Abstract
Background and Purpose
Radiomics provides opportunities to quantify the tumor phenotype non-invasively by applying a large number of quantitative imaging features. This study evaluates computed-tomography (CT) radiomic features for their capability to predict distant metastasis (DM) for lung adenocarcinoma patients.
Material and Methods
We included two datasets: 98 patients for discovery and 84 for validation. The phenotype of the primary tumor was quantified on pre-treatment CT-scans using 635 radiomic features. Univariate and multivariate analysis was performed to evaluate radiomics performance using the concordance index (CI).
Results
Thirty-five radiomic features were found to be prognostic (CI > 0.60, FDR < 5%) for DM and twelve for survival. It is noteworthy that tumor volume was only moderately prognostic for DM (CI=0.55, p-value=2.77 × 10−5) in the discovery cohort. A radiomic-signature had strong power for predicting DM in the independent validation dataset (CI=0.61, p-value=1.79 ×10−17). Adding this radiomic-signature to a clinical model resulted in a significant improvement of predicting DM in the validation dataset (p-value=1.56 × 10−11).
Conclusions
Although only basic metrics are routinely quantified, this study shows that radiomic features capturing detailed information of the tumor phenotype can be used as a prognostic biomarker for clinically-relevant factors such as DM. Moreover, the radiomic-signature provided additional information to clinical data.
INTRODUCTION
Lung cancer is the most deadly cancer worldwide for both men and women[1]. Nonsmall cell lung cancer (NSCLC) is the most common type of lung cancer (85–90% of all lung cancers) and adenocarcinoma is the most common subtype (about 40% of all lung cancers) of NSCLC. Patients with locally advanced (stage II-III) lung adenocarcinomas are typically treated with combined modality therapy including chemotherapy with local therapy including radiation therapy and/or surgery, but overall survival remains low due to a high risk of local recurrence and distant metastasis (DM) after treatment. Despite the use of concurrent chemotherapy with local therapy, the incidence of DM after combined modality therapy is as high as 30–40% in prospective trials [2–4]. However, large randomized trials studying consolidation chemotherapy after concurrent chemotherapy and radiation therapy have not shown improvement in overall survival with additional chemotherapy[5, 6] likely because there was no selection of patients at the highest risk of DM. Therefore, developing better biomarkers to predict patients at highest risk for DM may help identify sub-groups who benefit from intensification of systemic therapy and is crucial for improving outcomes.
Due to recent technological advances in medical imaging it is possible to capture tumor phenotypic characteristics non-invasively. The most widely used imaging modality is Computed-Tomography (CT), which can quantify tissue density. In lung cancer, CT imaging is routinely used for patient management, including diagnosis, radiation treatment planning and surveillance.
Tumor phenotypic differences (e.g. shapes irregularity, infiltration, heterogeneity or necrosis) can be quantified in CT images using radiomic features. Radiomics [7–9] aims to provide a comprehensive quantification of the tumor phenotype by analyzing robustly [10–12] a large set of quantitative data characterization algorithms . Biomarkers based on quantitative features have demonstrated strong prognostic performance across a range of cancer types and investigators have reported that these features are associated with clinical outcomes and underlying genomic patterns [13–26]. Radiomics has significant clinical potential, as it can be applied to routinely acquired medical imaging data at low costs.
In this manuscript we present a radiomic analysis to identify biomarkers of DM in patients treated with chemoradiation (chemoRT) for locally advanced lung adenocarcinoma. In a discovery dataset, we extracted 635 radiomics features to identify the optimal features for predicting metastasis. Only a limited number of features with high performance for predicting DM were tested in the independent validation dataset. We evaluated the ability of radiomic features to predict DM or overall survival, and how these features compare with basic metrics (e.g. volume, diameter) as prognostic factors [27–30].
MATERIALS AND METHODS
Patient characteristics
This study is an Institutional Review Board-approved analysis of CT for treatment simulation from North-American NSCLC patients receiving chemoRT at our institution from 2001 to 2013. We limited the patient population to pathologically-confirmed lung adenocarcinoma with locally advanced disease (overall stage II-III)[30]. Patients with surgery or chemotherapy before the scheduled radiation therapy planning CT date were excluded from the study. Patients treated before July 2009 were included in the discovery Dataset1 (n=98), and after July 2009 in an independent validation Dataset2 (n=84). In total 182 patients were included in our analysis.
Clinical endpoints
Patients were followed up every three to six months after treatment, and surveillance chest CT scans with contrast (unless patient’s contraindication, e.g. allergy or renal dysfunction) were performed to assess treatment response or tumor progression based on US national guidelines (NCCN). The primary endpoint of this study was distant metastasis (DM), which was defined as progression of disease to other organs as assessed in surveillance scans, and time to DM was defined as time from start of radiation to date of DM or censoring (date of last scan). Overall survival was analyzed as a secondary endpoint, and was defined as the time between the start of radiation treatment and last day of follow up or date of death.
Clinical variables
The conventional clinical prognostic factors (CPFs) used for this study included tumor grade (1-Well differentiated, 2-Moderately differentiated, 3-Poorly differentiated and 4-Not available), Eastern Cooperative Oncology Group (ECOG) performance status (PS)[31], TNM stage per the American Joint Committee on Cancer (AJCC) staging system (7th edition)[30]; CT-based measurements commonly utilized in the clinic (e.g. tumor volume and maximal tumor diameter measured on single axial slice), and treatment characteristics. Sub-group analyses of clinical variables were performed (e.g. overall stage II vs IIIA vs IIIB) and can be found in Table S1 (Supplement II.1).
CT acquisition and segmentation
Planning CT was performed according to standard clinical scanning protocols at our institution with a GE “LightSpeed” CT scanner (GE Medical System, Milwaukee, WI, USA). The most common pixel spacing was (0.93mm, 0.93mm, 2.5mm) for CT. The primary lung tumor was delineated manually on Eclipse (Varian Medical System, Palo Alto, CA, USA). It was first contoured in the abdomen window to identify the boundaries with the chest wall or other soft tissues, then in the lung window to capture the maximum extent in the lung parenchyma. All contours were reviewed by an experienced radiation oncologist (R.H.M).
Radiomic features extraction
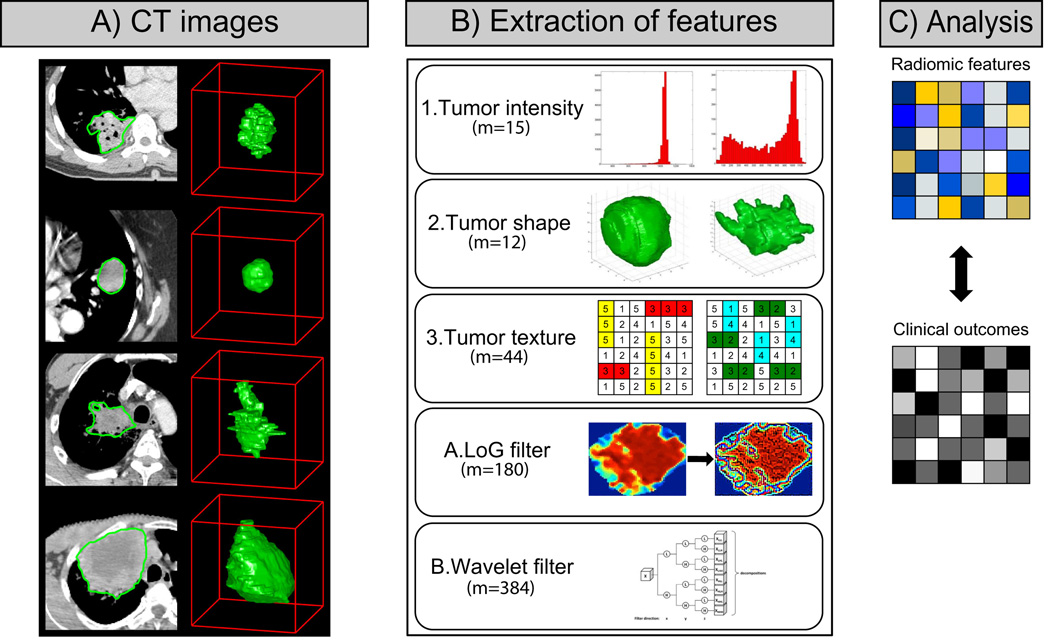
Radiomic features have the capacity to capture tumor phenotype differences by examining a large set of quantitative features (Figure 1). The feature extraction was performed in MATLAB 2013b (Mathworks, Natick, MA, USA) using an in-house developed toolbox running on the Computational Environment for Radiotherapy Research (CERR)[32]. DICOMs files (CT images + tumor contours) were imported into CERR to extract the radiomic features. The radiomic features set included is described in detail in the Supplement I.
Figure 1.
A) Differences between lung primary tumors with a same histology are apparent on CT images (3D model on the right and CT contours on the left). CT images of primary tumors contain critical information that can be used to predict outcomes or assess the RT treatment response. B) To quantify this information, a large set of features (m=635) is used to capture the tumor phenotype. It includes 1| intensity, 2| shape and 3| texture based features. Also, A| Laplacian of Gaussian (LoG) and B| Wavelet filtered features were investigated. C) The final step is to link radiomic information to clinical data.
Feature selection
Feature selection for the radiomic signature was performed with the minimum redundancy maximum relevance (mRMR) algorithm implemented in the mRMRe[33] package version 2.0.4 in R. The mRMR algorithm is an entropy based feature selection method, which starts by calculating the mutual information (MI) between a set of features and an outcome variable. MRMR ranks the input features by maximizing the MI with respect to outcome and minimizing the average MI of higher ranked features. Here, survival objects as implemented in R with “Survcomp” package[34] were used as outcome to select complementary features with respect to DM or survival.
Among available clinical covariates, those with p < 0.1 on univariate analysis of DM using a Log-Rank test were included into a multivariate clinical prognostic model.
Data analysis
Univariate and multivariate analyses were performed for this study. All analysis were performed on Dataset1, leaving Dataset2 as an independent validation cohort for evaluating the radiomic signature.
Statistical analysis was conducted using the survcomp[34] package version 1.12 and rmeta[35] package version 2.16 in Bioconductor[36]. Prognostic performances were evaluated by the concordance index[37] (CI), which is the probability that among two randomly drawn samples, the sample with the higher risk value has also the higher chance of experiencing an event (e.g. death or development of DM). CIs were either directly computed for continuous variables or on the predictions of a univariate Cox model with clinical categorical variables. Kaplan-Meier and Log-Rank statistics were used to analyze the univariate discrimination of survival and DM groups by imaging features and clinical covariates. To build the multivariate radiomic signature for DM, Cox regression models were trained on Dataset1 for selected prognostic variables and the predictions by these models were validated on Dataset2. Features were incrementally added to the model according to the relevance rank calculated by mRMR[33]. Intermediate models were tested by repeated random sub-sampling cross validation with 1,000 iterations on Dataset1. Once the mean CI of the growing model dropped, the corresponding feature set was retained selected as the final model. Only this selected model was and validated on Dataset2. Significance of CIs was assessed by bootstrapping subsamples of size 100 with 100 repetitions for A) true survival data and B) random permutations of survival data, and comparing the empirical distributions of A) and B) by an one-sided Wilcoxon signed rank test. The same procedure was used to assess if a CI was higher than another CI. To correct for multiple comparisons, we additionally adjusted P-values by the false-discovery-rate (FDR) procedure according to Benjamini and Hochberg[38]. All statistical analysis was performed using the R software[39] version 3.0.2.
RESULTS
The majority of all patients were female (62.6%) and the median age at start of treatment was 64 years (range: 35–93 years). The median follow-up time was 23.7 months (range: 1.8–119.2 months) and the median survival time was 24.7 months (range: 1.8–119.2 months). The median time to distant metastasis (DM) was 13.4 months (range: 0.3–117.5 months). Patient characteristics, clinical outcomes are shown in Table 1.
Table 1.
Patient characteristics and outcomes are reported for each datasets. For categorical variables, actual numbers are reported for each category (format A/B/C). Statistical comparison between dataset 1 and 2 was computed using Chi Square (categorical variables) or Wilcoxon rank sum test (continuous variables).
| Overall dataset (n=182) |
Dataset 1 (n=98) | Dataset 2 (n=84) |
P- value |
|
|---|---|---|---|---|
| Median (range) | Median (range) | Median (range) | ||
| Age [years] | 64 (35–93) | 62 (41–86) | 65 (35–93) | 0.63 |
| Gender [F/M] | 114(62.6%)/ 68(37.4%) |
66(67.3%)/ 32(32.7%) |
48(57.1%)/ 36(42.9%) |
0.29 |
| Overall stage [IIA/IIB/IIIA/IIIB] | 6/3/101/72 | 2/1/55/40 | 4/2/46/32 | 0.65 |
| T-stage [T1a/T1b/T2a/T2b/T3/T4] | 19/23/50/19/39/32 | 14/10/30/10/17/17 | 5/13/20/9/22/15 | 0.26 |
| N-stage [N0/N1/N2/N3] | 13/17/97/55 | 5/9/53/31 | 8/8/44/24 | 0.70 |
| Performance status [0/1/2/3] | 81/91/8/2 | 36/57/5/0 | 45/34/3/2 | 0.04 |
| Tumor grade [1/2/3/X] | 4/28/92/58 | 3/11/47/37 | 1/17/45/21 | 0.12 |
| Follow-up [months] | 23.7 (1.8–119.2) | 28.9 (1.8–119.2) | 19.5 (3.1–54.9) | 0.007 |
| Survival [months] | 24.7 (1.8–119.2) | 29.7 (1.8–119.2) | 21.4 (3.4–54.9) | 0.005 |
| Time to distant metastasis [months] | 13.4 (0.3–117.5) | 13.6 (0.3–117.5) | 13.3 (0.7–49.6) | 0.36 |
| Distant metastasis [No/Yes] | 69(37.9%)/ 113(62.1%) |
34(34.7%)/ 64(65.3%) |
35 (41.7%)/ 49(58.3%) |
0.45 |
|
Radiation dose delivered ≤ 54/≤ 60/≤ 66/> 66 [Gray] |
60(32.97%)/ 30(16.48%)/ 70(38.45%)/ 22(12.1%) |
28(28.57%)/ 17(17.35%)/ 33(33.67%)/ 20(20.41%) |
32(38.10%)/ 13(15.48%)/ 37(44.04%)/ 2(2.38%) |
0.002 |
|
Chemotherapy sequence [concurrent/adjuvant/induction] |
175/79/28 | 95/38/22 | 80/41/6 | 0.024 |
Time to DM was similar between Dataset1 and Dataset2 (p-value < 0.36), as for the numbers of DM (p-value < 0.45). However, survival (p-value < 0.005) and follow-up times (p-value < 0.007) were significantly different in Dataset1.
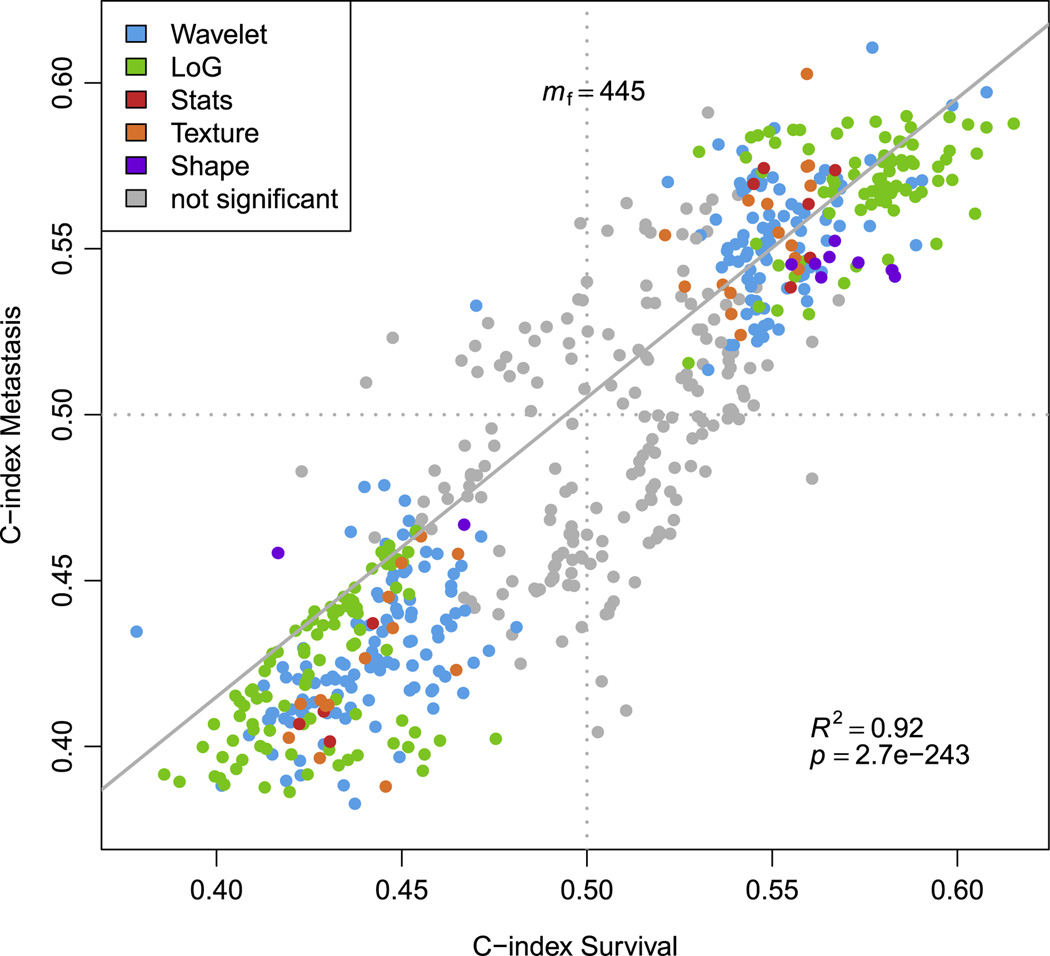
We investigated the association of radiomics data with DM and overall survival. In Figure 2 the association of the imaging features with DM and survival in the discovery Dataset1 is shown. Of the complete radiomic features set (m=635), a total of 520 (81.88%) and 582 (91.65%) features were significant from random (FDR < 5%) for DM and survival, respectively. A total of 445 radiomic features were significant for both DM and survival. A high linear relationship was observed (R2 =0.92, p-value < 2.7 × 10−243), for the features significant for both DM and survival. It is noteworthy that LoG features had the highest performance compared to the other features groups.
Figure 2. Univariate performances of prognostic features for Distant Metastasis (DM) and survival.
Each point refers to the CI of a feature evaluating the power of feature to predict metastasis, respectively, survival. Colors refer to the type of feature. Features whose CI estimation was not significant (FDR < 5%) for both DM and survival are shown in gray. Overall, 445 of these pairs of CIs are considered to be significant estimates. Linear regression for all significant pairs of CIs yielded an R-squared value of 0.92 (F-test, p-value < 2.7e-243).
Among all features, thirty-five radiomics features were strongly prognostic (CI > 0.60 and FDR < 5%) for DM (Table S2 in the Supplement II.3). Twelve features were found prognostic for survival. Specific details on statistic values of these features can be found in Table S3 in Supplement II.3. Between these two top performing feature sets there were four common prognostic features for both DM and survival. All of them were LoG based features (3 entropy and 1 standard deviation).
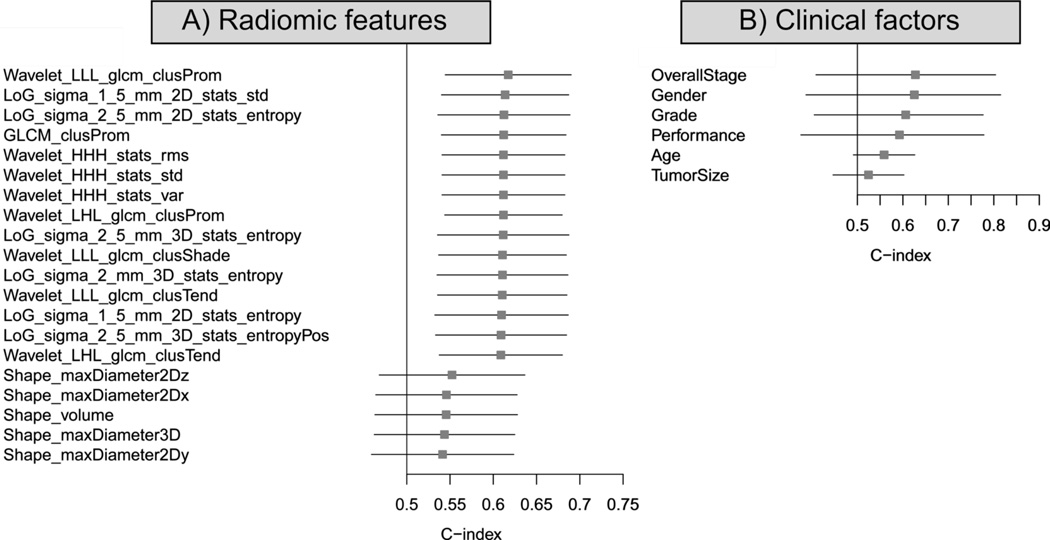
We compared the top 15 features that had the highest CIs (Top15), with tumor volume and diameter (equivalent to basic metrics). The Top15 radiomic features had notably higher CIs compared to tumor volume and diameter (Figure 3.A).
Figure 3.
A) Forest plot of the 15 best performing radiomic features for Distant Metastasis on univariate analysis (Dataset1, n=98). Radiomics equivalent of basic metrics (diameter and volume) was added for comparison. B) Forest plot of the clinical factors. The absolute C-indices and their 95% confidence interval are shown.
We also investigated the association of CPFs with DM in our data set. Three clinical parameters appeared to be significant univariate prognostic factors: Overall Stage (CI=0.63, p-value < 6.78 × 10−14), Gender (CI=0.63, p-value < 2.35 × 10−11) and tumor grade (CI=0.61, p-value < 2.35 × 10−11). Clinical parameters, ranked by their CI are displayed in Figure 3.B. Overall stage and gender yielded a higher CI than the radiomic features, although their 95% confidence interval is wider compared to the radiomic features.
An mRMR based feature selection on all features on Dataset1 (n=98) was performed to reduce redundancy and select a potential set of complementary and prognostic features. From this new ranking, the 15 highest mRMR-ranked features were kept after feature selection to build the radiomic signature. A multivariate Cox regression model to predict DM was developed. Features were iteratively added in order of high to low mRMR rank on Dataset1, and Dataset2 was used for independent validation. The combination that yielded the maximum CI on the discovery Dataset1 before dropping was defined as the optimal radiomic signature for predicting DM. This signature consists of three features: 1) Wavelet HHL – Skewness, 2) Gray-Level Co-occurrence Matrix – Cluster shade, and 3) LoG 5mm 2D – Skewness. Cluster shade is a textural feature sensitive to tumor heterogeneities. Skewness is a first-order feature that measures the asymmetry of the histogram from the mean, which here is associated with two different filters LoG and Wavelet.
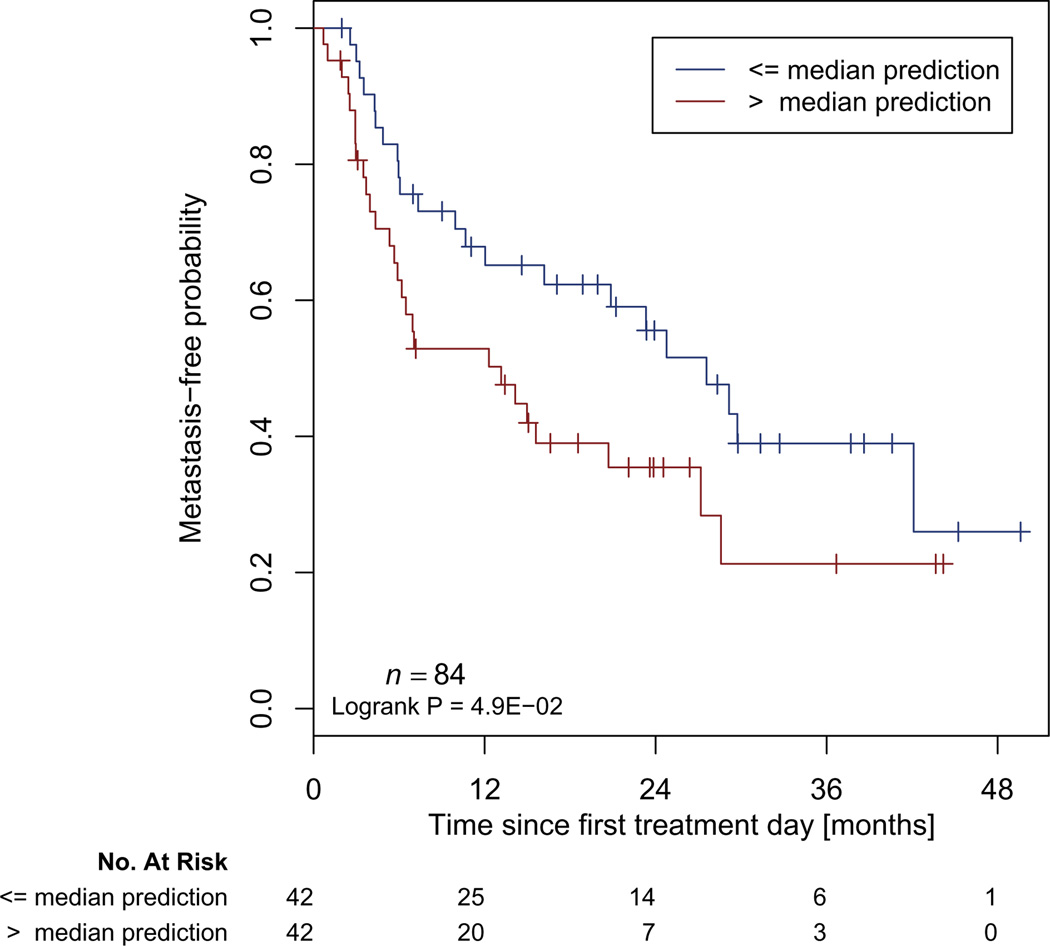
As a final step, we compared the radiomic signature to a clinical Cox regression model containing covariates that significantly discriminated between patients with and without DM in Dataset1 in univariate analysis. The final model contained overall stage and tumor grade. This clinical model showed moderate prognostic power when applied to Dataset2 with coefficients trained on Dataset1 (CI=0.57, p-value < 1.03 × 10−7). Combining the clinical and radiomic signature (trained on Dataset1) showed a significantly (p-value < 1.56 × 10−11) higher association with DM when applied to Dataset2 (CI=0.60, p-value < 3.57 × 10−16), compared to the clinical model. A median split of the patient prediction scores from applying the combined model on Dataset2 yielded a significant difference (p-value = 0.049) for metastasis-free probability estimates (Figure 4).
Figure 4. Kaplan Meier curves according to the combined model predicting score to predict metastasis-free probability in an independent dataset.
A significant survival difference appears between patients with a high or low risk of Distant Metastasis (Dataset2, n=84, Log-Rank test, p-value < 0.049).
DISCUSSION
Medical imaging gives valuable information for diagnostic, treatment planning or surveillance of cancer patients. Routinely, basic metrics are extracted from these images to utilize as a prognostic factor [27–30], or to assess treatment response. However, there is much more tumor phenotypic information captured in these images. Radiomics are able to quantify tumor phenotypical differences from medical images by using a large set of imaging features that can be linked to clinical factors of the tumors. In this study we extracted 635 radiomic features from a total of 182 lung cancer patients treated with chemoRT to assess the ability of radiomic features as a prognostic biomarker for distant metastasis (DM), and we validated a radiomic-based signature on an independent validation dataset. Since DM remains a major cause of mortality in 30–40% of patients with locally advanced lung adenocarcinoma, early identification of patients at highest risk of developing DM would allow clinicians to adapt treatment such as incorporating consolidation chemotherapy to improve outcomes. Moreover, the theoretical benefit of consolidation chemotherapy has not been shown in large randomized studies to date. It is likely because there was no selection of patients at the highest risk of distant metastases (i.e. patients who were at low risk of distant metastases were included in these trials and would not need additional treatment). Future trial design to demonstrate benefits of consolidation chemotherapy will likely require stratification to identify those at the highest risk of distant metastases and may benefit most from additional treatment.
We observed strong individual correlations between clinical outcomes and quantitative imaging features. A large number of features were significant from random to predict DM (91%) and survival (82%) in univariate analysis after correction for multiple testing. Moreover, a high linear correlation was found among those 445 features that were significant factors of both DM and survival (R2 =0.92, p-value < 2.7 × 10−243). This high linear correlation is expected as there is a high correlation between DM and survival (DM greatly impact patient survival, see Table S4 in Supplement II.4). Only a small number of features, 35 for DM and for 12 survival, were prognostic, as defined by a CI > 0.6 and FDR < 5%.
Although we tested a large number of features, to minimize any risk of over-fitting or bias, we performed a robust validation approach: all analysis steps, mRMR feature selection, and model fitting were performed on Dataset1 (n=98) and the results validated on an independent validation Dataset2 (n=84). With this approach we found a multivariate radiomic DM signature consisting of three features that yielded a high prognostic performance for DM in Dataset1 (CI=0.61). Combining the radiomics signature to clinical model predictors showed significant improvement (p-value < 1.56 × 10−11), compared to the clinical predictors alone.
A recent study from Fried et al.[22] investigated DM prediction for NSCLC patients. They found a significant model DM (P-value=0.005) using both texture features and CPFs. The model used consisted of eight parameters (two CPFs and six textures). In another study, Ganeshan et al.[15] applied textural analysis to find univariate prognostic factors for survival. They focused on two imaging features (uniformity, associated with two LoG filter). In our analysis, these features were significant from random but lowly ranked by their CI value (184th and 146th CI-ranked features in Dataset1). However, major differences in studies design and implementation made it difficult to compare them objectively. Fields et al.[22] used leave-one out cross validation to validate their model instead of an independent validation dataset. Ganeshan et al.[15] only used one CT image slice (presenting the largest cross section) to calculate their features when we used the whole primary tumor. Finally, both these studies have a smaller patient cohort, n=54[15] and n=91[22], and had mixed histology patients. Our analysis calculated the features from the complete 3D tumor volume, contained only a single histology of NSCLC (adenocarcinoma), and is based on larger cohorts (n=182) with an independent validation dataset for the radiomic signature.
A complementary point of the study was to compare basic metrics [27–29] to radiomic features as prognostic factors for DM. The first observation made was that Shape-Maximum diameter (in every direction x/y/z) is a better univariate prognostic factors than the maximal tumor diameter on an axial slice reported by a radiologist. The advantage of the radiomic shape features is that they can be automatically acquired, reproducible[10–12], and take into account the whole tumor volume, whereas clinically assessed tumor diameters are manually drawn on a CT slice and are therefore limited to one dimension of the tumor. Furthermore, shape or size-based features were not in the top ranked features in our study. Total tumor volume, has been associated with survival in stage I-III NSCLC patients treated with radiation therapy in a study from Etiz et al.[28], and a prior study from our institution by Alexander et al.[29] also demonstrated an association between primary tumor volume and overal survival, but not risk of distant metastasis. In our study, volume was ranked only the 405th (CI=0.55) and 224th (CI=0.56) best univariate prognostic factor for DM and survival respectively in Dataset1. Thus, while basic metrics such as size and volume have historically been used as used in the clinical setting because such data are easily acquired, radiomic shape and size measurements can provide stronger prognostic factors.
A short-coming of our study is the variability in CT acquisition and reconstruction parameters. Our dataset includes patients from 2001 to 2013. During this time period, the standard of care for CT acquisition has evolved, differences appeared between our cohorts for some factors (Table 1). However, despite this variability in the imaging data (evolution of hardware, progress in informatics), radiomics was able to detect a strong signal to predict DM despite a temporal split. Additionally, clinical outcomes are provided by one center, which makes it hard to evaluate the generalizability of outcomes to other institutions. However, in comparison with a recent study[20] investigating clinical outcomes from another center, patient characteristics or outcomes were comparable. Future work would therefore involve studying the DM signature in other histologies and in independent validation sets from other institutions, assessing its generalizability to all NSCLC.
In conclusion, this study demonstrated strong association between radiomic features and DM for patients with locally advanced adenocarcinoma; and presented an independently validated radiomics signature for DM. This signature would allow early identification of patients with locally advanced lung adenocarcinoma at risk of developing DM, allowing clinicians to individualize treatment (such as intensification of chemotherapy) to reduce the risk of DM and improve survival.
Supplementary Material
Highlights.
Early prediction of patients that will develop distant metastasis (DM) is crucial for improving overall treatment and patient outcomes.
This study demonstrated an association between radiomic features and DM for lung cancer patients.
A combined signature with clinical and radiomic features was able to predict DM in an independent validation dataset.
Acknowledgments
Source of support: Authors acknowledge financial support from the National Institute of Health (NIH-USA U01CA190234). Authors acknowledge financial support from the QuIC-ConCePT project, which is partly funded by EFPI A companies and the Innovative Medicine Initiative Joint Undertaking (IMI JU) under Grant Agreement No. 115151. This research is also supported by the Dutch technology Foundation STW (grant n° 10696 DuCAT), which is the applied science division of NWO, and the Technology Programme of the Ministry of Economic Affairs. Authors also acknowledge financial support from EU 7th framework program (EURECA, ARTFORCE), NGI Pre-Seed grant (n° 93612005), Kankeronderzoekfonds Limburg from the Health Foundation Limburg and the Dutch Cancer Society (KWF UM 2011-5020, KWF UM 2009-4454,KWF MAC 2013-6089).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclaimer: none
Conflict of interest: none
Bibliography
- 1.Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014: Cancer Statistics: 2014. CA Cancer J Clin. 2014;64:9–29. doi: 10.3322/caac.21208. [DOI] [PubMed] [Google Scholar]
- 2.Albain KS, Swann RS, Rusch VW, Turrisi AT, Shepherd FA, Smith C, Chen Y, Livingston RB, Feins RH, Gandara DR, Fry WA, Darling G, Johnson DH, Green MR, Miller RC, Ley J, Sause WT, Cox JD. Radiotherapy plus chemotherapy with or without surgical resection for stage III non-small-cell lung cancer: a phase III randomised controlled trial. Lancet. 2009;374:379–386. doi: 10.1016/S0140-6736(09)60737-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Curran WJ, Paulus R, Langer CJ, Komaki R, Lee JS, Hauser S, Movsas B, Wasserman T, Rosenthal SA, Gore E, Machtay M, Sause W, Cox JD. Sequential vs. concurrent chemoradiation for stage III non-small cell lung cancer: randomized phase III trial RTOG 9410. J Natl Cancer Inst. 2011;103:1452–1460. doi: 10.1093/jnci/djr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradley JD, Paulus R, Komaki R, Masters GA, Forster K, Schild SE, Bogart J, Garces YI, Narayan S, Kavadi V, Nedzi LA, Michalski JM, Johnson D, MacRae RM, Curran WJ, Choy H Radiation Therapy Oncology Group. A randomized phase III comparison of standard-dose (60 Gy) versus high-dose (74 Gy) conformal chemoradiotherapy with or without cetuximab for stage III non-small cell lung cancer: Results on radiation dose in RTOG 0617. ASCO Meet Abstr. 2013;31:7501. [Google Scholar]
- 5.Hanna N, Neubauer M, Yiannoutsos C, McGarry R, Arseneau J, Ansari R, Reynolds C, Govindan R, Melnyk A, Fisher W, Richards D, Bruetman D, Anderson T, Chowhan N, Nattam S, Mantravadi P, Johnson C, Breen T, White A, Einhorn L. Phase III Study of Cisplatin, Etoposide, and Concurrent Chest Radiation With or Without Consolidation Docetaxel in Patients With Inoperable Stage III Non–Small-Cell Lung Cancer: The Hoosier Oncology Group and U.S. Oncology. J Clin Oncol. 2008;26:5755–5760. doi: 10.1200/JCO.2008.17.7840. [DOI] [PubMed] [Google Scholar]
- 6.Kelly K, Chansky K, Gaspar LE, Albain KS, Jett J, Ung YC, Lau DHM, Crowley JJ, Gandara DR. Phase III trial of maintenance gefitinib or placebo after concurrent chemoradiotherapy and docetaxel consolidation in inoperable stage III non-small-cell lung cancer: SWOG S0023. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:2450–2456. doi: 10.1200/JCO.2007.14.4824. [DOI] [PubMed] [Google Scholar]
- 7.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RGPM, Granton P, Zegers CML, Gillies R, Boellard R, Dekker A, Aerts HJWL. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48:441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kumar V, Gu Y, Basu S, Berglund A, Eschrich SA, Schabath MB, Forster K, Aerts HJWL, Dekker A, Fenstermacher D, Goldgof DB, Hall LO, Lambin P, Balagurunathan Y, Gatenby RA, Gillies RJ. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lambin P, van Stiphout RGPM, Starmans MHW, Rios-Velazquez E, Nalbantov G, Aerts HJWL, Roelofs E, van Elmpt W, Boutros PC, Granone P, Valentini V, Begg AC, De Ruysscher D, Dekker A. Predicting outcomes in radiation oncology--multifactorial decision support systems. Nat Rev Clin Oncol. 2013;10:27–40. doi: 10.1038/nrclinonc.2012.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rios Velazquez E, Aerts HJWL, Gu Y, Goldgof DB, De Ruysscher D, Dekker A, Korn R, Gillies RJ, Lambin P. A semiautomatic CT-based ensemble segmentation of lung tumors: Comparison with oncologists’ delineations and with the surgical specimen. Radiother Oncol. 2012;105:167–173. doi: 10.1016/j.radonc.2012.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parmar C, Rios Velazquez E, Leijenaar R, Jermoumi M, Carvalho S, Mak RH, Mitra S, Shankar BU, Kikinis R, Haibe-Kains B, Lambin P, Aerts HJWL. Robust Radiomics Feature Quantification Using Semiautomatic Volumetric Segmentation. PLoS ONE. 2014;9:e102107. doi: 10.1371/journal.pone.0102107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leijenaar RTH, Carvalho S, Velazquez ER, van Elmpt WJC, Parmar C, Hoekstra OS, Hoekstra CJ, Boellaard R, Dekker ALAJ, Gillies RJ, Aerts HJWL, Lambin P. Stability of FDGPET Radiomics features: An integrated analysis of test-retest and inter-observer variability. Acta Oncol. 2013;52:1391–1397. doi: 10.3109/0284186X.2013.812798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganeshan B. Non_Small Cell Lung Cancer: Histopathologic Correlates for Texture Parameters at CT. Radiology. doi: 10.1148/radiol.12112428. [DOI] [PubMed] [Google Scholar]
- 14.Davnall F, Yip CSP, Ljungqvist G, Selmi M, Ng F, Sanghera B, Ganeshan B, Miles KA, Cook GJ, Goh V. Assessment of tumor heterogeneity: an emerging imaging tool for clinical practice? Insights Imaging. 2012;3:573–589. doi: 10.1007/s13244-012-0196-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ganeshan B, Panayiotou E, Burnand K, Dizdarevic S, Miles K. Tumour heterogeneity in non-small cell lung carcinoma assessed by CT texture analysis: a potential marker of survival. Eur Radiol. 2012;22:796–802. doi: 10.1007/s00330-011-2319-8. [DOI] [PubMed] [Google Scholar]
- 16.Ganeshan B, Abaleke S, Young RCD, Chatwin CR, Miles KA. Texture analysis of nonsmall cell lung cancer on unenhanced computed tomography: initial evidence for a relationship with tumour glucose metabolism and stage. Cancer Imaging. 2010;10:137–143. doi: 10.1102/1470-7330.2010.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He X, Sahiner B, Gallas BD, Chen W, Petrick N. Computerized characterization of lung nodule subtlety using thoracic CT images. Phys Med Biol. 2014;59:897–910. doi: 10.1088/0031-9155/59/4/897. [DOI] [PubMed] [Google Scholar]
- 18.Skogen K, Ganeshan B, Good C, Critchley G, Miles K. Measurements of heterogeneity in gliomas on computed tomography relationship to tumour grade. J Neurooncol. 2013;111:213–219. doi: 10.1007/s11060-012-1010-5. [DOI] [PubMed] [Google Scholar]
- 19.Ravanelli M, Farina D, Morassi M, Roca E, Cavalleri G, Tassi G, Maroldi R. Texture analysis of advanced non-small cell lung cancer (NSCLC) on contrast-enhanced computed tomography: prediction of the response to the first-line chemotherapy. Eur Radiol. 2013;23:3450–3455. doi: 10.1007/s00330-013-2965-0. [DOI] [PubMed] [Google Scholar]
- 20.Aerts HJWL, Velazquez ER, Leijenaar RTH, Parmar C, Grossmann P, Cavalho S, Bussink J, Monshouwer R, Haibe-Kains B, Rietveld D, Hoebers F, Rietbergen MM, Leemans CR, Dekker A, Quackenbush J, Gillies RJ, Lambin P. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5 doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chae H-D, Park CM, Park SJ, Lee SM, Kim KG, Goo JM. Computerized Texture Analysis of Persistent Part-Solid Ground-Glass Nodules: Differentiation of Preinvasive Lesions from Invasive Pulmonary Adenocarcinomas. Radiology. 2014:132187. doi: 10.1148/radiol.14132187. [DOI] [PubMed] [Google Scholar]
- 22.Fried DV, Tucker SL, Zhou S, Liao Z, Mawlawi O, Ibbott G, Court LE. Prognostic Value and Reproducibility of Pretreatment CT Texture Features in Stage III Non-Small Cell Lung Cancer. Int J Radiat Oncol. 2014 doi: 10.1016/j.ijrobp.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaidya M, Creach KM, Frye J, Dehdashti F, Bradley JD, El Naqa I. Combined PET/CT image characteristics for radiotherapy tumor response in lung cancer. Radiother Oncol. 2012;102:239–245. doi: 10.1016/j.radonc.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 24.Aerts HJWL, Bussink J, Oyen WJG, van Elmpt W, Folgering AM, Emans D, Velders M, Lambin P, De Ruysscher D. Identification of residual metabolic-active areas within NSCLC tumours using a pre-radiotherapy FDG-PET-CT scan: A prospective validation. Lung Cancer. 2012;75:73–76. doi: 10.1016/j.lungcan.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 25.Van Elmpt W, Das M, Hüllner M, Sharifi H, Zegers CML, Reymen B, Lambin P, Wildberger JE, Troost EGC, Veit-Haibach P, De Ruysscher D. Characterization of tumor heterogeneity using dynamic contrast enhanced CT and FDG-PET in non-small cell lung cancer. Radiother Oncol. 2013;109:65–70. doi: 10.1016/j.radonc.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balagurunathan Y, Gu Y, Wang H, Kumar V, Grove O, Hawkins S, Kim J, Goldgof DB, Hall LO, Gatenby RA, Gillies RJ. Reproducibility and Prognosis of Quantitative Features Extracted from CT Images. Transl Oncol. 2014;7:72–87. doi: 10.1593/tlo.13844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ball DL, Fisher RJ, Burmeister BH, Poulsen MG, Graham PH, Penniment MG, Vinod SK, Krawitz HE, Joseph DJ, Wheeler GC, McClure BE. The complex relationship between lung tumor volume and survival in patients with non-small cell lung cancer treated by definitive radiotherapy: A prospective, observational prognostic factor study of the Trans-Tasman Radiation Oncology Group (TROG 99.05) Radiother Oncol. 2013;106:305–311. doi: 10.1016/j.radonc.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Etiz D, Marks LB, Zhou S-M, Bentel GC, Clough R, Hernando ML, Lind PA. Influence of tumor volume on survival in patients irradiated for non—small-cell lung cancer. Int J Radiat Oncol Biol Phys. 2002;53:835–846. doi: 10.1016/s0360-3016(02)02814-6. [DOI] [PubMed] [Google Scholar]
- 29.Alexander BM, Othus M, Caglar HB, Allen AM. Tumor Volume Is a Prognostic Factor in Non–Small-Cell Lung Cancer Treated With Chemoradiotherapy. Int J Radiat Oncol • Biol • Phys. 2011;79:1381–1387. doi: 10.1016/j.ijrobp.2009.12.060. [DOI] [PubMed] [Google Scholar]
- 30.Mirsadraee S. The 7th lung cancer TNM classification and staging system: Review of the changes and implications. World J Radiol. 2012;4:0. doi: 10.4329/wjr.v4.i4.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oken MM, Creech RH, Tormey DC, Horton J, Davis TE, McFadden ET, Carbone PP. Toxicity and response criteria of the Eastern Cooperative Oncology Group. Am J Clin Oncol. 1982;5:649–655. [PubMed] [Google Scholar]
- 32.Deasy JO, Blanco AI, Clark VH. CERR: a computational environment for radiotherapy research. Med Phys. 2003;30:979–985. doi: 10.1118/1.1568978. [DOI] [PubMed] [Google Scholar]
- 33.De Jay N, Papillon-Cavanagh S, Olsen C, El-Hachem N, Bontempi G, Haibe-Kains B. mRMRe: an R package for parallelized mRMR ensemble feature selection. Bioinformatics. 2013;29:2365–2368. doi: 10.1093/bioinformatics/btt383. [DOI] [PubMed] [Google Scholar]
- 34.Schröder MS, Culhane AC, Quackenbush J, Haibe-Kains B. survcomp: an R/Bioconductor package for performance assessment and comparison of survival models. Bioinformatics. 2011;27:3206–3208. doi: 10.1093/bioinformatics/btr511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lumley T. Rmeta. 2012 [Google Scholar]
- 36.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harrell FE, Califf RM, Pryor DB, Lee KL, Rosati RA. Evaluating the yield of medical tests. JAMA J Am Med Assoc. 1982;247:2543–2546. [PubMed] [Google Scholar]
- 38.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 39.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. ISBN 3-900051-07-0. 2013 URL http://www.R-Project.org/.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.