several reasons: (i) it can be the result of misassembly of the plasmid, as transformation of the backbone without insert or repair oligo already resulted in ∼2000 colonies when making single deletions; (ii) a cell does not need to assemble all three plasmids, as one plasmid is enough to restore prototrophy; and (iii) errors present in the gRNA inserts affecting its targeting efficiency. Indeed, sequencing of in vivo assembled plasmids of three false positive colonies showed that the gRNA sequence contained nucleotidic insertions and deletions, impairing cas9 restriction in the target ORF. These mutations of the gRNA sequence likely derived from imperfect annealing of the two complementary oligonucleotides used to form the in vivo assembled gRNA fragment.
Figure 2.
Efficiency of gene deletion obtained after transformation with a single gRNA plasmid. (a) Quantification of the number of colonies and corresponding gene deletion efficiencies, obtained after transformation of S. cerevisiae IMX672 (ura3–52 trp1–289 leu2–3,112 his3Δ, can1Δ::cas9) with 100 ng pMEL10 backbone, 300 ng of gRNA insert DNA and 2 μg of the corresponding 120 bp dsDNA repair fragment. The transformation targeting MCH1, MCH2 and MCH5 simultaneously was performed using 300 ng of each insert and 2 μg of each repair fragment. For the transformations with repair fragments, the exact number of transformants could not be determined, but exceeded 5000 colonies per plate. The data represent average and standard deviation of transformants of three independent transformation experiments. The estimated total number of colonies carrying gene deletions was based on colony PCR results of 24 randomly picked colonies. In red: percentage of colonies without gene deletions, in blue: percentage of colonies containing one or two but not all deletions, green: percentage of colonies containing all desired gene deletions. No transformants with all three genes deleted were identified. (b) Diagnostic primers were designed outside of the target ORFs to differentiate between successful and non-successful colonies via PCR. In this colony PCR example, primers 6862 & 6863 were used to amplify the MCH1 locus. (c) Example of a diagnostic gel from the transformation targeting the MCH1 locus. The first lane (L) contains the GeneRuler DNA Ladder Mix. Lane 1–8 show the PCR results of eight randomly picked colonies. Successful deletion of MCH1 results in a PCR fragment with a length of 729 bp (lane 1, 3 and 7), when MCH1 is still present a band is observed at 2190 bp (lane 2, 4, 5, 6 and 8).
All collectively these results showed that single gene deletions could be easily performed by this method; however, when more than one locus has to be altered a more effective strategy is needed as is detailed in the next section.
Multiplexing seamless gene deletions using in vitro assembled plasmids containing two gRNAs
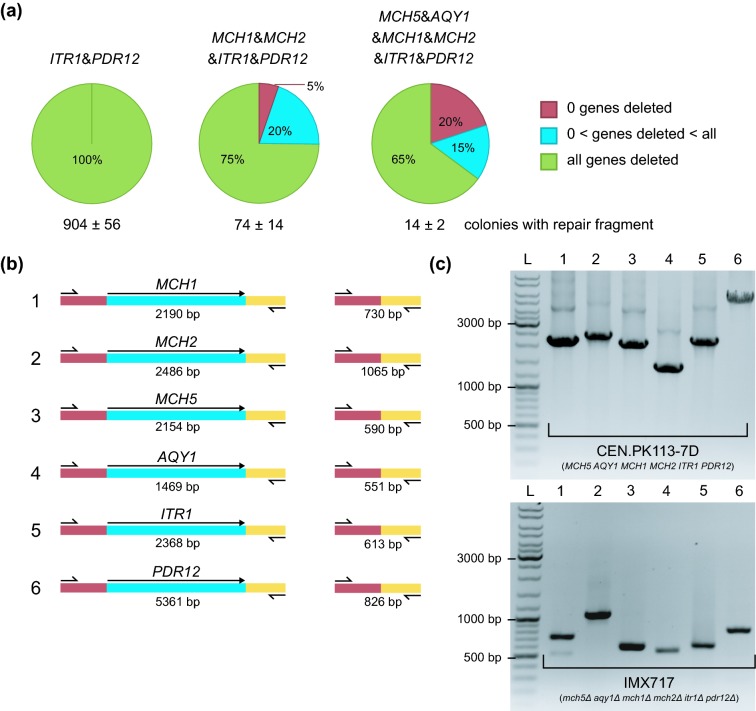
While in vivo assembly of the gRNA plasmid and CRISPR-assisted genetic modification can be combined in a single transformation, the relatively high incidence of false positive colonies limits the use of this system for simultaneous introduction of multiple chromosomal modifications. Since some of the false positives could be the result of misassembly of the gRNA containing plasmid, it was expected that pre-assembly of the plasmid using in vitro Gibson assembly would result in a lower number of false positives. Therefore, a double gRNA plasmid was constructed, expressing guide RNAs designed to target ITR1 and PDR12. First, the pROS10 backbone and the 2 μm fragment flanked by the gRNAs containing the target sequences for ITR1 and PDR12 were amplified using PCR (Fig. 1). The resulting DNA fragments were assembled and the plasmid (pUDR005) was verified by digestion with restriction enzymes specifically targeting the ITR1 and PDR12 target sequences (Table 3). Co-transformation of the resulting plasmid with the corresponding repair fragments into the cas9-expressing strain IMX672 yielded 900 colonies per plate. In contrast, a single transformation in which repair fragments were omitted yielded only two colonies. 24 colonies were randomly picked and PCR verification determined that all of these contained both gene disruptions (IMX715, Fig. 3). The transformation of an already assembled plasmid might enable immediate transcription of the gRNA, while the other method required assembly of a correct plasmid prior to transcription of the gRNA. This might explain the significantly higher observed efficiency of this approach compared to the method using in vivo assembled plasmids. Most likely, pre-assembly and confirmation of the plasmid containing two gRNAs greatly reduced the number of false positives.
Figure 3.
Efficiency of gene deletion obtained after transformation with double gRNA plasmids. (a) Quantification of the number of colonies and corresponding gene deletion efficiencies after transformation of S. cerevisiae IMX672 (ura3–52 trp1–289 leu2–3,112 his3Δ, can1Δ::cas9) with 2 μg of various double gRNA plasmids with 1 μg of the appropriate repair fragment(s). When multiple plasmids were transformed simultaneously, 2 μg of each plasmid was added. The data represent average and standard deviation of transformants of three independent transformation experiments. In red: percentage of colonies containing no gene deletions, in blue: percentage of colonies containing some but not all targeted gene deletions (1, 1–3 and 1–5 respectively), in green: percentage of colonies containing all targeted simultaneous gene deletions (2, 4 and 6 respectively). (b) Diagnostic primers were designed outside of the target ORFs to differentiate between successful and non-successful colonies via PCR. In this colony PCR example, primers 6862 & 6863, 6864 & 6865, 6866 & 6867, 6868 & 6870, 6869 & 6870 and 253 & 3998 were used to amplify the MCH1, MCH2, MCH5, AQY1, ITR1 and PDR12 loci, respectively. The expected sizes of the PCR products obtained when the gene is present (left) or deleted (right) are indicated. (c) Example of a diagnostic gel from the transformation introducing six simultaneous gene deletions, resulting in IMX717. The first lane (L) contains the GeneRuler DNA Ladder Mix. Lane 1–6 show the PCR results of the reference strain CEN.PK113–7D (top) and a randomly picked colony of IMX717 (bottom) with primers 6862 & 6863 for MCH1 (lane 1), 6864 & 6865 for MCH2 (lane 2), 6866 & 6867 for MCH5 (lane 3), 6868 & 5593 for AQY1 (lane 4), 6869 & 6870 ITR1 (lane 5) and 253 & 3998 for PDR12 (lane 6).
Encouraged by these extremely high efficiencies, transformations were performed using multiple plasmids that carried different genetic markers. To this end, two new plasmids were constructed, each targeting two different genetic loci [MCH1 & MCH2 (pUDR002) and MCH5 & AQY1 (pUDR004)]. Two and three plasmids were co-transformed, containing either the KlURA3 and TRP1 or the KlURA3, TRP1 and HIS3 selectable markers. In contrast to the transformations with a single plasmid, which yielded over 900 colonies per transformation plate, only 74 colonies and 14 colonies were obtained when transforming two and three plasmids, respectively. These lower numbers of colonies might reflect the decreasing probability that a single cell successfully takes up multiple plasmids and, subsequently, performs the corresponding gene deletions. The characterization of the colonies from the transformation with two plasmids revealed that out the 20 tested clones, 14 (70%) harboured all 4 deletions (IMX716). Similarly, out of 20 randomly tested colonies obtained from the transformations with 3 plasmids, 13 (65%) clones contained all 6 deletions (IMX717, Fig. 3).
These results unequivocally demonstrate the efficiency of CRISPR/Cas9-mediated genetic modification of yeast in simultaneously generating multiple deletions in a single transformation step. Recently, CRISPR/Cas9 was successfully applied to simultaneously disrupt all homozygous alleles in the polyploid ATCC4124 strain. In four transformation iterations, a quadruple ura3 trp1 leu2 his3 auxotrophic strain was constructed (Zhang et al. 2014). A design similar to the native polycistronic CRISPR array consisting of a tracrRNA and crRNA instead of the chimeric gRNA was used to achieve three concurrent deletions using a single crRNA array (Bao et al. 2014). To the best of our knowledge, the present study is the first to demonstrate generation of a sextuple deletion strain of S. cerevisiae in a single transformation event.
Multiplexing deletion, multigene integration and introduction of single nucleotide mutations
Hitherto, reported applications of CRISPR/Cas9 in S. cerevisiae focused on gene inactivation. I-SceI-mediated introduction of double-stranded breaks has previously been shown to facilitate simultaneous integration of several gene expression cassettes at chromosomal loci (Kuijpers et al. 2013a). To explore the potential of CRISPR to combine gene deletion with the simultaneous in vivo assembly and chromosomal integration of multiple DNA fragments, we attempted to construct a S. cerevisiae strain with a double ACS1 ACS2 deletion that also overexpresses the E. faecalis pyruvate dehydrogenase (PDH) complex (Kozak et al. 2014b) in a single transformation. To this end, IMX585 was transformed with a plasmid expressing the gRNAs targeting the ACS genes (pUDR022). A 120 bp repair fragment was co-transformed for the deletion of ACS1, and the ORF of ACS2 was replaced via integration of six gene cassettes expressing the genes of the E1α, E1β, E2 and E3 subunits of E. faecalis PDH encoded by pdhA, pdhB, aceF, and lpd, as well as the lplA and lplA2 genes required for PDH lipoylation (Kozak et al. 2014b) (Fig. 4A). Cytosolic acetyl-CoA is essential and a double acs1Δ acs2Δ mutant is not viable (van den Berg et al. 1996) unless acetyl-CoA is provided by an alternative route (Kozak et al. 2014a,b). The transformation targeting the two ACS genes yielded 11 colonies, with 10 out of 10 picked colonies showing the desired genotype (Fig. 4B). From one colony, the gRNA plasmid was removed by growing on non-selective YP medium, yielding strain IMX719. This strain showed only growth on SM with 2% glucose and lipoate and failed to grow on SM with 2% glucose and on YP with 2% ethanol. This phenotype is consistent with the phenotype reported by Kozak et al. (2014b) for an acs1 acs2 strain of S. cerevisiae that expresses the E. faecalis PDH subunits and lipoylation genes, thereby further confirming the genotype of the strain IMX719.
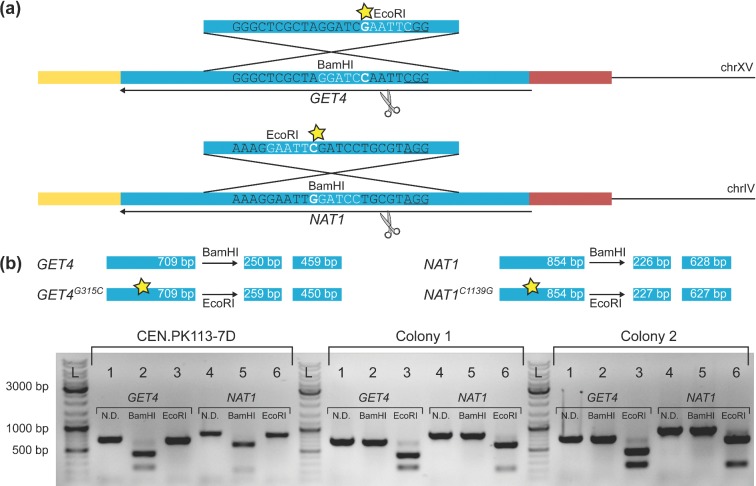
To explore the potential of CRISPR/Cas9 to introduce specific single-nucleotide mutations in genomic DNA, a plasmid was constructed with gRNAs targeting the GET4 and NAT1 loci (pUDR020). Both of the target sequences contained a BamHI (G|GATCC) restriction site. The corresponding repair oligonucleotides were designed to introduce a single-nucleotide change in the genomic target sequence, which at both sites resulted in the introduction of a restriction site for EcoRI (G|AATTC) and simultaneously disrupted the BamHI restriction site (Fig. 5A). Transformation of the cas9-bearing strain IMX672 with pUDR020 (NAT1 and GET4 gRNA, KlURA3) and the corresponding repair fragments resulted in ∼1500 colonies, while omitting both repair fragments did not result in any colonies. Eight colonies were randomly picked and a part of the ORF containing the target sequence was PCR amplified for both GET4 and NAT1. Digestion of the amplified PCR product with BamHI and EcoRI, followed by gel electrophoresis, showed that four of the eight colonies contained both mutations after transformation (Fig. 5B). To confirm whether these four colonies indeed contained the desired mutations, a fragment of 120 bp around the target site was sequenced. All colonies contained the desired mutations, although two of the four colonies also showed additional, undesired, mutations at the GET4 locus (supplementary data S1, colony 2 and 4). Although these results show that one mutation can already abolish restriction, Cas9-induced DSBs might still occur, as long as the gRNA cassette is expressed. We therefore advise to (re)sequence mutated sites after gRNA plasmid removal and/or use the two-step strategy discussed below.
Figure 5.
Simultaneous introduction of different single-nucleotide mutations in S. cerevisiae. (a) Transformation of IMX581 (ura3–52, can1Δ::CAS9) was performed with pUDR020, resulting in the introduction of two mutations in NAT1 and GET4. Underlined: the PAM sequences associated with the gRNA targets. In white: restriction sites present in the original gRNA targeting sequence (BamHI) and in the repair fragment used to correct the double-strand break (EcoRI). (b) Introduction of the double-strand break and subsequent repair using the mutagenic repair fragment resulted in a change of restriction site from BamHI to EcoRI. In this colony PCR example, primers 7030 & 7031 and 7036 & 7037 were used to amplify a part of the GET4 and NAT1 locus (lane 1 and 4, respectively) from CEN.PK113–7D and two colonies of IMX581, transformed with pUDR020 and mutagenic repair fragments. In lanes labelled 2 and 5: digestion of the PCR fragments with BamHI, which only results in digestion fragments of sizes 250 bp & 459 bp and 226 bp & 628 bp when the original restriction sites in GET4 and NAT1 are still present (CEN.PK113–7D). In lanes labelled 3 and 6: digestion of the PCR fragments with EcoRI, which only results in digestion fragments of sizes 259 bp & 450 bp and 227 bp & 627 bp when this new restriction site has been introduced via the mutagenic repair fragment (colony 1 and 2).
The ease with which specific single-nucleotide mutations can be simultaneously introduced makes the CRISPR/Cas9 system a highly valuable tool for analysing the biological significance of mutations identified by whole-genome resequencing of strains obtained by mutagenesis (e.g. UV, X-rays) (Jung et al. 2011) or evolutionary engineering (Hong et al. 2011; González-Ramos et al. 2013; Oud et al. 2013; Caspeta et al. 2014). Reverse engineering of such mutations essentially encompasses restoration of the reference nucleotide in the mutant strain and/or introduction of the mutated nucleotide position in a naïve (non-mutated or non-evolved) genetic background. Even laboratory evolution experiments performed in the absence of mutagenesis typically yield multiple mutations, not all of which contribute to the phenotype of interest (see e.g. de Kok et al. 2012; González-Ramos et al. 2013). Therefore, availability of methods that enabled reintroduction of multiple point mutations at various genomic loci is invaluable for rapid identification of relevant mutations. Similar to the YOGE (Yeast Oligo-Mediated Genome Engineering) method (DiCarlo et al. 2013a), the CRISPR approach enables multiplexing and offers flexibility. In contrast to the YOGE method, which requires a specific background (mlh1Δ msh2Δ RAD51K342E↑ RAD54↑) (DiCarlo et al. 2013a), the CRISPR approach should be applicable to any S. cerevisiae strain background that expresses a functional cas9 gene. The two examples used in this study (GET4 and NAT1) were designed for easy verification. If the desired point mutation is not present in a suitable target sequence, it may be possible to introduce (multiple) single nucleotide variation(s) (SNV) in two rounds of CRISPR/Cas9-mediated genome modification. In the first round, a larger part surrounding one or several SNVs could be deleted while repairing the DSBs with repair fragments that contain a generic synthetic target sequence. In a second round, these generic target sequences can then be cut by Cas9, combined with the repair of the DSBs with 120 bp sequences that contain the desired SNVs.
Outlook
The use of HR for the assembly of multigene constructs (Gibson et al. 2008; Shao, Zhao and Zhao 2009) had a tremendous impact on genetic engineering strategies (Annaluru et al. 2014; Casini et al. 2014). Even before the advent of CRISPR/Cas9, these developments have immensely decreased strain construction time in our group and enabled us to express complicated multistep pathways and multicomponent enzymes (Koopman et al. 2012; Guadalupe-Medina et al. 2013; Kuijpers et al. 2013a; Beekwilder et al. 2014; Kozak et al. 2014b). Until recently, the deletion of multiple genes and insertions at multiple loci remained a cumbersome and time-intensive process. Here, building on the pioneering conceptual proof of CRISPR functionality in lower eukaryotes (DiCarlo et al. 2013b), we show that CRISPR/Cas9 will further improve and accelerate yeast strain construction, not only by allowing multiplexed gene deletions (DiCarlo et al. 2013b; Zhang et al. 2014) but also by allowing simultaneous introduction of gene deletions, chromosomal integration of multigene constructs and the introduction of specific mutations. Although it is difficult to quantify the impact on the overall time requirements for complex pathway engineering, the examples presented here suggest that a 3- to 4-fold acceleration is unlikely to be exaggerated.
This paper focuses on S. cerevisiae, a microbial species that is already known for its easy genetic accessibility. Therefore, it is logical to speculate that this methodology should have an even higher impact on species of yeasts and filamentous fungi that are notoriously more difficult to alter genetically. Indeed, a very recent example described the benefit of the introduction of CRISPR/Cas9 in Schizosaccharomyces pombe (Jacobs et al. 2014). While the methodology reported in this study streamlines the use of CRISPR in S. cerevisiae, the method can be further improved. The RNA polymerase III-dependent promoter SNR52 is not a broadly recognized promoter (Ryan et al. 2014). Recently, it has been shown that guideRNAs, flanked by the hammerhead and hepatitis delta virus ribozymes, can be expressed using polymerase II promoters (Gao and Zhao 2014). A promising promoter would then be the TEF1 promoter from Blastobotrys adeninivorans (Terentiev et al. 2004) as this is a strong, constitutive promoter, recognized in different species, like S. cerevisiae, Hansenula polymorpha and Pichia pastoris.
We hope that by providing the easy-to-use Yeastriction design tool, two versatile plasmid series for gRNA expression, a set of S. cerevisiae CEN.PK strains harbouring the cas9 expression cassette and standardized protocols (Supplementary data), this paper will help colleagues to facilitate and accelerate yeast strain engineering.
SUPPLEMENTARY DATA
Supplementary data is available at FEMSYR online.
Acknowledgments
We thank James Dykstra for the construction of IMX719, Rudy Brinkman for constructing IMX581 and Arthur Dobbe for sharing the growth rates of IMX585.
FUNDING
PDL and NK were financially supported by the Technology Foundation STW (Vidi Grant 10776). HMvR, RM, AJAvM, JTP and JMD were supported by the BE-Basic R&D Program, which was granted an FES subsidy from the Dutch Ministry of Economic Affairs, Agriculture and Innovation (EL&I).
Conflict of interest statement. None declared.
REFERENCES
- Annaluru N, Muller H, Mitchell LA, et al. Total synthesis of a functional designer eukaryotic chromosome. Science. 2014;344:55–8. doi: 10.1126/science.1249252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao Z, Xiao H, Liang J, et al. Homology-Integrated CRISPR − Cas (HI-CRISPR) system for one-step multigene disruption in Saccharomyces cerevisiae. ACS Synth Biol. 2014 doi: 10.1021/sb500255k. DOI: 10.1021/sb500255k. [DOI] [PubMed] [Google Scholar]
- Barrangou R, Fremaux C, Deveau H, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–12. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- Beekwilder J, van Rossum HM, Koopman F, et al. Polycistronic expression of a β-carotene biosynthetic pathway in Saccharomyces cerevisiae coupled to β-ionone production. J Biotechnol. 2014;192:383–92. doi: 10.1016/j.jbiotec.2013.12.016. [DOI] [PubMed] [Google Scholar]
- Botstein D, Fink GR. Yeast: an experimental organism for 21st century biology. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braglia P, Percudani R, Dieci G. Sequence context effects on oligo(dT) termination signal recognition by Saccharomyces cerevisiae RNA polymerase III. J Biol Chem. 2005;280:19551–62. doi: 10.1074/jbc.M412238200. [DOI] [PubMed] [Google Scholar]
- Brouns SJJ, Jore MM, Lundgren M, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–4. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Genome engineering with zinc-finger nucleases. Genetics. 2011;188:773–82. doi: 10.1534/genetics.111.131433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casini A, MacDonald JT, De Jonghe J, et al. One-pot DNA construction for synthetic biology: the Modular Overlap-Directed Assembly with Linkers (MODAL) strategy. Nucleic Acids Res. 2014;42:e7. doi: 10.1093/nar/gkt915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspeta L, Chen Y, Ghiaci P, et al. Altered sterol composition renders yeast thermotolerant. Science. 2014;346:75–8. doi: 10.1126/science.1258137. [DOI] [PubMed] [Google Scholar]
- Chee MK, Haase SB. New and redesigned pRS Plasmid shuttle vectors for genetic manipulation of Saccharomyces cerevisiae. G3 (Bethesda) 2012;2:515–26. doi: 10.1534/g3.111.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–61. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, et al. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–22. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- De Kok S, Nijkamp JF, Oud B, et al. Laboratory evolution of new lactate transporter genes in a jen1Δ mutant of Saccharomyces cerevisiae and their identification as ADY2 alleles by whole-genome resequencing and transcriptome analysis. FEMS Yeast Res. 2012;12:359–74. doi: 10.1111/j.1567-1364.2012.00787.x. [DOI] [PubMed] [Google Scholar]
- De Kok S, Yilmaz D, Suir E, et al. Increasing free-energy (ATP) conservation in maltose-grown Saccharomyces cerevisiae by expression of a heterologous maltose phosphorylase. Metab Eng. 2011;13:518–26. doi: 10.1016/j.ymben.2011.06.001. [DOI] [PubMed] [Google Scholar]
- Delneri D, Tomlin GC, Wixon JL, et al. Exploring redundancy in the yeast genome: an improved strategy for use of the cre-loxP system. Gene. 2000;252:127–35. doi: 10.1016/s0378-1119(00)00217-1. [DOI] [PubMed] [Google Scholar]
- Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–7. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Conley AJ, Penttilä M, et al. Yeast oligo-mediated genome engineering (YOGE) ACS Synth Biol. 2013a;2:741–9. doi: 10.1021/sb400117c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCarlo JE, Norville JE, Mali P, et al. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013b;41:4336–43. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entian K-D, Kötter P. 25 Yeast genetic strain and plasmid collections. Method Microbiol. 2007;36:629–66. [Google Scholar]
- Feng Z, Zhang B, Ding W, et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013;23:1229–32. doi: 10.1038/cr.2013.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhao Y. Self-processing of ribozyme-flanked RNAs into guide RNAs in vitro and in vivo for CRISPR-mediated genome editing. J Integr Plant Biol. 2014;56:343–9. doi: 10.1111/jipb.12152. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Benders G a, Andrews-Pfannkoch C, et al. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215–20. doi: 10.1126/science.1151721. [DOI] [PubMed] [Google Scholar]
- Gietz DR, Woods RA. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Method Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- González F, Zhu Z, Shi Z-D, et al. An iCRISPR platform for rapid, multiplexable, and inducible genome editing in human pluripotent stem cells. Cell Stem Cell. 2014;15:215–26. doi: 10.1016/j.stem.2014.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ramos D, van den Broek M, van Maris AJ, et al. Genome-scale analyses of butanol tolerance in Saccharomyces cerevisiae reveal an essential role of protein degradation. Biotechnol Biofuels. 2013;6:48. doi: 10.1186/1754-6834-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz SJ, Cummings AM, Nguyen JN, et al. Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics. 2013;194:1029–35. doi: 10.1534/genetics.113.152710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guadalupe-Medina V, Wisselink HW, Luttik MA, et al. Carbon dioxide fixation by Calvin-Cycle enzymes improves ethanol yield in yeast. Biotechnol Biofuels. 2013;6:125. doi: 10.1186/1754-6834-6-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueldener U, Heinisch J, Koehler GJ, et al. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fielder T, et al. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–24. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegemann JH, Heick SB. Delete and repeat: a comprehensive toolkit for sequential gene knockout in the budding yeast Saccharomyces cerevisiae. Methods Mol Biol. 2011;765:189–206. doi: 10.1007/978-1-61779-197-0_12. [DOI] [PubMed] [Google Scholar]
- Hong K-K, Vongsangnak W, Vemuri GN, et al. Unravelling evolutionary strategies of yeast for improving galactose utilization through integrated systems level analysis. P Natl Acad Sci USA. 2011;108:12179–84. doi: 10.1073/pnas.1103219108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Lander ES, Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–78. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PD, Scott DA, Weinstein JA, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–32. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31:227–9. doi: 10.1038/nbt.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JZ, Ciccaglione KM, Tournier V, et al. Implementation of the CRISPR-Cas9 system in fission yeast. Nat Commun. 2014;5:5344. doi: 10.1038/ncomms6344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WS, Yoo YJ, Park JW, et al. A combined approach of classical mutagenesis and rational metabolic engineering improves rapamycin biosynthesis and provides insights into methylmalonyl-CoA precursor supply pathway in Streptomyces hygroscopicus ATCC 29253. Appl Microbiol Biotechnol. 2011;91:1389–97. doi: 10.1007/s00253-011-3348-6. [DOI] [PubMed] [Google Scholar]
- Koopman F, Beekwilder J, Crimi B, et al. De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb Cell Fact. 2012;11:155. doi: 10.1186/1475-2859-11-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak BU, van Rossum HM, Benjamin KR, et al. Replacement of the Saccharomyces cerevisiae acetyl-CoA synthetases by alternative pathways for cytosolic acetyl-CoA synthesis. Metab Eng. 2014a;21:46–59. doi: 10.1016/j.ymben.2013.11.005. [DOI] [PubMed] [Google Scholar]
- Kozak BU, van Rossum HM, Luttik MAH, et al. Engineering acetyl coenzyme A supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae. mBio. 2014b;5:e01696–14. doi: 10.1128/mBio.01696-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers NGA, Chroumpi S, Vos T, et al. One-step assembly and targeted integration of multigene constructs assisted by the I-SceI meganuclease in Saccharomyces cerevisiae. FEMS Yeast Res. 2013a;13:769–81. doi: 10.1111/1567-1364.12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijpers NGA, Solis-Escalante D, Bosman L, et al. A versatile, efficient strategy for assembly of multi-fragment expression vectors in Saccharomyces cerevisiae using 60 bp synthetic recombination sequences. Microb Cell Fact. 2013b;12:47. doi: 10.1186/1475-2859-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunes S, Botstein D, Fox SM. Transformation of yeast with linearized plasmid DNA. Formation of inverted dimers and recombinant plasmid products. J Mol Biol. 1985;184:375–87. doi: 10.1016/0022-2836(85)90288-8. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, et al. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Cradick TJ, Brown MT, et al. CRISPR/Cas9 systems have off-target activity with insertions or deletions between target DNA and guide RNA sequences. Nucleic Acids Res. 2014;42:7473–85. doi: 10.1093/nar/gku402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner Zu Siederdissen C, et al. ViennaRNA Package 2.0. Algorithms Mol Biol. 2011;6:26. doi: 10.1186/1748-7188-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marraffini LA, Sontheimer EJ. CRISPR interference limits horizontal gene transfer in Staphylococci by targeting DNA. 2008;322:1843–5. doi: 10.1126/science.1165771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–8. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Montague TG, Cruz JM, Gagnon JA, et al. CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–7. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lütge F, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39:9283–93. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Larsson C, van Maris A, et al. Metabolic engineering of yeast for production of fuels and chemicals. Curr Opin Biotechnol. 2013;24:398–404. doi: 10.1016/j.copbio.2013.03.023. [DOI] [PubMed] [Google Scholar]
- Nijkamp JF, van den Broek M, Datema E, et al. De novo sequencing, assembly and analysis of the genome of the laboratory strain Saccharomyces cerevisiae CEN.PK113–7D, a model for modern industrial biotechnology. Microb Cell Fact. 2012;11:36. doi: 10.1186/1475-2859-11-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW. Yeast recombination: the association between double-strand gap repair and crossing-over. P Natl Acad Sci USA. 1983;80:4417–21. doi: 10.1073/pnas.80.14.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver TL, Szostak JW, Rothstein RJ. Genetic applications of yeast transformation with linear and gapped plasmids. Method Enzymol. 1983;101:228–45. doi: 10.1016/0076-6879(83)01017-4. [DOI] [PubMed] [Google Scholar]
- Oud B, Guadalupe-Medina V, Nijkamp JF, et al. Genome duplication and mutations in ACE2 cause multicellular, fast-sedimenting phenotypes in evolved Saccharomyces cerevisiae. P Natl Acad Sci USA. 2013;110:E4223–31. doi: 10.1073/pnas.1305949110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddon CJ, Westfall PJ, Pitera DJ, et al. High-level semi-synthetic production of the potent antimalarial artemisinin. Nature. 2013;496:528–32. doi: 10.1038/nature12051. [DOI] [PubMed] [Google Scholar]
- Pronk JT. Auxotrophic yeast strains in fundamental and applied research. Appl Environ Microb. 2002;68:2095–210. doi: 10.1128/AEM.68.5.2095-2100.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ro D-K, Paradise EM, Ouellet M, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–3. doi: 10.1038/nature04640. [DOI] [PubMed] [Google Scholar]
- Roberts RJ. How restriction enzymes became the workhorses of molecular biology. P Natl Acad Sci USA. 2005;102:5905–8. doi: 10.1073/pnas.0500923102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan OW, Skerker JM, Maurer MJ, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. Elife. 2014;3:e03703. doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Z, Zhao H, Zhao H. DNA assembler, an in vivo genetic method for rapid construction of biochemical pathways. Nucleic Acids Res. 2009;37:e16. doi: 10.1093/nar/gkn991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewers V. An overview on selection marker genes for transformation of Saccharomyces cerevisiae. Methods Mol Biol. 2014;1152:3–15. doi: 10.1007/978-1-4939-0563-8_1. [DOI] [PubMed] [Google Scholar]
- Solis-Escalante D, Kuijpers NGA, Bongaerts N, et al. amdSYM, a new dominant recyclable marker cassette for Saccharomyces cerevisiae. FEMS Yeast Res. 2013;13:126–39. doi: 10.1111/1567-1364.12024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis-Escalante D, Kuijpers NGA, van der Linden FH, et al. Efficient simultaneous excision of multiple selectable marker cassettes using I-SceI-induced double-strand DNA breaks in Saccharomyces cerevisiae. FEMS Yeast Res. 2014a;14:741–54. doi: 10.1111/1567-1364.12162. [DOI] [PubMed] [Google Scholar]
- Solis-Escalante D, van den Broek M, Kuijpers NGA, et al. The genome sequence of the popular hexose transport deficient Saccharomyces cerevisiae strain EBY.VW4000 reveals LoxP/Cre-induced translocations and gene loss. FEMS Yeast Res. 2014b;15 doi: 10.1093/femsyr/fou004. doi.org/10.1093/femsyr/fou004. [DOI] [PubMed] [Google Scholar]
- Storici F, Coglievina M, Bruschi CV. A 2-micron DNA-based marker recycling system for multiple gene disruption in the yeast Saccharomyces cerevisiae. Yeast. 1999;15:271–83. doi: 10.1002/(SICI)1097-0061(19990315)15:4<271::AID-YEA371>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Storici F, Lewis LK, Resnick MA. In vivo site-directed mutagenesis using oligonucleotides. Nat Biotechnol. 2001;19:773–6. doi: 10.1038/90837. [DOI] [PubMed] [Google Scholar]
- Terentiev Y, Pico AH, Böer E, et al. A wide-range integrative yeast expression vector system based on Arxula adeninivorans-derived elements. J Ind Microbiol Biot. 2004;31:223–8. doi: 10.1007/s10295-004-0142-9. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, et al. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–46. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Van den Berg MA, de Jong-Gubbels P, Kortland CJ, et al. The two acetyl-coenzyme A synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem. 1996;271:28953–9. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- Verduyn C, Postma E, Scheffers WA, et al. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–17. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila CS, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153:910–8. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Wang L. New methods enabling efficient incorporation of unnatural amino acids in yeast. J Am Chem Soc. 2008;130:6066–7. doi: 10.1021/ja800894n. [DOI] [PubMed] [Google Scholar]
- Wieczorke R, Krampe S, Weierstall T, et al. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999;464:123–8. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- Zhang F. CRISPR-Cas systems and methods for altering expression of gene products. 2014 US patent no. 8,697,359 B1. [Google Scholar]
- Zhang G, Kong II, Kim H, et al. Construction of a quadruple auxotrophic mutant of an industrial polyploidy Saccharomyces cerevisiae using RNA-guided Cas9 nuclease. Appl Environ Microb. 2014;80:7694–701. doi: 10.1128/AEM.02310-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.