Summary
Phylogenetic reconstructions provide a means of inferring the branching relationships among members of multigene families that have diversified via successive rounds of gene duplication and divergence. Such reconstructions can illuminate the pathways by which particular expression patterns and protein functions evolved. For example, phylogenetic analyses can reveal cases in which similar expression patterns or functional properties evolved independently in different lineages, either through convergence, parallelism, or evolutionary reversals. The purpose of this paper is to provide a robust phylogenetic framework for interpreting experimental data and for generating hypotheses about the functional evolution of globin proteins in chordate animals. To do this we present a consensus phylogeny of the chordate globin gene superfamily. We document the relative roles of gene duplication and whole-genome duplication in fueling the functional diversification of vertebrate globins, and we unravel patterns of shared ancestry among globin genes from representatives of the three chordate subphyla (Craniata, Urochordata, and Cephalochordata). Our results demonstrate the value of integrating phylogenetic analyses with genomic analyses of conserved synteny to infer the duplicative origins and evolutionary histories of globin genes. We also discuss a number of case studies that illustrate the importance of phylogenetic information when making inferences about the evolution of globin gene expression and protein function. Finally, we discuss why the globin gene superfamily presents special challenges for phylogenetic analysis, and we describe methodological approaches that can be used to meet those challenges.
Keywords: Cytoglobin, gene family evolution, hemoglobin, myoglobin, neuroglobin
INTRODUCTION
During the last half century, hemoglobin (Hb) and myoglobin (Mb) played starring roles in research efforts to understand relationships between protein structure and function, and in efforts to identify molecular mechanisms of adaptation and pathophysiology. In the past decade, investigations into the structure, function, and evolution of globin proteins have been reinvigorated by the discovery of new members of the globin protein superfamily in humans and other vertebrates. Of these recently characterized globin proteins, neuroglobin (Ngb) and cytoglobin (Cygb) have received the most attention (1-9). These two proteins are highly conserved, and are possessed by all jawed vertebrates (gnathostomes) examined to date. In gnathostomes, the physiological functions of Ngb and Cygb are not yet clearly understood, but investigations into their structures, ligand reactivities, biochemical activities, and expression patterns are gradually yielding clues (6-10). In jawless fishes (cyclostomes, represented by lampreys and hagfish), homologs of the Ngb, Mb, and Hb genes appear to have been secondarily lost, and the Cygb homolog evolved a specialized respiratory function in blood-oxygen transport (11).
In addition to Ngb and Cygb, recent surveys of vertebrate genome assemblies and sequence data bases have led to the discovery of several other globin genes with much more restricted phyletic distributions. The Globin X (GbX) gene is the product of a duplication event that predates the divergence between protostomes and deuterostomes, and among vertebrates it has only been found in the genomes of teleost fish and Xenopus (12). The Globin Y (GbY) gene appears to be restricted to gnathostome vertebrates, and has thus far only been found in the genomes of Xenopus, anole lizard, bearded dragon lizard, and platypus (13-16). Finally, the Globin E (GbE) gene has thus far only been found in the genome of birds (16-19). Expression patterns of these recently discovered globin genes have been characterized, but the physiological functions of the encoded proteins remain a mystery. Ongoing experimental studies can be expected to yield new and surprising insights in coming years.
To interpret the results of functional experiments, and to generate informed hypotheses about the functional evolution of globin proteins, it is important to have a correct understanding of phylogenetic relationships. Phylogenetic reconstructions allow us to infer the branching relationships among members of a multigene family that have diversified via successive rounds of duplication and divergence. In comparisons among different species, phylogenetic reconstructions provide a means of distinguishing between paralogous genes (which trace their common ancestry to duplication events) and orthologous genes (which trace their common ancestry to speciation events – that is, they descend from a common ancestral gene by phylogenetic splitting at the organsimal level). The reconstruction of phylogenetic relationships among homologous members of a multigene family is also essential for understanding the pathways by which various functional properties evolved. For example, it is possible to reconstruct the history of evolutionary change in particular structural and functional features of proteins by “mapping” character states onto a phylogenetic tree that is estimated using independent data. This mode of inference can reveal whether certain physiological functions of modern-day globins represent derived, “repurposed” modifications of distinct ancestral functions. Phylogenetic reconstructions are also essential for identifying cases in which similar features evolved independently in different lineages, either through convergence, parallelism, or evolutionary reversals.
The purpose of the present paper is to provide a robust phylogenetic framework for interpreting experimental data and for generating hypotheses about the functional evolution of globin proteins in chordate animals. To do this we present a consensus phylogeny of the globin gene superfamily in chordates, based on genomic sequence data that were available as of December 2010. Our results demonstrate the value of integrating phylogenetic analyses with genomic analyses of conserved synteny to infer the duplicative origins and evolutionary histories of globin genes. We also discuss a number of case studies that illustrate the importance of phylogenetic information when making inferences about the evolution of globin gene expression and protein function. Finally, we discuss why the globin gene superfamily presents special challenges for phylogenetic analysis, and we describe methodological approaches that can be used to meet those challenges.
CONSENSUS PHYLOGENY OF CHORDATE GLOBINS
To reconstruct the phylogeny of chordate globins we assembled a data set that included the complete set of globins from representatives of all major vertebrate lineages, in addition to representatives of the two other chordate subphyla: the sea squirt (Ciona intestinalis, a urochordate, (20)) and amphioxus (Branchiostoma floridae, a cephalochordate, (21)). To compile the vertebrate globin dataset, we interrogated the genome assemblies of eleven gnathostome taxa (including teleost fish, amphibians, squamate reptiles, birds, and mammals), and we used bioinformatic tools to annotate the entire globin gene repertoire of each species. In addition, we compiled globin sequences from three representative cartilaginous fish (red stingray, gummy houndshark, and Port Jackson shark) and three representative cyclostomes: Arctic lamprey and sea lamprey (subclass Hyperartia) and hagfish (subclass Myxini). Even though most of the gnathostome species included in this study possess multiple paralogous copies of the α- and β-like globin genes, we only included a representative subset of sequences from each species in our analyses because monophyly of the α- and β-like globin genes has been well established (22, 23).
Amino acid sequences were aligned using the E-INS-i, G-INS-i, and L-INS-i strategies from Mafft version 6.8 (24, 25), and for each alignment we estimated phylogenetic relationships using maximum likelihood and Bayesian approaches. Maximum likelihood searches were carried out using Treefinder version October 2008 (26) using a mixed model of amino acid substitution, which was identified as the best-fitting model of amino acid substitution with the “propose model” subroutine from Treefinder. Support for the nodes was evaluated with 1,000 bootstrap pseudoreplicates. Bayesian analyses were conducted in Mr. Bayes v.3.1.2 (27) running six simultaneous chains for 10,000,000 generations, sampling every 1,000 generations, under a mixed model of amino acid substitution and using default priors. Support for the nodes and parameter estimates were derived from a majority rule consensus of the last 2,500 trees. The average standard deviation of split frequencies remained less than 0.01 after the burn-in threshold.
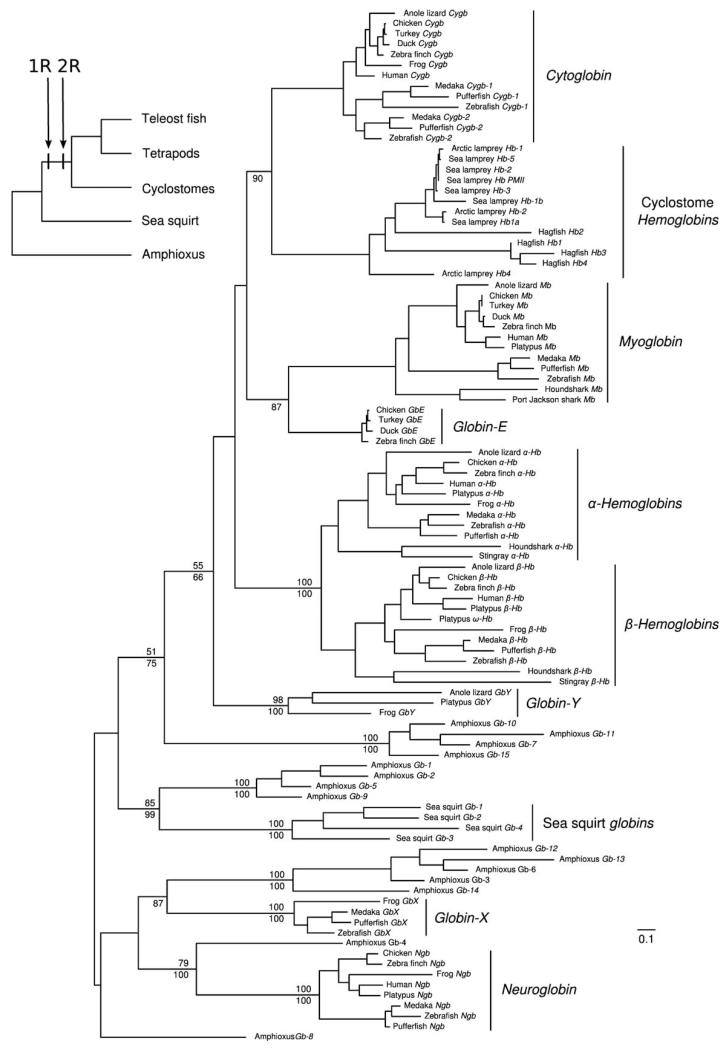
Bayesian and maximum likelihood phylogenies arranged chordate globins into 4 distint clades (Fig. 1). The first clade contains the complete set of vertebrate-specific globins (Cygb, Cyclostome Hbs, Mb, GbE, the α- and β-Hbs of gnathostomes, and GbY) plus Gbs-7, 10, 11, and 15 from amphioxus. The second clade includes the complete globin repertoire of the sea squirt plus Gbs-1, 2, 5, and 9 from amphioxus, the third clade includes vertebrate GbX plus Gbs-3, 6, 12, 13 and 14 from amphioxus, and the fourth clade includes vertebrate Ngb and its putative ortholog in amphioxus, Gb-4 (21). The correct phylogenetic placement of an additional amphioxus globin, Gb-8, is unclear. Consistent with results of previous studies (11-13, 18, 21), our phylogenetic reconstruction reveals that two of the vertebrate globin paralogs, GbX and Ngb, derive from independent duplication events that occurred prior to the deueterostome/protostome divergence. The remaining members of the vertebrate globin gene repertoire are all products of vertebrate-specific duplication events (18). Amphioxus possesses an especially diverse repertoire of globin genes, as sequences from this taxon are represented in each of the different globin gene lineages that trace back to the chordate common ancestor. In the absence of gene losses, each clade of orthologous sequences should independently recapitulate the expected organismal phylogeny. For example, a hypothetical clade of orthologous globin genes from representatives of the three chordate subphyla (Craniata, Urochordata, and Cephalochordata) would be expected to group the sea squirt with vertebrates to the exclusion of amphioxus (28, 29). Contrary to these expectations, the phylogenetic reconstruction shown in Figure 1 indicates that orthologs of GbX, Ngb, and the pro-ortholog of vertebrate-specific globins have all been secondarily lost from the sea squirt genome (18). Likewise, the pro-ortholog of the sea squirt globins was lost in the stem lineage of vertebrates.
Figure 1.
Maximum likelihood phylogram describing relationships among globin genes from representative chordates: 11 jawed vertebrates (Gnathostomata), 3 jawless fishes (Cyclostomata), the sea squirt (Ciona intestinalis [Urochordata]) and amphioxus (Branchiostoma floridae [Cephalochordata]). Numbers above the nodes correspond to maximum likelihood bootstrap support values, and those below the nodes correspond to Bayesian posterior probabilities. The inset tree depicts the organismal phylogeny and the timing of two successive whole-genome duplications (the “1R” and “2R” duplications) in the stem lineage of vertebrates.
The set of vertebrate-specific globin genes fall into four main clades: (i) Cygb and cyclostome Hbs; (ii) Mb + GbE; (iii) the α- and β-chain Hbs of gnathostomes; and (iv) GbY (Fig. 1; (11, 18)). As reported previously (11), the “Hb” genes of cyclostomes are clearly orthologous to the Cygb gene of gnathostome vertebrates. The relationships depicted in Fig. 1 indicate that progenitors of the four main vertebrate-specific globin lineages were present in the vertebrate common ancestor (11, 18). At face value, the fact that four clades of vertebrate-specific globins are sister to a single clade of amphioxus globins is consistent with the hypothesis that those four clades represent the paralogous products of two successive rounds of whole-genome duplication (WGD) in the stem lineage of vertebrates (the “1R” and “2R” WGDs). If the four main clades of vertebrate-specific globins represent products of the 1R/2R WGDs in the vertebrate common ancestor, then representatives of the four vertebrate-specific gene lineages should be embedded in unlinked chromosomal regions that share similar, interdigitated arrangements of paralogous genes (“paralogons”). The flanking tracts of paralogous duplicates may not contain identical subsets of genes, but the globin-defined paralogons should be united by gene families that trace their duplicative origins to the stem lineage of vertebrates. For example, we would expect that a number of globin-linked genes are members of “4:1” gene families – that is, quartets of paralogs that co-duplicated with the globin genes such that each of the four duplicate copies are located on a different globin-defined paralogon. The problem with identifying 4:1 gene families is that only a small subset of gene families would be expected to retain all four of the resultant paralogs following two rounds of WGD, and subsequent gene turnover via small-scale duplications and deletions would further obscure the signal of WGD (30). However, the same globin-defined paralogons that are united by 4:1 gene families would also be united – in various combinations – by 3:1 and 2:1 gene families. Members of such gene families are located on either three or two of the four globin-defined paralogons, respectively (the implication is that the missing members of the expected gene quartet were deleted after the first or second rounds of WGD). Finally, if the four clades of vertebrate-specific globins are products of two rounds of WGD in the vertebrate common ancestor, then the globin-defined paralogons should all derive from a single linkage group of the ancestral chordate proto-karyotype (29).
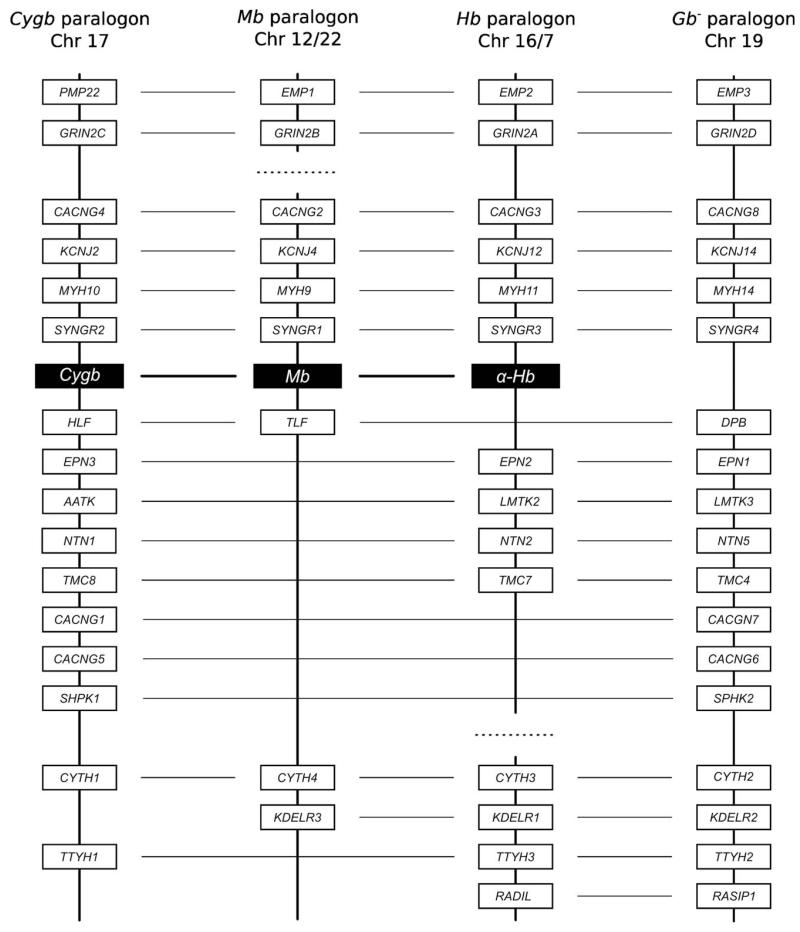
We tested each of the above predictions by examining the genomic map positions of the vertebrate-specific globin genes, by characterizing large-scale patterns in the physical locations of paralogous gene duplicates in the flanking chromosomal regions, and by reconstructing the phylogenetic relationships of the globin-linked genes (18). Results of this analysis revealed that the Cygb gene, the Mb/GbE gene pair, and the Hb/GbY gene pair are each embedded in clearly identifiable paralogons (18, 19). The Hb paralogon is defined by the α-globin gene cluster of amniotes, and is defined by the tandemly linked α- and β-globin gene clusters in teleost fishes and amphibians. In the human genome, the “Cygb” and “Hb” paralogons correspond to large segments of Chromosome 17 and 16, respectively, and the “Mb” paralogon is partitioned among segments of Chromosomes 7, 12, and 22. We also identified a fourth set of linked genes on human Chromosome 19 that co-duplicated with the Cygb, Mb, and Hb paralogons, but the associated globin paralog has been secondarily lost (Fig. 2). Synteny comparisons revealed that this segment of Chromosome 19 represents the fourth WGD-derived paralogon – the inference is that it once harbored a globin gene that was co-paralogous to the proto Cygb, Mb, and Hb genes (and it is possible that this globin gene lineage has been retained in the genomes of early-branching vertebrate lineages like cartilaginous fish). We henceforth refer to this 4th paralogon as the “globin minus” (Gb−) paralogon. As predicted by the genome duplication model, the 4:1, 3:1, and 2:1 gene families that unite the Cygb, Mb, Hb, and Gb− paralogons all trace their duplicative origins to the stem lineage of vertebrates (Fig. 3; (18)).
Figure 2.
Graphical depiction of gene duplicates that are shared between the three globin-defined paralogons (Cygb, Mb, and Hb) and the “Gb−” paralogon in the human genome. There are seven 4:1 gene families that unite the 4th paralogon with the Cygb, Mb, and Hb paralogons, there are seven 3:1 gene families that unite the Gb− paralogon with two of the three globin-defined paralogons, and there are four 2:1 gene families that unite the Gb− paralogon with a single globin-defined paralogon. On each chromosome, annotated genes are depicted as grey bars. The “missing” globin gene on the Gb− paralogon is denoted by an “X”. The shared paralogs are depicted in colinear arrays for display purposes only, as there is substantial variation in gene order among the four paralogons. For clarity of presentation, genes that are not shared between the Gb− paralogon and any of the three globin-defined paralogons are not shown. In the human genome, the Gb− paralogon on Chromosome 19 shares multiple gene duplicates with fragments of the Hb paralogon on Chromosomes 16 and 7.
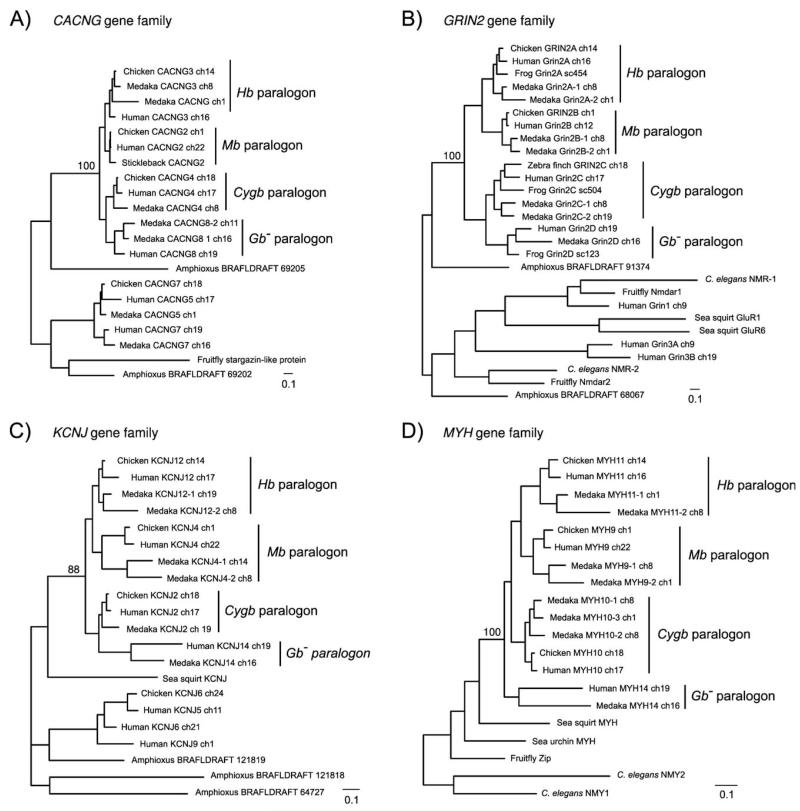
Figure 3.
Maximum likelihood phylogenies of representative 4:1 gene families that unite the Cygb, Mb, Hb, and Gb− paralogons. Individual members of the CACNG, Grin2, KCNJ, and MYH gene families (panels A-D, respectively) are located on each of the four globin-defined paralogons (see Fig. 2 for their chromosomal locations in the human genome). As the tree topologies indicate, paralogous members of the same gene family always form a monophyletic group relative to the putative ortholog in non-vertebrate chordates (amphioxus or sea squirt). In each of the four maximum likelihood trees, bootstrap support values are shown for the node uniting all vertebrate-specific gene as a monophyletic group. These phylogenies (and those for many other globin-linked gene duplicates; Hoffmann et al. 2010a) are consistent with the genome-duplication hypothesis, and indicate that each of the gene families diversified prior to the divergence between tetrapods and teleost fish.
Integrating the phylogenetic reconstruction with synteny comparisons revealed that the GbY gene and the proto-Hb gene represent paralogous products of an ancient tandem gene duplication that occurred prior to the two rounds of WGD in the stem lineage of vertebrates (18). The ancestral linkage arrangement of these two genes is still retained in the genomes of Xenopus, anole lizard, bearded dragon lizard, and platypus, as GbY is located at the 3′ end of the α-globin gene cluster in each of these taxa (13-16). Similarly, the GbE and Mb genes represent the paralogous products of a tandem gene duplication that occurred in the common ancestor of gnathostome vertebrates, and their ancestral linkage arrangement is still retained in the genomes of all birds examined to date (chicken, turkey, mallard duck, and zebra finch; (18, 19)).
By comparing the complete sequences of vertebrate genomes to those of non-vertebrate chordates like amphioxus, it is possible to reconstruct the proto-karyotpe of the chordate ancestor (29). As a result of the 1R/2R WGDs, each of the proto-chromosomes in the chordate ancestor would have been quadruplicated in the stem lineage of vertebrates. This is reflected by the well-documented “tetra-paralogon” structure that is detectable in the genomes of contemporary vertebrates (30). Consistent with the genome-duplication hypothesis, the synteny analysis of Hoffmann et al. (18) revealed that the Cygb, Mb, Hb, and Gb− paralogons all descend from a single ancestral linkage group in the reconstructed chordate proto-karyotpe (= linkage group 15 of Putnam et al. (29)). This provides conclusive evidence that the Cygb, Mb, and Hb gene lineages represent the paralogous products of WGD, not just large-scale segmental duplications (18).
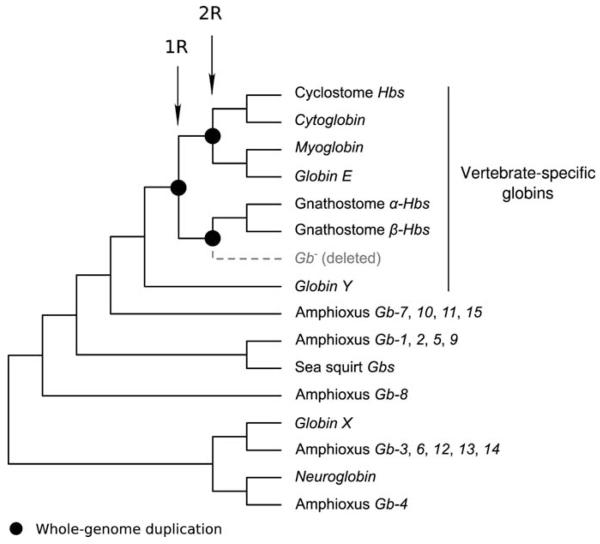
The traditional textbook account of the origins of vertebrate globins is that the proto Mb and Hb genes were produced by duplication of an ancestral, single-copy globin gene in the vertebrate common ancestor. In the Hb gene lineage, a subsequent tandem gene duplication gave rise to the progenitors of the α- and β-globin gene subfamilies. This scenario is correct in broad outline, but we now know that the progenitors of the Cygb/GbE/Mb gene lineage and the α/β-Hb gene lineage originated via WGD in the stem lineage of vertebrates (Fig. 4; 18, 19).
Figure 4.
Cladogram describing phylogenetic relationships among chordate globins. The products of whole-genome duplications (the 1R and 2R duplications) are indicated in the clade of vertebrate-specific globins.
CONVERGENT EVOLUTION OF GLOBIN GENE EXPRESSION AND PROTEIN FUNCTION
Phylogenetic analyses of the vertebrate α- and β-chain Hb genes have revealed a number of examples of convergent evolution in the developmental timing of gene expression. In all tetrapod vertebrates that have been examined to date, developmentally regulated members of the α- and β-globin gene families direct the synthesis of functionally distinct Hb isoforms in primitive (embryonic) erythrocytes derived from the yolk sac and in definitive (adult) erythroid cells derived from the bone marrow (31-35). In the α-globin gene cluster, the physiological division of labor between early- and late-expressed genes was established in the common ancestor of tetrapod vertebrates and it appears to have been retained in nearly all descendant lineages. The ancestral arrangement of the tetrapod α-globin gene cluster is 5′-αE-αD-αA-3′ (16, 36). In the stem lineage of tetrapods, the αE- and αA-globin genes originated via tandem duplication of an ancestral proto α-globin gene and the αD-globin gene originated subsequently via tandem duplication of the proto αE-globin gene (36). In all tetrapods that have been examined, the αE-globin gene is exclusively expressed in larval/embryonic erythroid cells and the αA-globin gene is expressed in definitive erythroid cells during later stages of prenatal development and postnatal life. The αD-globin gene does not appear to be expressed in mammalian erythrocytes, but it is expressed in both primitive and definitive erythroid cells in all birds and nonavian reptiles that have been examined to date (34, 35).
In contrast to the ancient functional diversification of the α-like globin genes, the developmental regulation of gene expression in the β-globin gene cluster evolved independently in several different tetrapod lineages (16). For example, in mammals and birds, the β-like globin genes that are expressed during the earliest stages of embryogenesis are not 1:1 orthologs, as they are independently derived from lineage-specific duplications of the same proto β-globin gene (16, 23, 37). Even within mammals, embryonic β-like globin genes appear to have originated independently as the products of lineage-specific duplication events in monotremes (platypus and echidnas) and in the common ancestor of marsupials and placental mammals (37). Likewise, fetally expressed β-like globin genes originated independently in anthropoid primates and in some artiodactyls such as goats and cows. In anthropoid primates (including humans), duplicate copies of the embryonic γ-globin genes have been co-opted for fetal expression, whereas in artiodactyls, duplicate copies of the adult β-globin gene were co-opted for fetal expression (23). These observations indicate that genes with similar stage-specific expression patterns do not necessarily descend from a homologous gene copy that was inherited from a common ancestor. Instead, the genes represent the paralogous products of independent, lineage-specific duplication events, and their similar patterns of stage-specific expression are attributable to convergent evolution.
An especially remarkable case of convergence involves the independent evolution of erythroid-specific, oxygen-transport Hbs from different ancestral precursor proteins in gnathostomes and cyclostomes (11). A comprehensive phylogenetic analysis of vertebrate globins revealed that the erythroid Hbs of cyclostomes are orthologous to the Cygb protein of gnathostome vertebrates, a hexacoordinate globin that has no oxygen-transport function. The independent evolution of oxygen-transport Hbs in cyclostomes and gnathostomes represents an example of “co-optive convergence” where paralogous members of the same gene family independently evolve the same specialization of function in different lineages. During their independent acquisition of oxygen-transport functions, the two paralogous globins convergently evolved distinct forms of both homotropic and heterotropic cooperativity from different ancestral precursor proteins that lacked cooperativity. In both cases, multisubunit quaternary structures provided the basis for cooperative oxygen-binding and allosteric regulation, but the underlying structural mechanisms are quite distinct.
It has been suggested that globins in the common ancestor of eukaryotes may have performed functions unrelated to oxygen-transport (31, 38, 39). It is not known whether the progenitor of eukaryotic globins had a “2-on-2” or “3-on-3” tertiary structure, or whether it had penta- or hexacoordinate heme chemistry. In pentacoordinate globins like Hb and Mb, the heme iron is coordinated by four nitrogen atoms in the porphyrin ring and the proximal histidine in the F helix (HisF8). In the deoxy state, the sixth coordination site of the ferrous (Fe2+) iron atom is accessible to oxygen-binding. In hexacoordinate globins like Ngb and Cygb, the distal histidine in the E helix (HisE7) is bound to the sixth coordination position of both Fe2+ and Fe3+ (4). The binding of oxygen or other exogenous ligands therefore requires the displacement of the distal histidine. Given that heme coordination chemistry is such an important determinant of ligand-binding dynamics (and hence, physiological function) it might be supposed that penta- and hexacoordinate globins represent two highly distinct and ancient phylogenetic lineages. However, this is not the case, as phylogenetic reconstructions have revealed that the alternative heme coordination chemistries have evolved multiple times independently (8, 10, 12). In fact, phylogenetic reconstructions of metazoan globins suggest that hexacoordination actually evolved multiple times from different pentacoordinate ancestral states (10).
CHALLENGES ASSOCIATED WITH THE PHYLOGENETIC ANALYSIS OF GLOBIN GENE FAMILY EVOLUTION
Historically, much of the uncertainty about the phylogenetic relationships among vertebrate globins was attributable to inadequate sampling of taxa and/or paralogous gene lineages. For example, it was previously assumed that Hb and Mb originated via duplication of an ancestral, single-copy globin gene prior to the cyclostome/gnathostome divergence, such that each of these two vertebrate lineages inherited orthologous copies of the same “proto-Hb” gene (22, 23). According to this scenario, the Hbs of cyclostomes would be sister to the clade of gnathostome α- and β-chain Hbs: (Mb (cyclostomes Hb, gnathostome Hb)). Until gnathostome Cygb sequences were included in phylogenetic reconstructions – which revealed the (gnathostome Hb (cyclostome Hb, gnathostome Cygb)) topology (11) – the phylogenetic affinities between cyclostome and gnathostome Hbs could not be correctly inferred.
The availability of complete genome assemblies for an exponentially growing list of taxa not only enhances the density of “taxon” sampling in phylogenetic analysis – it also increases the sampling of paralogous gene lineages because some globin genes have a very limited phyletic distribution. Despite the increasing availability of sequence data, reconstructing the phylogeny of chordate globins remains a challenging proposition. Phylogenetic uncertainty can stem from several sources, including ambiguities in the sequence alignment (40-42) and uncertainty about the best-fitting model of nucleotide or amino acid substitution (43). And of course there is a vast literature on the computational and statistical challenges associated with exploring the vast universe of all possible phylogenetic tree topologies (44). These inherent problems in phylogeny estimation are exacerbated by the fact that globin proteins are typically less than 200 amino acids in length, so there is a rather limited number of potentially informative sites. Finally, as discussed above, many globin gene lineages trace their origins to extremely ancient duplication events. For example, all vertebrate-specific globins shown in Fig. 1 originated via WGDs or subsequent small-scale duplication events that occurred prior to the split between cartilaginous fish and the common ancestor of teleost fish and tetrapods, over 500 million years ago. Over a broad range of timescales, lineage-specific gene duplications and deletions can also complicate phylogenetic inference (see below). Finally, because of the complex duplicative history of animal globins, the choice of appropriate outgroups is not always straightforward.
There are a number of steps that can be taken to minimize sources of uncertainty in phylogenetic reconstructions. First, it is highly advisable to conduct sensitivity analyses in which the same set of sequences is aligned using different algorithms, and the resultant alignments are then analyzed using different methods of phylogeny estimation under a range of different substitution models. For example, statistically robust inferences about the relationship between cyclostome and gnathostome Hbs required a comprehensive sensitivity analysis to evaluate how phylogeny estimates were affected by the use of different alignment algorithms, the use of different amino acid substitution models, and the use of different outgroup sequences (11). In the study by Hoffmann et al. (11), phylogenetic searches were performed on 10 alternative alignments under three different substitution models, and topology tests were then used to test alternative phylogenetic hypotheses.
In the case of Bayesian analyses, it is necessary to confirm the convergence of Markov chain Monte Carlo simulations to ensure an efficient exploration of parameter space. In the case of maximum likelihood analyses, competing phylogenetic hypotheses can be tested statistically using topology tests such as the Kishino-Hasegawa test (45), the Shimodaira-Hasegawa test (46), the approximately unbiased (AU) test (47), and the SOWT test (48, 49). These approaches have been used to test alternative phylogenetic hypotheses in a number of studies of globin gene family evolution (11, 19, 36, 37). Finally, it is often useful to incorporate additional sources of information to resolve phylogenetic relationships. As illustrated by the diversification of vertebrate-specific globin genes discussed above, tree topologies and patterns of conserved synteny provide reciprocally illuminating sources of information about the duplicative origins of globin genes.
CONCERTED EVOLUTION AND BIRTH-AND-DEATH EVOLUTION
In addition to the methodological issues discussed above, there are also certain modes of gene family evolution that can greatly complicate efforts to decipher the correct branching history of gene duplication and species divergence. Within the α- and β-globin subfamilies of vertebrates, tandemly linked genes are often identical or nearly identical in sequence. For example, most mammals possess 2-3 tandemly linked copies of the adult α-globin gene that have identical coding sequences (50, 51). This pattern is typically attributable to a history of gene conversion – a form of nonreciprocal recombinational exchange between duplicated genes. Recurrent gene conversion results in the gradual homogenization of sequence variation among paralogous members of the same gene family, giving rise to a pattern referred to as “concerted evolution”. Concerted evolution complicates phylogenetic reconstructions because the homogenization of sequence variation between paralogous genes erases phylogenetic history and creates the false appearance of recent common ancestry. Specifically, concerted evolution can create a confusing situation where paralogous genes within the genome of a single species are more similar to one another than they are to their orthologous counterparts in closely related species.
In the α- and β-globin gene clusters of mammals, there are a number of cases where the sequence similarity between tandemly duplicated globin genes is only partly attributable to concerted evolution between pre-existing paralogs – instead, it is often attributable to recent ancestry between the products of de novo gene duplications that occurred independently in different lineages (37, 50-54). To distinguish between the effects of concerted evolution and gene turnover (“birth-and-death evolution”) it is necessary to integrate phylogenetic information from multiple partitions of genomic sequence alignments. Because interparalog gene conversion is largely restricted to the coding regions of globin genes (55-58), orthologous and paralogous relationships can typically be determined by analyzing variation in flanking sequence and/or intronic sequence (37, 50, 52-54, 59).
CONCLUSIONS
The phylogenetic reconstruction presented in Figure 4 provides a framework for interpreting experimental data and for generating hypotheses about the functional evolution of globin proteins in chordates. As more complete genome sequences become available in coming years it will be possible to trace the ancestry of some chordate globins back to more ancient branch points in the metazoan phylogeny. To reconstruct these ancient pathways of gene family evolution, it will be necessary to use rigororous, model-based methods of phylogeny estimation that account for sources of uncertainty in the sequence alignments, the substitution models, and the choice of outgroup sequences that are used to root the tree. As it becomes possible to probe deeper into the evolutionary history of metazoan globin genes, it may become especially important to complement phylogenetic analyses with comparisons of conserved synteny. Unraveling phylogenetic relationships among the diverse globin genes in protostome taxa – and documenting their affinities with deuterostome globins – may require a combined approach that integrates molecular phylogenetics, alignment-free computational methods, and genomic analyses of conserved synteny.
ACKNOWELDGMENTS
We thank S. Vinogradov and one anonymous reviewer for helpful comments and suggestions. Our work on globin gene family evolution is funded by grants to JFS from the National Institutes of Health and the National Science Foundation.
REFERENCES
- 1.Burmester T, Weich B, Reinhardt S, Hankeln T. A vertebrate globin expressed in the brain. Nature. 2000;407:520–523. doi: 10.1038/35035093. [DOI] [PubMed] [Google Scholar]
- 2.Burmester T, Ebner B, Weich B, Hankeln T. Cytoglobin: a novel globin type ubiquitously expressed in vertebrate tissues. Mol. Biol. Evol. 2002;19:416–421. doi: 10.1093/oxfordjournals.molbev.a004096. [DOI] [PubMed] [Google Scholar]
- 3.Trent JT, Hargrove MS. A ubiquitously expressed human hexacoordinate hemoglobin. J. Biol. Chem. 2002;277:19538–19545. doi: 10.1074/jbc.M201934200. [DOI] [PubMed] [Google Scholar]
- 4.Pesce A, et al. Neuroglobin and cytoglobin - Fresh blood for the vertebrate globin family. Embo Reports. 2002;3:1146–1151. doi: 10.1093/embo-reports/kvf248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fago A, Hundahl C, Malte H, Weber RE. Functional properties of neuroglobin and cytoglobin. Insights into the ancestral physiological roles of globins. IUBMB Life. 2004;56:689–696. doi: 10.1080/15216540500037299. [DOI] [PubMed] [Google Scholar]
- 6.Hankeln T, et al. Neuroglobin and cytoglobin in search of their role in the vertebrate globin family. J. Inorg. Biochem. 2005;99:110–119. doi: 10.1016/j.jinorgbio.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Hankeln T, Burmester T. Neuroglobin and cytoglobin. In: Ghosh A, editor. The Smallest Biomolecules: Diatomics and their Interactions with Heme Proteins. Elsevier B. V.; 2008. pp. 203–218. [Google Scholar]
- 8.Burmester T, Hankeln T. Neuroglobin and other nerve globins. In: Bolognesi M, di Prisco G, Verde C, editors. Protein Reviews: Dioxygen Binding and Sensing Proteins. Vol. 9. Springer; Milan: 2008. pp. 211–222. [Google Scholar]
- 9.Burmester T, Hankeln T. What is the function of neuroglobin? J. Exp. Biol. 2009;212:1423–1428. doi: 10.1242/jeb.000729. [DOI] [PubMed] [Google Scholar]
- 10.Kakar S, Hoffmann FG, Storz JF, Fabian M, Hargrove MS. Structure and reactivity of hexacoordinate hemoglobins. Biophys. Chem. 2010;152:1–14. doi: 10.1016/j.bpc.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoffmann FG, Opazo JC, Storz JF. Gene cooption and convergent evolution of oxygen transport hemoglobins in jawed and jawless vertebrates. Proc. Natl. Acad. Sci. U. S. A. 2010;107:14274–14279. doi: 10.1073/pnas.1006756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roesner A, Fuchs C, Hankeln T, Burmester T. A globin gene of ancient evolutionary origin in lower vertebrates: evidence for two distinct globin families in animals. Mol. Biol. Evol. 2005;22:12–20. doi: 10.1093/molbev/msh258. [DOI] [PubMed] [Google Scholar]
- 13.Fuchs C, Burmester T, Hankeln T. The amphibian globin gene repertoire as revealed by the Xenopus genome. Cytogenet. Genome Res. 2006;112:296–306. doi: 10.1159/000089884. [DOI] [PubMed] [Google Scholar]
- 14.Patel VS, et al. Platypus globin genes and flanking loci suggest a new insertional model for β-globin evolution in birds and mammals. BMC Biol. 2008;6:22. doi: 10.1186/1741-7007-6-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patel VS, Ezaz T, Deakin JE, Marshall Graves JA. Globin gene structure in a reptile supports the transpositional model for aminote α- and β-globin gene evolution. Chromosome Research. 2010;18:897–907. doi: 10.1007/s10577-010-9164-5. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann FG, Storz JF, Gorr TA, Opazo JC. Lineage-specific patterns of functional diversification in the α- and β-globin gene families of tetrapod vertebrates. Mol. Biol. Evol. 2010;27:1126–1138. doi: 10.1093/molbev/msp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kugelstadt D, Haberkamp M, Hankeln T, Burmester T. Neuroglobin, cytoglobin, and a novel, eye-specific globin from chicken. Biochem. Biophys. Res. Commun. 2004;325:719–725. doi: 10.1016/j.bbrc.2004.10.080. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann FG, Opazo JC, Storz JF. Key innovations in the vertebrate oxygen transport system were fueled by whole-genome duplication.(in revision) 2011. [Google Scholar]
- 19.Hoffmann FG, Opazo JC, Storz JF. Differential loss and retention of cytoglobin, myoglobin, and globin E during the radiation of vertebrates. Genome Biol. Evol. 2011 doi: 10.1093/gbe/evr055. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebner B, Burmester T, Hankeln T. Globin genes are present in Ciona intestinalis. Mol. Biol. Evol. 2003;20:1521–1525. doi: 10.1093/molbev/msg164. [DOI] [PubMed] [Google Scholar]
- 21.Ebner B, et al. The globin gene family of the cephalochordate amphioxus: implications for chordate globin evolution. BMC Evolutionary Biology. 2010;10:370. doi: 10.1186/1471-2148-10-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goodman M, Moore GW, Matsuda G. Darwinian evolution in the genealogy of haemoglobin. Nature. 1975;253:603–608. doi: 10.1038/253603a0. [DOI] [PubMed] [Google Scholar]
- 23.Goodman M, Czelusniak J, Koop BF, Tagle DA, Slightom JL. Globins: a case study in molecular phylogeny. Cold Spring Harb Symp QuantBiol. 1987;52:875–890. doi: 10.1101/sqb.1987.052.01.096. [DOI] [PubMed] [Google Scholar]
- 24.Katoh K, Toh H. Recent developments in the mafft multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 25.Katoh K, Asimenos G, Toh H. Multiple alignment of DNA sequences with mafft. Methods Mol Biol. 2009;537:39–64. doi: 10.1007/978-1-59745-251-9_3. [DOI] [PubMed] [Google Scholar]
- 26.Jobb G, von Haeseler A, Strimmer K. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol Biol. 2004;4:18. doi: 10.1186/1471-2148-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Ronquist F, Huelsenbeck JP. Mrbayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- 28.Delsuc F, Brinkmann H, Chourrout D, Philippe H. Tunicates and not cephalochordates are the closest living relatives of vertebrates. Nature. 2006;439:965–968. doi: 10.1038/nature04336. [DOI] [PubMed] [Google Scholar]
- 29.Putnam NH, et al. The amphioxus genome and the evolution of the chordate karyotype. Nature. 2008;453:1064–1071. doi: 10.1038/nature06967. [DOI] [PubMed] [Google Scholar]
- 30.Dehal P, Boore JL. Two rounds of whole genome duplication in the ancestral vertebrate. PloS Biology. 2005;3:1700–1708. doi: 10.1371/journal.pbio.0030314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hardison R. Hemoglobins from bacteria to man: evolution of different patterns of gene expression. J. Exp. Biol. 1998;201:1099–1117. doi: 10.1242/jeb.201.8.1099. [DOI] [PubMed] [Google Scholar]
- 32.Hardison R. Organization, evolution and regulation of the globin genes. In: Steinberg MH, Forget BG, Higgs DR, Nagel RL, editors. Disorders of Hemoglobin: Genetics, Pathophysiology, and Clinical Management. Cambridge University Press; Cambridge: 2001. [Google Scholar]
- 33.Brittain T. Molecular aspects of embryonic hemoglobin function. Mol Asp Med. 2002;23:293–342. doi: 10.1016/s0098-2997(02)00004-3. [DOI] [PubMed] [Google Scholar]
- 34.Alev C, Shinmyozu K, McIntyre BAS, Sheng G. Genomic organization of zebra finch α and β globin genes and their expression in primitive and definitive blood in comparison with globins in chicken. Dev.Genes Evol. 2009;219:353–360. doi: 10.1007/s00427-009-0294-8. [DOI] [PubMed] [Google Scholar]
- 35.Storz JF, Hoffman FG, Opazo JC, Sanger TJ, Moriyama H. Developmental regulation of hemoglobin synthesis in the green anole lizard, Anolis carolinensis. J. Exp. Biol. 2011;214:575–581. doi: 10.1242/jeb.050443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoffmann FG, Storz JF. The αD-globin gene originated via duplication of an embryonic α-like globin gene in the ancestor of tetrapod vertebrates. Mol. Biol. Evol. 2007;24:1982–1990. doi: 10.1093/molbev/msm127. [DOI] [PubMed] [Google Scholar]
- 37.Opazo JC, Hoffmann FG, Storz JF. Genomic evidence for independent origins of β-like globin genes in monotremes and therian mammals. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1590–1595. doi: 10.1073/pnas.0710531105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vinogradov SN, et al. A model of globin evolution. Gene. 2007;398:132–142. doi: 10.1016/j.gene.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 39.Vinogradov SN, Moens L. Diversity of globin function: enzymatic, transport, storage, and sensing. J. Biol. Chem. 2008;283:8773–8777. doi: 10.1074/jbc.R700029200. [DOI] [PubMed] [Google Scholar]
- 40.Phillips A, Janies D, Wheeler W. Multiple sequence alignment in phylogenetic analysis. Mol. Phylogenet. Evol. 2000;16:317–330. doi: 10.1006/mpev.2000.0785. [DOI] [PubMed] [Google Scholar]
- 41.Loytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc. Natl. Acad. Sci. U. S. A. 2005;102:317–330. doi: 10.1073/pnas.0409137102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wong KM, Suchard MA, Huelsenbeck JP. Alignment uncertainty and genomic analysis. Science. 2008;319:473–476. doi: 10.1126/science.1151532. [DOI] [PubMed] [Google Scholar]
- 43.Sullivan J, Joyce P. Model selection in phylogenetics. Ann. Rev. Ecol. Evol. Syst. 2005;36:445–466. [Google Scholar]
- 44.Felsenstein J. Inferring Phylogenies. Sinauer Associates; Sunderland, MA: 2004. [Google Scholar]
- 45.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in hominoidea. J. Mol. Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 46.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol. Biol. Evol. 1999;16:1114–1116. [Google Scholar]
- 47.Shimodaira H. An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 2002;51:492–508. doi: 10.1080/10635150290069913. [DOI] [PubMed] [Google Scholar]
- 48.Swofford DL, Olsen GJ, Waddell PJ, Hillis DM. Phylogenetic inference. In: Hillis DM, Moritz C, Mable B, editors. Molecular Systematics. 2nd Ed. Sinauer Associates; Sunderland, MA: 1996. pp. 407–514. [Google Scholar]
- 49.Goldman N, Anderson JP, Rodrigo AG. Likelihood-based tests of topologies in phylogenetics. Syst. Biol. 2000;49:652–670. doi: 10.1080/106351500750049752. [DOI] [PubMed] [Google Scholar]
- 50.Hoffmann FG, Opazo JC, Storz JF. Rapid rates of lineage-specific gene duplication and deletion in the α-globin gene family. Mol. Biol. Evol. 2008;25:591–602. doi: 10.1093/molbev/msn004. [DOI] [PubMed] [Google Scholar]
- 51.Storz JF, Hoffmann FG, Opazo JC, Moriyama H. Adaptive functional divergence among triplicated α-globin genes in rodents. Genetics. 2008;178:1623–1638. doi: 10.1534/genetics.107.080903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hoffmann FG, Opazo JC, Storz JF. New genes originated via multiple recombinational pathways in the β-globin gene family of rodents. Mol. Biol. Evol. 2008;25:2589–2600. doi: 10.1093/molbev/msn200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Opazo JC, Hoffmann FG, Storz JF. Differential loss of embryonic globin genes during the radiation of placental mammals. Proc. Natl. Acad. Sci. U. S. A. 2008;105:12950–12955. doi: 10.1073/pnas.0804392105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Opazo JC, Sloan AM, Campbell KL, Storz JF. Origin and ascendancy of a chimeric fusion gene: The β/δ-globin gene of paenungulate mammals. Mol. Biol. Evol. 2009;26:1469–1478. doi: 10.1093/molbev/msp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Storz JF, et al. Complex signatures of selection and gene conversion in the duplicated globin genes of house mice. Genetics. 2007;177:481–500. doi: 10.1534/genetics.107.078550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Storz JF, et al. Evolutionary and functional insights into the mechanism underlying high-altitude adaptation of deer mouse hemoglobin. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14450–14455. doi: 10.1073/pnas.0905224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Storz JF, Runck AM, Moriyama H, Weber RE, Fago A. Genetic differences in hemoglobin function between highland and lowland deer mice. J. Exp. Biol. 2010;213:2565–2574. doi: 10.1242/jeb.042598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Runck AM, Weber RE, Fago A, Storz JF. Evolutionary and functional properties of a two-locus β-globin polymorphism in Indian house mice. Genetics. 2010;184:1121–1131. doi: 10.1534/genetics.109.113506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Runck AM, Moriyama H, Storz JF. Evolution of duplicated β-globin genes and the structural basis of hemoglobin isoform differentiation in Mus. Mol. Biol. Evol. 2009;26:2521–2532. doi: 10.1093/molbev/msp165. [DOI] [PMC free article] [PubMed] [Google Scholar]