Abstract
Due to recent improvements in mass spectrometry (MS), there is an increased interest in data independent acquisition (DIA) strategies in which all peptides are systematically fragmented using wide mass isolation windows (“multiplex fragmentation”). DIA-Umpire (http://diaumpire.sourceforge.net/), a comprehensive computational workflow and open-source software for DIA data, detects precursor and fragment chromatographic features and assembles them into pseudo MS/MS spectra. These spectra can be identified using conventional database searching and protein inference tools, allowing sensitive untargeted analysis of DIA data without the need for a spectral library. Quantification is obtained using both precursor and fragment ion intensities. Furthermore, DIA-Umpire enables targeted extraction of quantitative information based on peptides initially identified in only a subset of the samples, resulting in more consistent quantification across multiple samples. We demonstrate the performance of the method using control samples of varying complexity, and publicly available glycoproteomics and affinity purification - mass spectrometry data.
Introduction
The combination of liquid chromatography (LC) and tandem mass spectrometry (MS/MS) is a powerful technology frequently applied to high-throughput peptide and protein identification and quantification. The most common strategy for peptide identification remains the data-dependent acquisition (DDA) approach1, in which the instrument sequentially surveys all the peptide ions that elute from the LC column at a particular time (MS1 scans), followed by isolation and fragmentation of selected peptide ions (usually the top few most intense) to generate MS/MS (MS2) spectra. Peptides are identified from these MS/MS spectra, most often through database searching2 (spectrum-centric approach; Fig. 1a). However, mass spectrometers are not able to reliably isolate and acquire high quality MS/MS spectra for all peptides present in typical samples, introducing stochasticity in the process3-7.
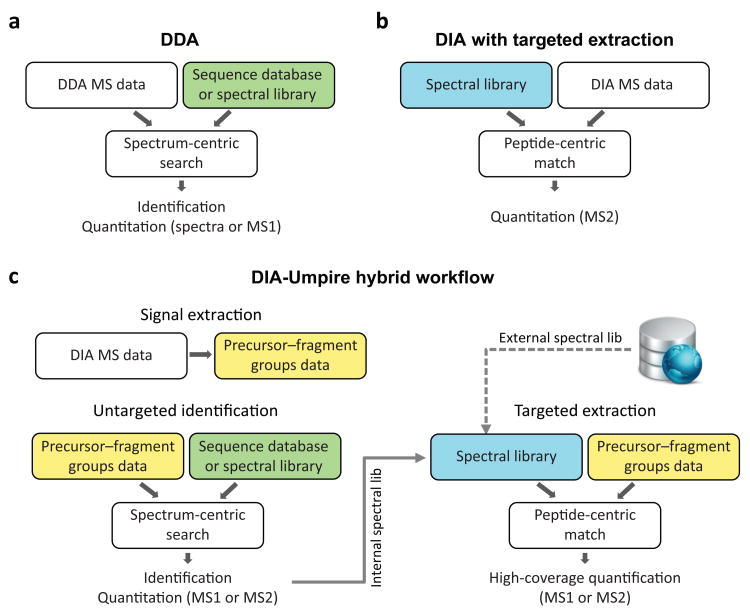
Fig. 1. Untargeted and targeted data analysis strategies and DIA-Umpire hybrid framework.
(a)Conventional analysis of DDA data is based on matching MS/MS spectra against a proteome-wide sequence database or a spectral library (spectrum-centric search). Peptides (and then proteins) are quantified using MS1 signal intensity or spectral counts (label-free quantification) (b) Current methods for DIA analysis are based on targeted data extraction, in which peptide ions from a spectral library are queried against experimental data (peptide-centric search) to find the best matching fragment ion signals and their intensities (MS2 based quantification). (c) DIA-Umpire hybrid workflow performs signal extraction from DIA MS1 and MS2 spectra to construct precursor–fragment groups (see Fig. 2 and Online Methods for details). Each precursor–fragment group is then analyzed using spectrum-centric searching to identify the peptides, as in (a). Peptide-centric matching is then performed to query unidentified precursor–fragment groups against a spectral library, as in (b). The spectral library can be built from the initial untargeted (spectrum-centric) results using the same DIA data, or can be combined (replaced) with an external spectral library built using DDA data. Quantification can be done from either MS1 precursor- or MS2 fragment-ion intensities.
Recent improvements in MS instrumentation have enabled alternative workflows to DDA, namely data-independent acquisition (DIA) methods4, 6, 8-14, now supported on multiple vendor platforms. These DIA strategies are based on acquiring fragment ion information for all precursor ions within a certain range of m/z values (DIA MS2 spectra), as exemplified by the Sequential Window Acquisition of all THeoretical Mass Spectra (SWATH)6 approach. The prevalent approach for DIA analysis is currently the targeted extraction of quantitative information from the acquired DIA data using libraries containing retention time and fragmentation information for the desired peptide species15, 16 (Fig. 1b; peptide-centric matching approach). Library generation is a current limitation of this strategy: either time and sample must be consumed to generate the libraries using the same samples and instrument, or libraries can be obtained independently17, but with the issues that fragmentation patterns and retention times may differ across experimental conditions. Additionally, DIA MS1 information (precursor peptide measurement scans) has not been systematically incorporated into DIA scoring so far, and the lack of accurate precursor mass leads to ambiguity in data interpretation, especially for peptides co-isolated in the same DIA window and sharing fragment ion peaks. Only a few computational tools4, 18, 19 have been developed so far for untargeted peptide identification from DIA and they have not been tested on SWATH-like DIA methods nor are they capable of performing both identification and quantification.
Here, we developed DIA-Umpire, a new computational approach that takes full advantage of DIA strategies such as SWATH. Our approach allows untargeted peptide identification directly from DIA data without the dependence on a spectral library for the data extraction: this enables us to readily employ tools developed for DDA data2 such as database search engines and post-identification analysis tools, facilitating incorporation of DIA into existing workflows. DIA-Umpire also reports DIA MS1- and MS2-based quantification results. Furthermore, DIA-Umpire is able to generate spectral libraries directly from the peptides it identifies, which can then be used to extract quantitative information in a targeted way in the samples where a particular peptide was not identified at the initial untargeted stage, increasing sensitivity in this hybrid approach (Fig. 1c). DIA-Umpire is neither vendor-specific nor limited to a particular DIA strategy, and is available as an open source pipeline.
Results
DIA-Umpire workflow
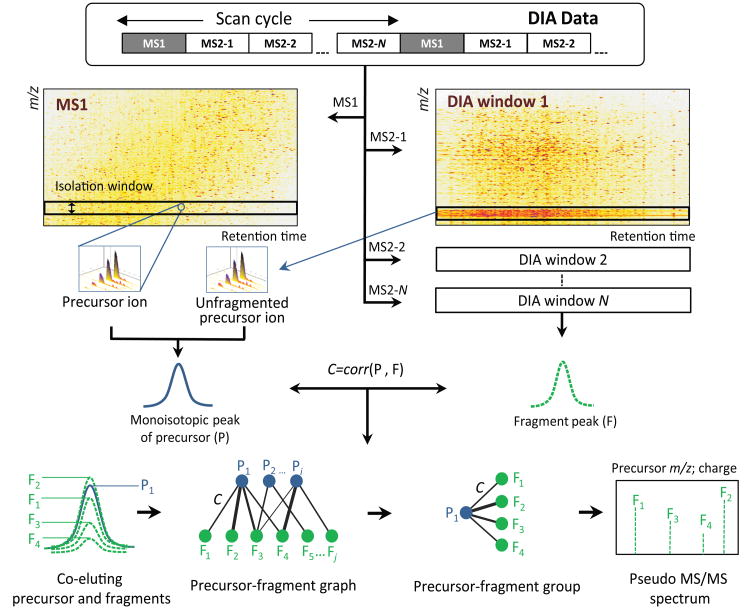
DIA-Umpire incorporates a number of computational algorithms for DIA analysis (see Online Methods for detail). It begins with a two dimensional (m/z – retention time) feature detection algorithm that discovers all possible precursor and fragment ion signals in DIA MS1 and MS2 data, respectively, and also possible unfragmented precursor ions in the MS2 data (Fig. 2). Because DIA usually employs wider isolation m/z range (e.g. 25 Da) than DDA, co-eluting peptides are more frequently co-fragmented, generating complex MS2 spectra. In order to measure the likelihood that a detected fragment signal is derived from a particular precursor peptide ion, the algorithm calculates the Pearson correlation coefficient of LC elution peaks and retention time differences of LC elution peak apexes between all detected precursor features and all co-eluting fragment ions. Reflecting the complex nature of precursor–fragment relationships, all precursor–fragment pairs are represented as a bipartite graph (Fig. 2). After filtering by a combination of thresholds, sets of fragment peaks are grouped with precursor features and stored as precursor–fragment groups (Fig. 2).
Fig. 2. DIA-Umpire signal processing algorithms.
The feature detection algorithm is applied to DIA MS1 and MS2 spectra to detect all possible MS1 peptide precursor ions and MS2 fragment signals. Each detected precursor feature is grouped with corresponding co-eluting fragment ion features based on Pearson correlation of LC elution peaks and retention times of peak apexes to form precursor-fragments groups. These precursor–fragment groups are used to construct pseudo MS/MS spectra (separated into different quality tiers based on the quality of detected precursor ion signal) for untargeted spectrum-centric database search and identification. The precursor–fragment groups are stored and are again queried during the second, peptide-centric targeted data extraction stage.
For direct untargeted analysis, DIA-Umpire generates a pseudo MS/MS spectrum (Supplementary Fig. 1) for each precursor–fragment group. The pseudo MS/MS spectra can be searched by any conventional database search engine. Here we used X! Tandem20 Comet21, and MSGF+22 followed by PeptideProphet23 or iProphet24 and ProteinProphet25 analysis. The resulting peptide and protein identification lists are filtered using computed peptide and protein probabilities controlling the false discovery rate (FDR) via, e.g., the target-decoy approach2. Identified peptides and proteins are quantified using either the MS1 precursor ion intensity or the MS2 fragment ion intensities (Fig. 1c).
DIA-Umpire is also compatible with a targeted quantification strategy in which the library is generated based on untargeted identification from DIA as described above (Fig. 1c). A peptide-centric spectral matching algorithm queries unidentified precursor–fragment groups against a spectral library built from spectrum-centric search results, allowing more consistent quantification of peptide ions across multiple experiments. Exact retention time is known for peptides identified in a given experiment, and the commonly identified peptides are used to perform retention time alignment across all the runs, negating the need for external retention time calibration peptides. An additional advantage of this approach over previously-described targeted extraction strategies6, 12, 16, 26 is that the precursor peptide m/z value is used to constrain the search space, enabling to distinguish between peptides with multiple shared fragments (e.g. modified and unmodified peptides).
Untargeted protein identification using DIA-Umpire
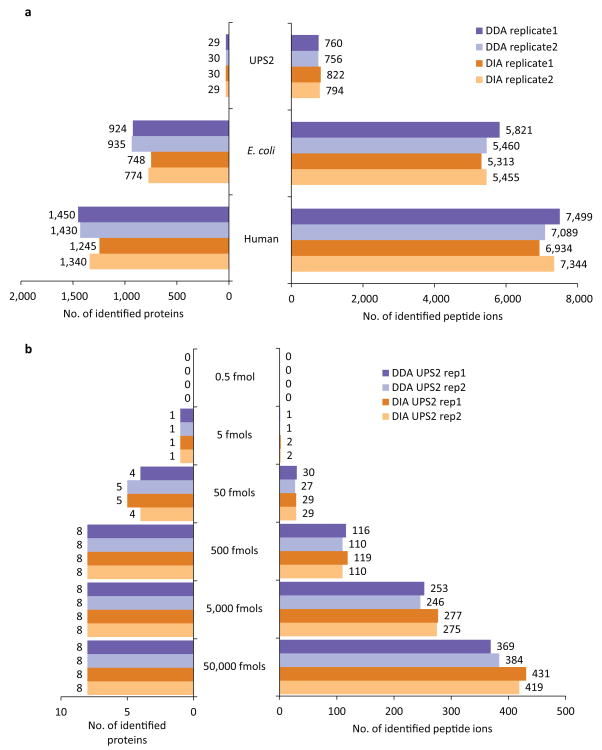
We first evaluated the performance of DIA-Umpire for untargeted protein identification using samples ranging from low complexity (48 Universal Protein Standard (UPS) proteins) to high complexity (E. coli and human cell lysates) by performing parallel DDA and DIA runs in at least duplicates on an AB SCIEX TripleTOF 5600. We acquired DIA data using 250 ms ion accumulation time for MS1 survey scans instead of the 50 ms SWATH setting used in earlier reports6, which improved the MS1 signal quality and detectability of precursor ion signals in complex samples (Supplementary Fig. 2). We identified close numbers of peptide ions and proteins in the DDA and DIA runs for all samples and search engines tested (Fig. 3a; Supplementary Table 1). As previously shown for DDA data, combining DIA pseudo MS/MS search results from multiple search engines with iProphet24 led to a consistent increase in the number of peptide ions and proteins identified at a given FDR (Supplementary Table 1). However, for the sake of clarity and because single search engine analyses are still prevalent in the field, the remainder of the manuscript, unless noted otherwise, is based on peptide and protein identifications using X! Tandem only (Fig. 3a; Supplementary Tables 2-4).
Fig. 3. Untargeted peptide and protein identification using DDA and DIA data from UPS2, E. coli, and human cell lysate samples.
(a) The number of peptide ions and proteins identified by X! Tandem search engine at 1% FDR in DDA and in DIA (SWATH) data from UPS2, E. coli, and human cell lysate samples. (b) The number of peptide ions and protein identifications (X! Tandem) in each replicate of the UPS2 sample DDA and DIA data plotted separately for proteins of different abundance (in UPS2 samples 48 proteins span 5 orders of magnitude of abundance ranging from 0.5 to 50,000 fmoles with 8 proteins in each abundance range).
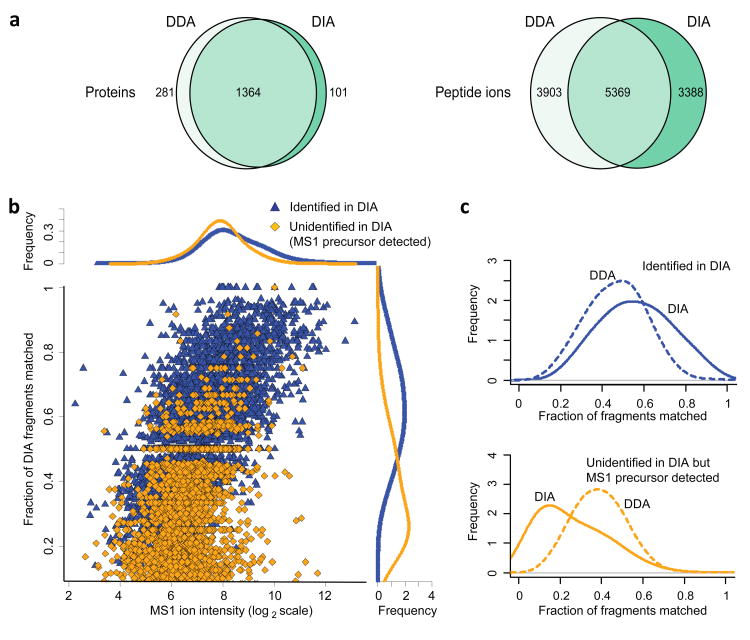
In low complexity UPS2 samples (48 proteins spanning 5 orders of magnitude in abundance), DIA and DDA identified similar numbers of peptide ions and proteins, with DIA identifying more peptide ions than DDA for higher abundance proteins (Fig 3b; Supplementary Table 1), and with the identification success depending on the abundance of each protein in the sample. In complex samples, such as human cell lysates (Fig. 4a; Supplementary Table 1), DDA slightly outperformed DIA at both peptide ion (9,272 vs. 8,757) and protein levels (1,645 vs. 1,465). Interestingly, the overlap between the peptide ions identified with high confidence (1% FDR) by both methods was relatively low (42% compared to 78% overlap at the protein level). While some of these differences were simply due to a detected peptide ion not passing the 1% FDR threshold in one or the other approach, DIA was also able to identify peptide ions where no MS/MS spectrum was acquired in DDA (2,326 peptide ions). The lack of an acquired MS/MS spectrum in DDA was observed even for some high intensity ions, possibly due to a combination of dynamic exclusion settings and co-elution of a different (more abundant) peptide. On the other hand, DDA was more successful in identifying peptide ions for which the pseudo MS/MS spectra extracted by DIA-Umpire from DIA data did not contain enough fragment ions, many of which were of low intensity (Fig. 4b,c). The loss of fragment ions in DIA can be attributed to a number of factors, including suppression of fragment ions by higher intensity species in the same DIA window, which is further compounded by computational challenges such as the imperfect de-multiplexing of co-eluting peptide ions. Similar results and trends were observed when the results from all three search engines were combined (Supplementary Fig. 3), and in the E. coli dataset (Supplementary Figs. 4 and 5).
Fig. 4. Comparative analysis of peptide identifications from DDA and DIA data from human cell lysate samples.
(a) The numbers of proteins and peptide ions identified at 1% FDR by X! Tandem search engine in DDA and in DIA (SWATH) data. Left: the number of protein identifications. Right: the number of peptide ion identifications (9,272 peptide ions identified from DDA data, 8,757 from DIA, 12,660 in total). Of the peptide ions identified by DIA and not DDA at 1% FDR (3,388), the majority were not identified by DDA because no MS/MS spectrum was acquired (2,326). Of the peptide ions identified from DDA data and not from DIA at 1% FDR (3,903), DIA-Umpire was able to detect precursor features for 3,338 of these peptide ions. (b) Fraction of fragment ions matched in pseudo MS/MS spectra extracted from DIA data as a function of MS1 peptide ion intensity in DDA data. Data points (peptide ions) and the summary density plots (“Frequencies”) are colored according to the two categories of peptide ions: those identified from DIA data at 1% FDR (high scoring in DIA, blue), and unidentified in DIA (orange; these ions were found in DIA data as described in Online Methods). (c) Comparison between DDA and DIA in terms of fraction of fragments matched among the two categories of peptide ions described in (b), showing that peptide ions identified with confidence from DDA but not from DIA have fewer matched fragments.
Comparison between untargeted and targeted DIA analysis
To investigate the differences between the untargeted approach described above and targeted data extraction strategies previously applied to SWATH data, we processed the human and E. coli datasets using OpenSWATH16. We used SpectraST27 to build spectral libraries by taking the union of DDA-identified spectra (9,272 peptide ions) from two replicates of human cell lysate data, and adding the same number of shuffled decoy spectra (Online Methods and Supplementary Fig. 6). In these data, OpenSWATH detected 7,372 peptide ions at 1% FDR according to mProphet28 (Supplementary Fig. 6a; Supplementary Table 5). In comparison, the untargeted analysis using DIA-Umpire (i.e. searching against the whole proteome database) identified 8,757 peptide ions at the same FDR. OpenSWATH had a better overlap with the identifications from the target library than DIA-Umpire, 79% vs. 58% (Supplementary Fig. 6b). This, to a large degree, can be explained by a smaller search space for OpenSWATH data extraction than for DIA-Umpire (Supplementary Fig. 6a). Indeed, when the pseudo MS/MS spectra were searched against a smaller database containing only the peptide sequences included in the spectral library used for OpenSWATH analysis (i.e. DDA-identified peptides), the overlap between the DIA-Umpire identified peptides and the DDA-identified peptides improved to 69% (Supplementary Fig. 6b). Similar results were obtained for E. coli samples (Supplementary Fig. 7; Supplementary Table 6). The use of the entire database however provided us with the opportunity to identify peptide ions not present in the DDA-constructed library.
Furthermore, when we built the spectral library for OpenSWATH from the pseudo-MS/MS spectra confidently identified by DIA-Umpire (8,757 peptide ions for the human cell lysate dataset), OpenSWATH confidently identified 8,650 (98.8%) of the library peptides, providing additional validation of the peptides identified using untargeted DIA-Umpire approach. We observed similar results (96.2% confirmation rate) for E. coli samples. The small percentage of peptide ions not identified by OpenSWATH was in part due to OpenSWATH's internal filtering of spectra from the input library.
We further assessed the performance of untargeted DIA-Umpire approach on a publicly available SWATH N-glycopeptide dataset from prostate cancer, which was already processed with OpenSWATH using a spectral reference library containing deamidated asparagine peptides built from a large number of DDA runs29. At 1% FDR, DIA-Umpire and OpenSWATH identified 1,821 and 1,383 deamidated asparagine peptide sequences (2,933 and 1,537 peptide ions) respectively (Online Methods; Supplementary Fig. 8; Supplementary Table 7). Among the additional identifications introduced by DIA-Umpire, more than 80% had a N-glycosylation (NX-S/T) motif, indicating high site specificity of the additional identifications (non-consensus identifications could be due to standard deamidation of non-glycosylated peptides, as we have not restricted our analysis to a library enriched in glycosylation sites, in contrast to the OpenSWATH library).
An additional drawback of current targeted extraction approaches (e.g. OpenSWATH) for DIA analysis is that they have difficulties resolving ambiguities for peptide ions that share many MS2 fragments (e.g. unmodified and post-translationally modified peptides co-fragmenting in the same isolation window), especially since the exact precursor mass is not used for scoring. We present examples (Supplementary Figs. 9–13) in which OpenSWATH was not able to distinguish deamidated peptide ions from unmodified ones. In contrast, DIA-Umpire constructs pseudo MS/MS spectra according to detected high mass accuracy MS1 precursor features, enabling better differentiation of peptide species.
Targeted extraction and protein quantification
Accuracy and coverage of protein quantification is of critical importance for downstream analysis of proteomic data. Following the initial untargeted analysis, DIA-Umpire fills in the missing peptide ion intensities across samples by creating an internal spectral library from all the identified peptides, followed by re-extraction of quantitative information across all precursor–fragment groups, including those which were not identified in the untargeted manner in some samples (Fig. 1c). In the human cell lysate data, targeted re-extraction improved the number of peptide ions and proteins identified across both replicate runs (estimated FDR less than 1%; see Online Methods), with the overlap between the replicates at the peptide ion and protein levels increasing from 63% to 80% and from 84% to 93%, respectively, compared to the initial untargeted identification results (Supplementary Fig. 14).
The same human cell lysate data was used to investigate the quantification performance of the algorithm. DIA-Umpire computes two iBAQ30 protein abundance measures (from MS1 and MS2 data) as well as “Top N peptides” (MS1)9 and “Top N peptides/ Top M fragments” (MS2)31 metrics (Supplementary Fig. 15 and 16; Online Methods). Using the reproducibility of protein quantification across the two replicate runs as a benchmark measure, the MS2-based method with a stringent peptide and fragment selection procedure (“MS2 Top6pep/Top6fra, Freq > 0.5”) outperformed the other methods considered (Supplementary Fig. 16). A similar MS1-based quantification metric (“MS1 Top6pep, Freq > 0.5”) performed almost equally well, but with fewer (1,310 vs. 1,341) proteins quantified across both replicates (Supplementary Fig. 15). A good agreement was observed between these two (MS1 and MS2-based) abundance measures (Supplementary Fig 17), further demonstrating the reliability of the feature detection and quantification algorithms in DIA-Umpire. In UPS2 standard protein sample, both MS1 and MS2 quantification recovered the expected trend of differential abundance, suggesting that these measures are suitable for estimation of absolute protein abundances (Supplementary Fig. 18).
Application of DIA-Umpire to interactome data
A popular application of MS based proteomics is interactome analysis which involves in most cases the use of quantitative MS to monitor the relative abundance of a given protein in a bait purification experiment in comparison to negative controls. The coupling of affinity purification (AP) with targeted extraction strategies for SWATH analysis (AP-SWATH) was recently described26, 32. At the same time, a large number of scoring tools have been previously developed to assist identification of true interaction partners over background contaminants in DDA data33, including Significance Analysis of INTeractome (SAINT)34, 35. We reasoned that AP-SWATH data would provide a good test case for a complete analytical pipeline by demonstrating that DIA-Umpire in combination with SAINT can detect true interactors from DIA data, without the need for spectral libraries. We analyzed a dataset consisting of three biological replicates of the baits EIF4A2 and MEPCE and the negative GFP control analyzed by DIA26 (Fig. 5a).
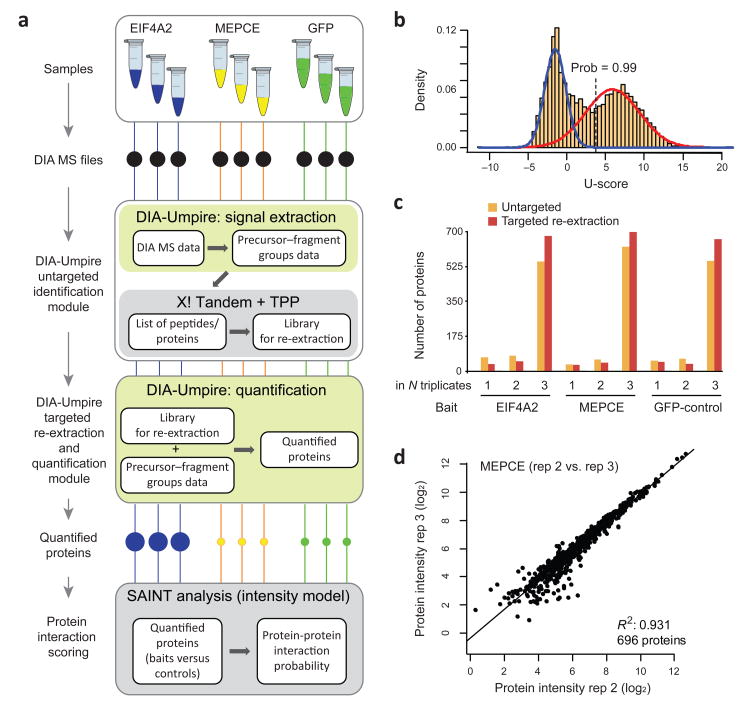
Fig. 5. Illustration of the entire DIA-Umpire workflow using affinity purification – SWATH interactome dataset.
(a) Two bait proteins (EIF4A2 and MEPCE) and the negative control (GFP) samples were analyzed in biological triplicates using AP-SWATH. The complete DIA-Umpire pipeline was applied to quantify proteins in these samples. The quantified proteins were further analyzed using SAINT (intensity model) to compute protein-protein interaction probabilities. (b) The distribution of scores (U-score) computed by the targeted re-extraction algorithm of DIA-Umpire. Data shown are from one biological replicate of MEPCE AP-SWATH run. The observed distribution was modeled using the mixture modeling approach (blue curve: false identification model; red curve: correct identifications) to compute the posterior probability for each match. Peptide ions with a computed probability above 0.99 were considered confidently identified and contributed, together with the peptide ions identified at the initial untargeted identification stage, to protein quantification for their corresponding protein. (c) The numbers of proteins identified in only one, two, or all three biological replicates for each experiment after the initial untargeted search and after targeted data re-extraction. Comparison between the sets of proteins identified in each experiment (all biological replicates combined) after the untargeted search and after targeted data re-extraction, showing an increase in the number of proteins quantified across all three samples. (d) High reproducibility of protein intensities between two MEPCE AP-SWATH biological replicates computed by DIA-Umpire using the “MS2 Top6pep/Top6fra, Freq>0.5” quantification approach.
In the first step, we processed DIA data through DIA-Umpire in an untargeted manner, leading to identification and quantification of 3,900 – 4,900 peptide ions (600 – 700 proteins) in each AP-SWATH run (Supplementary Table 8). As expected, using targeted re-extraction with a stringent 0.99 peptide-centric identification probability threshold (Fig 5b; Supplementary Fig. 19), we could identify and quantify additional peptide ions (1,300 – 2,300) and proteins (60 – 100) in each AP-SWATH run (Supplementary Fig. 20). Importantly, targeted re-extraction reduced the stochasticity issue, with an increase (by 19 – 23%) in the number of proteins quantified across all three replicates for the same bait (Fig. 5c). Protein abundances were estimated using the “MS2 Top6pep/Top6fra, Freq>0.5” approach, with an excellent quantification reproducibility observed across the biological replicates for each bait and the GFP controls (Fig. 5d; Supplementary Fig. 21).
Using SAINT35 we recovered 45 significant interactors (SAINT probability above 0.95) for the EIF4A2 bait, a translation initiation factor implicated in the association of mRNAs to the ribosome (Supplementary Table 9). These proteins included 19 associated translation initiation factors (specifically the multi-subunit factors eIF3 and eIF4), the poly(A) binding protein which binds eIF4 and, as expected, several RNA helicases and RNA-binding proteins that are likely recruited via the mRNA. The ubiquitin protease USP10 (previously reported as an interaction partner for the eIF4A2 direct interactor eIF4G1) and casein kinase II subunits (which interact with eIF3) were also detected, alongside a known eIF4A inhibitor, PDCD436. A similar result was observed for the MEPCE bait – a protein that methylates the cap of the 7SK snRNA, leading to its stabilization37: 54 proteins were confidently scored as interactors, including well-characterized partners such as CDK9, Cyclin T, HEXIM1, METTL16, LARP7, SART1 and 3, several splicing components, and multiple components of the large, but not small, ribosomal subunit26, 36, 37 (Supplementary Table 9). In summary, DIA-Umpire allows sensitive protein identification from DIA data, extraction of accurate quantitative information with less missing data, and is fully compatible with the existing interaction scoring methods such as SAINT, leading to the recovery of biologically-meaningful interactions.
Discussion
We demonstrated that by using DIA-Umpire we could identify a comparable number of peptides and proteins from DIA and DDA data. However, we have also observed the complementary nature of these two data acquisition strategies. As both the DDA and DIA technologies and the underlying instrumentation are rapidly improving, future work should include a comprehensive comparative analysis of different workflows as they are applied to a variety of biological problems. We also showed that reproducible and reliable quantification is possible using both DIA MS1 and MS2 data. Furthermore, DIA-Umpire is compatible with different DIA strategy variants including implementations on other instruments such as the Q Exactive Plus (Supplementary Fig. 22), and alternative approaches such as the MSX method12. The highly flexible design of the DIA-Umpire computational framework (importantly, with a full support for MS1 feature detection and quantification) should allow us to quickly adapt the algorithms to take advantage of new approaches and technological improvements, including emerging hybrid DIA/DDA strategies38. Finally, the pseudo MS/MS spectra generated by DIA-Umpire can be used to build spectral libraries for use with external tools, e.g. for visualization of spectra and precursor and fragment chromatograms in Skyline39 (Supplementary Fig. 23), or for targeted quantification using OpenSWATH. The internal (i.e. study-specific, DIA-derived) libraries can also be combined with external (e.g. DDA-derived) libraries for more complete analysis, a strategy that we are currently exploring in DIA-Umpire.
Accession Codes
All mass spectrometry files (Supplementary Table 11) along with DIA-Umpire results presented in this paper have been deposited to the ProteomeXchange Consortium40 (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001587.
Online Methods
Sample preparation
Proteomics Dynamic Range Standard (UPS2) sample was acquired from Sigma-Aldrich (St. Louis, MO), the MassPREP E. coli Digest Standard was acquired from Waters (Milford, MA) and the MS compatible human protein extract digest was from Promega (Madison, WI). The UPS2 samples were reduced with 5 mM TCEP (tris(2-carboxyethyl)phosphine), alkylated with 50 mM iodoacetamide, and digested overnight with 1 μg trypsin (Promega, Madison, WI) in 100 mM Tris pH 8 at 37°C. UPS2, E. coli, and human peptides were acidified with formic acid and loaded at various concentrations, alone or in combination, onto an in-house made 75 μm x 12 cm analytical column emitter packed with 3 μm ReproSil-Pur C18-AQ (Dr. Maisch HPLC GmbH, Germany). A NanoLC-Ultra 1D plus (Eksigent, Dublin CA) nano-pump was used to deliver a 90 minute gradient from 2% to 35% acetonitrile with 0.1% formic acid, followed by a 30 minute wash with 80% acetonitrile prior to re-equilibration to 2% acetonitrile with 0.1% formic acid.
Mass spectrometric analysis
Each sample was analyzed in duplicate (1 ug E. coli lysate, 500 ng Human lysate) or in triplicate (UPS2, UPS2 plus E. coli; affinity purified samples previously reported26) on a TripleTOFTM 5600 instrument (AB SCIEX, Concord, Ontario, Canada) once using DDA and once using DIA (SWATH) with an extended ion accumulation time of 250 ms for MS1 scans. UPS2 samples were also analyzed using SWATH with the previously-reported MS1 survey scan ion accumulation time of 50 ms6, 26. The DDA run consisted of one 250 ms MS1 TOF survey scan covering 400–1300 Da followed by ten data dependent 100 ms MS/MS scans (1 Da isolation window, scan range 100–2000 Da) with precursors excluded for 15 s after being selected for fragmentation once (dynamic exclusion option). The SWATH run consisted of one 250 ms or 50 ms MS1 TOF survey scan followed by 34 sequential MS2 windows of 25 Da covering a mass range of 400–1250 Da at 95 ms per each SWATH scan. The DIA run (Thermo Q Exactive Plus) consisted of one MS survey scan (17500 resolution, target 3e6, max fill time 50 ms) every 10 scans, and 24 sequential MS2 windows of 26 amu (17500 resolution, target 5e5, max fill time 80 ms) covering a mass range from 400–1000 Da. The DDA run (Thermo QE Plus) consisted of one MS survey scan (70000 resolution, target 1e6, max fill time 30 ms) followed by fifteen MS/MS scans (2 Da isolation, 17500 resolution, target 1e5, max fill time 125 ms), with former precursors excluded for 20 s after being selected once.
mzXML File conversion
The .wiff raw files from AB SCIEX 5600 TripleTOF were converted to mzML format with the AB MS Data Converter (AB SCIEX version 1.3 beta) using “centroid” option, and the resulting mzML files were further converted into mzXML format by msconvert.exe from the ProteoWizard package (version 3.0.4462)41 using the default parameters. The .raw files from Thermo Q Exactive Plus were directly converted to mzXML files by msconvert.exe.
Precursor and fragment ion 2D peak detection in DIA-Umpire
A two-dimensional feature detection algorithm was developed to locate precursor and fragment ion signals in MS1 and MS2 data (Fig. 2). Feature detection analysis starts with the LC elution profile (“peak curve”) detection step. A peak curve represents a mass trace continuous in time, and a peak must be present in at least three consecutive scans (for data presented in this study, >9 second on average). It is stored as three vectors of m/z values MZ = (m1, m2, …, mn), intensities INTraw = (i1, i2, … in), and retention times RTraw = (t1, t2, … tn), where n is the number of consecutive scans and ti+1>ti. For detected features the algorithm reports m/z value, retention time span (elution start and end times, t1 and tn) and extracted ion chromatograms (XICs). The m/z value M of a peak curve is calculated as a weighted average (by intensity) of detected m/z values in the retention time span,
Each peak curve is then smoothed by B-spline interpolation (using the 2nd degree basis function). XICs are represented as two vectors of interpolated retention times RT = (t1, t2, … tk) and intensities INT = (i1, i2, … ik), where k is the total number of interpolated points per peak (we used 150 points per minute, making k = 150(tn-t1)). As a peak curve might have multiple maxima, we apply a Continuous Wavelet Transform (CWT)-based approach42 for splitting it into several separate peak curves using Mexican-hat wavelet. For each unimodal peak curve, the apex intensity is determined as Imax = max(INT).
In MS1 data generated using high-resolution instruments, several isotope peaks for each peptide precursor ion can usually be detected (referred to as precursor ion features; see Fig. 2 and Supplementary Fig. 24), helping to distinguish true precursor signals from noise. Single peak curves detected in MS1 scans are grouped together to form isotopic clusters based on RT apex distance and m/z spacing, which should fit the spacing for a given charge state (in this work, +2, +3, and +4 only). In complex samples, however, the presence of multiple co-eluting peptides having similar m/z values results in overlapping signals, leading to multiple alternative possibilities for isotope peak grouping (see Supplementary Fig. 25 for an illustration). In such cases, the algorithm intentionally over predicts the number of precursor ion features by first considering the m/z of each peak curve as a possible monoisotope, and then attempting to find heavier isotope peaks for that presumed monoisotopic m/z value. In doing so, the algorithm maximizes the sensitivity with respect to finding true precursor ion features at the cost of introducing some redundant features with incorrectly assigned monoisotopic m/z values.
In general, the higher the number of isotope peaks detected for an MS1 feature, the more likely it is to be a true precursor ion signal. Thus, the algorithm uses the number of isotope peaks as a measure of quality of precursor ion features. Features with three or more isotope peaks are labeled as Quality Tier 1 (QT = 1 or Q1) precursors, i.e. the precursors that are most likely to represent true precursor peptides with the correctly determined monoisotopic m/z values. MS1 features with only two detected isotope peaks are labeled as Quality Tier 2 (QT = 2 or Q2). All single peaks observed in MS1 scans (i.e. peaks with no isotopic envelope detected) are discarded.
In addition to detection of precursor ion features in MS1 scans, unfragmented precursor ions can sometimes be observed in DIA MS2 spectra. This is likely due to the collision energy not being universally suitable for complete fragmentation of all the precursor ions within a particular DIA isolation window. To take advantage of this, all peaks in MS2 spectra having m/z values within the corresponding DIA isolation window are considered as potential unfragmented precursors (see Fig. 2). Unfragmented precursor ion features are detected as described above for MS1 data, requiring at least two isotope peaks. These features are added to the precursor list as Quality Tier 3 (QT = 3 or Q3). Note that some peptide precursor ions can be detected in both DIA MS1 and MS2 spectra, and their corresponding features thus may be included in both Quality Tier 3 and Quality Tier 1 (or 2) sets.
Fragment ion peak detection in MS2 data is performed similarly, with one modification. It is generally more difficult to detect multiple isotopic peaks for low intensity fragment ions. Relaxed stringency of feature detection for fragment ions (compared to MS1 precursor ions feature detection described above) resulted in improved sensitivity of peptide identification and reduced the computational time (data not shown). Thus, isotope peak grouping and charge state determination for fragment ions is not performed at this stage. Instead, each possible fragment peak is treated independently, and isotope detection and charge state determination is performed at a later stage (after the precursor–fragment grouping step described below).
Precursor-fragment grouping in DIA-Umpire
“Co-elution” is an important characteristic of the data that reveals relationships between a precursor ion and its fragments9. The algorithm takes advantage of this characteristic by calculating the Pearson correlation coefficient and the retention time difference of LC elution peak apexes between all detected precursors (P) and all possible fragment ions (F) (see Fig. 2). This pairing is naturally restricted to fragment ions in the DIA isolation window corresponding to the m/z value of the precursor. For a precursor Pq and a fragment Fr the Pearson correlation coefficient Cq,r = corr(Pq, Fr) is computed using the LC profiles (XICs) of monoisotopic precursor and fragment ion features. All precursor–fragment pairs are represented as a bipartite graph (see Fig. 2). In this representation, one fragment ion can have multiple precursors and several precursors can share the same fragment.
To better connect precursor ions to their most likely fragment ions, the following parameters are calculated based on the correlation scores for each possible Pq, Fr pair. First, given a fragment ion Fr, RP(Pq, Fr) score is calculated as the rank of the precursor ion Pq based on Pearson correlation Cq,r between that fragment and all candidate precursors. Second, given a precursor ion Pq, RF(Pq, Fr) score is calculated as the rank of the fragment Fr based on Pearson correlation between that precursor and all possible fragments. For a precursor ion with many co-eluting fragments, a higher-ranking fragment is more likely to be derived from it. Similarly, for a fragment ion, a higher-ranking precursor ion is more likely to be its true precursor. These two metrics, as well as the retention time difference of LC profile apexes, ΔT(Pq, Fr), are used to assemble precursor–fragment groups (see Fig. 2).
Generation of pseudo MS/MS spectra using DIA-Umpire
To generate a pseudo MS/MS spectrum for a precursor ion Pq, the algorithm first detects the charge state of each fragment peak (if only a single isotopic peak is detected, charge state +1 is assumed). It then detects all likely complementary y- and b-ions in the spectrum (detected as pairs of fragments summing up to the precursor peptide mass 43). For non-complementary ion peaks, only those fragments Fr are kept that pass the following set of thresholds: RF(Pq, Fr) ≤ RFmax, RP(Pq, Fr) ≤ RPmax, and ΔT(Pq, Fr) ≤ ΔTmax. These threshold parameters are implemented as user-specified options in the software, allowing re-evaluation and adjustment of the default thresholds (described below), if necessary.
Charge state and precursor m/z for each pseudo MS/MS spectrum are determined by precursor ion features. Fragment ion intensities are computed in three steps. For fragment Fr, the intensity is taken as LC apex intensity of the corresponding elution peak curve, Ir. Then for each complementary b-, y- fragment pair Fr1, Fr2, the intensity of the less intense fragment is boosted to match that of the more intense one, Ir1 = Ir2 = max(Ir1, Ir2). At the last step, intensities are adjusted by weighting according to the square of correlation with the precursor peak curve, Ir′ = Ir×C2q,r. The presence of complementary ions is a positive sign of a connection between the precursor and fragment ions, and boosting the intensities of complementary ions has been shown to improve the sensitivity of peptide identification44. Note that this fragment intensity adjustment step can optionally be skipped for other applications, e.g. to use a spectral library search engine for searching pseudo MS/MS spectra or to build a spectral library from the pseudo MS/MS spectra. Also note that the adjusted (boosted) intensities are not used for quantitation, only for identification. An example of a pseudo MS/MS spectrum (before and after complementary ion boosting), the underlying precursor ion and fragment ion elution profiles in DIA MS1 and MS2 data, and the DDA MS/MS spectrum for the same peptide are shown in Supplementary Fig. 1.
The performance of the DIA-Umpire algorithm for different combinations of the threshold parameters described above was evaluated using a subset of the data (Supplementary Table 10). When the pseudo MS/MS spectra extracted under different settings were searched using X! Tandem, the following threshold values resulted in the highest number of identifications (at 1% FDR) and were selected as default values in the software: allow the top 25 ranked precursors for each fragment (RPmax= 25), the top 300 ranked fragments for each precursor (RFmax = 300) and 0.6 minutes apex elution time difference (ΔTmax = 0.6). Note that the best performance was achieved by allowing the possibility of an MS2 fragment to be included in multiple MS/MS spectra (RPmax= 25). Because the algorithm takes the square of a peak shape correlation coefficient between the precursor and fragment signals as the weighting factors for calculation of adjusted fragment intensities in pseudo MS/MS spectra, true high intensity fragments can still contribute to the identification of their corresponding peptide even if they have a relatively poor correlation with the precursor (e.g., due to ion suppression effects affecting either the precursor ion or the fragment ion elution peak shape). The overall robustness of the pseudo MS/MS spectrum generation process was also evident from similar numbers of peptide ion identifications obtained by searching the spectra with three different database search engines (X! Tandem, Comet, and MSGF+, Supplementary Table 1). These results indicate that inclusion of more fragment ions in a pseudo MS/MS spectrum does not hamper the identification rate. On the contrary, by doing so the algorithm increases the chance of true fragments to be included, thus improving the number of confident identifications. An additional analysis was also carried out for E. coli and human cell lysate datasets by removing fragments from pseudo MS/MS spectra that were also matched in other pseudo MS/MS spectra identified with high confidence. Repeating X! Tandem search with those fragments removed did not change the number of identified peptide ions in either dataset.
Peptide and protein identification using pseudo MS/MS spectra
In this study, we used X! Tandem20, Comet21, and MSGF+22 as search engines to identify peptides from pseudo MS/MS spectra (however, any database search engine developed for searching DDA spectra can be used). Because of the similar characteristics of DDA and DIA pseudo MS/MS spectra, all downstream analysis of the database search results, including protein inference and estimation of posterior probabilities of correct identification and FDR, can also be performed using conventional strategies developed for DDA data. Database search output files were processed by PeptideProphet23 via the Trans-Proteomic Pipeline (TPP)45, followed by ProteinProphet25 analysis to assemble peptides into proteins/protein groups and to determine protein probabilities. The final protein and peptide identification lists were filtered to achieve a desired FDR (here – 1%) estimated using the target-decoy approach2. The only modification was to compute posterior peptide probabilities by PeptideProphet separately for each of the three quality categories of MS/MS spectra (Quality Tiers QT = 1, 2 or 3) because of very different ratios of correct vs. incorrect identifications among them. Further analysis of the model parameters and the distributions of scores reported by PeptideProphet (Supplementary Fig. 26) did not show any evidence indicating that pseudo MS/MS spectra extracted using DIA-Umpire behaved any different than conventional DDA spectra with respect to the basic assumptions in PeptideProphet or the target-decoy FDR estimation strategy.
Targeted extraction in DIA-Umpire using spectral libraries
The targeted extraction module of DIA-Umpire (peptide-centric matching) was developed as an optional second step in the DIA-Umpire workflow to increase the quantification coverage across multiple samples. Given a set of peptides identified by the initial spectrum-centric untargeted database search, the algorithm builds an internal consensus spectral library using confident identifications from all DIA runs. In addition, DIA runs are aligned (in retention time) using commonly identified peptides between the DIA runs as pivot points for non-linear regression46. For a peptide ion not identified in a particular DIA run in untargeted way, DIA-Umpire calculates its retention time (via retention time alignment) and m/z (via mass calibration model, described below), and performs targeted data re-extraction. This is achieved by matching the library spectrum of that peptide ion against precursor–fragment groups previously extracted from the experimental data within a narrow retention time window (in this work, ±1 minute of the calculated retention time) and a narrow precursor mass window (±30 ppm of the calibrated precursor mass). The details of this targeted data extraction algorithm are described below.
1. Spectral library generation
A consensus spectral library is built using confident identifications (here, 1% FDR at the peptide level) from the initial untargeted analysis of the DIA data. First, for each confident pseudo MS/MS spectrum match, the matched fragment intensities are normalized to the most intense matched fragment. For a peptide ion which has multiple spectra identified across samples, the intensity of a fragment in the consensus spectrum is computed as the average fragment intensity across all corresponding identified spectra. Decoy spectra are created by the “shuffle-and-reposition” method47, and such a decoy is generated for each peptide ion in a consensus spectral library.
2. Retention time prediction and mass calibration for target peptide ions
DIA-Umpire adopts a previously described46 nonlinear regression-based method for retention time calculation and a mass calibration model48 for adjusting precursor m/z values of a peptide ion in a DIA run. To generate the retention time model for a pair of DIA-runs, retention times of commonly identified peptide ions from the initial spectrum-centric search are used and a non-linear regression model is built based on these retention times. For mass calibration, mass errors are represented as a function of the retention time, and a non-linear LOWESS regression is done for calculation of peptide ion mass errors given the peptide retention time in a DIA run.
3. Peptide-centric matching
To find the best matching precursor–fragment group for a peptide ion from a spectral library, all precursor–fragment groups within the range of ± 30 ppm (user-defined parameter) of the calculated precursor m/z and ± 1 minute of calculated retention time are considered as candidates. A library spectrum S is denoted as
where NS is the number of fragment peaks in the spectrum, and IrS and MrS are the intensity and the theoretical m/z value, respectively, of each fragment Fr that belongs to spectrum S. A precursor-fragment group G is represented as
where NG is the number of fragment peaks, IrG and MrG are the intensity and m/z value, respectively, of each fragment Fr that belongs to precursor-fragment group G, and CrG is the Pearson correlation coefficient between the fragment Fr and the precursor anchoring group G. Given a library spectrum S and a precursor–fragment group G, matched fragment peaks from the precursor–fragment group are extracted using a predefined mass tolerance (e.g. ± 40 ppm for AB SCIEX 5600 instrument). The algorithm then calculates five sub-scores for the match between S and G. In addition to the number of matched fragments (L), it calculates a spectral similarity score as follows. Consider an intensity vector INTG-S = (I1G, I2G, …, INSG) of length NS, with IrG taken as the intensity of the fragment peak Fr in G that matches to a fragment in S, and as zero if no fragment peak can be found in G within the specified mass tolerance window around MrS. The spectral similarity is then calculated by Pearson correlation between the vector INTG-S and the library spectrum intensity vector (I1S, I2S, …, INSS).
Three more scores, Mass Error Score (MES), Intensity Score (IS), and Correlation Score (CS), are calculated using matched fragments Fj only as follows:
The final match score (U-score) between S and G is calculated as a linear combination of these five sub-scores (Supplementary Figure 19a), with the score weights determined using the linear discriminant analysis (LDA)28. For LDA model training, 50% of all matches in which S is a decoy spectrum are randomly selected and labeled as negative training data (the other 50% are held away from the training; these can be used at the final stage to assess the quality of the model fitted using the mixture modeling algorithm described below). Positively labeled training dataset is composed of likely true matches, i.e. matches between S and G that were identified with high scores at the initial untargeted (spectrum-centric search) stage.
4. Peptide-centric match probability and FDR
The U-score is computed for all targets considered in a particular DIA run. Targets are defined here as peptide ions represented in the spectral library (created, as described above, from all DIA runs in the experiment) that were not identified in that particular DIA run by the untargeted (spectrum-centric) search. The U-score distribution for these target matches computed as described above is assumed to be a bimodal distribution representing populations of correct and false matches (Supplementary Fig. 19c). This distribution is modeled as a mixture of two normal distributions and is de-convoluted using the expectation maximization (EM) algorithm23. The probability that a match is correct, given the U-score U, is determined as
Here f1(U) and f0(U) are Gaussian density functions (from the mixture model above) for correct and false matches, respectively. The parameters of the distributions and their mixing weights, π0 and π1, are determined directly from the data using the EM. FDR can then be estimated using computed probabilities2, 23. In this study, a probability threshold of 0.99 (estimated FDR of less than 1% in these data) was applied as the final filter.
Quantification in DIA data using DIA-Umpire
The quantification module of DIA-Umpire computes peptide and protein intensities estimated either from MS1 precursor ion intensities or from MS2 fragment ion intensities. We use the LC apex intensity of the smoothed MS1 monoisotopic peak to determine the MS1 precursor ion intensity. For MS2 fragment ion intensity, we use the raw LC apex intensity of the fragment signal. The MS2 fragment-based intensity for a protein can be computed by summing the intensities of all matched fragments of all identified peptide ions from that protein (or only using selected peptide ions and fragments, as described below). In rare cases, the same peptide ion is identified from multiple precursor ion features (i.e. at different retention times). Such peptides are excluded by default (optionally, such peptide ions can be used for quantification by selecting the precursor ion feature with the highest MS1 intensity). When computing protein-level intensities, the analysis can be based on all peptides or based only on peptides unique to a particular protein group (e.g. with ProteinProphet computed group weight above 0.9; default option).
For fragment-based quantification, DIA-Umpire computes two protein intensity measures. The MS2 iBAQ intensity is computed for each protein by summing the intensities of all matched fragments from all identified peptide ions divided by the number of expected tryptic peptides (similar to the iBAQ score30 commonly used for DDA MS1 intensity data). This intensity measure can be computed for all proteins identified in the dataset. In addition, DIA-Umpire also computes a protein intensity measure using selected fragments and peptide ions consistently identified across multiple samples within the whole dataset, as described below.
1. Fragment selection
For a peptide ion which is identified in Npep DIA runs within the experiment, only fragments detected in more than MinFreq × Npep DIA runs are kept. For each remaining fragment Fr, the fragment quality score
is calculated using the Pearson correlation Crj between fragment Fr and its precursor peak in DIA run j, and the apex intensity of a fragment Fr in DIA run j, Irj. For a peptide ion, its top TF best (i.e. with highest QF scores) fragments (e.g. TF =6, denoted as Top6fra option) are selected for quantification. Peptide ion intensity in a DIA run is then determined by summing the intensities of all selected fragments.
2. Peptide ion selection
To select peptide ions for protein quantification, only peptide ions identified in more than MinFreq × Nprot runs are kept, where Nprot is the number of DIA runs in which the protein was identified. The peptide ion intensity in each DIA run is computed using the intensities of its fragments selected as described above (e.g. Top6fra option). The peptide ion quality score is then computed as a sum of peptide ion intensities across all runs in the dataset. The protein intensity in then calculated in each DIA run by summing the intensities of the top TP highest quality peptide ions (e.g. TP =6, denoted asTop6pep option).
The thresholds described above were implemented as input parameter options. The following parameters were selected in this work based on the analysis of variability between two replicate human cell lysate runs: TP = 6, TF = 6, and MinFreq = 0.5 (denoted as “Top6pep/Top6fra Freq>0.5”; Supplementary Fig. 16). Note that this selection procedure may lead to a loss of a small number of identified proteins that cannot be quantified due to lack of reproducible peptide ions passing the filters described above. Out of 1,653 proteins identified in both replicates of the human cell lysate data (Supplementary Table 3), only 12 were not quantified by the “Top6pep/Top6fra Freq > 0.5” approach.
DIA-Umpire also reports two MS1-based quantification scores. The MS1 iBAQ protein intensity is computed as previously described30, with peptide ion intensities determined at the apex of the LC elution monoisotopic peak. Note that MS1-based peptide quantification is only available for peptide ions identified from QT = 1 or 2 category pseudo MS/MS spectra (no MS1 feature is detected for QT = 3 spectra). In the human cell lysate data, 1,324 out of 1,353 proteins were quantified by MS1 iBAQ in both replicates (Supplementary Table 3; Supplementary Fig. 15). In addition, DIA-Umpire reports a second MS1 quantification score computed as a sum of intensities of top TP peptide ions, selecting peptide ions for quantification in a similar manner as described above for MS2-based quantification (e.g. top 6 most intense peptide ions, with an additional MinFreq = 0.5 filter: “Top6pep, Freq > 0.5” option; Supplementary Fig. 15).
To demonstrate the importance of selecting the most reliable peptide ions and fragments across all samples for quantification, we also implemented a selection procedure applied independently within each DIA run (“MS1 Top3pep (indep. selection)” in Supplementary Fig. 15; “MS2 Top3pep/Top2fra (indep. selection)” in Supplementary Fig. 16), which produced significantly worse results.
Peptide and protein identification parameters
For UPS2, E. coli, and human cell lysate datasets, DDA MS/MS spectra and the DIA pseudo MS/MS spectra were searched by X! Tandem, Comet, and MSGF+ using the following parameters: allow tryptic peptides only, up to one missed cleavage, oxidation of methionine and cysteine alkylation as variable modifications. The glycoproteomics SWATH dataset was searched by X! Tandem only, with cysteine alkylation specified as a fixed modification and with deamidation of asparagine as a variable modification. The instrument-specific parameters – the precursor ion mass tolerance and the fragment ion mass tolerance – were set to 30 ppm and 40 ppm for AB SCIEX 5600 TripleTOF, and 10 ppm and 20 ppm for Thermo Q Exactive Plus, respectively. In X! Tandem, the analysis was limited to 140 most intense peaks which gave the best results based on the same subset of the data that was used to select the parameters for the DIA-Umpire pseudo MS/MS extraction algorithm (see above). However, the search results were not very sensitive to the choice of this parameter (which is also evident from the fact that similar results were obtained using Comet and MSGF+ search tools that do not provide an option to restrict the number of peaks in the spectra). The sequence database for the UPS2 experiment was compiled from the UPS sequences (total 50 sequences: 48 UPS1 proteins and 48 UPS2 proteins, www.sigmaaldrich.com). For the E. coli experiments, E. coli proteome sequences (4,431 proteins) were extracted from UniProtKB. The non-redundant human protein sequence FASTA file from the UniProt/SwissProt database (release of 09-Jan-2013), appended with common contaminant proteins, was used for the human cell lysate experiment, AP-SWATH interactome, and the glycoproteomics datasets. For all sequence databases, reversed sequences were added as decoys for target–decoy analysis. The initial search results from the search engines were first converted into pepXML format, followed by analysis using PeptideProphet23 via the Trans-Proteomic Pipeline (TPP)45 (v4.7). For DIA derived pseudo MS/MS spectra, PeptideProphet was run separately for each of the three quality categories of MS/MS spectra (Quality Tiers QT = 1, 2 or 3). The iProphet24 tool was used when merging the search results from all three search engines. Unless noted otherwise, peptide ion identification lists for each DDA or DIA run were filtered at 1% FDR, estimated by target–decoy approach based on PeptideProphet probability for each search engine (or iProphet peptide ion probability when using iProphet).
Protein inference for different analyses was performed as follows. To report the numbers of protein identification for individual DIA/DDA runs (Fig. 3 and Supplementary Table 1), PeptideProphet output files (individual search engine analysis) or iProphet output files (when combining the search results) were analyzed by ProteinProphet25 for protein inference. For the comparison between DDA and DIA (Fig. 4, Supplementary Figs 3–5, Supplementary Tables 2–4) or between DIA-Umpire and OpenSWATH (Supplementary Figs 6, 7) results at the protein level, PeptideProphet output files (based on X! Tandem results) for both DIA and DDA were processed together by ProteinProphet. For the AP-SWATH dataset, ProteinProphet analysis was done by taking all PeptideProphet output files (X! Tandem results) from all SWATH runs, i.e. EIF4A2 and MEPCE bait data (biological triplicates for each bait) and the three GFP negative controls. The final protein lists for each ProteinProphet analysis were determined using an 1% FDR threshold, estimated by target–decoy approach.
Quantification in DDA data
We used elution apex intensity of the MS1 precursor feature when performing peptide quantification for DDA MS1 data. For each MS/MS spectrum identified in a DDA experiment, all precursor features observed in the MS1 data with close monoisotopic m/z (same precursor m/z tolerance as used in the database search), close retention time (within ±1 minute), and same charge state were considered as candidates. Among these candidates, the MS1 feature with the closest retention time was considered as the precursor ion for the identified MS/MS spectrum. As with DIA MS1 data in DIA-Umpire, peptide ion intensity and its retention time in DDA MS1 were determined from the intensity and the retention time at the LC apex of the monoisotopic peak.
Comparison of ion intensities between DDA and DIA
To compare the fragment ions observed for the same peptide in DDA and in DIA experiments, the compomics-utilities library49 was used to generate theoretical peptide fragments. To find the signal of a peptide ion which was only identified in either DDA or DIA data, the retention time observed for that ion in the run where it was identified was used to detect the corresponding peptide ion feature in the other run. This was done without the need for retention time alignment between the runs because of the excellent retention time and ion intensity reproducibility between DDA and DIA runs on the same samples (see Supplementary Fig. 27 and 28). An MS1 precursor feature m/z window of ±30 ppm and a retention time window of ±1 minute were used. For DDA data, the highest intensity candidate was selected among multiple possible ones. For DIA data, the best candidate (precursor–fragment group) was selected based on the number of matched fragments between the corresponding pseudo MS/MS spectrum and the DDA identified peptide sequence. The number of matched fragments was calculated as follows: for each DDA MS/MS spectrum, or pseudo MS/MS spectrum in DIA, only the top 140 highest intensity peaks were considered. The mass tolerance for peak matching was set to 40 ppm for AB SCIEX 5600 TripleTOF and 10 ppm for Thermo Q Exactive Plus. The analysis was restricted to b- and y-fragment ions only. A peak in an experimental spectrum was allowed to be matched to only one theoretical fragment. The number of matched fragments for each spectrum was counted and then normalized by the total number of theoretical fragments for that peptide.
Targeted extraction analysis using OpenSWATH
The E. coli and human cell lysate experiments from AB SCIEX 5600 TripleTOF were also processed with OpenSWATH to identify proteins and peptides using the fully targeted approach. The two DDA replicates acquired for each sample were used to build the spectral library using SpectraST27 with the following options: best replicate; union; 0 minimum peaks for exclusion; 0 minimum amino acids for exclusion. Only the DDA non-decoy identification spectra that passed 1% FDR threshold were used for building the library. The probability thresholds were: 0.6979 for DDA E. coli replicate 1; 0.7877 for DDA E. coli replicate 2; 0.8075 for DDA human replicate 1; 0.8233 for DDA human replicate 2. This resulted in a total of 12,820 and 17,402 peptide ions including decoys represented in the “transition lists” used by OpenSWATH for E. coli and human, respectively. For OpenSWATH analysis using DIA-derived libraries, the libraries were built with SpectraST using the pseudo MS/MS spectra (without complementary b- and y-ion boosting) from peptide ions identified by the DIA-Umpire's untargeted workflow and filtered at 1% FDR (8,757 peptide ions for human and 6,364 for E. coli samples).
OpenSWATH was run using the following parameters: extraction elution time window (seconds): 60; minimum transitions: 2; maximum transitions: 6; unique ion signature threshold: -1; retention time normalization factor (seconds): 7200 (i.e. the whole LC-MS run duration in our case). Our dataset did not contain iRT50 peptides for retention time normalization because all the experiments were performed using the same instrumentation setup and the retention times were highly reproducible (within one minute) between the DDA/DIA runs (Supplementary Fig. 29). Peptide ion identification lists were filtered using mProphet28 at 1% FDR. The number of candidate peptide ions used for scoring against the extracted peak groups in OpenSWATH analysis was estimated as the number of ions in the DDA-derived library falling within the corresponding 25 Da SWATH isolation window and within the specified retention time tolerance (1 minute).
DIA-Umpire analysis using reduced database
In order to demonstrate how search space affects peptide identification, in addition to searching DIA pseudo MS/MS spectra against the proteome-wide sequence database (all E. coli or human proteome sequences plus decoys), we also used a smaller database of peptide sequences (5,997 and 8,784 sequences for E. coli and human cell lysate experiments, respectively) identified from the corresponding DDA data. Reverse versions of these sequences were also appended to the database for target-decoy analysis. All other search parameters and settings were the same as described above.
Isotopic pattern validation of glycopeptide identifications
Identification of N-linked glycopeptides relies on detection of asparagine deamidation due to PNGase F treatment which causes a small mass shift (0.984 Da). The mass shift is close to the mass difference between the isotopic peaks which could lead to false identification of a peptide as deamidated if an “M+1” isotopic peak is mis-recognized as a true monoisotopic peak. In another scenario, if there is a noise signal at “M-1” Da of a deamidated ion that is mis-recognized as the monoisotopic peak, the deamidated peptide would be mis-identified as an unmodified peptide ion. To remove these erroneous identifications, we applied a two-step filtering strategy. All confident identifications from DIA-Umpire were first grouped if their precursor features shared an isotopic peak at same retention time (see Supplementary Fig. 25 for one such example). We then removed grouped precursor features if the observed MS1 isotopic peak distribution did not fit the theoretical isotopic pattern (chi-squared goodness of fit probability < 0.8). This first stage filtering was able to remove misidentifications in the second scenario. To remove the cases in the first scenario, the precursor masses of peptides identified in each group were compared, and only the identification with the smallest mass in the group was kept.
SAINT interaction scoring for AP-SWATH interactome dataset
AP-SWATH interactome dataset was processed using the entire DIA-Umpire pipeline including feature detection, untargeted identification, targeted re-extraction, peptide ion and fragment selection, and protein quantification (Figure 5a). Protein and peptide identifications were filtered at 1% and 5% FDRs, respectively. Missing identification across replicates and samples were re-extracted by the peptide-centric matching with a 0.99 probability threshold as the filter. For protein quantitation, we used the “Top6pep/Top6fra, Freq > 0.5” approach to determine protein intensity using only peptides unique to a particular protein group (ProteinProphet computed group weights above 0.9). Protein quantification data from EIF4A2 and MEPCE bait experiments with GFP negative controls were analyzed using SAINT (intensity model; v2.3.4)35 to determine high confidence protein-protein interactions (here, SAINT probability above 0.95).
Code and data availability
The program was developed in the cross-platform Java language and is available at http://diaumpire.sourceforge.net/. All mass spectrometry files (Supplementary Table 11) along with DIA-Umpire results presented in this paper have been deposited to the ProteomeXchange Consortium40 (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD001587.
Supplementary Material
Acknowledgments
We thank Brendan MacLean for help with Skyline, Hannes Röst and Ben Collins for help with OpenSWATH, and Stephen Tate and for useful discussions. We also thank Steven Danielson at Thermo Scientific for access to the Q Exactive Plus and Zhen-Yuan Lin for the acquisition of the DIA samples for MEPCE, EIF4A2 and GFP. This work was supported by the US National Institutes of Health grants 5R01GM94231 (to A.I.N and A.-C.G), R01GM107148 and U24DK097153 (to A.I.N.), the Canadian Institutes of Health Research (to A.-C.G.; MOP-84314), a Genome Canada Bioinformatics and Computational Biology grant (to A.-C.G and A.I.N) and Singapore Ministry of Education grant (to H.C.; R-608-000-088-112).
Abbreviations
- LC
Liquid chromatography
- MS
Mass spectrometry
- DDA
Data Dependent Acquisition
- DIA
Data Independent Acquisition
- SWATH
Sequential Window Acquisition of all THeoretical Mass Spectra
- FDR
False Discovery Rate
- AP
Affinity Purification
- SAINT
Significance Analysis of INTeractome
- XIC
Extracted Ion Chromatogram
- UPS
Universal Protein Standard
Footnotes
Editorial summary: The computational workflow of DIA-Umpire allows untargeted peptide identification directly from DIA data without the dependence on a spectral library for the data extraction.
Author Contributions: A.I.N. and C.-C.T. conceived the project and developed the algorithm; C.-C.T. implemented the software; D.A. assisted with the OpenSWATH analysis and contributed to the algorithm and software development; B.L. and M.T. acquired mass spectrometry data; H.C. assisted with SAINT scoring and contributed to the development of protein quantification strategies, A.-C.G, C.-C.T., B.L., and A.I.N designed experiments and analyzed data; A.I.N., A.-C.G. supervised the project; C.-C.T., A.I.N. and A.-C.G. wrote the manuscript with input from B.L. and D.A.
Competing Financial Interests: The authors declare no competing financial interests.
References
- 1.Bantscheff M, Lemeer S, Savitski MM, Kuster B. Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal Bioanal Chem. 2012;404:939–965. doi: 10.1007/s00216-012-6203-4. [DOI] [PubMed] [Google Scholar]
- 2.Nesvizhskii AI. A survey of computational methods and error rate estimation procedures for peptide and protein identification in shotgun proteomics. Journal of Proteomics. 2010;73:2092–2123. doi: 10.1016/j.jprot.2010.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bailey DJ, McDevitt MT, Westphall MS, Pagliarini DJ, Coon JJ. Intelligent data acquisition blends targeted and discovery methods. J Proteome Res. 2014;13:2152–2161. doi: 10.1021/pr401278j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weisbrod CR, Eng JK, Hoopmann MR, Baker T, Bruce JE. Accurate Peptide Fragment Mass Analysis: Multiplexed Peptide Identification and Quantification. J Proteome Res. 2012;11:1621–1632. doi: 10.1021/pr2008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Michalski A, Cox J, Mann M. More than 100,000 Detectable Peptide Species Elute in Single Shotgun Proteomics Runs but the Majority is Inaccessible to Data-Dependent LC-MS/MS. J Proteome Res. 2011;10:1785–1793. doi: 10.1021/pr101060v. [DOI] [PubMed] [Google Scholar]
- 6.Gillet LC, et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol Cell Proteomics. 2012;11:O111 016717. doi: 10.1074/mcp.O111.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tate S, Larsen B, Bonner R, Gingras AC. Label-free quantitative proteomics trends for protein-protein interactions. Journal of proteomics. 2013;81:91–101. doi: 10.1016/j.jprot.2012.10.027. [DOI] [PubMed] [Google Scholar]
- 8.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 9.Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics. 2006;5:144–156. doi: 10.1074/mcp.M500230-MCP200. [DOI] [PubMed] [Google Scholar]
- 10.Panchaud A, et al. Precursor acquisition independent from ion count: how to dive deeper into the proteomics ocean. Anal Chem. 2009;81:6481–6488. doi: 10.1021/ac900888s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiger T, Cox J, Mann M. Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol Cell Proteomics. 2010;9:2252–2261. doi: 10.1074/mcp.M110.001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Egertson JD, et al. Multiplexed MS/MS for improved data-independent acquisition. Nat Methods. 2013;10:744–746. doi: 10.1038/nmeth.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Distler U, et al. Drift time-specific collision energies enable deep-coverage data-independent acquisition proteomics. Nat Methods. 2014;11:167–170. doi: 10.1038/nmeth.2767. [DOI] [PubMed] [Google Scholar]
- 14.Purvine S, Eppel JT, Yi EC, Goodlett DR. Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics. 2003;3:847–850. doi: 10.1002/pmic.200300362. [DOI] [PubMed] [Google Scholar]
- 15.Colangelo CM, Chung L, Bruce C, Cheung KH. Review of software tools for design and analysis of large scale MRM proteomic datasets. Methods. 2013;61:287–298. doi: 10.1016/j.ymeth.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rost HL, et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat Biotech. 2014;32:219–223. doi: 10.1038/nbt.2841. [DOI] [PubMed] [Google Scholar]
- 17.Rosenberger G, et al. A repository of assays to quantify 10,000 human proteins by SWATH-MS. Scientific Data. 2014;1:140031. doi: 10.1038/sdata.2014.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li GZ, et al. Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics. 2009;9:1696–1719. doi: 10.1002/pmic.200800564. [DOI] [PubMed] [Google Scholar]
- 19.Pak H, et al. Clustering and filtering tandem mass spectra acquired in data-independent mode. J Am Soc Mass Spectrom. 2013;24:1862–1871. doi: 10.1007/s13361-013-0720-z. [DOI] [PubMed] [Google Scholar]
- 20.Craig R, Cortens JP, Beavis RC. Open source system for analyzing, validating, and storing protein identification data. J Proteome Res. 2004;3:1234–1242. doi: 10.1021/pr049882h. [DOI] [PubMed] [Google Scholar]
- 21.Eng JK, Jahan TA, Hoopmann MR. Comet: an open-source MS/MS sequence database search tool. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, et al. The generating function of CID, ETD, and CID/ETD pairs of tandem mass spectra: applications to database search. Mol Cell Proteomics. 2010;9:2840–2852. doi: 10.1074/mcp.M110.003731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 24.Shteynberg D, et al. iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol Cell Proteomics. 2011;10:M111 007690. doi: 10.1074/mcp.M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nesvizhskii AI, Keller A, Kolker E, Aebersold R. A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- 26.Lambert JP, et al. Mapping differential interactomes by affinity purification coupled with data-independent mass spectrometry acquisition. Nat Methods. 2013;10:1239–1245. doi: 10.1038/nmeth.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam H, et al. Building consensus spectral libraries for peptide identification in proteomics. Nat Methods. 2008;5:873–875. doi: 10.1038/nmeth.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reiter L, et al. mProphet: automated data processing and statistical validation for large-scale SRM experiments. Nat Methods. 2011;8:430–435. doi: 10.1038/nmeth.1584. [DOI] [PubMed] [Google Scholar]
- 29.Liu Y, et al. Glycoproteomic analysis of prostate cancer tissues by SWATH mass spectrometry discovers N-acylethanolamine acid amidase and protein tyrosine kinase 7 as signatures for tumor aggressiveness. Mol Cell Proteomics. 2014;13:1753–1768. doi: 10.1074/mcp.M114.038273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwanhausser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 31.Ludwig C, Claassen M, Schmidt A, Aebersold R. Estimation of absolute protein quantities of unlabeled samples by selected reaction monitoring mass spectrometry. Mol Cell Proteomics. 2012;11:M111 013987. doi: 10.1074/mcp.M111.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Collins BC, et al. Quantifying protein interaction dynamics by SWATH mass spectrometry: application to the 14-3-3 system. Nat Methods. 2013;10:1246–1253. doi: 10.1038/nmeth.2703. [DOI] [PubMed] [Google Scholar]
- 33.Nesvizhskii AI. Computational and informatics strategies for identification of specific protein interaction partners in affinity purification mass spectrometry experiments. Proteomics. 2012;12:1639–1655. doi: 10.1002/pmic.201100537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi H, et al. SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods. 2011;8:70–73. doi: 10.1038/nmeth.1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi H, Glatter T, Gstaiger M, Nesvizhskii AI. SAINT-MS1: protein-protein interaction scoring using label-free intensity data in affinity purification-mass spectrometry experiments. J Proteome Res. 2012;11:2619–2624. doi: 10.1021/pr201185r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2013;41:D816–823. doi: 10.1093/nar/gks1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeronimo C, et al. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prakash A, et al. Hybrid data acquisition and processing strategies with increased throughput and selectivity: pSMART analysis for global qualitative and quantitative analysis. J Proteome Res. 2014;13:5415–5430. doi: 10.1021/pr5003017. [DOI] [PubMed] [Google Scholar]
- 39.MacLean B, et al. Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vizcaino JA, et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol. 2014;32:223–226. doi: 10.1038/nbt.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chambers MC, et al. A cross-platform toolkit for mass spectrometry and proteomics. Nature biotechnology. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tautenhahn R, Bottcher C, Neumann S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinformatics. 2008;9:504. doi: 10.1186/1471-2105-9-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nesvizhskii AI, et al. Dynamic spectrum quality assessment and iterative computational analysis of shotgun proteomic data: toward more efficient identification of post-translational modifications, sequence polymorphisms, and novel peptides. Mol Cell Proteomics. 2006;5:652–670. doi: 10.1074/mcp.M500319-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Kryuchkov F, Verano-Braga T, Hansen TA, Sprenger RR, Kjeldsen F. Deconvolution of mixture spectra and increased throughput of peptide identification by utilization of intensified complementary ions formed in tandem mass spectrometry. J Proteome Res. 2013;12:3362–3371. doi: 10.1021/pr400210m. [DOI] [PubMed] [Google Scholar]
- 45.Deutsch EW, et al. A guided tour of the Trans-Proteomic Pipeline. Proteomics. 2010;10:1150–1159. doi: 10.1002/pmic.200900375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsou CC, et al. IDEAL-Q, an automated tool for label-free quantitation analysis using an efficient peptide alignment approach and spectral data validation. Mol Cell Proteomics. 2010;9:131–144. doi: 10.1074/mcp.M900177-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam H, Deutsch EW, Aebersold R. Artificial decoy spectral libraries for false discovery rate estimation in spectral library searching in proteomics. J Proteome Res. 2010;9:605–610. doi: 10.1021/pr900947u. [DOI] [PubMed] [Google Scholar]
- 48.Cox J, Michalski A, Mann M. Software lock mass by two-dimensional minimization of peptide mass errors. J Am Soc Mass Spectrom. 2011;22:1373–1380. doi: 10.1007/s13361-011-0142-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barsnes H, et al. compomics-utilities: an open-source Java library for computational proteomics. BMC Bioinformatics. 2011;12:70. doi: 10.1186/1471-2105-12-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Escher C, et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12:1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.