Abstract
A significant limitation of fiber bundle endomicroscopy systems is that the field of view tends to be small, usually only several hundred micrometers in diameter. Image mosaicking techniques can increase the effective image size, but require careful manipulation of the probe to ensure sufficient overlap between adjacent frames. For confocal endomicroscopes, which typically have frame rates on the order of 10 fps, this is particularly challenging. In this paper we demonstrate that line-scanning confocal endomicroscopy can, by use of a high speed linear CCD camera, achieve a frame rate of 120 fps while maintaining sufficient resolution and signal-to-noise ratio to allow imaging of topically stained gastrointestinal tissues. This leads to improved performance of a cross-correlation based mosaicking algorithm when compared with lower frame-rate systems.
OCIS codes: (170.2150) Endoscopic imaging, (110.0180) Microscopy
1. Introduction
Confocal endomicroscopy is an endoscopic technique for obtaining fluorescence microscopy images in vivo [1, 2]. It serves as a real-time alternative to conventional biopsy and histology, and is beginning to find routine clinical applications. Typically, tissue is first stained with intravenous or topical fluorescent contrast agents, and the endomicroscopy probe is then introduced through the working channel of a flexible endoscope. When the probe is placed in gentle contact with the stained tissue, live confocal microscopy images are displayed.
For the probe to be compatible with a conventional endoscope channel, its maximum diameter must not exceed (typically) 3.2 mm. Building such a compact probe for confocal imaging is made difficult by the need to scan a focused laser spot over the tissue in two dimensions. While miniaturized confocal laser scanners have been demonstrated [3], and indeed commercialized [4], the images they generate tend to be limited to a few frames per second (fps). Higher frames rates (on the order of 10 fps) can be more readily achieved by generating the scanning pattern in bulk optics and then relaying it to the tissue by a flexible fiber imaging bundle and (optionally) a distal micro-objective [5, 6]. However, in this case the number of useful ‘pixels’ of information is limited by the number of fiber cores (typically 30,000), necessitating a trade-off between lateral image size and resolution.
The magnification factor of the probe’s distal optics is typically selected so as to provide an image diameter of between a quarter and three quarters of a millimeter. This is relatively small compared to a typical histology slide, thus imposing difficulties on image interpretation. This drawback can be mitigated to some extent by mosaicking, whereby images acquired from adjacent areas of tissue are fused to synthesize a larger field of view. Such mosaics can be assembled in real-time through simple frame-by-frame rigid registration [7], or by using computationally intensive algorithms to achieve more faithful reconstructions retrospectively [8]. In the latter case, improvements in resolution and signal-to-noise ratio may also be obtained.
Nevertheless, obtaining large mosaics is difficult. In particular, the speed by which the operator can translate the probe across the tissue is constrained by the need for sufficient overlap between adjacent images. The viability of mosaicking is therefore directly linked to the image acquisition rate. For commercial and research confocal endomicroscopes this is typically 10-20 fps, limited primarily by the maximum speed with which a laser spot can be scanned. If we take, as an example, the Cellvizio Coloflex UHD probe with a field of view of 240 µm and a frame rate of 12 fps, then sufficient frame overlap for reliable mosaicking limits the translational speed to around 0.5 mm/s [9]. Given the obvious difficulties in achieving this consistently, an increase in the frame rate is clearly desirable.
In bench-top microscopes, spinning disk illumination is commonly used for high speed, live cell studies, but has only been reported for endomicroscopy on one occasion [10], and then only for the higher signal conditions of reflectance-mode imaging. Higher frame rate imaging is also possible with non-confocal endomicroscopy, such as in the widefield fluorescence endomicroscopy technique known as high resolution microendoscopy (HRME) [11, 12]. Here, frame rates of 15 fps have been reported, but this can readily be improved upon by using a higher frame rate camera. However, as HRME is non-sectioning, the range of applications is more limited [13]. Some degree of optical sectioning can be introduced using structured illumination [14, 15], but this tends to produce noisier images from thick tissue, and as a multi-shot approach is subject to motion artefacts and a reduced net frame rate.
A further approach, and the one which is the subject of this paper, is to use line or slit scanning illumination. In this embodiment, rather than scanning a laser point in a 2D pattern across the tissue, a line is scanned in 1D, with the fluorescent emission from the entire line recorded simultaneously. This leads to a slight broadening of the axial sectioning profile (the full-width half-maximum increases by a factor of 1.4), as well as introducing a tail which falls off roughly linearly with distance [16]. The optical sectioning strength is therefore reduced, but remains sufficient for less demanding applications.
Sabharwal et al. first demonstrated this approach to fiber bundle endomicroscopy in 1999 [17]. In their implementation, a line was scanned across the proximal face of the fiber bundle, and hence ultimately the tissue, and the fluorescent emission was de-scanned, passed through a confocal slit, and then rescanned over a 2D camera to form an image. This system has been developed further [18–20], and demonstrated in vivo [21], but the frame rate of 30 fps did not fully realize the potential of line-scanning.
In this paper, we report a high speed, line-scanning, fiber bundle endomicroscope which uses a 1D line-scan camera. We achieve a frame rate 120 fps (for 500 lines per image), an order of magnitude improvement over commercially available point-scanning endomicroscopy systems. We compare the mosaicking performance to lower-frame rate systems, and thus assess the advantages of high speed imaging in general, and of line-scanning in particular.
2. Methods
2.1 Endomicroscopy system
The line-scan endomicroscopy system is shown schematically in Fig. 1 . While the system can be used with any fiber bundle, for this work we used a Coloflex UHD probe taken from a Cellvizio endomicroscope. This consisted of a packaged fiber bundle and distal micro-lens assembly. The fiber bundle was a 30,000 core Fujikura fused bundle, with an inter-core spacing of 2.7 µm and an image circle diameter of 600 µm. The distal lens provided a 2.5X magnification, resulting in a field of view of 240 µm and an effective core spacing of 1.1 µm when projected onto the tissue. This suggests a sampling-limited spatial resolution of 2.2 µm when the Nyquist criterion is applied.
Fig. 1.
Schematic of line-scanning endomicroscopy system, shown in the plane in which the cylindrical lens has power. Telescope 1 expands the beam, while telescopes 2 and 3 are relays with unity magnification.
The output from a 488 nm laser diode (50 mW) was expanded to 5 mm by a telescope, and focused by an f = 5 cm cylindrical lens to a line on a dichroic mirror. The reflected beam from the dichroic mirror was imaged onto a galvo scanner and then onto the back aperture of a X10 microscope objective via two relay 4f telescopes (using f = 5 cm achromatic doublets). This generated a line on the proximal face of the fiber bundle which could be scanned over its full extent by driving the galvo mirror.
When placed in contact with the distal tip of the probe, the tissue was illuminated with a time-averaged power of 2.5 mW. Fluorescence emission returned along the bundle, where it was de-scanned by the galvo mirror and transmitted by the dichroic and an emission filter to remove reflected 488 nm light. The de-scanned line was then imaged onto a linear CCD array (Basler Sprint spL2048-70km with 45-50% quantum efficiency in the 500-600 nm range). The magnification between the proximal face of the bundle and the camera was approximately 8, resulting in an image diameter of approximately 5 mm on the camera. The camera had a 10 µm pitch, providing 500 pixels across the fiber bundle, or approximately 2.3 pixels per core spacing. This was sufficient sampling to allow individual cores to be visualized in the images.
In the vertical direction, the camera had two lines of pixels, each 10 µm high. We averaged the signal from both lines to give an effective vertical pixel size of 20 µm. Projected onto the proximal face of the bundle, this provided a detector slit width of 2.4 µm, approximately equal to the core spacing. There would be no benefit to having a slit much smaller than the mode field diameter of the cores, since the returning fluorescence is homogenised over each core. A smaller slit would therefore result in a reduction in light collection efficiency for no gain in sectioning strength. We note, therefore, the advantage of having an effective 2:1 aspect ratio of pixel size on the camera, as this allows the detector slit to be the desired 1 core spacing high while still allowing for Nyquist sampling of the cores in the horizontal direction. This avoided the need to image a physical slit onto the detector.
An input/output board sent a ramp voltage to the galvo scanner, providing the vertical scan, while also triggering the acquisition of a frame on the linear CCD. By adjusting the galvo scanner speed and the exposure time for each line, the imaging frame rate could be varied. The camera could be run at up to a line-rate of 70 kHz, which allowed a frame rate of fps, where is the number of lines per image, and is the number of unusable lines due to fly-back of the galvo scanner. The line exposure time, , was therefore a function of the desired frame rate, , set such that . For images in this paper, we collected = 500 lines per image (with = 50 fly-back lines), and achieved a maximum frame rate of = 120 fps with a line exposure time of 15.2 µs and a frame acquisition time of 7.6 ms.
2.2 Image reconstruction and mosaicking system
Raw images were filtered by convolution with a 2D Gaussian function of 1.4 pixels standard deviation (equivalent to 0.7 µm in object space) to remove the core pattern. A background subtraction was then performed to remove background signal from the bundle. The background measurement was made by averaging 10 frames with the fiber tip placed inside a dark tube; we found that ambient light was otherwise sufficient to disturb this measurement. Finally, a circular window was applied to crop the edges of the bundle, leading to a final image diameter of approximately 240 µm. Due to the high frame rate of the system, data was streamed directly to disk and image processing and mosaicking were performed off-line. A preview of processed images at 20 fps was shown on screen to guide the operator.
The mosaicking algorithm, which is similar to a previously reported real-time confocal endomicroscopy mosaicking approach [7], estimated the shift between successive pairs of images using template matching. For images and , a central sub-region, , of size was extracted from for use as the template. The normalized cross-correlation, , was then calculated between this template and . The situation was then reversed, with a template extracted from used to calculate the normalized cross correlation with image , . The estimated shift between the two images was then taken to be the location of the largest peak across the two cross correlations. The absolute position of any frame could be calculated by summing all of the prior pairwise shifts. To assemble the mosaic, image frames were inserted at their estimated positions by merging with the existing pixels using distance-weighted alpha blending.
It should be noted that the template size used during registration was a manually tunable parameter. In general, increasing the size of the template reduces the size of the valid region of the cross-correlation, and hence reduces the maximum shift that can be permitted between images. However, a larger template size also improves the template matching, since it increases the probability of a high fidelity feature being present. The performance of the mosaicking algorithm is also highly dependent on the features of the tissue, as well as on the consistency of the probe-tissue contact force, but further discussion of these effects is beyond the scope of this paper.
3. Results
3.1. Optical system characterization
We measured an approximation to the axial depth sectioning strength by removing the emission filter and measuring the reflectance signal from a mirror. The distal probe tip was initially placed in contact with the mirror, and then driven away using a motorized translation stage. Images were acquired for every 0.1 µm step. The average intensities from a 25 x 25 µm region in the center of each image are plotted as a function of distance in Fig. 2(a) . For comparison, we performed the same experiment using a fully confocal (i.e. point-scanning) and a fully widefield endomicroscope, of which details are provided in the appendix. For the confocal system, the pinhole was chosen to be approximately the same size as the detector slit height for the line-scanning system (3.1 µm at the proximal bundle face). For this plot, the peaks of the profiles were manually aligned to the nominal working distance.
Fig. 2.

(a) Axial sectioning profiles for the line-scanning system (red), shown in comparison with point-scanning (green) and widefield illumination (blue). The line-scanning profile exhibits a full-width half-maximum only slightly larger than the point-scanning system, but with a significantly increased tail. The gentle fall-off in the widefield profile is due to simple geometric effects. (b) USAF resolution target, showing group 7 elements 2-6 and group 6 element 1, imaged with the line-scanning system. The target was back-illuminated by a green LED. The insets show a zoom on part of the target, with (top) and without (bottom) fiber-pattern removal by spatial filtering. Scale bar is 50 µm.
As expected, the line-scan system is an intermediate case between the widefield and confocal endomicroscopes. Averaged across 10 experimental measurements, the full-width half-maximum of the line-scanning profile peak is 6.7 ± 0.3 µm as opposed to 5.8 ± 0.1 µm for the point-scanning. While the change in the 3 dB fall-off point is relatively modest, there is a significant tail to the line-scanning profile which appears to fall off approximately linearly with depth. For example, the single sided 10 dB drop-off point is 23 ± 3 µm for the line-scanning, as opposed to only 10.0 ± 0.4 µm for the point-scanning. This will result in some additional background signal in the images, although clearly much reduced in comparison to the case of widefield illumination. There is also some drop in sensitivity with depth for widefield system (3 dB in 142 ± 2 µm); this is simply due to geometry, with the maximum collection angle being reduced as the bundle moves further from the mirror.
To illustrate the transverse resolution of the system, we placed the distal tip of the probe against a USAF resolution target which was back-illuminated by a green LED. We then imaged as normal, with the emission filter in place, but with the laser turned off. The resulting raw image, centered on group 7, is shown in Fig. 2(b). The inset, showing part of a ‘6’ character, demonstrates that individual fiber cores can be identified prior to spatial filtering, and hence that the resolution is limited by the fiber bundle rather than the scanning system.
3.2 Ex vivo tissue imaging results
Figure 3 shows images of (previously frozen) ex vivo porcine tissue from the oesophagus, stomach and colon, acquired with the line-scanning system at three different frame rates (10 fps, 30 fps, 120 fps) and also with the point-scanning and widefield systems (which ran at 10 fps and 30 fps respectively). Each system imaged the same piece of tissue, although the images are not exactly co-located. True co-location was not possible because of the need to switch the probe between systems and to wipe clean the probe tip to acquire a background correction.
Fig. 3.
Example images of porcine colon (left column), stomach (centre column) and oesophagus (right column) mucosa, taken with the line-scan system running at 10 fps, 30 fps and 120 fps, shown with similar images from a point-scanning and widefield endomicroscope. Scale bar is 50 µm.
Each sample was stained with topical proflavine (0.02%) for 2 minutes before washing with water. This acts as a nuclear stain, and has been previously been used for in vivo studies [12, 22]. Videos of typically 1 minute in length were then acquired from each sample, using each imaging system. For each video, a single frame was selected to be representative of the best images that could be acquired within a reasonable timescale. In addition to the standard processing described above, each image was resized to 300 x 300 pixels by cubic spline interpolation, and the image contrast and brightness was adjusted automatically so that 1% of pixels were saturated (high and low).
The line-scan images show improved sectioning over the widefield images, but do not provide the full confocal sectioning seen in the point-scan images. The effect is to reduce somewhat the contrast, although features are still clearly visible. There is also some degradation in image quality apparent in the 120 fps line-scan images which can be attributed to the short exposure time reducing the signal-to-noise ratio. However, tissue features remain clearly visible.
3.3 In vivo tissue imaging results
The practicality of the high speed imaging system for use in vivo was evaluated in a live porcine model during a simulated transanal surgical procedure. The experiment was conducted under UK Home Office animal project license PPL 70/7940. The bowel was stained with proflavine using a custom spraying catheter, and irrigated shortly afterwards. The probe was introduced through a 5 mm port, and held by a laparoscopic gripper deployed through a second 5 mm port. Visual guidance was provided by a rigid endoscope, from which an example frame can be seen in Fig. 4(a) . Example endomicroscope image frames are shown in Fig. 4(b) and Fig. 4(c), while a small mosaic is shown in Fig. 4(d).
Fig. 4.

Example results from in vivo validation in a porcine model. (a) Endoscope view of probe in rectum; (b,c) single image frames acquired at 120 fps; (c) short mosaic showing field of view expansion. The apparent deformation of the crypts is also present in the video, and is not an artefact of the mosaicking algorithm. Scale bars are 50 µm.
3.4 Mosaicking study
To verify that the line-scan system could generate mosaics at high velocities, and that this represents improved performance over lower frame rate systems, we first performed experiments using a known velocity. The probe tip was mounted on a translation stage (Standa 8MT173), and driven 3 mm across the surface of porcine colon tissue at varying velocities, with the system frame rate set at 10, 30 and then 120 fps. For each combination, we automatically built mosaics using the algorithm described above, with the additional criteria that a normalized cross correlation of at least 0.85 was required between each image frame, and that a minimum length of 10 images was required to build a mosaic. While these parameters are somewhat arbitrary, we found that in general mosaics that look visually correct have a cross-correlation value > 0.85 for each image pair. The cross-correlation template size was chosen as half the image diameter (120 µm in object space). Altering the template size would change the absolute performance but should not change the relative performance at different frame rates. We considered a given combination of frame rate and velocity to produce viable mosaics if at least one mosaic of 1 mm or more in length could be produced from three experimental trials.
For the system running at 10 fps, 0.6 mm/s was the largest velocity for which mosaics could be reliably assembled. This velocity corresponds to a frame overlap of 75%, whereas a velocity of 0.8 mm/s would have provided an overlap of only 67%. This is broadly consistent with a previous study that suggested a maximum velocity of 0.5 mm/s for a 12 fps system [9]. For the 30 fps runs, the maximum velocity was 1.8 mm/s, while the 120 fps runs were able to produce mosaics at the maximum translation stage velocity of 5 mm/s.
We then collected a set of 7 videos at 120 fps while the probe was moved by hand across the surface of the colon tissue at various velocities and in various directions. The combined data set contained 19648 image frames, collected over 164 s. We down-sampled the videos to 10 and 30 fps and then processed the three resulting data sets to extract all viable mosaics using the same procedure as above (i.e. requiring a length of at least 10 frames with an NCC greater than 0.85). Unlike in the study using the translation stage, it should be stressed here that as the 10 fps and 30 fps videos were created from the 120 fps video, all three share the same noise and depth-sectioning characteristics. In addition, a true lower frame-rate system would generally also have a longer frame integration time, and less of a ‘step’ between each frame.
The results are summarized in Table 1 , which shows the number of viable mosaics created, the mean and total length of the mosaics, and the percentage of all image frames which formed part of a mosaic. The length of a mosaic was defined as the integrated distance from the first frame to the final frame of each, and the total and mean length of the mosaics were calculated by summing the lengths of all viable mosaics, and then by dividing by the number of viable mosaics. There is a clear improvement with frame rate, as expected, with the 120 fps videos providing more and longer mosaics.
Table 1. Statistical results of mosaicking experiments using freehand and endoscope scanning.
| Freehand
|
Endoscope
|
||||||
|---|---|---|---|---|---|---|---|
| Video Frame Rate | 10 fps | 30 fps | 120 fps | 10 fps | 30 fps | 120 fps | |
| Number of mosaics | 6 | 58 | 120 | 20 | 57 | 51 | |
| Mean length of mosaics (mm) | 0.7 | 1.3 | 2.3 | 0.7 | 1.2 | 2.7 | |
| Total length of mosaics (mm) | 4.6 | 76.0 | 274.0 | 14.2 | 68.6 | 140.0 | |
| % image frames in mosaic | 5 | 36 | 86 | 25 | 63 | 88 | |
Finally, to more closely simulate clinical usage, we passed the probe through the working channel of a flexible endoscope. Videos were acquired (17588 frames in 147 s) while the probe was manipulated using the standard endoscopic steering controls and processed as above. The results are again shown in Table 1, showing a clear improvement in mosaicking performance with higher frames rates. Example mosaics from the 120 fps videos (both freehand and endoscope) are shown in Fig. 5 . These images demonstrate an apparently higher signal-to-noise ratio than the single frames of Fig. 3 due to the ‘averaging’ effect of the mosaicing algorithm.
Fig. 5.
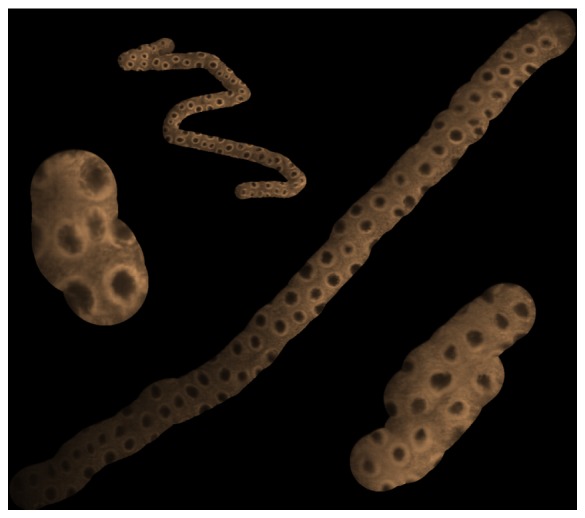
Several examples of mosaics created using freehand and endoscope scanning. The averaging effect of the mosaicking algorithm tends to reduce the noise when compared with single image frames.
4. Discussion
The results presented above demonstrate that a higher frame rate improves the performance of a simple mosaicking algorithm. These experiments were conducted using a line-scan system, but in principle this result should be applicable to any high frame rate endomicroscope. A further advantage of shorter frame acquisition times is that motion artifacts within image frames will tend to be reduced, increasing the number of useable frames within any video sequence.
However, it should be noted that the efficacy of mosaicking is not only limited by frame rate, but also affected by factors such as the clarity of image features, the consistency of probe-tissue contact, and the behavior of the tissue under deformation, all of which may be influenced by the velocity of the probe. Therefore, the full potential of the higher frame rate may not always be realized in practice. We also stress that the comparison was performed only for a simple cross-correlation based mosaicking algorithm. The absolute and relative performance may change when using algorithms that take into account non-rigid deformation, that perform a global optimization, or that filter the estimated probe motion. However, regardless of the algorithm used, image overlap is required in order to register images, and so we would still expect an improvement with higher frame rates.
While line-scanning allows for higher speeds, it also has several disadvantages over point-scanning. Most obviously, depth sectioning is weaker, which in general tends to lead to a reduction in useful image contrast. While this may not present a major limitation when topical nuclear staining agents such as proflavine are used, it is more likely to be problematic with intravenous stains. The line-scanning system will therefore need to be trialed in vivo using intravenous fluorescein sodium in order for this to be fully evaluated. The second limitation is that a higher imaging speed necessarily means shorter integration times, reducing the photon budget and generally leading to noisier images.
However, these problems are partly countered by other advantages of line-scanning. Firstly, point-scanning confocal using a resonant scanner for the fast axis is inefficient, as the motion of the scanner is sinusoidal, and hence only the relatively linear central portion of each sweep can be used. In comparison, the line-scanning system reported here has a duty cycle approaching 90%. Secondly, a thicker optical section also tends to increase the amount of light collected, leading to a brighter image. These factors help to make very high frame rates feasible. If higher signal-to-noise ratio is required for a particular application, such as imaging endogenous fluorescence of elastin fibers in the lung, the frame rate can be reduced on the fly.
In terms of cost and system complexity, the line-scanning system is comparable to point-scanning, with the resonant scanner and photodetector replaced with a line-scan camera. It is arguably slightly less amenable to compact packaging, since it is not possible to use a multimode fiber to collect fluorescence. Further, simultaneous multi-wavelength imaging would be more difficult to implement with line-scanning, as the cost of employing a line-scan camera in each detection arm would be prohibitive. However, by using multiband filters, it would be possible to either use a color line-scan camera for simultaneous multicolor imaging, or a monochrome camera and triggered lasers for sequential imaging where fluorophore bleed-through is possible. Indeed, line-scan has an advantage in the latter case, as the higher line-rate would reduce color motion artefacts.
5. Conclusion
We have demonstrated that a 120 fps line-scanning fiber bundle endomicroscope can obtain good quality fluorescence microscope images from ex vivo tissue samples and during an in vivo rectal mucosal study when using a topical contrast agent. While there is some degradation in optical sectioning strength, and a reduction in signal-to-noise ratio at the highest frame rates, this is countered by the improved performance of mosaicking algorithms.
Acknowledgments
Thank you to Petros Giataganas and Khushi Vyas for useful discussions and assistance with mosaicing algorithms, and to Dr. Nisha Patel, Dr. James Clark, Dr. Siyang Zuo and other members of the Hamlyn Centre and Northwick Park Institute for Medical Research for assistance. This work was supported by EPSRC Grant EP/I027769/1: SMART Endomicroscopy.
Appendix
The point-scanning confocal and widefield epi-fluorescence endomicroscopy systems used for comparison are similar to previously reported designs. They are therefore described here only in brief. The confocal endomicroscope, shown in Fig. 6
Fig. 6.

Schematic of confocal point-scanning endomicroscopy system used for comparison. Telescope 1 is a relay, telescope 2 provides a x2 beam expansion. L1 has a focal length of 75 cm. The mid-point between the two scanning mirrors is imaged onto the back focal plane of the objective.
, used a 488 nm laser (the same model as the line-scan system) which was scanned over the distal end of the probe using a two axis scanning system consisting of a galvo mirror and an 8 kHz resonant scanning mirror. After the returning fluorescence signal was de-scanned, a dichroic filter was used to direct it through a pinhole and onto an avalanche photodiode (APD). The physical pinhole diameter was 25 µm, which corresponded to 1.25 µm at the tissue plane. Signal from the APD was digitized by a high speed digitizer, and synchronized with the scanning mirrors via an input/output board to allow assembly of completed image frames. The frame rate was 10 fps for a 500 x 500 pixel image. Images were processed as for the line-scan system.
The widefield endomicroscopy system, shown in Fig. 7
Fig. 7.

Schematic of widefield endomicroscopy system used for comparison.
, was based on design for high resolution endomicroscopy by Pierce et al. [23]. A 450 nm LED was low-pass filtered by a 450 nm edge filter and directed into the bundle by a dichroic mirror and a x10 objective. Returning florescence was imaged on a monochrome CCD camera via the dichroic mirror, a tube lens, and a high pass 500 nm emission filter. The bundle was imaged to a diameter of 2.3 mm on the camera, covering approximately 500 pixels. The frame rate, determined by the camera read-out speed, was 30 fps. Image processing was as for the confocal endomicroscope.
References and links
- 1.Jabbour J. M., Saldua M. A., Bixler J. N., Maitland K. C., “Confocal endomicroscopy: instrumentation and medical applications,” Ann. Biomed. Eng. 40(2), 378–397 (2012). 10.1007/s10439-011-0426-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shukla R., Abidi W. M., Richards-Kortum R., Anandasabapathy S., “Endoscopic imaging: How far are we from real-time histology?” World J Gastrointest. Endosc. 3(10), 183–194 (2011). 10.4253/wjge.v3.i10.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dickensheets D. L., Kino G. S., “Micromachined scanning confocal optical microscope,” Opt. Lett. 21(10), 764–766 (1996). 10.1364/OL.21.000764 [DOI] [PubMed] [Google Scholar]
- 4.Polglase A. L., McLaren W. J., Skinner S. A., Kiesslich R., Neurath M. F., Delaney P. M., “A fluorescence confocal endomicroscope for in vivo microscopy of the upper- and the lower-GI tract,” Gastrointest. Endosc. 62(5), 686–695 (2005). 10.1016/j.gie.2005.05.021 [DOI] [PubMed] [Google Scholar]
- 5.Gmitro A. F., Aziz D., “Confocal microscopy through a fiber-optic imaging bundle,” Opt. Lett. 18(8), 565–567 (1993). 10.1364/OL.18.000565 [DOI] [PubMed] [Google Scholar]
- 6.Goualher G. L., Perchant A., Genet M., Cav C., Viellerobe B., Abrat B., Ayache N., “Towards Optical Biopsies with an Integrated Fibered Confocal Fluorescence Microscope,” in Medical Imaging Computing and Computer Aided Intervention, (Springer, 2004), 761–768. [Google Scholar]
- 7.Vercauteren T., Meining A., Lacombe F., Perchant A., “Real time autonomous video image registration for endomicroscopy: fighting the compromises,” Proc. SPIE 6082, 60820C (2008). [Google Scholar]
- 8.Vercauteren T., Perchant A., Malandain G., Pennec X., Ayache N., “Robust mosaicing with correction of motion distortions and tissue deformations for in vivo fibered microscopy,” Med. Image Anal. 10(5), 673–692 (2006). 10.1016/j.media.2006.06.006 [DOI] [PubMed] [Google Scholar]
- 9.M. S. Erden, B. Rosa, J. Szewczyk, and G. Morel, “Mechanical design of a distal scanner for confocal microlaparoscope: a conic solution,” in Robotics and Automation (ICRA),2013IEEE International Conference on, (IEEE, 2013), 1205–1212. 10.1109/ICRA.2013.6630725 [DOI] [Google Scholar]
- 10.Juškattis R., Wilson T., Watson T. F., “Real-time white light reflection confocal microscopy using a fibre-optic bundle,” Scanning 19(1), 15–19 (1997). 10.1002/sca.4950190102 [DOI] [Google Scholar]
- 11.Muldoon T. J., Pierce M. C., Nida D. L., Williams M. D., Gillenwater A., Richards-Kortum R., “Subcellular-resolution molecular imaging within living tissue by fiber microendoscopy,” Opt. Express 15(25), 16413–16423 (2007). 10.1364/OE.15.016413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pierce M. C., Vila P. M., Polydorides A. D., Richards-Kortum R., Anandasabapathy S., “Low-cost endomicroscopy in the esophagus and colon,” Am. J. Gastroenterol. 106(9), 1722–1724 (2011). 10.1038/ajg.2011.140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koucky M. H., Pierce M. C., “Axial response of high-resolution microendoscopy in scattering media,” Biomed. Opt. Express 4(10), 2247–2256 (2013). 10.1364/BOE.4.002247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bozinovic N., Ventalon C., Ford T., Mertz J., “Fluorescence endomicroscopy with structured illumination,” Opt. Express 16(11), 8016–8025 (2008). 10.1364/OE.16.008016 [DOI] [PubMed] [Google Scholar]
- 15.Santos S., Chu K. K., Lim D., Bozinovic N., Ford T. N., Hourtoule C., Bartoo A. C., Singh S. K., Mertz J., “Optically sectioned fluorescence endomicroscopy with hybrid-illumination imaging through a flexible fiber bundle,” J. Biomed. Opt. 14(3), 030502 (2009). 10.1117/1.3130266 [DOI] [PubMed] [Google Scholar]
- 16.Sheppard C. J. R., Mao X. Q., “Confocal microscopes with slit apertures,” J. Mod. Opt. 35(7), 1169–1185 (1988). 10.1080/09500348814551251 [DOI] [Google Scholar]
- 17.Sabharwal Y. S., Rouse A. R., Donaldson L., Hopkins M. F., Gmitro A. F., “Slit-scanning confocal microendoscope for high-resolution in vivo imaging,” Appl. Opt. 38(34), 7133–7144 (1999). 10.1364/AO.38.007133 [DOI] [PubMed] [Google Scholar]
- 18.Rouse A. R., Gmitro A. F., “Multispectral imaging with a confocal microendoscope,” Opt. Lett. 25(23), 1708–1710 (2000). 10.1364/OL.25.001708 [DOI] [PubMed] [Google Scholar]
- 19.Rouse A. R., Kano A., Udovich J. A., Kroto S. M., Gmitro A. F., “Design and demonstration of a miniature catheter for a confocal microendoscope,” Appl. Opt. 43(31), 5763–5771 (2004). 10.1364/AO.43.005763 [DOI] [PubMed] [Google Scholar]
- 20.Makhlouf H., Gmitro A. F., Tanbakuchi A. A., Udovich J. A., Rouse A. R., “Multispectral confocal microendoscope for in vivo and in situ imaging,” J. Biomed. Opt. 13(4), 044016 (2008). 10.1117/1.2950313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanbakuchi A. A., Udovich J. A., Rouse A. R., Hatch K. D., Gmitro A. F., “In vivo imaging of ovarian tissue using a novel confocal microlaparoscope,” Am. J. Obstet. Gynecol. 202(1), e1–e9 (2010). 10.1016/j.ajog.2009.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muldoon T. J., Roblyer D., Williams M. D., Stepanek V. M., Richards-Kortum R., Gillenwater A. M., “Noninvasive imaging of oral neoplasia with a high-resolution fiber-optic microendoscope,” Head Neck 34(3), 305–312 (2012). 10.1002/hed.21735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pierce M., Yu D., Richards-Kortum R., “High-resolution fiber-optic microendoscopy for in situ cellular imaging,” J. Vis. Exp. 47(47), e2306 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]




