Abstract
Under physiologically relevant conditions, the levels of non-viral gene transfer are low at best. The reason for this is that many barriers exist for the efficient transfer of genes to cells, even before any gene expression can occur. While many transfection strategies focus on DNA condensation and overcoming the plasma membrane, events associated with the intracellular trafficking of the DNA complexes have not been as extensively studied. Once internalized, plasmids must travel potentially long distances through the cytoplasm to reach their next barrier, the nuclear envelope. This review summarizes the current progress on the cytoplasmic trafficking and nuclear transport of plasmids used for gene therapy applications. Both of these processes utilize specific and defined mechanisms to facilitate movement of DNA complexes through the cell. The continued elucidation and exploitation of these mechanisms will lead to improved strategies for transfection and successful gene therapy.
Keywords: Transfection, plasmid, microtubules, nuclear pore complex, nuclear localization signal, nuclear transport, actin, cytoskeleton
BARRIERS TO GENE THERAPY
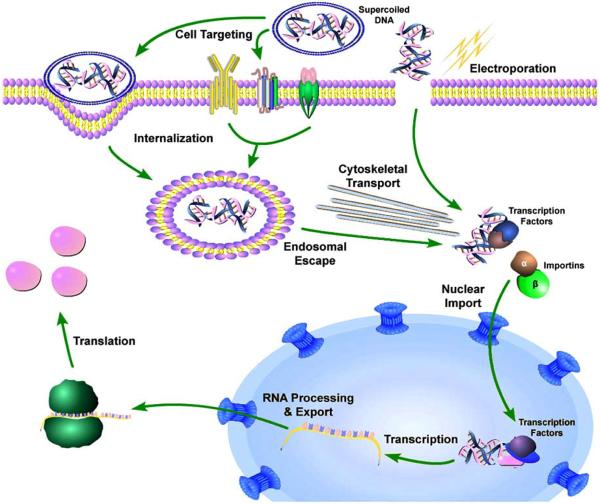
In order for gene therapy to be successful, many extracellular and intracellular barriers must be overcome (Fig. 1). Outside of the cell, a number of host systems can significantly impair gene delivery and result in sequestration of vectors by first pass organs (e.g. lung and liver), destruction of nucleic acids by serum nucleases, opsonization, and immune and inflammatory responses. Other barriers include the extracellular matrix, which can greatly affect how vectors traffic in and between tissues and into their target cells. Following successful targeting to any individual cell, the DNA must enter the cell, traverse the cytoplasm, and enter the nucleus prior to transcription, translation, and modification. Multiple chemical and physical approaches are routinely used to deliver DNA into the cell, but all result in the delivery of the DNA into the cytoplasm. However, getting into the cell is not the end. Once inside the cell, the DNA must still navigate through the cytoplasm to reach the nucleus and then gain access to the nuclear compartment and be expressed. In this review, we will focus on the intracellular events associated with DNA trafficking.
Fig. (1). Pathways and cellular barriers to gene delivery.
Supercoiled DNA can be internalized via receptor mediated pathways, liposomal endocytosis, or through transient pores created in the plasma membrane by physical forces such as electroporation. Once inside the cell, the DNA needs to traverse the cytoplasm to be imported into the nucleus where it can be transcribed.
CYTOPLASMIC DIFFUSION?
Until recently, the cytoplasm was viewed largely as a black box: most research focused first on how to get DNA into the cell and second, once inside the cell, how to get the DNA into the nucleus. Yet, how the DNA moves through the cytoplasm and how the DNA moves once inside the nucleus are largely unknown. Recent research focused on how plasmids traffic throughout the cell is imperative to enhancing gene delivery and gene therapy.
There are two basic mechanisms by which plasmids can reach the nucleus once they enter the cell: by diffusion or by active transport. It had previously been shown by spot-photobleaching that small solutes could diffuse freely and rapidly in the cytoplasm and the nucleus [Bicknese et al. 1993; Kao et al. 1993]. This suggested that diffusion of large macromolecules like DNA may be possible in the cytoplasm. However, Alan Verkman and colleges elegantly demonstrated that it is highly improbable that DNA is able to freely diffuse in the cytoplasm. After microinjection of fluorescein-labeled linear DNAs of varying lengths into cells, the diffusional mobilities were determined by fluorescence recovery after photobleaching (FRAP) and it was shown that movement of DNA larger than 250 base-pairs was greatly slowed (>17-fold) compared to that seen in water [Lukacs et al. 2000]. Further, DNA fragments larger than 2,000 base-pairs were effectively unable to diffuse in the cytoplasm in any reasonable physiological time frame [Lukacs et al. 2000]. It has been shown that the primary mechanism for the size-dependent reduced mobility of DNA in the cytoplasm is likely due to the highly cross-linked actin cytoskeleton, since its disruption by cytochalasin D eliminated the size-dependent reduction in DNA diffusion [Dauty & Verkman 2005]. Interestingly, this data is supported by previous observations that the length of actin filaments is estimated to be between 100 and 500 nm with a spacing of around 100 nm between filaments [Niederman et al. 1983; Stossel 1984]. Since a 250 bp fragment of double-stranded DNA has an extended linear length of approximately 85 nm, it would be expected to be sterically hindered in its movement through this network. Thus, it appears highly unlikely that DNA is able to diffuse in the cytoplasm. However, since transfections do work, there must be mechanisms whereby DNA can navigate through the dense meshwork of the cytoskeleton to reach the nucleus. Consequently, if diffusion cannot explain the successful movement of DNA to the nucleus, the major alternative is directed trafficking along cytoskeletal elements.
VIRAL VECTORS
In order to understand how plasmids, plasmid-polymer, and protein-DNA complexes traffic in the cytoplasm, it is useful to first understand how viruses have evolved to efficiently target their genomes to the nucleus. Many viruses have the ability to enter cells and effectively deliver their DNA to the nucleus. Further, they have devised a number of approaches to exploit normal cellular processes for their successful trafficking. The most common viral vectors used include those derived from retroviruses, adenoviruses, and herpes simplex virus, all of which have evolved unique mechanisms for intracellular trafficking.
Multiple cytoskeletal networks appear to be involved in the intracellular trafficking of retroviruses. Retroviral vectors are RNA viruses that replicate via a DNA intermediate. Conversion of the viral RNA to double-stranded DNA occurs in the cytoplasm and the resulting pre-integration complex consists of the reverse-transcribed DNA and a number of viral proteins that are present in the incoming virus. How a retrovirus traffics depends on how the virus enters the cell. Since the retroviral envelope protein determines how the virus enters and controls cell entry, it also affects intracellular trafficking [Burns et al. 1993; Anderson & Hope 2005]. When viruses are pseudotyped with the HIV-1 envelope protein Env, fusion between the cell membrane and the viral envelope occurs and the viral core then enters the cytoplasm directly, bypassing endocytosis and endosomal escape. In this case, the viral protein Nef is required for intracellular trafficking [Tobiume et al. 2003; Campbell et al. 2004]. Nef is a multifunctional viral protein that appears to have a role in depolymerizing cortical actin [Chazal et al. 2001]. It has been shown that depolymerization of the actin cytoskeleton can mimic the effects of Nef in its absence [Campbell et al. 2004], supporting the idea that initially the cortical actin cytoskeleton immediately beneath the plasma membrane impedes trafficking of the virus. However, some studies suggest that the actin cytoskeleton actually enhances intracellular trafficking of other viruses (e.g., vaccinia and HIV)[Bukrinskaya et al. 1998; Komano et al. 2004]. By contrast, when a virus is pseudotyped with the VSV-G or the Ebola envelope proteins, the virus enters by endocytosis and the viral core then must exit the endosome to enter the cytoplasm [Chazal et al. 2001]. Viruses that are pseudotyped with these proteins do not require the virally-encoded Nef protein for efficient cell entry. This suggests that when a virus enters by endocytosis, disruption of the cortical actin cytoskeleton is not required.
After traversing the cortical actin network or bypassing it by endocytosis and endosomal escape, the retroviral core and reverse-transcribed pre-integration complex must traffic through the rest of the cytoplasm. Early studies demonstrated that when the microtubule network was depolymerized by nocodazole, infection by HIV was reduced 2-fold [Bukrinskaya et al. 1998], suggesting the possibility that HIV may traffic toward the nucleus on the microtubule network. This is supported by work from Tom Hope's lab who imaged GFP-tagged HIV particles [McDonald et al. 2002]. Viruses were labeled with a GFP-Vpr fusion protein (part of the viral core which remains with the pre-integration complex) and real-time fluorescence microscopy of living cells demonstrated that cytoplasmic movement occurred in curvilinear paths. These GFP-Vpr particles co-localized with microtubules, and accumulated around the microtubule organizing center. Additional experiments have shown that this trafficking appears to use the microtubule-based, molecular motor dynein for movement towards the nucleus [Saib et al. 1997; McDonald et al. 2002; Petit et al. 2003]. Thus, while entry and initial trafficking may rely on disruption of the actin cytoskeleton, it is the microtubule network that is required for movement to the nucleus.
DNA viruses, like retroviruses, can enter the cell in a variety of ways, including endocytosis and fusion with the cell membrane. Adenoviruses enter the cell via the classical clathrin-mediated endocytic pathway [Medina-Kauwe 2003]. Acidification of the endosome is then thought to introduce conformational changes in the adenovirus capsid which results in endosomal escape and entry into the cytoplasm [Seth et al. 1985]. Unlike retroviruses which uncoat in the cytoplasm, the adenoviral capsid remains largely associated with the DNA genome and docks at the nuclear pore to release its DNA to the nucleus [Trotman et al. 2001]. Leopold et al. have shown that once in the cytoplasm, GFP-labeled adeno-virus particles use the microtubule network to reach the nucleus [Leopold et al. 2000]. They found that depolymerization of the microtubule network with nocodazole inhibited nuclear localization of the virus whereas pharmacological disruption of the actin network with cytochalasin D did not alter nuclear localization. Further, using direct video observation, they found that when nocodazole was used to disrupt the microtubules, they no longer observed linear movement of the virus, suggesting that viruses could not travel via directed movement towards the nucleus without the microtubule network. In addition, microinjection of blocking antibodies to the microtubule retrograde motor dynein resulted in the inhibition of nuclear accumulation of the viruses, whereas blocking kinesin did not affect nuclear accumulation [Leopold et al. 2000]. Since the environment in which the Adenovirus particles were translocating was at a neutral pH, as determined by fluorescence ratio imaging, they concluded that the movement was occurring in the cytoplasm and not inside endosomes. Later studies used an in vitro microtubule assay to show that adenovirus is indeed able to interact with dynein and microtubules [Kelkar et al. 2006]. Recent data also suggests that adeno-associated viruses, as well as different serotypes of adenoviruses, bind to dynein by a shared mechanism [Kelkar et al. 2006].
Other viruses, such as herpes simplex virus can enter the cell by membrane fusion, resulting in the tegument and DNA-containing capsid being deposited at the periphery of the cytoplasm, where it has been shown to localize with dynein [Sodeik et al. 1997; Dohner et al. 2002]. This interaction between HSV capsids and dynein results in retrograde transport along microtubules to the microtubule organizing center, which is often located with close proximity to the nucleus [Kristensson et al. 1986; Sodeik et al. 1997; Mabit et al. 2002]. Indirect immunofluorescence has been used to analyze the transport of individual capsids and it was found that within four hours of infection, most of the virus was located at the nuclear rim [Sodeik et al. 1997]. Most of the capsids not associated with the nucleus were associated with microtubules and it was shown that these capsids contained viral DNA. Additionally, using immunoelectron microscopy they showed that incoming cytosolic viral capsids were associated with dynein [Sodeik et al. 1997].
NON-VIRAL VECTORS
Although viral vectors have been regarded as the most efficient delivery vehicles for gene therapy, they have a number of limitations, including the generation of host immune and inflammatory reactions, size limitations of cargo genes, and random integration into the host genome, in the case of retroviruses. Therefore, much research has focused on the development of non-viral vectors. However, without the viral machinery to facilitate cellular uptake, alternate approaches have been developed since free DNA is not readily taken up by cells. A number of current reviews have addressed the issues associated with overcoming the barriers presented by the cell membrane [El-Andaloussi et al. 2005; Patil et al. 2005; Khalil et al. 2006]. Briefly, to enable up-take, the DNA can be complexed with cationic polymers (polyplex), cationic lipids (lipoplex) or a mixture (lipopolyplex). These complexes impart a positive charge on the DNA and the complex is able to enter the cell. Originally, it was thought that the complexed DNA was able to freely enter the cell [Felgner et al. 1987; Smith et al. 1993]. More recent data suggests that the complex enters by endocytosis [Zabner et al. 1995; Clark & Hersh 1999]. This is supported by confocal image-assisted three-dimensionally integrated quantification, which was used to determine that one hour post lipofectamine treatment, a large percentage of the transfected plasmid was in the endosome/lysosome fraction [Hama et al. 2006]. Zabner and colleagues demonstrated that much of the DNA remains in the endosome and that the fusion of these endosomes results in the formation of large perinuclear aggregates of lipoplexes [Zabner et al. 1995]. This, in addition to their observation that lipid-DNA complexes microinjected directly into the nucleus are unable to express, suggests that the DNA not only needs to escape from the endsome but also must dissociate from the lipoplex, polyplex, or lipopolyplex to be transcribed.
As an alternative to these chemical approaches, physical methods also work efficiently to deliver DNA into the cytoplasm. Physical methods such as hydroporation, electroporation, biolistic delivery (gene gun), or ultrasound are all used to deliver DNA across the plasma membrane and result in naked DNA being deposited into the cytoplasm [Dean 2005]. Thus, regardless of the method of transfection, it appears that plasmids must enter the cytoplasm prior to nuclear localization. As discussed earlier, diffusion does not appear to be a feasible mechanism to explain DNA cytoplasmic transport. Thus, whether the DNA enters directly via a physical force or after endosomal/lysomal escape there must be a means whereby the free DNA can reach the nucleus. Since retrograde movement of many viruses and endocytosed materials, including DNA/lipoplex complexes and polyethyleneimine (PEI)/DNA polyplexes [Bausinger et al. 2006], occurs via the microtubule network, it is reasonable to assume that movement of plasmids towards the nucleus could utilize the microtubule network as well. Indeed, our lab and others have shown that this is the case [Mesika et al. 2005; Vaughan & Dean 2006].
When GFP expressing plasmids were cytoplasmically microinjected into cultured cells, a significant level of expression was detected within four hours [Vaughan & Dean 2006]. If the microtubule network was depolymerized with nocodazole, expression levels were greatly reduced. By contrast, depolymerization of microtubules had no effect on gene expression following microinjection of the DNA directly into the nucleus, suggesting that the differences observed in the cytoplasmic injections are likely due to trafficking on microtubules. Similarly, co-injections of a GFP-expressing plasmid and a dynein inhibitory antibody resulted in decreased expression levels, whereas control nuclear injections and co-injection of DNA with a control antibody did not affect expression levels. To determine if DNA can interact with microtubules, a spin-down assay was used [Vaughan & Dean 2006]. After high speed centrifugation, microtubules pelleted along with interacting proteins. When microtubules and DNA were incubated and subjected to this assay, the DNA was found in the supernatant, suggesting that there is not a direct interaction between microtubules and the DNA. This was not surprising, since the experiments in cells suggested that dynein and perhaps other cellular proteins are mediating the interaction. When cell extract was added to the reactions, the DNA was found almost exclusively in the pellet, demonstrating that in the presence of cellular proteins, DNA can bind indirectly to microtubules [Vaughan & Dean 2006].
Thus, plasmids, like many viruses, use the microtubule network to reach the nucleus. However, the mechanism whereby DNA attaches to dynein, and hence the microtubule network, remains unknown. What is known is that covalent or non-covalent attachment of a nuclear localization sequence to plasmids have been shown to improve transfection efficiency (Collas and Alestrom 1997; Aronsohn and Hughes 1998; Sebestyen, et al. 1998; Branden, et al. 1999; Ciolina, et al. 1999; Ludtke, et al. 1999; Subramanian, et al. 1999; Zanta, et al. 1999; Chan and Jans 2001; Balicki, et al. 2002). Further, Mesika et al. have shown that the addition of an NLS improves transfection efficiency not only by enhancing nuclear entry of the plasmid but also by facilitating transport in the cytoplasm [Mesika et al. 2005]. They demonstrated an increased transfection efficiency when plasmids containing repetitive binding sites for the p50 subunit of the NF-κB transcription factor were co-injected with p50. p50 contains an NLS, thus suggesting that the NLS-containing protein may facilitate cytoplasmic transport. They further demonstrated that pharmacological disruption of the microtubule network abrogated the increased expression observed and that co-injection of an anti-dynein antibody also resulted in diminished transfection efficiency [Mesika et al. 2005]. This suggests that the retrograde microtubule-based transport of DNA may be due to the presence of NLSs on proteins that bind to the DNA. Indeed, using direct single-particle tracking in a cell-free assay, another group was able to demonstrate that NLSs facilitate active transport of proteins along microtubules [Salman et al. 2005]. It is known that for classical NLS-meditated nuclear entry, the NLS must bind to a heterodimer of importins and [Gorlich 1997]. Interestingly, Mesika et al. were able to pull down plasmids with importin [Mesika et al. 2005]. It was originally assumed that importins were found in the perinuclear area because of their function, but some studies suggest that they may interact with microtubules to funnel proteins to the nuclear pore [Smith & Raikhel 1998; Lam et al. 2002; Mavlyutov et al. 2002]. Moreover, in neuronal axoplasm, importin α has been found to be constitutively associated with dynein and importin β has been shown to be located throughout the axoplasm [Hanz et al. 2003], allowing for the possibility that the NLS is able to bind the importins in the cytoplasm for dynein-mediated transport on the microtubule network towards the nucleus.
NUCLEAR ENVELOPE AS A BARRIER
The nuclear envelope has been proposed to be one of the most substantial barriers for DNA delivery to cells. This was first demonstrated in 1980 when pBR322-based plasmids were injected into the cytoplasm and showed no gene expression, while nuclear injected plasmids showed expression in 50–100% of cells [Capecchi 1980]. Additional studies confirmed that when 1000 to 3000 copies of plasmid were microinjected into the cytoplasm, the levels of gene expression were approximately 3% of that seen when the plasmids were injected into the nucleus [Graessman et al. 1989]. Zabner and colleagues demonstrated in Xenopus oocytes that nuclear injected DNA resulted in high level gene expression, but the same DNA injected into the cytoplasm gave no expression [Zabner et al. 1995]. Later studies in several other mammalian cell types further support the dependence of gene expression on plasmid nuclear localization [Mirzayans et al. 1992; Thornburn & Alberts 1993].
This is not to say that DNA in the cytoplasm does not reach the nucleus. Rather, the amount of DNA that gains access to the nuclear compartment is small, regardless of how it is delivered to a cell or whether the cells are actively dividing. It has been estimated that between 2000 and 10,000 plasmids are delivered per cell following lipofection, but that only between 20 and 1000 are detected in the nucleus by 24–36 hours following DNA addition [Coonrod et al. 1997; Tseng et al. 1997; James & Giorgio 2000]. Other studies have placed the value between 30 and 60% of the input DNA reaching the nucleus [James & Giorgio 2000], but due to the use of labeled plasmid and the methods of detection [Sebestyén et al. 1998; Ludtke et al. 2002], this may be an overestimation. However, these studies reveal that a large percentage of the input DNA never reaches the nucleus. Indeed, two studies have shown that even in actively dividing cells, it takes between 30 and 100 times more plasmid microinjected into the cytoplasm compared to the nucleus to give equivalent levels of gene expression [Dean et al. 1999; Ludtke et al. 2002]. One reason for this is that trafficking through the cytoplasm is inefficient. Second, DNA degradation likely plays a major role in limiting the amount of DNA that can reach the nucleus. It has been estimated that the half-life of naked plasmid DNA in the cytoplasm of cells ranges between 50 minutes and 5 hours. If an average half-life of 3 hours is assumed, during a typical 24 hour transfection experiment, unless the DNA reaches the nucleus quickly, less than 0.4% of the input DNA would remain by 24 hours [Tseng et al. 1997; Escriou et al. 1998; Lechardeur et al. 1999; Zelphati et al. 1999; James & Giorgio 2000; Pollard et al. 2001; Banks et al. 2003].
MITOSIS OR NUCLEAR PORE MEDIATED?
Access to the nucleus in many cell types, including most primary cells and many cellular targets for gene delivery in vivo, remains highly restricted by the nuclear envelope [Capecchi 1980; Zabner et al. 1995; Dean et al. 1999; Zupan et al. 2000; Escriou et al. 2001; Li et al. 2001; Young & Dean 2002]. In the absence of mitosis, the nuclear membrane remains largely impermeable to plasmids. However, during mitosis, the nuclear envelope breaks down and plasmid in the cytoplasm can gain access to the newly-formed nuclei of daughter cells prior to nuclear envelope formation. This is the case for most routine transfections in cell culture. Indeed, one study demonstrated that actively dividing cells, identified by BrdU incorporation, were ten times more likely to express the transferred gene product than BrdU-negative cells [Fasbender et al. 1997]. It has been shown that transfections are largely dependent on the cell cycle, with a 50- to 300- fold higher level of gene expression obtained when cells were exposed to lipoplexes just before (G2 phase) or during mitosis (G2-M Phase)[Tseng et al. 1999; Brunner et al. 2000]. Similarly, gene transfer and nuclear entry of DNAPEI complexes are highly dependent on mitotic events [Grosse et al. 2006]. This demonstrates that nuclear envelope breakdown is critical for efficient DNA nuclear entry and gene expression. Though certain physical methods such as electroporation may show greater cell-cycle independent gene transfer, [Brunner et al. 2002], in non-dividing or growth arrested cells, the only access plasmid DNA (or viral genomes) will have to the nucleus is through the nuclear pore complex, or NPC.
The NPC is an aqueous channel in the nuclear envelope through which proteins and ribonucleoproteins can traffic [Gorlich 1997]. Indeed, all exchange of macromolecules between the nucleus and cytoplasm during interphase occurs through NPCs. The pores are large ( ~ 125 Mdal) multiprotein complexes that are composed of upwards of 100 distinct proteins present in multiple copies. Transport across the nuclear envelope occurs by either signal-independent diffusion in the case of proteins less than 50 Kdal at rates inversely proportional to their size or by signal-mediated import which requires the presence of a nuclear localization signal (NLS) within the imported protein. Proteins containing an NLS interact with a set of cytoplasmic receptor proteins, termed importins, which bind to both the NLS and subunits of the NPC itself to facilitate translocation across the pore. The directionality of nuclear transport is controlled by a small GTP-binding protein, RAN, which in its GTP-bound state is localized exclusively to the nucleus and promotes disassembly of the NLS-importin complex on the inner face of the nuclear envelope. Since a typical plasmid is between 2 and 10 Mdal, passive import is expected to be almost nonexistent. Thus, some sort of facilitated import must account for translocation of DNA across the NPC.
Wolff and colleagues demonstrated that plasmids can indeed enter the nuclei of terminally differentiated cells in the absence of cell division [Dowty et al. 1995]. They showed that plasmids localized to the nucleus following cytoplasmic microinjection in cultured myotubes, based on detection of reporter gene expression. Nuclear import was dose- and energy-dependent and was inhibited by agents that block transport through the NPC. Further, they demonstrated that greater levels of gene expression (hence trafficking) were obtained when the plasmids were injected near the nuclei as opposed to far away from the nuclei [Dowty et al. 1995]. Using a similar approach, our laboratory demonstrated that plasmids do indeed traffic into the nucleus through the nuclear pore complex by localizing the injected DNA by in situ hybridization [Dean 1997] or directly using plasmids labeled with fluorescent peptide nucleic acid clamps [Wilson et al. 1999]. Following microinjection of plasmids between 4 and 14 kb into the cytoplasm, we were able to detect movement of the DNA into the nuclei of injected cells within 8 hours. Further, this import was energy-dependent and utilized the NPC, since co-injection of lectins or antibodies that block nucleocytoplasmic trafficking of proteins and mRNA through the NPC also blocked import of plasmids.
SEQUENCE-SPECIFIC NUCLEAR IMPORT OF PLASMID DNA
Although it has been shown in vivo that numerous plasmids can lead to productive expression, a number of reports have suggested that certain specific sequences of DNA can enhance nuclear accumulation of plasmid DNA. Indeed, in cultured cells, it appears that DNA nuclear import is dependent on the presence of these sequences in the absence of cell division. When plasmid DNA from the DNA tumor virus SV40 was microinjected into the cytoplasm of growth-arrested cells, the majority of cells showed nuclear localization of the plasmid within 6 to 8 hours [Dean 1997]. By contrast, plasmids lacking SV40 sequences failed to be imported into the nuclei of cells and in synchronized cells, remained in the cytoplasm until after cell division [Dean 1997; Dean et al. 1999]. It was further shown that when included on one of these cytoplasmically localizing plasmids, as little as 50 bp of the SV40 enhancer was able to promote plasmid entry into the nucleus with the same kinetics as the full SV40 genome [Dean et al. 1999]. Previous studies in transfected and microinjected cells pointed to this nuclear import activity, since when plasmids lacking the enhancer sequence were injected into the cytoplasm of dividing cells, they showed 30-fold lower levels of expression than when delivered directly into the nucleus, whereas plasmids containing the SV40 enhancer showed only modestly increased expression after nuclear injection versus that found with cytoplasmic injection [Graessman et al. 1989]. This SV40 sequence, termed a DNA targeting sequence (DTS), has been shown to be active in cell lines derived from monkey, rat, mouse, hamster, chicken, and human origin [Dean et al. 1999]. Other sequences having similar ability to promote plasmid nuclear import have also been identified (see below) [Langle-Rouault et al. 1998; Vacik et al. 1999; Zennou et al. 2000; Mesika et al. 2001; Vaysse et al. 2004; Mesika et al. 2005; Arhel et al. 2006]. However, it should be stressed that such sequence-specific nuclear import appears to be important mainly in the absence of cell division. Thus, when cells go through mitosis, any DNA can enter the nuclear space, and as such, these sequences will play little, if any, role in enhancing nuclear import. It is interesting to note that many of the studies that first identified the nuclear envelope as a barrier to gene transfer used plasmids that lacked this SV40 sequence [Capecchi 1980; Graessman et al. 1989; Thornburn & Alberts 1993; Zabner et al. 1995]. Since most of these studies employed plasmids expressing reporter genes driven from the Cytomegalovirus (CMV) immediate early promoter, the Rous sarcoma virus long terminal repeat (LTR) promoter, or the herpes thymidylate kinase (TK) promoter, all of which have been shown to have no plasmid nuclear import activity [Dean et al. 1999; Vacik et al. 1999], it is not surprising that the plasmids failed to enter the nucleus.
The SV40 DTS has shown significant effects on gene transfer and subsequent expression in vitro and in vivo. In synchronized cells microinjected with GFP-expressing plasmids, it was shown that the presence of the SV40 DTS allowed for gene expression within 2 to 4 hours from the CMV promoter when as few as 10 plasmids were microinjected into the cytoplasm, whereas injection of 1000 similar plasmids lacking the SV40 sequence failed to show any expression until cell division [Dean et al. 1999]. Similarly, in transfected cells, the presence of the SV40 sequence has been shown to increase gene expression, presumably due to its import activity [Reddy et al. 1999; Vacik et al. 1999; Yanai et al. 2006] In mouse muscle, plasmids containing the SV40 DTS have shown a 20-fold increase in reporter gene expression when compared to plasmids lacking the DTS [Li et al. 2001; Blomberg et al. 2002]. Similarly, plasmids carrying the SV40 DTS increased gene expression 40 to 200 fold in the vasculature of living rats following delivery by electro-poration, and in situ hybridization confirmed that the increased expression was a result of increased nuclear import of the plasmids [Young et al. 2003]. The central caveat to these experiments is that the cells and tissues are largely non-dividing. Indeed, in a recent study using electroporation to deliver genes to the cornea, Zhou and Dean found that while the presence of the SV40 DTS was required for DNA nuclear import in cultured corneal epithelial cells and fibro-blasts [Dean et al. 1999], it had little impact on gene delivery to the injured cornea, presumably due to the fact that the cells in the cornea were actively dividing in a wound repair process [Zhou & Dean 2006].
The mechanism of nuclear import of the SV40 DTS is currently not known. It is known, however, that the SV40 enhancer contains binding sites for numerous transcription factors, such as AP1, AP2, AP3, NF- B, Oct-1, TEF-I and TEF-II [Dynan & Tjian 1983; Wildeman 1988; Dynan & Chervitz 1989]. The main function of transcription factors is to activate transcription of target genes in the nucleus. Since they are translated in the cytoplasm, they require NLS motifs to translocate into the nucleus and bind their respective DNA regulatory elements. However, when their binding sites are presented on a plasmid in the cytoplasm, it is hypothesized that these transcription factors can bind the exogenous DNA in the cytoplasm to form a DNA-protein complex and mediate interactions between DNA and importin proteins via their NLS motifs. Indeed, in modified ChIP assays and in purification assays, plasmids are indeed found to bind to a number of cellular proteins, including transcription factors, in the cytoplasm and in cytoplasmic extracts (F. Munkonge, E. Vaughan, A. Miller, and D. A. Dean, unpublished observations). Further, it has been demonstrated that such DNA-transcription factor-importin complexes can be formed in vitro and in cells [Chan & Jans 1999]. Interestingly, other strong viral promoters and enhancers, such as the cytomegalovirus promoter, Rous sarcoma virus and Moloney murine leukaemia virus long terminal repeats, and the herpes simplex virus thymidylate kinase promoter do not facilitate nuclear import of plasmid DNA in the absence of cell division [Dean et al. 1999; Vacik et al. 1999]. Since these elements also contain numerous binding sites for multiple transcription factors, this suggests that a specific tertiary protein-DNA complex is likely responsible for the NLS-importin interactions.
Based on this model, it was hypothesized that certain promoters may bind cell-specific transcription factors and mediate plasmid nuclear import in selected cell types. Several sequences have been identified, including a DTS active only in endothelial cells, and a DTS active only in smooth muscle cells [Vacik et al. 1999; Dean 2002]. Upon microinjection into the cytoplasm, plasmids containing the smooth muscle gamma actin (SMGA) promoter were imported into the nuclei of smooth muscle cells, but not in non-smooth muscle cell types, such as endothelial or epithelial cells [Vacik et al. 1999]. This promoter is transcriptionally regulated by serum response factor (SRF) and Nkx3 [Browning et al. 1998; Kovacs & Zimmer 1998; Carson et al. 2000], and mutation of either of these DNA binding elements abrogates SMGA DTS nuclear import activity.
Other groups have identified additional DTS elements that enhance exogenous gene expression when included on a plasmid. For example, it is well known that the transcription factor NF-κB is translocated to the nucleus when activated by the cytokine TNF-α [Hayden & Ghosh 2004]. Based on this data, a series of five NF-κB binding sites were included on a plasmid and transfected into cells with the intent of enhancing plasmid nuclear import. It was observed that upon treating cells with TNF-α, nuclear import and subsequent gene expression increased significantly when compared with parent plasmids [Mesika et al. 2001]. Though the SV40 DTS was also included on this plasmid [Mesika et al. 2001], subsequent reports using plasmids containing only NF-κB sites have confirmed that this sequence provides enhanced DNA nuclear import [Mesika et al. 2005]. Another sequence that can act as a DTS is the oriP sequence from the Epstein-Barr virus (EBV). When EBV nuclear antigen (EBNA)-1 expressing cells were cytoplasmically microinjected with plasmids containing the oriP DTS, increased gene expression was detected when compared to plasmids lacking oriP [Langle-Rouault et al. 1998]. More recently, combinations of the tet operator and a modified tetracycline repressor containing an NLS (tetO and TetR-NLS, respectively) have been used in cis and trans to show that nuclear import can be controlled and enhanced by protein-DNA interactions [Vaysse et al. 2004; Vaysse et al. 2006]. When multiple copies of the tetO sequence were cloned into a plasmid and transfected into cells expressing TetR-NLS, gene expression increased almost 20-fold in growth-arrested cells, and nuclear localization increased by 4-fold.
Although a number of specific sequences have been identified and shown to increase DNA nuclear import both in vitro and in vivo, using a number of experimental approaches from multiple laboratories, other experiments suggest that while these sequences may enhance transport, they may not be “required” for DNA nuclear import in the absence of cell division. For example, a number of different plasmids lacking specific DNA nuclear import sequences have been delivered by direct injection or electroporation to mouse muscle and have shown robust gene expression [Wolff et al. 1990; Wolff et al. 1992; Wolff et al. 1992; Manthorpe et al. 1993; Hartikka et al. 1996; Doh et al. 1997], despite the fact that myotubes are terminally differentiated and do not divide. In these experiments, both the CMV promoter and the RSV LTR showed robust expression in muscle. This implies that in certain tissues, especially skeletal muscle, plasmids lacking the SV40 enhancer are still able to enter the nucleus, although other studies have reported that the SV40 enhancer can increase gene expression in mouse muscle [Li et al. 2001; Blomberg et al. 2002]. Another recent study used a series of plasmids containing or lacking various segments of the SV40 DTS and found that upon liposome-mediated transfection of several cell types, there were no beneficial effects of the import sequence on gene expression in cultured cells [Prasad & Rao 2005]. However, in this study, effects on nuclear import of the DNA were not evaluated. One possibility is that these DNA nuclear import sequences simply increase the rates or nuclear import and that this can shift the balance between productive nuclear targeting and expression and degradation in the cytoplasm. If this is the case, it could be possible to flood the cytoplasm with DNA and drive the nuclear import reaction independent of specific DNA sequences. In support of this, it has been shown that when 105 plasmids (lacking SV40 sequences) are injected into the cytoplasm of a single mouse myotube in vivo, no gene expression is seen, but when 106 plasmids are injected, gene expression is detected [Utvik et al. 1999]. Another possible reason for these differing results is that many studies on intracellular trafficking of DNA have used a labeling technique that has been shown to alter DNA trafficking properties, perhaps leading to misinterpretations [Gasiorowski & Dean 2005].
NLS:DNA CONJUGATES
Since nuclear localization signals were identified and shown to direct protein nuclear localization [Kalderon et al. 1984; Kalderon et al. 1984], their use to promote and enhance nuclear import of plasmids has been proposed and studied [Cartier & Reszka 2002; Escriou et al. 2003; Hebert 2003]. In early studies, increased nuclear localization and transgene expression was observed in zebrafish embryos after cytoplasmic injection of plasmid DNA complexed (non-covalently) with SV40 T-antigen NLS peptides [Collas & Alestrom 1996; Collas et al. 1996; Collas & Alestrom 1997; Collas & Alestrom 1997]. This process has been elucidated as a two-step mechanism, involving peptide binding to DNA and their subsequent translocation across the NPC [Collas & Alestrom 1997]. Using in vitro assembled sea urchin nuclei, they were able to demonstrate that DNA nuclear import was energy-dependent, inhibited by agents that block the NPC, and required cytoplasmic extracts [Collas & Alestrom 1996]. When this approach was taken in mammalian cells, only a 3-fold increase in gene expression was seen following cationic liposome-mediated transfection [Aronsohn & Hughes 1998]. A similar effect was observed by utilizing a non-classical NLS, the M9 sequence of the human hnRNP A1 protein, to enhance gene expression by an order of magnitude in non-dividing endothelial cells [Subramanian et al. 1999]. NLS peptides have also been covalently attached to plasmids and other forms of DNA to facilitate nuclear import, but the enhancement of gene delivery/expression has been modest at best [Sebestyén et al. 1998; Ciolina et al. 1999; Neves et al. 1999]. Perhaps the greatest improvements in nuclear targeting and gene expression were seen when a single NLS peptide fused to a linear piece of DNA that was capped at both ends by DNA hairpins [Zanta et al. 1999]. In this study, the authors reported that gene expression levels increased by nearly 1000-fold when a wild type NLS was fused to the DNA. Though the condensation of the plasmid DNA by the peptide may have contributed to the effect, the involvement of the NLS was implicated by the finding that a single amino acid substitution within the NLS peptide abolished increased gene expression [Zanta et al. 1999].
Peptide nucleic acids (PNAs) have also been used to link plasmid DNA to NLS peptides. Peptide nucleic acids are nucleic acid analogs in which the phosphodiester backbone is replaced with a polyamide backbone made up of repeating N-(2-aminoethyl)glycine units [Dean 2000; Koppelhus & Nielsen 2003]. PNAs can form triplex structures with specific sequences of DNA, and bind with a high affinity (10−6 – 10−9 M) [Dean 2000]. These PNAs have been explored as a tool to attach peptides and other molecules, and are especially useful due to their ability to maintain supercoiled conformation when bound to plasmid DNA [Zelphati et al. 1999; Zelphati et al. 2000]. Moreover, when bound to plasmids, they do not alter the trafficking or transcriptional properties of the DNA, whereas a number of commercially available kits either destroy the ability of a plasmid to move within the cell or even cause it to mislocalize [Gasiorowski & Dean 2005]. When PNA-NLS peptides were complexed with plasmids and transfected into cells with polyethylenimine (PEI), nuclear localization and gene expression was enhanced up to eightfold when compared to uncomplexed plasmid DNA [Branden et al. 1999]. Further, when similar PNA-NLS/plasmid and PNA-NLS/oligonucleotide conjugates were injected into mouse organs in vivo, increased levels of gene expression and nuclear localization of the conjugates were observed in skin, skeletal muscle, and liver [Branden et al. 2001].
As an alternative to linking individual NLS peptides by various means to plasmids, others have taken the approach of forming NLS-containing protein complexes with DNA in attempts to increase gene transfer and expression. Unlike highly charged NLS peptides which may electrostatically interact with the plasmid backbone rather than be accessible to the NLS-dependent nuclear import machinery, an advantage to using NLS-containing proteins is that the DNA binding domains of the proteins and the NLS may be spatially separated and structurally fixed so that the NLS is free to interact with the importins. Complexes of plasmids with high mobility group-1 (HMG-1) proteins or the nuclear protein nucleoplasmin have been shown to increase nuclear localization of plasmids by about 3-fold within 4 to 8 hours of transfection of cells and to increase gene expression by a factor of 5 within the first 12 hours in rat liver, compared to plasmids complexed with IgG or BSA [Kaneda et al. 1989]. Wolff and colleagues found that addition of histone H1 to plasmids gave a 20-fold stimulation of gene expression versus DNA without H1 when the complexes were transfected into cells using cationic [Fritz et al. 1996] or anionic [Hagstrom et al. 1996] liposomes. More recently, as discussed above, Mesika et al. have demonstrated that complexation of plasmids with the p50 subunit of the NF-kB transcription factor can faciliate both cytoplasmic trafficking and nuclear import [Mesika et al. 2001; Mesika et al. 2005].
Use of viral proteins coupled to plasmid DNA has gained momentum recently as alternative to viral delivery systems. The human immunodeficiency virus-1 (HIV) has the ability to transport its cDNA through an intact nuclear envelope, and consequently has the ability to infect non-dividing cells [Hearps & Jans 2006]. Thus, the proteins involved in this mechanism are being investigated as potential candidates for enhancement of DNA nuclear import. Recently, one study analyzed the ability of a tetracycline repressor protein TetR fused to the TAT peptide to enhance nuclear import and gene expression of minicircle DNA in vivo. It was found that there was more than a 6-fold increase in gene expression in mouse lung [Vaysse et al. 2006]. Another HIV protein, the integrase IN, has shown nuclear accumulation and DNA binding activity in vitro, and may be a candidate for enhancement of nonviral gene delivery [Hearps & Jans 2006].
Several recent studies have used approaches that bypass use of an NLS but still try to increase NLS-dependent nuclear import. Daniel Scherman's group has taken an intriguing, although unfortunately, not particularly successful approach of bypassing the NLS and fusing plasmids directly to an importinβ peptide directly. During transport of classic NLS-containing proteins, the NLS binds to importinα which in turn binds to importinβ through its importinβ binding domain (IBB), and this IBB then facilitates translocation into the nucleus [Gasiorowski & Dean 2003]. When the IBB peptide was covalently conjugated to plasmids, a 20-fold increase in gene expression was seen in inefficiently transfected cells [Carriere et al. 2003]. However, when transfection conditions were optimized, the peptide had no effect, leading the authors to conclude that the improvement was not due to nuclear import, but rather altered physiochemical properties of the peptide-DNA complex. In a similar approach, another study linked the recombinant importinβ protein to a plasmid using biotinylation. It was observed that, when this DNA conjugate was microinjected into NIH3T3 fibroblasts, the nuclear localization and gene expression efficiency was markedly higher than plasmid DNA alone [Nagasaki et al. 2005]. Another approach has exploited the glucocorticoid receptor which normally is cytoplasmic but upon ligand binding, translocates into the nucleus. Rebuffet and colleagues complexed dexamethasone, a glucocorticoid receptor ligand, to plasmids by either a direct linkage via a psoralen linker or using a PNA clamp [Rebuffat et al. 2002]. When these complexes were transfected into glucocorticoid receptor-expressing cells, gene transfer and expression was 20- to 40-fold greater than that seen with unmodified DNA. Finally, another study observed that when vimentin, a cytoplasmic intermediate filament protein, was bound to oligonucleotides or plasmids (via a cryptic DNA binding domain), the protein caused rapid migration of the DNAs into the nucleus, although the mechanisms for this remain unclear [Hartig et al. 1998; Hartig et al. 1998].
CONCLUSIONS
The limitations of gene therapy are many while the improvements have been few and incremental. Even though we can now deliver a higher amount of transgene into the cells of many different tissues with new generation transfection reagents and methods for in vivo delivery, there are many intracellular barriers that impede intracellular DNA trafficking, degrade DNA in transit, and silence gene expression once delivery has ceased. This often results in plasmids that never reach the nucleus and are subsequently degraded. Thus, it is vital that we understand how plasmids move inside the cell in order to develop novel ways to enhance trafficking and improve gene delivery and expression. Although the intracellular trafficking of exogenous DNA may not be a normal event in the cell, it occurs everyday in and out of the laboratory, and as such must be understood if gene therapy is to be an effective strategy to eventually treat disease.
ACKNOWLEDGEMENTS
We would like to thank Drs. Jennifer Young and Joshua Z. Gasiorowski, and all the members of the Dean laboratory for intriguing discussions and critical reading of the manuscript. Work in the authors' lab was supported in parts by grants HL59956, HL71643, and HL81148 from the NIH and predoctoral fellowships from the Midwest Affiliate of the American Heart Association (EEV and JVD).
Footnotes
Copyright of Current Gene Therapy is the property of Bentham Science Publishers Ltd. and its content may not be copied or emailed to multiple sites or posted to a listserv without the copyright holder's express written permissin. However, users may print, download, or email articles for individual use.
REFERENCES
- Anderson JL, Hope TJ. Intracellular trafficking of retroviral vectors: obstacles and advances. Gene Ther. 2005;12:1667–1678. doi: 10.1038/sj.gt.3302591. [DOI] [PubMed] [Google Scholar]
- Arhel NJ, Souquere-Besse S, Charneau P. Wild-type and central DNA flap defective HIV-1 lentiviral vector genomes: intracellular visualization at ultrastructural resolution levels. Retrovirology. 2006;3:38. doi: 10.1186/1742-4690-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsohn AI, Hughes JA. Nuclear localization signal peptides enhance cationic liposome-mediated gene therapy. J. Drug Target. 1998;5:163–169. doi: 10.3109/10611869808995871. [DOI] [PubMed] [Google Scholar]
- Banks GA, Roselli RJ, Chen R, Giorgio TD. A model for the analysis of nonviral gene therapy. Gene Ther. 2003;10:1766–1775. doi: 10.1038/sj.gt.3302076. [DOI] [PubMed] [Google Scholar]
- Bausinger R, von Gersdorff K, Braeckmans K, Ogris M, Wagner E, Brauchle C, Zumbusch A. The transport of nanosized gene carriers unraveled by live-cell imaging. Angew Chem. Int. Ed. Engl. 2006;45:1568–1572. doi: 10.1002/anie.200503021. [DOI] [PubMed] [Google Scholar]
- Bicknese S, Periasamy N, Shohet SB, Verkman AS. Cytoplasmic viscosity near the cell plasma membrane: measurement by evanescent field frequency-domain microfluorimetry. Biophys. J. 1993;65:1272–1282. doi: 10.1016/S0006-3495(93)81179-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomberg P, Eskandarpour M, Xia S, Sylven C, Islam KB. Electroporation in combination with a plasmid vector containing SV40 enhancer elements results in increased and persistent gene expression in mouse muscle. Biochem. Biophys. Res. Commun. 2002;298:505–510. doi: 10.1016/s0006-291x(02)02486-5. [DOI] [PubMed] [Google Scholar]
- Branden LJ, Christensson B, Smith CI. In vivo nuclear delivery of oligonucleotides via hybridizing bifunctional peptides. Gene Ther. 2001;8:84–87. doi: 10.1038/sj.gt.3301345. [DOI] [PubMed] [Google Scholar]
- Branden LJ, Mohamed AJ, Smith CI. A peptide nucleic acid-nuclear localization signal fusion that mediates nuclear transport of DNA. Nat. Biotechnol. 1999;17:784–787. doi: 10.1038/11726. [DOI] [PubMed] [Google Scholar]
- Browning CL, Culberson DE, Aragon IV, Fillmore RA, Croissant JD, Schwartz RJ, Zimmer WE. The developmentally regulated expression of serum response factor plays a key role in the control of smooth muscle-specific genes. Dev. Biol. 1998;194:18–37. doi: 10.1006/dbio.1997.8808. [DOI] [PubMed] [Google Scholar]
- Brunner S, Furtbauer E, Sauer T, Kursa M, Wagner E. Overcoming the nuclear barrier: cell cycle independent nonviral gene transfer with linear polyethylenimine or electroporation. Mol. Ther. 2002;5:80–86. doi: 10.1006/mthe.2001.0509. [DOI] [PubMed] [Google Scholar]
- Brunner S, Sauer T, Carotta S, Cotten M, Saltik M, Wagner E. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000;7:401–407. doi: 10.1038/sj.gt.3301102. [DOI] [PubMed] [Google Scholar]
- Bukrinskaya A, Brichacek B, Mann A, Stevenson M. Establishment of a functional human immunodeficiency virus type 1 (HIV-1) reverse transcription complex involves the cytoskeleton. J. Exp. Med. 1998;188:2113–2125. doi: 10.1084/jem.188.11.2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns JC, Friedmann T, Driever W, Burrascano M, Yee JK. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc. Natl. Acad. Sci. USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell EM, Nunez R, Hope TJ. Disruption of the actin cytoskeleton can complement the ability of Nef to enhance human immunodeficiency virus type 1 infectivity. J. Virol. 2004;78:5745–5755. doi: 10.1128/JVI.78.11.5745-5755.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capecchi MR. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell. 1980;22:479–488. doi: 10.1016/0092-8674(80)90358-x. [DOI] [PubMed] [Google Scholar]
- Carriere M, Escriou V, Savarin A, Scherman D. Coupling of importin beta binding peptide on plasmid DNA: transfection efficiency is increased by modification of lipoplex's physico-chemical properties. BMC Biotechnol. 2003;3:14. doi: 10.1186/1472-6750-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JA, Fillmore RA, Schwartz RJ, Zimmer WE. The smooth muscle gamma-actin gene promoter is a molecular target for the mouse bagpipe homologue, mNkx3-1, and serum response factor. J. Biol. Chem. 2000;275:39061–39072. doi: 10.1074/jbc.M006532200. [DOI] [PubMed] [Google Scholar]
- Cartier R, Reszka R. Utilization of synthetic peptides containing nuclear localization signals for nonviral gene transfer systems. Gene Ther. 2002;9:157–167. doi: 10.1038/sj.gt.3301635. [DOI] [PubMed] [Google Scholar]
- Chan CK, Jans DA. Synergy of importin alpha recognition and DNA binding by the yeast transcriptional activator GAL4. FEBS Lett. 1999;462:221–224. doi: 10.1016/s0014-5793(99)01515-x. [DOI] [PubMed] [Google Scholar]
- Chazal N, Singer G, Aiken C, Hammarskjold ML, Rekosh D. Human immunodeficiency virus type 1 particles pseudotyped with envelope proteins that fuse at low pH no longer require Nef for optimal infectivity. J. Virol. 2001;75:4014–4018. doi: 10.1128/JVI.75.8.4014-4018.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciolina C, Byk G, Blanche F, Thuillier V, Scherman D, Wils P. Coupling of nuclear localization signals to plasmid DNA and specific interaction of the conjugates with importin alpha. Bioconjug. Chem. 1999;10:49–55. doi: 10.1021/bc980061a. [DOI] [PubMed] [Google Scholar]
- Clark PR, Hersh EM. Cationic lipid-mediated gene transfer: current concepts. Curr. Opin. Mol. Ther. 1999;1:158–176. [PubMed] [Google Scholar]
- Collas P, Alestrom P. Nuclear localization signal of SV40 T antigen directs import of plasmid DNA into sea urchin male pronuclei in vitro. Mol. Reprod. Dev. 1996;45:431–438. doi: 10.1002/(SICI)1098-2795(199612)45:4<431::AID-MRD4>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- Collas P, Alestrom P. Nuclear localization signals: a driving force for nuclear transport of plasmid DNA in zebrafish. Biochem. Cell Biol. 1997;75:633–640. [PubMed] [Google Scholar]
- Collas P, Alestrom P. Rapid targeting of plasmid DNA to zebrafish embryo nuclei by the nuclear localization signal of SV40 T antigen. Mol. Mar. Biol. Biotechnol. 1997;6:48–58. [PubMed] [Google Scholar]
- Collas P, Husebye H, Alestrom P. The nuclear localization sequence of the SV40 T antigen promotes transgene uptake and expression in zebrafish embryo nuclei. Transgenic Res. 1996;5:451–458. doi: 10.1007/BF01980210. [DOI] [PubMed] [Google Scholar]
- Coonrod A, Li FQ, Horwitz M. On the mechanism of DNA transfection: efficient gene transfer without viruses. Gene Ther. 1997;4:1313–1321. doi: 10.1038/sj.gt.3300536. [DOI] [PubMed] [Google Scholar]
- Dauty E, Verkman AS. Actin cytoskeleton as the principal determinant of size-dependent DNA mobility in cytoplasm: a new barrier for non-viral gene delivery. J. Biol. Chem. 2005;280:7823–7828. doi: 10.1074/jbc.M412374200. [DOI] [PubMed] [Google Scholar]
- Dean BS, Byrd JN, Jr., Dean DA. Nuclear targeting of plasmid DNA in human corneal cells. Cur. Eye Res. 1999;19:66–75. doi: 10.1076/ceyr.19.1.66.5344. [DOI] [PubMed] [Google Scholar]
- Dean DA. Import of plasmid DNA into the nucleus is sequence specific. Exp. Cell Res. 1997;230:293–302. doi: 10.1006/excr.1996.3427. [DOI] [PubMed] [Google Scholar]
- Dean DA. Peptide nucleic acids: versatile tools for gene therapy strategies. Adv. Drug Deliv. Rev. 2000;44:81–95. doi: 10.1016/s0169-409x(00)00087-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean DA. Nucleocytoplasmic trafficking. In: Mahato RI, editor. Pharmaceutical perspectives of nucleic acid-based therapeutics. Harwood Academic Publishers; London: 2002. pp. 229–260. [Google Scholar]
- Dean DA. Nonviral gene transfer to skeletal, smooth, and cardiac muscle in living animals. Am. J. Physiol. Cell Physiol. 2005;289:C233–245. doi: 10.1152/ajpcell.00613.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean DA, Dean BS, Muller S, Smith LC. Sequence requirements for plasmid nuclear entry. Exp. Cell Res. 1999;253:713–722. doi: 10.1006/excr.1999.4716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doh SG, Vahlsing HL, Hartikka J, Liang X, Manthorpe M. Spatial-temporal patterns of gene expression in mouse skeletal muscle after injection of lacZ plasmid DNA. Gene Ther. 1997;4:648–663. doi: 10.1038/sj.gt.3300460. [DOI] [PubMed] [Google Scholar]
- Dohner K, Wolfstein A, Prank U, Echeverri C, Dujardin D, Vallee R, Sodeik B. Function of dynein and dynactin in herpes simplex virus capsid transport. Mol. Biol. Cell. 2002;13:2795–2809. doi: 10.1091/mbc.01-07-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowty ME, Williams P, Zhang G, Hagstrom JE, Wolff JA. Plasmid DNA entry into postmitotic nuclei of primary rat myotubes. Proc. Natl. Acad. Sci. USA. 1995;92:4572–4576. doi: 10.1073/pnas.92.10.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan WS, Chervitz SA. Characterization of a minimal simian virus 40 late promoter: enhancer elements in the 72-base-pair repeat not required. J. Virol. 1989;63:1420–1427. doi: 10.1128/jvi.63.3.1420-1427.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynan WS, Tjian R. The promoter-specific transcription factor Sp1 binds to upstream sequences in the SV40 early promoter. Cell. 1983;35:79–87. doi: 10.1016/0092-8674(83)90210-6. [DOI] [PubMed] [Google Scholar]
- El-Andaloussi S, Holm T, Langel U. Cell-penetrating peptides: mechanisms and applications. Curr. Pharm. Des. 2005;11:3597–3611. doi: 10.2174/138161205774580796. [DOI] [PubMed] [Google Scholar]
- Escriou V, Carriere M, Bussone F, Wils P, Scherman D. Critical assessment of the nuclear import of plasmid during cationic lipid-mediated gene transfer. J. Gene Med. 2001;3:179–187. doi: 10.1002/jgm.174. [DOI] [PubMed] [Google Scholar]
- Escriou V, Carriere M, Scherman D, Wils P. NLS bioconju-gates for targeting therapeutic genes to the nucleus. Adv. Drug Deliv. Rev. 2003;55:295–306. doi: 10.1016/s0169-409x(02)00184-9. [DOI] [PubMed] [Google Scholar]
- Escriou V, Ciolina C, Helbling-Leclerc A, Wils P, Scherman D. Cationic lipid-mediated gene transfer: analysis of cellular up-take and nuclear import of plasmid DNA. Cell Biol. Toxicol. 1998;14:95–104. doi: 10.1023/a:1007425803756. [DOI] [PubMed] [Google Scholar]
- Fasbender A, Zabner J, Zeiher BG, Welsh MJ. A low rate of cell proliferation and reduced DNA uptake limit cationic lipid-mediated gene transfer to primary cultures of ciliated human airway epithelia. Gene Ther. 1997;4:1173–1180. doi: 10.1038/sj.gt.3300524. [DOI] [PubMed] [Google Scholar]
- Felgner PL, Gadek TR, Holm M, Roman R, Chan HW, Wenz M, Northrop JP, Ringold GM, Danielsen M. Lipofection: a highly efficient, lipid-mediated DNA-transfection procedure. Proc. Natl. Acad. Sci. USA. 1987;84:7413–7417. doi: 10.1073/pnas.84.21.7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz JD, Herweijer H, Zhang G, Wolff JA. Gene transfer into mammalian cells using histone-condensed plasmid DNA. Hum. Gene Ther. 1996;7:1395–1404. doi: 10.1089/hum.1996.7.12-1395. [DOI] [PubMed] [Google Scholar]
- Gasiorowski JZ, Dean DA. Mechanisms of nuclear transport and interventions. Adv. Drug Deliv. Rev. 2003;55:703–716. doi: 10.1016/s0169-409x(03)00048-6. [DOI] [PubMed] [Google Scholar]
- Gasiorowski JZ, Dean DA. Postmitotic Nuclear Retention of Episomal Plasmids Is Altered by DNA Labeling and Detection Methods. Mol. Ther. 2005;12:460–467. doi: 10.1016/j.ymthe.2005.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorlich D. Nuclear protein import. Curr. Opin. Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- Graessman M, Menne J, Liebler M, Graeber I, Graessman A. Helper activity for gene expression, a novel function of the SV40 enhancer. Nucleic Acids Res. 1989;17:6603–6612. doi: 10.1093/nar/17.16.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosse S, Thevenot G, Monsigny M, Fajac I. Which mechanism for nuclear import of plasmid DNA complexed with polyethylenimine derivatives? J. Gene Med. 2006;8:845–851. doi: 10.1002/jgm.915. [DOI] [PubMed] [Google Scholar]
- Hagstrom JE, Sebestyen MG, Budker V, Ludtke JJ, Fritz JD, Wolff JA. Complexes of non-cationic liposomes and histone H1 mediate efficient transfection of DNA without encapsulation. Biochim. Biophys. Acta. 1996;1284:47–55. doi: 10.1016/0005-2736(96)00106-x. [DOI] [PubMed] [Google Scholar]
- Hama S, Akita H, Ito R, Mizuguchi H, Hayakawa T, Harashima H. Quantitative comparison of intracellular trafficking and nuclear transcription between adenoviral and lipoplex systems. Mol. Ther. 2006;13:786–794. doi: 10.1016/j.ymthe.2005.10.007. [DOI] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hartig R, Shoeman RL, Janetzko A, Grub S, Traub P. Active nuclear import of single-stranded oligonucleotides and their complexes with non-karyophilic macromolecules. Biol. Cell. 1998;90:407–426. [PubMed] [Google Scholar]
- Hartig R, Shoeman RL, Janetzko A, Tolstonog G, Traub P. DNA-mediated transport of the intermediate filament protein vimentin into the nucleus of cultured cells. J. Cell Sci. 1998;111:3573–3584. doi: 10.1242/jcs.111.24.3573. [DOI] [PubMed] [Google Scholar]
- Hartikka J, Sawdey M, Cornefert-Jensen F, Margalith M, Barnhart K, Nolasco M, Vahlsing HL, Meek J, Marquet M, Hobart P, Norman J, Manthorpe M. An improved plasmid DNA expression vector for direct injection into skeletal muscle. Hum. Gene Ther. 1996;7:1205–1217. doi: 10.1089/hum.1996.7.10-1205. [DOI] [PubMed] [Google Scholar]
- Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- Hearps AC, Jans DA. HIV-1 integrase is capable of targeting DNA to the nucleus via an importin alpha/beta dependent mechanism. Biochem. J. 2006;398:475–484. doi: 10.1042/BJ20060466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert E. Improvement of exogenous DNA nuclear importation by nuclear localization signal-bearing vectors: a promising way for nonviral gene therapy? Biol. Cell. 2003;95:59–68. doi: 10.1016/s0248-4900(03)00007-8. [DOI] [PubMed] [Google Scholar]
- James MB, Giorgio TD. Nuclear-associated plasmid, but not cell-associated plasmid, is correlated with transgene expression in cultured mammalian cells. Mol. Ther. 2000;1:339–346. doi: 10.1006/mthe.2000.0054. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE. Seqeuence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 1984;311:33–38. doi: 10.1038/311033a0. [DOI] [PubMed] [Google Scholar]
- Kalderon D, Richardson WD, Markham AF, Smith AE. A short amino acid sequence able to specify nuclear location. Cell. 1984;39:499–509. doi: 10.1016/0092-8674(84)90457-4. [DOI] [PubMed] [Google Scholar]
- Kaneda Y, Iwai K, Uchida T. Increased expression of DNA cointroduced with nuclear protein in adult rat liver. Science. 1989;243:375–378. doi: 10.1126/science.2911748. [DOI] [PubMed] [Google Scholar]
- Kao HP, Abney JR, Verkman AS. Determinants of the translational mobility of a small solute in cell cytoplasm. J. Cell Biol. 1993;120:175–184. doi: 10.1083/jcb.120.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar S, De BP, Gao G, Wilson JM, Crystal RG, Leopold PL. A common mechanism for cytoplasmic dynein-dependent microtubule binding shared among adeno-associated virus and adenovirus serotypes. J. Virol. 2006;80:7781–7785. doi: 10.1128/JVI.00481-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil IA, Kogure K, Akita H, Harashima H. Uptake pathways and subsequent intracellular trafficking in nonviral gene delivery. Pharmacol. Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- Komano J, Miyauchi K, Matsuda Z, Yamamoto N. Inhibiting the Arp2/3 complex limits infection of both intracellular mature vaccinia virus and primate lentiviruses. Mol. Biol. Cell. 2004;15:5197–5207. doi: 10.1091/mbc.E04-04-0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koppelhus U, Nielsen PE. Cellular delivery of peptide nucleic acid (PNA) Adv. Drug Deliv. Rev. 2003;55:267–280. doi: 10.1016/s0169-409x(02)00182-5. [DOI] [PubMed] [Google Scholar]
- Kovacs AM, Zimmer WE. Cell specific transcription of the smooth muscle g-actin gene requires both positive and negative acting cis-elements. Gene Exp. 1998;7:115–129. [PMC free article] [PubMed] [Google Scholar]
- Kristensson K, Lycke E, Roytta M, Svennerholm B, Vahlne A. Neuritic transport of herpes simplex virus in rat sensory neurons in vitro. Effects of substances interacting with microtubular function and axonal flow [nocodazole, taxol and erythro-9-3-(2-hydroxynonyl) adenine] J. Gen. Virol. 1986;67:2023–2028. doi: 10.1099/0022-1317-67-9-2023. [DOI] [PubMed] [Google Scholar]
- Lam MH, Thomas RJ, Loveland KL, Schilders S, Gu M, Martin TJ, Gillespie MT, Jans DA. Nuclear transport of parathyroid hormone (PTH)-related protein is dependent on microtubules. Mol. Endocrinol. 2002;16:390–401. doi: 10.1210/mend.16.2.0775. [DOI] [PubMed] [Google Scholar]
- Langle-Rouault F, Patzel V, Benavente A, Taillez M, Silvestre N, Bompard A, Sczakiel G, Jacobs E, Rittner K. Up to 100-fold increase of apparent gene expression in the presence of Epstein-Barr virus oriP sequences and EBNA1: implications of the nuclear import of plasmids. J. Virol. 1998;72:6181–6185. doi: 10.1128/jvi.72.7.6181-6185.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechardeur D, Sohn K-J, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O'Brodovich H, Lukacs GL. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6:482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- Leopold PL, Kreitzer G, Miyazawa N, Rempel S, Pfister KK, Rodriguez-Boulan E, Crystal RG. Dynein- and microtubule-mediated translocation of adenovirus serotype 5 occurs after endosomal lysis. Hum. Gene Ther. 2000;11:151–165. doi: 10.1089/10430340050016238. [DOI] [PubMed] [Google Scholar]
- Li S, MacLaughlin FC, Fewell JG, Gondo M, Wang J, Nicol F, Dean DA, Smith LC. Muscle-specific enhancement of gene expression by incorporation of the SV40 enhancer in the expression plasmid. Gene Ther. 2001;8:494–497. doi: 10.1038/sj.gt.3301419. [DOI] [PubMed] [Google Scholar]
- Ludtke JJ, Sebestyen MG, Wolff JA. The effect of cell division on the cellular dynamics of microinjected DNA and dextran. Mol. Ther. 2002;5:579–588. doi: 10.1006/mthe.2002.0581. [DOI] [PubMed] [Google Scholar]
- Lukacs GL, Haggie P, Seksek O, Lechardeur D, Freedman N, Verkman AS. Size-dependent DNA mobility in cytoplasm and nucleus. J. Biol. Chem. 2000;275:1625–1629. doi: 10.1074/jbc.275.3.1625. [DOI] [PubMed] [Google Scholar]
- Mabit H, Nakano MY, Prank U, Saam B, Dohner K, Sodeik B, Greber UF. Intact microtubules support adenovirus and herpes simplex virus infections. J. Virol. 2002;76:9962–9971. doi: 10.1128/JVI.76.19.9962-9971.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthorpe M, Cornefert-Jensen F, Hartikka J, Felgner J, Rundell A, Margalith M, Dwarki V. Gene therapy by intramuscular injection of plasmid DNA: studies on firefly luciferase gene expression in mice. Hum. Gene Ther. 1993;4:419–431. doi: 10.1089/hum.1993.4.4-419. [DOI] [PubMed] [Google Scholar]
- Mavlyutov TA, Cai Y, Ferreira PA. Identification of RanBP2- and kinesin-mediated transport pathways with restricted neuronal and subcellular localization. Traffic. 2002;3:630–640. doi: 10.1034/j.1600-0854.2002.30905.x. [DOI] [PubMed] [Google Scholar]
- McDonald D, Vodicka MA, Lucero G, Svitkina TM, Borisy GG, Emerman M, Hope TJ. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 2002;159:441–452. doi: 10.1083/jcb.200203150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina-Kauwe LK. Endocytosis of adenovirus and adenovirus capsid proteins. Adv. Drug Deliv. Rev. 2003;55:1485–1496. doi: 10.1016/j.addr.2003.07.010. [DOI] [PubMed] [Google Scholar]
- Mesika A, Grigoreva I, Zohar M, Reich Z. A regulated, NFkappaB-assisted import of plasmid DNA into mammalian cell nuclei. Mol. Ther. 2001;3:653–657. doi: 10.1006/mthe.2001.0312. [DOI] [PubMed] [Google Scholar]
- Mesika A, Kiss V, Brumfeld V, Ghosh G, Reich Z. Enhanced intracellular mobility and nuclear accumulation of DNA plasmids associated with a karyophilic protein. Hum. Gene Ther. 2005;16:200–208. doi: 10.1089/hum.2005.16.200. [DOI] [PubMed] [Google Scholar]
- Mirzayans R, Remy AA, Malcom PC. Differential expression and stability of foreign genes introduced into human fibroblasts by nuclear versus cytoplasmic microinjection. Mutation Res. 1992;281:115–122. doi: 10.1016/0165-7992(92)90045-j. [DOI] [PubMed] [Google Scholar]
- Nagasaki T, Kawazu T, Tachibana T, Tamagaki S, Shinkai S. Enhanced nuclear import and transfection efficiency of plasmid DNA using streptavidin-fused importin-beta. J. Control Rel. 2005;103:199–207. doi: 10.1016/j.jconrel.2004.11.024. [DOI] [PubMed] [Google Scholar]
- Neves C, Byk G, Scherman D, Wils P. Coupling of a targeting peptide to plasmid DNA by covalent triple helix formation. FEBS Lett. 1999;453:41–45. doi: 10.1016/s0014-5793(99)00674-2. [DOI] [PubMed] [Google Scholar]
- Niederman R, Amrein PC, Hartwig J. Three-dimensional structure of actin filaments and of an actin gel made with actin-binding protein. J. Cell Biol. 1983;96:1400–1413. doi: 10.1083/jcb.96.5.1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil SD, Rhodes DG, Burgess DJ. DNA-based therapeutics and DNA delivery systems: a comprehensive review. AAPS J. 2005;7:E61–77. doi: 10.1208/aapsj070109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Giron ML, Tobaly-Tapiero J, Bittoun P, Real E, Jacob Y, Tordo N, De The H, Saib A. Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J. Cell Sci. 2003;116:3433–3442. doi: 10.1242/jcs.00613. [DOI] [PubMed] [Google Scholar]
- Pollard H, Toumaniantz G, Amos JL, Avet-Loiseau H, Guihard G, Behr JP, Escande D. Ca2+-senssitive cytosolic nucleases prevent efficient delivery to the nucleus of injected plasmids. J. Gene Med. 2001;3:153–164. doi: 10.1002/jgm.160. [DOI] [PubMed] [Google Scholar]
- Prasad TK, Rao NM. The role of plasmid constructs containing the SV40 DNA nuclear-targeting sequence in cationic lipid-mediated DNA delivery. Cell Mol. Biol. Lett. 2005;10:203–215. [PubMed] [Google Scholar]
- Rebuffat AG, Nawrocki AR, Nielsen PE, Bernasconi AG, Bernal-Mendez E, Frey BM, Frey FJ. Gene delivery by a steroid-peptide nucleic acid conjugate. FASEB J. 2002;16:1426–1428. doi: 10.1096/fj.01-0706fje. [DOI] [PubMed] [Google Scholar]
- Reddy JA, Dean D, Kennedy MD, Low PS. Optimization of Folate-Conjugated Liposomal Vectors for Folate Receptor-Mediated Gene Therapy. J. Pharm. Sci. 1999;88:1112–1118. doi: 10.1021/js990169e. [DOI] [PubMed] [Google Scholar]
- Saib A, Puvion-Dutilleul F, Schmid M, Peries J, de The H. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J. Virol. 1997;71:1155–1161. doi: 10.1128/jvi.71.2.1155-1161.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman H, Abu-Arish A, Oliel S, Loyter A, Klafter J, Granek R, Elbaum M. Nuclear localization signal peptides induce molecular delivery along microtubules. Biophys. J. 2005;89:2134–2145. doi: 10.1529/biophysj.105.060160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestyén MG, Ludtke JL, Bassik MC, Zhang G, Budker V, Lukhtanov EA, Hagstrom JE, Wolff JA. DNA vector chemistry: the covalent attachment of signal peptides to plasmid DNA. Nat. Biotech. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- Seth P, Willingham MC, Pastan I. Binding of adenovirus and its external proteins to Triton X-114. Dependence on pH. J. Biol. Chem. 1985;260:14431–14434. [PubMed] [Google Scholar]
- Smith HM, Raikhel NV. Nuclear localization signal receptor importin alpha associates with the cytoskeleton. Plant Cell. 1998;10:1791–1799. doi: 10.1105/tpc.10.11.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JG, Walzem RL, German JB. Liposomes as agents of DNA transfer. Biochim. Biophys. Acta. 1993;1154:327–340. doi: 10.1016/0304-4157(93)90004-8. [DOI] [PubMed] [Google Scholar]
- Sodeik B, Ebersold MW, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J. Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stossel TP. Contribution of actin to the structure of the cytoplasmic matrix. J. Cell Biol. 1984;99:15s–21s. doi: 10.1083/jcb.99.1.15s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Ranganathan P, Diamond SL. Nuclear targeting peptide scaffolds for lipofection of nondividing mammalian cells. Nat. Biotechnol. 1999;17:873–877. doi: 10.1038/12860. [DOI] [PubMed] [Google Scholar]
- Thornburn AM, Alberts AS. Efficient expression of miniprep plasmid DNA after needle micro-injection into somatic cells. Biotechniques. 1993;14:356–358. [PubMed] [Google Scholar]
- Tobiume M, Lineberger JE, Lundquist CA, Miller MD, Aiken C. Nef does not affect the efficiency of human immunodeficiency virus type 1 fusion with target cells. J. Virol. 2003;77:10645–10650. doi: 10.1128/JVI.77.19.10645-10650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotman LC, Mosberger N, Fornerod M, Stidwill RP, Greber UF. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 2001;3:1092–1100. doi: 10.1038/ncb1201-1092. [DOI] [PubMed] [Google Scholar]
- Tseng W, Haselton F, Giorgio T. Transfection by cationic liposomes using simultaneous single cell measurements of plasmid delivery and transgene expression. J. Biol. Chem. 1997;272:25641–25647. doi: 10.1074/jbc.272.41.25641. [DOI] [PubMed] [Google Scholar]
- Tseng WC, Haselton FR, Giorgio TD. Mitosis enhances transgene expression of plasmid delivered by cationic liposomes. Biochim. Biophys. Acta. 1999;1445:53–64. doi: 10.1016/s0167-4781(99)00039-1. [DOI] [PubMed] [Google Scholar]
- Utvik JK, Nja A, Gundersen K. DNA injection into single cells of intact mice. Hum. Gene Ther. 1999;10:291–300. doi: 10.1089/10430349950019075. [DOI] [PubMed] [Google Scholar]
- Vacik J, Dean BS, Zimmer WE, Dean DA. Cell-specific nuclear import of plasmid DNA. Gene Ther. 1999;6:1006–1014. doi: 10.1038/sj.gt.3300924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan EE, Dean DA. Intracellular trafficking of plasmids during transfection is mediated by microtubules. Mol. Ther. 2006;13:422–428. doi: 10.1016/j.ymthe.2005.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaysse L, Gregory LG, Harbottle RP, Perouzel E, Tolmachov O, Coutelle C. Nuclear-targeted minicircle to enhance gene transfer with non-viral vectors in vitro and in vivo. J. Gene Med. 2006;8:754–763. doi: 10.1002/jgm.883. [DOI] [PubMed] [Google Scholar]
- Vaysse L, Harbottle R, Bigger B, Bergau A, Tolmachov O, Coutelle C. Development of a self-assembling nuclear targeting vector system based on the tetracycline repressor protein. J. Biol. Chem. 2004;279:5555–5564. doi: 10.1074/jbc.M311894200. [DOI] [PubMed] [Google Scholar]
- Wildeman AG. Regulation of SV40 early gene expression. Biochem. Cell Biol. 1988;66:567–577. doi: 10.1139/o88-067. [DOI] [PubMed] [Google Scholar]
- Wilson GL, Dean BS, Wang G, Dean DA. Nuclear import of plasmid DNA in digitonin-permeabilized cells requires both cytoplasmic factors and specific DNA sequences. J. Biol. Chem. 1999;274:22025–22032. doi: 10.1074/jbc.274.31.22025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff JA, Dowty ME, Jiao S, Repetto R, Berg RK, Ludtke JJ, Williams P, Slautterback DB. Expression of naked plasmids by cultured myotubes and entry into T tubules and caveolae of mammalian skeletal muscle. J. Cell Sci. 1992;103:1249–1253. doi: 10.1242/jcs.103.4.1249. [DOI] [PubMed] [Google Scholar]
- Wolff JA, Ludtke JJ, Acsadi G, Williams P, Jani A. Long-term persistence of plasmid DNA and foreign gene expression in mouse muscle. Hum. Mol. Genetics. 1992;1:363–369. doi: 10.1093/hmg/1.6.363. [DOI] [PubMed] [Google Scholar]
- Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science. 1990;247:1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- Yanai H, Hayashi Y, Watanabe Y, Ohtaki N, Kobayashi T, Nozaki Y, Ikuta K, Tomonaga K. Development of a novel Borna disease virus reverse genetics system using RNA polymerase II promoter and SV40 nuclear import signal. Microbes Infect. 2006;8:1522–1529. doi: 10.1016/j.micinf.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Young JL, Benoit JN, Dean DA. Effect of a DNA nuclear targeting sequence on gene transfer and expression of plasmids in the intact vasculature. Gene Ther. 2003;10:1465–1470. doi: 10.1038/sj.gt.3302021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JL, Dean DA. Non-Viral Gene Transfer Strategies for the Vasculature. Microcirculation Res. 2002;9:35–50. doi: 10.1038/sj/mn/7800120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabner J, Fasbender AJ, Moninger T, Poellinger KA, Welsh MJ. Cellular and molecular barriers to gene transfer by a cationic lipid. J. Biol. Chem. 1995;270:18997–19007. doi: 10.1074/jbc.270.32.18997. [DOI] [PubMed] [Google Scholar]
- Zanta MA, Belguise-Valladier P, Behr JP. Gene delivery: A single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc. Natl. Acad. Sci. USA. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelphati O, Liang X, Hobart P, Felgner PL. Gene chemistry: functionally and conformationally intact fluorescent plasmid DNA. Hum. Gene Ther. 1999;10:15–24. doi: 10.1089/10430349950019156. [DOI] [PubMed] [Google Scholar]
- Zelphati O, Liang X, Nguyen C, Barlow S, Sheng S, Shao Z, Felgner PL. PNA-dependent gene chemistry: stable coupling of peptides and oligonucleotides to plasmid DNA. Biotechniques. 2000;28:304–316. doi: 10.2144/00282rr01. [DOI] [PubMed] [Google Scholar]
- Zennou V, Petit C, Guetard D, Nerhbass U, Montagnier L, Charneau P. HIV-1 genome nuclear import is mediated by a central DNA flap. Cell. 2000;101:173–185. doi: 10.1016/S0092-8674(00)80828-4. [DOI] [PubMed] [Google Scholar]
- Zhou R, Dean DA. Gene transfer of IL-10 to the murine cornea using electroporation. Exp. Biol. Med. 2006 in press. [PMC free article] [PubMed] [Google Scholar]
- Zupan J, Muth TR, Draper O, Zambryski P. The transfer of DNA from agrobacterium tumefaciens into plants: a feast of fundamental insights. Plant J. 2000;23:11–28. doi: 10.1046/j.1365-313x.2000.00808.x. [DOI] [PubMed] [Google Scholar]