Summary
Naked mole-rats (Heterocephalus glaber), the longest-lived rodents, live 7-10 times longer than similarly–sized mice and exhibit normal activities for ~75% of their lives. Little is known about the mechanisms that allow them to delay the aging process and live so long. Neuregulin-1 (NRG-1) signaling is critical for normal brain function during both development and adulthood. We hypothesized that long-lived species will maintain higher levels of NRG-1 and that this contributes to their sustained brain function and concomitant maintenance of normal activity. We monitored the levels of NRG-1 and its receptor ErbB4 in H. glaber at different ages ranging from 1 day to 26 years and found that levels for NRG-1 and ErbB4 were sustained throughout development and adulthood. In addition, we compared seven rodent species with widely divergent (4-32y) maximum lifespan potential (MLSP) and found that at a physiologically-equivalent age, the longer-lived rodents had higher levels of NRG-1 and ErbB4. Moreover, phylogenetic independent contrast analyses revealed that this significant strong correlation between MLSP and NRG-1 levels was independent of phylogeny. These results suggest that NRG-1 is an important factor contributing to divergent species MLSP through its role in maintaining neuronal integrity.
Keywords: Aging, neurotrophic factors, naked mole-rat, maximum lifespan, Spalax, cerebellum, Neuregulin-1, ErbB4
INTRODUCTION
“The reasons for some animals being long-lived and others short-lived, and, in a word, causes of the length and brevity of life call for investigation”
-Aristotle, On Longevity and Shortness of Life. (Aristotle, 350 B.C.E)
Human awareness that different animal species show divergent rates of aging predates modern science. Indeed it has been known for millennia that mice and rats are shorter-lived than domestic livestock such as sheep and cows. This relationship is largely due to species differences in body size. Species maximum lifespan potential (MLSP) correlates with body mass such that for every doubling of body mass, MLSP increases by 16% (Hulbert et al 2007). However, only 35% of the variability in species’ lifespan can be explained by body mass and it is still unknown why some species (e.g., humans and naked mole-rats) age more slowly than do others (e.g., shrews and mice). Maximum lifespan varies by ~40,000-fold across the animal kingdom and even within a single mammalian order, Rodentia, there is a 10-fold range in lifespan among similarly-sized species with the shortest-lived rodents living less than three years and the longest-lived rodents, naked mole-rats, (Heterocephalus glaber) with a MLSP of ~32 years; this MLSP is based upon data from an adult-mass wild-caught male that was maintained in captivity for 31 years. These differences in rodent longevity are much greater than those that can be experimentally induced through caloric restriction (Masoro, 2005), drug intervention (Harrison et al., 2009) or even genetic mutations (Hulbert et al., 2007). The central nervous system obviously plays a vital role in sustaining life and here we explore one link between disparate species MLSP and a key component of brain function, namely growth factors which may play a pivotal role in longevity among species (Cho and Leahy, 2002; Kraus et al., 1989). These factors help maintain cellular integrity, promote repair and provide a putative mechanism for replacing cells that may have either died through necrosis/ apoptosis or may have senesced. Growth factors are critically important in brain development and although expression commonly declines with age, expression of these neurotrophic factors can persist throughout the lifetime of an organism, albeit at a lower level than during development, (Hwang et al., 2006) thereby maintaining neuronal integrity in mature animals. As such, mature adult levels potentially may serve as a determinant of lifespan itself.
Neuregulins are growth and differentiation factors that play critical roles in neuron development, protection and survival (Esper et al., 2006) and are critical for normal brain function. A key neurotrophic factor within this family is Neuregulin-1 (NRG-1, previously known as Heregulin, NDF, or ARIA). NRG-1 has two isoforms (α and β) both of which are present in human brains as well as those of young and old rats (Bernstein et al., 2006). NRG-1 receptors (ErbB2, ErbB3 and ErbB4), like NRG-1, are present in the nervous system throughout life (Esper et al., 2006; Gerecke et al., 2001) and widely distributed throughout the brain, although a decrease in both NRG-1 and its receptors is evident in the adult brain (Bernstein et al., 2006; Law et al., 2004). This ligand-receptor system initiates signaling pathways responsible for a variety of biological activities including the development of the cerebellum and is involved in cell differentiation, survival and repair of mature neurons as well as cell migration and synaptic formation during neurogenesis (Bernstein et al., 2006). NRG-1 signaling in the adult brain has been studied extensively (reviewed in (Buonanno, 2010); It is critically important in synaptic transmission being involved in both GABA and AMPAR-mediated synaptic transmission (reviewed in (Fischbach, 2007)). Moreover, recent studies have highlighted the importance of NRG-1 signaling in sustaining neuronal function especially under toxic conditions. In an elegant set of studies, (Min et al., 2011) highlight the neuroprotective role of NRG-1 signaling. The adverse effects of amyloid beta (Aβ1-42) excitotoxicity on long-term potentiation (LTP) in hippocampal slices were rescued by the treatment with NRG-1. Furthermore, these actions were mediated via the ErbB4 receptor but not via ErbB2. Clearly NRG-1 prevents impairment of hippocampal function by both endogenous and exogenous neurotoxins and as such this signaling pathway may play a pivotal role in the maintenance of neuronal integrity over the lifespan of long-lived species.
In humans, perturbations in the activity and distribution of NRG-1 and/or its receptors are associated with neurological diseases such as schizophrenia (Hahn et al., 2006), multiple sclerosis (Esper et al., 2006) and Alzheimer’s Disease (Chaudhury et al., 2003). NRG-1 levels are regulated by several transcription factors including hypoxia-inducible transcription factors (HIF-1α). For example, when hypoxia is induced in neonatal rats, the change in HIF-1α levels lead to increased NRG-1α levels in their brains (Nadri et al., 2007).
We hypothesize that since neurotrophic growth factors contribute to the maintenance of neuronal integrity, long-lived species maintain higher levels of NRG-1 than do short-lived species. This therefore facilitates their observed prolonged and sustained brain function. Since H. glaber live an order of magnitude longer than do similar-sized mice, we predict that naked mole-rats do not show the typical age-related decline in NRG-1 levels reported in mice. We therefore determined the expression levels of NRG-1 and its receptor, ErbB4, throughout the long-life of H. glaber (from 1 day to 26 years). Since NRG-1 and its receptors have the potential to mediate both neurogenesis and neuronal survival, we assessed if adult H. glaber brains have higher expression levels of NRG-1 and its receptor than six shorter-lived rodent species diverging in MLSP from 3.5-32 years (Figure 1; Table 1). These include C57Bl/6 Mice (Mus musculus), spiny mice (Acomys cahirinus), guinea pigs (Cavia porcellus), damaraland mole-rats (Fukomys damarensis), blind mole-rats (Spalax carmeli and Spalax judaei, both belonging to the Spalax ehrenbergi superspecies) and naked mole-rats (H. glaber). Moreover, we specifically tested if levels of NRG-1 in adult cerebella correlate with species MLSP. These species fall into two distinct rodent suborders (Myomorpha; Hystricomorpha sensus (Wilson and Reeder, 2005); and two distinct lifestyles (above-ground dwellers and subterranean rodents). Given that subterranean rodents naturally encounter a more hypoxic gaseous atmosphere below ground (Arieli, 1979) we also measured hypoxia-inducible factor 1 alpha (HIF-1α) a transcription factor induced by hypoxia. We specifically chose to use the cerebellum for this study because it is involved in movement control, an important aspect of healthy aging. Moreover, cerebella are large, discrete, easily identifiable tissues that can be readily isolated in all rodent species. Furthermore, cerebella contain high levels of both NRG-1 (Bernstein et al., 2006) and its receptor, ErbB4 (Gerecke et al., 2001) in humans, primates and rodents.
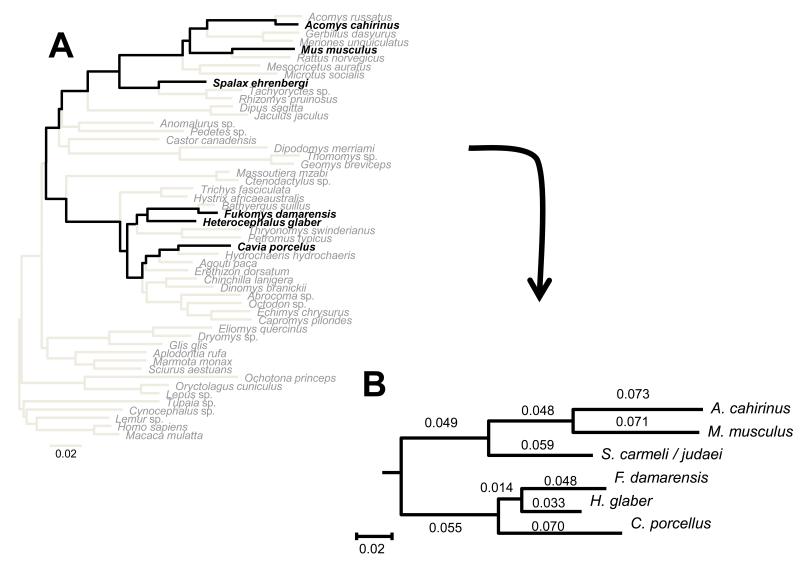
Fig. 1. Phylogenetic trees for rodent species.
Panel A: Maximum likelihood tree reconstructed based on five nuclear genes under the GTR model of sequence evolution. Panel B: phylogenetic tree and branch lengths inferred, for the species under study, based on the larger tree. Species included in this study appear in black.
Table 1. Rodent species in this study.
All rodents included in this study were physiologically aged-matched adults which had lived at least 50% of their lives. Body mass, MLSP and phylogeny differed between these species, and these variables were taken into account in our analyses. Levels of NRG-1 and ErbB4 were assessed by immunoblot, normalized to actin and appear in arbitrary units (A.U).
| Species | Common name | Suborder | Family | Average body mass (g) |
Average brain mass (g) |
MLSP (years) |
NRG-1 (A.U) |
ErbB4 (A.U) |
MLSP Reference |
|---|---|---|---|---|---|---|---|---|---|
| Mus musculus | Lab mouse (C57Bl/6) |
Myomorpha | Muridae | 30 | 0.4 | 3.5 | 0.84 | 1.70 | Jax.org |
| Acomys cahirinus | Spiny mouse | Myomorpha | Muridae | 50 | 0.5 | 5.9 | 0.79 | 1.71 | Weigl 2005 |
| Cavia porcellus | Guinea pig | Hystricognathi | Caviidae | 970 | 3.1 | 12 | 1.00 | 1.74 | Kiklevich, VJ (AnAge website) |
| Spalax carmeli | Blind mole-rat | Myomorpha | Spalacidae | 130 | 1.7 | 19 | 1.17 | 2.20 | Nevo, per. obs. |
| Spalax judaei | Blind mole-rat | Myomorpha | Spalacidae | 100 | 1.4 | 19 | 0.87 | 2.30 | Nevo, per. obs. |
| Fukomys damarensis | Damaraland mole-rat |
Hystricognathi | Bathyergidae | 140 | 1.4 | 16 | 1.23 | 3.47 | Goldman B, per. comm. |
|
Heterocephalus
glaber |
Naked mole-rat | Hystricognathi | Bathyergidae | 40 | 0.4 | 32 | 1.63 | 2.61 | Buffenstein, per. obs. |
RESULTS
NRG-1, ErbB4 and HIF1α epitope regions show high percentage of identity among rodent species
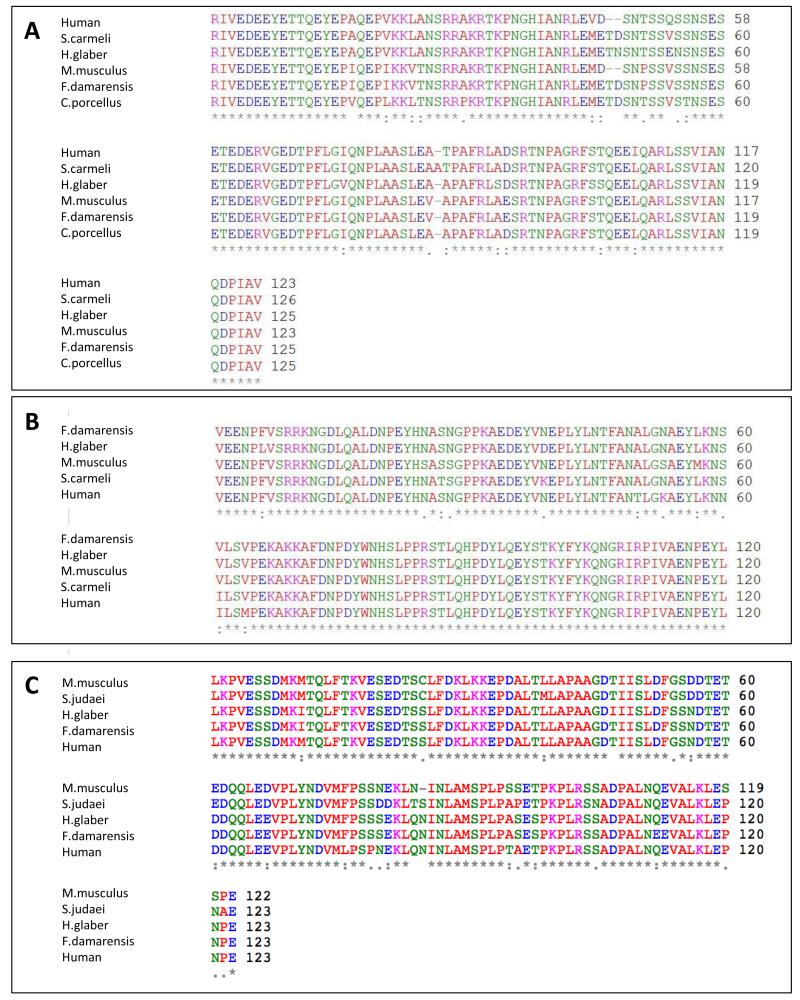
For species whose sequence was not found in Genbank, we sequenced the epitopes for NRG-1, ErbB4 and/or HIF1α. These sequences were submitted to GenBank under the following accession numbers: For NRG-1: Fukomys damarensis HQ014873.1; H. glaber HM438944.1; Spalax carmeli HM438945.1; Cavia porcellus HQ014872.1. For ErbB4: F. damarensis HQ014868.1; S. carmeli HQ014869.1; H. glaber HQ014870.1. For HIF1: F. damarensis HQ014863.1; H. glaber HQ014864.1. To determine if the antibodies raised against human proteins would recognize rodent antigens, we aligned the human and rodent sequences and determined the percentage of identity among these sequences. The percentage of identity between two NRG-1 sequences in the epitope region was at least 91% in all cases, which corresponds to the percent of identity between the human and mouse NRG-1 epitopes (Fig. 2A). For ErbB4, the percentage of identity was lowest between F. damarensis and human (90%) whereas mouse and human aligned with 93% identity in the epitope region (Fig. 2B). For HIF1α the percentage identity between humans and H. glaber or F. damarensis was 92%, 90% between M. musculus and H. glaber or F. damarensis, 91% between S. judaei and M. musculus and 87% between S. judaei and humans (Fig. 2C). Since the antibodies for NRG-1, ErbB4 and HIF1α are routinely used to test expression levels in mice as well as humans, similar homology among the exotic species and mice/humans reveal that the use of these antibodies also reflect quantitative assessments of protein levels in the novel species. Furthermore, bands were evident in all species tested and had identical molecular weights. In cases where blocking peptide for the proteins was available, these were used to serve as an additional indicator for the specificity of the binding recognition for all novel species. Finally, it is likely that each species has its own unique protein milieu that could introduce recognition artifacts and interfere with antibody recognition and/or binding. However, in all cases the novel species exhibited higher expression bands for all three proteins under examination. We therefore deduce that the patterns may represent an underestimation rather than an overestimation of the actual levels of proteins in these species.
Fig. 2. NRG-1, ErbB4 and HIF1α protein sequence alignment for novel species used in this study.
In order to verify proper recognition of the antibodies used to the non-traditional species of rodents in this study, in cases where the sequences were not available, the areas containing the epitopes were sequenced. These were then compared to human sequences for which the antibodies are raised against, and to M.musculus which are routinely used in research. Panel A: NRG-1 alignments show 97% between humans and S.carmeli, 91% between humans and M.musculus or F.damarensis and 92% between humans and H.glaber and C.porcellus. Panel B: ErbB4 alignments show 93% between humans and M.musculus, 94% between humans and H.glaber and S.carmeli and 95% between humans and F.damarensis. Panel C: HIF1α alignment show 92% humans and H.glaber or F.damrensis, 90% between M.musculus and H.glaber or F.damarensis, 91% between S.judaei and M.musculus and 87% between S.judaei and humans.
NRG-1 and ErbB4 expression levels do not change throughout the naked mole-rat’s long life
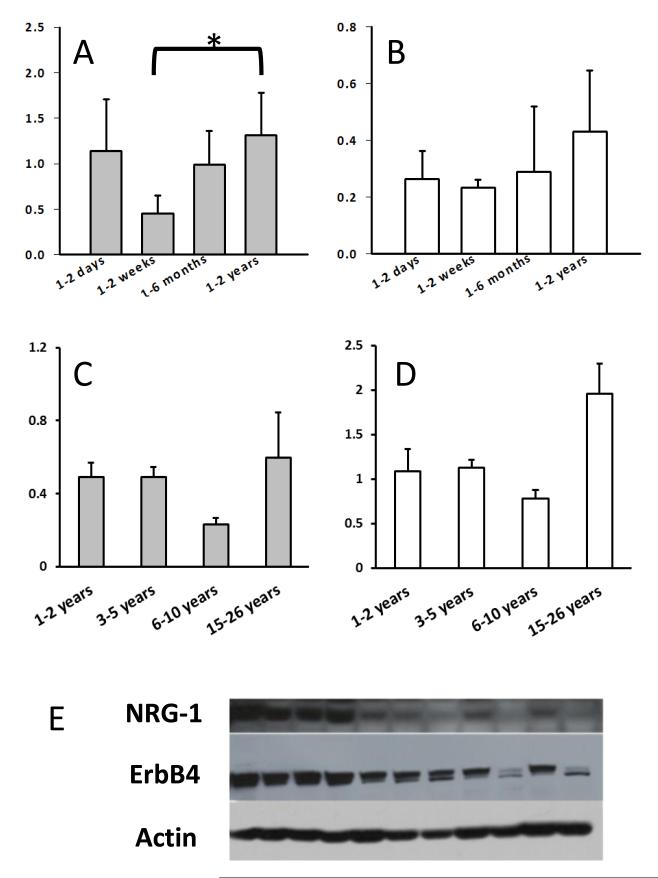
We compared the age-related (1 day old to 26 years) profile of NRG-1 expression in the cerebella of H. glaber during development and throughout adulthood. Expression levels were highest in the first days of life, but surprisingly not significantly different from young adults (1-2 years). Though interestingly, the 1-2 weeks of age cohort were significantly lower than the young adult group (Fig.3A; ANOVA DF=3, F=3.54, p=0.034). Expression levels of the receptor ErbB4 were unchanged during development and similar to that of young adults (Fig. 3B; ANOVA D=3, F=1.638, p=0.214).
Fig. 3. Levels of NRG1 and ErbB4 throughout the life of H.glaber.
Panel A: NRG-1 levels of 1-day old individuals were higher to those of young adults but did not reach statistical significance. NRG-1 levels were significantly lower at 1-2 weeks of age (ANOVA DF=3, F=3.54, p=0.034). Panel B: Levels of the receptor ErbB4 were unchanged from young pups to young adults (ANOVA D=3, F=1.638, p=0.214). Panel C: Levels of NRG-1 did not change from the youngest adult cohort to that of the oldest group (ANOVA DF=3, F=1.48, p=2.60). Levels of ErbB4 during adulthood did not change significantly (ANOVA DF=3, F=2.78, p=0.077). Panel D: Data are presented as SEM. Sample sizes 1-2 days (n= 8), 1-2 weeks (n=5), 1-6 months (n=5), 1-2 years (n=5). Sample sizes for 2 yrs. (n=9), 3-5 yrs. (n=5), 10yrs. (n=3), 15-26 yrs. (n=3). Panel E: Representative immunoblot for NRG-1 and ErbB4 containing all ages of H.glaber used in this study. Data are normalized by actin.
Young adult (1-2 years) NRG-1 levels did not significantly differ from those of the oldest age group (15-26 years) (Fig. 3C; ANOVA DF=3, F=1.48, p=2.60) and neither did protein expression of the ErbB4 receptor (Fig. 3D; ANOVA DF=3, F=2.78, p=0.077). These findings suggest that unlike laboratory rodents and humans NRG-1 and ErbB4 expression levels do not decline with age in adult H. glaber (Law et al., 2004).
NRG-1 and ErbB4 expression correlate with maximum lifespan in rodent species but not with species body mass
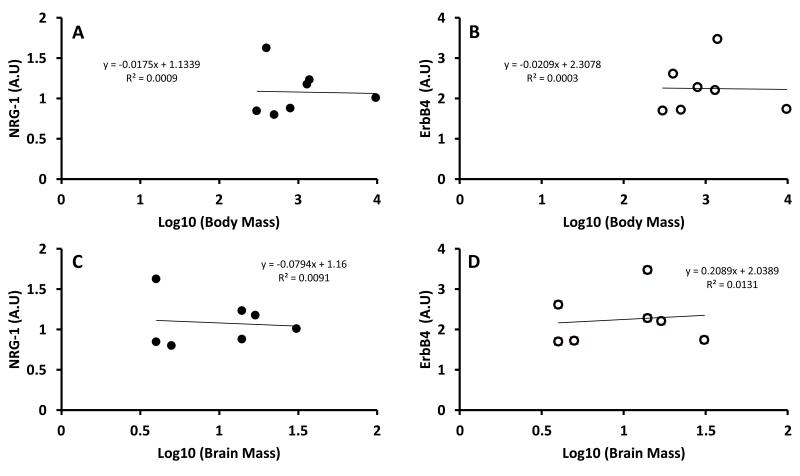
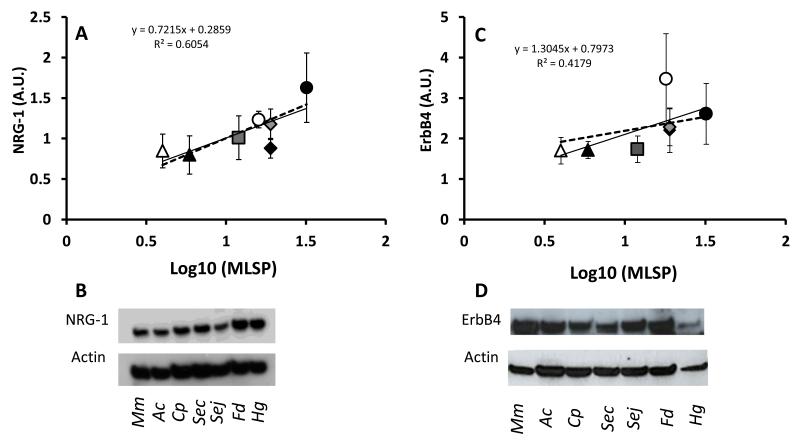
Both NRG-1 and ErbB4 expression in cerebella showed a two-fold difference among species (Table 1; p<0.001). We assessed if NRG-1 and ErbB4 levels were correlated with species average brain mass (Fig. 4A-B), body mass (Fig. 4C-D) and/or MLSP. Given the strong allometric relationship between species’ body size and longevity, we were surprised that neither NRG-1, nor ErbB4 were correlated with species brain mass (NRG-1, R2=0.00915, p=0.85; ErbB4, R2=0.0131, p=0.90, Fig 4A, B) or with species body mass (NRG-1, R2=0.00092, p=0.85; ErbB4, R2=0.00027, p=0.56, Fig 5C, D). However, there was a highly significant correlation between NRG-1 expression and MLSP (R2=0.605, p=0.008, Fig.5A) but not between ErbB4 levels and MLSP (R2=0.418, p=0.193, Fig. 5B). The lack of significance of the correlation between the receptor levels and longevity was primarily due to one species, F. damarensis, that had higher expression levels than three longer-lived species (two Spalax species and H. glaber).
Fig. 4. Levels of NRG-1 and ErbB4 in rodent species do not correlate with brain mass or body mass.
NRG-1 (Panel A) and ErbB4 (Panel B) expression levels did not correlate with brain mass (p=0.85, p=0.56, respectively). Similarly, levels of NRG-1 (Panel C) and ErbB4 (Panel D) did not correlate with body mass (p=0.85, p=0.90) in the rodent species under study (See Table 1).
Fig. 5. Expression levels of NRG1 and its receptor ErbB4 correlate with MLS in 7 rodent species.
Expression levels were assessed in cerebella from 6 individuals per species by immunoblots. The comparisons were performed on physiologically age-matched adults that had lived at least 50% of their lives. The species include short-lived M.musculus (white triangles) and A.cahirinus (black triangles). The longer-lived species include C.porcellus (rectangles), F.damarensis (black circles), S.carmeli (grey diamonds) and S.judaei (black diamonds). The longest living rodent is the H.glaber (black circles). Panel A: NRG-1 expression levels correlate with MLS (R2=0.605). Panel B: Representative immunoblot of 46kDa NRG-1 in all of the species in this study. To test whether the observed correlations of NRG-1/ ErbB4 and MLSP were confounded by the phylogenetic distances between these species, an independent contrast analysis was employed. Taking the phylogenetic distances between species into account, the high correlation between NRG-1 levels was statistically significant (p=0.045, Panel A, dashed line). Panel C: ErbB4 expression levels correlate with MLS (R2=0.418). Panel D: Representative immunoblot of 180kDa ErbB4 for all species in this study. The correlation between ErbB4 and MLSP was lower and was not significant once the phylogenetic lineage was included in the analysis (p=0.57, Panel B, dashed line). This suggests that for ErbB4, the initial correlation with MLSP was biased due to a shared trait by phylogenetic distance, and does not truly reflect the MLSP property. Immunoblots include from left to right: M. musculus, A. cahirilus, C. porcellus, S. carmeli, S. judaei, F. damarensis and H. glaber. Data are presented as mean ± SEM. Arbitrary units (A.U) are used as normalized by actin.
To remove the potentially confounding effects of shared phylogenetic lineages among species that could influence our interpretation, we calculated the maximum likelihood phylogenetic distances between species and implemented these into a Generalized Least Squares (GLS) analysis. We found a statistically significant correlation between MLSP and NRG-1 levels (p=0.045, Fig.5A). This suggests that the observed correlation between NRG-1 levels and MLSP are phylogenetically independent and as such NRG-1 may serve as a determinant of maximum lifespan in rodents. In contrast, phylogenetic independent contrast analysis revealed that ErbB4 expression levels are not directly involved in species longevity (p=0.57, Fig. 5B). It is possible that NRG-1 activity does not depend upon a stoichiometric relationship with ErbB4, and that the other receptors ErbB2 and ErbB3 may also facilitate NRG-1 activity.
HIF1α levels are higher in species that have evolved underground
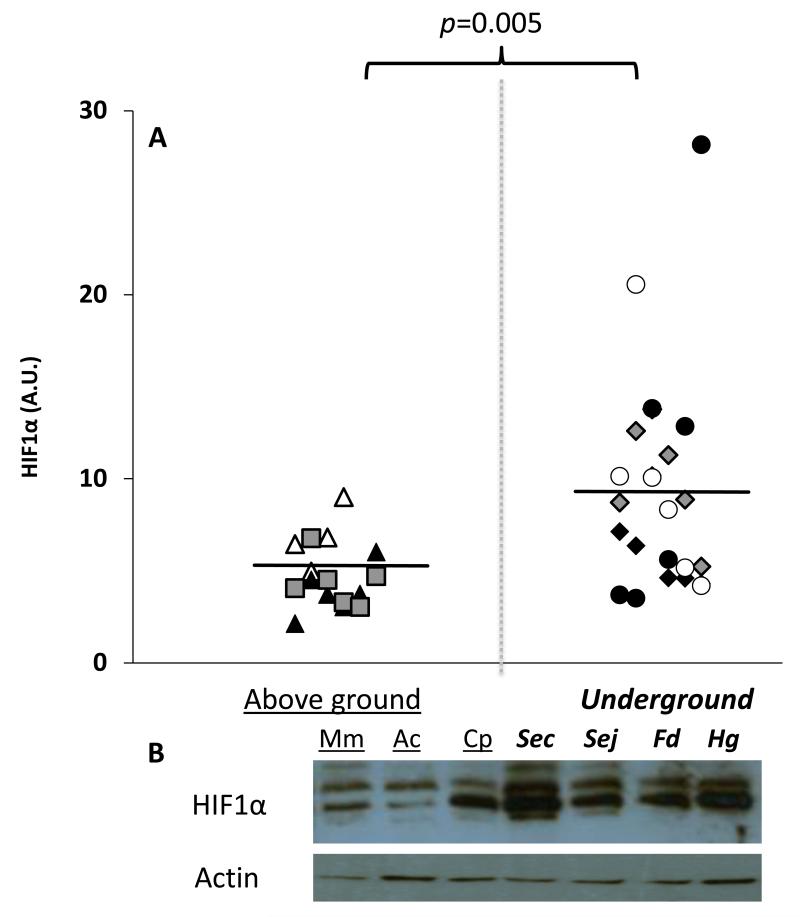
All four of the subterranean species (H. glaber, F. damarensis, S. carmeli, S. judaei) had similar levels of HIF1α (ANOVA, DF=7, F=0.122, p=0.996) and had on average 1.8-fold higher than those of animals that naturally reside above ground (t-test p<0.005, Fig. 6). These results are not surprising given the fact that these species have resided underground for millennia and have evolved a suite of adaptations well suited to life under hypoxic conditions reviewed in (Edrey et al., 2011).
Fig. 6. HIF1α expression levels are higher in species that live underground.
Expression levels were assessed in cerebella from 6 individuals per species by immunoblots. The comparisons were performed on physiologically age-matched adults that had lived at least 50% of their lives. The species include short-lived M. musculus (white triangles) and A.cahirinus (black triangles). The longer-lived species include C. porcellus (rectangles), F.damarensis (white circles), S. carmeli (grey diamonds), S. judaei (black diamonds) and H.glaber (black circles). Panel A: Individuals of species that live above ground were significantly different from those that have evolved to live underground (t-test p=0.005). All individual animals in this study are represented on the graphs. Panel B: Representative images of the immunoblot with corresponding species appear below. Data appear in arbitrary units (A.U) and are normalized by actin.
DISCUSSION
In this study we asked if the levels of NRG-1 and its receptor ErbB4 change during both development and aging in H. glaber and if this may contribute to the more than three-decades of sustained cognition of these eusocial rodents. Furthermore, we asked if adult levels of NRG-1 may serve as a determinant of species lifespan, given that this neuroprotective growth factor helps maintain neuronal integrity and brain function. Here we show for the first time that, unlike laboratory rodents and humans both NRG-1 and its receptor, ErbB4 are maintained at high levels throughout life in H. glaber. Furthermore, cerebella from this species exhibits the highest NRG-1 levels among the seven species of rodents examined and NRG-1 levels correlate with species MLSP. These findings suggest that NRG-1 may be integral to sustained neuron integrity in species that have long lifespans and that neuroprotective growth factor expression may be an important component of species longevity.
Little is known about the mechanisms contributing to the observed prolonged healthspan, sustained activity and exceptional longevity of H. glaber (Buffenstein, 2005). These eusocial animals also show many signs of well-maintained cognitive longevity as indicated by their ability to carry out complex behavioral and social routines such as communal foraging, care of younger siblings and burrow maintenance in their vast maze-like underground habitat throughout their lives (per. obs.). High levels of NRG-1 and concomitant maintenance of cognitive function goes hand in hand with the abrogated decline in physiological function, reproduction, body composition and biochemical traits (Andziak and Buffenstein, 2006; Ungvari et al., 2008). Sustained physiological and molecular function may also contribute to the pronounced resistance of H. glaber to both spontaneous and induced neoplasia (Liang et al., 2010). It is speculated that because H. glaber have low extrinsic mortality, living in large communal groups in a thermally buffered and protected underground habitat, they have evolved protective mechanisms to maintain cellular and organ-based function well into old age (Edrey et al., 2011).
One such protective mechanism is the high and sustained levels of neuroprotective NRG-1 (Min et al., 2011) which is critical for synaptic function (reviewed in (Buonanno, 2010). This may contribute to the prolonged survival and maintenance of neuronal function. Expression of both NRG-1 and ErbB4 in adult H. glaber is maintained at similar high levels to those observed in 1 day-old pups that are still undergoing brain development. As such it is highly likely that NRG-1 is used to maintain neuronal integrity and phenotypic plasticity in this species. For example, although we only examined non-breeding animals, any subordinate is capable of changing its status to a breeder and when this occurs undergoing marked changes in brain morphology and function in keeping with altered social status (reviewed in Edrey et al., 2011).
Levels of NRG-1 are altered in schizophrenia patients, and this finding has spurred the use of genetically modified mice with either elevated or decreased levels of NRG-1 to examine behaviors associated with this growth factor. Studies in these animals have demonstrated the role of NRG-1 signaling in social interactions and in sensory and locomotor activity (Ehrlichman et al., 2009; O’Tuathaigh et al., 2007). For example, a recent study on NRG-1 heterozygous mutant mice revealed that NRG-1 is critical for social interactions (O’Tuathaigh et al., 2007) in addition to its known roles in neural maintenance and development. H. glaber are the most social rodent species known, living in colonies of up to 300 individuals with a diverse array of social interactions and a repertoire of various modes of communication (Edrey et al., 2011). Given the role of NRG-1 in social interactions in humans, it is possible that sustained high levels of NRG-1 are needed to maintain neuronal integrity and the highly developed social skills of naked mole-rats. However, a careful balance of NRG-1 expression levels is needed for normal synaptic activity and brain function, because synaptic activity can be impaired if NRG-1 levels are either reduced (i.e., knocked out) or overexpressed, a phenomenon referred to as an ‘inverted U model’ in schizophrenia research (Role and Talmage, 2007).
Interestingly, increased NRG-1 signaling reportedly promotes cancer in humans (reviewed in (Montero et al., 2008)), yet despite sustained high levels of these potential oncogenes, naked mole-rats appear impervious to spontaneous cancer induction (Buffenstein, 2005). It is most likely therefore that the NRG-1 levels found in a healthy adult animal reflect the optimal level to both sustain neuronal function and avoid cancer induction in this species.
High levels of NRG-1 may reflect the observed high levels of the transcription factor, HIF-1α. HIF-1α regulates transcription of growth factors, particularly in response to hypoxic conditions. Since naked mole-rats have evolved to inhabit deep underground hypoxic burrows since the early Miocene, one would anticipate high levels of both HIF-1α and NRG-1 reflect the evolutionary history of H. glaber rather than are a response to captive environmental conditions. Subterranean rodents may be particularly sensitive to HIF-1α for all the subterranean animals assessed in this study showed similar high levels of HIF-1α. Both NRG-1 and HIF-1α expression are inversely associated with low thyroid hormone concentration (Hulbert and Else, 2004). Given the low thyroid hormone profile of H. glaber (Woodley and Buffenstein, 2002), we speculate that the reported longevity enhancement by low levels of thyroid hormone may indeed work through this inverse relationship. Future studies may address the interactions and roles of these important variables in aging and longevity.
There are a myriad of neurobiological changes that accompany aging. The decline in availability of growth factors (Hayakawa et al., 2008) and neurotransmitters (Kelly et al., 2006) to neurons and glia result in loss of select neuronal populations, synaptic alteration in neuronal circuits, changes in dendritic densities as well as the soma itself. These changes are markers of the aging brain (Reviewed in (Dickstein et al., 2007). The observation that a neurotrophic factor does not decline with age in the long-lived H. glaber suggests a potential mechanism that protects this species from age-related impairments in neuronal function. The mechanisms responsible for enhanced NRG-1 signaling in long-lived species may yield novel insights for sustaining neuronal integrity, and cognitive function well into old age.
The significant positive link between NRG-1 expression levels and MLSP in rodents suggests that this growth factor, either through its specific effects upon brain function or throughout the body, is an important contributor to species longevity. Understanding how NRG-1 levels are regulated and how these impact upon species longevity may yield important insights into the mechanism that may delay age-related declines in brain function, and thereby prolong both healthspan and lifespan.
Experimental procedures
Rodent Species
We assessed expression levels of NRG-1 and ErbB4 in cerebella of different physiologically age-matched healthy mature individuals of different species (Table 1), which had lived at least 50% of their maximal lifespan. In addition, age-related changes in levels of NRG-1, ErbB4 and HIF1α were monitored in H. glaber beginning on the day of birth and at regular intervals until 26 years of age. In all cases, the abundance of NRG-1 and ErbB4 in the cerebellum is sufficient to detect and quantify by immunoblot analyses.
Tissue collection and processing
Animal procurement and euthanasia were carried out in adherence to NIH, Federal, State, and Institutional guidelines at The City College of New York (M. musculus, C. porcellus, F. damarensis and H. glaber), Haifa University (S. carmeli, S. judaei), and Rider University (A. cahirinus) by the same investigators. Cerebella were collected from six animals of each species within 10 minutes of euthanasia and flash frozen prior to storage at −80°C. Cerebella were sonicated and homogenized in lysis buffer containing protease inhibitors (PMSF, aprotinin and leupeptin), insoluble material was pelleted by centrifugation and samples were alliquoted and stored at −80°C until use. Cerebella were collected at two time points: the first contained all samples that were used in the species comparison. A second, separate collection was used for comparisons made within the Heterocephalus glaber. Samples were processed within 4 months of eachother as the samples became available.
Phylogenetic reconstructions
In order to assess whether the differences in growth factor expression levels were independent of the species’ phylogenetic lineage, we first computed phylogenetic distances between all species used in this study. These distances were based on sequence divergence from five nuclear genes: alpha 2B adrenergic receptor (ADRA2B); cannabinoid receptor 1 (CNR1); growth hormone receptor (GHR); recombination activating gene 2 (RAG2); and von Willebrand factor (vWF). Nuclear genes were chosen because they are less saturated than mitochondrial genes (Springer et al., 2001). These five genes were specifically chosen because they have been sequenced for numerous rodent species, (e.g., (Blanga-Kanfi et al., 2009; Huchon et al., 2007) and because large sampling improves the estimation of evolutionary distance (e.g., (Heath et al., 2008). The IRBP gene which is a commonly sequenced gene was not included in the analysis since this gene is not functional and fast-evolving in blind species like Spalax (Stanhope et al., 1996). Three species were newly sequenced A. cahirinus, F. damarensis and Meriones unguiculatus. Sequences were determined as follows: total DNA was extracted from ethanol-preserved samples following (Sambrook et al., 1989) with slight modifications. Amplification of ADRA2B and vWF were performed following (Huchon et al., 2007) and amplifications of CB1, GHR and RAG2 were performed following (Blanga-Kanfi et al., 2009). Purification of the PCR products and sequencing were performed according to (Huchon et al., 2007) and sequences were submitted to EMBL under accession numbers FN984740 -FN984757.
DNA sequences of ADRA2B, CNR1, GHR, RAG2 and vWF from the three novel species were added to the alignment of (Blanga-Kanfi et al., 2009). This DNA sequence alignment included 52 species and 5016 characters. We performed tree reconstruction on the concatenated nucleotide data set using the maximum likelihood (ML) criterion. Following (Blanga-Kanfi et al., 2009) the GTR+G+I model was selected as a model of sequence evolution. ML searches for the best trees were executed with PAUP* (Swofford, 2003). The parameters of the model and the ML tree were determined by successive approximation (Sullivan et al., 2005). The initial parameter values were estimated based on the Neighbor-Joining (NJ) topology reconstructed using p-distances. These values were then used for a first round of heuristic search starting with a Neighbor-Joining (NJ) tree and TBR branch-swapping. Parameters were then estimated on the resulting tree and used for a second round of heuristic search. This process was repeated twice until all parameter values were stable. As expected, the resulting phylogenetic tree was identical to the tree published by (Blanga-Kanfi et al., 2009) with A. cahirinus and F. damarensis placed respectively as sister clades to Acomys russatus (another Acomys) and to Bathyergus suillus (another Bathyergidae; Fig 1A). Pairwise distances between H. glaber, F. damarensis, C. porcellus, M. musculus, A. cahirinus and Spalax (Fig 1B), were inferred based on the patristic tree distances obtained from the larger tree (Fig 1A).
Data Analysis- Interspecies comparison
The NRG-1, ErbB4 and HIF1α values were averaged for the blind mole-rat Spalax ehrenbergi superspecies (S. carmeli and S. judaei). For NRG-1 and ErbB4, a covariance matrix was derived from the phylogenetic tree assuming a Brownian model of trait evolution using the Analyses of Phylogenetics and Evolution (ape) R package (Paradis et al., 2004). Generalized Least Squares (GLS) regression of log10 Maximum lifespan (MLS) was performed onto NRG-1 and ErbB4 levels using the PHYSIG matlab program written by Theodore Garland (Garland and Ives, 2000).
NRG-1, ErbB4 and HIF1α protein sequences
NRG-1 and ErbB4 antibodies SC-348 and SC-283 respectively (Santa Cruz Biotechnology) and HIF1α antibodies NB100-134 (Novus Biologicals) were tested for novel species which are not routinely used in biomedical studies. To ensure that there were no recognition artifacts, the epitope-containing regions of NRG-1 and ErbB4 were sequenced based on information obtained from the manufacturer (Santa Cruz). For each mole-rats species (F. damarensis, S. judaei, S .carmeli and H. glaber), RNA was extracted from two brains by homogenization in TRI reagent (Sigma) according to the manufacturer’s protocol. Integrity of RNA was verified by agarose gel electrophoresis. cDNA was synthesized using the Masterscript kit (5 PRIME). Following manufacturer’s instructions, 2ug of RNA were used per reaction. Semi-degenerate primers were designed based on the availability of species’ sequences for NRG-1, ErbB4 and HIF1α in GenBank. NRG-1 Forward: 5′-AGGATAGTGGAGGATGAGG-3′; NRG-1 Reverse: 5′-TACAGCRATAGGGTCTTGG-3′; ErbB4 Forward: 5′-AAGGYTACATGACYCCHATG-3′; ErbB4 Reverse: 5′-TYACACCACAGTATTYCG-3′. HIF1a Forward: 5′-ACAGAATGTGTCCTCAAACCAG-3′; HIF1a Reverse: 5′-TCTCTACCAGTTCCAACTTGAATAC-3′.
PCR reactions were performed using the following conditions: One denaturation cycle at 94°C for 2 minutes; followed by 40 cycles for NRG-1/ HIF1α or 35 cycles for ErbB4, each consisting of 1 minute at 94°C, 30 seconds at 53°C for NRG-1/ ErbB4 and 54 °C for HIF1α and 1 minute at 72°C; the program ended with a final extension step at 72°C for 10 minutes. The amplicons obtained were cloned using One Shot TOPO10 chemically potent E.coli cells (Invitrogen). Plasmids were prepared by QIAprep Miniprep kits (Qiagen) and sequenced (Genewiz, NJ). To verify that the sequences did not contain errors, PCR fragments from each sample were sequenced for both strands and these were compared to one another. In cases where mismatches occurred, the entire process was repeated until 100% homology was reached.
Assessment of NRG-1, ErbB4 and HIF1α protein levels
This study focuses on 46kDa band for NRG-1α, which was prominent across all species under examination. Blocking peptide (Santa Cruz Biotechnology (C-20)) was used to ensure specificity of the antibody to the growth factor. ErbB4 was chosen because unlike ErbB2 and ErbB3, ErbB4 both homo- and heterodimerizes to ErbB2-ErbB4, and carries out complete tyrosine kinase activity (Peles and Yarden, 1993; Plowman et al., 1993; Tzahar and Yarden, 1998) of the full length receptor observed at ~180kDa. For HIF1α we quantified the bands at ~100kDa which were apparent in all species tested.
Immunoblots were performed on 50 μg protein aliquots on 10% acrylamide pre-cast gels (Pierce) such that all species were represented on each gel in the interspecies comparison and were repeated in triplicates. Similarly, to compare expression levels of NRG-1, ErbB4 and HIF1α within H. glaber, each age group was represented on the same gel. One sample was randomly loaded onto every gel for control purposes. Proteins were electro-transferred onto PVDF 0.45 μm pore membranes (Millipore) followed by incubation in PBS-T with 5% non-fat dry milk. Immunoblotting was carried out with rabbit anti-NRG-1 (Santa Cruz SC-348; 1:500), rabbit anti-ErbB4 (Santa Cruz SC-283; 1:500) or rabbit anti-HIF1α (Novus Biologicals NB100-134; 1:1000) primary antibodies, and horseradish peroxidase-conjugated anti-rabbit secondary antibody (DCO3L; Calbiochem; 1:2000). Membranes were incubated with a chemiluminescent substrate (Pierce) and exposed to film (Kodak Bio Max Light film) for visualization. Molecular weights of resulting bands were determined by interpolation with the migration distances of the molecular weight markers. Protein levels were normalized by dividing signal intensity of the bands of interest by actin values obtained from the same membranes to control for variations in protein content, and are presented as arbitrary units (A.U.). Bands were quantified by densitometry with ImageJ (NIH) software.
Acknowledgments
We gratefully acknowledge Anne Pidel, Rukmani Lekhraj, Shani Blanga-Kanfi, Dr. Tomáš Pavlíček, Pavel Tovbin and Dr. Robert Bases for their help. We also gratefully acknowledge two anonymous reviewer’s for their helpful critiques. E.N. thanks the Ancell-Teicher Research Foundation for Genetics and Molecular Evolution for financial support. D.H. thanks the support of the National Evolutionary Synthesis Center (NESCent, NSF #EF-215 0905606). This work was supported by grants to R.B. from NIA/NIH AG022891.
Footnotes
“And in the end, it’s not the years in your life that count. It’s the life in your years.”
-Abraham Lincoln.
Dedicated to my husband Itai Zilberschmid who died young and forever lives. -YHE
Author contributions
YHE, DC and RB designed the experiments and wrote the paper. DH produced the phylogenetic data set. JG analyzed the data for the phylogenetic independent contrast analysis. EN provided tissues and information. YHE, DMK and RB collected the tissues. YHE performed the experiments and analyzed data. JM provided technical support.
References
- Andziak B, Buffenstein R. Disparate patterns of age-related changes in lipid peroxidation in long-lived naked mole-rats and shorter-lived mice. Aging Cell. 2006;5:525–532. doi: 10.1111/j.1474-9726.2006.00246.x. [DOI] [PubMed] [Google Scholar]
- Arieli R. The atmospheric environment of the fossorial mole-rat (Spalax ehrenbergi): effects of season, soil texture, rain, temperature and activity. Comp Biochem PhysioZ. 1979;63A:569–575. [Google Scholar]
- Aristotle Ross T.b.G.R.T., editor. On Longevity and Shortness of Life. 350 B.C.E http://classics.mit.edu/Aristotle/longev_short.html
- Bernstein H, Lendeckel U, Bertram I, Bukowska A, Kanakis D, Dobrowolny H, Stauch R, Krell D, Mawrin C, Budinger E, et al. Localization of neuregulin-1alpha (heregulin-alpha) and one of its receptors, ErbB-4 tyrosine kinase, in developing and adult human brain. Brain Res Bull. 2006;69:546–559. doi: 10.1016/j.brainresbull.2006.02.017. [DOI] [PubMed] [Google Scholar]
- Blanga-Kanfi S, Miranda H, Penn O, Pupko T, DeBry R, Huchon D. Rodent phylogeny revised: analysis of six nuclear genes from all major rodent clades. BMC Evol Biol. 2009;9:71. doi: 10.1186/1471-2148-9-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffenstein R. The naked mole-rat: a new long-living model for human aging research. J Gerontol A Biol Sci Med Sci. 2005;60:1369–1377. doi: 10.1093/gerona/60.11.1369. [DOI] [PubMed] [Google Scholar]
- Buonanno A. The neuregulin signaling pathway and schizophrenia: from genes to synapses and neural circuits. Brain Res Bull. 2010;83:122–131. doi: 10.1016/j.brainresbull.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A, Gerecke K, Wyss J, Morgan D, Gordon M, Carroll S. Neuregulin-1 and erbB4 immunoreactivity is associated with neuritic plaques in Alzheimer disease brain and in a transgenic model of Alzheimer disease. J Neuropathol Exp Neurol. 2003;62:42–54. doi: 10.1093/jnen/62.1.42. [DOI] [PubMed] [Google Scholar]
- Cho H, Leahy D. Structure of the extracellular region of HER3 reveals an interdomain tether. Science. 2002;297:1330–1333. doi: 10.1126/science.1074611. [DOI] [PubMed] [Google Scholar]
- Dickstein D, Kabaso D, Rocher A, Luebke J, Wearne S, Hof P. Changes in the structural complexity of the aged brain. Aging Cell. 2007;6:275–284. doi: 10.1111/j.1474-9726.2007.00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edrey Y, Park T, Kang H, Biney A, Buffenstein R. Endocrine function and neurobiology of the longest-living rodent, the naked mole-rat. Exp Gerontol. 2011;46:116–123. doi: 10.1016/j.exger.2010.09.005. [DOI] [PubMed] [Google Scholar]
- Ehrlichman R, Luminais S, White S, Rudnick N, Ma N, Dow H, Kreibich A, Abel T, Brodkin E, Hahn C, et al. Neuregulin 1 transgenic mice display reduced mismatch negativity, contextual fear conditioning and social interactions. Brain Res. 2009;1294:116–127. doi: 10.1016/j.brainres.2009.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esper R, Pankonin M, Loeb J. Neuregulins: versatile growth and differentiation factors in nervous system development and human disease. Brain Res Rev. 2006;51:161–175. doi: 10.1016/j.brainresrev.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Fischbach G. NRG1 and synaptic function in the CNS. Neuron. 2007;54:495–497. doi: 10.1016/j.neuron.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Garland JT, Ives A. Using the past to predict the present: Confidence intervals for regression equations in phylogenetic comparative methods. Am Nat. 2000;155:346–364. doi: 10.1086/303327. [DOI] [PubMed] [Google Scholar]
- Gerecke K, Wyss J, Karavanova I, Buonanno A, Carroll S. ErbB transmembrane tyrosine kinase receptors are differentially expressed throughout the adult rat central nervous system. J Comp Neurol. 2001;433:86–100. doi: 10.1002/cne.1127. [DOI] [PubMed] [Google Scholar]
- Hahn C, Wang H, Cho D, Talbot K, Gur R, Berrettini W, Bakshi K, Kamins J, Borgmann-Winter K, Siegel S, et al. Altered neuregulin 1-erbB4 signaling contributes to NMDA receptor hypofunction in schizophrenia. Nat Med. 2006;12:824–828. doi: 10.1038/nm1418. [DOI] [PubMed] [Google Scholar]
- Harrison D, Strong R, Sharp Z, Nelson J, Astle C, Flurkey K, Nadon N, Wilkinson J, Frenkel K, Carter C, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa N, Abe M, Eto R, Kato H, Araki T. Age-related changes of NGF, BDNF, parvalbumin and neuronal nitric oxide synthase immunoreactivity in the mouse hippocampal CA1 sector. Metab Brain Dis. 2008;23:199–211. doi: 10.1007/s11011-008-9084-7. [DOI] [PubMed] [Google Scholar]
- Heath T, Hedtke S, Hillis D. Taxon sampling and the accuracy of phylogenetic analyses. Journal of Systematics and Evolution. 2008;46:239–257. [Google Scholar]
- Huchon D, Chevret P, Jordan U, Kilpatrick C, Ranwez V, Jenkins P, Brosius J, Schmitz J. Multiple molecular evidences for a living mammalian fossil. Proc Natl Acad Sci U S A. 2007;104:7495–7499. doi: 10.1073/pnas.0701289104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert A, Pamplona R, Buffenstein R, Buttemer W. Life and death: metabolic rate, membrane composition, and life span of animals. Physiol Rev. 2007;87:1175–1213. doi: 10.1152/physrev.00047.2006. [DOI] [PubMed] [Google Scholar]
- Hwang I, Yoo K, Jung B, Cho J, Kim D, Kang T, Kwon Y, Kim Y, Won M. Correlations between neuronal loss, decrease of memory, and decrease expression of brain-derived neurotrophic factor in the gerbil hippocampus during normal aging. Exp Neurol. 2006;201:75–83. doi: 10.1016/j.expneurol.2006.02.129. [DOI] [PubMed] [Google Scholar]
- Kelly K, Nadon N, Morrison J, Thibault O, Barnes C, Blalock E. The neurobiology of aging. Epilepsy Res. 2006;68(Suppl 1):S5–20. doi: 10.1016/j.eplepsyres.2005.07.015. [DOI] [PubMed] [Google Scholar]
- Kraus M, Issing W, Miki T, Popescu N, Aaronson S. Isolation and characterization of ERBB3, a third member of the ERBB/epidermal growth factor receptor family: evidence for overexpression in a subset of human mammary tumors. Proc Natl Acad Sci U S A. 1989;86:9193–9197. doi: 10.1073/pnas.86.23.9193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law AJ, Shannon Weickert C, Hyde TM, Kleinman JE, Harrison PJ. Neuregulin-1 (NRG-1) mRNA and protein in the adult human brain. Neuroscience. 2004;127:125–136. doi: 10.1016/j.neuroscience.2004.04.026. [DOI] [PubMed] [Google Scholar]
- Liang S, Mele J, Wu Y, Buffenstein R, Hornsby J. Resistance to experimental tumorigenesis in cells of a long-lived mammal, the naked mole-rat (Heterocephalus glaber) Aging Cell. 2010;9:626–635. doi: 10.1111/j.1474-9726.2010.00588.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoro E. Overview of caloric restriction and ageing. Mech Ageing Dev. 2005;126:913–922. doi: 10.1016/j.mad.2005.03.012. [DOI] [PubMed] [Google Scholar]
- Min S, An J, Lee J, Seol G, Im J, Kim H, Baik T, Woo R. Neuregulin-1 prevents amyloid beta-induced impairment of long-term potentiation in hippocampal slices via ErbB4. Neurosci Lett. 2011 doi: 10.1016/j.neulet.2011.05.246. [DOI] [PubMed] [Google Scholar]
- Montero J, Rodríguez-Barrueco R, Ocaña A, Díaz-Rodríguez E, Esparís-Ogando A, Pandiella A. Neuregulins and cancer. Clin Cancer Res. 2008;14:3237–3241. doi: 10.1158/1078-0432.CCR-07-5133. [DOI] [PubMed] [Google Scholar]
- Nadri C, Belmaker R, Agam G. Oxygen restriction of neonate rats elevates neuregulin-1alpha isoform levels: possible relationship to schizophrenia. Neurochem Int. 2007;51:447–450. doi: 10.1016/j.neuint.2007.03.013. [DOI] [PubMed] [Google Scholar]
- O’Tuathaigh C, Babovic D, O’Sullivan G, Clifford J, Tighe O, Croke D, Harvey R, Waddington J. Phenotypic characterization of spatial cognition and social behavior in mice with ‘knockout’ of the schizophrenia risk gene neuregulin 1. Neuroscience. 2007;147:18–27. doi: 10.1016/j.neuroscience.2007.03.051. [DOI] [PubMed] [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: analyses of phylogenetics and evolution in R language. Bioinformatics. 2004:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Peles E, Yarden Y. Neu and its ligands: from an oncogene to neural factors. Bioessays. 1993;15:815–824. doi: 10.1002/bies.950151207. [DOI] [PubMed] [Google Scholar]
- Plowman G, Culouscou J, Whitney G, Green J, Carlton G, Foy L, Neubauer M, Shoyab M. Ligand-specific activation of HER4/p180erbB4, a fourth member of the epidermal growth factor receptor family. Proc Natl Acad Sci U S A. 1993;90:1746–1750. doi: 10.1073/pnas.90.5.1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Role L, Talmage D. Neurobiology: new order for thought disorders. Nature. 2007;448:263–265. doi: 10.1038/448263a. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch E, Maniatis T. Molecular cloning: a laboratory manual. 1989 [Google Scholar]
- Springer M, DeBry R, Douady C, Amrine H, Madsen O, de Jong W, Stanhope M. Mitochondrial versus nuclear gene sequences in deep level mammalian phylogeny reconstruction. Mol Biol Evol. 2001;18:132–143. doi: 10.1093/oxfordjournals.molbev.a003787. [DOI] [PubMed] [Google Scholar]
- Stanhope M, Smith M, Waddell V, Porter C, Shivji M, Goodman M. Mammalian evolution and the interphotoreceptor retinoid binding protein (IRBP) gene: convincing evidence for several superordinal clades. J Mol Evol. 1996:2. doi: 10.1007/BF02337352. [DOI] [PubMed] [Google Scholar]
- Sullivan J, Abdo Z, Joyce P, Swofford D. Evaluating the performance of a successive-approximations approach to parameter optimization in maximum-likelihood phylogeny estimation. Mol Biol Evol. 2005;22:1386–1392. doi: 10.1093/molbev/msi129. [DOI] [PubMed] [Google Scholar]
- Swofford D. PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods) Sinauer Associates; Sunderland, Massachusetts: 2003. [Google Scholar]
- Tzahar E, Yarden Y. The ErbB-2/HER2 oncogenic receptor of adenocarcinomas: from orphanhood to multiple stromal ligands. Biochim Biophys Acta. 1998;1377:M25–37. doi: 10.1016/s0304-419x(97)00032-2. [DOI] [PubMed] [Google Scholar]
- Ungvari Z, Buffenstein R, Austad S, Podlutsky A, Kaley G, Csiszar A. Oxidative stress in vascular senescence: lessons from successfully aging species. Front Biosci. 2008;13:5056–5070. doi: 10.2741/3064. [DOI] [PubMed] [Google Scholar]
- Wilson D, Reeder D. Mammal Species of the World. A Taxonomic and Geographic Reference. 3rd edn Johns Hopkins University Press; 2005. [Google Scholar]
- Woodley R, Buffenstein R. Thermogenic changes with chronic cold exposure in the naked mole-rat (Heterocephalus glaber) Comparative biochemistry and physiology Part A, Molecular & integrative physiology. 2002;133:827–834. doi: 10.1016/s1095-6433(02)00199-x. [DOI] [PubMed] [Google Scholar]