Abstract
The catalytic (C) subunit of PKA was the first protein kinase structure to be solved, and it continues to serve as the prototype for the protein kinase superfamily. In contrast, by comparing many active and inactive kinases, we developed a novel ‘spine’ concept where every active kinase is composed of two hydrophobic spines anchored to a hydrophobic F-helix. The R-spine (regulatory spine) is dynamically assembled, typically by activation loop phosphorylation, whereas the C-spine (catalytic spine) is completed by the adenine ring of ATP. In the present paper, we show how the spine concept can be applied to B-Raf, specifically to engineer a kinase-dead pseudokinase. To achieve this, we mutated one of the C-spine residues in the N-lobe (N-terminal lobe), Ala481, to phenylalanine. This mutant cannot bind ATP and is thus kinase-dead, presumably because the phenylalanine ring fills the adenine-binding pocket. The C-spine is thus fused. However, the A481F mutant is still capable of binding wild-type B-Raf and wild-type C-Raf, and dimerization with a wild-type Raf leads to downstream activation of MEK [MAPK (mitogen-activated protein kinase)/ERK (extracellular-signal-regulated kinase) kinase] and ERK. The mutant requires dimerization, but is independent of Ras and does not require enzymatic activity. By distinguishing between catalytic and scaffold functions of B-Raf, we define kinases as being bifunctional and show that, at least in some cases, the scaffold function is sufficient for downstream signalling. Since this alanine residue is one of the most highly conserved residues in the kinome, we suggest that this may be a general strategy for engineering kinase-dead pseudokinases and exploring biological functions that are independent of catalysis.
Keywords: ATP, GTP, protein kinase, protein kinase A (PKA), pseudokinase, Raf
History and evolution of protein kinases
Protein kinases represent one of the largest gene families in eukaryotic organisms [1]. They have evolved to be dynamic molecular switches, similar to the G-proteins, and they regulate much of biology [2]. Because of their widespread importance, dysfunctional protein kinases are also associated with many diseases, especially cancers, where a single mutation can create a driver of tumorigenesis. The importance of protein phosphorylation as a regulatory mechanism was first discovered with the pioneering work of Krebs and Fischer and the regulation of glycogen phosphorylase by phosphorylase kinase [3,4]. Glycogen phosphorylase was the first regulatory protein kinase to be discovered and PKA (cAMP-dependent protein kinase) was the second [5]. Protein kinase C was discovered shortly thereafter by Nishazuka and colleagues [6,7]. With the advent of cloning, it quickly became clear not only that that this was a large family, but also that it included kinases that could phosphorylate tyrosine as well as serine and threonine [8–10].
On the basis of just a handful of kinases, Hanks, Quinn and Hunter [11] aligned the different sequence motifs that were shared by a kinase core and classified them into 11 subdomains. Our understanding of the protein kinase family made another major advance when the first protein kinase structure was solved [12]. Our structure of the PKA catalytic subunit not only showed the fold that would be conserved by all members of the family, but also gave functional significance to the subdomains and to the conserved sequence motifs that mostly clustered around the active-site cleft between two lobes: the N-lobe (N-terminal lobe) and C-lobe (C-terminal lobe) [13]. The adenine ring of ATP is buried at the base of the cleft between the two lobes, allowing the phosphates to extend out towards the edge of the cleft where the substrate is docked [14]. These first structures of PKA also showed the structural importance of the AL (activation loop) phosphate since they represented a fully active protein kinase that was phosphorylated on the AL and locked into a closed conformation. The subsequent structure of a ternary complex with a pseudosubstrate inhibitor peptide provided a glimpse of what a transition state complex might look like [15].
Although these crystal structures provide a static picture of a protein kinase ternary complex, they do not tell us about dynamics or flexibility. For this we need NMR, and results from Veglia and colleagues [16–19] have defined a conformational range of dynamics that extend from a catalytically uncommitted state for the apoenzyme, to a ‘committed’ state that results when MgATP and/or peptide is added [18]. Although the complex is more closed in the ternary complex, the backbone motions in the millisecond–microsecond range are much more dynamic. In the presence of PKI (protein kinase inhibitor), ATP and two Mg2+ ions, the dynamic properties of the pseudosubstrate complex are almost completely quenched.
Two hydrophobic spines define the core architecture of all protein kinases
Because of the widespread correlation between disease and dysfunctional protein kinases, the protein kinases have become major therapeutic targets, and, as a result, many protein kinase structures have been solved by academics, by structural genomics consortia, and by the biotechnology community. By having many kinase structures to compare (in contrast with delving deeply into the structure and function of one protein kinase, as we have done with PKA), we could explore common structural features in addition to just the conserved sequence motifs. One of the most important features of these enzymes is their dynamic regulation, which is frequently achieved by phosphorylation of the AL. By comparing active and inactive kinases, we discovered that there is a conserved hydrophobic core architecture that is shared by all protein kinases in addition to the conserved sequence motifs [20–22].
A fundamental feature of this core architecture is best described in terms of a ‘spine’ model where two hydrophobic spines are anchored to the long hydrophobic αF-helix which spans the entire C-lobe. This buried hydrophobic helix is an unusual feature for a globular proteins such as the protein kinases. Usually such a hydrophobic helix is associated with membranes. The two spines are referred to as the C-spine (catalytic spine) and the R-spine (regulatory spine). The C-spine is assembled by the binding of ATP where the adenine ring is lodged between two N-lobe spine residues (Ala70 and Val57 in PKA) and one C-spine residue (Leu173 in PKA) from the C-lobe (Figure 1). In contrast with the C-spine, the R-spine is typically assembled and disassembled, or at least stabilized, by phosphorylation of the AL. A fundamental feature that emerged from the initial computational analysis of active and inactive kinases is that the R-spine is dynamically regulated and typically broken in inactive kinases. Phosphorylation of the AL stabilizes the R-spine and prevents its ‘melting’ back into the inactive conformation, which tends to be more stable. This leaves most kinases also sensitive to nearby phosphatases which in part explains why the kinases function as such powerful and dynamically regulated ‘molecular switches’.
Figure 1. Internal hydrophobic core architecture defines every active kinase.
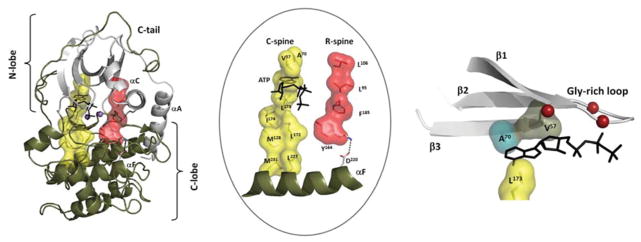
On the left, using PKA as a prototype, we show the overall hydrophobic core that consists of two hydrophobic spines that are anchored to the F-helix. The core architecture, highlighted in the centre panel, shows the R-spine in red and the C-spine in yellow. The R-spine is dynamically assembled, whereas the C-spine is completed by the binding of ATP. The right-hand panel shows how the adenine ring is sandwiched between Ala70 (in β-strand 3) and Val57 (in β-strand 2) on the top and Leu173 on the bottom. WT, wild-type
Pseudokinases versus active kinases
An analysis of the initial kinome revealed a curious thing. In addition to the conventional kinases, which shared all of the essential catalytic residues, approximately 10% of the kinome were found to be missing an essential catalytic residue [23–26]. These were referred to as ‘pseudokinases’ and were predicted to be devoid of catalytic activity. However, this prediction proved to be incorrect when the structure of WNK1 (with no lysine kinase 1) was solved [27,28]. This kinase lacked the highly conserved lysine residue in β-strand 3 which binds to the α- and β-phosphates of ATP and to the conserved glutamate residue in the αC-helix. The structure showed that WNK1 had evolved a novel mechanism whereby another basic amino acid filled the same space as the catalytic lysine residue and apparently can carry out the same function. It was thus a fully active kinase, even though it lacked an essential residue. Another interesting kinase that was predicted initially to be a pseudokinase was CASK (Ca2+/calmodulin-activated serine kinase) because it lacked both the residues that bind to the Mg2+ ions that position the ATP phosphates (Asp185 in the DFG motif and Asn171 in the catalytic loop, using PKA nomenclature). However, it was later demonstrated that CASK could transfer the γ-phosphate from ATP to a protein substrate, neurexin, in an Mg2+-independent manner [24,29]. This is not necessarily true for other pseudokinases. In some cases such as VRK3 (vaccinia-related kinase 3) (Figure 2) the kinase is completely dead because a hydrophobic side chain fills the space that is usually occupied by the adenine ring of ATP [25,30].
Figure 2. Mechanism for ‘fusing’ the C-spine.
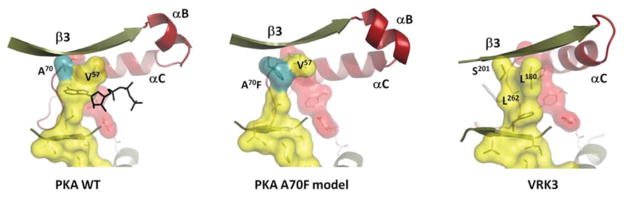
Two elements of the endogenous C-spine are fused by the adenine ring of ATP (left). We predict that the phenylalanine side chain in the A70F mutant will fill in the adenine-binding pocket (centre). In VRK3, several residues (Leu180, Ser201 and Leu262) fill in the adenine-binding pocket making it unable to bind ATP. Leu180 and Ser201 are equivalent to Val57 and Ala70 respectively in PKA, whereas Leu262 corresponds to Val123 from the hinge.
Functional properties of the pseudokinases
Although classified as pseudokinases because they lack critical catalytic residues, increasing numbers of pseudokinases such as KSR (kinase suppressor of Ras) and HER3 (human epidermal growth factor receptor 3) have been shown to retain some residual kinase activity [31,32]. Whether this level of kinase activity is important for their function, however, is controversial. Mutations in catalytic residues in general do not impair ATP binding. For example, kinases that lack the Lys72, Asp166 or Asp184 equivalents can still bind ATP with an affinity similar to that of the wild-type protein, but cannot correctly position the γ-phosphate for efficient transfer to a substrate [33]. In the case of CASK or KSR, this low level of kinase activity may be sufficient for phosphotransfer to a very specific substrate that is co-localized in close proximity to the kinase. In other cases, the binding of ATP alone may be important or sufficient to convey a functional property to the kinase even if transfer of the phosphate is not essential. One has only to look at small G-proteins to appreciate how ATP or GTP binding is sufficient to mediate a biological response [34]. This suggests that some pseudokinases may function as switches using ATP binding (or ATP hydrolysis) to oscillate between an active and inactive conformations, but may not have to actually transfer the phosphate to a protein substrate.
How do we then establish whether a true kinase-dead pseudokinase can still mediate a biological response? An important function is indicated when knocking out the gene gives a biological phenotype. A chemical validation would require strategies that would fix the pseudokinase in either the active or inactive conformation and comparing their functions. This function may not be restricted to pseudokinases and could also be part of the function of traditional kinases. Are, in fact, all kinases bifunctional? To address this, we turn to the Rafs.
Raf activation
In humans and other higher eukaryotes, there are three Raf homologues: A-Raf, B-Raf and C-Raf. Epistasis screens in fruitflies and nematodes identified KSR1 and KSR2 as proteins highly similar to the Raf family members and part of the pathway, either in a position that is parallel to or upstream of Raf. For many years, it was assumed that KSR was a pseudokinase because it lacked the equivalent of Lys72, although Lys72 is present in KSRs from lower eukaryotes such as Drosophila [35–37].
The process for activation of B-Raf and C-Raf in cells is complex and highly regulated by a series of events, some of which are dependent on catalytic activity and others which are not. Basically, B-Raf and C-Raf are maintained in an inactive state by interactions of the NTD (N-terminal domain) with the kinase domain [38,39]. This probably represents the most stable state of B-Raf and C-Raf, although no structures are available of a full-length kinase. Activation is transient and dynamic. The first step is the binding of Ras-GTP to the NTD. This releases the kinase domain rendering it much more dynamic. What follows next is dimerization with another Raf, which then leads to autophosphorylation of the AL. This ‘scaffold’ function of the Rafs has been well documented in crystal structures [40]. Whereas dimerization alone seems able to induce the active conformation and the assembly of the R-spine, the spine is subsequently stabilized by phosphorylation of the AL, which then supposedly leads to the release of the active kinase (Figure 3). This process is reversible due to phosphatases, which remove the phosphates from the AL. This mechanism for activation of Raf, coupled with inactivation by phosphatases, which are localized in close proximity to the kinase and typically constitutively active, creates a highly dynamic ‘molecular switch’.
Figure 3. The A70F mutation short-circuits the activation mechanism for B-Raf.
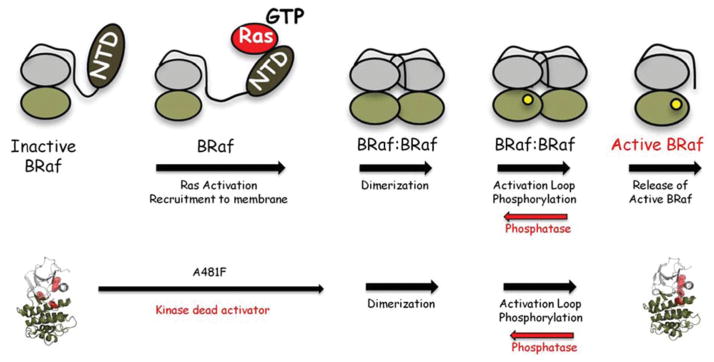
The general steps of the activation process are summarized on the top. Mutating Ala481 to phenylalanine creates a dead kinase that short-circuits part of the activation process, but can still dimerize and lead to downstream activation of ERK.
Discriminating between the catalytic and scaffold functions of the Raf family members
To discriminate between the scaffold and catalytic functions of the Raf homologues, we developed a new strategy that was based on the C-spine residues. Ala70 in PKA is a C-spine residue that sits on top of the adenine ring of ATP. This alanine is one of the most highly conserved residues in the kinase core. Could we abolish ATP binding by replacing this residue with a large hydrophobic residue? To test this hypothesis, we replaced the alanine equivalent in B-Raf (Ala481) with a series of hydrophobic residues. Replacing it with a large hydrophobic residue such as isoleucine or methionine did not abolish ATP binding, but replacing it with phenylalanine was sufficient to abolish ATP binding [41]. We then replaced the equivalent alanine residue in C-Raf and KSR with phenylalanine, and in each case the mutant protein could no longer bind to ATP. All three were thus catalytically ‘dead’ (Figure 2).
To determine whether this kinase-dead form of B-Raf was still capable of activating downstream signalling in cells, we expressed the mutant in HEK (human embryonic kidney)-293 cells. The B-Raf(A418F) mutant, although no longer able to bind ATP, was able to activate downstream ERK (extracellular-signal-regulated kinase) in a Ras-independent manner. To determine whether dimerization was still required for downstream activation by the dead B-Raf, we replaced Arg509 at the dimer interface with histidine, a mutation that is known to reduce dimerization [40]. This double mutant was no longer able to active MEK [MAPK (mitogen-activated protein kinase)/ERK kinase] and ERK. Thus, by engineering a kinase-dead version of B-Raf, we demonstrated that it is perfectly capable of activating wild-type C-Raf or wild-type B-Raf. The mutation thus short-circuits the first part of the activation process (Figure 3). Once the dead mutant forms a dimer with a wild-type Raf, it can lead to the activation of the wild-type Raf. It is a stable scaffold that lacks kinase activity.
Dynamic bifunctional molecular switches
In 2006, we first identified the hydrophobic R-spine as a conserved feature of every active protein kinase and hypothesized that it would be a driving force for kinase activation [20]. The subsequent description of the C-spine that, along with the R-spine, is anchored to the hydrophobic F-helix, defined a new conceptual way to look at protein kinases. This hydrophobic core hypothesis has subsequently been validated as a new framework for understanding protein kinase activation, drug design and drug resistance [42–44]. Assembly of the R-spine is the driving force for the molecular switch mechanism that defines this enzyme family. Our subsequent work with B-Raf allowed us to create a kinase-dead protein that was still capable of functioning as an activator of downstream MEK and ERK. This strategy provides a general tool for creating a catalytically dead kinase that is still correctly folded and capable of serving as a scaffold or as an allosteric activator. It is a strategy that can be used, in principle, to analyse any kinase, but, in particular, the pseudokinases where activity may be compromised. In some cases, the actual transfer of the phosphate may be required for function, whereas in others such as VRK3, the ‘scaffold’ function is sufficient. We must now therefore consider all kinases as bifunctional molecular switches. By modifying critical C-spine residues that appear to be capable of ‘fusing’ the C-spine, we provide a strategy for resolving this question.
Acknowledgments
Funding
This work was funded by the National Institutes of Health [grant numbers GM19301 (to S.S.T.) and AI57966 (to A.S.)]. H.S.M. was supported by the National Science Foundation [grant number DGE1144086].
Abbreviations used
- AL
activation loop
- CASK
Ca2+/calmodulin-activated serine kinase
- C-lobe
C-terminal lobe
- C-spine
catalytic spine
- ERK
extracellular-signal-regulated kinase
- KSR
kinase suppressor of Ras
- MEK
MAPK (mitogen-activated protein kinase)/ERK kinase
- N-lobe
N-terminal lobe
- NTD
N-terminal domain
- PKA
cAMP-dependent protein kinase
- R-spine
regulatory spine
- VRK3
vaccinia-related kinase 3
- WNK1
with no lysine kinase 1
References
- 1.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. The protein kinase complement of the human genome. Science. 2002;298:1912–1934. doi: 10.1126/science.1075762. [DOI] [PubMed] [Google Scholar]
- 2.Taylor SS, Keshwani MM, Steichen JM, Kornev AP. Evolution of the eukaryotic protein kinases as dynamic molecular switches. Philos Trans R Soc London Ser B. 2012;367:2517–2528. doi: 10.1098/rstb.2012.0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krebs EG, Fischer EH. The phosphorylase b to a converting enzyme of rabbit skeletal muscle. Biochim Biophys Acta. 1956;20:150–157. doi: 10.1016/0006-3002(56)90273-6. [DOI] [PubMed] [Google Scholar]
- 4.Krebs EG, Graves DJ, Fischer EH. Factors affecting the activity of muscle phosphorylase b kinase. J Biol Chem. 1959;234:2867–2873. [PubMed] [Google Scholar]
- 5.Walsh DA, Perkins JP, Krebs EG. An adenosine 3′,5′-monophosphate-dependant protein kinase from rabbit skeletal muscle. J Biol Chem. 1968;243:3763–3765. [PubMed] [Google Scholar]
- 6.Takai Y, Kishimoto A, Inoue M, Nishizuka Y. Studies on a cyclic nucleotide-independent protein kinase and its proenzyme in mammalian tissues. I Purification and characterization of an active enzyme from bovine cerebellum. J Biol Chem. 1977;252:7603–7609. [PubMed] [Google Scholar]
- 7.Castagna M, Takai Y, Kaibuchi K, Sano K, Kikkawa U, Nishizuka Y. Direct activation of calcium-activated, phospholipid-dependent protein kinase by tumor-promoting phorbol esters. J Biol Chem. 1982;257:7847–7851. [PubMed] [Google Scholar]
- 8.Collett MS, Erikson RL. Protein kinase activity associated with the avian sarcoma virus src gene product. Proc Natl Acad Sci USA. 1978;75:2021–2024. doi: 10.1073/pnas.75.4.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckhart W, Hutchinson MA, Hunter T. An activity phosphorylating tyrosine in polyoma T antigen immunoprecipitates. Cell. 1979;18:925–933. doi: 10.1016/0092-8674(79)90205-8. [DOI] [PubMed] [Google Scholar]
- 10.Hunter T, Sefton BM. Transforming gene product of Rous sarcoma virus phosphorylates tyrosine. Proc Natl Acad Sci USA. 1980;77:1311–1315. doi: 10.1073/pnas.77.3.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanks SK, Quinn AM, Hunter T. The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science. 1988;241:42–52. doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 12.Knighton DR, Zheng JH, ten Eyck LF, Ashford VA, Xuong NH, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 13.Knighton DR, Zheng JH, Ten Eyck LF, Xuong NH, Taylor SS, Sowadski JM. Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:414–420. doi: 10.1126/science.1862343. [DOI] [PubMed] [Google Scholar]
- 14.Zheng J, Knighton DR, ten Eyck LF, Karlsson R, Xuong N, Taylor SS, Sowadski JM. Crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MgATP and peptide inhibitor. Biochemistry. 1993;32:2154–2161. doi: 10.1021/bi00060a005. [DOI] [PubMed] [Google Scholar]
- 15.Madhusudan, Akamine P, Xuong NH, Taylor SS. Crystal structure of a transition state mimic of the catalytic subunit of cAMP-dependent protein kinase. Nat Struct Biol. 2002;9:273–277. doi: 10.1038/nsb780. [DOI] [PubMed] [Google Scholar]
- 16.Masterson LR, Mascioni A, Traaseth NJ, Taylor SS, Veglia G. Allosteric cooperativity in protein kinase A. Proc Natl Acad Sci USA. 2008;105:506–511. doi: 10.1073/pnas.0709214104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masterson LR, Cheng C, Yu T, Tonelli M, Kornev A, Taylor SS, Veglia G. Dynamics connect substrate recognition to catalysis in protein kinase A. Nat Chem Biol. 2010;6:821–828. doi: 10.1038/nchembio.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masterson LR, Shi L, Metcalfe E, Gao J, Taylor SS, Veglia G. Dynamically committed, uncommitted, and quenched states encoded in protein kinase A revealed by NMR spectroscopy. Proc Natl Acad Sci USA. 2011;108:6969–6974. doi: 10.1073/pnas.1102701108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masterson LR, Cembran A, Shi L, Veglia G. Allostery and binding cooperativity of the catalytic subunit of protein kinase A by NMR spectroscopy and molecular dynamics simulations. Adv Protein Chem Struct Biol. 2012;87:363–389. doi: 10.1016/B978-0-12-398312-1.00012-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kornev AP, Haste NM, Taylor SS, Ten Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci USA. 2008;105:14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor SS, Kornev AP. Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem Sci. 2011;36:65–77. doi: 10.1016/j.tibs.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR. Emerging roles of pseudokinases. Trends Cell Biol. 2006;16:443–452. doi: 10.1016/j.tcb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 24.Kannan N, Taylor SS. Rethinking pseudokinases. Cell. 2008;133:204–205. doi: 10.1016/j.cell.2008.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kornev AP, Taylor SS. Pseudokinases: functional insights gleaned from structure. Structure. 2009;17:5–7. doi: 10.1016/j.str.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zeqiraj E, van Aalten DM. Pseudokinases: remnants of evolution or key allosteric regulators? Curr Opin Struct Biol. 2010;20:772–781. doi: 10.1016/j.sbi.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu B, English JM, Wilsbacher JL, Stippec S, Goldsmith EJ, Cobb MH. WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem. 2000;275:16795–16801. doi: 10.1074/jbc.275.22.16795. [DOI] [PubMed] [Google Scholar]
- 28.Min X, Lee BH, Cobb MH, Goldsmith EJ. Crystal structure of the kinase domain of WNK1, a kinase that causes a hereditary form of hypertension. Structure. 2004;12:1303–1311. doi: 10.1016/j.str.2004.04.014. [DOI] [PubMed] [Google Scholar]
- 29.Mukherjee K, Sharma M, Urlaub H, Bourenkov GP, Jahn R, Südhof TC, Wahl MC. CASK functions as a Mg2+-independent neurexin kinase. Cell. 2008;133:328–339. doi: 10.1016/j.cell.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G. Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure. 2009;17:128–138. doi: 10.1016/j.str.2008.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taylor SS, Kornev AP. Yet another “active” pseudokinase, Erb3. Proc Natl Acad Sci USA. 2010;107:8047–8048. doi: 10.1073/pnas.1003436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brennan DF, Dar AC, Hertz NT, Chao WC, Burlingame AL, Shokat KM, Barford D. A Raf-induced allosteric transition of KSR stimulates phosphorylation of MEK. Nature. 2011;472:366–369. doi: 10.1038/nature09860. [DOI] [PubMed] [Google Scholar]
- 33.Iyer GH, Garrod S, Woods VL, Jr, Taylor SS. Catalytic independent functions of a protein kinase as revealed by a kinase-dead mutant: study of the Lys72His mutant of cAMP-dependent kinase. J Mol Biol. 2005;351:1110–1122. doi: 10.1016/j.jmb.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 34.Londos C, Salomon Y, Lin MC, Harwood JP, Schramm M, Wolff J, Rodbell M. 5′-Guanylylimidodiphosphate, a potent activator of adenylate cyclase systems in eukaryotic cells. Proc Natl Acad Sci USA. 1974;71:3087–3090. doi: 10.1073/pnas.71.8.3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kornfeld K, Hom DB, Horvitz HR. The ksr-1 gene encodes a novel protein kinase involved in Ras-mediated signaling in C. elegans. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 36.Sundaram M, Han M. The C. elegans ksr-1 gene encodes a novel Raf-related kinase involved in Ras-mediated signal transduction. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 37.Therrien M, Chang HC, Solomon NM, Karim FD, Wassarman DA, Rubin GM. KSR, a novel protein kinase required for RAS signal transduction. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 38.Dhillon AS, Kolch W. Untying the regulation of the Raf-1 kinase. Arch Biochem Biophys. 2002;404:3–9. doi: 10.1016/s0003-9861(02)00244-8. [DOI] [PubMed] [Google Scholar]
- 39.Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, Kolch W. Raf family kinases: old dogs have learned new tricks. Genes Cancer. 2011;2:232–260. doi: 10.1177/1947601911407323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rajakulendran T, Sahmi M, Lefrançois M, Sicheri F, Therrien M. A dimerization-dependent mechanism drives RAF catalytic activation. Nature. 2009;461:542–545. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 41.Hu J, Yu H, Kornev AP, Zhao J, Filbert EL, Taylor SS, Shaw AS. Mutation that blocks ATP binding creates a pseudokinase stabilizing the scaffolding function of kinase suppressor of Ras, CRAF and BRAF. Proc Natl Acad Sci USA. 2011;108:6067–6072. doi: 10.1073/pnas.1102554108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bossi RT, Saccardo MB, Ardini E, Menichincheri M, Rusconi L, Magnaghi P, Orsini P, Avanzi N, Borgia AL, Nesi M, et al. Crystal structures of anaplastic lymphoma kinase in complex with ATP competitive inhibitors. Biochemistry. 2010;49:6813–6825. doi: 10.1021/bi1005514. [DOI] [PubMed] [Google Scholar]
- 44.Jura N, Zhang X, Endres NF, Seeliger MA, Schindler T, Kuriyan J. Catalytic control in the EGF receptor and its connection to general kinase regulatory mechanisms. Mol Cell. 2011;42:9–22. doi: 10.1016/j.molcel.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]


