Abstract
Gelatinase B/matrix metalloproteinase-9 (MMP-9) (EC 3.4.24.35) cleaves many substrates and is produced by most cell types as a zymogen, proMMP-9, in complex with the tissue inhibitor of metalloproteinases-1 (TIMP-1). Natural proMMP-9 occurs as monomers, homomultimers, and heterocomplexes, but our knowledge about the overall structure of proMMP-9 monomers and multimers is limited. We investigated biochemical, biophysical, and functional characteristics of zymogen and activated forms of MMP-9 monomers and multimers. In contrast to a conventional notion of a dimeric nature of MMP-9 homomultimers, we demonstrate that these are reduction-sensitive trimers. Based on the information from electrophoresis, atomic force microscopy (AFM) and transmission electron microscopy (TEM), we generated a 3Dstructure model of the proMMP-9 trimer. Remarkably, the proMMP-9 trimers possessed a 50-fold higher affinity for TIMP-1 than the monomers. In vivo, this finding was reflected in a higher extent of TIMP-1 inhibition of angiogenesis induced by trimers versus monomers. Our results show that proMMP-9 trimers constitute a novel structural and functional entity that is differentially regulated by TIMP-1.
Keywords: MMP-9, TIMP-1, angiogenesis, homotrimer, protease, model
INTRODUCTION
At present, matrix metalloproteinase-9 (MMP-9/gelatinase B) is becoming one of the most studied enzymes in biology, immunology and medicine [1]. MMP-9 has been defined as a molecular trigger of the angiogenic switch in cancer progression [2] and is the most complex member of the entire MMP family in terms of protein domain structure. In contrast to the structure of MMP-2 (MMP-2/gelatinase A), which is fully resolved [3], the structure of full-length MMP-9 zymogen (proMMP-9) remains elusive in part because proMMP-9 naturally occurs as monomers, heterocomplexes and homomultimers.
MMP-9 monomers are highly flexible entities. This flexibility is attributed to a central 54-residue proline- and glycine-rich O-glycosylated (OG) domain that bridges the catalytic enzyme core and the hemopexin domain, allowing the terminal domains to act independently of each other (Figures 1A and 1B) [1, 4]. Moreover, it was suggested that this feature is crucial for the translocation of MMP-9 along collagen fibrils [5].
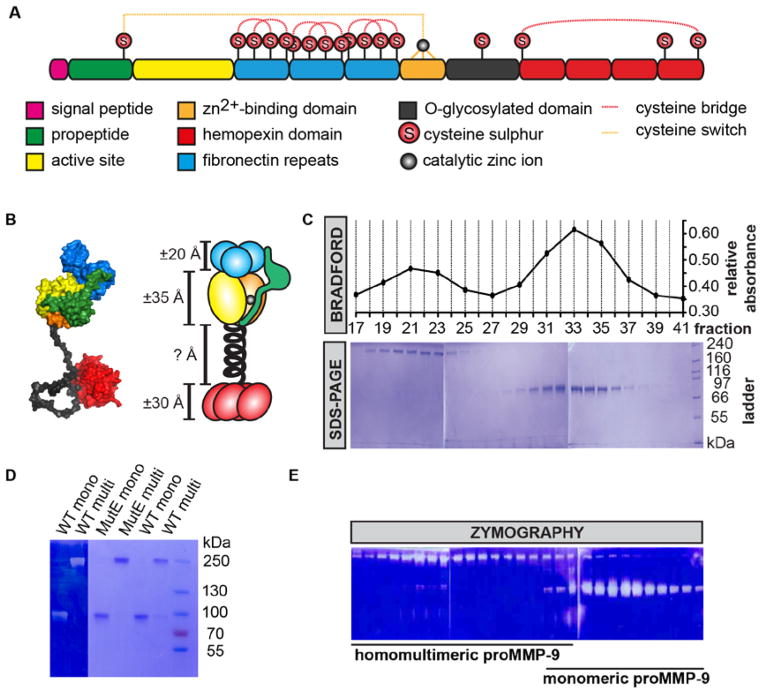
Figure 1. Separation of proMMP-9 monomers and multimers.
(A) Primary structure of full-length human proMMP-9 with indication of protein domains, cysteine sulfhydryl (S) groups, the propeptide cysteine switch (orange dotted line) and cysteine bridges (red dotted lines). (B) Scaled, schematic model representation of the proMMP-9 monomer, including the propeptide (green), the fibronectin like domain (blue), the Zn2+-binding domain (orange), the catalytic domain (yellow), the flexible OG domain (black) and the hemopexin-like domain (red). In the atomic model the central cysteine in the OG-domain is bridged to the free cysteine in the hemopexin domain [12]. (C) Separation of proMMP-9 MutE monomers and multimers by ultracentrifugation. The graph shows the relative absorbance obtained for each fraction collected from a single tube after ultracentrifugation (top). Two absorbance peaks were detected, consistent with the separation of proMMP-9 MutE multimers and monomers. The purity of each fraction was determined by SDS-PAGE (bottom). (D) Zymography (left) and SDS-PAGE (right) analysis of the WT proMMP-9 and MutE proMMP-9 separated by ultracentrifugation. The right lane contains a set of standard proteins with known molecular masses, indicated in kDa. (E) Separation of recombinant human WT proMMP-9 and zymography analysis of each fraction collected from a single tube after ultracentrifugation.
The neutrophil gelatinase B-associated lipocalin (NGAL)-MMP-9 heterocomplex is a typical product of neutrophils, a unique cell type that does not synthesize MMP-2 or TIMP-1 [6–7]. Interestingly, tissue macrophages can alter their phenotype: whereas M1-polarized macrophages produce proMMP-9 in complex with TIMP-1, M2-polarized macrophages and tumor-associated macrophages produce TIMP-1-deficient proMMP-9, which renders it with high angiogenic capacity [8,9].
Neutrophils, macrophages and other cell types often produce homomultimers of MMP-9. Homomultimerization of MMP-9 involves cysteine bridge formation as indicated by the disintegration of multimers into monomers after chemical reduction [6, 10]. Two supposedly free cysteines in the monomeric proMMP-9 are located in the center of the OG domain and the fourth blade of the hemopexin domain (Figure 1A) [1]. Based on the crystal structure of the recombinant hemopexin domain of proMMP-9, it has been assumed that the latter cysteine is buried and not accessible for cysteine bridging [11]. This theoretical rationale has provided the basis for a common view that homomultimers are dimers (vide infra). Molecular modeling based on sedimentation data, suggests that two free cysteines in a proMMP-9 monomer form an intramolecular cysteine bridge between the OG and hemopexin domains [12] (Figure 1B). In several studies, proMMP-9 homomultimers have been designated as dimers [11, 13–15]. However, experimental evidence of the actual structure of MMP-9 homomultimers is only circumstantial. There is also a paucity of studies addressing the possible functions of MMP-9 monomers versus homomultimers in physiological processes [13–15]. This study was conducted to identify the molecular structure of proMMP-9 homomultimers and to establish their unique in vitro and in vivo biological functions.
MATERIALS AND METHODS
Protein production and purification
A catalytically inactive mutant of proMMP-9 with the catalytic Glu402 mutated into Ala (proMMP-9 MutE) and wild-type (WT) proMMP-9 were expressed in Sf9 insect cells and purified by gelatin-Sepharose chromatography as previously described [12, 16]. In contrast to catalytically active WT proMMP-9, proMMP-9 MutE remained intact after prolonged incubation at room temperature, thereby facilitating structure-function studies. When necessary, WT proMMP-9 was activated with the catalytic domain of MMP-3 as previously described [17]. Recombinant human TIMP-1 was purchased and handled according to the manufacturer’s instructions (R&D Systems, Minneapolis, MN, USA).
Separation of proMMP-9 monomers and multimers
The proMMP-9 monomers and multimers were separated by glycerol gradient ultracentrifugation. Prior to centrifugation, 12 ml polyallomer tubes (Thermo Scientific, Waltham, MA, USA) were coated with Sigmacote® (Sigma, St. Louis, MO, USA). Dilutions of 20%, 25% and 30% glycerol were prepared in assay buffer (0.1M Tris-HCl pH 7.4, 0.1M NaCl, 10 mM CaCl2) and an equal volume of each solution was applied per polyallomer tube. Subsequently, 100 μg of proMMP-9 was loaded on each gradient. Ultracentrifugation (Sorvall WX Ultra 80 ultracentrifuge and TH-641 swinging bucket rotor, Thermo Scientific, Waltham, MA, USA) was executed for 46 h at 300 000 × g. After centrifugation, the fractions were collected (from bottom to top) with the use of a peristaltic pump (Minipuls 3, Gilson, Middleton, WI, USA) connected to a fraction collector (model 2128, Bio-Rad, Hercules, CA, USA). The fractions were analyzed for protein content and the positions of the proMMP-9 MutE monomer and multimer peak fractions were determined with the method of Bradford [18]. Next, the fractions containing monomeric or trimeric proMMP-9 were pooled and subjected to ultrafiltration in Vivaspin 4 concentrators with a cut-off-value of 30 kDa (Sartorius Stedim Biotech, Göttingen, Germany). Glycerol buffer was replaced by assay buffer. Finally, the concentration and purity of proMMP-9 preparations were determined by absorbance at 280 nm and SDS-PAGE analysis.
SDS-PAGE, zymography and Western blot analysis
Where indicated, proMMP-9 was analyzed by PAGE. We used a discontinuous buffer system with a 5% PAA stacking gel and a 7.5% polyacrylamide resolving gel in Tris-HCl/pH 6.8 and Tris-HCl/pH 8.8 buffer, respectively. For the analysis of high-molecular weight proMMP-9 forms, the ProSieve® 50 gel solution (Lonza Group Ltd, Basel, Switzerland) with a Tris/glycine buffer system was used. The protein bands were detected by Coomassie brilliant blue staining. The gels were further analyzed with ImageJ software [19]. The retention factors for individual marker-proteins (HiMark, Invitrogen Ltd, Paisley, UK) were plotted against the logarithm of their corresponding molecular weights [20], allowing then to determine the retention factor (Rf) for each of analyzed proteins. Next, zymography analysis was performed as detailed elsewhere [21]. Western blot analysis was performed with the use of an antibody against human MMP-9, as previously described [8].
Redox experiments
Purified proMMP-9 MutE monomers, at a concentration of 2 μM, were incubated at 37°C with increasing concentrations (0.03 mM, 0.3 mM, 3 mM, and 30 mM) of GSH and GSSG. All dilutions were prepared in 150 mM Tris buffer (pH 8.6) based on the conditions previously established for the correct folding of chemokines [22]. At multiple time points, samples were analyzed by SDS-PAGE on non-reducing 7.5% Prosieve® 50 gels stained with Coomassie brilliant blue.
Glycan analysis
N-glycans were released from gelatinase B samples by in situ digestion with N-glycosidase F (PNGase F, Prozyme) in-gel-blocks as previously described [23]. Briefly, samples were reduced and alkylated, set into SDS-gel blocks and washed. Following N-glycans release by PNGase F, O-glycans were released by ammonia-based β-elimination [24], and fluorescently labelled with 2-aminobenzamide (2AB) by reductive amination [25] (LudgerTag 2-AB labeling kit, LudgerLtd., Abingdon, UK). All enzymes were purchased from Prozyme. The 2AB-labelled glycans were digested for 18 h at 37°C in 10 μl of 50 mM sodium acetate buffer, pH 5.5, supplemented with the following enzymes: ABS-Arthrobacter ureafaciens sialidase (EC 3.2.1.18), 1 U/ml; BTG-bovine testes β-galactosidase (EC 3.2.1.23), 1 U/ml; and BKF – bovine kidney α-fucosidase (EC 3.2.1.51), 1 U/ml. After incubation, the enzymes were removed by filtration through protein binding EZ filters (Millipore Corporation, Bedford, MA, USA), and the N-glycans were analyzed by HILIC. HILIC was performed using a TSK-Gel Amide-80 4.6 × 250 mm column (Anachem, Luton, UK) on a 2695 Alliance separations module (Waters, Milford, MA, USA) equipped with a Waters temperature control module and a Waters 2475 fluorescence detector. Solvent A was 50 mM formic acid adjusted to pH 4.4 with ammonia solution. Solvent B was acetonitrile. The column temperature was set to 30°C. Conditions used were as follows (180-min method): a linear gradient of 20 to 58% solvent A at a flow rate of 0.4 ml/min over 152 min, followed by 3 min at 58–100% A, then increasing the flow rate to 1 ml/min for 5 min at 100% A, returning to 20% A over 1 min, running at 20% A for 14 min at 1 ml/min, then returning to 0.4 ml/min over 1.5 min, finishing with 1.5 min flow at 20% A, giving a total run time of 180 min [26]. Fluorescence was measured at 420 nm with excitation at 330 nm. The system was calibrated using an external standard of hydrolyzed and 2AB-labeled glucose oligomers to create a dextran ladder, as described previously [23].
Atomic force microscopy (AFM)
Freshly cleaved Mica (Ted Pella, Inc., USA) was incubated with 0.5 % (3-aminopropyl)triethoxysilane (APTES, 99 %; Sigma Aldrich, Israel) in toluene (analytical grade; Frutarom, Israel) for 10 min and successively rinsed with 3× 200 μl of absolute ethanol (Bio-Lab, Israel), 3× 200 μl of acetone (99.5 %; Mallinckrodt Chemicals, USA) and 3× 200 μl of ethanol. The amine functionalized Mica surface was gently dried in air flow for 1 min and activated by incubation with 6 % glutaraldehyde in 100 mM Tris-HCl buffer, pH 9.0, for 30 min [27]. The Mica substrate was rinsed with water (10× 200 μl) and gently dried for 1 min. 10 μl aliquots (2.8 mg ml−1) of purified proMMP-9 multimers in 100 mM Tris-HCl buffer, pH 7.4, supplemented with 100 mM NaCl and 10 mM CaCl2, were incubated on activated Mica for 1 h at room temperature. The Mica substrate was rinsed with water (10× 200 μl) and dried under a gentle flow of nitrogen gas for 3 min. Five separate areas of the Mica surface were examined by AFM to ensure that a representative sample of the structures on the Mica was obtained. Images were collected at a MultiMode AFM with a Nanoscope V controller (Veeco Metrology LLC, USA) and acquired with an E-scanner in soft tapping mode at room temperature (22–24°C). All images were recorded at a scan rate of 1–3 Hz using silicon microcantilevers (OMCL-AC160TS-W2, Olympus, Japan) with a spring constant of ~42 Nm−1 and a resonant frequency of ~300 kHz (manufacturer specified). The target amplitude was 300 mV with a set point of 220–250 mV. Images were acquired in different scan directions and at different length scales to verify the consistency of the observed structures. Image processing was performed across the whole image. AFM images were processed by a first order flattening. Thereby, all protein molecules were excluded prior to background flattening. Profiles of proMMP-9 multimers were acquired from the AFM images and the dimensions of the multimers were evaluated. The heights and extensions were calculated, binned, normalized, and plotted using OriginPro 9 (OriginLab Co., USA). For accurate height determination, the proMMP-9 multimers were cropped from the AFM images and the image was brought to a uniform level by a plane fit. The height of proMMP-9 molecules was determined relatively to the height of the background and indicated by a color scale bar.
Transmission electron microscopy (TEM)
5 μL of proMMP-9 multimer (2.8 mg ml−1) in 100 mM Tris-HCl buffer, pH 7.4, containing 100 mM NaCl and 10 mM CaCl2, were deposited onto carbon coated copper grids, incubated for 1 min and stained with 1% (w:v) aqueous uranyl acetate for 30 s. The samples were observed in a Tecnai 12 Transmission Electron Microscope (FEI, Eindhoven, The Netherlands), operated at 120 kV. Micrographs were taken with a MegaView III CCD camera (SIS, Münster, Germany).
Molecular modeling
For the molecular modeling of trimeric proMMP-9 we used a previous model established for monomeric proMMP-9 [12]. Based on the TEM measurements, four hexagons with different diameters were applied to encompass correspondingly, the minimal and maximal diameters of small conformers and the minimal and maximal diameters of large conformers. By using the assisted model building with energy refinement (AMBER) software, the N-terminal and C-terminal lobes of three molecules of monomeric proMMP-9 were placed on the corners of these hexagons. Next, the flexible O-glycosylated domain was bent so that intermolecular bridges could be formed between Cys468and Cys 674.
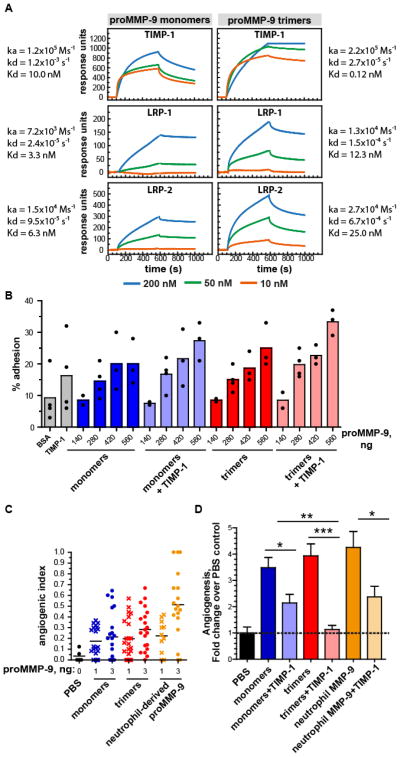
Surface plasmon resonance (SPR)
A 1:1 mixture of 0.2 M N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide and 0.05 M N-hydroxysuccimide in water was used to activate Biacore sensor chips type CM513. TIMP-1 was immobilized at 10 μg/ml in 10 mM sodium acetate, pH 4.5. LRP-1 was immobilized at 15 μg/ml in 10 mM sodium acetate, pH 3.0. LRP-2/Megalin was immobilized at 10 μg/ml in 10 mM sodium acetate, pH 4.5. The remaining binding sites were blocked with 1 M ethanolamine, pH 8.5. The resulting receptor densities were 70 fmol TIMP-1/mm2, 31 fmol LRP-1/mm2 and 25 fmol LRP-2/megalin/mm2. A control chip underwent only the activation and blocking procedures. ProMMP-9 monomer and trimer samples were diluted and analyzed at various concentrations in 100 mM Tris/HCl, 100 mM NaCl, 10 mM CaCl2 and 0.005% Tween-20 pH 7.4. After each analysis the sensory chips were regenerated with 1.6 M glycine-HCl pH 3.0. The analyses were performed on a Biacore 3000 instrument (Biacore, Sweden), and the response was expressed in relative response units [12].
Cell adhesion assay
96-Well plates were coated with proMMP-9 monomers or multimers, complexed with or without TIMP-1. The proMMP-9 monomers or multimers were used at increasing quantities (from 140 ng to 560 ng per well). When TIMP-1 was added (45 ng per well), the samples were first pre-incubated (30min, 37°C) to allow the formation of proMMP-9/TIMP-1 complexes. Control plates were coated with 0.5% BSA. Bone marrow cells (1 × 105), freshly collected from MMP-9 knockout (KO) mice [28], were fluorescently labeled with 1.4 ng/ml 2,7-bis(carboxyethyl)-5(6)-carboxyfluorescein-acetoxymethyl ester (BCECF-AM, Molecular Probes, CA, USA) for 20 min. The labeled cells were washed to eliminate free dye, suspended in adhesion medium (RPMI 1640, 0.5% BSA), and added to the coated wells at 1×105 cells per well. After 60 min at 37 °C, attached cells were lysed with 0.1% SDS in PBS and fluorescence was quantified using an FL600 Microplate fluorescence reader (Biotek, Winooski, USA) [29].
Chick embryo chorioallantoic membrane (CAM) angiogenesis model
The CAM angiogenesis assays were performed as previously described in detail [30–31]. An angiogenic index was determined within 72 to 96 h for each 3D-collagen graft as the fraction of grids with newly formed blood vessels over the total number of grids scored.
RESULTS
Production and fractionation of proMMP-9 monomers and multimers
We investigated various forms of recombinant proMMP-9 and found that a catalytically inactive mutant of proMMP-9, proMMP-9 MutE, was optimal for structural studies because of enhanced protein stability and absence of autocatalysis. Analogous to human MMP-9 forms in vivo, when expressed in insect cells, proMMP-9 is produced as both monomers and homomultimers [12]. Following optimization of preparative isolation protocols, we achieved a good separation of monomers and multimers by glycerol gradient ultracentrifugation (Figure 1C) and produced and purified proMMP-9 monomeric and multimeric forms for further structural, biochemical and biological characterization (Figure 1D). Separation of WT species confirmed that production and purification of proMMP-9 were reproducible and yielded consistent ratios of monomers and multimers as judged by SDS-PAGE and gelatin zymography (Figures 1D and 1E).
Electrophoretic analysis of proMMP-9 monomers and multimers
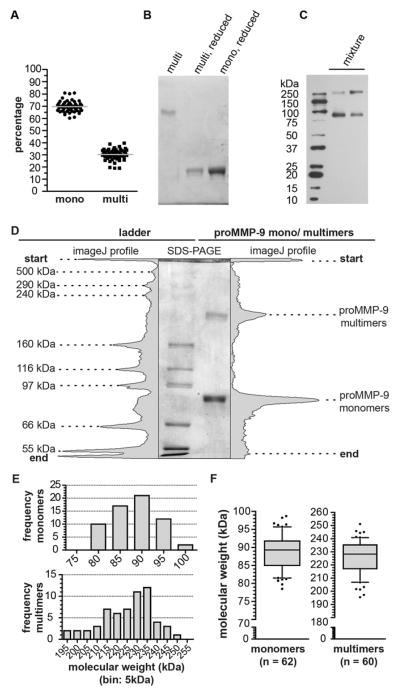
The original proMMP-9 preparations contained approximately 70% monomers and 30% multimers (Figure 2A). In addition, when subjected to reducing conditions, multimeric proMMP-9 was completely reduced into monomers (Figure 2B). Because no reliable protein size markers exceeding the apparent molecular weights (>200 kDa) of dimers were included in previous studies [6, 15], we performed a regression analysis of electrophoretic separation data based on protein standards, which included 250 and 500-kDa mol. wt. markers (Figures 2C and 2D). Although some variations were observed among 60 independent, weekly-produced batches of proMMP-9, the apparent molecular weight of multimers repeatedly situated between dimers and trimers. The frequency distribution of monomers was less skewed than that of the multimers (Figure 2E). Statistical analysis (Figure 2F) yielded 89.3 ± 6.78 kDa for the monomers and 228.3 ± 18.1 kDa for the multimers [median ± interquartile range (IQR)]. Based on the specific molecular weight of the marker mixture, the analysis yielded a molecular weight ratio of 2.5 for multimers versus monomers, which was exactly between trimers and dimers.
Figure 2. Electrophoretic analysis of proMMP-9 monomers and multimers.
(A) Percentage (mean ± SD) of monomers (69.7 ± 4.6%) and multimers (30.3 ± 4.6%) in independent batches (n = 50) of proMMP-9, purified from Sf9 cells cultures. Relative amounts of monomers and multimers were determined by Coomassie brilliant blue staining and expressed as percentage of total protein content. Grey lines indicate the mean values. (B) Electrophoretic separation and Coomassie brilliant blue staining of a proMMP-9 trimer preparation. Upon chemical reduction with β-mercaptoethanol, proMMP-9 trimers migrated to a similar position as monomers. (C) Western blot analysis of two recombinant proMMP-9 samples (mixture of proMMP-9 monomers and proMMP-9 trimers). The proMMP-9 multimers were resolved at the position higher than the 250-kDa molecular weight marker. (D) An example of an SDS-PAGE gel with the HiMark® ladder (left) and a mixture of proMMP-9 monomers and multimers (right). The gel is complemented with the corresponding scanning densitometry analysis performed by using ImageJ software. (E) Frequency distribution histograms of proMMP-9 monomers and multimers as determined from multiple SDS-PAGE analyses. (F) Box-whisker-plots for the molecular weight distribution profiles of proMMP-9 monomers and multimers (90 percentile, 75 percentile, median, 25 percentile, 10 percentile and outliers, represented by dots). The median values (± IQR) for the monomers and multimers were 89.3 ± 6.8 kDa and 228.3 ± 18.1 kDa, respectively. Statistical analysis by two-tailed, non-paired Student’s t-test (P<0.0001), also indicates a clear separation of the fractions.
In vitro stability of monomeric and multimeric proMMP-9
We examined whether proMMP-9 multimers can be generated in vitro from the purified monomer preparations. No multimeric forms were generated from proMMP-9 monomers following a series of oxido-reduction pair reaction cycles known to induce proper folding and double cysteine bridge formation of chemokines [22]. Hence, it is plausible to conclude that multimerization occurs naturally during cell-mediated production of proMMP-9 and is not artificially triggered during biochemical purifications. In line with previous findings [10], chemical reduction of pure multimers with β-mercaptoethanol yielded exclusively monomers (Figure 2B), attesting that the multimer is formed by cysteine bridging.
Structural analysis of monomeric and multimeric proMMP-9
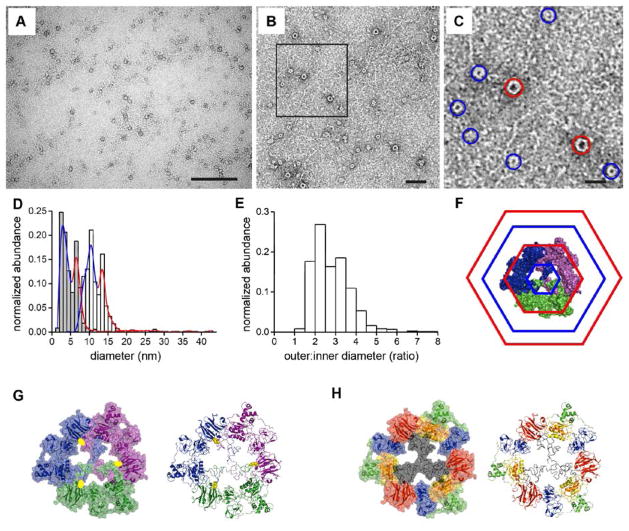
We investigated whether monomeric proMMP-9 molecules, having certainly one and possibly two accessible free cysteine sulfhydryl groups, form dimers, trimers or other multimers. AFM was used to visualize individual multimers (Figures 3A to 3D). The majority of proMMP-9 multimers appeared as circular structures with a height of 4.75 ± 0.63 nm (Figure 3E), similar to the known height of proMMP-9 monomers [4]. AFM images indicated the existence of two distinct populations of circular proMMP-9 multimers with different sizes. Since the diameter of the AFM probe (ØAFM-tip ~15 nm) has a size similar to that of the diameter of the proMMP-9 multimer, the diameter of the latter can be artificially broadened in the x-y-plane [32–34]. Thus, we analyzed the large proMMP-9 multimers characterized by an inner diameter of 18.8 ± 5.7 nm and an outer diameter of 52 ± 10 nm (Figures 3F and 3G). Previously, AFM analysis of proMMP-9 monomers yielded a two-lobed structure with a flexible linker, the larger lobe representing the catalytic part and smaller lobe being the hemopexin domain [4]. Herein, the high-resolution AFM images of proMMP-9 multimers indicated multi-lobed ring structures (Figures 2B to 2D), consistent with a trimeric composition of the proMMP-9 multimers. We also analyzed the size of large proMMP-9 multimers pre-complexed with TIMP-1 and found that the inner and outer diameters of the proMMP-9 multimers in complex with TIMP-1 did not significantly differ from the non-complexed trimers (douter ± SE = 58 ± 13 nm, dinner ± SE = 25.8 ± 4.6 nm, n = 21), probably due to dissociation of the proMMP-9/TIMP-1 complex during the AFM preparation procedure.
Figure 3. Structural analysis of proMMP-9 trimers by AFM.
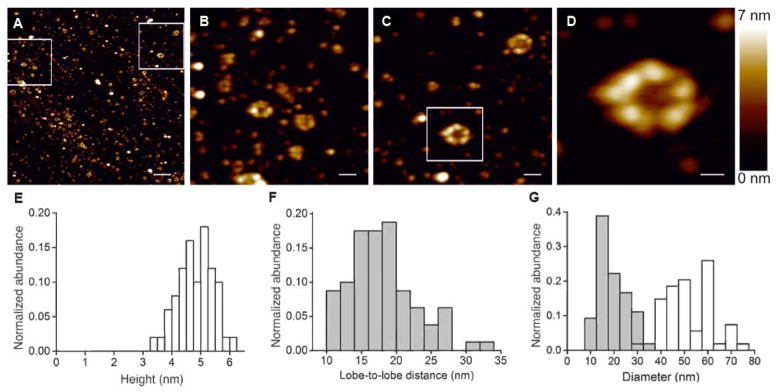
(A) Overview image of purified proMMP-9 trimers. Scale bar = 200 nm. Two boxed areas are enlarged and presented in images panels B and C. (B – C) zoomed areas. Scale bar = 50 nm. The boxed area is magnified and presented in panel D. (D) The proMMP-9 trimer demonstrating a multi-lobed composition with a hollow center. The colors have been adjusted for contrast enhancement. Scale bar = 20 nm. (E) Histogram depicts distribution of proMMP-9 multimers according to their height determined from AFM data (n = 50). The calculated height of 4.75 ± 0.63 nm (mean ± SD) is similar to the height of proMMP-9 monomers established previously [4]. (F) Histogram depicts distribution of lobe-to-lobe distances within monomeric subunits (n = 160), determined from AFM analysis of proMMP-9 trimers. (G) Histogram depicts distribution of inner diameters (mean = 18.8 ± 5.7 nm; grey) and outer diameters (mean = 52 ± 10 nm; white), determined for proMMP-9 trimers (n = 54).
In accordance with the AFM findings, TEM analysis of proMMP-9 trimers also demonstrated that the trimers were represented by large and small circular structures (Figures 4A to 4C). Quantitative processing of a total of 868 individual trimers revealed that the large trimers had outer diameters of 14.1 ± 2.2 nm and inner diameters of 6.9 ± 0.8 nm, whereas the outer and inner diameters of small trimers were respectively 10.8 ± 2.1 nm and 2.8 ± 0.7 nm (Figures 4D and 4E). The statistical analysis of TEM data indicated that the small and large trimers were present at a ratio of 3:1(see also Figure 4C).
Figure 4. TEM analysis and molecular model of proMMP-9 trimers.
(A) TEM overview image. The proMMP-9 trimers occur as negatively stained round structures. Scale bar, 200 nm. (B) Representative TEM image of proMMP-9 trimers at higher magnification. Scale bar, 50 nm. (C) Magnified area from panel B. Two populations of proMMP-9 trimers of different sizes are indicated by blue (small conformers) and red (large conformers) circles. Scale bar, 20 nm. (D) Statistical analysis of TEM images (n = 868) revealed two distinct maxima for the outer (white histogram) and inner (grey histogram) diameters, indicating the presence of two size populations of the proMMP-9 trimers. For the large conformers of proMMP-9 trimers (red circles in C), an inner diameter of 6.9 ± 0.8 nm (mean ± SD) and an outer diameter of 14.1 ± 2.2 nm have been obtained. For the small conformers (blue circles in C), an inner diameter of 2.8 ± 0.7 nm and an outer diameter of 10.8 ± 2.1 nm have been obtained. The colored curves indicate distribution of small (blue) and large (red) conformers. (E) Histogram, distribution of the ratios between the outer and inner diameters of proMMP-9 trimers. Mean = 2.84 ± 0.82. (F–H) Molecular models of trimeric proMMP-9. The models assume that the Cys468 and Cys674 are involved in trimerization. (F) Model of trimeric proMMP-9 based on the AFM and TEM data. Hexagons indicate the inner minimal and the outer maximal diameters of the large (red) and small (blue) proMMP-9 conformers. (G) 3D model of a proMMP-9 trimer. A space filling model (left panel) and a cartoon model (right panel) with the three monomers in different colors, illustrate the intermolecular cysteine bridges (in yellow) between the monomers. (H) A space filling model (left panel) and a cartoon model (right panel) depicting the functional domains of proMMP-9 in the trimeric structure. The same color code was adopted as used in Figure 1. The molecular orientations are the same as in panel G.
Trimeric hydrolases are rare in nature and their formation, mediated by protein disulfide isomerase, would require at least two free cysteine residues per monomeric form. Fulfilling this requirement, there are two possibly free cysteines in the primary structure of proMMP-9 (Figure 1A). In addition, the cysteine in the OG-domain is located in the very center of this highly flexible domain [1], which provides a link between the globular N-terminal catalytic segment and the C-terminal hemopexin domain (Figure 1A). Considering structural aspects, namely the presence of two free cysteines, flexibility of the OG domain [4], central location of the cysteine (Cys468) within the OG domain and the circular structures observed by AFM and TEM, we generated a model for trimeric proMMP-9 (Figures 4F to 4H). This model depicts how three proMMP-9 monomers can form a circular structure with globular domains at the periphery and a central hollow. We indicate how three intermolecular bridges are formed between the cysteine (Cys468) in the center of the one OG domain and cysteine (Cys674) in the blade of the hemopexin domain of the adjacent molecule, resulting in the formation of a trimer (Figure 4G).
We next attempted to identify the exact cysteines involved in trimerization of proMMP-9 and to quantify free cysteines present in monomeric proMMP-9 compared to trimeric proMMP-9. However, despite implementation of several independent techniques, including mass spectrometry following tryptic digestion used previously [12], we were not able to determine the arrangements of disulfide bridges and free cysteines in proMMP-9 monomers and trimers, mostly due to extensive N- and O-glycosylation of proMMP-9 molecules. Furthermore, we also attempted to determine the amount of free SH groups in monomeric proMMP-9 using urea and Ellman’s reagent or Alexa Fluor® 633 C5 Maleimide [35–36]. However, these approaches which are suitable for free cysteine quantification in peptides, failed due to relatively high molecular weight and complexity of the proMMP-9 molecule.
Glycan composition of monomeric and multimeric proMMP-9
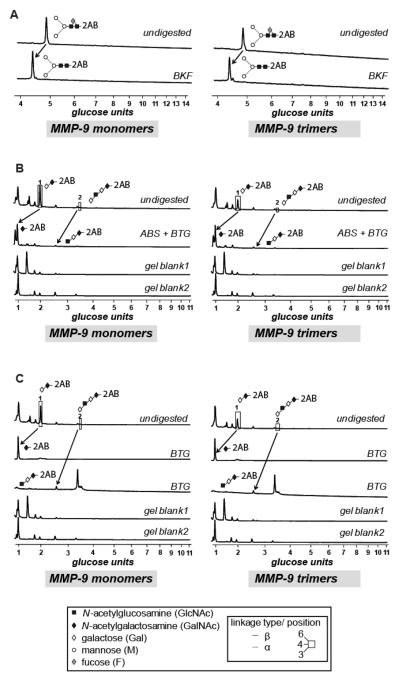
The proMMP-9 molecule is decorated with both N- and O-linked oligosaccharides [37], which define the overall structure of glycoproteins [38, 39]. Therefore, we investigated the possibility that differential glycosylation of individual proMMP-9 monomers may affect their ability to form multimers. To test this hypothesis, N- and O-linked glycans present on proMMP-9 monomers versus trimers were analyzed by hydrophilic interaction liquid chromatography (HILIC) in combination with sequential digestion by an array of exoglycosidases. Both forms of proMMP-9 contained the N-linked glycan F(6)M3, as indicated by bovine kidney alpha-fucosidase (BKF) digestion (Figure 5A). O-linked glycans were subjected to a range of exoglycosidases. For precise assignments, individual peaks containing O-glycans were collected and digested with bovine testes β-galactosidase (BTG). Both monomeric and trimeric proMMP-9 contained the same core 1 glycans Galβ1–3GalNAc and Galβ1–3GlcNAcβ1–3Galβ1–3GalNAc (Figures 5B, 5C and Table 1), and therefore, no structural differences were observed between monomer and trimer O-linked glycans.
Figure 5. Glycan analysis of proMMP-9 monomers and trimers.
(A) N-linked glycans of separated proMMP-9 monomer and trimer preparations were isolated and analyzed by HILIC. Typical profiles of undigested and BKF-digested monomeric and trimeric proMMP-9 N-glycomes are shown. (B, C) Typical HILIC profiles of monomeric and trimeric proMMP-9 O-glycomes before and after digestion with a range of exoglycosidases. Peaks were collected and each digested with BTG and/or ABS. Structural symbols for the glycans and their linkages are shown in the box situated at the bottom of the figure. These symbols are also the keys to the structures shown in Table 1.
Table 1.
N-and O-glycans identified in monomeric and trimeric proMMP-9
| peak | abbreviations | structure | glucose units |
|---|---|---|---|
| N-glycans | |||
|
| |||
| F(6)M3 |
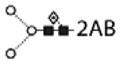
|
4.84 | |
|
| |||
| O-glycans | |||
|
| |||
| 1 | Galβ1–3GalNAc |
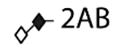
|
1.96 |
| 2 | Galβ1–3GlcNAcβ1–3Galβ1–3GalNAc |
|
3.5 |
Binding of monomeric and trimeric proMMP-9 to LRP-1, LRP-2 and TIMP-1
In a physiological environment, the majority of different cell types, except of neutrophilic granulocytes [6, 7], produce proMMP-9 in complex with its inhibitor, TIMP-1, bound to the hemopexin domain of the zymogen. In addition, the bioavailability of proMMP-9 is also regulated by the cargo receptors, namely low density lipoprotein receptor-related protein-1 (LRP-1) and LRP-2/megalin, through their binding to the hemopexin domain of proMMP-9 [12]. Since the hemopexin domain would be involved in trimerization via its cysteine residue, we compared proMMP-9 monomers and trimers for their efficiency to bind TIMP-1, LRP-1 and LRP-2.
Monomers and trimers of proMMP-9 MutE at increasing concentrations ranging from 10 nM to 200 nM, were tested for their binding efficiency to TIMP-1, LRP-1, or LRP-2 immobilized on sensor chips (Figure 6A). The global fit analysis of surface plasmon resonance (SPR) data demonstrated a substantial decrease in dissociation rates of TIMP-1 from proMMP-9 trimers, indicating a significantly higher affinity between TIMP-1 and proMMP-9 trimers than TIMP-1 and proMMP-9 monomers, exhibiting the dissociation constants (Kd) of 0.12 nM and 10 nM, respectively (Figure 6A). A similar differential in affinity to TIMP-1 was also characteristic of WT proMMP-9, which demonstrated a dissociation constant (Kd) of 10 nM for monomers and a Kd of 0.4 nM for trimers. Importantly, there were no differences in this order of magnitude between proMMP-9 monomers versus trimers in their binding strengths to LRP-1 or LRP-2/megalin (Figure 6A). We have previously shown that the LRP-1 and LRP-2 docking sites do not overlap with the TIMP-1 binding site in the MMP-9 molecule, although all these sites are located in the hemopexin domain [12]. Therefore, it is plausible to suggest that the binding of proMMP-9 to LRPs would not be influenced by pre-complexing of proMMP-9 with TIMP-1 and that the uptake of monomeric and trimeric proMMP-9 by cell surface LRPs would not be regulated differentially by TIMP-1.
Figure 6. Functional analysis of trimeric proMMP-9.
(A) SPR binding of proMMP-9 monomers and trimers to TIMP-1, LRP-1 and LRP-2 (megalin). The y-axis indicates the difference between the sample and control runs in relative response units. The functional affinity of proMMP-9 monomers to TIMP-1 was approximately 50 times less than that of the proMMP-9 trimers. The binding affinity of monomeric proMMP-9 MutE to LRP-1 and LRP-2 was slightly higher than the binding affinity of the trimers. (B) Cell adhesion assays. Binding of bone marrow cells isolated from MMP-9 KO mice to proMMP-9 monomers and trimers. Increasing quantities of monomers and trimers (140 ng to 560 ng) were tested in the presence or absence of TIMP-1 (45 ng). (C) Angiogenic capacity of purified proMMP-9 monomers (blue), proMMP-9 trimers (red) and neutrophil-derived proMMP-9 (orange). Monomers and trimers of proMMP-9 were compared in the CAM angiogenesis model. 3-D collagen grafts, containing gridded meshes, were supplemented with proMMP-9 monomers, proMMP-9 trimers, or neutrophil-derived proMMP-9 (1 or 3 ng per graft). In the negative control, the grafts were supplemented with PBS only (black). Collagen grafts were placed on the CAM of chick embryos (5–6 grafts per embryo; 5–6 embryos per variant). The levels of angiogenesis were quantified as angiogenic index (the ratio of grids containing newly formed blood vessels versus total number of evaluated grids). Horizontal lines indicate medians. (D) TIMP-1 inhibition of angiogenesis induced by proMMP-9 monomers (blue, n=43), trimers (red, n=44) and neutrophil-derived MMP-9 (orange, n=20) preparations. CAM angiogenesis assay was performed as described in C. Monomers and trimers of proMMP-9 were supplemented at 3 ng per graft, alone or with 10 ng of recombinant TIMP-1. Negative control contained PBS only (black, n=24). *, ** and ***, P<0.05, 0.005 and 0.0001, respectively; two-tailed Student’s t-test.
Cell binding properties of monomeric and trimeric proMMP-9
ProMMP-9 binds to a number of cell surface molecules (e.g. LRP-1, LRP-2, CD44 and integrins) and thereby controls cell surface proteolysis and activates outside-in cell signaling pathways. Since these interactions are mediated by the hemopexin domain of MMP-9 [40], we hypothesized that the efficiency of hemopexin-mediated binding to cell surfaces might be different for proMMP-9 trimers.
To test this hypothesis, we compared the efficiency of cell binding to proMMP-9 MutE trimers and monomers immobilized on plastic surfaces. In a cell adhesion assay employing bone marrow cells from MMP-9 KO mice to avoid interference with endogenously produced proMMP-9, we found that proMMP-9 MutE trimers and monomers induced similar levels of cell adhesion. The cells bound to both proMMP-9 monomers and multimers in a dose-dependent manner, and this binding was insensitive to TIMP-1 complexed to proMMP-9 (Figure 6B). In agreement with the findings that TIMP-1 can interact with cell surface molecules, e.g. β1 integrins [41–42], the plastic-bound TIMP-1 also induced cell adhesion, albeit at relatively low levels. Since the proMMP-9/TIMP-1 complex was shown to interact with CD44 via the hemopexin domain of the MMP-9 molecule [43], the cell binding properties of free TIMP-1 and TIMP-1 bound to MMP-9 may vary.
Induction of angiogenesis by monomeric and trimeric proMMP-9
Since proMMP-9 triggers the angiogenic switch [2] and induces angiogenesis [44], we evaluated the angiogenic potential of proMMP-9 monomers and trimers. We previously established that in its TIMP-free form, recombinant and neutrophil proMMP-9 (both containing mixtures of multimeric and monomeric forms of the zymogen) exhibited a high angiogenic activity in vivo [7, 30, 45]. Both the recombinant and neutrophil TIMP-free proMMP-9 are proteolytically activated in the tissue microenvironment by either endogenous activators of proMMP-9 such as stromal MMP-3 [46] or by conformational activation of highly concentrated proMMP-9 bound to stromal matrix components and undergoing autoactivation [47]. Then, the proteolytically or conformationally activated MMP-9 enzyme could liberate extracellular matrix-bound angiogenic factors such as VEGF and FGF-2, making them bioavailable to induce angiogenesis [30].
Herein, we analyzed and compared the angiogenesis-inducing potency of proMMP-9 monomers and trimers and their sensitivity to TIMP-1 inhibition in vivo. By employing a chick embryo angiogenesis model [31], we found that TIMP-free trimers were capable of inducing angiogenesis and that the efficiency of proMMP-9 trimers was comparable with that of monomers (Figure 6C). However, the angiogenic response induced by multimers was significantly more sensitive to TIMP-1 inhibition than that induced by monomers (Figure 6D). This finding indicates that the induction of angiogenesis by MMP-9 trimers could be regulated differentially from that by MMP-9 monomers.
DISCUSSION
Metalloproteinases and their inhibitors mediate many biological and pathological processes [48–50] and, within this family of enzymes, MMP-9 is an outstanding example. Not only did the present study provide the first experimental evidence for the molecular nature of MMP-9 multimers, it also revealed the trimeric structure of MMP-9 as a specific entity that exhibits high affinity to TIMP-1. Herein, trimeric MMP-9 was shown to participate in a number of biological activities, including binding to cell surfaces and the induction of angiogenesis. The following important conclusions can be drawn from our findings: (i) The previously described flexibility of monomers [4] now comes into the context of bending and assembly into trimers via disulfide bridges. (ii) The ongoing debate about which individual free cysteines, namely in the OG domain or in the hemopexin domain, are involved in multimerization [11–15], might be resolved. In the MMP-9 trimerization process, the cysteines from both the OG and hemopexin domains would be involved, analogous to trimeric haptoglobin 2-2, where 2 free cysteines in each monomer bridge with 2 free cysteines in the adjacent molecule to form a circular multimer [51]. (iii) The finding of a trimeric MMP-9 might shed light on how triple helical collagens are being processed in nature. Triple helical collagens are hydrolyzed by collagenases (e. g. MMP-1, MMP-8, and MMP-13), into 1/4 and 3/4 length fragments. Subsequently, these fragments are further degraded by MMP-2/gelatinase A and MMP-9/gelatinase B [52, 53]. At the molecular level it is still difficult to understand how such a large triple helical substrate can be processed since molecular studies showed that triple helical collagens do not fit into the active site of monomeric MMPs [54]. Additionally, whether or not MMP-9 digests type I, II and III collagens has been a point of debate [55]. The existence of the MMP-9 trimer might better explain how MMP-9 molecules are able to accommodate cleaved triple helical collagen fragments. (iv) Our findings are particularly interesting for leukocyte biology, because neutrophils [6, 7] and M2-polarized macrophages and tumor-associated macrophages [8,9] uniquely produce TIMP-free proMMP-9. However, only neutrophils produce copious quantities of proMMP-9 abundant in trimers without TIMP-1, which could uniquely serve as quick TIMP-1 trap, allowing the less sensitive MMP-9 monomers to induce angiogenesis. In addition, from the moment that TIMP-1 is secreted into the micro-environment by cell types different from neutrophils, the catalysis by trimers would be shut down first, leaving the monomers still active. With more TIMP-1 being produced at later stages, all MMP-9 activity would become inhibited. This aspect could be critical for all physiological and pathological conditions such as cancer [49, 56], inflammation [48, 57] and angiogenesis [7, 45], where neutrophils play key roles associated with the delivery of TIMP-free proMMP-9.
Summary statement.
We have discovered a homotrimeric protease form of gelatinase B/matrix metalloproteinase-9 (MMP-9) and showed that the angiogenic activity of MMP-9 trimers is more tightly controlled by the natural inhibitor TIMP-1 than that of monomers. These findings open new roads for selective inhibition of MMP-9.
Acknowledgments
The authors thank the Irving and Cherna Moskowitz Center for Nano and Bio-Nano Imaging for the electron microscopy studies and Sidney Cohen and Arkady Bitler for support with the AFM measurements at the Weizmann Institute of Science.
Funding: The research leading to these results has received funding from the European Union Seventh Framework Programme [FP7/2007-2013] under grant agreement n° 263307 and from The Fund for Scientific Research of Flanders (FWO-Vlaanderen) and the Geconcerteerde OnderzoeksActies [GOA 2013 014] of the Regional Government of Flanders. The authors acknowledge support of the Israel Science Foundation and the German-Israel Minerva Foundation. BB acknowledges financial support from the Human Frontier Science Program [LT000336/2011]. RS acknowledges funding from the European Union Seventh Framework Programme [FP7/2007-2013] under grant agreement n°260600 (“GlycoHIT”). MS is a fellow of the Verband der Chemischen Industrie e.V. EUB was supported by a fellowship from the Spanish Ministry of Economy and Competiveness (MINECO) and AGP was supported by grants SAF2012-31613 (MINECO) and S2010/BMD-2314 (Comunidad de Madrid/European Union). The research by EZ, JPQ and EID was supported by the NIH grant [1R01CA157792].
Abbreviation
- 2AB
2-aminobenzamide
- ABS
Arthrobacter ureafaciens sialidase
- AFM
atomic force microscopy
- AMBER
assisted model building with energy refinement
- BKF
bovine kidney α-fucosidase
- BTG
bovine testes β-galactosidase
- CAM
chick embryo chorioallantoic membrane
- HILIC
hydrophilic interaction liquid chromatography
- IQR
interquartile range
- LRP
low density lipoprotein receptor-related protein
- MMP
matrix metalloproteinase
- NGAL
neutrophil gelatinase B-associated lipocalin
- OG
O-glycosylated
- Rf
retention factor
- SPR
surface plasmon resonance
- TEM
transmission electron microscopy
- TIMP-1
tissue inhibitor of metalloproteinases-1
- WT
wild-type
Footnotes
The authors declare no conflict of interest.
References
- 1.Vandooren J, Van den Steen PE, Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): The next decade. Crit Rev Biochem Mol Biol. 2013;48:222–272. doi: 10.3109/10409238.2013.770819. [DOI] [PubMed] [Google Scholar]
- 2.Bergers G, Brekken R, McMahon G, Vu TH, Itoh T, Tamaki K, Tanzawa K, Thorpe P, Itohara S, Werb Z, Hanahan D. Matrix metalloproteinase-9 triggers the angiogenic switch during carcinogenesis. Nat Cell Biol. 2000;2:737–744. doi: 10.1038/35036374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Morgunova E, Tuuttila A, Bergmann U, Isupov M, Lindqvist Y, Schneider G, Tryggvason K. Structure of human pro-matrix metalloproteinase-2: activation mechanism revealed. Science. 1999;284:1667–1670. doi: 10.1126/science.284.5420.1667. [DOI] [PubMed] [Google Scholar]
- 4.Rosenblum G, Van den Steen PE, Cohen SR, Grossmann JG, Frenkel J, Sertchook R, Slack N, Strange RW, Opdenakker G, Sagi I. Insights into the structure and domain flexibility of full-length pro-matrix metalloproteinase-9/gelatinase B. Structure. 2007;15:1227–1236. doi: 10.1016/j.str.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 5.Overall CM, Butler GS. Protease yoga: extreme flexibility of a matrix metalloproteinase. Structure. 2007;15:1159–1161. doi: 10.1016/j.str.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Opdenakker G, Van den Steen PE, Dubois B, Nelissen I, Van Coillie E, Masure S, Proost P, Van Damme J. Gelatinase B functions as regulator and effector in leukocyte biology. J Leukoc Biol. 2001;69:851–859. [PubMed] [Google Scholar]
- 7.Ardi VC, Kupriyanova TA, Deryugina EI, Quigley JP. Human neutrophils uniquely release TIMP-free MMP-9 to provide a potent catalytic stimulator of angiogenesis. Proc Natl Acad Sci U S A. 2007;104:20262–20267. doi: 10.1073/pnas.0706438104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zajac E, Schweighofer B, Kupriyanova TA, Juncker-Jensen A, Minder P, Quigley JP, Deryugina EI. Angiogenic capacity of M1- and M2-polarized macrophages is determined by the levels of TIMP-1 complexed with their secreted proMMP-9. Blood. 2013;122:4054–4067. doi: 10.1182/blood-2013-05-501494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deryugina E, Zajac E, Juncker-Jensen A, Kupriyanova TA, Welter L, Quigley JP. Tissue-infiltrating neutrophils constitute the major in vivo source of angiogenesis-inducing MMP-9 in the tumor microenvironment. Neoplasia. 2014 doi: 10.1016/j.neo.2014.08.013. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van den Steen PE, Proost P, Wuyts A, Van Damme J, Opdenakker G. Neutrophil gelatinase B potentiates interleukin-8 tenfold by aminoterminal processing, whereas it degrades CTAP-III, PF-4, and GRO-alpha and leaves RANTES and MCP-2 intact. Blood. 2000;96:2673–2681. [PubMed] [Google Scholar]
- 11.Cha H, Kopetzki E, Huber R, Lanzendorfer M, Brandstetter H. Structural basis of the adaptive molecular recognition by MMP9. J Mol Biol. 2002;320:1065–1079. doi: 10.1016/s0022-2836(02)00558-2. [DOI] [PubMed] [Google Scholar]
- 12.Van den Steen PE, Van Aelst I, Hvidberg V, Piccard H, Fiten P, Jacobsen C, Moestrup SK, Fry S, Royle L, Wormald MR, Wallis R, Rudd PM, Dwek RA, Opdenakker G. The hemopexin and O-glycosylated domains tune gelatinase B/MMP-9 bioavailability via inhibition and binding to cargo receptors. J Biol Chem. 2006;281:18626–18637. doi: 10.1074/jbc.M512308200. [DOI] [PubMed] [Google Scholar]
- 13.Collier IE, Legant W, Marmer B, Lubman O, Saffarian S, Wakatsuki T, Elson E, Goldberg GI. Diffusion of MMPs on the surface of collagen fibrils: the mobile cell surface-collagen substratum interface. PLoS ONE. 2011;6:e24029. doi: 10.1371/journal.pone.0024029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dufour A, Zucker S, Sampson NS, Kuscu C, Cao J. Role of matrix metalloproteinase-9 dimers in cell migration: design of inhibitory peptides. J Biol Chem. 2010;285:35944–35956. doi: 10.1074/jbc.M109.091769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Olson MW, Bernardo MM, Pietila M, Gervasi DC, Toth M, Kotra LP, Massova I, Mobashery S, Fridman R. Characterization of the monomeric and dimeric forms of latent and active matrix metalloproteinase-9. Differential rates for activation by stromelysin 1. J Biol Chem. 2000;275:2661–2668. doi: 10.1074/jbc.275.4.2661. [DOI] [PubMed] [Google Scholar]
- 16.Masure S, Proost P, Van Damme J, Opdenakker G. Purification and identification of 91-kDa neutrophil gelatinase. Release by the activating peptide interleukin-8. Eur J Biochem. 1991;198:391–398. doi: 10.1111/j.1432-1033.1991.tb16027.x. [DOI] [PubMed] [Google Scholar]
- 17.Vandooren J, Geurts N, Martens E, Van den Steen PE, De Jonghe S, Herdewijn P, Opdenakker G. Gelatin degradation assay reveals MMP-9 inhibitors and function of O-glycosylated domain. World J Biol Chem. 2011;2:14–24. doi: 10.4331/wjbc.v2.i1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 19.Abramoff MD, Magalhaes PJ, Ram SJ. Image Processing with ImageJ. Biophotonics International. 2004;11:36–42. [Google Scholar]
- 20.Weber K, Osborn M. The reliability of molecular weight determinations by dodecyl sulfate-polyacrylamide gel electrophoresis. J Biol Chem. 1969;244:4406–4412. [PubMed] [Google Scholar]
- 21.Vandooren J, Geurts N, Martens E, Van den Steen PE, Opdenakker G. Zymography methods for visualizing hydrolytic enzymes. Nat Methods. 2013;10:211–220. doi: 10.1038/nmeth.2371. [DOI] [PubMed] [Google Scholar]
- 22.Proost P, Van Leuven P, Wuyts A, Ebberink R, Opdenakker G, Van Damme J. Chemical synthesis, purification and folding of the human monocyte chemotactic proteins MCP-2 and MCP-3 into biologically active chemokines. Cytokine. 1995;7:97–104. doi: 10.1006/cyto.1995.1013. [DOI] [PubMed] [Google Scholar]
- 23.Royle L, Radcliffe CM, Dwek RA, Rudd PM. Detailed structural analysis of N-glycans released from glycoproteins in SDS-PAGE gel bands using HPLC combined with exoglycosidase array digestions. Methods Mol Biol. 2006;347:125–143. doi: 10.1385/1-59745-167-3:125. [DOI] [PubMed] [Google Scholar]
- 24.Huang Y, Mechref Y, Novotny MV. Microscale nonreductive release of O-linked glycans for subsequent analysis through MALDI mass spectrometry and capillary electrophoresis. Anal Chem. 2001;73:6063–6069. doi: 10.1021/ac015534c. [DOI] [PubMed] [Google Scholar]
- 25.Bigge JC, Patel TP, Bruce JA, Goulding PN, Charles SM, Parekh RB. Nonselective and efficient fluorescent labeling of glycans using 2-amino benzamide and anthranilic acid. Anal Biochem. 1995;230:229–238. doi: 10.1006/abio.1995.1468. [DOI] [PubMed] [Google Scholar]
- 26.Royle L, Campbell MP, Radcliffe CM, White DM, Harvey DJ, Abrahams JL, Kim YG, Henry GW, Shadick NA, Weinblatt ME, Lee DM, Rudd PM, Dwek RA. HPLC-based analysis of serum N-glycans on a 96-well plate platform with dedicated database software. Anal Biochem. 2008;376:1–12. doi: 10.1016/j.ab.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 27.Vandenberg E, Elwing H, Askendal A, Lundstrom I. Protein Immobilization to 3-Aminopropyl Triethoxy Silane Glutaraldehyde Surfaces - Characterization by Detergent Washing. J Colloid Interface Sci. 1991;143:327–335. [Google Scholar]
- 28.Dubois B, Masure S, Hurtenbach U, Paemen L, Heremans H, van den Oord J, Sciot R, Meinhardt T, Hämmerling G, Opdenakker G, Arnold B. Resistance of young gelatinase B-deficient mice to experimental autoimmune encephalomyelitis and necrotizing tail lesions. J Clin Invest. 1999;104:1507–1515. doi: 10.1172/JCI6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ugarte-Berzal E, Bailón E, Amigo-Jiménez I, Vituri CL, Hernández del Cerro M, Terol MJ, Albar JP, Rivas G, García-Marco JA, García-Pardo A. A 17-residue sequence from the matrix metalloproteinase-9 (MMP-9) hemopexin domain binds alpha4beta1 integrin and inhibits MMP-9-induced functions in chronic lymphocytic leukemia B cells. J Biol Chem. 2012;287:27601–27613. doi: 10.1074/jbc.M112.354670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ardi VC, Van den Steen PE, Opdenakker G, Schweighofer B, Deryugina EI, Quigley JP. Neutrophil MMP-9 proenzyme, unencumbered by TIMP-1, undergoes efficient activation in vivo and catalytically induces angiogenesis via a basic fibroblast growth factor (FGF-2)/FGFR-2 pathway. J Biol Chem. 2009;284:25854–25866. doi: 10.1074/jbc.M109.033472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deryugina EI, Quigley JP. Chapter 2. Chick embryo chorioallantoic membrane models to quantify angiogenesis induced by inflammatory and tumor cells or purified effector molecules. Methods Enzymol. 2008;444:21–41. doi: 10.1016/S0076-6879(08)02802-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Müller DJ, Fotiadis D, Scheuring S, Müller SA, Engel A. Electrostatically balanced subnanometer imaging of biological specimens by atomic force microscope. Biophys J. 1999;76:1101–1111. doi: 10.1016/S0006-3495(99)77275-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim D, Blanch HW, Radke CJ. Direct imaging of lysozyme adsorption onto mica by atomic force microscopy. Langmuir. 2002;18:5841–5850. [Google Scholar]
- 34.Meinander K, Jensen TN, Simonsen SB, Helveg S, Lauritsen JV. Quantification of tip-broadening in non-contact atomic force microscopy with carbon nanotube tips. Nanotechnology. 2012;23:405705. doi: 10.1088/0957-4484/23/40/405705. [DOI] [PubMed] [Google Scholar]
- 35.Steinert PM, Baliga BS, Munro HN. Estimation of protein sulfhydryl groups with 5,5′-(35S) dithio-bis(2-nitrobenzoate) Anal Biochem. 1974;59:416–425. doi: 10.1016/0003-2697(74)90294-2. [DOI] [PubMed] [Google Scholar]
- 36.Sekine T, Ando K, Machida M, Kanaoka Y. Fluorescent thiol reagents. V. Microfluorometry of thiol compounds with a fluorescent-labeled maleimide. Anal Biochem. 1972;48:557–568. doi: 10.1016/0003-2697(72)90111-x. [DOI] [PubMed] [Google Scholar]
- 37.Mattu TS, Royle L, Langridge J, Wormald MR, Van den Steen PE, Van Damme J, Opdenakker G, Harvey DJ, Dwek RA, Rudd PM. O-glycan analysis of natural human neutrophil gelatinase B using a combination of normal phase-HPLC and online tandem mass spectrometry: implications for the domain organization of the enzyme. Biochemistry. 2000;39:15695–15704. doi: 10.1021/bi001367j. [DOI] [PubMed] [Google Scholar]
- 38.Rudd PM, Dwek RA. Glycosylation: heterogeneity and the 3D structure of proteins. Crit Rev Biochem Mol Biol. 1997;32:1–100. doi: 10.3109/10409239709085144. [DOI] [PubMed] [Google Scholar]
- 39.Van den Steen P, Rudd PM, Dwek RA, Opdenakker G. Concepts and principles of O-linked glycosylation. Crit Rev Biochem Mol Biol. 1998;33:151–208. doi: 10.1080/10409239891204198. [DOI] [PubMed] [Google Scholar]
- 40.Redondo-Muñoz J, Ugarte-Berzal E, Terol MJ, Van den Steen PE, Hernández del Cerro M, Roderfeld M, Roeb E, Opdenakker G, García-Marco JA, Garcia-Pardo A. Matrix metalloproteinase-9 promotes chronic lymphocytic leukemia B cell survival through its hemopexin domain. Cancer Cell. 2010;17:160–172. doi: 10.1016/j.ccr.2009.12.044. [DOI] [PubMed] [Google Scholar]
- 41.Jung KK, Liu XW, Chirco R, Fridman R, Kim HR. Identification of CD63 as a tissue inhibitor of metalloproteinase-1 interacting cell surface protein. EMBO J. 2006;25:3934–3942. doi: 10.1038/sj.emboj.7601281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Redondo-Muñoz J, Ugarte-Berzal E, García-Marco JA, Hernández del Cerro M, Van den Steen PE, Opdenakker G, Terol MJ, García-Pardo A. Alpha4beta1 integrin and 190-kDa CD44v constitute a cell surface docking complex for gelatinase B/MMP-9 in chronic leukemic but not in normal B cells. Blood. 2008;112:169–178. doi: 10.1182/blood-2007-08-109249. [DOI] [PubMed] [Google Scholar]
- 43.Lambert E, Bridoux L, Devy J, Dassé E, Sowa ML, Duca L, Hornebeck W, Martiny L, Petitfrère-Charpentier E. TIMP-1 binding to proMMP-9/CD44 complex localized at the cell surface promotes erythroid cell survival. Int J Biochem Cell Biol. 2009;41:1102–1115. doi: 10.1016/j.biocel.2008.10.017. [DOI] [PubMed] [Google Scholar]
- 44.Deryugina EI, Quigley JP. Pleiotropic roles of matrix metalloproteinases in tumor angiogenesis: contrasting, overlapping and compensatory functions. Biochim Biophys Acta. 2010;1803:103–120. doi: 10.1016/j.bbamcr.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bekes EM, Schweighofer B, Kupriyanova TA, Zajac E, Ardi VC, Quigley JP, Deryugina EI. Tumor-recruited neutrophils and neutrophil TIMP-free MMP-9 regulate coordinately the levels of tumor angiogenesis and efficiency of malignant cell intravasation. Am J Pathol. 2011;179:1455–1470. doi: 10.1016/j.ajpath.2011.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramos-DeSimone N, Hahn-Dantona E, Sipley J, Nagase H, French DL, Quigley JP. Activation of matrix metalloproteinase-9 (MMP-9) via a converging plasmin/stromelysin-1 cascade enhances tumor cell invasion. J Biol Chem. 1999;274:13066–13076. doi: 10.1074/jbc.274.19.13066. [DOI] [PubMed] [Google Scholar]
- 47.Bannikov GA, Karelina TV, Collier IE, Marmer BL, Goldberg GI. Substrate binding of gelatinase B induces its enzymatic activity in the presence of intact propeptide. J Biol Chem. 2002;277:16022–16027. doi: 10.1074/jbc.M110931200. [DOI] [PubMed] [Google Scholar]
- 48.Hu J, Van den Steen PE, Sang QX, Opdenakker G. Matrix metalloproteinase inhibitors as therapy for inflammatory and vascular diseases. Nat Rev Drug Discov. 2007;6:480–498. doi: 10.1038/nrd2308. [DOI] [PubMed] [Google Scholar]
- 49.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khokha R, Murthy A, Weiss A. Metalloproteinases and their natural inhibitors in inflammation and immunity. Nat Rev Immunol. 2013;13:649–665. doi: 10.1038/nri3499. [DOI] [PubMed] [Google Scholar]
- 51.Kristiansen M, Graversen JH, Jacobsen C, Sonne O, Hoffman HJ, Law SK, Moestrup SK. Identification of the haemoglobin scavenger receptor. Nature. 2001;409:198–201. doi: 10.1038/35051594. [DOI] [PubMed] [Google Scholar]
- 52.Rosenblum G, Van den Steen PE, Cohen SR, Bitler A, Brand DD, Opdenakker G, Sagi I. Direct visualization of protease action on collagen triple helical structure. PLoS One. 2010;5:e11043. doi: 10.1371/journal.pone.0011043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Van den Steen PE, Proost P, Brand DD, Kang AH, Van Damme J, Opdenakker G. Generation of glycosylated remnant epitopes from human collagen type II by gelatinase B. Biochemistry. 2004;43:10809–10816. doi: 10.1021/bi0493665. [DOI] [PubMed] [Google Scholar]
- 54.Chung L, Dinakarpandian D, Yoshida N, Lauer-Fields JL, Fields GB, Visse R, Nagase H. Collagenase unwinds triple-helical collagen prior to peptide bond hydrolysis. EMBO J. 2004;23:3020–3030. doi: 10.1038/sj.emboj.7600318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bigg HF, Rowan AD, Barker MD, Cawston TE. Activity of matrix metalloproteinase-9 against native collagen types I and III. FEBS J. 2007;274:1246–1255. doi: 10.1111/j.1742-4658.2007.05669.x. [DOI] [PubMed] [Google Scholar]
- 56.Piccard H, Muschel RJ, Opdenakker G. On the dual roles and polarized phenotypes of neutrophils in tumor development and progression. Crit Rev Oncol Hematol. 2012;82:296–309. doi: 10.1016/j.critrevonc.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 57.Opdenakker G, Van den Steen PE, Van Damme J. Gelatinase B: a tuner and amplifier of immune functions. Trends Immunol. 2001;22:571–579. doi: 10.1016/s1471-4906(01)02023-3. [DOI] [PubMed] [Google Scholar]