Abstract
Salix L. is the largest genus in the family Salicaceae (450 species). Several classifications have been published, but taxonomic subdivision has been under continuous revision. Our goal is to establish the phylogenetic structure of the genus using molecular data on all American willows, using three DNA markers. This complete phylogeny of American willows allows us to propose a biogeographic framework for the evolution of the genus. Material was obtained for the 122 native and introduced willow species of America. Sequences were obtained from the ITS (ribosomal nuclear DNA) and two plastid regions, matK and rbcL. Phylogenetic analyses (parsimony, maximum likelihood, Bayesian inference) were performed on the data. Geographic distribution was mapped onto the tree. The species tree provides strong support for a division of the genus into two subgenera, Salix and Vetrix. Subgenus Salix comprises temperate species from the Americas and Asia, and their disjunction may result from Tertiary events. Subgenus Vetrix is composed of boreo-arctic species of the Northern Hemisphere and their radiation may coincide with the Quaternary glaciations. Sixteen species have ambiguous positions; genetic diversity is lower in subg. Vetrix. A molecular phylogeny of all species of American willows has been inferred. It needs to be tested and further resolved using other molecular data. Nonetheless, the genus clearly has two clades that have distinct biogeographic patterns.
Introduction
Salix L. is the largest genus of family Salicaceae with about 450 species [1–4]. The genus is distributed across the temperate to arctic regions of the Northern Hemisphere, entering tropical regions along montane ranges; willows also have been introduced worldwide. Over half the willow species, 275 are found in China [2], 107 in the former Soviet Union [5], 65 in Europe [3], and 103 in North America north of Mexico [4]. In Canada, 30% of the woody species are willows. Willows are mostly shrubs that play an important role in riparian habitats, wetlands and in shrub tundra. Willows contribute socially and economically to human societies [6–8]. During the last century, interest in environmental applications of Salix has grown, notably for biomass production and bioremediation [8–12].
Chase et al. [13] characterized the relationships among the genera of an expanded family Salicaceae, and Alford et al. [14] studied more closely the relationships of Salix and Populus Populus to their closest tropical and subtropical relatives. Salix and Populus are sister to each other and form a monophyletic group. In Chase et al. [13], Itoa and Poliothyrsis are successive sister to Salix-Populus. In Alford et al. [14], the genera Idesia, Bennettiodendron and Olmediella are sister to Populus-Salix, with Itoa, Poliothyrsis, Carrierea and Macrohasseltia sister to this clade.
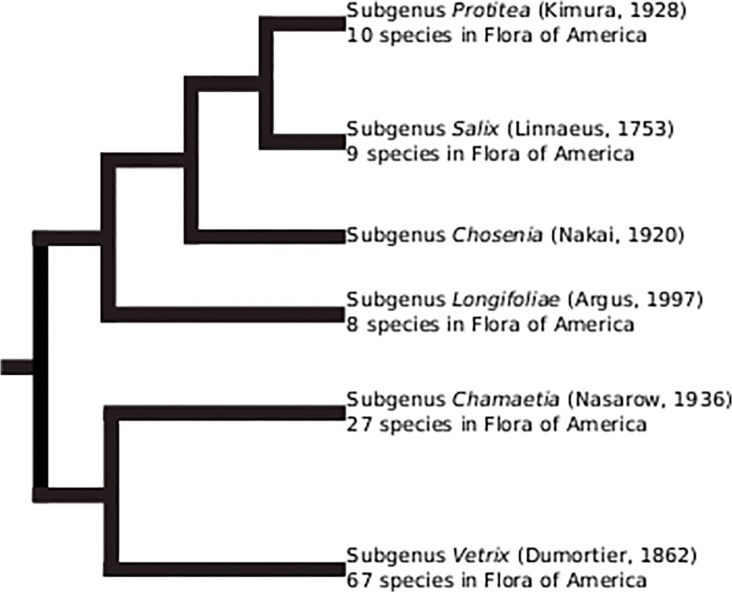
Several classifications of Salix have been published and the subdivision of the genus has been under continuous revision. Argus [3] reviewed the history of Salix classifications and showed that the genus has been divided into 35 genera since its description by Linnaeus, each author using different morphologic characters to justify these divisions. For instance, some Asian treatments recognized the genera Chosenia Nakai [15] (Salix arbutifolia) and Toisusu Kimura [16] (Salix cardiophylla). Argus [3] showed that subgenus Chosenia (including both species above) is sister to subgenus Salix (Fig 1). The Angiosperm Phylogeny Group [17] included Chosenia and Toisusu within Salix. Several subgeneric classifications of Salix have been proposed. Most recently, Skvortsov [18] divided the species of the former Soviet Union and Asia into three subgenera, Salix, Chamaetia and Vetrix. Dorn [19] divided the American species into two subgenera, Salix and Vetrix. Based on morphology, Argus [3–4] suggested five subgenera for American willows (Longifoliae, Protitea, Salix, Chamaetia and Vetrix), Fig 1 illustrating the relationships between these subgenera.
Fig 1. Relationships between the five subgenera of Salix in North America based on morphological characters (Argus, 1997).
The number of species per subgenus is indicated.
Five molecular phylogenies have addressed the relationships between willow species [20–24]. Table 1 summarizes the number of species and the molecular markers used in these studies. They affirm the monophyly of Salix, membership of Chosenia (S. arbutifolia) and Toisusu (S. cardiophylla) within Salix and the presence of two major clades within the genus. These studies all included a small number of willow species relative to the total number of species in the genus.
Table 1. Summary of molecular phylogenies of the genus Salix.
| Reference | Molecular region | Number of species | Main native area | |
|---|---|---|---|---|
| Used in study | Sequenced for study | |||
| Leskinen and Alström-Rapaport [20] | ITS region | 13 | 13 | Europe |
| Azuma et al. [21] | rbcL gene | 19 | 19 | Asia |
| Chen et al. [22] | rbcL gene; atpB-rbcL spacer; trnD-T spacer | 46 a | 32 b | Asia |
| Hardig et al. [23] | ITS region; matK gene | 25 | 25 c | North America |
| Abdollahzadeh et al. [24] | ITS region; trnL-F region | 57 d | 26 e | Iran |
| This study (2014) | ITS region; matK gene; rbcL gene | 123 | 122 | North America |
The molecular regions investigated are indicated. The number of species has been evaluated for each study (total number used and number specifically sequenced). The main native area of the species involved is indicated.
a: 45 species used with rbcL analysis. 31 species used in strict consensus of combined rbcL, atpB-rbcL and trnD-T analysis. Salix babylonica f. rokkaku was excluded from the count.
b: For 4 species, the sequences of rbcL were from Azuma et al. [21] and the spacer region was sequenced from the same specimens.
c: The sequences of matK for S. exigua and S. interior were from Brunsfeld et al. [76]. The ITS sequences of S. arctica and S. discolor are not available in GenBank.
d: trnL-F only for 14 species. Salix alba f. alba and Salix sp. (unidentified) were excluded from the count.
e: For ITS; only 6 species were sequenced for trnL-F (no analysis published). Salix sp. (unidentified) was excluded from the count.
The plastid genes matK and rbcL, and the nuclear ribosomal ITS region have been used extensively in molecular phylogenetic studies (e.g., [25–27]). Their widespread use and ease of amplification has led to their selection as the main DNA regions in the barcoding program [28–30] to be used for the identification of plants (e.g., [31]).
Our objectives are to determine the phylogenetic relationship among all American Salix species (107 species), using ITS, matK and rbcL, in order to evaluate current willow classifications and distribution patterns. We show that Salix is subdivided into two major clades, the first composed of temperate and the second of boreo-arctic species.
Materials and Methods
Plant material
This study includes all Salix species from America (107 species) plus the species introduced in North America (14 species: 7 from Europe, 5 from Eurasia, and 3 from Asia) [4]. The specimens were obtained from G. Argus' personal collection, the Marie Victorin Herbarium (MT), live collections of the Montreal Botanical Garden, the Canadian Museum of Nature (CAN), the Herbarium of the University of Texas (TEX), the University of Arizona Herbarium (ARIZ), and the Missouri Botanical Garden Herbarium (MO). Chosenia arbutifolia (= Salix arbutifolia) was sampled from the live collection of the Montreal Botanical Garden. A total of 213 specimens (122 species) of Salix were used in this study, with 1 to 3 specimens per species (S1 Table). The identity of a majority of specimens has been confirmed by G. Argus. We verified other specimens using Argus (2014). We downloaded sequences from GenBank for Toisusu cardiophylla (= Salix cardiophylla) and two outgroup genera, Idesia and Populus [13–14, 17].
DNA extraction, amplification and sequencing
Genomic DNA was extracted from herbarium specimens or fresh leaves dried in silica gel. The CTAB method [32] was used, as modified in Lauron-Moreau et al. [33]. Three molecular regions were used in this study: ITS, matK (partial) and rbcL (partial). They were amplified using the specific primers detailed in Table 2. PCRs were carried out in a 20 μL solution containing 1 μL of genomic DNA (approximately 50–70 ng), 0.75X of PCR buffer (BIO BASIC, Markham, ON, Canada), 0.25 μM of each primer, 0.25 mM of dNTPs, 2.25 mM of MgCl2, and 1 U Taq DNA polymerase (BIO BASIC). PCRs were performed using an Eppendorf Mastercycler pro Thermal Cyclers (Eppendorf Canada, Mississauga, ON, Canada) under the following cycling parameters: initial denaturation at 94°C for 3 min followed by 33–35 cycles (Ca) of 30 s at 94°C, 30 s at 52°C, 45–70 s at 72°C; and followed by a final extension at 72°C for 5 min. PCR products were sequenced by the group McGill University and Génome Québec Innovation Centre. Over half the sequences of matK and rbcL were obtained with the help of the Barcode of Life Data Systems (BOLD) following standard protocols at the Canadian Centre for DNA barcoding (CCDB) for plants, as described in Kuzmina et al. [31].
Table 2. Primers and PCR cycle characteristics for the three genes used in the study, indicating: source of primers; number of sequences obtained and their length (bp); percentage of polymorphic sites (including and excluding outgroup).
| ene | Source | Primer name | Sequence 5'-3' | Number of cycles (Ca) | Elongation time (s) | Number of sequences | Alignment length (bp) | Polymorphic sites | |
|---|---|---|---|---|---|---|---|---|---|
| With outgtoup | Without outgroup | ||||||||
| ITS | this paper | ALM-P001 | F: CGTAACAAGGTTTCCGTAGG | 35 | 60 | 210 | 608 | 125 (21%) | 95 (16%) |
| ALM-P002 | R: TGCTTAAACTCAGCGGGTAG | ||||||||
| matK | Ford et al. [77] | matK X | F: TAATTTACGATCAATTCATTC | 33 | 70 | 212 | 874 | 106 (12%) | 50 (6%) |
| Kew Barcoding | matK_Equisetum | R: GTACTTTTATGTTTACGAGC | |||||||
| rbcL | Levin et al. [78] | P1630 | F: ATGTCACCACAAACAGAGACTAAAGC | 33 | 45 | 212 | 553 | 44 (8%) | 20 (4%) |
| Kress et al. [79] | rbcLa-R | R: GTAAAATCAAGTCCACCRCG | |||||||
Sequence alignment and phylogenetic analyses
Sequences were assembled using Geneious Pro version 4.8.5 created by Biomatters (http://www.geneious.com). Alignments were done in SeaView version 4.2.6 [34] using Muscle parameters [35], followed by manual correction. Parsimony, maximum likelihood (ML), and Bayesian (BA) analyses were performed to determine the phylogenetic relationships on four datasets: ITS, matK, rbcL, and the concatenated matK-rbcL sequences. The program jModelTest2 [36–37] was used to select the model of sequence evolution for ML and BA analyses. Data matrices from this study are available on TreeBase (website: http://treebase.org) by searching for study ID14313.
Parsimony analyses were performed using PAUP version 4.0b10 [38]. We selected the optimal trees using a heuristic search following these parameters: 100 random additions of sequences followed by tree bisection and reconnection (TBR) branchswapping, retaining at most 100 trees at each replicate. Branch support was estimated using 10,000 bootstrap replicates with the same heuristic settings.
Maximum likelihood analyses were performed using PhyML 3.0 [39]. ML heuristic searches and bootstrap analysis (10,000 replicates) were conducted to obtain the best trees under the parameters of the evolution model selected by jModelTest2. The adequate evolution models were GTR+G+I for ITS, GTR+G for matK and matk-rbcL, and K80+I for rbcL.
Bayesian analyses were performed using MrBayes version 3.1.2 [40] and BEAST v1.7.5 [41]. In MrBayes, two independent runs were performed, each consisting of four parallel Markov chain Monte Carlo (MCMC) of 100 million generations (the average standard deviation of split frequencies being lower than 0.01). Trees were sampled every 10,000 generations. The evolution models used were identical with those in the ML analyses. Tree parameters reached stationarity after a burn-in period of 250,000 generations. Optimal trees were then sampled every 1,000 generations to obtain the final consensus tree and associated posterior probabilities. For the BEAST analysis, each molecular region was analyzed separately and the species tree was developed concurrently. Two independent runs of 100 million generations were performed, each with sampling every 10,000 generations. We used the same evolution models as above, with four gamma categories, a coalescent tree prior and a strict clock model for each partition. After analysis, the software Tracer [42] was used to evaluate the convergence after the first 20% of generations had been discounted as burn-in. The software TreeAnnotator v1.7.5 (available in BEAST package) was used to estimate the maximum-clade-credibility using the Bayesian posterior probabilities.
On the BEAST species tree, we illustrated the main native area of each species following Argus ([43]; for North America), using seven zones: four in North America (western temperate, western boreo-arctic, eastern temperate, eastern boreo-arctic) and three representing Europe, Asia and Mexico (including Central and South America). Finally, we constrained a BEAST analysis to conform with the morphological classification of Argus [3].
Results and Discussion
Success rate of the amplifications and DNA sequences
We obtained 211 sequences for ITS (including 2 partial sequences) and 213 sequences for matK and rbcL (all sequences are available in GenBank) (S1 Table). For the ITS region, amplification of two specimens of S. atrocinerea was not a successful, and amplification was partial only for S. jaliscana and S. prolixa. New DNA extractions and a modification to the PCR protocol did not give better results. Fifteen sequences were downloaded from GenBank and aligned with our data. We did not find intra-species variation within our data. The alignment of ITS, matK and rbcL resulted in 608, 874 and 553 aligned nucleotides, respectively (Table 2). Including the GenBank data, we had 215 sequences for ITS, and 217 sequences for matK and rbcL. The ITS region had a higher proportion of polymorphic sites (21%) when compared with matK (12%) and rbcL (8%) (Table 2).
Polymorphisms
We observed many polymorphic sites in the ITS region. Sixteen species (S. arbusculoides, S. arctica, S. arctophila, S. barclayi, S. cana, S. columbiana, S. discolor, S. exigua, S. famelica, S. floridana, S. humboldtiana, S. jejuna, S. monticola, S. raupii, S. richardsonii, S. rotundifolia) had polymorphisms at 17 nucleotide sites (1–5 polymorphic sites per species). We also found polymorphisms in the plastid genes. Salix aeruginosa and S. jaliscana are polymorphic at four (34, 367–368, 398) and two sites (80, 514), respectively, in matK. Six species had polymorphic sites on rbcL: S. jaliscana (396–397); S. pedicellaris, S. pseudomyrsinites (285); and S. argyrocarpa, S. cascadensis, S. orestera (286).
Phylogenetic analyses
We compared the resolution and branch support of four analytical approaches (PhylML, MrBayes, BEAST and PAUP) on all datasets. The topologies were similar and we are presenting the results from BEAST because its support values were higher (Figs 2 and 3). Bayesian posterior probabilities and ML bootstrap values are provided on the trees shown.
Fig 2. BEAST gene tree of matK and rbcL.
Branch support is Bayesian posterior probabilities and ML bootstrap values; subgenera are identified using colors; Idesia and Populus are outgroups.
Fig 3. BEAST gene tree of ITS.
Branch support is Bayesian posterior probabilities and ML bootstrap values; subgenera are identified using colors; Idesia and Populus are outgroups.
The phylogenetic trees obtained for matK and rbcL were identical except for the position of Salix petrophila, and we are presenting the consensus tree of these two plastid genes (Fig 2). Two major clades are apparent on the cp DNA tree. Clade A1 includes 32 Salix species and clade A2, 88. The two clades are well supported. The relationships within each clade are not well resolved, however, and branches with a posterior probability lower than 0.7 were collapsed. Clade A1 comprises the majority of species from subgenera Longifoliae, Protitea and Salix. Clade A2 includes most species of the subgenera Chamaetia and Vetrix. Salix arbutifolia and S. cardiophylla belong to clade A2.
In the ITS tree, four different crown are well supported (Fig 3). Clade B1 comprises most species of subgenus Protitea, clade B2 most species of subgenus Salix, clade B3 most species of subgenus Longifoliae, and clade B4 most species of subgenera Chamaetia and Vetrix. Overall, the species of clades B1, B2 and B3 (Fig 3) are present in clade A1 of the cp trees (Fig 2), while B4, S. arbutifolia and S. cardiophylla are in clade A2. Fourteen taxa have incongruent positions on the two trees, however.
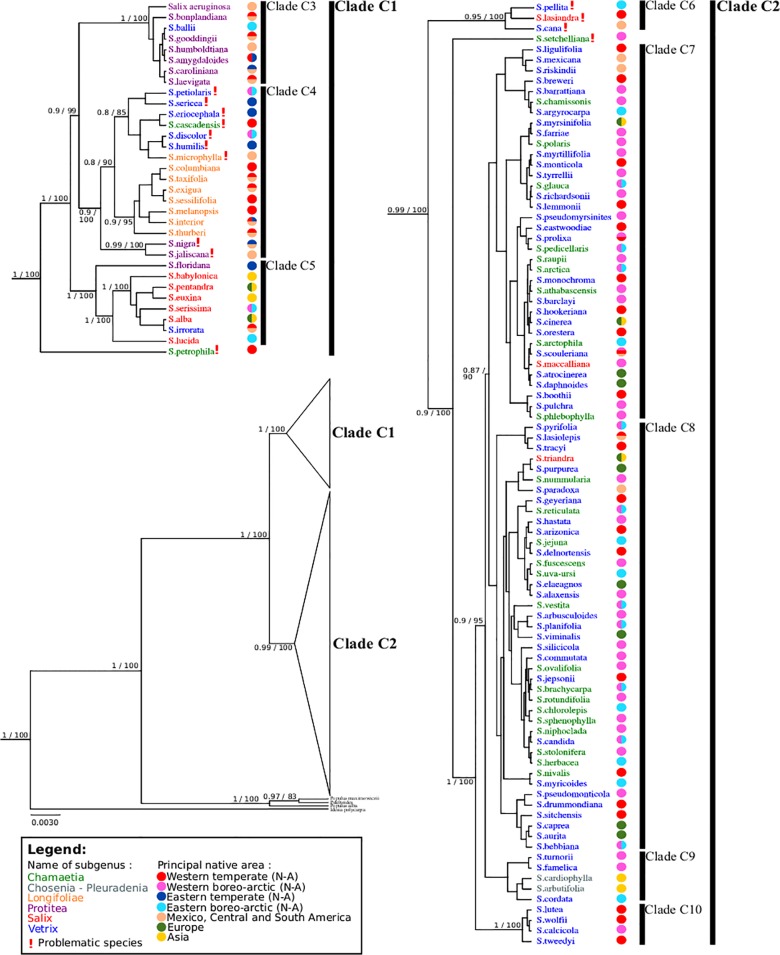
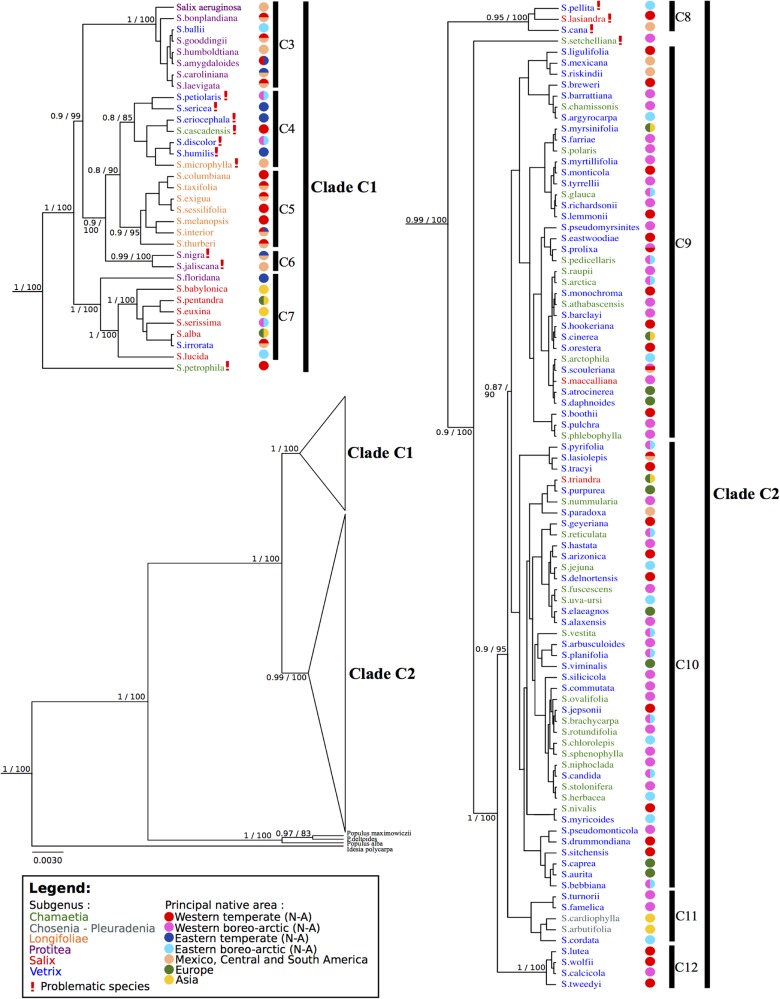
Fig 4 presents the species tree of the simultaneous BEAST analysis of the three markers. The tree exhibits two major clades, C1 and C2. Many subclades are shown in clade C1 and in clade C2. Subclades C9 or C10 have low support. The distribution of species is presented on the tree.
Fig 4. BEAST species tree generated with ITS, matK and rbcL.
Branch support is Bayesian posterior probabilities and ML bootstrap values; subgenera are identified using colors; native areas (Argus 2007) are indicated by colored circles; Idesia and Populus are outgroups.
S1 Fig shows a BEAST analysis where the subgenera were constrained according to Argus [4]. Support is slightly lower than Fig 2. In a manner similar to the unconstrained BEAST tree (Fig 4), one clade includes the species of the subgenera Longifoliae, Protitea and Salix, and a second one, the species of subgenera Chamaetia and Vetrix plus S. arbutifolia and S. cardiophylla. Species that in Fig 4 did not group with the subgenus in which Argus classified it all diverge early in the clades of the constrained tree.
Sequence polymorphism
As in Leskinen and Alström-Rapaport [20], we observed intra-individual polymorphic nucleotide sites in the ITS region. Ribosomal sequences are present in thousands of copies in the nuclear genome [44]. Usually, sequences within individuals are uniformized due to concerted evolution [45–46]. However, in cases of recent hybridization or homoploid speciation, sequence homogenization is often not achieved in the short period of time involved [45–46]. For instance, Leskinen and Alström-Rapaport [20] hypothesized that S. schwerinii could result from homoploid speciation after hybridization between S. viminalis and a second, unidentified species. In our study, S. exigua, a diploid species, shows polymorphic sites. It could be due either to introgression from recent hybridization or the species could be the result of homoploid speciation. Other species, however, are polyploid and polymorphism may merely result from a lack of homogenization, particularly in allopolyploid taxa. Conversely, Salix alba, a tetraploid, is without polymorphic sites in the ITS region. Data are currently insufficient to explain the presence of polymorphic sites in the ITS of American Salix.
Polymorphic nucleotide sites in plastid sequences may seem surprising but are not new. Few studies have reported this [47–48]. One hypothesis to explain such polymorphism would be the inclusion of cp DNA fragments in nuclear DNA [48–49]. A second hypothesis would be an error occurring during plastid division [50]. Our data are insufficient to determine what mechanism is acting. It would require, among others, detailed population and genomic studies of the species concerned.
Phylogenetic relationships between American species of willows
Plastid trees
The plastid tree (Fig 2), using Populus and Idesia as outgroups, affirms the monophyly of genus Salix and the inclusion of S. arbutifolia (Chosenia) and S. cardiophylla (Toisusu) within the genus, as was shown by Chen et al. [22]. This tree also shows the separation of American Salix species into two major clades, as was also found by Azuma et al. [21] and Chen et al. [22] on Asian species (see Table 1). As in our study, one clade included species of subgenera Chamaetia and Vetrix (our A2 clade), and the other (our clade A1) subgenus Salix (no representative of subg. Longifoliae and Protitea were included). Twenty species were shared between our study and that of Chen et al. [22], 18 of which are found in the same clade in both analyses (shown by black stars in Fig 2). Two taxa, S. discolor and S. maccaliana, were found in different clades, however. This could be explained by the fact that the two species are polyploid, 4x and 10x, respectively [4]; it could also be the result of intra-specific variability or chloroplast capture following hybridization, or of an error of identification or manipulation. Despite differences possibly caused by the taxonomy used (see below), Hardig et al. [23], working on American species (Table 1), also retrieved two similar clades. Resolution within clades is low and poorly supported, resulting in polytomies. Low rates of evolution of the plastid genome or recency of speciation in willow species could explain this. The positions of S. petrophila (clade A1) and S. lasiandra (clade A2) are surprising, both being early divergent in each clade; this cannot be readily explained with current data.
ITS tree
The ITS region shows more variation than the cp DNA markers, but resolution of the tree was not greatly improved. ITS trees in Leskinen and Alström-Rapaport [20], Hardig et al. [23] and Abdollahzedeh et al. [24], built respectively using parsimony, maximum likelihood and MrBayes, were similar to our analyses (not shown) carried out with the same approaches: a large polytomy is retrieved, with a single small clade comprised of species belonging to subg. Salix, Longifolia and Protitea. Our BEAST tree (Fig 3), however, provided greater resolution, identifying four clades: species of subgenus Protitea in clade B1, subgenus Salix in B2, and subgenus Longifoliae in B3, with subgenera Chamaetia and Vetrix species intermixed in the large clade B4. All North American species used by Hardig et al. [23] were also included in our study (shown by black stars in Fig 3). Differences were observed in the placement of a few species. For instance, Hardig et al. included specimens of S. eriocephala and S. lucida from Idaho, species that Argus [4] do not report for this area; this may result from the taxonomy used, since varieties sometimes attributed to S. eriocephala in western North America, for instance, are considered distinct species by Argus.
Incongruence between the chloroplastic and ITS trees
Overall, clades A2 (cp) and B4 (ITS) include species of subg. Chamaetia and Vetrix, while the species of clade A1 (cp) coincide with those in clades B1, B2 and B3 (ITS) (Figs 2 and 3). The global structure of the trees is similar. There is significant incongruence however, for 14 taxa. Ten species of clade A1 in the cp tree (Fig 2) were found in clade B4 on the ITS tree (Fig 3). Conversely, four species of clade A2 (cp) were retrieved in clade B4. These taxa are highlighted in our trees. Five hypotheses could explain these incongruences. Firstly, plastid or rDNA capture following hybridization could have occurred. For instance, S. pellita (clade B4) can form natural hybrids in nature with S. alaxensis (clade B1) (Argus, 2014). Secondly, part of the chloroplast genome of one parent could have migrated to the nucleus in allopolyploid taxa, a rare but not impossible phenomenon [51]. Thirdly, horizontal gene transfer from another species is possible [52]. Fourth, plastid fusion may occur, though it is rarely documented [53]. And finally, field or laboratory errors could have happened, which seems improbable given the number of taxa involved.
Species tree
The species tree (Fig 4) results from the simultaneous BEAST analysis of the cpDNA and ITS datasets. More resolution is apparent on this tree. The topology affirms the presence of two major clades (C1 and C2), such as described above, which mirrors the tree inferred from plastid data but is incongruent (in part) with the tree inferred from nuclear data. Within clade C1, species are mostly grouped according to the subgenera where they are assigned by Argus [4], i.e., subg. Longifoliae (subclade C5), Protitea (subclade C3), and Salix (subclade C7, also retrieved in Chen et al. [22]), which are well supported. A few species (discussed below) appear in novel positions with respect to Argus [4]. Subgenera Chamaetia and Vetrix (clade C2) form one group, which corresponds to the observations of Chen et al. [22]. There is no clear pattern of subgeneric segregation in clade C2. All subclades within C2 have low support. Skvortsov [18], discussing Russian material, also indicated that the distinction between these subgenera was difficult, while Dorn [19] only recognized subg. Vetrix. The branching order and groupings observed in this analysis are similar to those obtained in a morphology-based, numerical analysis of Salix by Argus [3], if one excepts the position of subg. Chosenia, which groups in C2 in our analysis and the equivalent of C1 in that of Argus.
Fourteen species had different positions in the ITS and cp DNA analyses. They are indicated by a! in Figs 2–4. Seven species, Salix cascadensis, S. discolor, S. eriocephala, S. humilis, S. microphylla, S. petiolaris and S. sericea, form a subclade (C4) within clade C1 in the analysis. Yet, the morphology of these species is heterogeneous [4] and no morphologic character appears to support such a group. Also in clade C1, S. jaliscana and S. nigra are grouped, both of which belong to subg. Protitea (Argus [4]), which would make subg. Protitea paraphyletic to subg. Longifoliae and the artificial subclade C4. All these species were in clade A1 in the cp tree and in clade B2 in the ITS tree. The grouping of species in subclade C4 suggests a random grouping of species with similar behaviors. Subclade C4 appears artificial. Salix petrophila (subg. Chamaetia) appears to be an early diverging branch of clade C1. This species occupied different positions in the matK and rbcL trees (not shown). Similarly, clade C6, comprised of S. cana, S. lasiandra, and S. pellita, form an early diverging group sister to clade C2. Salix setcheliana is an early diverging branch within clade C2. All these species were in clade A2 in the cp tree and in clade B1 in the ITS tree. In all instances, it appears as if the position in the species tree is determined primarily by the cp DNA.
Tree constrained by subgenera
A BEAST analysis of all datasets was done while constraining species to their subgeneric affiliation (fide Argus, [4]) (S1 Fig). Support for the subgenera in this tree is lower than for clades and subclades in the unconstrained species tree, where some subclades roughly correspond to subgenera. The constrained tree is divided into two clades, as in the unconstrained species tree. Subgenera Chamaetia and Vetrix are sisters within one clade, and subg. Longifoliae, Protitea and Salix are grouped in the second. Five species, however, that belong to a particular subgenus in the constrained tree, occupy a different position in the species tree. This cannot be explained readily by chloroplast capture, introgression or hybridization between Salix species. The 14 problematic taxa considered above, when constrained to group with their subgenus, acquire a basal position in their subgenus, which casts doubt as to their membership. Constraining the analysis to respect the subgenera defined by Argus [3–4] assumes that the morphological characters used are proper to classify willow species. The species tree (Fig 4), however, indicates that this is not valid for all characters and species, particularly when one considers the distinction between subg. Chamaetia and Vetrix.
Subgenus attribution of three species
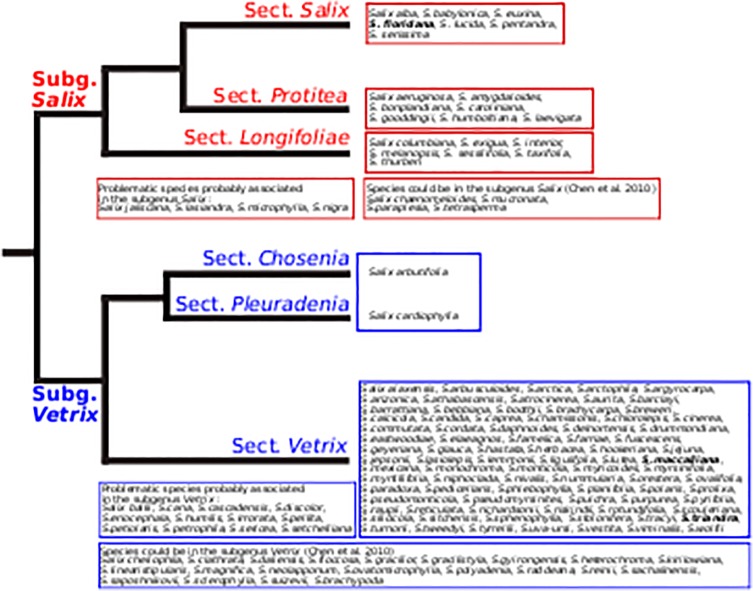
The species tree (Fig 4) shows that three species, Salix floridana (subg. Protitea), S. maccalliana and S. triandra (both subg. Salix), probably are assigned to the wrong subgenus. Our data suggest that S. floridana belongs to subgenus Salix, where it would be sister to the other species. Chen et al. [22] also found a similar position for S. floridana. The composition of subg. Protitea (S. amygdaloides, S. bonplandiana, S. caroliniana, S. gooddingii, S. humboltiana) has been discussed repeatedly [54–56], without consensus. Dorn [19] proposed the exclusion of S. floridana from this subgenus, placing it instead in either subg. Salix or Vetrix. He hypothesized that the morphological similarities (bud scales distinct, flowers with 3 to 7 stamens) of this species to subgenus Protitea was the result of hybridization [19]. Argus [4, 57] classified species of subgenus Protitea together because they share many morphological traits. The branching of the species tree (Fig 4) suggests that the morphological similarities highlighted by Dorn could be symplesiomorphic and not the result of hybridization. Chmelar [58] proposed that ovule number could be taxonomically significant. Salix floridana and S. babylonica have 2 ovules per carpel, S. alba 3 to 6, and S. amygdaloides, S. caroliniana and S. nigra 6 to 9 [57]. Low ovule number could be a feature of sect. Salix.
Our data suggest that S. maccalliana and S. triandra belong in a large subgenus Vetrix (see below). Salix maccalliana is decaploid or dodecaploid [4], which indicates a complex origin. Its morphology is similar to that of S. lucida (subg. Salix) [4]. The staminate flowers with abaxial nectaries and tawny, persistent bracts, and the villous ovaries, however, suggest relationship with S. glauca (subg. Chamaetia). Dorn [19] placed this species in subgenus Vetrix. Chen et al. [22] included S. maccalliana in subgenus Salix. The provenance of their sample appears geographically suspect, however. In the case of S. triandra, both our study and those of Leskinen and Alström-Rapaport [20] and Chen et al. [22] that it belongs with subg. Vetrix. The latter fully discussed this issue and indicated that S. triandra could be considered to belong to a distinct subgenus. In our tree (Fig 4), however, S. triandra falls fully within subg. Vetrix.
Placement of Salix ballii and Salix irrorata
Salix ballii and S. irrorata are assigned to subgenus Vetrix by Argus [4]. Our data, however, show that S. ballii is related to subg. Protitea, and S. irrorata to subg. Salix. Two distinct specimens were sequenced for each species with the same result. One hypothesis would be the capture of plastid or ribosomal DNA after hybridization. A laboratory error cannot be excluded. At this time, data are insufficient to explain these placements.
Biogeography of Salix
Formal biogeographic analyses (DEC [59], or DIVA [60]) could not be carried out with our tree due notably to the lack of resolution in clade C2 (Fig 4). Doing a calibrated datation was not feasible at this time due to a lack of verified Salix fossil material that could be accurately placed on the tree topology. Nonetheless, the patterns observed on Fig 4 allow the formulation of biogeographic hypotheses.
Overall, most species of clade C1 are found in temperate regions of both North America (western and eastern) and Eurasia, while those of clade C2 include mostly species from boreo-arctic regions or montane areas southward. Globally, distribution patterns in Salix reflect well the biogeographic regions delimited by Takhtajan [61] within the Holarctic kingdom, Boreal subkingdom: C1 taxa are predominantly in the Eastern Asiatic, North American Atlantic and Rocky Mountain regions (= subg. Salix, see below), and C2 taxa in the Circumboreal region (= subg. Vetrix, see below).
Within C1, species of subclade C3 are western and eastern North American, with extensions to Mexico and Central to South America (S. humboldtiana only). Most species of subclade C5 are temperate western North American and Mexican (S. interior spreading to eastern North America). Within subclade C7, Salix floridana is in southeastern temperate America (mostly Florida) and is sister to a clade comprised of temperate Eurasian and American elements. Subclade C4, which includes problematic elements, is predominantly eastern North American with some elements more boreal and therefore more widespread. We hypothesize that inter-continental migrations between temperate regions during the Tertiary may explain the pattern observed. During the Tertiary, the North Atlantic and Bering land bridges [62–64] allowed inter-continental exchanges at high latitudes because of warmer climates [65]; such a disjunction pattern appears to fit distributions in clade C1, and has been documented and dated for other taxa in the Northern Hemisphere (e.g. [66–69]).
Clade C2 comprises more northern, boreo-arctic and montane to alpine species of both Eurasia and North America; all strictly European species appear to belong here. North American and Eurasian elements are intermixed throughout the tree, as are western and eastern North American boreal species. A few Mexican species (S. cana, S. mexicana, S. riskindii, and S. paradoxa), closely related to western North American montane species, are found here, as are species found in both Mexico and western North America (S. lasiolepis, S. scouleriana). In Asia, S. cardiophylla (Toisusu) is montane and cool temperate, and S. arbutifolia (Chosenia) is montane and boreo-temperate; biogeographically, they fit well within this clade. We hypothesize that the large radiation within clade C2 is the result of events that occurred during the Pleistocene. Several species in subg. Vetrix are circumarctic, widespread at boreal latitudes, amphi-Beringian or amphi-Atlantic. During this period, the Northern Hemisphere was subjected to several glaciation periods [70–71].
Notably, lowering of the sea during glacial events repeatedly opened the Bering land bridge for long periods of time, allowing migration at high latitudes in a corridor of steppe and tundra. Ice extension also forced boreo-arctic species to seek refuges south of the ice. Both phenomena impacted species distributions [72]. These migrations brought species in contact and favored hybridization and the formation of polyploid species (e.g., [73]), two factors that appear to have played an important role in the rapid diversification of subg. Vetrix.
Classification of Salix
Skvortsov [18] divided the species of the former Soviet Union and Asia into three subgenera, Dorn [19] the American species into two subgenera, and Argus [3–4] American taxa into five subgenera (Longifoliae, Protitea, Salix, Chamaetia and Vetrix). Our molecular phylogenetic study and that of Chen et al. [22] show a primary subdivision of Salix into two clades (Fig 4), the latter pointing out that the number of subgenera proposed for Salix was too high. The studies by Leskinen and Alström-Rapaport [20], Azuma et al. [21], Hardig et al. [23] and Abdollahzedeh et al. [24] also suggest such a division. We are proposing to divide Salix into two subgenera, Salix and Vetrix. Three sections may be recognized within subgenus Salix: Salix, Protitea, and Longifoliae, the latter American only. Within subgenus Vetrix, lack of resolution prevents the definition of sections at this time. Salix arbutifolia (Chosenia) and S. cardiophylla (Toisusu) are definitely members of subg. Vetrix, possibly as an early branch including other willows, a group that may deserve sectional recognition. Another unresolved issue is the definitive position of the 16 problematic species discussed above, notably the clade within subg. Salix.
The three molecular regions used in our study are the markers selected for the barcoding of plants. The degree of variation of these molecular markers in Salix, however, is insufficient to provide species identification in subg. Vetrix, and as other studies have shown [74], other regions will need to be developed for full barcoding of willows.
Conclusions
We present the first complete phylogeny of willows for the Americas, based on three molecular markers from plastid and nuclear ribosomal DNA. We affirm the subdivision of genus Salix species into two clades that correspond to two subgenera proposed earlier on the basis of morphologic and molecular studies. Nonetheless, relationships among species remain tentative due to a lack of resolution within subg. Vetrix and to the unusual relationships exhibited by a 16 problematic species. Further phylogenetic analyses using low-copy nuclear genes should help address this lack of resolution and membership issues, and help in obtaining a tree that could be the object of formal biogeographic analyses. The challenge presented in this genus by hybridization and polyploidy may be resolved by phylogeographic analyses of species complex, such as was done by Tsai and Carstens [75]. In a recent report (2014), Percy et al. described the difficulty of DNA fingerprinting for willow species when using only plastid regions because these markers are unable to delineate possible widespread hybridization events, thus supporting our choice to include a nuclear marker [80].
Our phylogenetic analysis provides a framework to interpret data from other fields of study, such as eco-physiology and the development of willows for economic usages, such as biomass production.
Supporting Information
Branch support is Bayesian posterior probabilities and ML bootstrap values.
(TIF)
We indicated species name (according in Argus [4] and IPNI), their status in America (native or introduced), their principal native area, their subgenus (Argus [4]), the herbarium informations: live collections of the Montreal Botanical Garden (MBG), the Canadian Museum of Nature (CAN), the Herbarium of the University of Texas (TEX), the University of Arizona Herbarium (ARIZ), the Missouri Botanical Garden Herbarium (MO), or unmounted. Finally, we have indicated the GenBank number.
(DOCX)
Acknowledgments
The authors thank the following herbaria for the use of material in this study: Marie Victorin Herbarium (MT), Canadian Museum of Nature (CAN), Herbarium of the University of Texas (TEX), Herbarium of the Missouri Botanical Garden (MO), and the University of Arizona Herbarium (ARIZ); and the Montreal Botanical Garden for the use of their live collection. We also thank the RQCHP (Réseau Québécois de Calcul de Haute Performance) for access to their facilities. We thank the program BOLD (The Barcode of Life Data Systems) for their help with sequencing.
Data Availability
All DNA sequences are available from the Genbank database (accession numbers: see S1 Table).
Funding Statement
This study was supported by the Centre de recherche sur les grains (CEROM) and the Conseil pour le développement de l’agriculture au Québec (CDAQ). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Newsholme C. Willows: the genus Salix Portland: Timber Press; 1992. [Google Scholar]
- 2. Fang ZF, Zhao SD, Skvortsov AK. Salicaceae In: Wu Z, Raven PH, editors. Flora of China vol. 4 St. Louis, MO: Missouri Botanical Garden Press; 1999. pp. 139–274. [Google Scholar]
- 3. Argus GW. Infrageneric classification of New World Salix L. (Salicaceae). Syst Bot Monogr. 1997;52: 1–121. [Google Scholar]
- 4. Argus GW. Salix In: Flora of North America Editorial Committee, editors. Flora of North America, vol. 7: Magnoliophyta: Salicaceae to Brassicaceae. New York: Oxford University Press; 2010. pp. 23–51. [Google Scholar]
- 5. Skvortsov AK. Willows of Russia and adjacent countries Joensuu, Finland: University of Joensuu; 1999. [Google Scholar]
- 6. Sneader W. The discovery of aspirin: a reappraisal. Br Med J. 2000;321: 1591–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kuzovkina YA, Weih M, Abalos Romero M, Charles J, Hurst S, et al. Salix: botany and global horticulture. Hortic Rev. 2008;34: 447–489. [Google Scholar]
- 8. Karp A, Hanley SJ, Trybush SO, Macalpine W, Pei M, et al. Genetic improvement of willow for bioenergy and biofuels. J Integr Plant Biol. 2011;53: 151–165. 10.1111/j.1744-7909.2010.01015.x [DOI] [PubMed] [Google Scholar]
- 9. Stott KG. Improving the biomass potential of willow by selection and breeding In: Perttu KL, editor. Ecology and management of forest biomass production systems. Uppsala: Swedish University of Agricultural Sciences; 1984. pp. 233–260 [Google Scholar]
- 10. Lindegaard KN, Barker JHA. Breeding willows for biomass. Asp Appl Biol. 1997;49: 155–162. [Google Scholar]
- 11. Volk TA, Abrahamson LP, Nowak CA, Smart LB, Tharakan PJ, et al. The development of short-rotation willow in the northeastern United States for bioenergy and bioproducts, agroforestry and phytoremediation. Biomass & Bioenergy 2006;30: 715–727. [Google Scholar]
- 12. Guidi W, Pitre FE, Labrecque M. Short-rotation coppice of willows for the production of biomass in Eastern Canada In: Matovic MD, editor. Biomass Now—Sustainable Growth and Use, Chapter 17. Rijeka, Croatia: InTech; 2013. pp. 421–448. Available: http://www.intechopen.com/books/biomass-now-sustainable-growth-and-use. [Google Scholar]
- 13. Chase MW, Zmarzty S, Lledo MD, Wurdack KJ, Swensen SM, et al. When in doubt, put it in Flacourtiaceae: a molecular phylogenetic analysis based on plastid rbcL DNA sequences. Kew Bull. 2002;57: 141–181. [Google Scholar]
- 14. Alford MH, Brantley RJ, Hernandez CL, Samarakoon T. What are the closest relatives of Salix and Populus? Snowbird, Utah: Botany and Mycology; 2009. Available: http://2009.botanyconference.org/engine/search/index.php?func=detail&aid=786. Accessed 30 October 2014. [Google Scholar]
- 15. Nakai T. Chosenia, a new genus of Salicaceae. Bot Mag (Tokyo) 1920;34: 66–69. [Google Scholar]
- 16. Kimura A. Über Toisusu—eine neue Salicaceen-Gattung und die systematische Stellung derselben. Bot Mag (Tokyo) 1928;42: 287–290. [Google Scholar]
- 17. The Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc. 2009;161: 105–121. [Google Scholar]
- 18. Skvortsov AK. Willows of the URSS. Moscow: Nauka; 1968. [In Russian]. [Google Scholar]
- 19. Dorn RD. A synopsis of American Salix . Can J Bot. 1976;54: 2769–2789. [Google Scholar]
- 20. Leskinen E, Alstrom-Rapaport C. Molecular phylogeny of Salicaceae and closely related Flacourtiaceae: evidence from 5.8 S, ITS 1 and ITS 2 of the rDNA. Plant Syst Evol 1999;215: 209–227. [Google Scholar]
- 21. Azuma T, Kajita T, Yokoyama J, Ohashi H. Phylogenetic relationships of Salix (Salicaceae) based on rbcL sequence data. Am J Bot. 2000;87: 67–75. [PubMed] [Google Scholar]
- 22. Chen JH, Sun H, Wen J, Yang YP. Molecular phylogeny of Salix L. (Salicaceae) inferred from three chloroplast datasets and its systematic implications. Taxon 2010;59: 29–37. [Google Scholar]
- 23.Hardig TM, Anttila CK, Brunsfeld SJ. A phylogenetic analysis of Salix (Salicaceae) based on matK and ribosomal DNA sequence data. J Bot. 2010: 1–12.
- 24. Abdollahzadeh A, Osaloo K, Maassoumi AA. Molecular phylogeny of the genus Salix (Salicaceae) with an emphasize to its species in Iran. Iran J Bot. 2010;17: 244–253. [Google Scholar]
- 25. Hilu KW, Borsch T, Muller K, Soltis DE, Soltis PS, et al. Angiosperm phylogeny based on matK sequence information. Am J Bot. 2003;90: 1758–1776. 10.3732/ajb.90.12.1758 [DOI] [PubMed] [Google Scholar]
- 26. Davis CC, Anderson WR. A complete generic phylogeny of Malpighiaceae inferred from nucleotide sequence data and morphology. Am J Bot. 2010;97: 2031–2048. 10.3732/ajb.1000146 [DOI] [PubMed] [Google Scholar]
- 27. Vijaykumar A, Saini A, Jawali N. Phylogenetic analysis of subgenus Vigna species using nuclear ribosomal RNA ITS: Evidence of hybridization among Vigna unguiculata subspecies. J Hered. 2010;101: 177–188. 10.1093/jhered/esp084 [DOI] [PubMed] [Google Scholar]
- 28. Chase MW, Cowan RS, Hollingsworth PM, Van Den Berg C, Madrinan S, et al. A proposal for a standardised protocol to barcode all land plants. Taxon 2007;56: 295–299. 17464884 [Google Scholar]
- 29. Ausubel JH. A botanical macroscope. Proc Natl Acad Sci USA. 2009;106: 12569–12570. 10.1073/pnas.0906757106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yao H, Song J, Liu C, Luo K, Han J, et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLOS ONE 2010;5(10): e13102 10.1371/journal.pone.0013102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kuzmina ML, Johnson KL, Barron HR, Hebert PDN. Identification of the vascular plants of Churchill, Manitoba, using a DNA barcode library. BMC Ecology 2012;12: 25 10.1186/1472-6785-12-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19: 11–15. [Google Scholar]
- 33. Lauron-Moreau A, Pitre FE, Brouillet L, Labrecque M. Markers of willow species and characterization of 11 polymorphic microsatellites for Salix eriocephala (Salicaceae), a potential native species for biomass production in Canada. Plants 2013;2(2): 203–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gouy M, Guindon S, Gascuel O. SeaView Version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol Biol Evolut. 2010;27: 221–224. [DOI] [PubMed] [Google Scholar]
- 35. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52: 696–704. [DOI] [PubMed] [Google Scholar]
- 37. Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nature Methods 2012;9: 772–772. 10.1038/nmeth.2109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Swofford DL. Paup—a Computer-Program for Phylogenetic Inference Using Maximum Parsimony. J Gen Physiol. 1993;102: A9–A9. [Google Scholar]
- 39. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59: 307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- 40. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003;19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 41. Drummond AJ, Suchard MA, Xie D, Rambaut A. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 2012;29: 1969–1973. 10.1093/molbev/mss075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rambaut A, Drummond AJ. Tracer v1.4. 2007. Available: http://beast.bio.ed.ac.uk/Tracer. Accessed 15 January 2013.
- 43. Argus GW. Salix (Salicaceae) distribution maps and a synopsis of their classification in North America, north of Mexico. Harvard Pap Bot. 2007;12: 335–368. [Google Scholar]
- 44. Rogers SO, Bendich AJ. Ribosomal-rna genes in plants—Variability in copy number and in the intergenic spacer. Plant Mol Biol. 1987;9: 509–520. 10.1007/BF00015882 [DOI] [PubMed] [Google Scholar]
- 45. Alvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Mol Phylogenet Evol, 2003;29: 417–434. [DOI] [PubMed] [Google Scholar]
- 46. Bailey CD, Carr TG, Harris SA, Hughes CE. Characterization of angiosperm nrDNA polymorphism, paralogy, and pseudogenes. Mol Phylogenet Evol 2003;29: 435–455. [DOI] [PubMed] [Google Scholar]
- 47. McCauley DE, Stevens JE, Peroni PA, Raveill JA. The spatial distribution of chloroplast DNA and allozyme polymorphisms within a population of Silene alba (Caryophyllaceae). Am J Bot. 1996;83: 727–731. [Google Scholar]
- 48. Oliver MJ, Murdock AG, Mishler BD, Kuehl JV, Boore JL, et al. Chloroplast genome sequence of the moss Tortula ruralis: gene content, polymorphism, and structural arrangement relative to other green plant chloroplast genomes. BMC Genomics 2010;11: 143 10.1186/1471-2164-11-143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Guo X, Ruan S, Hu W, Cai D, Fan L. Chloroplast DNA insertions into the nuclear genome of rice: the genes, sites and ages of insertion involved. Funct Integr Genomics 2008;8: 101–108. [DOI] [PubMed] [Google Scholar]
- 50. Renzaglia KS, Maden AR, Duckett JG, Whittier DP. Monoplastidy in spermatogenesis of Lycopodium obscurum. Can J Bot. 1994;72: 1436–1444. [Google Scholar]
- 51. Timmis JN, Ayliffe MA, Huang CY, Martin W. Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nature Rev Genet. 2004;5: 123–U16. [DOI] [PubMed] [Google Scholar]
- 52. Stegemann S, Keuthe M, Greiner S, Bock R. Horizontal transfer of chloroplast genomes between plant species. Proc Natl Acad Sci USA. 2012;109: 2434–2438. 10.1073/pnas.1114076109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Birky CW. The inheritance of genes in mitochondria and chloroplasts: Laws, mechanisms, and models. Annu Rev Genet. 2001;35: 125–148. [DOI] [PubMed] [Google Scholar]
- 54. Schneider C. Notes on American willows. IV. Species and varieties of section Longifoliae . Bot Gaz 1919;67: 309–346. [Google Scholar]
- 55. Rehder A. Bibliography of cultivated trees and shrubs hardy in the cooler temperate regions of the northern hemisphere Jamaica Plain, Massachusetts: Arnold Arboretum; 1949. [Google Scholar]
- 56. Ball CR. Salix In: Lundell CL, et al. , editors. Flora of Texas, vol. 3(6). Renner, TX: Texas Research Foundation; 1961. pp. 369–392. [Google Scholar]
- 57. Argus GW. The genus Salix (Salicaceae) in the southeastern United States. Syst Bot Monogr. 1986;9: 1–170. [Google Scholar]
- 58. Chmelar J. Taxonomic importance of bud scale in the Salix genus. Folia Dendrologica 1978;4: 5–21. [Google Scholar]
- 59. Ree RH, Smith SA. Maximum likelihood inference of geographic range evolution by dispersal, local extinction, and cladogenesis. Syst Biol. 2008;57: 4–14. 10.1080/10635150701883881 [DOI] [PubMed] [Google Scholar]
- 60. Nylander JAA, Olsson U, Alstrom P, Sanmartin I. Accounting for phylogenetic uncertainty in biogeography: A Bayesian approach to dispersal-vicariance analysis of the thrushes (Aves: Turdus). Syst Biol. 2008;57: 257–268. 10.1080/10635150802044003 [DOI] [PubMed] [Google Scholar]
- 61. Takhtajan A, Crovello TJ. Floristic regions of the world Berkeley, CA: University of California Press; 1986. [Google Scholar]
- 62. Tiffney BH. The Eocene North Atlantic land bridge: its importance in Tertiary and modern phytogeography of the Northern Hemisphere. J Arnold Arbor. 1985;66: 243–273. [Google Scholar]
- 63. Milne RI. Northern hemisphere plant disjunctions: A window on Tertiary land bridges and climate change? Ann Bot. 2006;98: 465–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Condamine FL, Sperling FAH, Kergoat GJ. Global biogeographical pattern of swallowtail diversification demonstrates alternative colonization routes in the Northern and Southern hemispheres. J Biogeogr. 2013;40: 9–23. [Google Scholar]
- 65. Wen J. Evolution of eastern Asian-Eastern North American biogeographic disjunctions: A few additional issues. Int J Plant Sci. 2001;162: S117–S122. [Google Scholar]
- 66. Manchester SR. Biogeographical relationships of North American Tertiary floras. Ann Missouri Bot Gard. 1999;86: 472–522. [Google Scholar]
- 67. Xiang QY, Soltis DE, Soltis PS, Manchester SR, Crawford DJ. Timing the eastern Asian–eastern North American floristic disjunction: molecular clock corroborates paleontological estimates. Mol Phylogenet Evol. 2000;15: 462–472. [DOI] [PubMed] [Google Scholar]
- 68. Manchester SR, Tiffney BH. Integration of paleobotanical and neobotanical data in the assessment of phytogeographic history of holarctic angiosperm clades. Int J Plant Sci. 2001;162: S19–S27. [Google Scholar]
- 69. Xiang QYJ, Soltis DE. Dispersal-vicariance analyses of intercontinental disjuncts: Historical biogeographical implications for angiosperms in the Northern Hemisphere. Int J Plant Sci. 2001;162: S29–S39. [Google Scholar]
- 70. Roy M, Clark PU, Barendregt RW, Glasmann JR, Enkin RJ. Glacial stratigraphy and paleomagnetism of late Cenozoic deposits of the north-central United States. Geol Soc Am Bull. 2004;116: 30–41. [Google Scholar]
- 71. Ehlers J, Gibbard PL. The extent and chronology of Cenozoic global glaciation. Quat Int. 2007;164: 6–20. [Google Scholar]
- 72. Holderegger R, Stehlik I, Abbott RJ. Molecular analysis of the Pleistocene history of Saxifraga oppositifolia in the Alps. Mol Ecol. 2002;11: 1409–1418. [DOI] [PubMed] [Google Scholar]
- 73. Turgeon J, Stoks R, Thum RA, Brown JM, Mcpeek MA. Simultaneous Quaternary radiations of three damselfly clades across the Holarctic. Am Nat 2005;165: E78–E107. [DOI] [PubMed] [Google Scholar]
- 74. Hollingsworth PM. Refining the DNA barcode for land plants. Proc Natl Acad Sci USA. 2011;108: 19451–19452. 10.1073/pnas.1116812108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Tsai YHE, Carstens BC. Assessing model fit in phylogeographical investigations: an example from the North American sandbar willow Salix melanopsis . J Biogeogr. 2013;40: 131–141. [Google Scholar]
- 76. Brunsfeld SJ, Miller TR, Carstens BC. Insights into the biogeography of the Pacific Northwest of North America: Evidence from the phylogeography of Salix melanopsis . Syst Bot. 2007;32: 129–139. [Google Scholar]
- 77. Ford CS, Ayres KL, Toomey N, Haider N, Stahl JV, et al. Selection of candidate coding DNA barcoding regions for use on land plants. Bot J Linn Soc. 2009;159: 1–11. [Google Scholar]
- 78. Levin RA, Wagner WL, Hoch PC, Nepokroeff M, Pires JC, et al. Family-level relationships of Onagraceae based on chloroplast rbcL and ndhF data. Am J Bot. 2003;90: 107–115. 10.3732/ajb.90.1.107 [DOI] [PubMed] [Google Scholar]
- 79. Kress WJ, Erickson DL, Jones FA, Swenson NG, Perez R, et al. Plant DNA barcodes and a community phylogeny of a tropical forest dynamics plot in Panama. Proc Natl Acad Sci USA. 2009;106: 18621–6. 10.1073/pnas.0909820106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Percy DM, Argus GW, Cronk QC, Fazekas AJ, Kesanakurti PR, et al. Understanding the spectacular failure of DNA barcoding in willows (Salix): Does this results from a trans-specific selective sweep? Mol Ecol. 2014;23: 4737–4756. 10.1111/mec.12837 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Branch support is Bayesian posterior probabilities and ML bootstrap values.
(TIF)
We indicated species name (according in Argus [4] and IPNI), their status in America (native or introduced), their principal native area, their subgenus (Argus [4]), the herbarium informations: live collections of the Montreal Botanical Garden (MBG), the Canadian Museum of Nature (CAN), the Herbarium of the University of Texas (TEX), the University of Arizona Herbarium (ARIZ), the Missouri Botanical Garden Herbarium (MO), or unmounted. Finally, we have indicated the GenBank number.
(DOCX)
Data Availability Statement
All DNA sequences are available from the Genbank database (accession numbers: see S1 Table).