Abstract
Low back pain is a major physical and socioeconomic problem. Degeneration of the intervertebral disc and especially that of nucleus pulposus (NP) has been linked to low back pain. In spite of much research focusing on the NP, consensus among the research community is lacking in defining the NP cell phenotype. A consensus agreement will allow easier distinguishing of NP cells from annulus fibrosus (AF) cells and endplate chondrocytes, a better gauge of therapeutic success, and a better guidance of tissue-engineering-based regenerative strategies that attempt to replace lost NP tissue. Most importantly, a clear definition will further the understanding of physiology and function of NP cells, ultimately driving development of novel cell-based therapeutic modalities. The Spine Research Interest Group at the 2014 Annual ORS Meeting in New Orleans convened with the task of compiling a working definition of the NP cell phenotype with hope that a consensus statement will propel disc research forward into the future. Based on evaluation of recent studies describing characteristic NP markers and their physiologic relevance, we make the recommendation of the following healthy NP phenotypic markers: stabilized expression of HIF-1α, GLUT-1, aggrecan/collagen II ratio >20, Shh, Brachyury, KRT18/19, CA12, and CD24.
Keywords: nucleus pulposus, intervertebral disc, cell phenotype
Low back pain (LBP) is a major crippling problem, both physically and socioeconomically. The United States alone spends between $ 100 and $ 200 billion annually in health care costs treating LBP, a condition that affects up to 80% of adults at least once during their lifetime, and has a one-month prevalence of 23.2% of the global population.1–3 Degeneration of the intervertebral disc is associated with, and has been proposed to be a cause of, a large percentage of these cases of LBP.4 Disc degeneration is a chronic, progressive disease, and current clinical strategies, including both surgical as well as non-surgical approaches aimed at symptomatic relief, have done very little to improve mental, physical, or social health of those patients affected.5,6 As a result, the research community continues to focus on understanding the molecular nature of the healthy intervertebral disc and its degeneration, with the hope that novel therapeutic and regenerative strategies will eventually be developed to more adequately treat patients with the disabling condition of excessive and pathological disc degeneration.
The intervertebral disc contains three distinct tissue compartments. The outer fibrocartilaginous annulus fibrosus (AF) and the superior and inferior cartilaginous endplates completely enclose the gel-like nucleus pulposus (NP), an avascular, aggrecan-rich tissue. The NP is frequently implicated in disc degeneration and, as such, has been the focus of much research in the basic science and tissue engineering fields. In spite of this, a consensus is lacking in defining the young healthy, NP cell phenotype. We define the NP cell phenotype as a collection of “markers”—genes, proteins, and metabolic characteristics that are representative and distinguishing of NP cells. It is important to focus on defining a phenotype, rather than merely a genotype, since phenotypic characteristics likely have physiologic consequences and dictate NP cell function. With such a definition, we will be able to more easily distinguish NP cells from AF cells and other related cell types, better diagnose degenerate conditions and gauge therapies while comparing healthy and degenerate samples, and better guide stem cell and tissue engineering strategies that attempt to replace lost NP cells and tissue. Perhaps most important, a clear identification of phenotype will allow us to better understand the physiology and function of NP cells, ultimately driving development of novel cell-based therapeutic modalities. Phenotypic characterization of NP cells at all stages of development, growth, maturation, and disease warrant further characterization and identification. This study focused on defining the phenotype of the young healthy NP cell because regenerative strategies focused on recapitulating this healthy phenotype are likely to have greatest likelihood of successful clinical outcomes. The Spine Research Interest Group at the 2014 Annual ORS Meeting in New Orleans convened with the task of discussing the phenotype of the NP cell and the hope that a consensus statement will help propel disc research forward into the future. Presented here is the outcome of that meeting and discussions that followed.
ONTOGENY AND HETEROGENEITY OF NP CELLS
There is controversy and a long-held debate over the ontogeny and heterogeneity of the cells that make up the mature, human adult NP. Early anatomical studies recognized a morphological heterogeneity of cells within the NP.7 A population of larger (25–85 μm), vacuolated cells has been considered “notochordal,”, because of their resemblance to embryonic notochord cells, whereas a population of smaller (10 μm), rounder cells has been considered “chondrocyte-like”. Fluorescence-activated cell sorting (FACS) of these two different populations has revealed differential expression of certain genes.8 Although the human embryonic notochord and juvenile NP possess populations of “notochordal” cells, these cells gradually disappear or change their appearance and are replaced by the “chondrocyte-like” cells in the adult.9,10 Different animal models have been described as possessing either “notochordal” or “chondrocyte-like” mature NP cells, and as a result, some authors have argued against using animal models with distinctly “notochordal” cells (e.g., rat, mouse, non-chondrodystrophic dogs, porcine) and rather favor animal models with distinctly “chondrocyte-like” cells (e.g., bovine, ovine, chondrodystrophic dog), due to their resemblance to mature human NP. Although these cells exhibit different morphologies, recent fate-mapping studies in mice using notochord-specific Cre recombinase driven by either Sonic hedgehog (Shh)11 or Noto12 promoters have convincingly revealed that all cells of the adult NP derive from the embryonic notochord. Additionally, “notochordal” cells of porcine,13 rabbit,14 as well as mouse,15 NP have been shown to transition into “chondrocyte-like” cells both in vitro and in vivo. This transition was favored by both physiological stimuli, such as dynamic loading 13 and standard tissue culture conditions,14 as well as pathological stimulus like needle puncture injury15 (Fig. 1). Importantly, a robust expression of brachyury, an unequivocal marker of notochord, has been seen in adult human NP tissues.16 These results suggest that morphological differences between “notochordal” and “chondrocyte-like” cells represent different stages of cellular activity or differentiation, rather than a different lineage. Although some evidence exists suggesting that during degeneration, cells from the endplate or AF may migrate into the degenerate NP,17 most convincing evidence indicates that the overwhelming majority of cells within the NP at all points have differentiated along the notochordal lineage, and morphological differences seen represent different physiological or pathological stages of aging and degeneration.10–16 Accordingly, discounting certain animal models based solely on differences in cellular morphology may not be appropriate.
Figure 1.
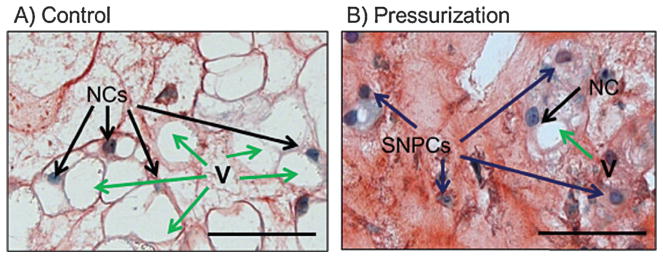
Morphology of large notochordal cells (NCs) versus small nucleus pulposus cells (SNPCs) from organ cultured porcine discs (A). Control group and (B) Pressurization group. Black arrows, NCs; blue arrows, SNPCs. A large NC was defined as nuclei in direct contact and surrounded by large vacuoles, and an SNPC was defined as nuclei surrounded by matrix with no contact with vacuoles. V, vacuoles (green arrows). Scale bar = 50 μm. Image licensed under Creative Commons Attribution License 4.0 and reproduced with permission from Purmessur D, et al. 2013. Dynamic pressurization induces transition of notochordal cells to a mature phenotype while retaining production of important patterning ligands from development. Arthritis Res. Ther. 15:R122.
This must be kept in mind when considering potential markers that define the NP phenotype. Characteristic markers with important physiological consequences and relevance would be most helpful to better understand NP cell function and subsequently guide regenerative strategies. Therefore, proposed markers that show certain interspecies variability are less likely to be physiologically important, and should perhaps not be considered critical, defining NP markers. Instead, we reviewed the literature to identify a list of markers that show no or very little interspecies differences and could be used to unequivocally describe the young healthy NP phenotype.
MARKERS THAT RELATE TO NP CELL PHYSIOLOGY
The NP resides in a unique hypoxic and hyperosmotic niche.18,19 Additionally, all cells of the NP appear to derive from the embryonic notochord, making them quite unlike any other cell type in the body.11,12,16 As a result, NP cells have developed distinguishing characteristics that adapt them well to their environment. These characteristics of distinctive protein expression, though perhaps not classified as traditional “markers” per se, should certainly not be overlooked when describing the NP phenotype. Such non-traditional markers reflect the function of the NP cell, and speak directly to its metabolism and physiology, which is critical when monitoring and assessing the success of stem cell and regenerative therapies.
Hypoxia-Inducible Factors
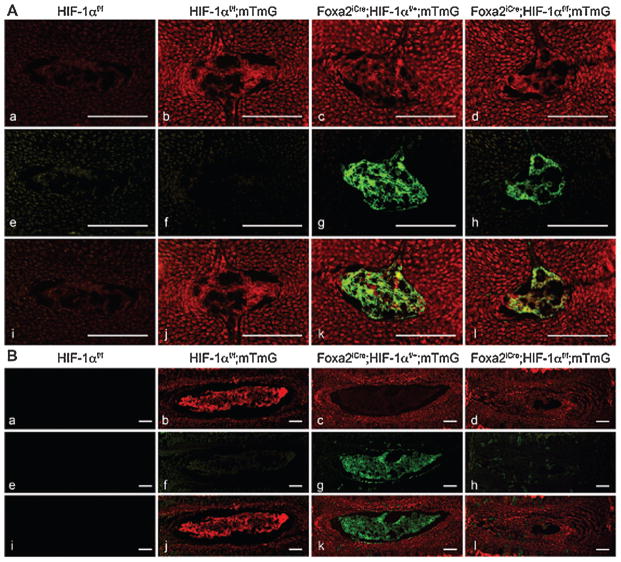
The NP is the largest avascular tissue in the body and is physiologically hypoxic.18 Cells of the body are attuned to survive hypoxic settings through expression of the transcription factors hypoxia-inducible factor-1α and -2α (HIF-1/2). In most cells of the body, HIF-α protein stability and activity are independently regulated by different molecular dioxygenases.20 HIF-α is normally degraded in the presence of oxygen via hydroxylation of specific proline residues (Pro 402 and Pro 564 in HIF-1α, and Pro 405 and Pro 531 in HIF-2α) by members of the iron-dependent prolyl-hydroxylase domain (PHD) family, and subsequent degradation through 26 S proteasome. Additionally, activity of HIF-α in the presence of oxygen is usually regulated by another molecular dioxygenase, factor inhibiting HIF-1 (FIH-1), which hydroxylates an asparagine residue (Asn 803 in HIF-1α and Asn 851 in HIF-2α) in the C-terminus transactivation domain of the protein, disrupting interaction with essential transcriptional co-activator p300/CBP. However, this standard model of HIF-α regulation does not apply to NP cells. NP cells constitutively and robustly express HIF-1α and HIF-2α, even when cultured under normoxia.21,22 Noteworthy, these HIF-α proteins are either partially or wholly refractory to PHD-mediated regulation in NP and show reliance on other non-oxemic pathways for their turnover.23–25 Of importance to the discussion of phenotypic markers, this constitutive expression of HIF-1α in NP is consistent across multiple species.22 Similarly, in NP cells FIH-1 does not control HIF-1α transcriptional activity.26 This constitutive HIF-1 activity in NP cells has important metabolic consequences - NP cells are obligate glycolytic, and rely very little on aerobic respiration, even when oxygen is abundant, e.g., under normoxic culture conditions,27 suggesting that HIF-1 strongly drives glycolytic metabolism. HIF-1/2 promotes survival and function of NP cells through upregulation of crucial genes including: glucose transporter (GLUT) 1 and 3, glyceraldehyde 3-phosphate dehydrogenase (GAPDH), aggrecan,27,28 β-1,3-glucuronyltransferase 1,29 galectin-3,30 and vascular endothelial growth factor-A (VEGF-A).21 Of note, higher expression of GLUT-1 in rat NP compared to AF and endplate chondrocytes has been shown at the protein level by Rajpurohit et al.,28 an observation supported by a later study in human tissues.31 HIF-1/2 has also been shown in NP to suppress expression of progressive ankylosis protein homolog (ANK), a pyrophosphate transporter, expression of which is sensitive to the disease process, possibly playing a role in preventing dystrophic mineralization of the NP.32 Importantly, recent work by Merceron and colleagues has conclusively demonstrated that HIF-1 is indispensable for NP cell survival.33 Conditional deletion of HIF-1α in mouse notochord by Foxa2-driven Cre recombinase results in early morphologic changes in the NP beginning at E15.5. Specifically, the mutant NP is smaller and comprises non-vacuolated cells. Postnatally, there is massive apoptotic cell death in the NP after HIF-1α deletion, likely due to energetic failure by decreased glycolysis. Eventually the NP is replaced with a fibrocartilaginous tissue with inferior biomechanical properties. Importantly, lineage-tracing studies clearly showed the notochordal lineage of the NP cells, and their replacement with non-notochordal cells, likely migrating into the tissue from the inner AF (Fig. 2). Thus, because of the absolute indispensability of HIF-1α in maintaining NP cell survival and function, the constitutive expression of HIF-α subunits should be considered a crucial and necessary marker of NP cell phenotype.
Figure 2.
Lineage study in control and mutant IVDs. A, B. Detection of fluorescence in frozen sections of NP isolated from E15.5 (A) and 1 month (B) HIF-1αf/f (a,e,i), HIF-1αf/f;mTmG (b,f,j), Foxa2iCre;HIF-1αf/+;mTmG (c,g,k) and Foxa2iCre;HIF-1αf/f;mTmG (d,h,l) mice, respectively. Red fluorescence (a–d), green fluorescence (e–h) and merged filters (i–l) are shown. Red signal is membrane-bound tdtomato fluorescent protein indicating cells in which recombination has not occurred. Green signal is membrane-bound EGFP indicating cells in which Cre mediated recombination has occurred. Bar = 100 μm. Image reproduced with permission from Merceron C, et al. 2014. Loss of HIF-1α in the Notochord Results in Cell Death and Complete Disappearance of the Nucleus Pulposus. PLoS One. 9:e110768.
Sonic Hedgehog
Sonic hedgehog (Shh) is a ligand of the hedgehog family that is expressed specifically in the developing notochord. Studies by Brian Harfe’s group using conditional deletion of both Smoothened (Smo) as well as Shh itself in Shh-expressing cells elegantly showed that this signaling pathway is required for the formation of notochordal sheath and patterning of the NP.34,35 Importantly, Shh expression remains active in the postnatal NP, at least for the first few weeks, and is required for proper functioning of NP cells.36,37 Although Shh expression and signaling are decreased with aging and degeneration, a very recent study has demonstrated that reactivation of Shh signaling in aged discs through activation of Wnt signaling can increase expression of brachyury and aggrecan, as well as accumulation of chondroitin sulfate.38 Because of its specificity in NP and promising results in reversing the aging and degenerative phenotype, Shh should be considered as a critical NP phenotypic marker.
Brachyury
Brachyury (T) is the founding-member of the T-box family of transcription factors. It is necessary for development of the embryonic mesoderm, in particular the morphogenesis of the notochord. Although previously considered only a notochordal gene and studied in a developmental context, the expression of brachyury has been recently reported in postnatal and mature NP cells using various techniques.16,39–41 Whereas one of these studies showed that, in comparison to non-chondrodystrophic dogs, in chondrodystrophic breeds “chondrocyte-like” cells showed decreased brachyury expression relative to “notochordal” cells with aging and degeneration,40 correlation with degeneration was not seen in other human studies.16,39 Of note, transition from “notochordal” to “chondrocyte-like” morphology in organ-cultured porcine discs by continuous pressurization did not involve a decrease in brachyury expression.13 Despite importance of this transcription factor in notochord development and function, direct causative studies establishing the role of brachyury exclusively in maintenance of postnatal NP have not been performed.
Relevant to this discussion of brachyury function, recent studies using chromatin-immunoprecipitation approaches in mouse embryonic stem cells42 and a human chordoma cell line43 have given insight into the direct gene targets of brachyury binding in mammals. Targets identified by these studies that are of potential importance to NP physiology include fibroblast growth factor 8 (FGF8), which has been implicated in pathogenesis of osteoarthritis44; Axin2, a negative regulator of the Wnt pathway with altered expression in degenerative NP40,45; pleiotrophin, which is expressed in NP (see below)46,47; and connective tissue growth factor (CTGF, also called CCN2), an extracellular matrix protein that is necessary for post-natal NP function48 and is also capable of suppressing the catabolic effects of IL-1β in NP cells.49 Although there are yet few studies specifically investigating the role of brachyury in the post-natal NP, it deserves attention and should nevertheless be considered as a physiologically relevant NP cell phenotype marker.
Aggrecan/Collagen II Ratio
The biomechanical role of the NP is to resist compressive loads of the spine. Hydration of the NP is responsible for resisting these compressive forces, and the extracellular matrix of the NP is responsible for maintaining hydration. During aging and degeneration, change in matrix composition is responsible for loss of hydration and subsequent exacerbation of degeneration.50–52 The NP matrix comprises proteoglycans and collagens—the major aggregating proteoglycan in the NP is aggrecan while the major collagen is collagen II.53 Although chondrocytes also express aggrecan and collagen II, the NP matrix can be distinguished from cartilage by a distinct glycosaminoglycan (GAG): hydroxyproline ratio. While the GAG:hydroxyproline ratio in juvenile cartilage is ~3:1 and decreases to ~2:1 with age, in NP the ratio is much higher at all ages. In juvenile (2–5 years) and young adult (15–25 years) NP, the ratio is ~25:1 and ~27:1, respectively. Again, the ratio decreases with age and degeneration, but not to the level of articular cartilage, reaching ~5:1 in the elderly (60–80 years).54 Because of the physiologic importance of extracellular matrix production by NP cells, aggrecan and collagen II, specifically at the high ratio (>20) seen in healthy juvenile and young adult NP, should be included in any discussion about NP phenotypic markers.
Matrix-Binding Proteins
Because of the importance of the extracellular matrix in the biological and biomechanical function of the NP, some groups have begun investigating matrix-binding proteins of NP cells, namely integrins. Studies have shown elevated expression in porcine and human NP cells compared to AF cells of integrins α3, α6, and β4, and laminin α5.55,56 Integrins α3β4 and α6β4 bind laminin, suggesting a high level of integrin-lamin binding in NP cells. Follow-up studies observed NP-specific expression of laminin-111, -511, and -322, and that NP cells adhere strongly to these laminins, suggesting their crucial role in cell-matrix interactions.57,58 Although in the porcine NP integrin α6β1 mediates laminin-111 attachment, in human NP this interaction is mediated by integrins α3β1 and α5β1.59 This interspecies variability suggests that although cell-matrix adhesion may play a major role in NP cell function and development, the exact underlying nature of this interaction maybe not be as critical.
Worth noting are observations that α5β1 expression increased in human lumbar herniated discs,60 and that the anti-catabolic role of CCN2 in suppressing effects of IL-1β requires CCN2 binding to α5β1 and αvβ3 integrins.49 Additionally, it is known that the vacuolated notochordal phenotype of NP cells is very sensitive to the surrounding microenvironment in culture,61 and differences in cell-matrix connectivity of the notochordal NP cells is culture functionally impact the cells they contact.62 As more physiological roles of integrins in NP cells are elucidated, especially those that exhibit an NP-distinctive expression pattern, the more important they will become in the discussion of NP phenotypic markers.
“-OMICS” APPROACHES AND PROGENITOR STUDIES
Because of the wealth of data microarrays provide, it would be prudent to use results of NP cell microarray profiling to help define the NP phenotype. We must be wary and recognize, however, that protein and mRNA expression do not always correlate, and all proposed markers described from transcript expression profiles should indeed be validated and informed by protein level measurements.
One of the earliest studies using microarray to analyze NP cells specifically looked for differential expression of cell surface markers and found CD24 as such a marker.63 Another important early use of microarray was reported by Lee et al.46 Comparing NP cells to AF cells and articular chondrocytes harvested from rats, the authors described genes with significantly higher mRNA expression in NP cells, including annexin A3, the cell-surface heparan sulfate proteoglycan glypican-3 (GPC3), keratin-19 (KRT19), and pleiotrophin (PTN). Authors did validate differences in GPC3 and KRT19 expression at the protein level through immunohistochemistry. Interestingly, Rutges et al. in a subsequent analysis of human tissues observed that pleiotrophin was more highly enriched in articular chondrocytes than NP cells.47
Noteworthy, a follow-up study by the same group using a dog model could not replicate the significant differential expression for GPC3 and KRT19 seen in rat.64 Instead, when comparing NP to AF, they found significantly higher expression of α2-macroglobulin (A2M), desmocollin-2 (DSC2), CD56, and keratin-18 (KRT18), all of which were confirmed at the protein level with immunohistochemistry. Of potential importance to disc physiology, α2-macroglobulin is a protease inhibitor, and has been shown to play a role in osteoarthritis prevention.65 Specifically, α2-macroglobulin has been proposed as an endogenous inhibitor of the aggrecanases ADAMTS-4 and -5, as well as MMP-13, all of which exhibit increased expression during degenerative disc disease.53 While an elevated A2M gene expression in NP compared to AF cells was also reproduced in human disc cells,47 it must be noted, however, that Güner et al. were unable to show protein expression of α2-macroglobulin in human surgical disc specimens using rate nephelometry techniques.66 This therefore suggests that expression and physiological relevance of α2-macroglobulin requires further investigation.
A microarray study by Minogue et al. using bovine NP cells39 found NP-specific expression of several genes compared to AF, including keratin-8, -18, and -19, the t-SNARE protein SNAP25, cadherin-2 (CDH2), brain acid soluble protein 1 (BASP1), and sclerostin domain-containing protein 1 (SOSTDC1). The authors were able to validate differential expression of CDH2 by qPCR in human NP cells compared to human AF and articular chondrocytes; expression at the protein level, however, was not investigated. Still worth noting is the observation that SNAP25, KRT-8, -18, and CDH2 mRNA expression decreased in degenerate human NP cells. A follow-up microarray study from the same group using human NP cells67 compared expression between NP cells and articular chondrocytes and found NP-specific mRNA expression of transcription factors PAX1 and FOXF1, hemoglobin β-chain (HBB), carbonic anhydrase XII (CA12), and ovostatin 2 (OVOS2).
A later study by Power et al. independently validated NP-specific expression of carbonic anhydrase XII by both microarray and immunohistochemistry and showed strong disease-dependence of expression levels.68 Carbonic anhydrase proteins are involved in acid-base balance. Because of the avascular and glycolytic nature of the NP, there is elevated lactic acid found in the tissue.69 It is therefore expected that acid-base physiology plays an important role in NP cell survival and function. Carbonic anhydrase XII may be a key player in buffering acidic environments, and, as such, should be considered an NP marker and should be investigated further for its contribution to NP cell survival. Also worth noting, an early study by Lyons et al. observed robust expression of carbonic anhydrase III in mouse notochord and postnatal NP, a CA family member previously considered to be tissue-specific with expression predominantly limited to skeletal muscle.70 Although the authors also found prenatal expression of CAIII transcript in a few other non-muscle tissues, post-partum expression was only reported in NP and brown fat, with protein expression in NP equal to that of hindlimb muscle at two days post-partum. Despite no recent gene profiling techniques recommending carbonic anhydrase III as an NP marker, its robust expression at the protein level, like carbonic anhydrase XII, suggests physiologic importance. As such, carbonic anhydrase III should perhaps enter into the NP marker discussion.
A recent study used immortalized human NP cell lines to uncover “subpopulations” within the NP -different groups of cells with different expression levels of NP-specific genes.71 The authors described two subpopulations of NP cells, based on their ability to respond to a chondrocytic differentiation protocol and increase expression of Sox9 and collagen II. The authors tested expression of six genes described above—KRT19, CA12, CD24, FoxF1, PAX1, and PTN. Importantly, both subpopulations of cells expressed all markers, supporting the argument that all NP cells have similar ontogeny, and morphological differences seen between different populations represent different stages of development. In spite of the fact that both subpopulations expressed all of these proposed markers, there were significant differences in CA12 and FoxF1 expression between the two subpopulations before differentiation, and between FoxF1 and PTN expression after differentiation. Although future studies may elucidate potential physiological consequences caused by these differences, it is again important to consider the expression of all genes at the protein level. Just as is true when comparing cells of different species, variability between subpopulations of cells within a single species may show differences at the transcript level but not at the protein level, which are much more tightly regulated.72
A study from Dick Heinegård’s lab used quantitative proteomics approaches to analyze musculoskeletal tissues, including NP, AF, and articular cartilage. Of note, they reported significantly higher lubricin (PRG4) expression in human NP compared to articular cartilage of the femoral and humeral head (~9-fold),73 supporting an earlier study reporting lubricin expression in human cadaveric NP tissue.74 Additionally, there was a trend of higher lubricin expression in NP when compared directly to AF (p = 0.09). Lubricin is a highly conserved proteoglycan that is often described in the context of synovial joints, implicated in reducing shear stress, inflammation, and apoptosis, and maintenance of joint health.75 The intervertebral disc shares many properties with synovial joints, to such an extent that some argue the spinal motion segment should be re-classified as a polyaxial diarthrosis, rather than as an amphiarthrosis, as it is often currently described.76 Therefore, while the physiologic role of lubricin is yet to be elucidated in the NP, it is likely to have substantial relevance, and is certainly worth future investigation.
An important study from Sakai et al. identified a population of progenitor cells within the NP compartment.77 The study observed that progenitor cells change expression of specific cell-surface markers sequentially from angiopoeitin-1 receptor (Tie2) positive, to disialoganglioside 2 (GD2) positive, to CD24 positive cells as they differentiate and lose proliferative capacity. Additionally, as reported earlier,78 NP cells at all stages of differentiation showed positivity for CD44, CD49f, CD56, CD73, CD90, CD105 and CD166, which is helpful for FACS applications. Importantly, although Tie2 positive progenitors were found in human discs, the number of Tie2 positive cells decreased with aging and degeneration. These markers will certainly have an impact on future regenerative strategies, since they help define and identify a specific precursor cell subpopulation within the NP.
This discussion would not be complete unless we consider the potential change in NP cell phenotype with age. Long has it been known that degeneration of the NP seen with aging is associated with a shift in balance from anabolism to catabolism, including decreased production of aggrecan and collagen II, increased production of several MMP and ADAMTS enzymes, and increased cytokine production.53,79–82 For tissue engineering and regenerative strategies, it is therefore important to achieve a young healthy NP cell phenotype, rather than an aged, degenerated phenotype, to allow for the optimal outcome. Recently, groups have focused on unbiased approaches to better understand the changes in gene expression seen with aging. Tang et al.83 demonstrated an increase in expression with age of BASP1 in rat NP, confirming its NP cell-specific expression as previously reported,39 as well as an increase in neurochondrin and CD155. Interestingly, the authors saw no difference in T expression between aged and immature rat NP cells. The authors additionally identified NP-cell specific markers neuropilin-1 and CD221 through their microarray analysis.
Very recent studies have used bioinformatics approaches to identify specific gene networks that change with aging. It was noted that differentially expressed genes with aging and degeneration are associated with membrane-bound vesicles, calcium-ion binding,84 MAPK and Rho families,85 and TGF-β and extracellular matrix networks, particularly focused around MMP2.86 While important for understanding the pathogenesis of degeneration, the usefulness of such studies in this discussion is limited due to the lack of NP-cell specificity of these proteins.
CONCLUSIONS
The current literature evaluates the NP cell phenotype using several techniques and a variety of species in development and aging in order to provide primary phenotypic markers (Table 1) with greater consensus and secondary phenotypic markers (Table 2) that have been less well validated. Until we validate more proposed targets at the protein level or employ more large-scale proteomics approaches, we must rely on proposed markers with physiologic importance that have not only been investigated through gene expression studies but also validated at the protein level using multiple methods. We therefore suggest that markers including stabilized expression of HIF-1α, GLUT-1, aggrecan/collagen II ratio >20, Shh, Brachyury, KRT18/19, carbonic anhydrase XII, and CD24 should be used to characterize and define the phenotype of the healthy NP cell. These markers are not an exhaustive list but are characteristic of and relevant to healthy NP cells. By focusing on these cellular markers as a starting point when attempting to repair a degenerate or aged NP, we can better hone therapeutic strategies to successfully develop and restore a healthy NP phenotype. Although few recent studies have used a collection of some of these markers to assess differentiation potential of iPSCs87,88 and MSCs89,90 into an NP-cell phenotype, due to the lack of a clear phenotypic definition of NP cells, some studies still focus only on expression of collagen II, aggrecan, and Sox9.91,92 While some studies have reported promising results, it is still unclear, given the unique notochordal origin of NP cells, whether mesoderm-derived MSCs will be fully capable of differentiating in the NP cell lineage. Furthermore, it is only after rigorous testing of these therapeutic strategies that we will better understand which phenotypic markers are sufficient for healthy NP function. Therefore, the phenotypic definition of NP markers presented here can help serve as an initial guideline to assess the success of MSC-driven cell therapies. We hope that a consensus among the research community for use of the above markers to characterize the NP cell phenotype will help drive NP cell research into the future.
Table 1.
Proposed Primary Markersa of Young Healthy NP Cells and Their Relevance to NP Cell Physiology
| Identified NP phenotypic marker | Relevance to NP physiology | Species | Method |
|---|---|---|---|
| Stabilized HIF-1/2α | Transactivate many pro-survival genes in NP; absolutely necessary for post-natal NP cell survival | Human, mouse rat, sheep21,22,27 | Western blot, IF, IHC, qPCR |
| GLUT-1 | Glucose transporter expressed in hypoxic tissues; expression controlled by HIF-1 | Human, rat28,31 | Western blot, qPCR, IHC |
| Shh | Signaling ligand necessary for post-natal function of NP cells | Human, mouse36–38 | Western blot, IF, in situ hybridization |
| Brachyury (T) | Transcription factor necessary for notochordal morphogenesis and patterning | Human, mouse, dog, bovine16,39–41 | Western blot, IHC, in situ hybridization, microarray, qPCR, flow cytometry |
| Aggrecan /collagen II ratio >20 | High PG content maintains hydration to resist loads | Human, many others53,54 | DMMB/DMAB, qPCR, Western blot, IHC |
| Carbonic anhydrase 3/12 | Acid-base balance | Human, mouse67,68,70 | Microarray, qPCR, IHC, Western blot |
| Microarray studies | |||
| CD24 | Unknown | Human, rat, mouse63,77,78 | Microarray, flow cytometry, IHC, qPCR |
| Cytokeratins 8, 18, and 19 | Cellular structural integrity and possibly signaling | Human, rat, bovine, dog39,46,64 | Microarray, IHC |
Primary markers were chosen based on criteria including: (i) specific expression in young healthy NP cells, (ii) requirement for proper NP cell function and relevance to NP cell physiology, and (iii) mRNA and protein expression validated across different species.
Table 2.
Proposed Secondary Markersa of Young Healthy NP and NP Progenitor Cells and Their Relevance to NP Cell Physiology
| Identified NP phenotypic marker | Relevance to NP physiology | Species | Method |
|---|---|---|---|
| Integrins α3, α6, β4 | Cell-matrix adhesion | Human, porcine55,56 | Flow cytometry, IHC |
| Microarray and proteomics studies | |||
| Annexin A3 | Unknown | Rat46 | Microarray |
| Glypican 3 | Rat46 | Microarray | |
| α2-macroglobulin | Human, dog47,64 | Microarray | |
| Desmocollin-2 | Dog64 | Microarray, IHC | |
| CD56 | Dog, mouse64,77 | Microarray, flow cytometry, IHC | |
| SNAP25 | Bovine, human39 | Microarray, qPCR | |
| CDH2 | Bovine, human39 | Microarray, qPCR | |
| BASP1 | Bovine, rat39,83 | Microarray | |
| SOSTDC1 | Bovine39 | Microarray | |
| PAX1 | Human67,71 | Microarray, qPCR | |
| FOXF1 | Human67,71 | Microarray, qPCR | |
| Hemoglobin β-chain | Human67 | Microarray | |
| Ovostatin | Human67 | Microarray | |
| Neurochondrin | Rat79 | Microarray, qPCR, IHC | |
| Neuropilin-1 | Rat79 | Microarray, IHC | |
| CD155 | Rat79 | Microarray | |
| CD221 | Rat79 | Microarray, IHC | |
| Lubricin (PRG4) | Human73,74 | IHC, LC-MS/MS | |
| Progenitor markers | |||
| Tie2 | Receptor for angiopoietin-1; drives proliferation of progenitor cells | Human, mouse77 | IF, flow cytometry |
| GD2 | Unknown | Human, mouse77 | IF, flow cytometry |
Secondary markers were identified in the literature but less well validated for young healthy NP cells than primary markers.
Acknowledgments
Grant sponsor: National Institutes of Arthritis and Musculoskeletal and Skin Diseases; Grant numbers: R01AR055655 (MVR), R01AR064733 (MVR), R01AR050087 (MVR), T32AR052273 (ZRS), R01AR064157 (JCI), R01AR057397 (JCI); Grant sponsor: CIHR (RAK).
Authors wish to acknowledge ORS for supporting Spine Research Interest Group at its annual meetings since 2011. We wish to thank all those who actively contributed and participated in discussions at the 2014 RIG and Robert Hartman, University of Pittsburgh, for his efforts in conducting pre-RIG surveys. The authors would additionally like to thank Devina Purmessur for her kind assistance providing histologic images. M.V.R is supported by grants from National Institutes of Arthritis and Musculoskeletal and Skin Diseases R01AR055655, R01AR064733, and R01AR050087. Z.R.S is supported by T32AR052273. R.A.K is supported by CIHR. J.C.I is supported by grants from National Institutes of Arthritis and Musculoskeletal and Skin Diseases R01AR064157 and R01AR057397.
References
- 1.Katz JN. Lumbar disc disorders and low-back pain: socioeconomic factors and consequences. J Bone Joint Surg Am. 2006;88 (Suppl 2):21–24. doi: 10.2106/JBJS.E.01273. [DOI] [PubMed] [Google Scholar]
- 2.Shvartzman L, Weingarten E, Sherry H, et al. Cost-effectiveness analysis of extended conservative therapy versus surgical intervention in the management of herniated lumbar intervertebral disc. Spine (Phila Pa 1976) 1992;17:176–182. doi: 10.1097/00007632-199202000-00010. [DOI] [PubMed] [Google Scholar]
- 3.Hoy D, Bain C, Williams G, et al. A systematic review of the global prevalence of low back pain. Arthritis Rheum. 2012;64:2028–2037. doi: 10.1002/art.34347. [DOI] [PubMed] [Google Scholar]
- 4.Freemont AJ, Peacock TE, Goupille P, et al. Nerve ingrowth into diseased intervertebral disc in chronic back pain. Lancet. 1997;350:178–181. doi: 10.1016/s0140-6736(97)02135-1. [DOI] [PubMed] [Google Scholar]
- 5.Martin BI, Deyo RA, Mirza SK, et al. Expenditures and health status among adults with back and neck problems. JAMA. 2008;299:656–664. doi: 10.1001/jama.299.6.656. [DOI] [PubMed] [Google Scholar]
- 6.Williams CM, Maher CG, Latimer J, et al. Efficacy of paracetamol for acute low-back pain: a double-blind, randomised controlled trial. Lancet. 2014 doi: 10.1016/S0140-6736(14)60805-9. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 7.Trout JJ, Buckwalter JA, Moore KC, et al. Ultrastructure of the human intervertebral disc. Tissue Cell. 1982;14:359–369. doi: 10.1016/0040-8166(82)90033-7. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Yan W, Setton LA. Molecular phenotypes of notochordal cells purified from immature nucleus pulposus. Eur Spine J. 2006;15(Suppl 3):S303–S311. doi: 10.1007/s00586-006-0088-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gilson A, Dreger M, Urban JP. Differential expression level of cytokeratin 8 in cells of the bovine nucleus pulposus complicates the search for specific intervertebral disc cell markers. Arthritis Res Ther. 2010;12:R24. doi: 10.1186/ar2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weiler C, Nerlich AG, Schaaf R, et al. Immunohistochemical identification of notochordal markers in cells in the aging human lumbar intervertebral disc. Eur Spine J. 2010;19:1761–1770. doi: 10.1007/s00586-010-1392-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi K-S, Cohn MJ, Harfe BD. Identification of nucleus pulposus precursor cells and notochordal remnants in the mouse: implications for disk degeneration and chordoma formation. Dev Dyn. 2008;237:3953–3958. doi: 10.1002/dvdy.21805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCann MR, Tamplin OJ, Rossant J, et al. Tracing notochord-derived cells using a Noto-cre mouse: implications for intervertebral disc development. Dis Model Mech. 2012;5:73–82. doi: 10.1242/dmm.008128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Purmessur D, Guterl CC, Cho SK, et al. Dynamic pressurization induces transition of notochordal cells to a mature phenotype while retaining production of important patterning ligands from development. Arthritis Res Ther. 2013;15:R122. doi: 10.1186/ar4302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim JH, Deasy BM, Seo HY, et al. Differentiation of intervertebral notochordal cells through live automated cell imaging system in vitro. Spine (Phila Pa 1976) 2009;34:2486–2493. doi: 10.1097/BRS.0b013e3181b26ed1. [DOI] [PubMed] [Google Scholar]
- 15.Yang F, Leung VYL, Luk KDK, et al. Injury-induced sequential transformation of notochordal nucleus pulposus to chondrogenic and fibrocartilaginous phenotype in the mouse. J Pathol. 2009;218:113–121. doi: 10.1002/path.2519. [DOI] [PubMed] [Google Scholar]
- 16.Risbud MV, Shapiro IM. Notochordal cells in the adult intervertebral disc: new perspective on an old question. Crit Rev Eukaryot Gene Expr. 2011;21:29–41. doi: 10.1615/critreveukargeneexpr.v21.i1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim K-W, Lim T-H, Kim JG, et al. The origin of chondrocytes in the nucleus pulposus and histologic findings associated with the transition of a notochordal nucleus pulposus to a fibrocartilaginous nucleus pulposus in intact rabbit intervertebral discs. Spine (Phila Pa 1976) 2003;28:982–990. doi: 10.1097/01.BRS.0000061986.03886.4F. [DOI] [PubMed] [Google Scholar]
- 18.Risbud MV, Schipani E, Shapiro IM. Hypoxic regulation of nucleus pulposus cell survival: from niche to notch. Am J Pathol. 2010;176:1577–1583. doi: 10.2353/ajpath.2010.090734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson ZI, Shapiro IM, Risbud MV. Extracellular Osmolarity Regulates Matrix Homeostasis in the Intervertebral Disc and Articular Cartilage: Evolving role of TonEBP. Matrix Biol. 2014 doi: 10.1016/j.matbio.2014.08.014. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semenza GL. Oxygen sensing, hypoxia-inducible factors, and disease pathophysiology. Annu Rev Pathol. 2014;9:47–71. doi: 10.1146/annurev-pathol-012513-104720. [DOI] [PubMed] [Google Scholar]
- 21.Agrawal A, Gajghate S, Smith H, et al. Cited2 modulates hypoxia-inducible factor-dependent expression of vascular endothelial growth factor in nucleus pulposus cells of the rat intervertebral disc. Arthritis Rheum. 2008;58:3798–3808. doi: 10.1002/art.24073. [DOI] [PubMed] [Google Scholar]
- 22.Risbud MV, Guttapalli A, Stokes DG, et al. Nucleus pulposus cells express HIF-1 alpha under normoxic culture conditions: a metabolic adaptation to the intervertebral disc microenvironment. J Cell Biochem. 2006;98:152–159. doi: 10.1002/jcb.20765. [DOI] [PubMed] [Google Scholar]
- 23.Fujita N, Chiba K, Shapiro IM, et al. HIF-1α and HIF-2α degradation is differentially regulated in nucleus pulposus cells of the intervertebral disc. J Bone Miner Res. 2012;27:401–412. doi: 10.1002/jbmr.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fujita N, Markova D, Anderson DG, et al. Expression of prolyl hydroxylases (PHDs) is selectively controlled by HIF-1 and HIF-2 proteins in nucleus pulposus cells of the intervertebral disc: distinct roles of PHD2 and PHD3 proteins in controlling HIF-1α activity in hypoxia. J Biol Chem. 2012;287:16975–16986. doi: 10.1074/jbc.M111.334466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gogate SS, Fujita N, Skubutyte R, et al. Tonicity enhancer binding protein (TonEBP) and hypoxia-inducible factor (HIF) coordinate heat shock protein 70 (Hsp70) expression in hypoxic nucleus pulposus cells: role of Hsp70 in HIF-1α degradation. J Bone Miner Res. 2012;27:1106–1117. doi: 10.1002/jbmr.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirose Y, Johnson ZI, Schoepflin ZR, et al. FIH-1-Mint3 Axis Does Not Control HIF-1α Transcriptional Activity in Nucleus Pulposus Cells. J Biol Chem. 2014;289:20594–20605. doi: 10.1074/jbc.M114.565101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Agrawal A, Guttapalli A, Narayan S, et al. Normoxic stabilization of HIF-1α drives glycolytic metabolism and regulates aggrecan gene expression in nucleus pulposus cells of the rat intervertebral disk. Am J Physiol Cell Physiol. 2007;293:C621–C631. doi: 10.1152/ajpcell.00538.2006. [DOI] [PubMed] [Google Scholar]
- 28.Rajpurohit R, Risbud MV, Ducheyne P, et al. Phenotypic characteristics of the nucleus pulposus: expression of hypoxia inducing factor-1, glucose transporter-1 and MMP-2. Cell Tissue Res. 2002;308:401–407. doi: 10.1007/s00441-002-0563-6. [DOI] [PubMed] [Google Scholar]
- 29.Gogate SS, Nasser R, Shapiro IM, et al. Hypoxic regulation of β-1,3-glucuronyltransferase 1 expression in nucleus pulposus cells of the rat intervertebral disc: role of hypoxia-inducible factor proteins. Arthritis Rheum. 2011;63:1950–1960. doi: 10.1002/art.30342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Y, Danielson KG, Albert TJ, et al. HIF-1 alpha is a regulator of galectin-3 expression in the intervertebral disc. J Bone Miner Res. 2007;22:1851–1861. doi: 10.1359/jbmr.070620. [DOI] [PubMed] [Google Scholar]
- 31.Richardson SM, Knowles R, Tyler J, et al. Expression of glucose transporters GLUT-1, GLUT-3, GLUT-9 and HIF-1alpha in normal and degenerate human intervertebral disc. Histochem Cell Biol. 2008;129:503–511. doi: 10.1007/s00418-007-0372-9. [DOI] [PubMed] [Google Scholar]
- 32.Skubutyte R, Markova D, Freeman TA, et al. Hypoxia-inducible factor regulation of ANK expression in nucleus pulposus cells: possible implications in controlling dystrophic mineralization in the intervertebral disc. Arthritis Rheum. 2010;62:2707–2715. doi: 10.1002/art.27558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merceron C, Mangiavini L, Robling A, et al. Loss of HIF-1a in the Notochord Results in Cell Death and Complete Disappearance of the Nucleus Pulposus. PLoS One. 2014;9:e110768. doi: 10.1371/journal.pone.0110768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi K-S, Harfe BD. Hedgehog signaling is required for formation of the notochord sheath and patterning of nuclei pulposi within the intervertebral discs. Proc Natl Acad Sci US A. 2011;108:9484–9489. doi: 10.1073/pnas.1007566108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi K-S, Lee C, Harfe BD. Sonic hedgehog in the notochord is sufficient for patterning of the intervertebral discs. Mech Dev. 2012;129:255–262. doi: 10.1016/j.mod.2012.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dahia CL, Mahoney E, Wylie C. Shh signaling from the nucleus pulposus is required for the postnatal growth and differentiation of the mouse intervertebral disc. PLoS One. 2012;7:e35944. doi: 10.1371/journal.pone.0035944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dahia CL, Mahoney EJ, Durrani AA, et al. Intercellular signaling pathways active during intervertebral disc growth, differentiation, and aging. Spine (Phila Pa 1976) 2009;34:456–462. doi: 10.1097/BRS.0b013e3181913e98. [DOI] [PubMed] [Google Scholar]
- 38.Winkler T, Mahoney EJ, Sinner D, et al. Wnt signaling activates shh signaling in early postnatal intervertebral discs, and reactivates shh signaling in old discs in the mouse. PLoS One. 2014;9:98444. doi: 10.1371/journal.pone.0098444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Minogue BM, Richardson SM, Zeef LA, et al. Transcriptional profiling of bovine intervertebral disc cells: implications for identification of normal and degenerate human intervertebral disc cell phenotypes. Arthritis Res Ther. 2010;12:R22. doi: 10.1186/ar2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smolders LA, Meij BP, Riemers FM, et al. Canonical Wnt signaling in the notochordal cell is upregulated in early intervertebral disk degeneration. J Orthop Res. 2012;30:950–957. doi: 10.1002/jor.22000. [DOI] [PubMed] [Google Scholar]
- 41.Maier JA, Lo Y, Harfe BD. Foxa1 and Foxa2 are required for formation of the intervertebral discs. PLoS One. 2013;8:e55528. doi: 10.1371/journal.pone.0055528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Evans AL, Faial T, Gilchrist MJ, et al. Genomic targets of Brachyury (T) in differentiating mouse embryonic stem cells. PLoS One. 2012;7:e33346. doi: 10.1371/journal.pone.0033346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nelson AC, Pillay N, Henderson S, et al. An integrated functional genomics approach identifies the regulatory network directed by brachyury (T) in chordoma. J Pathol. 2012;228:274–285. doi: 10.1002/path.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Uchii M, Tamura T, Suda T, et al. Role of fibroblast growth factor 8 (FGF8) in animal models of osteoarthritis. Arthritis Res Ther. 2008;10:R90. doi: 10.1186/ar2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Smolders LA, Meij BP, Onis D, et al. Gene expression profiling of early intervertebral disc degeneration reveals a down-regulation of canonical Wnt signaling and caveolin-1 expression: implications for development of regenerative strategies. Arthritis Res Ther. 2013;15:R23. doi: 10.1186/ar4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee CR, Sakai D, Nakai T, et al. A phenotypic comparison of intervertebral disc and articular cartilage cells in the rat. Eur Spine J. 2007;16:2174–2185. doi: 10.1007/s00586-007-0475-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rutges J, Creemers LB, Dhert W, et al. Variations in gene and protein expression in human nucleus pulposus in comparison with annulus fibrosus and cartilage cells: potential associations with aging and degeneration. Osteoarthritis Cartilage. 2010;18:416–423. doi: 10.1016/j.joca.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 48.Bedore J, Sha W, McCann MR, et al. Impaired intervertebral disc development and premature disc degeneration in mice with notochord-specific deletion of CCN2. Arthritis Rheum. 2013;65:2634–2644. doi: 10.1002/art.38075. [DOI] [PubMed] [Google Scholar]
- 49.Tran CM, Schoepflin ZR, Markova DZ, et al. CCN2 suppresses catabolic effects of interleukin-1β through α5β1 and αVβ3 integrins in nucleus pulposus cells: implications in intervertebral disc degeneration. J Biol Chem. 2014;289:7374–7387. doi: 10.1074/jbc.M113.526111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Murakami H, Yoon TS, Attallah-Wasif ES, et al. Quantitative differences in intervertebral disc-matrix composition with age-related degeneration. Med Biol Eng Comput. 2010;48:469–474. doi: 10.1007/s11517-010-0586-1. [DOI] [PubMed] [Google Scholar]
- 51.Antoniou J, Steffen T, Nelson F, et al. The human lumbar intervertebral disc: evidence for changes in the biosynthesis and denaturation of the extracellular matrix with growth, maturation, ageing, and degeneration. J Clin Invest. 1996;98:996–1003. doi: 10.1172/JCI118884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Iatridis JC, MacLean JJ, O’Brien M, et al. Measurements of proteoglycan and water content distribution in human lumbar intervertebral discs. Spine (Phila Pa 1976) 2007;32:1493–1497. doi: 10.1097/BRS.0b013e318067dd3f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Le Maitre CL, Pockert A, Buttle DJ, et al. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35:652–655. doi: 10.1042/BST0350652. [DOI] [PubMed] [Google Scholar]
- 54.Mwale F, Roughley P, Antoniou J. Distinction between the extracellular matrix of the nucleus pulposus and hyaline cartilage: a requisite for tissue engineering of intervertebral disc. Eur Cell Mater. 2004;8:58–63. doi: 10.22203/ecm.v008a06. [DOI] [PubMed] [Google Scholar]
- 55.Chen J, Jing L, Gilchrist CL, et al. Expression of laminin isoforms, receptors, and binding proteins unique to nucleus pulposus cells of immature intervertebral disc. Connect Tissue Res. 2009;50:294–306. [PMC free article] [PubMed] [Google Scholar]
- 56.Nettles DL, Richardson WJ, Setton LA. Integrin expression in cells of the intervertebral disc. J Anat. 2004;204:515–520. doi: 10.1111/j.0021-8782.2004.00306.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gilchrist CL, Francisco AT, Plopper GE, et al. Nucleus pulposus cell-matrix interactions with laminins. Eur Cell Mater. 2011;21:523–532. doi: 10.22203/ecm.v021a39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilchrist CL, Chen J, Richardson WJ, et al. Functional integrin subunits regulating cell-matrix interactions in the intervertebral disc. J Orthop Res. 2007;25:829–840. doi: 10.1002/jor.20343. [DOI] [PubMed] [Google Scholar]
- 59.Bridgen DT, Gilchrist CL, Richardson WJ, et al. Integrin-mediated interactions with extracellular matrix proteins for nucleus pulposus cells of the human intervertebral disc. J Orthop Res. 2013;31:1661–1667. doi: 10.1002/jor.22395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xia M, Zhu Y. Expression of integrin subunits in the herniated intervertebral disc. Connect Tissue Res. 2008;49:464–469. doi: 10.1080/03008200802325425. [DOI] [PubMed] [Google Scholar]
- 61.Spillekom S, Smolders LA, Grinwis GCM, et al. Increased Osmolarity and Cell Clustering Preserve Canine Notochordal Cell Phenotype in Culture. Tissue Eng Part C Methods. 2014;20:652–662. doi: 10.1089/ten.TEC.2013.0479. [DOI] [PubMed] [Google Scholar]
- 62.Purmessur D, Schek RM, Abbott RD, et al. Notochordal conditioned media from tissue increases proteoglycan accumulation and promotes a healthy nucleus pulposus phenotype in human mesenchymal stem cells. Arthritis Res Ther. 2011;13:R81. doi: 10.1186/ar3344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujita N, Miyamoto T, Imai J, et al. CD24 is expressed specifically in the nucleus pulposus of intervertebral discs. Biochem Biophys Res Commun. 2005;338:1890–1896. doi: 10.1016/j.bbrc.2005.10.166. [DOI] [PubMed] [Google Scholar]
- 64.Sakai D, Nakai T, Mochida J, et al. Differential phenotype of intervertebral disc cells: microarray and immunohistochemical analysis of canine nucleus pulposus and anulus fibrosus. Spine (Phila Pa 1976) 2009;34:1448–1456. doi: 10.1097/BRS.0b013e3181a55705. [DOI] [PubMed] [Google Scholar]
- 65.Wan R, Hu J, Zhou Q, et al. Application of co-expressed genes to articular cartilage: new hope for the treatment of osteoarthritis (review) Mol Med Rep. 2012;6:16–18. doi: 10.3892/mmr.2012.859. [DOI] [PubMed] [Google Scholar]
- 66.Güner A, Oktay G, Kerman M, et al. Immunoglobulins and alpha-1-proteinase inhibitor in human intervertebral disc material. Biochem Soc Trans. 1995;23:212S. doi: 10.1042/bst023212s. [DOI] [PubMed] [Google Scholar]
- 67.Minogue BM, Richardson SM, Zeef LAH, et al. Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum. 2010;62:3695–3705. doi: 10.1002/art.27710. [DOI] [PubMed] [Google Scholar]
- 68.Power KA, Grad S, Rutges JPHJ, et al. Identification of cell surface-specific markers to target human nucleus pulposus cells: expression of carbonic anhydrase XII varies with age and degeneration. Arthritis Rheum. 2011;63:3876–3886. doi: 10.1002/art.30607. [DOI] [PubMed] [Google Scholar]
- 69.Bartels EM, Fairbank JC, Winlove CP, et al. Oxygen and lactate concentrations measured in vivo in the intervertebral discs of patients with scoliosis and back pain. Spine (Phila Pa 1976) 1998;23:1–7. doi: 10.1097/00007632-199801010-00001. [DOI] [PubMed] [Google Scholar]
- 70.Lyons GE, Buckingham ME, Tweedie S, et al. Carbonic anhydrase III, an early mesodermal marker, is expressed in embryonic mouse skeletal muscle and notochord. Development. 1991;111:233–244. doi: 10.1242/dev.111.1.233. [DOI] [PubMed] [Google Scholar]
- 71.Van den Akker GG, Surtel DA, Cremers A, et al. Novel immortal human cell lines reveal subpopulations in the nucleus pulposus. Arthritis Res Ther. 2014;16:R135. doi: 10.1186/ar4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. 2012;13:227–232. doi: 10.1038/nrg3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Önnerfjord P, Khabut A, Reinholt FP, et al. Quantitative proteomic analysis of eight cartilaginous tissues reveals characteristic differences as well as similarities between subgroups. J Biol Chem. 2012;287:18913–18924. doi: 10.1074/jbc.M111.298968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shine KM, Simson JA, Spector M. Lubricin distribution in the human intervertebral disc. J Bone Joint Surg Am. 2009;91:2205–2212. doi: 10.2106/JBJS.H.01344. [DOI] [PubMed] [Google Scholar]
- 75.Jay GD, Waller KA. The biology of Lubricin: Near frictionless joint motion. Matrix Biol. 2014;39C:17–24. doi: 10.1016/j.matbio.2014.08.008. [DOI] [PubMed] [Google Scholar]
- 76.Shapiro IM, Vresilovic EJ, Risbud MV. Is the spinal motion segment a diarthrodial polyaxial joint: what a nice nucleus like you doing in a joint like this. Bone. 2012;50:771–776. doi: 10.1016/j.bone.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sakai D, Nakamura Y, Nakai T, et al. Exhaustion of nucleus pulposus progenitor cells with ageing and degeneration of the intervertebral disc. Nat Commun. 2012;3:1264. doi: 10.1038/ncomms2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Risbud MV, Guttapalli A, Tsai T-T, et al. Evidence for skeletal progenitor cells in the degenerate human intervertebral disc. Spine (Phila Pa 1976) 2007;32:2537–2544. doi: 10.1097/BRS.0b013e318158dea6. [DOI] [PubMed] [Google Scholar]
- 79.Buckwalter JA. Aging and degeneration of the human intervertebral disc. Spine (Phila Pa 1976) 1995;20:1307–1314. doi: 10.1097/00007632-199506000-00022. [DOI] [PubMed] [Google Scholar]
- 80.Zhao C-Q, Wang L-M, Jiang L-S, et al. The cell biology of intervertebral disc aging and degeneration. Ageing Res Rev. 2007;6:247–261. doi: 10.1016/j.arr.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 81.Roughley PJ. Biology of intervertebral disc aging and degeneration: involvement of the extracellular matrix. Spine (Phila Pa 1976) 2004;29:2691–2699. doi: 10.1097/01.brs.0000146101.53784.b1. [DOI] [PubMed] [Google Scholar]
- 82.Risbud MV, Shapiro IM. Role of cytokines in intervertebral disc degeneration: pain and disc content. Nat Rev Rheumatol. 2014;10:44–56. doi: 10.1038/nrrheum.2013.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tang X, Jing L, Chen J. Changes in the molecular phenotype of nucleus pulposus cells with intervertebral disc aging. PLoS One. 2012;7:e52020. doi: 10.1371/journal.pone.0052020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chen K, Wu D, Zhu X, et al. Gene expression profile analysis of human intervertebral disc degeneration. Genet Mol Biol. 2013;36:448–454. doi: 10.1590/S1415-47572013000300021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Chen Y, Chen K, Li M, et al. Genes associated with disc degeneration identified using microarray gene expression profiling and bioinformatics analysis. Genet Mol Res. 2013;12:1431–1439. doi: 10.4238/2013.April.26.5. [DOI] [PubMed] [Google Scholar]
- 86.Tang Y, Wang S, Liu Y, et al. Microarray analysis of genes and gene functions in disc degeneration. Exp Ther Med. 2014;7:343–348. doi: 10.3892/etm.2013.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu Y, Fu S, Rahaman MN, et al. Native nucleus pulposus tissue matrix promotes notochordal differentiation of human induced pluripotent stem cells with potential for treating intervertebral disc degeneration. J Biomed Mater Res A. 2014 doi: 10.1002/jbm.a.35243. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 88.Chen J, Lee EJ, Jing L, et al. Differentiation of mouse induced pluripotent stem cells (iPSCs) into nucleus pulposus-like cells in vitro. PLoS One. 2013;8:e75548. doi: 10.1371/journal.pone.0075548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Clarke LE, McConnell JC, Sherratt MJ, et al. Growth differentiation factor 6 and transforming growth factor-beta differentially mediate mesenchymal stem cell differentiation, composition and micromechanical properties of nucleus pulposus constructs. Arthritis Res Ther. 2014;16:R67. doi: 10.1186/ar4505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sun Z, Liu Z-H, Zhao X-H, et al. Impact of direct cell co-cultures on human adipose-derived stromal cells and nucleus pulposus cells. J Orthop Res. 2013;31:1804–1813. doi: 10.1002/jor.22439. [DOI] [PubMed] [Google Scholar]
- 91.Ni L, Liu X, Sochacki KR, et al. Effects of hypoxia on differentiation from human placenta-derived mesenchymal stem cells to nucleus pulposus-like cells. Spine J. 2014;14:2451–2458. doi: 10.1016/j.spinee.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 92.Allon AA, Butcher K, Schneider RA, et al. Structured coculture of mesenchymal stem cells and disc cells enhances differentiation and proliferation. Cells Tissues Organs. 2012;196:99–106. doi: 10.1159/000332985. [DOI] [PMC free article] [PubMed] [Google Scholar]