Abstract
The dopamine transporter (DAT) is a transmembrane protein belonging to the family of Neurotransmitter:Sodium Symporters (NSS). Members of the NSS are responsible for the clearance of neurotransmitters from the synaptic cleft, and for their translocation back into the presynaptic nerve terminal. The DAT contains long intracellular N- and C-terminal domains that are strongly implicated in the transporter function. The N-terminus (N-term), in particular, regulates the reverse transport (efflux) of the substrate through DAT. Currently, the molecular mechanisms of the efflux remain elusive in large part due to lack of structural information on the N-terminal segment. Here we report a computational model of the N-term of the human DAT (hDAT), obtained through an ab initio structure prediction, in combination with extensive atomistic molecular dynamics (MD) simulations in the context of a lipid membrane. Our analysis reveals that whereas the N-term is a highly dynamic domain, it contains secondary structure elements that remain stable in the long MD trajectories of interactions with the bilayer (totaling >2.2 µs). Combining MD simulations with continuum mean-field modeling we found that the N-term engages with lipid membranes through electrostatic interactions with the charged lipids PIP2 (phosphatidylinositol 4,5-Biphosphate) or PS (phosphatidylserine) that are present in these bilayers. We identify specific motifs along the N-term implicated in such interactions and show that differential modes of N-term/membrane association result in differential positioning of the structured segments on the membrane surface. These results will inform future structure-based studies that will elucidate the mechanistic role of the N-term in DAT function.
Keywords: Neurotransmitter transporter, sodium symporter, signaling, efflux, amphetamine, phosphorylation, molecular dynamics, electrostatics
Introduction
The dopamine transporter (DAT) is a membrane protein in the family of mammalian Neurotransmitter:Sodium Symporters (NSS) that also includes the closely related transporters for serotonin (SERT) and norepinephrine (NET)1–3. The NSS are responsible for the clearance of neurotransmitters (e.g. dopamine (DA), by DAT) from the synaptic cleft, and their translocation back into the cytoplasm of the presynaptic neuronal cell. The uptake of neurotransmitter is made possible by coupling the transmembrane (TM) Na+ gradient to the uphill transport of the respective substrate2. The functions of NSS proteins in neuronal signaling implicate them in a number of psychiatric and neurological disorders that include schizophrenia, and Parkinson’s disease4, and in the mechanisms of action of abused psychostimulants, such as cocaine and amphetamine (AMPH)5,6. Their essential neurophysiological roles have made these transporters primary targets for antidepressant medications.
The first X-ray structure of the NSS DAT from Drosophila (dDAT) was reported recently 7, and revealed 12 TM segments, a domain architecture that has been predicted as well from previous molecular modeling of human DAT (hDAT) based on sequence homology to the bacterial Leucine transporter (LeuT) 8–14 for which several crystal structures have existed since 200515–22. But in contrast to LeuT, DAT has much longer cytoplasmic N- and C-terminal segments. These intracellular segments possess numerous putative phosphorylation sites, and several protein kinases have been implicated in the regulation of DAT function 23–26.
The phosphorylation of the N-terminal segment at serine residues positioned in its distal portions (i.e., close to its starting residue, Met1, see Figure 1A), leads to the intriguing phenotype of efflux in which the substrate, DA, is transported via DAT in the reverse direction, out of the cell 27–31. Under physiological conditions, efflux can be triggered by the action of the psychostimulant amphetamine (AMPH), which apparently leads to a DAT conformation suitable for the phosphorylation of the N-terminus. Indeed, in studies of phosphomimetic S-to-D mutations substituting the distal serine residues of the N-terminus (N-term), DAT-mediated DA efflux was observed even in the absence of AMPH 27.
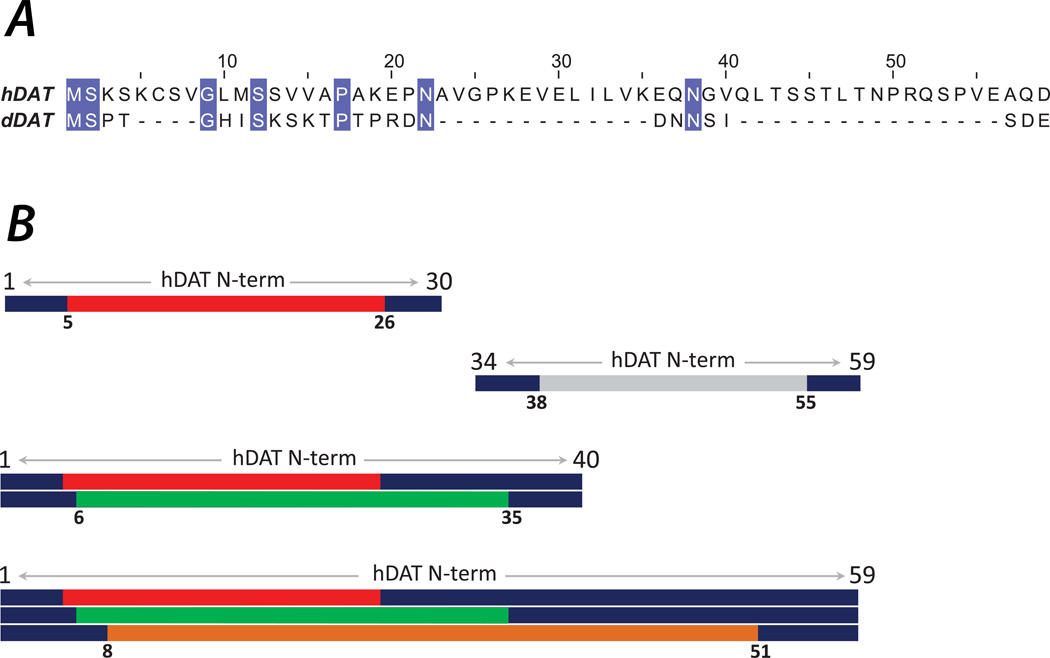
Figure 1.
(A) Sequence alignment of human (top row) and Drosophila (bottom row) DAT N-terminal segments illustrating low sequence conservation between the two species. The conserved residues are colored. The alignment has been performed with the BLAST tool74. (B) hDAT N-terminus constructs used in ab initio fold prediction. N-terminal 30, 40 and 59 residue segments, as well as the 34–59 stretch were considered. The predicted structures were clustered using various criteria for residue exclusion: For the 1–30 construct, only the 5–26 (red bar) segment was used for clustering; for the 34–59 construct, only 38–55 (silver bar); for the 1–40 construct, the 5–26 and 6–35 (green bar) segments were utilized; and for the 1–59 fragment, the clustering was performed based on the 5–26, 6–35, and 8–51 (orange bar) segments.
Interestingly, efflux can be regulated separately from the substrate uptake process. Thus, we have shown that for hDAT, charge-neutralizing substitution of K3 and K5 residues in the N-term with either Ala or Asn dramatically reduces AMPH-induced DA efflux while leaving the DA uptake unchanged 32. The same studies have suggested a central mechanistic role for direct binding of the N-term to highly charged anionic PIP2 (phosphatidylinositol 4,5-Biphosphate) lipids in the efflux process 32. The importance of the efflux process is underscored by results from recent studies showing that it is affected by specific de novo mutations in DAT linked to various neurological disorders 33–38.
Despite rapid progress in identifying key elements of the molecular machinery that regulates the efflux process in NSS proteins (e.g., involvement of specific components of the cell membrane, interactions with scaffolding proteins) 39–42, the molecular mechanism of DAT-mediated reverse transport remains unclear. This includes mechanistic questions about the role of the N-terminal segment (residues 1–59 in hDAT), for which structural information is lacking because this functionally important region in DAT is absent in the prototypical NSS, the bacterial transporter LeuT, and also had to be excised from the construct used to obtain the only available X-ray structure of the DAT protein from Drosophila (dDAT) 7. Moreover, the sequence of the DAT N-terminus is not homologous to any protein with known fold, and exhibits important variations among DAT proteins from different species (Figure 1A).
To overcome this difficulty, we sought a prediction of the three-dimensional (3D) conformation of the N-terminus from the human DAT (hDAT) by combining, as described here, ab initio structure prediction tools and extensive atomistic molecular dynamics (MD) simulations. The ab initio modeling, carried out with the Rosetta software 43, yielded predictions of structured regions within the first 57 residues of the N-term. Subsequent long (>2.2 µs in total) atomistic MD simulations of the N-term in complex with a lipid membrane showed that the identified secondary structure elements were stable on the simulation timescales, and were involved in specific interactions with pertinent models of the cell membrane. Notably, we found that the spatial orientation and position relative to the membrane of these structured elements is largely determined by specific modes of N-term/membrane association. Analysis of the MD trajectories and evaluation of membrane responses to interactions with the N-term, using quantitative self-consistent mean-field modeling 44–47, showed that the N-term engages in strong electrostatic interactions with the lipid bilayer through contacts between several basic residues and charged PIP2 or PS (phosphatidylserine) lipids. Notably, when the predicted N-terminus model was attached to the full TM bundle of the crystallographically solved dDAT structure (substituting for the incomplete dDAT terminus), the MD simulations (~600 ns in total) revealed similar modes of interactions with the lipid membrane. These constructs, and the molecular insights regarding the preferred modes of interaction of the DAT N-term with model membranes obtained from the study, provide a novel structural context for future explorations of the functional mechanisms of this NSS-family transporter.
Methods
Ab initio modeling of the hDAT N-terminus with Rosetta
The Rosetta (version 3.0) ab initio structure prediction tool 43 was used to obtain a 3D model of the 1–59 N-terminal region of hDAT (see Figure 1A). For comparison, fragments composed of residues 1–30, 1–40, 1–59, and 34–59 were also modeled separately (Figure 1B). Using Rosetta, 1000 different conformations were generated for each of these sequences. The predicted structures were then subjected to a root-mean-square clustering implemented in Rosetta, with a cluster radius set at 4Å and excluding the terminal residues that are highly dynamic (see Figure 1B). The structures in the top clusters were further evaluated for common secondary structure element conservation, using the RMSDTT algorithm developed in our lab and implemented in VMD 48,49. This algorithm performs pair-wise iterative fitting on a cluster of conformations in order to identify regions with common secondary structure elements within the cluster. With each iteration, the residues are weighted according to their residence in the motifs that are conserved in the iteration (structurally the most rigid ones), and at the end of the fitting routine each residue is assigned a score representing the result of residue-based root-mean-square deviation, RMSD. In this manner, common structural elements within each cluster are identified as the motifs containing residues with the lowest RMSD scores.
Application of RMSDTT to both the full-length N-term and its fragments enabled us to assess whether the preferred fold for the complete N-term (1–59) predicted from the Rosetta clustering and the scoring function, harbored the same secondary structure elements that were found in the different size N-term fragments (see Results for more details). The final conformation of the 1–59 N-term obtained from this rigorous computational framework, was used as the starting conformation of the N-term in all subsequent atomistic MD simulations described below.
We note that because a direct structural validation of the predicted fold for the N-term is currently not feasible, we carried out a qualitative validation test taking advantage of the structure reported for the relatively larger segment of the C-terminus of dDAT which is included in the latest X-ray structure 7. Thus, we used the exact same protocol and algorithms of the modeling approach described above for the hDAT N-term to predict the conformation of the C-terminus of dDAT (Figure S1 in the Supporting Material). The procedure yielded a structure with an RMSD <2Å to the backbone atoms of residues 586–596 (the C-terminal stretch resolved in the X-ray structure, see Figure S1), offering strong support for the appropriateness of the approach used to predict the structure of the hDAT N-term discussed in this work.
Atomistic MD simulation of the hDAT N-terminus
All MD simulations were carried out with the NAMD 2.8 package 50 and the CHARMM36 force field for proteins 51 and lipids 52. The simulations implemented rigidbonds all option, PME for electrostatics interactions 53, and were carried out in NPT ensemble under semi-isotropic (in the presence of a lipid membrane) or isotropic (for simulations in a water box) pressure coupling conditions, at 310 K temperature. The Nose-Hoover Langevin piston 50 algorithm was used to control the target P = 1 atm pressure with the LangevinPistonPeriod set to 100 fs and LangevinPistonDecay set to 50 fs. The van der Waals interactions were calculated applying a cutoff distance of 12 Å and switching the potential from 10 Å.
MD simulations of the N-term in a water box
Atomistic MD simulations of the 59-residue long hDAT N-term were carried out in a TIP3P water box of 82Å×82Å×82Å dimension. The peptide was acetylated on its N-terminal and methylamidated on the C-terminal. After initial minimization (for 5000 steps), a 670 ns long MD trajectory was accumulated with 2fs time-step.
Replica Exchange MD (REMD) simulations of the N-term and plectasin peptide
Conformational dynamics of the N-terminal segment and the structurally related plectasin peptide (see Results) were explored in water environment using REMD simulations. To minimize the number of solvent molecules in the simulation box, we employed a setup in which a solute molecule (the 1–59 residue N-term fragment or the 1–40 residue-long plectasin peptide, PDBID: 3E7U54) was placed in the center of a spherical water droplet (resulting in a simulation box containing overall ~22,000 atoms). The REMD simulations were then run using spherical boundary conditions, with sphericalBCk1 set to 0.002, sphericalBCexp1=2, sphericalBCk2=1.0, and sphericalBCexp2=2.
The REMD simulations were initiated on 84 replicas, exponentially spaced within the temperature range TL=290K and TH=500K with intervals between 2 and 3K. Each replica was run for total of 5 × 106 steps (10ns) resulting in overall REMD time of 840ns. The exchange trials between the replicas were attempted every 1000 steps. This setup resulted in an average exchange probability of 0.3, with the average and maximal temperature ranges spanned by replicas, being 115.25K and 210K, respectively (see Figure S2A in the Supporting Material).
To keep the solute in the water phase at all times during the REMD and to maintain proper water density across the simulation box at all temperatures 55, a combination of several restraining potentials was used: 1) A soft harmonic potential (with k=0.002 kcal/mole/Å2 force constant) restrained waters that were at a distance greater than (R-5)Å from the water droplet center, R being the radius of the water sphere; 2) A hard harmonic potential (with k=1 kcal/mole/Å2) was applied to waters at distances (R+5)Å or greater from the water droplet center to avoid water evaporation; 3) The center of mass of the solute was restrained to the center of the water drop using k=1 kcal/mole/Å2 as a force constant; 4) The solute molecules were restrained to a sphere of radius (R-5)Å by applying a harmonic force with k=0.25 kcal/mole/Å2 force constant to those solute atoms that were at a distance greater than (R-5)Å. The latter restraining potential was necessary only in rare cases and only at the highest temperatures considered (the restraint not active > 95% of the time at 500K), showing that the water droplet was large enough to accommodate the peptide as it underwent conformational changes during REMD. Importantly, we verified that MD simulations performed with the above set of restrains resulted in water densities that were in good agreement with those established from experiments 55 for the entire [290–500]K temperature range (see Figure S2B).
MD simulations of the N-term in complex with a lipid membrane
The stability of the N-term conformation, and especially of the secondary structure elements predicted with the Rosetta protocol (see above) was examined with extensive MD simulations in the environment of a lipid membrane. The N-term was anchored to different lipid bilayers by a palmitoyl (PALM) extension in the following way: the 59-residue model of the N-term was connected to a coiled 60–65 segment of hDAT (60-RETWGK-65 sequence, homologous to 5-REHWAT-10 stretch in LeuT that is available in several X-ray structures), and a single PALM tail was connected at position Lys65, where the transmembrane helix 1 in hDAT connects to the N-terminus.
The palmitoylated N-term peptide, acetylated on the N-terminal, was inserted into membranes of asymmetric lipid composition designed to mimic the lipid content of the neuronal plasma membrane56. The full specifications of the simulated systems are given in Tables 1 and Table 2, and Figure S3A shows the initial placement of the palmitoylated N-term model (from System 1 in Table 1) in the membranes.
TABLE 1.
Specification of the atomistic MD simulations (system size, total simulation time, number of waters per lipid) of the hDAT N-terminus palmitoylated at Lys65. System3 and 4 are replicas of System1 and 2, respectively, initiated with different random seeds. The lipid membranes were modeled with or without PIP2 lipid (see Table 2).
| name | PIP2 | ions | waters/lipid | system size |
time, ns |
|---|---|---|---|---|---|
| System1 | no | 15 K+ | 127 | 128,822 | 590 |
| System2 | yes | 90 K+ | 127 | 129,032 | 557 |
| System3 | no | 15 K+ | 127 | 128,822 | 520 |
| System4 | yes | 90 K+ | 127 | 129,032 | 556 |
CHARMM-GUI membrane builder57 was used to create bilayer models of desired lipid compositions. The membrane preparation protocol in the software takes advantage of experimentally determined values for the area per molecule for different lipids and makes use of the extensive library of lipid conformations to generate a lipid bilayer in a stress-free condition. Since at the time of these simulations, inositol lipids were not part of the CHARMM-GUI lipid library, the PIP2-containing mixtures (for Systems 2, 4 in Table 1) were constructed by first building a 451-lipid size bilayer that contained a 100:40:32:27:29 mixture of POPE/POPC/DAPC/POPS/Cholesterol in one layer (intracellular leaflet) and 176:29:18 mixture of POPC/DPPC/Cholesterol in another layer (extracellular leaflet, see Table 2 for lipid name abbreviations). Then, all DAPC (1,2-diarachidonoyl-sn-glycero-3-phosphocholine) lipids were replaced by PIP2 and the resulting membrane was equilibrated with MD for a short time (15ns). This pre-equilibrated bilayer patch was then trimmed to its final size (see Table 2) before the palmitoylated N-term constructs were added and the systems were solvated and electro-neutralized with K+ ions (see Table 1). For PIP2-depleted systems (Systems 1 and 3 in Table 1), PIP2 lipids from the trimmed bilayers (Table 2) were replaced by POPE lipids before inserting the palmitoylated N-terminus.
The N-term/membrane complexes were equilibrated using a previously established multi-step protocol (see, for example, Refs. 58,59): 1) minimization for 5,000–10,000 steps and running MD with 1fs integration time-step for 250ps by fixing all atoms in the system except for the lipid tails and the PALM chain; 2) minimization for 2,500 steps and performing MD with 1fs time-step for 500ps by constraining protein backbone and lipid headgroups with force constant of 1 kcal/ (mol Å2) and keeping water out of the membrane hydrophobic core; 3) gradual release of the constraints on the protein backbone and lipid headgroup atoms (force constant of 0.5 and 0.1 kcal/(mol Å2)) while still keeping water out of the membrane interior. At each value of the force constant, the system was minimized for 2,500 steps followed by 500ps MD (with 1fs time-step). After this initial phase, all the constraints were removed, the systems were minimized for 1,200 steps and subjected to long MD (Table 1), using 2fs time-step.
Simulations of the hDAT N-term attached to the transmembrane bundle of dDAT
The construct was obtained by integrating the predicted structure of the hDAT N-terminus (residues 1–59) with the transmembrane segment of dDAT 7 (PDB ID:4M48, see Figure S3B). In this X-ray model of dDAT, which includes the ligand Nortriptyline (NOR), two Na+ ions, and a single Cl− ion in the respective binding sites, the N-terminal region is resolved only from residue D25, which aligns with Q58 in hDAT (Figure 1A). To position the hDAT N-term onto the dDAT bundle, we took advantage of the conserved RETW sequence (60-RETW-63 in hDAT and 27-RETW-30 in dDAT) as the best transition point for an alignment using VMD 60. Therefore, in the resulting model of the chimera (hDAT/dDAT) construct, the hDAT N-term extends up to D59, and the dDAT TM domain starts at R27, which was then re-numbered to R60 (the numbering of the remainder of the dDAT sequence was adjusted accordingly). In addition to providing a reasonable anchoring for the N-term, the inclusion of the dDAT TM bundle makes the model consonant with the hDAT/dDAT chimera construct studied experimentally in our ongoing collaborative structure/function studies. Furthermore, we note that in such collaborative studies, the in vivo behavior of flies 32 was demonstrated to involve N-term/PIP2 interactions in the DA efflux through the endogenous (dDAT) transporter, just as observed with hDAT, suggesting that the mechanistic effects of the interaction of the N-term with PIP2 lipids are recapitulated by dDAT.
The extracellular loop 4, which is truncated in the X-ray model of dDAT, was left unaltered. Disulfide bonds were introduced between Cys180 and Cys189 (in hDAT numbering). The prediction of protonation states of ionizable residues in dDAT with PROPKA 3.0 61, suggested a protonated state for Glu490. The CHARMM36 compatible force field parameters for the ligand NOR were derived with the MATCH software 62.
The hDAT/dDAT chimera was inserted into the compositionally asymmetric pre-equilibrated membrane patch containing 451-lipids including PIP2 (see above), and the overlapping lipids were removed. The protein/membrane system was then solvated and ionized for electro-neutrality within VMD. The total atom count of the simulated box was ~150,000. During equilibration of the system the backbone of the protein was harmonically constrained, with gradually reduced force constants. Water was initially prevented from entering the lipid-water interface. The constraints were released gradually in three steps of 300 ps each, changing the force constants from 1, to 0.5, and 0.1 kcal/ (mol Å2), respectively. After this equilibration phase two independent trajectories of ~340 ns and ~245 ns time-length were run with 2 fs integration step.
Continuum mean-field model of hDAT N-terminus/membrane interactions
The extent of lipid segregation near the hDAT N-terminus was quantified with the application of the continuum self-consistent mean-field model (SCMFM) described previously 44–47. With SCMFM we evaluated the steady state distributions of charged lipid species (PIP2, in this case) under the influence of electrostatic forces from a membrane-adsorbing macromolecule (here, the hDAT N-terminus), and quantified the corresponding adsorption energies. Briefly (see Refs. 45,47 for details), the SCMFM is a mesoscale approach based on the non-linear Poisson-Boltzmann (NLPB) theory of electrostatics63 and Cahn-Hilliard dynamics64, in which the protein is considered in 3-dimensional full atomistic detail, and the lipid membrane is considered as a 2-dimensional, tensionless, incompressible low-dielectric slab in which the equilibrium distribution of different lipid species around the adsorbing protein is obtained from self-consistent minimization of the free energy. The governing free energy function contains contributions from electrostatic interactions, lipid mixing entropy, and mixing entropy of mobile salt ions in the solution. In the SCMFM calculations, the hDAT N-terminus (residues 1–59) was considered in full atomistic detail (with partial charge and atomic radii taken from the all-atom CHARMM36 force field for proteins), and was positioned 2Å away from the lipid surface with average surface charge density of −0.0031e (corresponding to a lipid mixture with ~5% PIP2). Maintaining a separation of 2Å between the protein and the membrane allows for a layer of water between the interacting charged surfaces and is required for numerical accuracy of the NLPB equation that is solved using the APBS package 65 as described before 44–47. The conditions were set by neutralizing the system with 0.1 M ionic solution of monovalent counterions (corresponding to λ=9.65Å Debye length), and a dielectric constant of 2 for membrane interior and protein, and 80 for the solution. All the analyses reported below were performed with combination of various routines of VMD60, OpenStructure 66,67, and in-house built analyses tools as mentioned.
Results
Ab initio structure modeling reveals folded motifs in the hDAT N-terminus
The N-terminal segment of hDAT (residues 1–59) is a functional unit whose 3D structure has not been determined. It is also a protein fragment with little sequence similarity to any known fold. To evaluate the existence of secondary structure motifs in the N-terminal, and the possible role of such structural elements in the mechanisms of transporter function, it is necessary to obtain a model of the N-term 3D structure and assess its dynamic rearrangements under conditions relevant to the function of the protein. To obtain the prediction we used the Rosetta ab initio tool following the extensive protocol and controls described in Methods. Various fragments of the N-term (shown in Figure 1B) were subjected to the Rosetta fold-prediction algorithm, and for each construct the predicted structures were clustered under various residue exclusion conditions (Figure 1B). Clusters containing a majority of the structures were identified (top 2–3 clusters), and the representative models for each cluster were chosen according to the best Rosetta scores. In the top clusters, the conformations of the best scoring models were evaluated with the RMSDTT fitting routine (see Methods) to identify regions with the highest structural similarity within each cluster. The results from these analyses are given in Figure 2–Figure 3.
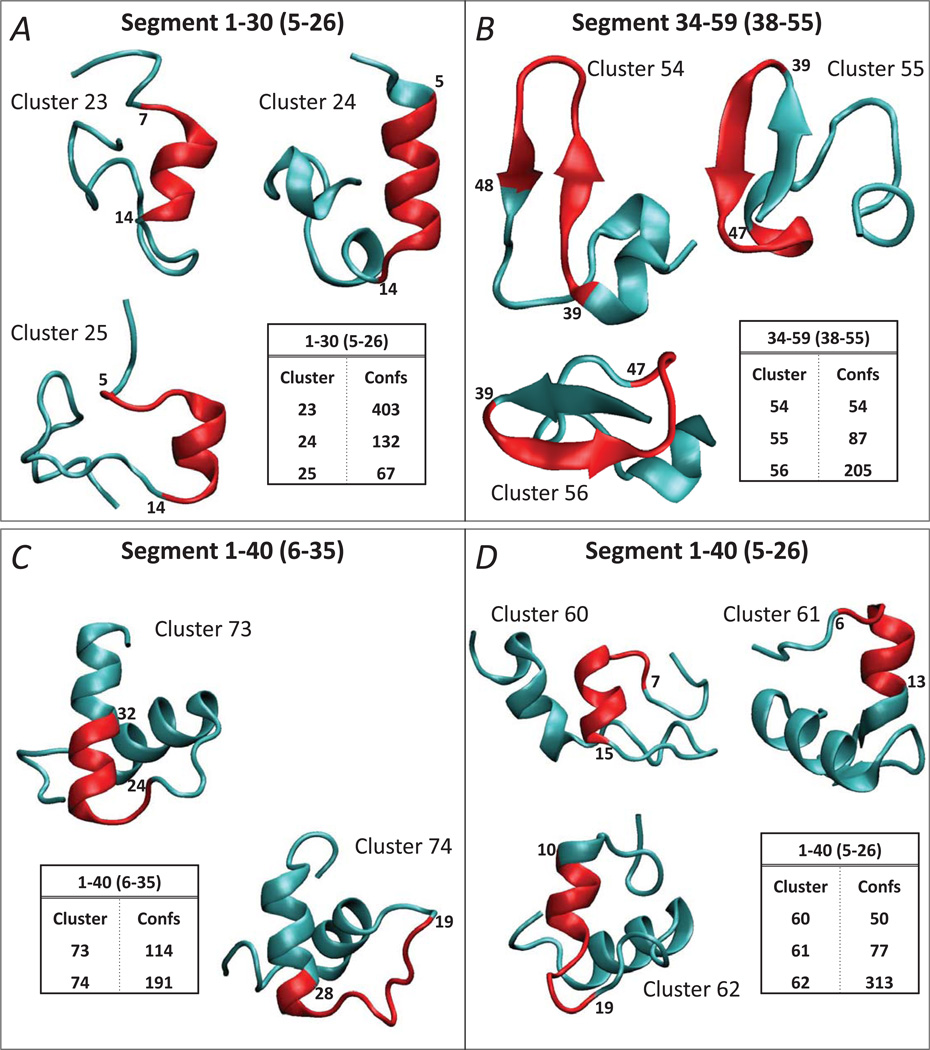
Figure 2.
Ab initio structure prediction for hDAT N-term segments 1–30 (A), 34–59 (B), and for hDAT N-term 1–40 segment using two different fragments for clustering, 6–35 (C) and 5–26 (D) (see also Figure 1B). The inserts identify the clusters with the largest number of structures for each construct; the structures with the best Rosetta energy scores in each cluster are shown in cartoon. The segments considered for clustering are given in brackets (see also Figure 1B). The structural elements with the highest degree of conservation within each cluster as identified by the RMSDTT analysis, are colored in red, and the residues flanking these conserved segments are labeled.
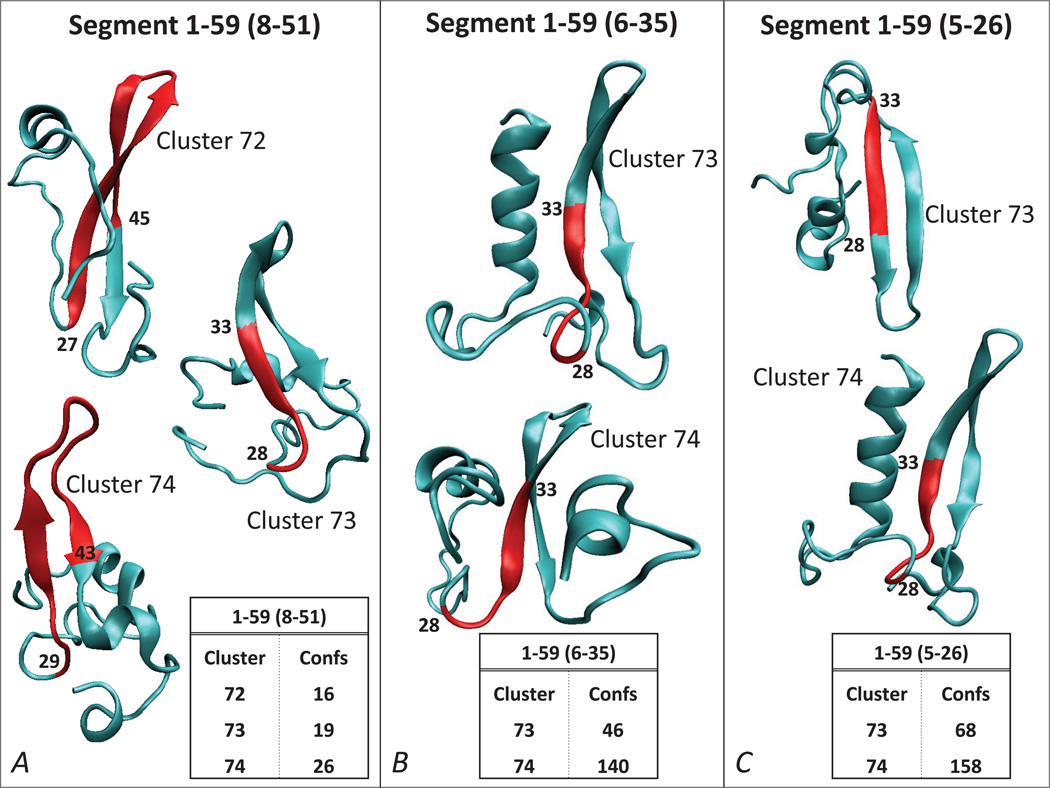
Figure 3.
Ab initio structure prediction for hDAT N-term 1–59 segment using three different segments for clustering (same representations as in Figure 2), for 8–51 (A), 6–35 (B) and 5–26 (C). The inserts identify the clusters with the largest number of structures for each construct, and the structures with the best Rosetta energy scores in each cluster are shown in cartoon. The segments considered for clustering are given in brackets (see also Figure 1B). The structural elements with the highest degree of conservation within each cluster as identified by the RMSDTT analysis, are colored in red, and the residues flanking these conserved segments are labeled.
Figure 2A shows the predicted structures with the best Rosetta energy scores in the top three clusters found for the 1–30 residue segment of the hDAT N-term. Using residues 5–26 for clustering, the calculations predict an α-helical secondary structure in the distal N-terminal part (spanning residues 8–12, 3–12, 8–13 in the top three clusters, respectively). The RMSDTT iterations show that this helical segment is a highly conserved structural element in all top clusters (see red colored cartoon in Figure 2A).
The application of the same set of approaches for the 34–59 fragment of the N-term (with residues 38–55 used for the Rosetta clustering) identified the presence of an extended anti parallel beta sheet motif (involving residue stretches 35–42, 35–43, 41–48 in the top three clusters, respectively, see Figure 2B). The RMSDTT analysis revealed that the beta sheet structure within 34–59 fragment belonged to the region with the highest structural similarity in all three top clusters.
To verify the validity of the structural elements identified from the analysis of the 1–30 and 34–59 segments of the N-term and establish their spatial relationship, we repeated the analysis for larger stretches of the N-term, spanning residues 1–40 and 1–59. Panels C and D on Figure 2 show the results from the ab initio structure prediction and clustering analysis for the 1–40 segment. To enable comparisons of the predicted structures for the 1–40 segment to those for the 1–30 segment described above, the clustering for the 1–40 stretch was done by implementing two different residue exclusion criteria: 1) excluding the terminal 5 residues (thus using the stretch 6–35 for clustering, Figure 2C); and 2) clustering on the same 5–26 residue segment as was done for the 1–30 segment (compare Figures 2D and 2A).
Clustering on the 5–26 segment for the 1–40 fragment (Figure 2D) again revealed the presence of the conserved α-helical motif in the distal N-terminal (red cartoon in Figure 2D), consistent with the predictions obtained for the 1–30 stretch. Furthermore, the results from the RMSDTT analysis on the top cluster (Cluster 62 in Figure 2D) suggested that the helix is followed at its C-terminal end by an unstructured region (extending to residue 19). This coil structure appears as well in the clustering based on the 6–35 segment (Figure 2C) followed by the RMSDTT routine. Thus, the most conserved region of the top cluster (Cluster 74, Figure 2C) includes a coil segment of residues 19 to 24. Together, the predictions from 1–30 and 1–40 segments suggest the presence of an α-helical motif in the distal N-terminal part followed by an unstructured coil region.
Because the analysis of the 1–40 segment did not reveal any extended structure in the N-term, contrary to the results we obtained for the 34–59 fragment (Figure 2B), we reasoned that the C-terminal part of the N-term may be required to stabilize the extended beta sheet motif. This hypothesis is supported by the results for the 1–59 segment (Figure 3). This fragment was investigated by clustering the predicted structures under three different conditions: 1) by excluding the terminal 7 residues from the clustering (Figure 3A); 2) clustering on the 6–35 segment (Figure 3B), which is the same fragment used for the clustering of the 1–40 stretch (in Figure 2C); and 3) clustering on the 5–26 segment (Figure 3C), which is the same fragment used for the clustering of the 1–30 and 1–40 segments (Figures 2A and 2D, respectively).
As shown in Figure 3, regardless of the clustering condition used, an antiparallel beta sheet was predicted in the 1–59 segment, positioned in the same region of the N-term where the extended structure was identified in the 34–59 fragment (Figure 2B). Furthermore, clustering on 5–26 and 6–35 revealed the presence of an α-helical structure in the distal part of the N-term, consistent with the predictions from 1–30 and 1–40 fragments (Figure 2). Collectively, the ab initio analysis on different parts of the N-term presented above supports the presence of the same structured motifs within the first 59 residues of the N-term, and an interdependence as identified in the largest segment, 1–59. Specifically, the most distal N-terminal part is predicted to contain an α-helical secondary structure (residues ~2–12), and the more distal C-terminal part of the N-term (residues ~30–45) includes the antiparallel beta sheet motif. These structured regions are connected by a stretch of coil, and the beta sheet is followed by another coil that connects to the TM bundle of the hDAT.
The predicted structure of the N-term resembles that of the peptide antibiotic plectasin, but is not well preserved in an aqueous environment
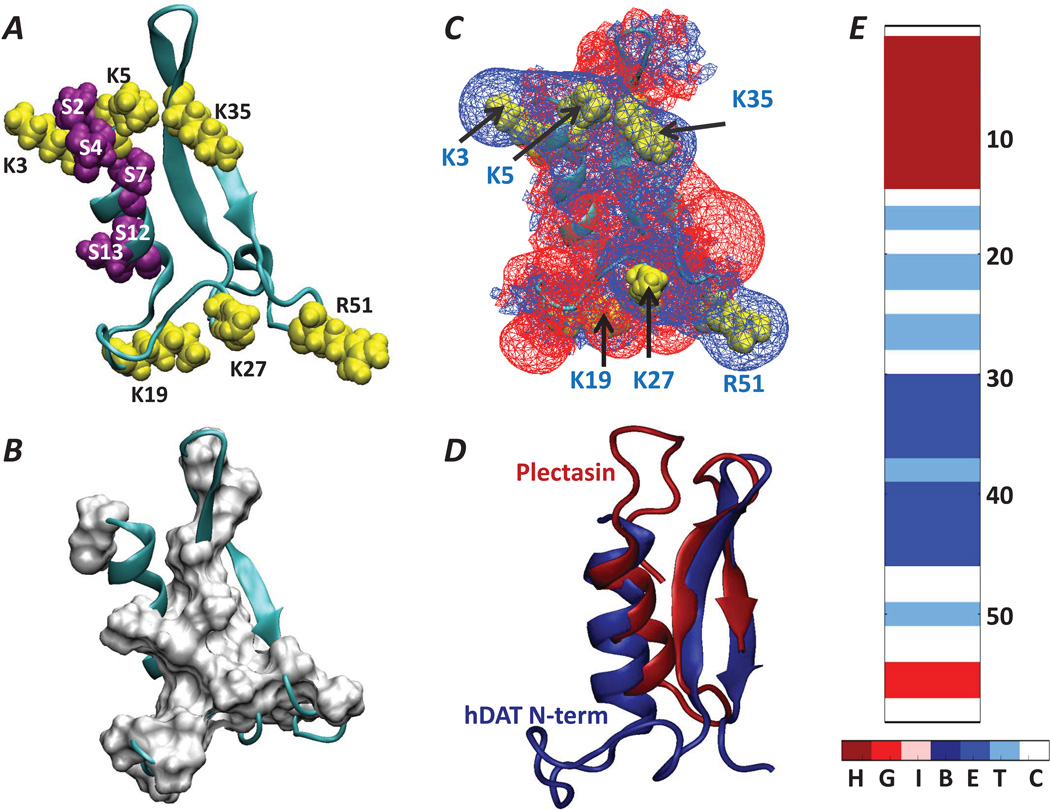
The best scoring structure from the top cluster of 1–59 (Cluster 74 in Figure 3C, clustered on 5–26 region) in the ab initio modeling described above (Figure 4A), was chosen to serve in subsequent analyses. Figure 4E shows the locations of the different secondary structure elements in this predicted structure of the N-term. Interestingly, this conformation positions the multiple basic R/K residues in a belt-like arrangement (Figure 4A) that generates an extended isosurface of positive electrostatic potential (Figure 4C). The overall fold of the N-term is largely stabilized by hydrophobic interactions between the α-helix and the beta sheet (Figure 4B). The surprising find of a 40-amino acid long cysteine-rich peptide antibiotic, plectasin, with a structure known from solution NMR and X-ray crystallography studies 54,68 that is very similar to the predicted fold of the N-term, suggests the importance of these interactions in the N-term. Thus, the remarkably similar fold (Figure 4D) of the two peptides (the backbone atom-based RMSD between the structurally aligned segments of the two peptides is 2.9Å) in spite of the lack of any sequence similarity between them, is stabilized in plectasin by several disulfide bonds not present in the hDAT N-term.
Figure 4.
(A) 3D fold of the hDAT N-terminal segment (in cartoon, residues 1–59) according to Rosetta ab initio structure prediction. The positions of the basic residues and serine residues are shown in yellow and purple space fill representation. (B) Surface representation of the hydrophobic residues (in white) overlaid on the cartoon of the N-term. (C) Electrostatic potential isosurfaces (EPIs) (+1kt/e shown as blue wireframes and −1kt/e as red wireframes) derived from the predicted structure in panel A. Locations of basic residues are highlighted in yellow space fill representations and are identified with corresponding labels and black arrows. Note the presence of extended EPI stemming from a belt-like arrangement of K/R residues. The electrostatic potential was calculated with the APBS software65. (D) Structural alignment (performed with FAST tool75) of the 59-residue long hDAT N-term (blue) and the 40-residue long plectasin, PDB ID: 1ZFU (red). The backbone atom-based RMSD between the structurally aligned segments of the two peptides (that includes the N-term segments 4–14, 34–37, 39–42) is 2.9Å. (E) Secondary structure elements in the hDAT N-terminus. Secondary structure elements are designated as: Turn (T), Extended conformation (E); Isolated bridge (B); α-helix (H); 3–10 helix (G); π-helix (I); Coil (C). The positions of selected residues along the sequence are labeled.
Given the apparent importance of the hydrophobic interactions for the 3D fold of the N-term predicted from Rosetta, its stability was first assessed in ~670ns long unbiased atomistic MD simulations in water. In this trajectory the peptide underwent significant conformational changes in the ~190–270 ns time interval, illustrated by large RMSD values in Figure S4A (in the Supporting Material). Closer inspection of the time sequence of the secondary structure elements revealed gradual loss of the beta sheet motif (blue shades in Figure S4B). Concomitantly, as shown in the time-evolution of pair-wise Cα distance difference matrices in Figure S4C, we observe the disruption of hydrophobic contacts between the α-helical and the beta sheet elements that stabilized the initial fold of the peptide (the average distance between the beta-sheet and α-helix in Figure S4C increases). Overall, the conformation of the N-term changes significantly, as indicated by the stabilization of the RMSD measure at a higher value (~11Å) within the last ~250ns of the trajectory (Figure S4A).
Instabilities in the fold and secondary structure elements in the aqueous environment were further confirmed with replica exchange MD (REMD) simulations performed on the hDAT N-term in water (see Methods). These computations revealed some loss of the α-helix and the beta sheet (see Figure S5 in the Supporting Material). Interestingly, in the similar REMD simulations performed on the structurally related plectasin (see above and Figure 4D) we observed relatively minor structural perturbations, even in the absence of the disulfide bridges that stabilize the plectasin fold (Figure S5).
When interacting with the membrane, the N-term is dynamic, maintains stable structural motifs, and is oriented by the PIP2 content of the bilayer
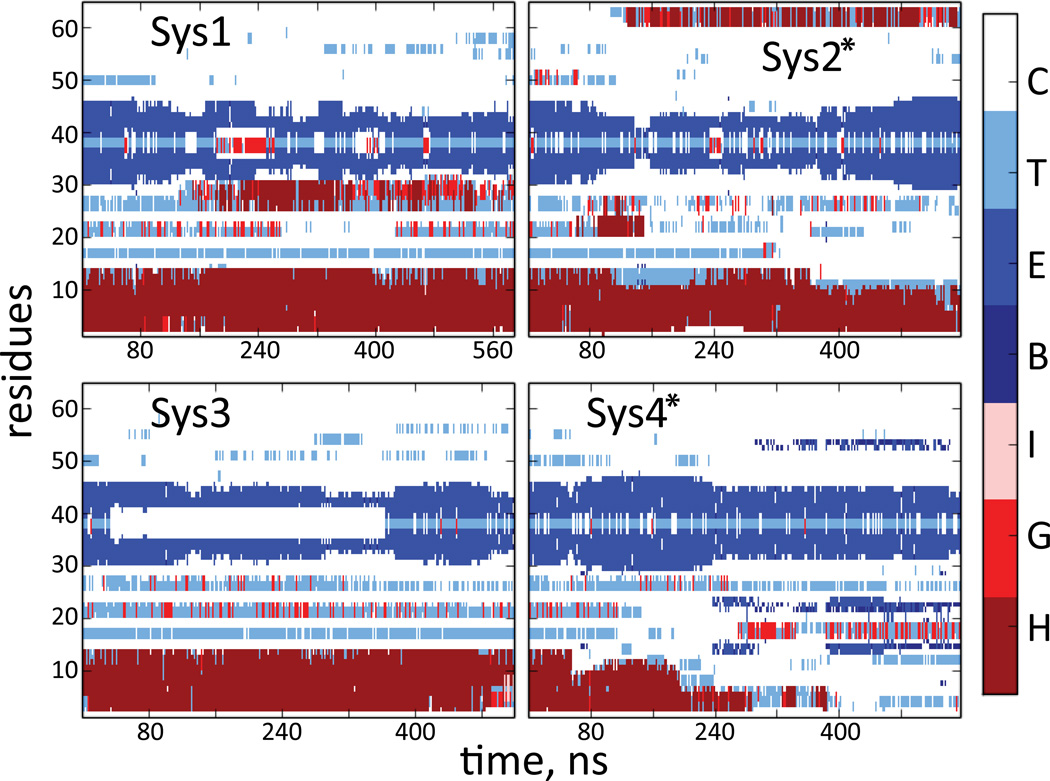
We have previously demonstrated that the hDAT N-term is involved in functional interactions with PIP2 lipids 32. To provide a detailed structural context for the N-term/PIP2 association, we examined the effect of membrane interactions on the predicted structure of the N-term peptide for both PIP2-enriched and PIP2-depleted lipid bilayers (Table 2). The N-term was anchored to the membrane systems by a palmitoyl chain (PALM) attached to its C-terminal end (see Methods, Figure S3).
In four independent MD trajectories totaling > 2.2µs in simulation time (Table 1), the membrane-bound N-term exhibited the same highly dynamic behavior as seen in the simulations in water, but in stark contrast to the aqueous system, the α-helix and beta sheet segments remained mostly stable on the simulation timescales (Figure 5). Among the runs performed with membrane-bound N-term, we observed some loss of the α-helical content only in System 4 (see more below).
Figure 5.
Time evolution of the secondary structure (SS) elements during the MD simulations of the hDAT N-term in lipid membranes (a star sign designates runs carried out in PIP2-enriched membranes). SS motifs are labeled as in Figure 4.
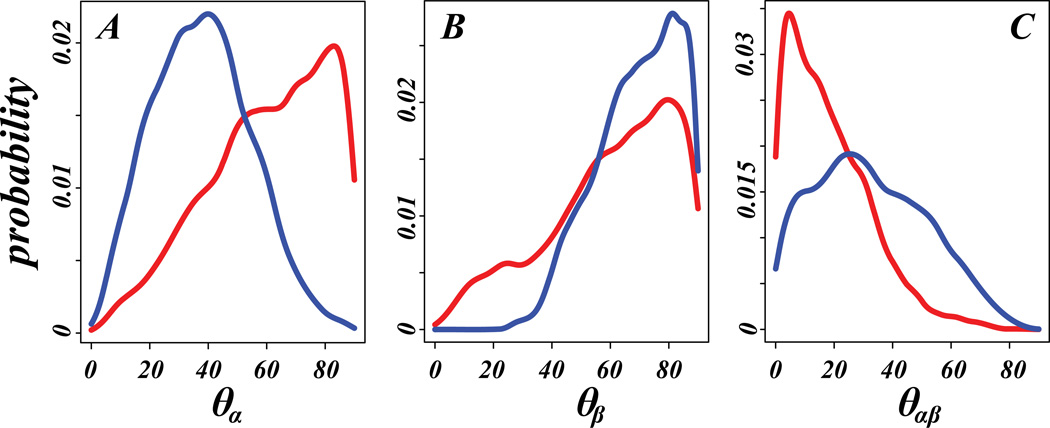
Most notable, however, is the observation that the positioning of the structured elements of the N-term depends on the PIP2 content of the membranes. The dynamics and orientations of the two structural segments in membrane-bound N-term are monitored in the trajectories with two “director vectors”, r⃗α (average of two vectors connecting Cα atoms of residues 4 and 8, and residue 5 and 9, respectively) and r⃗β (connecting Cα atoms of residues 39 and 43). These vectors define the orientations of the α-helix and the beta sheet, respectively, and the positions are defined by (i)-the angle between r⃗α and r⃗β (θαβ), and (ii)-the angle between the director vectors and the membrane normal axis (denoted by θα and θβ, correspondingly). In Figure 6, the probability densities of θα, θβ, and θαβ are compared for PIP2-containing and PIP2-depleted systems. The α-helical domain in PIP2-depleted membranes is seen to remain mostly co-planar with the lipid/water interface (red curve in Figure 6A peaks at high θα angle), but is mostly “upright” in PIP2-enriched bilayers (blue in Figure 6A, see below for more details). As seen in Figure 6C, the α-helical segment is oriented along the direction of the beta-sheet motif in the membranes lacking PIP2 (red in Figure 6C), but its orientation with respect to the beta-sheet is variable in PIP2-containing bilayers (the distribution of θαβ in Figure 6C is broad). The beta-sheet segment appears to prefer orientations co-planar with the lipid/water interface regardless of PIP2 content (Figure 6B). As described below, these trends in the arrangement of the structured segments in the two types of lipid compositions are related to differential modes of interactions of the N-term with PIP2-containing and PIP2-depleted lipid membranes.
Figure 6.
Probability density distributions of θα (A), θβ (B) and θαβ (C) angles (see text for the definitions of the angles) in trajectories of the hDAT N-terminus in PIP2-enriched (blue) and PIP2-depleted (red) simulations. The histograms of angles were combined for Systems 1 and 3, and for Systems 2 and 4 (see Table 1) before the distributions were calculated.
The electrostatic interactions with PIP2 lipids determine the N-term position relative to the lipid membranes
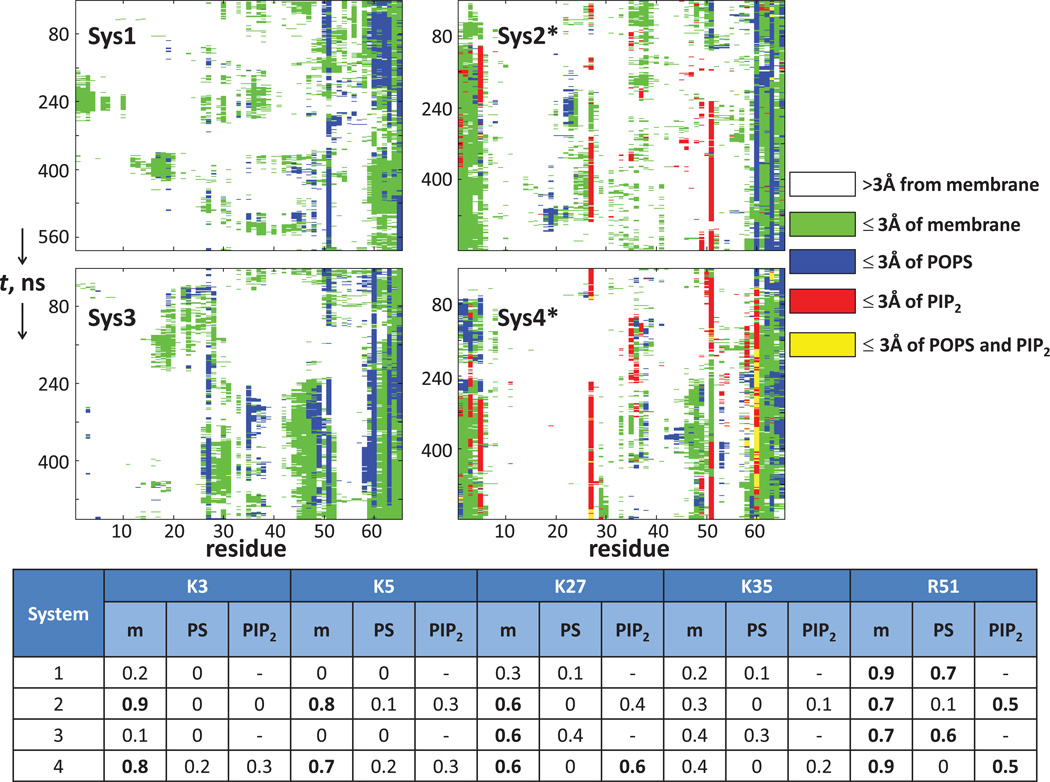
The contacts between the individual N-term residues and the membrane components were quantified in the four trajectories, as described in Figure 7 that shows the time-propagation of these contacts. The table in Figure 7 lists the contacts in terms of the fraction of time in an individual trajectory when several key basic residues of the N-term are within 3Å of any lipid or within 3Å of charged lipids (PIP2 or POPS). These data show extensive contacts between the specific regions of the N-term and PIP2 lipids (see Systems 2 and 4 in Figure 7). The positively charged residues K27 and R51 are seen to be simultaneously engaged with PIP2 lipids in the latter half of System 2 trajectory, as well as throughout the System 4 run. Figure 8A illustrates how this conformation positions the K27/R51 pair in coordination with two PIP2 molecules.
Figure 7.
N-term/membrane contacts in the MD simulations (the star symbol designates simulations carried out in PIP2-enriched membranes). N-term residues at instantaneous points along the trajectories are identified by the following color code: white - if a residue is farther than 3Å from the membrane; green - if a residue is within 3Å of the membrane; blue - if a residue is within 3Å of POPS; red - if a residue is within 3Å of PIP2; yellow - if a residue is within 3Å of both PIP2 and POPS. Numbers on the horizontal axis of each panel identify the N-term residues, and the vertical axis shows the progression of the trajectories. In all simulations the N-term is anchored to the lipid membranes via PALM attached to residue 65. The table shows time fractions, in the individual trajectories, when several key basic residues (K3, K5, K27, K35, and R51) were found within 3Å of any lipid (m) or within 3Å of charged lipids, PIP2 or POPS. Bold entries in the table highlight fractions ≥ 0.5.
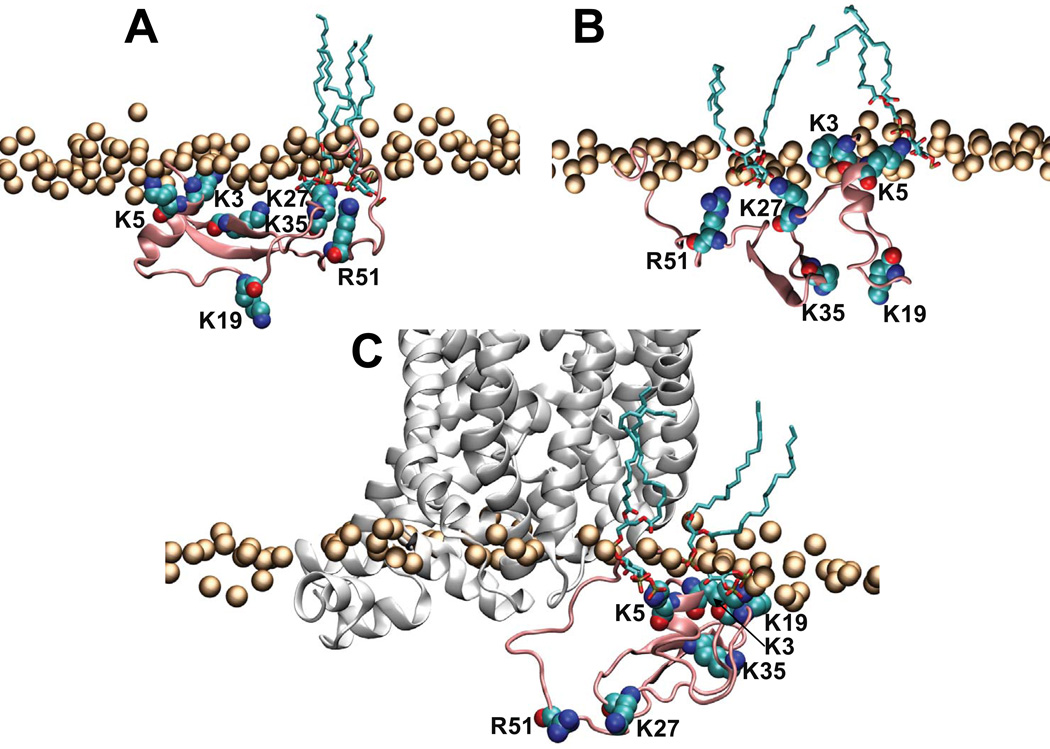
Figure 8.
Illustration of interactions of the hDAT N-term with PIP2 lipids in the membrane in selected snapshots from System4 (A–B), and from the simulations of the chimera construct composed of hDAT N-term and dDAT TM bundle (C). In all panels the hDAT N-term (residues 1–65) is shown in pink cartoon, and K3, K5, K19, K27, K35, and R51 residues shown in space fill and labeled. The panels also show PIP2 lipids within 3Å of the hDAT N-term (in licorice rendering) and the lipid head group phosphate atoms (orange spheres). In panel C the dDAT TM bundle is shown in white cartoon.
The region of the N-terminus containing the K3/K5 residue pair remains positioned in close proximity to PIP2 lipids in the System 2 and 4 trajectories (Figure 7; see also Figure 8B), but the N-term/membrane associations are dynamic in nature and different basic residues are positioned to interact with PIP2 lipids. For example, within the ~150–250ns time interval of the System 4 run, the K3 and K35 side chains jointly coordinate PIP2 (Figure 7), enabled by the close proximity of the K3, K5, and K35 residues in the 3D fold of the N-term (see Figures 4A–B). Later in the System 4 trajectory, the N-term is associated with PIP2 lipids through the K3/K5 pair, as well as through K27/R51 (see Figure 7).
The modes of interaction with PIP2 relate directly to the positioning of the structural motifs in the N-term. Thus, when the N-term interacts with PIP2 lipids through the K3/K5 residue pair, the N-terminal α-helical segment (harboring these residues) adopts orientations that distance it from the plane of the lipid/water interface (see Figure 6A, Figure 7 for Systems 2 and 4), while the beta sheet remains co-planar to this interface (Figure 6B). Even when the helix unfolds somewhat in the later stages of the System 4 trajectory (Figure 5), this segment retains its orientation due to the strong interactions of K3/K5 residues with PIP2 (Figure 7).
In the PIP2-depleted system, the extent of the N-term association with the membrane is significantly reduced relative to the PIP2-enriched bilayers (Figure 7). Indeed, the segment containing residues K3/K5, which actively participates in the interaction of the N-term with PIP2-containing membranes, remains distant from the lipid surface in Systems 1 and 3 (Figure 7 and also see Figure S6 in the Supporting Material). Interestingly, however, R51 maintains long-lasting contacts with the PIP2-depleted bilayers (mostly through interactions with charged POPS lipids present in these mixtures - see blue traces in Figure 7). It is noteworthy that the R51 residue is close to the membrane-anchoring C-terminal segment of the N-peptide (see residues 60–65 in Figure 7, also Figure S6).
Overall, in PIP2-depleted membranes the α-helical segment remains distant from the membrane surface, and the N-term associates with the lipids through the beta-sheet (Figure 7, Systems 1 and 3, see also Figure S6) that remains mostly parallel to the membrane surface (Figure 6B and Figure S6); the α-helix motif maintains an orientation co-planar with the beta sheet (Figure 6C red trace, Figure S6). Together, these results suggest that it is the association with the lipid membranes that determines the spatial positioning of the structural motifs in the N-terminus relative to the bilayer surface.
Strong segregation of PIP2 lipids near the N-term by the electrostatic interaction
Because the extent of PIP2 sequestration by the N-term appears to be an important element of the spatial organization of the peptide relative to the membrane, it is essential to obtain a reliable assessment of lipid segregation and redistribution in the bilayer. This is known to be a slow process that may not be captured completely during the several hundred nanosecond simulations 69. Therefore, we evaluated the relation between PIP2 segregation and protein-membrane interaction using the SCMFM approach (see Methods). Following a protocol described previously 44,46,47, the predicted N-term structure was positioned in specific poses near the membrane so as to place the largest positive electrostatic potential isosurfaces (EPIs) towards the membrane surface. These poses served as starting configurations in the SCMFM approach that evaluates the dynamics of PIP2 under the influence of electrostatic forces generated by the N-term. The reorganization of the lipid composition under these conditions is calculated from a self-consistent energy minimization protocol that includes the electrostatic terms as well as entropic contributions from the mixing of lipids in the membrane and from the mobile ions in the solution (see Methods for the SCMFM approach).
Figure 9A shows the position of the N-term found from the mean-field calculations to generate the largest EPI on the bilayer surface. In this configuration, the K3/K5 pair is interacting with the membrane, and the strong PIP2 lipid sequestration by the N-term peptide calculated from the SCMFM protocol results in 2.5-fold increase of PIP2 density near the K3/K5 pair of the N-term (blue shaded area). The extent of the aggregation of PIP2 lipids around the N-term obtained here is in line with observations that many juxtamembrane proteins (or protein domains) enriched in positively charged residues become anchored to lipid membranes through local concentrations of charge (for some specific examples see 45–47 and references therein).
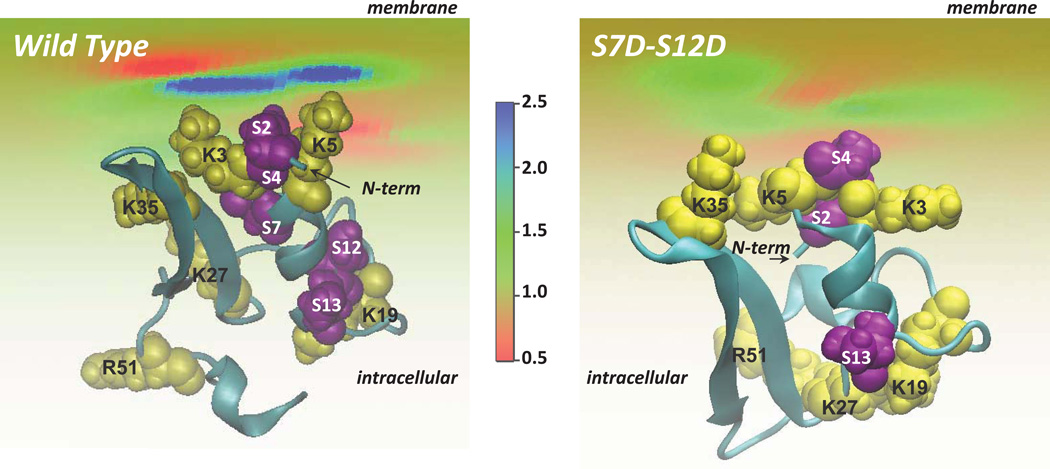
Figure 9.
View from the intracellular side of the wild type (left) and S7D–S12D (right) hDAT N-terminus (cartoon) adsorbing on the lipid membrane. The level of PIP2 segregation by the N-terminus is illustrated (as ratios of local and bulk lipid fraction values) in color code. The positive residues (yellow) and serine residues (purple) are highlighted in space fill and labeled. The membrane is composed of charged and neutral lipids generating a surface charge density of −0.0031e (corresponding to a lipid mixture with 5% PIP2). The 3D fold of the S7D–S12D N-term was predicted using the same set of Rosetta and RMSTT protocols as the ones used for the wild type N-peptide.
Importantly, the level of PIP2 segregation predicted from the SCMFM is consistent with that observed in the MD simulations. Thus, in Systems 2 and 4 we find at most 2 PIP2 lipids near K3/K5 pair (Figure 7). Assuming a 65Å2 area per lipid head group and taking into consideration an approximate membrane-exposed surface area of the N-term, two bound PIP2 lipids correspond to a local lipid composition around the N-term in which PIP2 concentration is elevated by ~2.4-fold over the bulk. Such consistency between results from the mean-field SCMFM and the all-atom MD simulation, demonstrated as well in our previous work 44, illustrates the convergence of the two approaches in a suitable quantification of PIP2 lipid sequestration around the N-term. Importantly, since the steady state lipid distribution in the SCMFM is derived through minimization of the free energy with only electrostatic and lipid mixing contributions, the reproducibility of the predictions from MD simulations in the SCMFM reinforces the conclusion that direct electrostatic interactions with PIP2 lipids play major role in anchoring the N-terminus to membranes.
Phospho-mimetic S-to-D mutations in silico alter the electrostatic reactivity features of the N-term
To test the role of electrostatic interactions between the N-term and PIP2 lipids in the positioning of the N-term we chose an approach that corresponds to experimental probing by introducing phospho-mimetic mutations at sites known to be phosphorylated in the N-terminus of hDAT 25–27, in the proximity of the R/K residues (Figure 4A). To model the S7D–S12D double mutant we used the same structure prediction protocol as for the wild type N-term (see Methods). The approach yielded a 3D structure of the S7D–S12D construct containing the same secondary structure elements (Figure S7A in the Supporting Material) as those identified in the wild type. However, the α-helix in the S7D–S12D mutant was somewhat shorter (spanning only residues 7–12, compare to Figure 4E). Importantly, quantification of PIP2 segregation around the S7D–S12D construct using the SCMFM approach (Figure 9B) revealed that the mutant N-term does not sequester PIP2 lipids as much as the wild-type peptide does (compare to Figure 9A). This difference is due to specific spatial positioning of the basic residues in the S7D–S12D mutant brought about by the two additional negative charges. The position is distinct from that observed in the wild type N-term, which disrupts the extended positive electrostatic potential isosurface seen in the wild type N-term (Figure 4C; cf. Figure S7B in the Supporting Material).
When attached to the complete transmembrane domain of the dDAT structure the N-term maintains the same modes of interaction with PIP2-containing membranes
To evaluate the modes of N-term/membrane interactions in the full-length DAT, we connected the atomistic model of the hDAT N-term to the TM bundle of dDAT determined from X-ray crystallography (see Methods). This construct (hDAT/dDAT) was subjected to atomistic MD simulations in the same PIP2-containing lipid membrane (see Figure S3B). Analysis of two separate MD trajectories (~340ns and ~245ns long) of hDAT/dDAT showed the secondary structure elements identified in the N-term to remain relatively stable on the simulation timescales (some transient changes are identifiable in the two panels of Figure S8A in the Supporting Material). Importantly, the analysis revealed the same mode of N-term binding to lipid membranes as detected throughout in Systems 2 and 4 described above for the palmitoylated N-term peptide. Thus, the K3/K5 pair is involved in strong electrostatic interactions with PIP2 lipids (see panels C and D in Figure S8, also compare panels B and C in Figure 8), while the segment containing K3/K5 maintains orientations with respect to the membrane normal that are similar to those observed in the simulations of membrane-anchored peptide, Systems 2 and 4 (see panel B in Figure S8, compare to Figure 6A). We found recently (Khelashvili et al, in preparation) that the stability of the secondary structure elements in the N-term, as well as the mode of interaction with PIP2 lipids that involves K3/K5 pair of residues occur as well in the full-length human DAT (hDAT) model obtained from homology modeling based on the dDAT structure. The corresponding atomistic MD simulations (an aggregate of >20 microseconds from individual 2–4 microsecond simulations), were carried out in the same complement of PIP2-containing/ PIP2-depleted membranes.
Discussion
The functional importance of the N-terminal segment of hDAT has been rigorously established through various in vitro and in vivo experiments 11,25–27,32,40,41,70–72. In particular, the first 33 residues of hDAT were also shown to have a critical role in the reverse transport process, efflux 40, in which the substrate DA is transported out of the nerve cell and into the synaptic cleft via hDAT. Consonant with these findings, we showed recently 32 that hDAT efflux requires direct interactions of the N-term with PIP2 lipids, and that these interactions were disrupted by mutations of the positively charged K3 and K5 residues to neutral residues (K-to-A or K-to-N). Notably, disrupting these interactions reduces AMPH-induced DA efflux without altering physiological uptake of the substrate through hDAT.
Structural information about the N-term is still lacking for the eukaryotic transporters in the NSS family73 in spite of the recently determined structure of dDAT (the termini were not elucidated in the structures), and there is no sequence homology to proteins of known fold. Therefore, we undertook the prediction of the 3D fold of the hDAT N-term computationally as described herein and concluded that it contains two secondary structure elements, an α-helical segment and a beta sheet motif connected by flexible segments that rearrange upon interaction of the N-term with the membrane. The predicted structured elements remained stable in long (>2.2µs) MD simulations of this peptide tethered to lipid membranes via C-terminal palmitoylation, whereas simulations of the same peptide in water led to rapid destabilization of the beta sheet motif and major changes in the overall structure (since, as described, the α-helix is in sustained contact with the beta sheet in the folded peptide). Biased MD simulations with the replica exchange method (REMD) carried out in water support the loss of stability of the N-term structure, especially in comparison to the structurally related plectasin peptide, for which the fold is known.
The functional relevance of the structural framework becomes evident from the role of direct electrostatic interactions of the K3/K5 pair of residues in the structured N-term, with negatively charged lipids in the membrane. Throughout the long MD simulations, we found the N-term to engage in long-lasting contacts with the PIP2 components of the membrane. As a result of this association, the N-terminal segment harboring K3/K5 residues becomes anchored to the lipid membrane and assumes orientations that are nearly perpendicular to the bilayer plane. But in the absence of PIP2, this N-terminal segment remains distant from the membrane surface and does not engage in the same type of interactions. Since PIP2, and the positively charged residues at positions 3 and 5 of the N-term, were shown to be required for the efflux phenotype, it is tempting to propose that the difference in orientation is involved in the mechanism eliciting the phenotype. This hypothesis gains support from the described observation that when the predicted N-term structure of the human DAT protein (hDAT) was incorporated in a chimera construct (hDAT/dDAT) containing the structurally known TM bundle of dDAT, the MD simulations showed a conservation of the key dynamic and structural features of the N-term alone interacting with PIP2-containing membranes, featuring the electrostatic interactions between K3/K5 residues and PIP2 lipids. Moreover, this N-term/membrane interaction is modulated by phosphorylation, which plays a key role in the efflux phenotype. The results from continuum mean-field level computations performed on the S7D–S12D double mutant showed lower levels of PIP2 lipid segregation around S7D–S12D mutant than around the wild type N-terminus, consonant with weaker binding to PIP2 membranes measured for the mutant construct than for the wild type N-term.
The findings based on the predicted structure of the N-term are remarkably consistent with the experimental results, for which they provide for the first time a detailed structural context. This includes not only the effects of phosphorylation, but also the distinct effects on efflux observed for the K-to-A or K-to-N mutations that were shown to disrupt PIP2 binding 32. Moreover, the specific conformations visited by the N-term region anchored to the membrane, and the manner in which they support the role of the K3/K5 residues and phosphorylation of the N-terminal serine residues in the efflux process, are also consonant with the proposed hypothesis that the N-term plays the role of a “lever” in regulating this function of DAT 70 and identify the portion of the N-term that is likely to function in this manner.
Based on the noted agreement with the inferences from experimental probing of efflux conditions and properties, we suggest that the presented findings showing that the N-term harbors structured elements that are reasonably stable during the course of the long time-scale MD simulations could inform further structure-guided experimental explorations of the N-term in the membrane environment. With that, our studies lay the foundation for future structure/function investigations of the role of the N-term in DAT function in general, and the efflux mechanism in particular, by following its spatially ordered interactions with the transmembrane domain of the DAT structure, and its C-terminus41.
Supplementary Material
Acknowledgments
We thank Drs. Jonathan A. Javitch, Aurelio Galli and Ulrik Gether for very helpful discussions. This work was supported by National Institute of Health Grants 2P01DA012408, 1R01DA035263, and R01DA017293. The following computational resources are gratefully acknowledged: XSEDE allocation at the Texas Advanced Computing Center at the University of Texas at Austin (Stampede supercomputer, projects TG-MCB090132, TG-MCB120008); allocation at the National Energy Research Scientific Computing Center (NERSC, repository m1710), supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231; and computational resources of the David A. Cofrin Center for Biomedical Information in the HRH Prince Alwaleed Bin Talal Bin Abdulaziz Alsaud Institute for Computational Biomedicine.
References
- 1.Kanner BI, Zomot E. Sodium-coupled neurotransmitter transporters. Chem Rev. 2008;108(5):1654–1668. doi: 10.1021/cr078246a. [DOI] [PubMed] [Google Scholar]
- 2.Broer S, Gether U. The solute carrier 6 family of transporters. Br J Pharmacol. 2012;167(2):256–278. doi: 10.1111/j.1476-5381.2012.01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen NH, Reith ME, Quick MW. Synaptic uptake and beyond: the sodium- and chloride-dependent neurotransmitter transporter family SLC6. Pflugers Arch. 2004;447(5):519–531. doi: 10.1007/s00424-003-1064-5. [DOI] [PubMed] [Google Scholar]
- 4.Sora I, Li B, Fumushima S, Fukui A, Arime Y, Kasahara Y, Tomita H, Ikeda K. Monoamine transporter as a target molecule for psychostimulants. Int Rev Neurobiol. 2009;85:29–33. doi: 10.1016/S0074-7742(09)85003-4. [DOI] [PubMed] [Google Scholar]
- 5.Sulzer D, Sonders MS, Poulsen NW, Galli A. Mechanisms of neurotransmitter release by amphetamines: a review. Prog Neurobiol. 2005;75(6):406–433. doi: 10.1016/j.pneurobio.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 6.Robertson SD, Matthies HJ, Galli A. A closer look at amphetamine-induced reverse transport and trafficking of the dopamine and norepinephrine transporters. Mol Neurobiol. 2009;39(2):73–80. doi: 10.1007/s12035-009-8053-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Penmatsa A, Wang KH, Gouaux E. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 2013;503(7474):85–90. doi: 10.1038/nature12533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beuming T, Shi L, Javitch JA, Weinstein H. A comprehensive structure-based alignment of prokaryotic and eukaryotic neurotransmitter/Na+ symporters (NSS) aids in the use of the LeuT structure to probe NSS structure and function. Mol Pharmacol. 2006;70(5):1630–1642. doi: 10.1124/mol.106.026120. [DOI] [PubMed] [Google Scholar]
- 9.Kantcheva AK, Quick M, Shi L, Winther AM, Stolzenberg S, Weinstein H, Javitch JA, Nissen P. Chloride binding site of neurotransmitter sodium symporters. Proc Natl Acad Sci U S A. 2013;110(21):8489–8494. doi: 10.1073/pnas.1221279110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shan J, Javitch JA, Shi L, Weinstein H. The substrate-driven transition to an inward-facing conformation in the functional mechanism of the dopamine transporter. PLoS One. 2011;6(1):e16350. doi: 10.1371/journal.pone.0016350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guptaroy B, Zhang M, Bowton E, Binda F, Shi L, Weinstein H, Galli A, Javitch JA, Neubig RR, Gnegy ME. A juxtamembrane mutation in the N terminus of the dopamine transporter induces preference for an inward-facing conformation. Mol Pharmacol. 2009;75(3):514–524. doi: 10.1124/mol.108.048744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beuming T, Kniazeff J, Bergmann ML, Shi L, Gracia L, Raniszewska K, Newman AH, Javitch JA, Weinstein H, Gether U, Loland CJ. The binding sites for cocaine and dopamine in the dopamine transporter overlap. Nat Neurosci. 2008;11(7):780–789. doi: 10.1038/nn.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kniazeff J, Shi L, Loland CJ, Javitch JA, Weinstein H, Gether U. An intracellular interaction network regulates conformational transitions in the dopamine transporter. J Biol Chem. 2008;283(25):17691–17701. doi: 10.1074/jbc.M800475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stockner T, Montgomery TR, Kudlacek O, Weissensteiner R, Ecker GF, Freissmuth M, Sitte HH. Mutational analysis of the high-affinity zinc binding site validates a refined human dopamine transporter homology model. PLoS Comput Biol. 2013;9(2):e1002909. doi: 10.1371/journal.pcbi.1002909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krishnamurthy H, Gouaux E. X-ray structures of LeuT in substrate-free outward-open and apo inward-open states. Nature. 2012;481(7382):469–474. doi: 10.1038/nature10737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang H, Elferich J, Gouaux E. Structures of LeuT in bicelles define conformation and substrate binding in a membrane-like context. Nat Struct Mol Biol. 2012;19(2):212–219. doi: 10.1038/nsmb.2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Piscitelli CL, Gouaux E. Insights into transport mechanism from LeuT engineered to transport tryptophan. EMBO J. 2012;31(1):228–235. doi: 10.1038/emboj.2011.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Gouaux E. Substrate binds in the S1 site of the F253A mutant of LeuT, a neurotransmitter sodium symporter homologue. EMBO Rep. 2012;13(9):861–866. doi: 10.1038/embor.2012.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Piscitelli CL, Krishnamurthy H, Gouaux E. Neurotransmitter/sodium symporter orthologue LeuT has a single high-affinity substrate site. Nature. 2010;468(7327):1129–1132. doi: 10.1038/nature09581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Singh SK, Piscitelli CL, Yamashita A, Gouaux E. A competitive inhibitor traps LeuT in an open-to-out conformation. Science. 2008;322(5908):1655–1661. doi: 10.1126/science.1166777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Singh SK, Yamashita A, Gouaux E. Antidepressant binding site in a bacterial homologue of neurotransmitter transporters. Nature. 2007;448(7156):952–956. doi: 10.1038/nature06038. [DOI] [PubMed] [Google Scholar]
- 22.Yamashita A, Singh SK, Kawate T, Jin Y, Gouaux E. Crystal structure of a bacterial homologue of Na+/Cl--dependent neurotransmitter transporters. Nature. 2005;437(7056):215–223. doi: 10.1038/nature03978. [DOI] [PubMed] [Google Scholar]
- 23.Giambalvo CT. Protein kinase C and dopamine transport--2. Effects of amphetamine in vitro. Neuropharmacology. 1992;31(12):1211–1222. doi: 10.1016/0028-3908(92)90049-u. [DOI] [PubMed] [Google Scholar]
- 24.Giambalvo CT. Differential effects of amphetamine transport vs. dopamine reverse transport on particulate PKC activity in striatal synaptoneurosomes. Synapse. 2003;49(2):125–133. doi: 10.1002/syn.10223. [DOI] [PubMed] [Google Scholar]
- 25.Foster JD, Pananusorn B, Cervinski MA, Holden HE, Vaughan RA. Dopamine transporters are dephosphorylated in striatal homogenates and in vitro by protein phosphatase 1. Brain Res Mol Brain Res. 2003;110(1):100–108. doi: 10.1016/s0169-328x(02)00645-9. [DOI] [PubMed] [Google Scholar]
- 26.Foster JD, Pananusorn B, Vaughan RA. Dopamine transporters are phosphorylated on N-terminal serines in rat striatum. J Biol Chem. 2002;277(28):25178–25186. doi: 10.1074/jbc.M200294200. [DOI] [PubMed] [Google Scholar]
- 27.Khoshbouei H, Sen N, Guptaroy B, Johnson L, Lund D, Gnegy ME, Galli A, Javitch JA. N-terminal phosphorylation of the dopamine transporter is required for amphetamine-induced efflux. PLoS Biol. 2004;2(3):E78. doi: 10.1371/journal.pbio.0020078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Foster JD, Cervinski MA, Gorentla BK, Vaughan RA. Regulation of the dopamine transporter by phosphorylation. Handb Exp Pharmacol. 2006;(175):197–214. doi: 10.1007/3-540-29784-7_10. [DOI] [PubMed] [Google Scholar]
- 29.Kahlig KM, Binda F, Khoshbouei H, Blakely RD, McMahon DG, Javitch JA, Galli A. Amphetamine induces dopamine efflux through a dopamine transporter channel. Proc Natl Acad Sci U S A. 2005;102(9):3495–3500. doi: 10.1073/pnas.0407737102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahlig KM, Javitch JA, Galli A. Amphetamine regulation of dopamine transport. Combined measurements of transporter currents and transporter imaging support the endocytosis of an active carrier. J Biol Chem. 2004;279(10):8966–8975. doi: 10.1074/jbc.M303976200. [DOI] [PubMed] [Google Scholar]
- 31.Thwar PK, Guptaroy B, Zhang M, Gnegy ME, Burns MA, Linderman JJ. Simple transporter trafficking model for amphetamine-induced dopamine efflux. Synapse. 2007;61(7):500–514. doi: 10.1002/syn.20390. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton PJ, Belovich AN, Khelashvili G, Saunders C, Erreger K, Javitch JA, Sitte HH, Weinstein H, Matthies HJ, Galli A. PIP2 regulates psychostimulant behaviors through its interaction with a membrane protein. Nat Chem Biol. 2014;10(7):582–589. doi: 10.1038/nchembio.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen FH, Skjorringe T, Yasmeen S, Arends NV, Sahai MA, Erreger K, Andreassen TF, Holy M, Hamilton PJ, Neergheen V, Karlsborg M, Newman AH, Pope S, Heales SJ, Friberg L, Law I, Pinborg LH, Sitte HH, Loland C, Shi L, Weinstein H, Galli A, Hjermind LE, Moller LB, Gether U. Missense dopamine transporter mutations associate with adult parkinsonism and ADHD. J Clin Invest. 2014;124(7):3107–3120. doi: 10.1172/JCI73778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hamilton PJ, Campbell NG, Sharma S, Erreger K, Herborg Hansen F, Saunders C, Belovich AN, Sahai MA, Cook EH, Gether U, McHaourab HS, Matthies HJ, Sutcliffe JS, Galli A. De novo mutation in the dopamine transporter gene associates dopamine dysfunction with autism spectrum disorder. Mol Psychiatry. 2013;18(12):1315–1323. doi: 10.1038/mp.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sakrikar D, Mazei-Robison MS, Mergy MA, Richtand NW, Han Q, Hamilton PJ, Bowton E, Galli A, Veenstra-Vanderweele J, Gill M, Blakely RD. Attention deficit/hyperactivity disorder-derived coding variation in the dopamine transporter disrupts microdomain targeting and trafficking regulation. J Neurosci. 2012;32(16):5385–5397. doi: 10.1523/JNEUROSCI.6033-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bowton E, Saunders C, Erreger K, Sakrikar D, Matthies HJ, Sen N, Jessen T, Colbran RJ, Caron MG, Javitch JA, Blakely RD, Galli A. Dysregulation of dopamine transporters via dopamine D2 autoreceptors triggers anomalous dopamine efflux associated with attention-deficit hyperactivity disorder. J Neurosci. 2010;30(17):6048–6057. doi: 10.1523/JNEUROSCI.5094-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazei-Robison MS, Bowton E, Holy M, Schmudermaier M, Freissmuth M, Sitte HH, Galli A, Blakely RD. Anomalous dopamine release associated with a human dopamine transporter coding variant. J Neurosci. 2008;28(28):7040–7046. doi: 10.1523/JNEUROSCI.0473-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ng J, Zhen J, Meyer E, Erreger K, Li Y, Kakar N, Ahmad J, Thiele H, Kubisch C, Rider NL, Morton DH, Strauss KA, Puffenberger EG, D'Agnano D, Anikster Y, Carducci C, Hyland K, Rotstein M, Leuzzi V, Borck G, Reith ME, Kurian MA. Dopamine transporter deficiency syndrome: phenotypic spectrum from infancy to adulthood. Brain. 2014;137(Pt 4):1107–1119. doi: 10.1093/brain/awu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cremona ML, Matthies HJ, Pau K, Bowton E, Speed N, Lute BJ, Anderson M, Sen N, Robertson SD, Vaughan RA, Rothman JE, Galli A, Javitch JA, Yamamoto A. Flotillin-1 is essential for PKC-triggered endocytosis and membrane microdomain localization of DAT. Nat Neurosci. 2011;14(4):469–477. doi: 10.1038/nn.2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Binda F, Dipace C, Bowton E, Robertson SD, Lute BJ, Fog JU, Zhang M, Sen N, Colbran RJ, Gnegy ME, Gether U, Javitch JA, Erreger K, Galli A. Syntaxin 1A interaction with the dopamine transporter promotes amphetamine-induced dopamine efflux. Mol Pharmacol. 2008;74(4):1101–1108. doi: 10.1124/mol.108.048447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fog JU, Khoshbouei H, Holy M, Owens WA, Vaegter CB, Sen N, Nikandrova Y, Bowton E, McMahon DG, Colbran RJ, Daws LC, Sitte HH, Javitch JA, Galli A, Gether U. Calmodulin kinase II interacts with the dopamine transporter C terminus to regulate amphetamine-induced reverse transport. Neuron. 2006;51(4):417–429. doi: 10.1016/j.neuron.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 42.Buchmayer F, Schicker K, Steinkellner T, Geier P, Stubiger G, Hamilton PJ, Jurik A, Stockner T, Yang JW, Montgomery T, Holy M, Hofmaier T, Kudlacek O, Matthies HJ, Ecker GF, Bochkov V, Galli A, Boehm S, Sitte HH. Amphetamine actions at the serotonin transporter rely on the availability of phosphatidylinositol-4,5-bisphosphate. Proc Natl Acad Sci U S A. 2013;110(28):11642–11647. doi: 10.1073/pnas.1220552110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Das R, Baker D. Macromolecular modeling with rosetta. Annu Rev Biochem. 2008;77:363–382. doi: 10.1146/annurev.biochem.77.062906.171838. [DOI] [PubMed] [Google Scholar]
- 44.Khelashvili G, Galli A, Weinstein H. Phosphatidylinositol 4,5-biphosphate (PIP(2)) lipids regulate the phosphorylation of syntaxin N-terminus by modulating both its position and local structure. Biochemistry. 2012;51(39):7685–7698. doi: 10.1021/bi300833z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khelashvili G, Harries D. Modelling signalling processes across cellular membranes using a mesoscopic approach. Annual Reports in Computational Chemistry. 2010;6:236–261. [Google Scholar]
- 46.Khelashvili G, Harries D, Weinstein H. Modeling membrane deformations and lipid demixing upon protein-membrane interaction: the BAR dimer adsorption. Biophys J. 2009;97(6):1626–1635. doi: 10.1016/j.bpj.2009.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Khelashvili G, Weinstein H, Harries D. Protein diffusion on charged membranes: a dynamic mean-field model describes time evolution and lipid reorganization. Biophys J. 2008;94(7):2580–2597. doi: 10.1529/biophysj.107.120667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gracia L. RMSDTT: RMSD Trajectory Tool. 2.5 ed. Weill Medical College of Cornell University, Department of Physiology and Biophysics; 2005. [Google Scholar]
- 49.Zhao C, Stolzenberg S, Gracia L, Weinstein H, Noskov S, Shi L. Ion-controlled conformational dynamics in the outward-open transition from an occluded state of LeuT. Biophys J. 2012;103(5):878–888. doi: 10.1016/j.bpj.2012.07.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. Journal of Computational Chemistry. 2005;26(16):1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Best RB, Zhu X, Shim J, Lopes PE, Mittal J, Feig M, Mackerell AD., Jr Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J Chem Theory Comput. 2013;8(9):3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Klauda JB, Venable RM, Freites JA, O'Connor JW, Tobias DJ, Mondragon-Ramirez C, Vorobyov I, MacKerell AD, Pastor RW. Update of the CHARMM All-Atom Additive Force Field for Lipids: Validation on Six Lipid Types. Journal of Physical Chemistry B. 2010;114(23):7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. Journal of Chemical Physics. 1995;103(19):8577–8593. [Google Scholar]
- 54.Mandal K, Pentelute BL, Tereshko V, Thammavongsa V, Schneewind O, Kossiakoff AA, Kent SB. Racemic crystallography of synthetic protein enantiomers used to determine the X-ray structure of plectasin by direct methods. Protein Sci. 2009;18(6):1146–1154. doi: 10.1002/pro.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sankararamakrishnan R, Konvicka K, Mehler E, Weinstein H. Solvation in simulated annealing and high-temperature molecular dynamics of proteins: A restrained water droplet model. International Journal of Quantum Chemistry. 2000;77(1):174–186. [Google Scholar]
- 56.Ariga T, Macala LJ, Saito M, Margolis RK, Greene LA, Margolis RU, Yu RK. Lipid composition of PC12 pheochromocytoma cells: characterization of globoside as a major neutral glycolipid. Biochemistry. 1988;27(1):52–58. doi: 10.1021/bi00401a010. [DOI] [PubMed] [Google Scholar]
- 57.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI Membrane Builder for Mixed Bilayers and Its Application to Yeast Membranes. Biophysical Journal. 2009;97(1):50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mondal S, Khelashvili G, Shi L, Weinstein H. The cost of living in the membrane: A case study of hydrophobic mismatch for the multi-segment protein LeuT. Chem Phys Lipids. 2013;169:27–38. doi: 10.1016/j.chemphyslip.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stolzenberg S, Khelashvili G, Weinstein H. Structural intermediates in a model of the substrate translocation path of the bacterial glutamate transporter homologue GltPh. J Phys Chem B. 2012;116(18):5372–5383. doi: 10.1021/jp301726s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14(1):33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 61.Olsson MHM, Sondergaard CR, Rostkowski M, Jensen JH. PROPKA3: Consistent Treatment of Internal and Surface Residues in Empirical pKa Predictions. Journal of Chemical Theory and Computation. 2011;7(2):525–537. doi: 10.1021/ct100578z. [DOI] [PubMed] [Google Scholar]
- 62.Yesselman JD, Price DJ, Knight JL, Brooks CL., 3rd MATCH: an atom-typing toolset for molecular mechanics force fields. J Comput Chem. 2011;33(2):189–202. doi: 10.1002/jcc.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sharp KA, Honig B. Electrostatic interactions in macromolecules: theory and applications. Annual review of biophysics and biophysical chemistry. 1990;19:301–332. doi: 10.1146/annurev.bb.19.060190.001505. [DOI] [PubMed] [Google Scholar]
- 64.Chaikin PM, Lubensky TC. Principles of Condensed Matter Physics. Cambridge: Cambridge university press; 2000. [Google Scholar]
- 65.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proceedings of the National Academy of Sciences of the United States of America. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biasini M, Schmidt T, Bienert S, Mariani V, Studer G, Haas J, Johner N, Schenk AD, Philippsen A, Schwede T. OpenStructure: an integrated software framework for computational structural biology. Acta Crystallogr D Biol Crystallogr. 2013;69(Pt 5):701–709. doi: 10.1107/S0907444913007051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Biasini M, Mariani V, Haas J, Scheuber S, Schenk AD, Schwede T, Philippsen A. OpenStructure: a flexible software framework for computational structural biology. Bioinformatics. 2010;26(20):2626–2628. doi: 10.1093/bioinformatics/btq481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mygind PH, Fischer RL, Schnorr KM, Hansen MT, Sonksen CP, Ludvigsen S, Raventos D, Buskov S, Christensen B, De Maria L, Taboureau O, Yaver D, Elvig-Jorgensen SG, Sorensen MV, Christensen BE, Kjaerulff S, Frimodt-Moller N, Lehrer RI, Zasloff M, Kristensen HH. Plectasin is a peptide antibiotic with therapeutic potential from a saprophytic fungus. Nature. 2005;437(7061):975–980. doi: 10.1038/nature04051. [DOI] [PubMed] [Google Scholar]
- 69.Hong C, Tieleman DP, Wang Y. Microsecond Molecular Dynamics Simulations of Lipid Mixing. Langmuir. 2014 doi: 10.1021/la502363b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sucic S, Dallinger S, Zdrazil B, Weissensteiner R, Jorgensen TN, Holy M, Kudlacek O, Seidel S, Cha JH, Gether U, Newman AH, Ecker GF, Freissmuth M, Sitte HH. The N terminus of monoamine transporters is a lever required for the action of amphetamines. J Biol Chem. 2010;285(14):10924–10938. doi: 10.1074/jbc.M109.083154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moritz AE, Foster JD, Gorentla BK, Mazei-Robison MS, Yang JW, Sitte HH, Blakely RD, Vaughan RA. Phosphorylation of dopamine transporter serine 7 modulates cocaine analog binding. J Biol Chem. 2012;288(1):20–32. doi: 10.1074/jbc.M112.407874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Foster JD, Yang JW, Moritz AE, Challasivakanaka S, Smith MA, Holy M, Wilebski K, Sitte HH, Vaughan RA. Dopamine transporter phosphorylation site threonine 53 regulates substrate reuptake and amphetamine-stimulated efflux. J Biol Chem. 2012;287(35):29702–29712. doi: 10.1074/jbc.M112.367706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fenollar-Ferrer C, Stockner T, Schwarz TC, Pal A, Gotovina J, Hofmaier T, Jayaraman K, Adhikary S, Kudlacek O, Mehdipour AR, Tavoulari S, Rudnick G, Singh SK, Konrat R, Sitte HH, Forrest LR. Structure and regulatory interactions of the cytoplasmic terminal domains of serotonin transporter. Biochemistry. 2014;53(33):5444–5460. doi: 10.1021/bi500637f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36(Web Server issue):W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu J, Weng Z. FAST: a novel protein structure alignment algorithm. Proteins. 2005;58(3):618–627. doi: 10.1002/prot.20331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.