Abstract
Poly(ADP-ribose) polymerases have been linked to several cellular functions, most of which being mediated through the dynamics of poly(ADP-ribose) (pADPr). In several pathways, pADPr is the effector molecule that regulates cellular signaling and dictates biological outcomes. pAPDr is a central molecule that is capable of promoting both cell survival through the maintenance of genome integrity and cell death that occurs by way of a signal-mediated apoptotic-like process. Thus, interactions with pADPr are extremely important in bringing about the balanced regulation that controls cell fate. Further clues regarding these functions are emerging from a growing list of proteins with which pADPr interacts. Here, we describe the current approaches for investigating noncovalent protein interactions with pADPr.
Keywords: Poly(ADP-ribose), pADPr, PARP, PARG, siRNA, Noncovalent binding, Consensus motif prediction, Immunoprecipitation, Mass spectrometry, Polymer-blot, Electrophoretic mobility shift assay, EMSA, Surface plasmon resonance, SPR
1. Introduction
Most of our understanding of the events associated with poly(ADP-ribose) (pADPr) metabolism is related to the maintenance of genome integrity and most aspects of nuclear metabolism (1). An abundant literature shows that DNA-dependent poly(ADP-ribose) polymerases (PARPs) are crucial sensors and effectors for the maintenance of nuclear architecture, chromatin structure, and DNA repair (2). This statement is in agreement with the increasing evidence that pADPr effects may be through modulation of several pathways involved in genome stability. In the current model, pADPr is considered as a chromatin remodeling and histone modifying molecule that promotes the unwinding and destabilization of histone–DNA interactions. Recent experimental findings also suggest that pADPr is a scaffolding polymer that facilitates efficient repair by recruiting several of the proteins that catalyze single- and double-strand break repair (3-5). Therefore, pADPr is a central component of the molecular cascades that detects and responds to several forms of DNA damage caused by genotoxic stress. Given its wide involvement in pathways affecting genomic stability, pADPr can be conceptualized as a nonproteinaceous tumor-suppressor (6).
Although pADPr metabolism has been mostly studied in the context of genomic integrity through PARP-1 and PARP-2 activation, recent evidence shows that it plays critical roles in the regulation of several other pathways, such as cell cycle progression (7), telomere stability (8), transcriptional control (9), or cell death (10). In the latter case, pADPr itself has been shown to act as an efficient apoptogenic molecule causing caspase-independent cell death (11). It is interesting to note that the cytotoxicity of pADPr is directly related to its complexity and molecular weight. The pADPr involved in PARP-dependent cell death (and likely DNA damage signaling) is characterized by a complex structure configuration mostly composed of long and branched chains of ADP-ribose units. This polymer is also thought to share some structural features with nucleic acids (12). In particular, a helical conformation of pADPr has been postulated based on its physicochemical properties (13, 14). A recent comparative study of the catalytic core structures of PARP family members put forward the concept of pADPr heterogeneity (15). Indeed, a reassessment of the molecular basis for the ADP-ribose transferase activity of PARP-like proteins brought Kleine and colleagues (15) to propose that only PARP-1, PARP-2, and PARP-3 possess a connecting loop that meets the requirements for the catalysis of long and branched pADPr polymers. Following this model, other bona fide PARPs, such as the vault-PARP and the telomeric tankyrase-1 and -2 are predicted to synthesize much shorter and probably unbranched polymers. The biological significance of these variations remains to be fully explored, but it is reasonable to think that it could modulate different cellular pathways. Indeed, tankyrase-1 was identified as the unique poly(ADP-ribosyl)ating enzyme required for spindle organization in mitosis (16), suggesting that structural proteins or mitotic regulators involved in cell cycle progression could be affected by pADPr. This pADPr is a crucial and global regulator of cell cycle progression despite its more simple configuration (short and unbranched polymer) (17). Although structural predictions provide a framework to further advance our understanding of the mechanisms related to poly(ADP-ribosyl)ation, extensive experimental validations are still required to evaluate the actual behavior of pADPr synthesized by the different PARP family members. For example, despite the high degree of similarity to the catalytic domains of PARP-1 and PARP-2 and the presence of structural requirements thought to be involved in pADPr elongation and branching, PARP-3 appears to be mainly a mono ADP-ribosyl transferase (18). The heterogeneity of pADPr molecules also arise from the dynamic interplay between pADPr-synthesizing enzymes and the poly(ADP-ribose) glycohydrolase (PARG) that regulates pADPr catabolism (19). We are just beginning to understand the signaling networks and the cross-talks based on specific structural features that might exist for different classes of pADPr (20).
Like most protein modifications, poly(ADP-ribosyl)ation can directly impart functional changes in target proteins. In this view, poly(ADP-ribosyl)ation can be seen as a highly dynamic posttranslational process, such as phosphorylation. Covalently poly(ADP-ribosyl)ated proteins at specific amino acid positions may undergo deep structural modifications which typically alter protein structure and function. The relaxation of chromatin topology after poly(ADP-ribosyl)ation of nucleosomes is one example of striking pADPr-dependent structural rearrangements (21, 22). However, with regards to size, steric hindrance and, most importantly, very high-negative charge density, pADPr characteristics extend outside of a solely posttranslational mechanism. Noncovalent pADPr interaction also provides an ability for proteins to adapt to new functional conditions. The identification of an increasing number of proteins that strongly interact with pADPr in a noncovalent fashion might indicate a full range of functional complexity and diversity in biological systems modulated by pADPr metabolism.
Protein domains are now emerging as modules that interact selectively with pADPr. The macro domain (23-25) and the PBZ zinc finger (26) are such protein folds for which specific noncovalent pADPr binding was demonstrated. Different mechanisms have been proposed to explain the interactions governing the binding to pADPr. While the PBZ zinc fingers seem to present structural determinants that could allow strong noncovalent interactions with multiple ADP-ribose residues along pADPr polymers (27), structural analysis of the macro domain suggest that its binding to pADPr would rather be limited to terminal ADP-ribose rings (28). It appears that conserved mechanisms with different structural adaptations to pADPr conformations can contribute to the regulation of specific cellular processes.
On the other hand, several DNA damage checkpoint and repair proteins strongly bind pADPr despite being devoid of the aforementioned binding modules (29). Instead, the interaction of these proteins with pADPr is mediated through a discrete binding motif characterized by a sequence pattern of alternating hydrophobic and basic amino acids (29). The pADPr-binding sequence was further defined with key amino acids at preferred positions within the motif (30). The noncovalent pADPr-binding motif often overlaps with other functional domains of proteins. This motif is also frequently associated with lysine- and arginine-rich regions of proteins (29). It is not surprising to find pADPr-binding motifs in a context of positively charged protein regions given the high negative charge of the pADPr and the electrostatic nature of the noncovalent interaction. The presence of a pADPr-binding motif in a nucleic acid-binding protein can alter its DNA/RNA-binding functions. For example, pADPr plays a role in regulating the DNA-binding properties of p53 (31). Table 1 summarizes the three noncovalent pADPr-binding modules previously reported in the literature.
Table 1.
Noncovalent pADPr-binding modules
| pADPr-binding modules | References |
|---|---|
| Poly(ADP-ribose)-binding zinc finger (PBZ) motif [e.g., APFL, CHFR] | (26, 27, 54, 57) |
| Macro domain (C-terminal domain of core histone macro-H2A) [e.g., PARP-9, PARP-14, PARP-15, CHD1L/ALC1] | (5, 23, 24, 28, 32, 58) |
| Poly(ADP-ribose)-binding sequence motif [e.g., p53, XRCC1, XPA, hnRNP A1, ASF/SF2, MRE11, ATM] | (29, 30, 41, 59, 60) |
The noncovalent pADPr-binding capacity of proteins can be verified through combined strategies. Computational in silico methods for the prediction of pADPr-binding proteins based on consensus motifs are the starting point for target identification. Affinity-based, proteome-wide target identification methods are also used to isolate pADPr-binding proteins. Affinity-purification mass spectrometry (AP-MS) approaches have been especially instrumental in identifying pADPr interactors and pADPr-containing multiprotein complexes (30, 32). Taking advantage of the broad specificity and high affinity of anti-pADPr antibodies, poly(ADP-ribosyl)ated and pADPr-binding proteins can be isolated in complex protein mixtures by immunoprecipitation (IP) assays. Predicted candidates or other protein of interest can be assayed for direct binding to pADPr through quantitative in vitro methods. Much of the published research on pADPr-binding proteins has been carried out using polymer-blot assays, a technique essentially derived from nitrocellulose filter-binding assays developed for protein-DNA interactions (33). This technique should be considered as a powerful tool for the identification of pADPr-binding proteins. Refined procedures based on polymer-blot assays now include synthetic peptide arrays covering putative pADPr-binding regions and the use of fractionated pADPr for the evaluation of size-specificity of the binding. Electrophoretic mobility shift assay (EMSA) with pADPr has also been adapted from common gel shift assays used to characterize interaction with nucleic acids (34). Indeed, nucleic acid-like pADPr shares several chemical features with DNA/RNA such that several variants of these methods have been published in the context of pADPr studies. More recently, surface plasmon resonance (SPR) was used to precisely quantify the binding affinity for pADPr, which interaction appeared to be surprisingly strong (34).
The current methods used so far to identify pADPr-binding proteins include various approaches from standard biochemical validation to computer-assisted modeling for the prediction of pADPr-binding motif. We recommend the following methods as a guide to evaluate whether a given protein binds pADPr and to define the region-mediating pADPr binding.
2. Materials
2.1. Cell Culture and PARG siRNA
Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS) (Wisent, St-Bruno, Canada). Pen-Strep (Invitrogen, Carlsbad, CA) containing penicillin (10,000 U/ml) and streptomycin (10,000 μg/ml) is added to culture media.
Solution of 0.5 g/l of trypsin (0.05%) and 0.2 g/l of EDTA•4Na in Hanks’ Balanced Salt Solution (Invitrogen, Carlsbad, CA).
100- and 150-mm cell culture dishes and cell scrapers (Sarstedt, Newton, NC).
GlutaMAX™ media (Invitrogen, Carlsbad, CA).
HiPerFect™ transfection reagent (Qiagen, Germantown, MD).
hPARG siRNA: AAGAUGAGAAUGGUGAGCGAAdTdT and hPARG control siRNA (mismatch): AAGAUGAG-AAUCCUGAGCGAAdTdT (Dharmacon, Lafayette, CO). Prepare 1 μM stock solution in 60 mM KCl, 6 mM HEPES-pH 7.5, 0.2 mM MgCl2 (resuspension and long-term storage buffer).
2.2. Immunoprecipitation
100 mM stock solution of N-methyl-N′-nitro-N-nitroso-guanidine (MNNG) in dimethyl sulfoxide (DMSO).
Phosphate-buffered saline (PBS). Prepare 10× stock with 1.37 M NaCl, 27 mM KCl, 100 mM Na2HPO4, 18 mM KH2PO4 (adjust to pH 7.4 with HCl if necessary).
Lysis buffer: 20 mM Tris–HCl pH 7.5, 150 mM NaCl, 0.5% NP-40, Complete™ protease inhibitor cocktail (Roche Applied Science, Indianapolis, IN). The lysis buffer can also be supplemented with 1 μM ADP-HPD [adenosine 5′-diphosphate hydroxymethyl pyrrolidinediol] (Calbiochem-EMD Biosciences, San Diego, CA) an analog of ADP-ribose that acts as a PARG inhibitor (35, 36).
0.1 M sodium acetate buffer, pH 5.0.
Dynabeads® magnetic beads covalently coupled with Protein G and DynaMag™ magnets (Invitrogen, Carlsbad, CA).
Anti-pADPr antibody clone 10H (Tulip Biolabs, West Point, PA) and equivalent amount of normal mouse IgGs (Calbiochem-EMD Biosciences, San Diego, CA).
PBS containing 1% (w/v) bovine serum albumin (BSA), fraction V (Sigma-Aldrich, Oakville, Canada).
3× Laemmli sample buffer (150 mM Tris–HCl pH 6.8, 6% SDS, 0.3% bromophenol blue, 30% glycerol) containing 5% β-mercaptoethanol.
4–12% Criterion™ XT Bis-Tris gradient gel (Bio-Rad, Hercules, CA).
Criterion™ Precast Electrophoresis System (Bio-Rad, Hercules, CA).
Gel fixation solution: 40% methanol, 10% acetic acid.
SYPRO Ruby® Protein gel stain (Invitrogen, Carlsbad, CA).
CCD-based imaging device, such as the Geliance Imaging System (PerkinElmer, Waltham, MA) equipped with emission filters for optimal visualization of the SYPRO® Ruby protein stain.
2.3. Polymer-Blot Detection of pADPr-Binding Proteins (Isotopic and Nonisotopic Assays)
Mini-PROTEAN® electrophoresis and Mini Trans-Blot® transfer system (Bio-Rad-Hercules, CA).
SDS-PAGE running buffer: 25 mM Tris–HCl, pH 7.4, 200 mM glycine, 0.1% SDS.
SDS-PAGE transfer buffer: 10 mM Tris–HCl, pH 7.4, 100 mM glycine, 20% methanol.
PROTRAN® 0.2 μm pore size nitrocellulose membrane (Whatman GmbH, Dassel, Germany).
SDS-PAGE prestained molecular weight markers (Bio-Rad-Hercules, CA).
Vacuum manifold, such as the Convertible® Filtration Manifold System (Whatman GmbH, Dassel, Germany).
Tris-buffered saline supplemented with Tween-20 (TBS-T): 10 mM Tris–HCl, pH 7.4, 150 mM NaCl and 0.05% Tween-20.
Blocking buffer (TBS-MT): TBS-T containing 5% nonfat dried milk.
Laboratory platform shaker.
Blot trays.
Peroxidase-conjugated secondary antibodies (Jackson Immunoresearch, West Grove, PA).
3 MM Chromatography/blotting paper (Whatman GmbH, Dassel, Germany).
Western Lightning® Plus-ECL, Enhanced Chemiluminescence Substrate (PerkinElmer, Waltham, MA).
Kodak BioMax® films.
Packard BioScience’s Instant-Imager™ or other quantitative radioactivity imaging device (such as Molecular Dynamics PhosphorImager).
2.4. Electrophoretic Mobility Shift Assay
Fractionated pADPr.
Acrylamide stock solution: 30% acrylamide, 0.8% bisacrylamide (150 g acrylamide, 4 g N,N′-methylene bis-acrylamide, deionized water to 500 ml). Mix well and filter through 0.2-μm filter. Store at 4°C.
10× TBE: 108 g Tris base, 55 g boric acid, 9.3 g EDTA, deionized water to 1 l. Store at room temperature.
10% (w/v) ammonium persulfate (APS) solution. Stable for 1 month at 4°C.
N,N,N′, N′-tetramethylethylenediamine (TEMED). Store at room temperature.
Monitoring dye: 10 mM Tris–HCl pH 8.0, 0.05% (w/v) bromophenol blue and 0.05% (w/v) xylene cyanol. Store at room temperature.
Vertical slab gel apparatus, semi-dry electrophoretic transfer cell and power supply.
Hybond N + nylon membrane (GE Healthcare Life Sciences, Piscataway, NJ).
Streptavidin–horseradish peroxidase conjugated antibody (GE Healthcare Life Sciences, Piscataway, NJ).
Vacuum gel dryer with heating unit.
X-ray film, cassettes, and developer.
3. Methods
3.1. Computational Predictions of pADPr-Binding Proteins
Using sequence alignments of DNA damage checkpoint proteins, Pleschke and colleagues (29) recognized a consensus pADPr-docking site pattern characterized by the presence of a cluster of basic amino acids and the conservation of an interspersed arrangement of basic and hydrophobic amino acid residues located in close proximity. Numerous protein pattern and profile search engines can be used for large-scale organism-specific predictions of pADPr-binding proteins or targeted protein sequences (see Note 1). Interestingly, evolutionarily conserved functional domains frequently overlap with the pADPr-binding motif, such as domains mediating protein–protein interactions, DNA binding, nuclear localization, nuclear export, and protein degradation (29). However, the tolerance for several amino acids at different positions within the sequence limits its ability to accurately predict matching motifs for protein database searches. Subsequent motif refinement was performed by Gagné and collaborators (30) on the basis of the original motif. Experimentation demonstrates that pADPr-binding motif refinements resulted in more matches to functionally related proteins, with significant enrichment in proteins involved in transactions with nucleic acids. The refined pADPr-binding motif may therefore be used as a more reliable guide to predict pADPr-binding proteins in this context. For a more comprehensive understanding of the sequence requirements for pADPr-binding, it will be important to extend similar studies to larger pADPr-binding motif datasets. The systematic use of both complementary pADPr-binding motifs is still mandatory (see Note 2). Together they may help users to locate a noncovalent pADPr-binding, but users should always keep in mind that searches should be performed at medium or low stringency to see weaker or divergent pADPr-binding sites. The frequency distribution of the motifs in protein databases varies according to the similarity level threshold applied to the database searches. The importance of the individual amino acids of the consensus motif can be assessed by site-directed mutagenesis, but computational predictions are usually first validated through direct pADPr-binding assays. The following methodologies were developed to identify pADPr-binding proteins, screen for the putative pADPr-binding regions and for quantitative analysis of pADPr-binding affinity.
3.2. Affinity Purification of pADPr-Binding Proteins
Anti-pADPr antibodies are characterized by a broad specificity as a consequence of being reactive with various pADPr structures (37). The high affinity and specificity of anti-pADPr-antibodies makes them highly suitable for specific immunoprecipitation from crude lysates. Because of the high turnover of pADPr, pull-down assays must be performed in conditions where hydrolysis is limited. For example, strategies can include the use of a PARG inhibitor or siRNA-mediated PARG knock-down. pADPr has notably been efficiently immunoprecipitated with a complex mixture of interacting proteins using the mouse monoclonal antibody clone 10H (Tulip Biolabs) (38) or the rabbit polyclonal antibody clone 96-10 (39). pADPr and its associated protein can be pulled down from protein extracts using regular buffer compositions usually employed for protein immunoprecipitation assays. Detergents that tend to preserve noncovalent interactions (e.g., Triton X-100, NP-40, CHAPS) are preferred. The ionic strength of the wash buffers (i.e., NaCl molarity) can also be modified to modulate the stringency of the pulled-down assay.
The pull-down assays do not distinguish whether the interaction between pADPr and proteins is mediated by free polymers or covalently attached polymers onto protein substrates or interaction complexes. However, pull-down and mass spectrometric strategies are very useful resources for target and pathway identification and for guiding future biological experiments.
3.2.1. Cell Culture and Endogenous PARG Knock-Down Using siRNA
Human neuroblastoma SK-N-SH (see Note 3) are cultured in DMEM medium supplemented with 10% FBS and Pen-Strep. SK-N-SH cells are passaged when approaching confluence with trypsin/EDTA to maintain stock cultures in 100-mm tissue dishes and experimental cultures in 150-mm dishes.
Experimental SK-N-SH cultures are grown until cells reached 50% confluence under air/CO2 ratio of 19:1 at 37°C. Before performing transfection, the medium is replaced with fresh DMEM containing GlutaMAX™ (1×) and 10% FBS (without antibiotics).
Prepare the siRNA transfection complexes as follow. For 150-mm plates, put 600 μl DMEM in a 1.5 ml tube. Add 100 μl of siRNA stock solution (for a final concentration of 5 nM in the culture plate). Mix well (vortex) and then add 40 μl of HiPerfect® reagent. Mix well by inverting the tube several times but without vortexing.
Leave the mixtures for 5–10 min at room temperature to allow the formation of transfection complexes.
Add the complexes drop-wise onto the cells. Gently swirl the plate to ensure uniform distribution of the transfection complexes.
Incubate the cells with the transfection complexes under their normal growth conditions for 48 h (cells will reach 100% confluency).
Trypsinize the cells and replate at 50% confluence in a medium containing freshly added siRNA complexes prepared as above.
When the cells become confluent, they are passaged and transfected again as described. To decrease PARG activity by more than 80%, silencing is conducted for over 6 days by passaging cells every 48 h as described (see Note 4). Examples of pADPr accumulation in PARG siRNA treated cells are shown in Fig. 1.
Fig. 1.
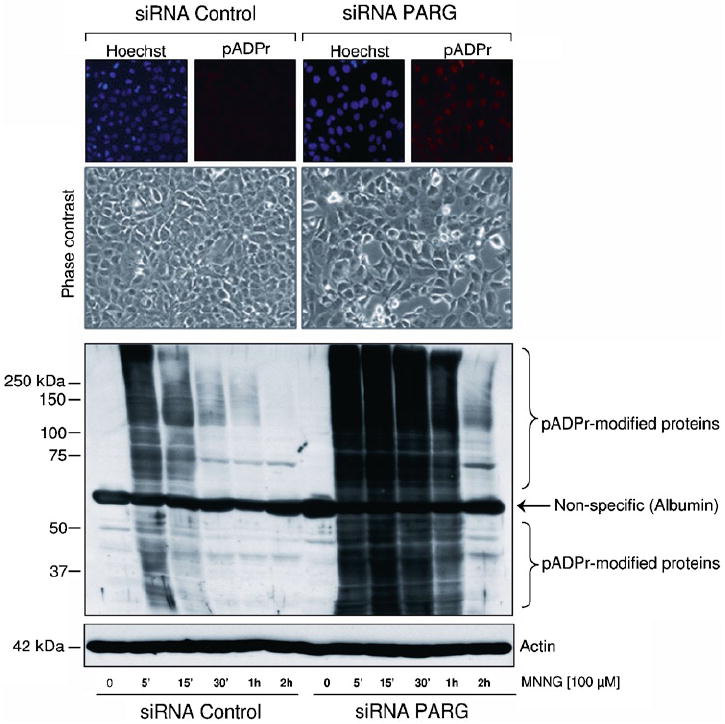
pADPr levels are increased and sustained in siRNA-mediated knock-down of PARG in SK-N-SH cells. Upper panel: immunostaining of pADPr in untreated and PARG knocked-down cells after 5 min of MNNG treatment. DNA was stained with Hoechst 33342. Phase contrast images of control and PARG siRNA-treated cells are shown. Lower panel: time course of protein poly(ADP-ribosyl)ation after MNNG treatment. Crude protein extracts were loaded and subjected to SDS-PAGE and immunoblotting. Western blots were revealed with anti-pADPr polyclonal antibody 96-10. Blots show the rapid accumulation and disappearance of pADPr in control cells while pADPr levels remain elevated in PARG siRNA knocked-down cells.
3.2.2. Immunoprecipitation of pADPr-Associated Proteins Following Alkylation-Induced DNA Damage
Neuroblastoma SK-N-SH cells are seeded onto three 150-mm cell-culture dishes and endogenous PARG is knocked down as described above (see Subheading 3.2.1).
The cells should reach 80–90% confluency prior to the treatment with an appropriate DNA-damaging agent. To induce sustained pADPr levels in PARG knocked-down SK-N-SH cell line, cells are incubated with 100 μM MNNG (see Note 5) freshly diluted in serum-free DMEM for 5 min and then rapidly rinsed twice with ice-cold PBS. All further steps should be performed on ice or at 4°C.
Immediately, 2 ml/plate of lysis buffer (see Note 6) is added. Cells are collected with a cell scraper and the extracts are pooled. The concentration of NaCl is raised to 2 M and placed on ice for 15–20 min (see Note 7).
The extracts are gently mixed for 30 s and extensively dialyzed against ice-cold lysis buffer.
During the dialysis steps, prepare the appropriate beads suspension for immunoprecipitation experiments. IP of pADPr-associated proteins is typically performed using Dynabeads® magnetic beads covalently coupled with protein G.
300 μl of the protein G beads suspension is washed twice with 1 ml of 0.1 M sodium acetate buffer, pH 5.0, before addition of 30 μg of mouse monoclonal anti-pADPr antibody clone 10H in 500 μl sodium acetate buffer. To prepare a negative control, add an equivalent amount of normal mouse IgGs to the washed protein G beads suspension (see Note 8). Incubate for 1 h at room temperature with gentle rotating agitation and wash twice with 2 ml of lysis buffer.
The antibody-coupled beads suspension is then incubated for 1 h with 1 ml of PBS containing 1% (w/v) BSA to block non-specific antibody binding sites.
Residual blocking solution is discarded and antibody-coupled beads washed three times with 1 ml of lysis buffer.
The pADPr/protein extract is then added to the bead suspension and incubated for 2 h with gentle rotating agitation.
The samples are washed at least three times with two volumes of lysis buffer for 5 min (with gentle rotating agitation).
Elution of protein complexes is performed using 100–150 μl of 3× Laemmli sample buffer containing 5% β-mercaptoethanol. Samples are heat denaturated at 95°C for 5 min and then resolved by SDS-PAGE using a 4–12% gradient gel.
After the electrophoresis, the gel is fixed for 30 min in 100 ml of gel fixation solution, rinsed three times with water and stained overnight with SYPRO® Ruby. Cover the incubation tray with aluminum foil to protect the stain from light.
Perform image acquisition on a CCD-based bioimaging system for fluorescence applications, such as PerkinElmer’s Geliance system. A typical pull-down result is shown in Fig. 2.
Immunoprecipitation extracts can be probed for pADPr or any targeted protein after transfer onto nitrocellulose and Western blot analysis (see Fig. 3).
Fig. 2.
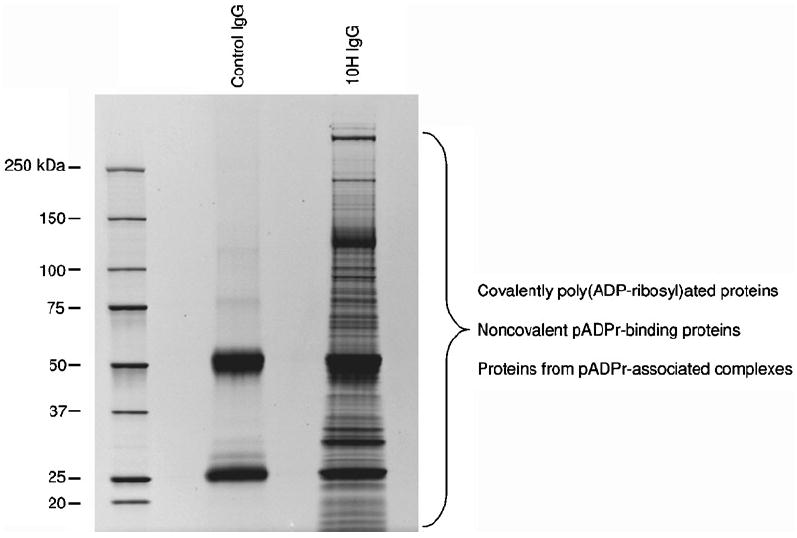
SDS-PAGE analysis of pADPr immunoprecipitation extracts from MNNG-treated and PARG-inhibited HEK 293 cells. pADPr-associated proteins were pulled-down using anti-pADPr antibody clone 10H (Tulip Biolabs) bound to protein G-coated magnetic beads (in the presence of 1 μM PARG inhibitor ADP-HPD). Immunoprecipitates were resolved by 4–12% SDS-PAGE and stained with SYPRO® Ruby fluorescent dye. Normal mouse IgGs were used to assess nonspecific binding.
Fig. 3.
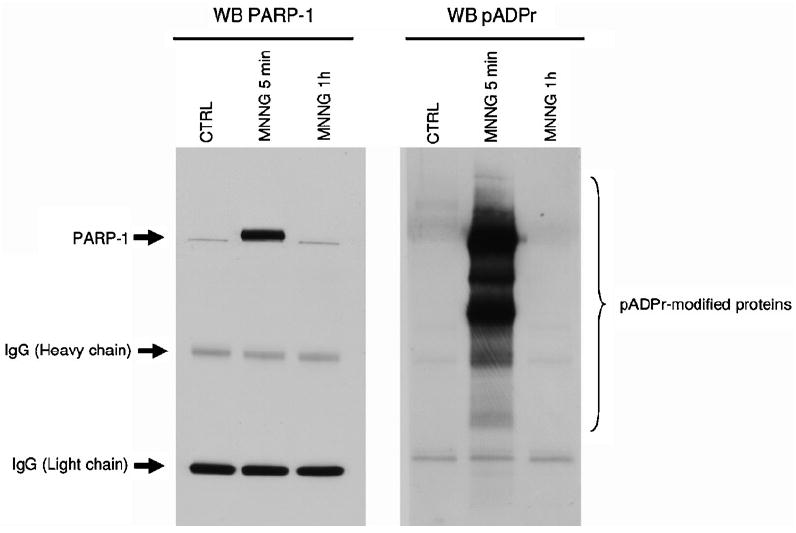
pADPr immunoprecipitation followed by Western blot analysis. HEK 293 cell extracts were subjected to pADPr immunoprecipitation with anti-pADPr antibody 10H (coupled to protein G-coated magnetic beads) under three conditions: untreated cells (CTRL), cells treated with 100 μM MNNG for 5 min (MNNG 5 min) and cells that have been allowed to recover for 1 h after the removal of the MNNG (MNNG 1 h). Cells were lysed in the presence of 1 μM PARG inhibitor ADP-HPD. Immunoprecipitation extracts were resolved by SDS-PAGE on a precast 4–12% gradient gel and transferred onto nitrocellulose membrane. Protein patterns were analyzed on immunoblots probed for either PARP-1 (anti-PARP-1 antibody C2-10) or pADPr (anti-pADPr antibody 96-10). Blots confirm the retention of poly(ADP-ribosyl)ated proteins in immunoprecipitation extracts and the accumulation of PARP-1 at the peak of pADPr levels following alkylation-induced DNA damage and PARP activation (see Fig.1).
3.3. Identification of Noncovalent pADPr-Binding Proteins by Polymer-Blot Analysis
Several novel methods assessing the noncovalent interaction between pADPr and specific binding proteins have been described. Yet, most of the pADPr-binding proteins have been identified biochemically by a polymer-blot-binding assay using synthetic peptides covering the putative pADPr-binding motif. In particular, the nitrocellulose-binding assay first described by Panzeter and colleagues (40) and modified by Gagné et al. (41) remains one of the leading techniques currently used. Because this in vitro pADPr-binding assay is quantitative, it also allows the measurement of the pADPr-binding properties of purified proteins. Polymer-blot analysis requires the immobilization of peptides or purified proteins onto nitrocellulose either by hand-spotting or transblot procedures. This method is basically an adaptation of blot overlays protocols to pADPr. Protein–pADPr interactions can be detected by using a radio-labeled pADPr probe or an anti-pADPr antibody (similar to the far-Western blotting technique). Automodified PARP-1 can also be used as a probe for detecting interactions with protein-bound pADPr. The benefit of this probe is that it does not need to be purified through dihydroxyboronyl BioRex (DHBB) chromatography (42) while providing, most of the time, equivalent end result as assays with protein-free pADPr. Investigators should, however, rule out the possibility that interactions with PARP-1 account for some of the observed binding.
3.3.1. Detection of pADPr-Binding Proteins Using Radio-Labeled Polymer-Blot Assay
These instructions assume the use of purified recombinant proteins or mimicking synthetic peptides corresponding to the predicted pADPr-binding domain of selected proteins. The ability of these proteins or peptides to interact with pADPr in a noncovalent manner is determined by polymer-blot assay using [32P]-labeled protein-free pADPr or [32P]-labeled auto-modified PARP-1 as probes (see Note 9).
For hand-spotted arrays, synthetic peptides or purified proteins are diluted in TBS-T. One microgram is hand-spotted onto a nitrocellulose membrane or dot-blotted using a vacuum manifold system. Hand-spotted peptides are air-dried for 15 min. Peptides or proteins blotted with a manifold system are washed once with TBS-T. The membrane is removed from the manifold and air-dried for at least 15 min.
Purified proteins can also be resolved by SDS-PAGE and transferred onto nitrocellulose membranes with the blotting system.
The following procedures apply for all spotting approaches (i.e., hand-spotting, vacuum-spotting, and transblot procedures).
The membranes are soaked in TBS-T and washed three times.
The membranes are then incubated for at least 1 h in TBS-T and placed on a platform or orbital shaker at room temperature to allow proper refolding of proteins (see Note 10).
The membranes are then incubated for 1 h in 25 ml of TBS-T containing 250 nM of [32P]-labeled probe at room temperature with constant shaking (see Note 11).
The membranes are extensively washed with TBS-T until no radioactivity could be detected in waste.
The membranes are then air-dried prior to analysis. Radioactivity is detected by using an Instant-Imager™ system which quantifies the distribution of radioactivity on flat samples. A hand-spotted protein array of histones is shown in Fig. 4 as an example.
For autoradiogram, expose the membranes to an X-ray film for an appropriate length of time.
Fig. 4.
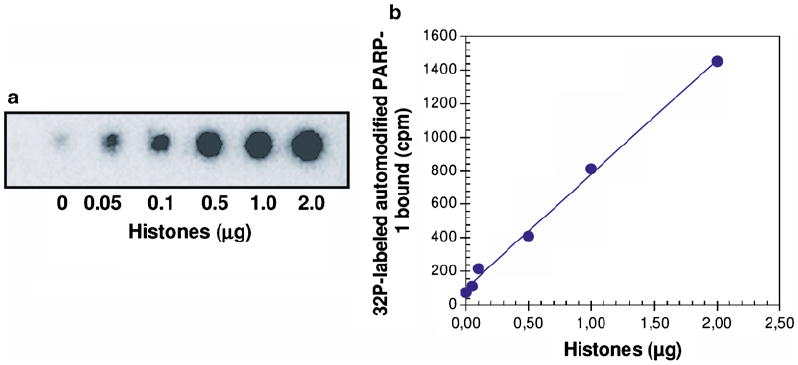
Analysis of pADPr-binding activity of purified chromatin histones. Noncovalent pADPr-binding was analyzed by polymer-blot with a [32P]-labeled automodified PARP-1 probe. (a) Autoradiogram of the polymer-blot assay in function of the amount of blotted histones. (b) Linearity dependence of [32P]-labeled automodified PARP-1 bound in function of increasing amount of blotted histones.
3.3.2. Identification of Noncovalent pADPr-Binding Proteins by Nonisotopic Polymer-Blot Analysis
Prepare arrays of hand- or vacuum-spotted peptides and nitrocellulose-transferred purified proteins as described for the isotopic detection of pADPr-binding (see Subheading 3.3.1).
The nitrocellulose membranes are soaked in TBS-T and washed three times.
The membranes are then incubated for at least 1 h in TBS-T and placed on a platform or orbital shaker at room temperature to allow proper refolding of proteins (see Note 10).
The incubation buffer is discarded and the membrane quickly rinsed in TBS-T prior to addition of 250 nM DHBB purified pADPr in 25 ml TBS-T. Incubate for 1 h at room temperature with gentle agitation.
The unbound pADPr is then removed by extensively washing the membrane with TBS-T.
Block nonspecific antibody-binding sites by incubating the membrane for 1 h in TBS-MT.
The presence of pADPr-binding proteins is detected with an appropriate anti-pADPr antibody (see Note 12). Dilute the anti-pADPr antibody in 25 ml of TBS-MT and incubate the membrane for 1 h at room temperature with gentle agitation. The primary antibody is then removed and the membrane washed at least three times for 15 min each with 50 ml TBS-MT.
The appropriate peroxidase-conjugated secondary antibody is freshly prepared for each experiment. A 1:5,000 dilution is prepared in 25 ml of TBS-MT and added to the membrane for 30 min at room temperature with gentle agitation.
The secondary antibody is discarded and the membrane is washed as follows: three times with 50 ml TBS-MT for 15 min each, three times with 50 ml of TBS-T, and then twice with 50 ml TBS without Tween-20 detergent.
The blot is revealed using ECL reagents for HRP-conjugated secondary antibody. For optimal results, the ECL reagents should be warmed at room temperature and used as soon as possible after mixing. Mix no more than is needed at one time and perform these steps preferably in a dark room under safe light condition.
Incubate the ECL reagents with the membrane for 1 min at room temperature. Use 0.125 ml per cm2 of membrane and incubate with gentle agitation.
Drain any excess of ECL reagents by depositing the membrane on a Whatman 3MM blotting paper. To keep the film dry, use a plastic transparent sheet protector or a thin plastic film typically used for sealing food to seal the wet membrane.
The membrane is then placed in an X-ray film cassette with BioMax® film for a suitable exposure time.
Repeat film exposure, varying the time as needed for optimal detection (see Note 13).
3.4. Electrophoretic Mobility Shift Assays
EMSA is a technique that was first developed to characterize the association of DNA-binding proteins with their substrates (43). The assay is based on the observation that complexes of protein and DNA migrate through a nondenaturing polyacrylamide gel more slowly than free DNA fragments. This technique can easily be adapted to study other type of associations, such as noncovalent binding to pADPr. This variant technique is carried out by first incubating a putative pADPr-binding protein with radiolabeled pADPr. The reaction products (i.e., complexes formation) are then analyzed on a nondenaturing polyacrylamide gel. This method can be used to obtain quantitative-binding parameters to pADPr, such as equilibrium and rate constants. However, the success of this technique depends on many factors which can affect the stability or the mobility of the protein-pADPr complexes (e.g., running buffer composition, temperature, molecular weight of protein). Therefore, factors influencing the electrophoresis must be taken into account and an optimal protocol has to be established according to the studied proteins. Moreover, in contrast to protein-DNA or -RNA studies, pADPr is generally not defined as a homogeneous component but rather as a complex mixture of pADPr polymers of various size and branching configurations which adds another challenge to understanding the observed interaction. In 2007, Fahrer et al. have assessed the binding affinity of XPA and p53 to biotinylated pADPr by EMSA (34). The authors developed an elegant and efficient method to circumvent the main problems associated with pADPr fractionation and detection. There are various advantages of using biotinylated pADPr. Instead of using anti-pADPr-specific antibodies, biotin end-labeled pADPr chains are readily accessible to high-affinity anti-biotin antibodies and the use of nonisotopic pADPr fractionation eliminates radioactive materials which might cause further contamination on chromatographic equipments. Using this approach, Fahrer et al. (34) demonstrated the size-dependent pADPr-binding of XPA and p53. This procedure is amenable to evaluate the pADPr-binding affinities of virtually any purified putative pADPr-binding protein.
3.4.1. Native Polyacrylamide Gel
Prepare a 6% native polyacrylamide gel in 0.25× TBE by mixing 8 ml of 30% acrylamide stock solution (see Note 14) with 1 ml of 10× TBE and adjusting the volume to 40 ml with deionized water. Then, add 0.4 ml of 10% APS, and mix by inverting the tube. This is followed by addition of 40 μl TEMED with mixing by inversion of the tube. The acrylamide solution is poured gently into a 15 × 17 cm (W × L), 1.5-mm thick slab gel.
Pre-run the gel in 0.25× TBE for ~1 h at 100 V. The binding reaction can be performed while the gel is being pre-run.
3.4.2. pADPr-Binding Reaction and EMSA
pADPr-binding reactions are performed in 20 μl reaction mixture containing 10 mM Tris–HCl pH 7.4 and 1 mM EDTA. Purified protein and fractionated pADPr (see Note 15) are incubated together at 25°C for 20 min to allow complex formation to reach equilibrium (see Note 16).
The mixture is adjusted to 10% glycerol and then applied immediately to the pre-run native polyacrylamide gel.
The samples are electrophoresed for 2.5 h at 160 V to separate free and bound pADPr. The monitoring dye can be loaded to visualize the progress of the electrophoretic run.
When using radiolabeled pADPr as a probe, the gel is transferred to a double layer of Whatman 3 MM filter paper. After applying an overlay of plastic wrap, the gel is dried under vacuum on a heated (80°C) platform gel dryer and autoradiographed.
For antibody-based detection of biotinylated pADPr, the gel is transferred to a positively charged nylon membrane with a semi-dry electrophoretic transfer system (20 V for 50 min followed by a heat-fixation at 90°C for 1 h). Then, the membrane is blocked with 2% BSA in TBS-T (137 mM NaCl, 20 mM Tris–HCl pH 7.6, 0.1% Tween-20) and biotinylated ADP-ribose chains are detected with Streptavidin–horseradish peroxidase conjugated antibody (1:2,000 in blocking solution for 1 h at room temperature) (see Note 17).
3.5. Other Techniques
A fairly recent development in the study of protein–pADPr interactions involves the measurements of biomolecular interactions by SPR (reviewed in (44)). This technique assesses changes in refractive index very close to a sensor surface. The noncovalent bond between a protein and pADPr results in a change in the refractive index measured in real time. This technology allows to measure association and dissociation constants with much more precision and reproducibility than EMSA (34). In 2007, Fahrer et al. (34) showed that XPA displayed high affinity for long pADPr chains with a KD value of 6.5 nM, whereas p53 displayed a strong binding to both short and long chains (34). Other groups also used SPR technology to study interactions of proteins with pADPr, such as the PBZ-containing proteins APLF and CHFR. As we only give a brief overview of the technique, we refer those readers who are interested in a greater understanding of SPR in the context of pADPr-binding studies to references (26, 27, 34).
Another method of interest reported by Panzeter et al. describes the synthesis of a stable and reusable pADPr-affinity resin for affinity studies (45). They observed that pADPr-agarose beads can efficiently isolate pADPr-binding proteins like histones. However, the noncovalent affinity purification of non-histone proteins from cell lysates with this approach is yet to be demonstrated. Nevertheless, this strategy may still be optimized and adapted to more recent technologies, such as magnetic agarose beads. We refer the readers interested in this technique to the original publication.
Acknowledgments
The authors are supported by research funds from a Canada research chair in proteomics, the Canadian Institutes of Health Research (CIHR grants MOP-74648 and IG1-14052), the Cancer Research Society, the Alberta Cancer Board, the National Institutes of Health (NIH grant P50 CA136393-01), Fonds de la Recherche en Santé du Québec (scholarship to JPG); Strategic Training Program grant in genomics, proteomics and bioinformatics (CIHR STP-53894 to JPG). The authors thank Michèle Rouleau for critical review of the manuscript and their colleagues who contributed to the methods described in this chapter.
Footnotes
We suggest the use of the Network Protein Sequence @nalysis (NPS@) Web server (46). It includes several tools dedicated to protein sequence analysis available for the biologist community (URL: http://npsa-pbil.ibcp.fr/). The Sequence Retrieval System (SRS) is particularly useful for organism-specific database searches. Extracted protein datasets can be scanned for one or several patterns using the PATTINPROT engine. The PATTINPROT algorithm is especially relevant to pADPr-binding motif searches because it allows divergence from the queried pattern. Errors toward the pattern are set by the number of mistmatch allowed or by a similarity threshold.
Computational analysis and prediction of pADPr-binding motifs are challenging because the sequence motif that mediates pADPr interaction is degenerated and poorly defined. In silico identifications also result from predictions made out of any structural context. Though variable in sequence, two motifs were experimentally proven to confer binding to pADPr: a motif derived from DNA damage checkpoint proteins (Pleschke et al. (29)): [AVILMFYW]-X-[KR]-X-[AVILMFYW]-[AVILMFYW]-[KR]-[KR]-[AVILMFYW]-[AVILMFYW]-[KR] and a refined, more stringent version with a bias toward proteins involved in DNA/RNA transactions (Gagné et al. (30)): [HKR]-X-X-[AIQVY]-[KR]-[KR]-[AILV]-[FILPV].
This protocol can be adapted for many other cell types. Affinity purification of pADPr-binding proteins has also been performed in cell extracts from HeLa, 293 F cell lines (16) and HEK 293 (Gagné et al. unpublished).
The optimal cell confluency for siRNA transfection should be determined for every new cell type to be transfected and kept constant in future experiments. Using this procedure, efficient knockdown without cytotoxicity has been observed up to 2 weeks after the initial transfection. To circumvent the lack of good antibodies for directly assessing endogenous PARG levels, the evaluation of PARG knockdown is performed using PARG thin layer chromatography (TLC) activity assays as described (47).
MNNG is a very potent mutagen. Wear gloves and safety glasses. Handle as a carcinogen. Heat, light, and moisture sensitive. Prepare in DMSO and store at 4°C. Stable at 4°C for 2 weeks. For alkylation-induced DNA damage, an optimal incubation time must be determined by the investigator. This time should be determined carefully, as pADPr can decrease rapidly at longer time points due to NAD depletion and PARG hydrolysis.
Alternative lysis buffers have been used to successfully pull-down pADPr-binding protein in affinity purification experiments: (1) 150 mM NaCl, 50 mM HEPES, pH 7.4, 1 mM MgCl2, 0.5% Triton-X-100, 1 mM DTT, 1 mM EGTA (16), (2) 50 mM Tris–HCl pH 8.0 containing 150 mM NaCl, 1% NP-40, 5 mg/ml deoxycholate and 0.1% SDS (39) and (3) 40 mM HEPES pH 7.5, 120 mM NaCl, 0.3% CHAPS, 1 mM EDTA, 1× Complete® protease inhibitors (Gagné et al. unpublished).
This salt extraction procedure was adapted from Cardenas-Corona et al. (48). High salt conditions typically result in the disruption of protein complexes. This procedure was performed to optimize the release of chromatin-bound pADPr-binding proteins especially abundant in the context of PARP hyperactivation with DNA damaging agents. To preserve multiprotein complexes integrity, skip the high salt extraction, centrifuge the cell lysate at 3,000 × g for 5 min to pellet any insoluble material/cellular debris and proceed to step 6.
Native IgG from mouse serum are used as a negative control in parallel with specific mouse IgG-type primary antibodies. A sample preclear can also be performed by first incubating the cell extracts with uncoupled beads to reduce unspecific background.
The pADPr synthesized by in vitro automodification of PARP-1 has a mean length of 40 ADP-ribose residues (49). The concentrations of pADPr used in these assays are within the range of in vivo pADPr concentrations found in MNNG-treated cells (50, 51). In vitro synthesis of pADPr is performed according to Ménard and Poirier (47), and pADPr is purified by DHBB chromatography (42). Alternatively, protein-free pADPr can be prepared using DNAse I and proteinase K followed by a phenol–chloroform extraction as described by Ahel et al. (26).
Proteins must fold to unique native structures in order to perform their functions. In this context, one can presume that it might be unlikely that a protein stays intact after SDS-PAGE sample preparation and nitrocellulose transfer. Nonetheless, it is well-known that many types of protein interactions do in fact still occur even after one of the partners has been reduced, denatured, run on SDS-PAGE, and Western blotted (52). Reducing agents, such as dithiothreitol (DTT) and β-mercaptoethanol can also be added to help the renaturation of some proteins (1 mM DTT or 5 mM β-mercaptoethanol). Reducing agents must be added fresh. This step is preferably performed in tightly closed container under a fume hood.
Another important consideration for this step is that the binding assay should also be performed at low concentration of ligand (pADPr down to 10 nM) to minimize the possibility of non-specific interactions. Under conditions of low concentrations of ligand, the binding is saturated when the system reaches the equilibrium. It was observed that, for all concentrations of ligand tested, the saturation of pADPr-binding is obtained within 3 h. Working under conditions of low concentration of ligand offers the advantage of being quantitative over a wide range of protein concentrations.
Generally, experiments are performed with mouse monoclonal antibody clone 10H diluted 1:250 or rabbit polyclonal antibody 96–10 diluted at 1:5,000.
Quantification of data may be desired, and this can be done using standard scanning densitometry of the films if care is taken to ensure that signal has not been saturated. Alternatively, the chemiluminescence signal can be captured digitally using a cooled CCD camera equipped with digital imaging software.
Acrylamide is a neurotoxin and therefore must be handled carefully. Wear a dust mask and rubber gloves.
pADPr is generally fractionated by anion exchange HPLC as reported by the Jacobson’s group (49, 53). A recent method using gel-filtration chromatography has also been reported to crudely fractionate pADPr (54). Fractions can be characterized by high resolution 20% native PAGE (42) based on improvements of the original method published by Alvarez-Gonzalez and Jacobson (49). Average polymer size can also be estimated from reversed-phase HPLC analysis of nucleosides generated by phosphodiesterase-digestion of pADPr (53). The described technique is based on Shuman’s EMSA protocol (55) and nonisotopic biotinylated pADPr prepared by Fahrer and colleagues (34).
Reaction mixtures need to be optimized empirically. Reported ranges of pADPr (125–250 fmol of size-fractionated pADPr) and target proteins (up to 1.6 μM) (34) must be modulated according to pADPr-binding strength and the method of detection.
Specific immunodetection of pADP-protein complexes transferred to nylon membrane can also be performed using anti-pADPr antibodies, such as the monoclonal antibody clone 10H or the polyclonal antibody 96–10 essentially by the method described by Affar et al. (39). Briefly, the gel is transferred onto Hybond N + membrane in 45 mM Tris–borate pH 8.3 and 1 mM EDTA using transblot electrophoresis transfer apparatus (0.4 A for 1.5 h at room temperature). After the transfer, the membrane is air-dried with a hair dryer and pADPr is cross-linked on the membrane for 5 min using a UV (312 nm) transilluminator apparatus (Fisher Scientific). After pADPr fixation, the membrane is blocked with PBS containing 0.1% Tween-20 and 5% milk for 1 h and processed essentially as described previously (56) with anti-pADPr antibody 96–10 at a dilution of 1:5000. The blot is revealed using ECL reagents for HRP-conjugated secondary rabbit antibody.
References
- 1.Hassa PO, Haenni SS, Elser M, Hottiger MO. Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev. 2006;70:789–829. doi: 10.1128/MMBR.00040-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schreiber V, Dantzer F, Ame JC, de Murcia G. Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol. 2006;7:517–528. doi: 10.1038/nrm1963. [DOI] [PubMed] [Google Scholar]
- 3.Malanga M, Althaus F. DNA damage signaling through poly(ADP-Ribose) In: Burkle A, editor. Poly(ADP-Ribosyl)ation. Landes Bioscience and Springer Science + Business Media; New York, NY: 2006. pp. 41–50. [Google Scholar]
- 4.Haince JF, McDonald D, Rodrigue A, Dery U, Masson JY, Hendzel MJ, Poirier GG. PARP1-dependent kinetics of recruitment of MRE11 and NBS1 proteins to multiple DNA damage sites. J Biol Chem. 2008;283:1197–1208. doi: 10.1074/jbc.M706734200. [DOI] [PubMed] [Google Scholar]
- 5.Gottschalk AJ, Timinszky G, Kong SE, Jin J, Cai Y, Swanson SK, Washburn MP, Florens L, Ladurner AG, Conaway JW, Conaway RC. Poly(ADP-ribosyl)ation directs recruitment and activation of an ATP-dependent chromatin remodeler. Proc Natl Acad Sci USA. 2009;106:13770–13774. doi: 10.1073/pnas.0906920106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beneke S, Burkle A. Poly(ADP-ribosyl) ation in mammalian ageing. Nucleic Acids Res. 2007;35:7456–7465. doi: 10.1093/nar/gkm735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang P, Jacobson MK, Mitchison TJ. Poly(ADP-ribose) is required for spindle assembly and structure. Nature. 2004;432:645–649. doi: 10.1038/nature03061. [DOI] [PubMed] [Google Scholar]
- 8.d’Adda di Fagagna F, Hande MP, Tong WM, Lansdorp PM, Wang ZQ, Jackson SP. Functions of poly(ADP-ribose) polymerase in controlling telomere length and chromosomal stability. Nat Genet. 1999;23:76–80. doi: 10.1038/12680. [DOI] [PubMed] [Google Scholar]
- 9.Kraus WL. Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol. 2008;20:294–302. doi: 10.1016/j.ceb.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hassa PO. The molecular “Jekyll and Hyde” duality of PARP1 in cell death and cell survival. Front Biosci. 2009;14:72–111. doi: 10.2741/3232. [DOI] [PubMed] [Google Scholar]
- 11.Andrabi SA, Kim NS, Yu SW, Wang H, Koh DW, Sasaki M, Klaus JA, Otsuka T, Zhang Z, Koehler RC, Hurn PD, Poirier GG, Dawson VL, Dawson TM. Poly(ADP-ribose) (PAR) polymer is a death signal. Proc Natl Acad Sci USA. 2006;103:18308–18313. doi: 10.1073/pnas.0606526103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.D’Amours D, Desnoyers S, D’Silva I, Poirier GG. Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J. 1999;342(Pt 2):249–268. [PMC free article] [PubMed] [Google Scholar]
- 13.Minaga T, Kun E. Probable helical conformation of poly(ADP-ribose). The effect of cations on spectral properties. J Biol Chem. 1983;258:5726–5730. [PubMed] [Google Scholar]
- 14.Minaga T, Kun E. Spectral analysis of the conformation of polyadenosine diphosphoribose. Evidence indicating secondary structure. J Biol Chem. 1983;258:725–730. [PubMed] [Google Scholar]
- 15.Kleine H, Poreba E, Lesniewicz K, Hassa PO, Hottiger MO, Litchfield DW, Shilton BH, Luscher B. Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell. 2008;32:57–69. doi: 10.1016/j.molcel.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 16.Chang P, Coughlin M, Mitchison TJ. Tankyrase-1 polymerization of poly(ADP-ribose) is required for spindle structure and function. Nat Cell Biol. 2005;7:1133–1139. doi: 10.1038/ncb1322. [DOI] [PubMed] [Google Scholar]
- 17.Rippmann JF, Damm K, Schnapp A. Functional characterization of the poly(ADP-ribose) polymerase activity of tankyrase 1, a potential regulator of telomere length. J Mol Biol. 2002;323:217–224. doi: 10.1016/s0022-2836(02)00946-4. [DOI] [PubMed] [Google Scholar]
- 18.Loseva O, Jemth AS, Bryant HE, Schuler H, Lehtio L, Karlberg T, Helleday T. PARP-3 is a mono-ADP-ribosylase that activates PARP-1 in the absence of DNA. J Biol Chem. 2010;285:8054–8060. doi: 10.1074/jbc.M109.077834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gagné JP, Hendzel MJ, Droit A, Poirier GG. The expanding role of poly(ADP-ribose) metabolism: current challenges and new perspectives. Curr Opin Cell Biol. 2006;18:145–151. doi: 10.1016/j.ceb.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 20.Andrabi SA, Dawson TM, Dawson VL. Mitochondrial and nuclear cross talk in cell death: parthanatos. Ann NY Acad Sci. 2008;1147:233–241. doi: 10.1196/annals.1427.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Murcia G, Huletsky A, Lamarre D, Gaudreau A, Pouyet J, Daune M, Poirier GG. Modulation of chromatin superstructure induced by poly(ADP-ribose) synthesis and degradation. J Biol Chem. 1986;261:7011–7017. [PubMed] [Google Scholar]
- 22.de Murcia G, Huletsky A, Poirier GG. Modulation of chromatin structure by poly(ADP-ribosyl)ation. Biochem Cell Biol. 1988;66:626–635. doi: 10.1139/o88-072. [DOI] [PubMed] [Google Scholar]
- 23.Egloff MP, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, Gruez A, Campanacci V, Cambillau C, Ziebuhr J, Ahola T, Canard B. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol. 2006;80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG. The macro domain is an ADP-ribose binding module. EMBO J. 2005;24:1911–1920. doi: 10.1038/sj.emboj.7600664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Till S, Ladurner AG. Sensing NAD metabolites through macro domains. Front Biosci. 2009;14:3246–3258. doi: 10.2741/3448. [DOI] [PubMed] [Google Scholar]
- 26.Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- 27.Li GY, McCulloch RD, Fenton AL, Cheung M, Meng L, Ikura M, Koch CA. Structure and identification of ADP-ribose recognition motifs of APLF and role in the DNA damage response. Proc Natl Acad Sci USA. 2010;107(20):9129–9134. doi: 10.1073/pnas.1000556107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EH, Scheffzek K, Hottiger MO, Ladurner AG. A macrodomain-containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol. 2009;16:923–929. doi: 10.1038/nsmb.1664. [DOI] [PubMed] [Google Scholar]
- 29.Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- 30.Gagné JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Malanga M, Pleschke JM, Kleczkowska HE, Althaus FR. Poly(ADP-ribose) binds to specific domains of p53 and alters its DNA binding functions. J Biol Chem. 1998;273:11839–11843. doi: 10.1074/jbc.273.19.11839. [DOI] [PubMed] [Google Scholar]
- 32.Dani N, Stilla A, Marchegiani A, Tamburro A, Till S, Ladurner AG, Corda D, Di Girolamo M. Combining affinity purification by ADP-ribose-binding macro domains with mass spectrometry to define the mammalian ADP-ribosyl proteome. Proc Natl Acad Sci USA. 2009;106:4243–4248. doi: 10.1073/pnas.0900066106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riggs AD, Bourgeois S, Newby RF, Cohn M. DNA binding of the lac repressor. J Mol Biol. 1968;34:365–368. doi: 10.1016/0022-2836(68)90261-1. [DOI] [PubMed] [Google Scholar]
- 34.Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slama JT, Aboul-Ela N, Goli DM, Cheesman BV, Simmons AM, Jacobson MK. Specific inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J Med Chem. 1995;38:389–393. doi: 10.1021/jm00002a021. [DOI] [PubMed] [Google Scholar]
- 36.Slama JT, Aboul-Ela N, Jacobson MK. Mechanism of inhibition of poly(ADP-ribose) glycohydrolase by adenosine diphosphate (hydroxymethyl)pyrrolidinediol. J Med Chem. 1995;38:4332–4336. doi: 10.1021/jm00021a023. [DOI] [PubMed] [Google Scholar]
- 37.Kawamitsu H, Hoshino H, Okada H, Miwa M, Momoi H, Sugimura T. Monoclonal antibodies to poly(adenosine diphosphate ribose) recognize different structures. Biochemistry. 1984;23:3771–3777. doi: 10.1021/bi00311a032. [DOI] [PubMed] [Google Scholar]
- 38.Kawamitsu H, Hoshino H, Miwa M, Sugimura T. Monoclonal antibodies against poly(ADP-ribose) recognize different structures of poly(ADP-ribose) Princess Takamatsu Symp. 1983;13:41–47. [PubMed] [Google Scholar]
- 39.Affar EB, Duriez PJ, Shah RG, Winstall E, Germain M, Boucher C, Bourassa S, Kirkland JB, Poirier GG. Immunological determination and size characterization of poly(ADP-ribose) synthesized in vitro and in vivo. Biochim Biophys Acta. 1999;1428:137–146. doi: 10.1016/s0304-4165(99)00054-9. [DOI] [PubMed] [Google Scholar]
- 40.Panzeter PL, Realini CA, Althaus FR. Noncovalent interactions of poly(adenosine diphosphate ribose) with histones. Biochemistry. 1992;31:1379–1385. doi: 10.1021/bi00120a014. [DOI] [PubMed] [Google Scholar]
- 41.Gagné JP, Hunter JM, Labrecque B, Chabot B, Poirier GG. A proteomic approach to the identification of heterogeneous nuclear ribonucleoproteins as a new family of poly (ADP-ribose)-binding proteins. Biochem J. 2003;371:331–340. doi: 10.1042/BJ20021675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah GM, Poirier D, Duchaine C, Brochu G, Desnoyers S, Lagueux J, Verreault A, Hoflack JC, Kirkland JB, Poirier GG. Methods for biochemical study of poly(ADP-ribose) metabolism in vitro and in vivo. Anal Biochem. 1995;227:1–13. doi: 10.1006/abio.1995.1245. [DOI] [PubMed] [Google Scholar]
- 43.Fried MG. Measurement of protein-DNA interaction parameters by electrophoresis mobility shift assay. Electrophoresis. 1989;10:366–376. doi: 10.1002/elps.1150100515. [DOI] [PubMed] [Google Scholar]
- 44.Willander M, Al-Hilli S. Analysis of biomolecules using surface plasmons. Methods Mol Biol. 2009;544:201–229. doi: 10.1007/978-1-59745-483-4_14. [DOI] [PubMed] [Google Scholar]
- 45.Panzeter PL, Zweifel B, Althaus FR. Synthesis of poly(ADP-ribose)-agarose beads: an affinity resin for studying (ADP-ribose)n-protein interactions. Anal Biochem. 1992;207:157–162. doi: 10.1016/0003-2697(92)90517-b. [DOI] [PubMed] [Google Scholar]
- 46.Combet C, Blanchet C, Geourjon C, Deleage G. NPS@: network protein sequence analysis. Trends Biochem Sci. 2000;25:147–150. doi: 10.1016/s0968-0004(99)01540-6. [DOI] [PubMed] [Google Scholar]
- 47.Ménard L, Poirier GG. Rapid assay of poly(ADP-ribose) glycohydrolase. Biochem Cell Biol. 1987;65:668–673. doi: 10.1139/o87-088. [DOI] [PubMed] [Google Scholar]
- 48.Cardenas-Corona ME, Jacobson EL, Jacobson MK. Endogenous polymers of ADP-ribose are associated with the nuclear matrix. J Biol Chem. 1987;262:14863–14866. [PubMed] [Google Scholar]
- 49.Alvarez-Gonzalez R, Jacobson MK. Characterization of polymers of adenosine diphosphate ribose generated in vitro and in vivo. Biochemistry. 1987;26:3218–3224. doi: 10.1021/bi00385a042. [DOI] [PubMed] [Google Scholar]
- 50.Wielckens K, Schmidt A, George E, Bredehorst R, Hilz H. DNA fragmentation and NAD depletion. Their relation to the turnover of endogenous mono(ADP-ribosyl) and poly(ADP-ribosyl) proteins. J Biol Chem. 1982;257:12872–12877. [PubMed] [Google Scholar]
- 51.Juarez-Salinas H, Sims JL, Jacobson MK. Poly(ADP-ribose) levels in carcinogen-treated cells. Nature. 1979;282:740–741. doi: 10.1038/282740a0. [DOI] [PubMed] [Google Scholar]
- 52.Hall RA. Studying protein-protein interactions via blot overlay or Far Western blot. Methods Mol Biol. 2004;261:167–174. doi: 10.1385/1-59259-762-9:167. [DOI] [PubMed] [Google Scholar]
- 53.Kiehlbauch CC, Aboul-Ela N, Jacobson EL, Ringer DP, Jacobson MK. High resolution fractionation and characterization of ADP-ribose polymers. Anal Biochem. 1993;208:26–34. doi: 10.1006/abio.1993.1004. [DOI] [PubMed] [Google Scholar]
- 54.Isogai S, Kanno S, Ariyoshi M, Tochio H, Ito Y, Yasui A, Shirakawa M. Solution structure of a zinc-finger domain that binds to poly-ADP-ribose. Genes Cells. 2010;15:101–110. doi: 10.1111/j.1365-2443.2009.01369.x. [DOI] [PubMed] [Google Scholar]
- 55.Shuman S. Analysis of topoisomerase-DNA interactions by electrophoretic mobility shift assay. Methods Mol Biol. 2001;95:65–74. doi: 10.1385/1-59259-057-8:65. [DOI] [PubMed] [Google Scholar]
- 56.Affar EB, Duriez PJ, Shah RG, Sallmann FR, Bourassa S, Kupper JH, Burkle A, Poirier GG. Immunodot blot method for the detection of poly(ADP-ribose) synthesized in vitro and in vivo. Anal Biochem. 1998;259:280–283. doi: 10.1006/abio.1998.2664. [DOI] [PubMed] [Google Scholar]
- 57.Eustermann S, Brockmann C, Mehrotra PV, Yang JC, Loakes D, West SC, Ahel I, Neuhaus D. Solution structures of the two PBZ domains from human APLF and their interaction with poly(ADP-ribose) Nat Struct Mol Biol. 2010;17:241–243. doi: 10.1038/nsmb.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, Owen-Hughes T, Boulton SJ. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Malanga M, Althaus FR. The role of poly(ADP-ribose) in the DNA damage signaling network. Biochem Cell Biol. 2005;83:354–364. doi: 10.1139/o05-038. [DOI] [PubMed] [Google Scholar]
- 60.Althaus FR, Kleczkowska HE, Malanga M, Muntener CR, Pleschke JM, Ebner M, Auer B. Poly ADP-ribosylation: a DNA break signal mechanism. Mol Cell Biochem. 1999;193:5–11. [PubMed] [Google Scholar]


