Abstract
We have used probe-based molecular dynamics (pMD) simulations to search for interaction hotspots on the surface of the therapeutically highly relevant oncogenic K-Ras G12D. Combining the probe-based query with an ensemble-based pocket identification scheme and an analysis of existing Ras-ligand complexes, we show that (i) pMD is a robust and cost-effective strategy for binding site identification, (ii) all four of the previously reported ligand binding sites are suitable for structure-based ligand design, and (iii) in some cases probe binding and expanded sampling of configurational space enable pocket expansion and increase the likelihood of site identification. Furthermore, by comparing the distribution of hotspots in non-pocket-like regions with known protein- and membrane-interacting interfaces, we propose that pMD has the potential to predict surface patches responsible for protein-biomolecule interactions. These observations have important implications for future drug design efforts and will facilitate the search for potential interfaces responsible for the proposed transient oligomerization or interaction of Ras with other biomolecules in the cellular milieu.
Keywords: Probe-based molecular dynamics simulation, allosteric sites, drug design
Introduction
K-Ras is a guanine triphosphate (GTP)-binding enzyme that regulates signaling pathways involved in cell division and proliferation1-3. It is made up of a soluble catalytic domain that is conserved in the Ras family and a variable C-terminus that undergoes post-translational farnesylation. K-Ras achieves high affinity binding to the plasma membrane, which is required for its cellular activity, through hydrophobic interactions via its farnesyl tail and electrostatic interactions via the proximal polybasic domain4. Malfunction of Ras proteins is associated with 15-20% of all human tumors and mutations in K-Ras account for over 85% of Ras-associated cancer cases5-7. These include some of the deadliest ones such as pancreatic and colorectal cancer8-10. Therefore, K-Ras remains a highly sought-after anti-cancer drug target11-13.
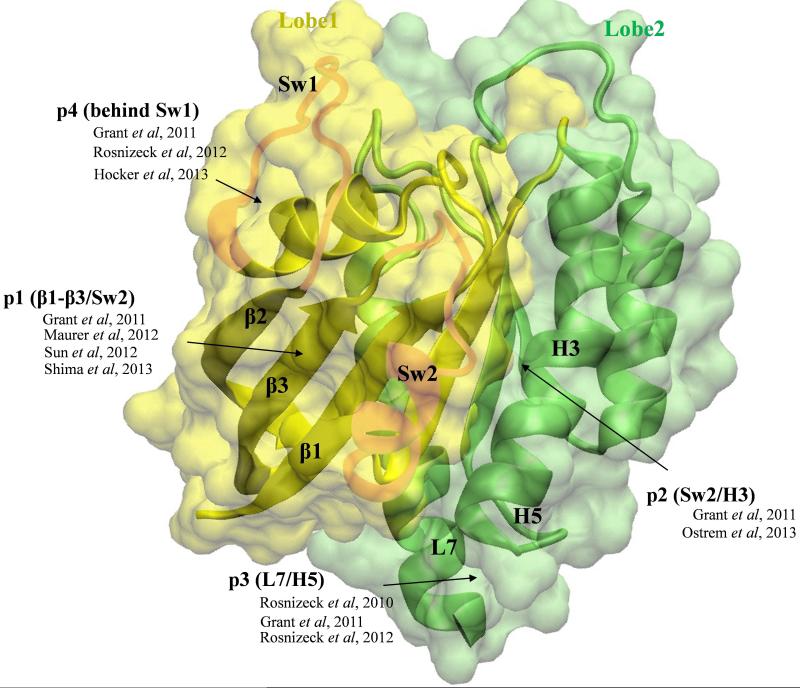
The catalytic domain of Ras (residues 1-166) has been structurally characterized by both X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy (e.g.,14-16). However, until recently, there was no known ligand-binding site on Ras aside from the nucleotide-binding site, which is an unsuitable drug target due to a variety of reasons17,18. Previous computational and crystallographic studies suggested that Ras is an allosteric enzyme19-23. This was followed by the prediction of four allosteric ligand-binding sites using FTMAP24,25 and ensemble docking24,26,27. These allosteric sites are now confirmed by the discovery of several small-molecule ligands that act as allosteric modulators of Ras28-33. The general location for each of the four well-characterized allosteric ligand-binding sites is shown in Fig 1.
Figure 1. The catalytic domain of K-Ras is bi-lobal and contains four allosteric ligand-binding sites.
The catalytic domain of K-Ras is shown in surface and ribbon representations with lobe1 (residues 1-86) in yellow and lobe2 (residues 87-167) in green. The functionally important switch regions sw1 and sw2 are highlighted. Arrows indicate the general location of the four allosteric ligand-binding sites discussed in this paper, along with the relevant previous reports that described them.
One of the key observations from the previous computational studies, particularly those based on molecular dynamics (MD) simulation24,26, was that pockets invisible in an average experimental structure could become accessible when the protein is allowed to flex. However, limitations of sampling inherent in MD and, following the simulation, the reliance on a few representative structures for binding site identification have left a number of important questions unanswered. One is whether there aren't additional ligand binding sites on Ras. The other is if there isn't a better way of more efficiently accounting for conformational flexibility to improve the chance of identifying new sites. Furthermore, all of the known non-covalent Ras binders have weak affinity28-32. One way to improve affinity is by (re)designing ligands that interact with the protein at multiple cavities34, which requires searching for trenches near known ligand binding sites or multiple novel cavities that could be simultaneously targeted by a ligand. Another important question is whether lobe2 of K-Ras (residues 87-17220) possesses hotspot surface patches capable of interacting with other proteins or membrane. This is important because (i) a number of studies have shown that a fraction of membrane-bound K-Ras exist as dimers or higher oligomers35-41, and (ii) the possibility that Ras may interact with some of its many partners via a region other than, or in addition to, the canonical switches sw1 and sw2 (e.g.,19).
As part of our initial effort toward addressing these issues, here we used probe-based MD (pMD)42-45 simulations in conjunction with structure-based pocket identification and analyses of experimental Ras-ligand complexes. pMD is a solvent mapping approach that can be regarded as the computational equivalent of multi-solvent crystallography46 and fragment-based nuclear magnetic resonance (NMR) spectroscopy47. It entails running a regular MD simulation in the presence of probe molecules as part of the solvent environment. Its premises are (i) hotspot residues, i.e., residues that contribute the most to binding free energy, govern protein-ligand interactions, and (ii) the frequency of contact between a target site and molecular probes is proportional to the druggability of the site, a notion supported by spectroscopic studies47. pMD has three main advantages over other binding site identification methods such as FTMap48 and various other approaches (for example ref.27,49,50). First, it is able to directly account for protein motion during the site identification process. Second, it does not depend on surface descriptors or training sets. Third, it allows for a rough estimation of binding affinities.
We applied pMD on oncogenic G12D K-Ras4B (here after K-Ras) and compared the results with data from ensemble-based pocket identification with MDpocket51. We show that the previously characterized ligand binding sites are robustly reproducible, and that they are suitable targets for docking small-molecule ligands. We also show that not only can pMD guide the search for bonafide ligand binding sites, but it can also be used to find clusters of small cavities that can be exploited to improve affinity of known ligands or to design new multivalent ligands. Our results also suggest that there exist hotspots on the surface of K-Ras that have the potential to interact with other biomolecules.
Computational Methods
Molecular dynamics simulation in mixed solvent
We ran six probe-based MD (pMD) simulations using the NAMD program52 and the CHARMM27 force field53 (Table 1). In three of these the CMAP dihedral correction54 was applied whereas in the other three no CMAP was used. The starting structure for the simulations was obtained from the RCSB protein data bank (PDB, id: 4DSO29). The protein was solvated in a 72 × 72 × 96 Å3 water box containing 13319 TIP3 waters and 698 isopropanol molecules (~20:1 ratio). Crystallographic waters were treated as part of the protein structure. The system was neutralized and 40 sodium and 41 chloride ions were added to achieve an ionic strength of 0.15 M. Isopropanol was parameterized using CGenFF version 2b755,56. Simulated annealing was used to homogenize the distribution of the mixed probe/water solvent and to prevent kinetic trapping of probe molecules. The annealing involved the application of a harmonic force constant of 4 kcal/mol/Å2 on the protein heavy atoms to prevent unfolding, raising the system temperature to 650 K at intervals of 50 K, and gradual cooling by 10 K every 5000 steps to 310 K. The system was then equilibrated for 1 ns while gradually decreasing the force constant to zero. Then, starting from the equilibrated configuration we ran three production simulations for 70, 100 and 100 ns with CMAP, assigning different initial velocities. Another three simulations were run for 30, 50 and 100 ns without CMAP. We used a non-bonded cutoff of 12 Å and particle mesh Ewald57 during both the equilibration and production phases of the simulations, and coordinates were saved every 10 ps for analysis. Notice that some of our simulations are significantly longer than in previous pMD studies (~10-40 ns-long42,43,45,58) in order to capture infrequently sampled conformations.
Table 1.
Average distance between pairs of selected atoms from the simulations performed in this work*.
| CMAP correction | Simulation type | T35:Oγ-Mg2+ | T35:Oγ-GTP:Pγ | G60:Cα-G10:Cα | Y32:Oη-GTP:Oγ2 | G60:N-GTP: Oγ2 |
|---|---|---|---|---|---|---|
| yes | pMDa | 2.1 ± 0.1 | 4.4 ± 0.1 | 5.5 ± 0.2 | 3.4 ± 1.9 | 3.0 ± 0.3 |
| yes | cMD | 2.4 ± 3.8 | 4.4 ± 0.1 | 5.6 ± 0.2 | 3.1 ± 0.8 | 2.9 ± 0.2 |
| no | pMDa | 8.5 ± 3.1 | 8.8 ± 3.3 | 8.4 ± 1.2 | 16.9 ± 4.1 | 6.9 ± 0.7 |
| no | cMDb | 7.5 ± 6.2 | 9.2 ± 5.6 | 7.3 ± 1.7 | 12.3 ± 6.1 | 5.4 ± 1.9 |
Total (aggregate) simulation length: pMD with CMAP = 270 ns; cMD with CMAP = 100 ns; pMD without CMAP = 180 ns; cMD without CMAP = 200 ns. pMD = probe-based molecular dynamics; cMD = classical molecular dynamics.
Average over three trajectories.
Average over two trajectories.
For reference, we conducted three 100 ns-long classical MD simulations in the absence of probe molecules (cMD); one of these was with CMAP and the other two without CMAP (Table 1). We also ran a 50 ns pMD simulation on an isopropanol/water system without protein. The former were used to check if the probes affect Ras dynamics while the latter was used to estimate expected probe occupancies to serve as a reference for grid binding free energy calculations (eq. 1).
Binding site and affinity prediction from pMD
Binding site identification with pMD builds on previous crystallographic and nuclear magnetic resonance (NMR) observations that small organic molecules bind to druggable sites with high probability46,47. Probe-binding probabilities can therefore be used to determine binding preferences. Based on this concept and aiming at developing computational druggability index, Seco et al42 demonstrated that probe-binding probabilities from binary solvent (water plus isopropanol) MD simulations could be converted to “achievable binding affinities” of drug-like molecules. Bakan et al45 expanded this approach to a diverse set of probes as well as probe mixtures and developed a simpler protocol to quantify “maximum achievable affinities”, which we have used here. We refer the reader to their work for important details, such as the concept of “ligand efficiency”. Below we briefly describe the procedure we have followed in this work.
A grid-based approach was utilized to determine druggable pockets and estimate their maximum ligand binding affinity, as follows. (i) Trajectories from the three pMD runs were combined and aligned on the initial structure based on backbone atoms, excluding the flexible switch regions and the termini. (ii) Probe molecules with any non-hydrogen atom within 4 Å from protein heavy atoms were selected and used to calculate occupancies of cubic grids of size 1 Å, referred to as voxels. Probe occupancy was then used to calculate time-averaged number densities per voxel using the Volmap and Volutil plugins of VMD59. The resulting number density per voxel (ni) was used to calculate grid-binding free energy (ΔGgrid) using the inverse Boltzmann relation42,45:
| (1) |
n0 is the expected number density per voxel obtained from the reference simulation (probe plus water in the absence of protein, described above), R is the gas constant and T = 300K is the temperature. (iii) Grid points with ΔGgrid≤−0.5 kcal/mol were retained and the rest eliminated. We used an upper limit of −0.5 kcal/mol instead of the more common −1 kcal/mol45 because all of the known Ras ligands bind in the micromolar (μM) to millimolar (mM) range (see Results). (iv) Starting from the voxel with the most favorable grid binding free energy (lowest ΔGgrid), we defined as central voxel the one with the lowest energy and discarded all voxels within 5 Å of it. This procedure was repeated until every voxel was accounted for, and the surviving voxels were defined as interaction spots. Since the 5 Å cutoff was based on the 2.5 Å radius of isopropanol (see ref45) a single interaction spot represents a molecule of isopropanol. (v) To convert ΔGgrid to the common measure of affinity (i.e., dissociation constant (Kd)), interaction spots were clustered based upon a distance cutoff of 6 Å. Clusters of two or more interaction spots were regarded as distinct binding sites, and their maximum binding affinity or Kd was estimated as (eq. 2):
| (2) |
where x is the number of interaction spots within the cluster. (vi) Since four interaction spots represent about 16 heavy atoms (equivalent to four isopropanol molecules or roughly the size of a small (~250 Da) ligand) we defined binding sites with four or more interaction spots as druggable sites. Assuming that a viable Ras inhibitor would have at least 55 mM affinity, we defined clusters of two or three interaction spots with Kd ≤ 55 mM as sub-sites. All other interaction spots were regarded as non-specific probe-protein interactions.
Additional analyses
MDpocket51, a fast and easy to use analysis tool to identify pockets and measure their volume in structural ensembles, was used to perform structure-based ligand binding site identification. Standard procedures and tools were used to monitor system equilibration as well as the structural integrity and equilibrium fluctuations of K-Ras during the simulations. These include root-mean-square deviation (RMSD) and root-mean-square fluctuation (RMSF) measurements. Structural superimposition was performed using backbone atoms excluding the flexible sw1, sw2 and C-terminus.
Results and Discussion
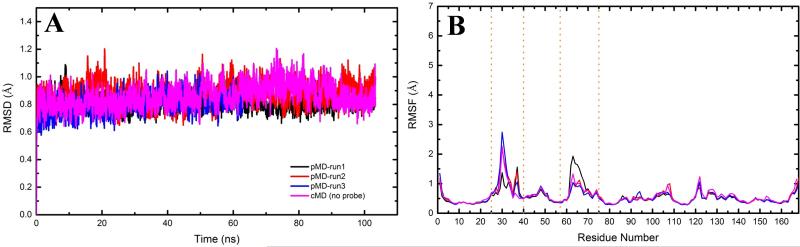
Time evolution of the backbone RMSD is nearly identical among the three pMD runs and between pMD and cMD simulations with CMAP (Fig 2A). For all of them the time-averaged RMS deviation is only ~0.8 Å excluding the flexible sw1 and sw2. The per-residue RMSFs derived from the trajectories are also essentially the same (Fig 2B). Similarly, no significant RMS differences were found among the no-CMAP simulations with and without probes (averages of 0.75-1.10 Å vs. 0.8 Å). Thus, the probes do not appear to significantly alter the overall structure of K-Ras. Moreover, comparison of the three independent pMD runs showed convergence in the sampling of protein-probe molecule interactions (discussed later). In what follows, we first describe results from the pMD simulations with CMAP and then compare cMD and pMD, followed by the simulations without CMAP.
Figure 2. K-Ras structure is stable during molecular dynamics simulations.
Shown are (A) backbone root-mean square deviation (RMSD) and (B) Cα atom root-mean square fluctuation (RMSF) for the CMAP simulations in the presence (pMD) and absence (cMD) of 20% isopropanol probe molecules in the solvent.
Identification of hotspot regions and druggable sites by pMD
We analyzed the combined trajectory of the three CMAP pMD runs to identify potential ligand binding sites and other reactive regions on the entire surface of K-Ras. Our analysis entailed dividing up the simulation box into 1 Å3 cubic grids to calculate grid binding free energies (ΔGgrid, see eq. 1). This yielded numerous grid points across the protein surface with favorable (negative) ΔGgrid of up to −2.03 kcal/mol. We used ΔGgrid to eliminate false positives, locate sites with high likelihood of binding ligands and predict their maximal affinities.
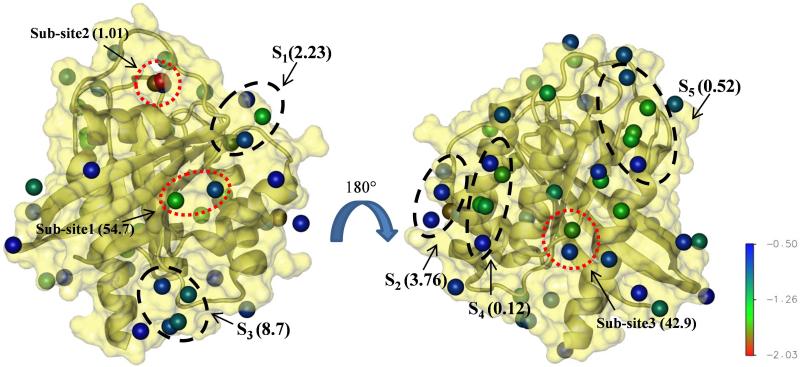
We considered all interaction spots with ΔGgrid ≤ −0.5 kcal/mol. As noted in Methods, the somewhat soft upper limit of −0.5 kcal/mol (typical is −1 kcal/mol, see ref45) was because (i) we are interested in sites even with weak ligand affinity and (ii) all Ras ligands known to date have high-μM to mM activity28-32. Applying the −0.5 kcal/mol cutoff, we ended up with 52 interaction spots. Clustering was applied on these interaction spots to determine druggable sites, and maximum affinity was calculated using eq. 2. This yielded five druggable sites (i.e., sites with four or more interaction spots, labeled as S1 to S5 in Fig 3). S1 is located in the conserved lobe1, S2 is between the lobes, and the rest are in the less conserved allosteric lobe (i.e., lobe 2). The maximum predicted binding affinities (see Fig 3) vary from high μM (S4 and S5) to low mM (S1-S3). As discussed later, S1, S2 and S3 overlap remarkably well with previously characterized allosteric ligand binding pockets (Figs 1 & 3 and, also Fig 4) whereas molecular probes have been found to bind at S425.
Figure 3. pMD predicts five potentially druggable sites on K-Ras.
Putative druggable sites S1-S5 and sub-sites 1-3 predicted by pMD are shown in black and red dotted circles, respectively. Numbers in bracket besides the labels are predicted Kd's in mM. The front view (left) is mainly lobe1 while the backside (right, after rotation by 180°) is the allosteric lobe2. Interaction spots with ΔGgrid between −2.03 and −0.5 kcal/mol are shown as balls and colored as indicated by the color bar.
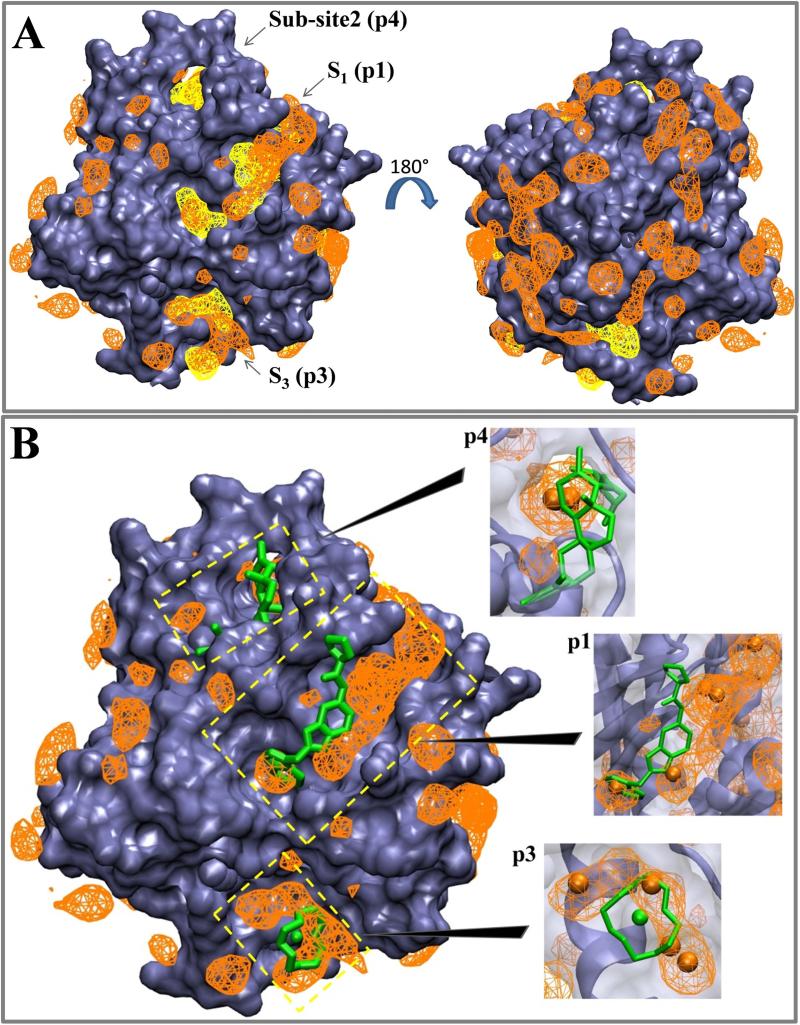
Figure 4. Structure-based pocket identification and experimental data are consistent with pMD predictions.
(A) Overlay of normalized probe number densities from pMD (orange wireframe) and binding sites derived from MDpocket (yellow). Probe number densities shown here correspond to a grid free energy range of −2.03 to −0.5 kcal/mol. For the MDpocket meshes we used an isovalue of 2.5, which is within the range recommended for detection of transient binding sites. Backbone atoms excluding the flexible switch regions were used for alignment onto the initial structure. (B) Superimposition of normalized probe number densities onto ligand-bound Ras structures. Ligands shown here include N-[2-(1H-indol-3-ylmethyl)-1H-benzimidazol- 5-yl]-L-prolinamide bound at p1 and Zn(II)1,4,7,10-tetraazaocylododecane bound at p3 (PDB ids 4EPY30 and 3L8Y28), as well as an Andrographolide derivative bound at p4 from Hocker et al, 201326. Interaction spots that make up druggable sites S1 and S3 as well as sub-site2 from Figure 3 are shown as orange balls in the close ups, which are shown as insets. We used the last snapshot of one of the pMD runs for superimposition to the ligand-bound structures.
In addition to these putative druggable sites, we also identified three sub-sites (defined as sites with two or three interaction spots and Kd ≤ 55 mM, see Methods) with predicted affinities of 54.7, 1.0 and 42.9 mM, respectively (Fig 3). The notion of sub-pockets is supported by the fact that some of the known binding sites on Ras accommodate ligands of diverse size and conformation28-33. Ras binders range from small molecules such as glycerol and metal-cyclens25,28,29 through medium-sized drug-like compounds30-33 to large macrocyclic compounds60. The latter interact across multiple sub-pockets. Even the interaction space of the drug-like Kobe family inhibitors involves the C-termini of both sw1 and sw232. Also, ligands reported by Ostrem et al33 occupy two sub-pockets within a trench broadly defined as p2 (Fig 1).
Assessing the ligand binding potential of pMD-predicted druggable sites based on geometric considerations
We used MDpocket51 to examine if the predicted druggable sites represent pockets large enough for ligand binding. As a control, MDpocket was able to correctly locate the nucleotide-binding site (see Fig 5). Note that here we used number density of probes (rather than grid free energies) for a more direct comparison with the isosurfaces/meshes generated by MDpocket. Superimposition of MDpocket-derived pockets whose volume was ≥100 Å3 in at least 50% of the structures with pMD-derived probe number densities yielded significant overlap in several places, including at S1, S3 and sub-site1 (Fig 4A). S2 was not captured by MDpocket because, first, it was open in less than 50% of the ensemble and, second, pMD-derived interaction spots near this site were mostly on the surface (see Fig 3). However, further analysis revealed that S2 becomes more accessible when the switches are more open (discussed later). Moreover, we believe S2 is a viable pocket because (i) there exist ligands that bind to this general area33 and (ii) transiently open pockets can be captured and stabilized by ligands61. In contrast, the fact that S4, S5 and sub-site3 are not captured by MDpocket (Fig 4A) suggests that they are probably on the surface of the protein and do not qualify as pockets.
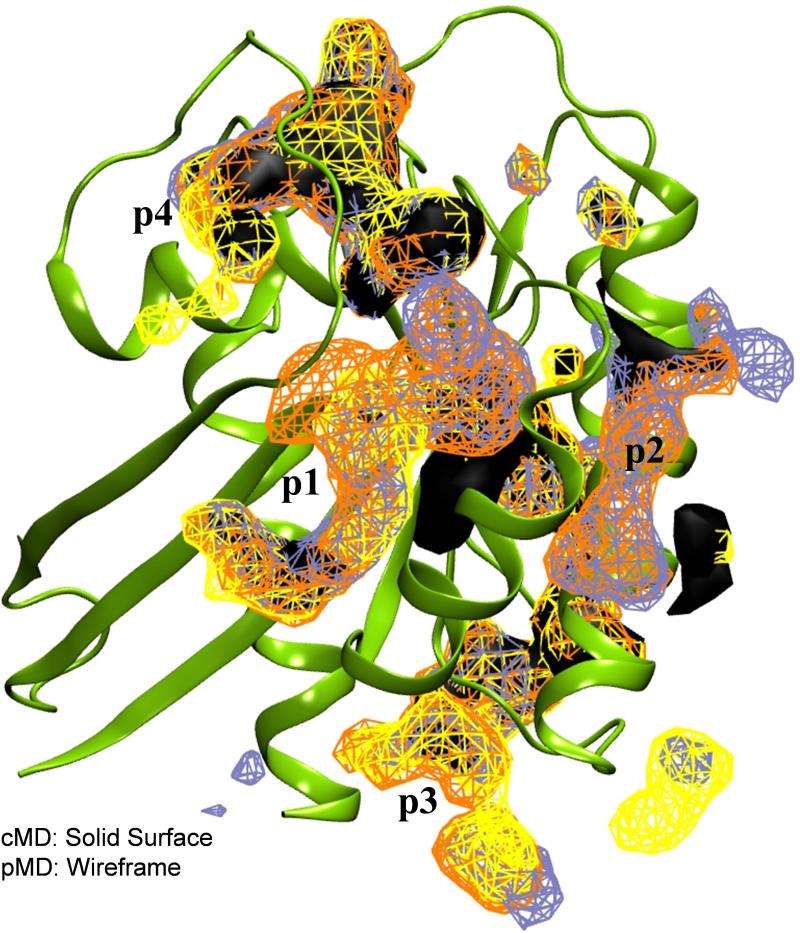
Figure 5. Probes facilitate pocket opening.
Shown are data from MDpocket analyses of cMD (black solid surface) and pMD (wireframe) trajectories. Yellow, orange and ice blue represent the three independent pMD runs. In each case, an isovalue of 2.02 was used.
Overall, we found very good overlap between druggable sites derived from the probe-based and structure-based pocket identification schemes. However, pMD appears to provide additional information regarding hotspot surface patches that MDpocket was not designed to locate.
Comparison between pMD-predicted and experimentally observed ligand binding sites
To further validate our predictions, we compared pMD-predicted druggable sites with the allosteric ligand binding sites p1 to p4 highlighted in Fig 1 (numbering based on Grant et al24). Fig 4B shows superposition of ligands bound to p1, p3 and p4 with probe number density meshes (main figure) and close ups with interaction spots (insets). The agreement between predicted druggable sites and experimentally observed (S1 + sub-site1 vs. p1; S2 vs. p2) or predicted (sub-site2 vs. p4) ligand poses is very good. Combining this with the results from MDpocket analysis, we conclude: (i) S1 + sub-site1 ≈ p1; (ii) S2 ≈ p2; (iii) S3 ≈ p3; (iv) sub-site2 ∈ p4. The current and next sections will thus focus on p1 to p4; all other sites or sub-sites are henceforth referred to as “other” interaction regions and discussed separately.
p1
Pocket p1 is located behind sw2 in the core β-sheet region. It is made up of a hydrophobic cavity involving V7, L56, M67, a polar region involving K5, D54, T74 and Y71 and an adjacent electronegative cleft near E37 and D3829,30,32. Typically, the primary binding site is found occupied by an indole or imidazole ring29,30 and the secondary binding cleft by other ring structures30. Our predicted sub-site1 overlaps exactly with the primary binding site while S1 lies in the secondary binding region (Figs 3 and 4B). The predicted Kd‘s for sub-site1 and S1 are ~55 mM and ~2 mM, respectively (Fig 3). The combined Kd of 0.12 mM is very close to the 1.5 mM Kd of benzimidazole and 4,6-dichloro-2-methyl-3-aminoethyl-indole29 or the 0.2-2 mM Kd of the ligands reported by Sun et al30.
p2
Primarily defined by V7, V9, G60, F78, M72, Q99 and I100, p2 stretches toward either of the two sub-pockets near sw2 and H3 (Figs 1, 3 and 4B). Ligands that target this general region and covalently interact with the Cys side chain of G12C Ras have been recently discovered33. However, there exist neither a p2-bound non-covalent ligand nor binding data to compare with our predicted affinity of ~4 mM (Fig 3).
p3
The polar pocket p3 is near the C-terminus involving residues from loop7 and H5, including D105, S106, D107, D108, M111, E162, Q165 and H166. There is an excellent overlap between calculated probe number densities/interaction spots and the pose of Zn(II)1,4,7,10-tetraazaocylododecane (Fig 4B). This and similar other metal cyclens have been shown to bind Ras at p3 with an apparent affinity of 2-6 mM28, which compares well with our predicted Kd of 8.7mM (Fig 3).
p4
p4 is located at the back of sw1 involving polar residues D30, D33, D38, S39 and Y40 as well as hydrophobic residues I21, I3626,31. No crystal structure of a ligand bound at this site is currently available. However, we previously used modeling and biochemical assays to show that Andrographolide derivatives target p426. Fig 4B shows that the predicted pose matches reasonably well with sub-site2 and surrounding probe densities. Moreover, NMR chemical shift perturbation and paramagnetic relaxation analyses suggested that Zn(II)-bis(2picolyl)amines bind at this general location with an apparent Kd of about 2 mM31, which is very close to the ~1 mM Kd we predict for sub-site2 (Fig 3). Note, however, that higher affinity could be found if an extended pocket was considered (see Fig 7B).
Figure 7. Predicting additional hotspot regions.
Overlay of interaction spots from pMD with (A) Ras-Sos complex (pdb: 1NVV67) and (B) membrane-bound H-Ras-GTP (from ref19). Membrane and Sos are in cyan and Ras is in yellow. Balls represent interaction spots from Fig 3.
Probe molecules facilitate pocket opening
Considering the role of solvent in protein dynamics (e.g., ref62), an important question regarding the utility of pMD is whether the probe molecules facilitate or hamper the probability of pocket opening relative to simulation in water. In other words, do the probe molecules play an active role in protein motion or are they passive spectators? The effect will likely vary with the physicochemical property of the co-solvent. In the case of isopropyl alcohol, which is small and uncharged, one would expect stabilization of both polar and non-polar surface cavities. This is because isopropanol is able to donate a hydrogen bond via its polar head and form vdW contacts via its apolar tail. In principle, therefore, it can have a measurable impact on the distribution and size of surface cavities as well as more buried pockets.
To examine this issue in some detail, we ran a 100 ns classical MD (cMD) simulation under identical condition as the pMD runs. As mentioned earlier, cMD and pMD simulations resulted in very similar overall structure and dynamics of the protein (Fig 2). We therefore decided to examine the trajectories in terms of pocket openings with MDpocket. As a control, MDpocket applied separately to the three CMAP pMD trajectories showed little variation in the location or size of the pockets despite the different lengths of the simulations (Fig 5, wireframes). This highlights the convergence of sampling for probe binding and the robustness of our analysis. Comparison of the data from pMD and cMD also shows substantial overlap in many regions (including an almost 100% overlap at the catalytic site, see Fig 5). However, there are significant differences, too. For instance, while pockets p1, p2 and p3 were visible in both cMD and pMD, p1 is fragmented in the former and does not cover the entire pocket occupied by the ligand shown in Fig 4B. Instead, cMD predicted an extension of the pocket deep into the core. Similar variations are apparent at p2 and p3 (Fig 5), where pMD yielded more expanded volumes than cMD. There also are several small pockets in the pMD but not the cMD run.
These data suggest that, while cMD/MDpocket analysis does remarkably well and is probably sufficient for most applications, pMD enables more robust pocket identification. We attribute this to the ability of the probe molecules to interact with and stabilize cavities and trenches that would otherwise be inaccessible. This is important not only because such information could aid in probe selection but also due to its fundamental implication that some drug-like ligands may exert a similar effect on their target.
Enhanced protein motion facilitates detection of hotspots by pMD
As noted earlier, each of the four allosteric ligand-binding sites on Ras were initially predicted by computational approaches24,25. A common feature of pMD and the previous computational analyses is their emphasis on protein motion, which is critical because most of the allosteric pockets on Ras become visible only upon relaxation of the protein by MD or NMR26,28. In particular, opening of p2 and p4 requires rearrangement of the switches and therefore they are accessible only in some GDP-bound conformations or in one of the two GTP-bound forms26,31,33. Perhaps as a result of this, no crystal structure is currently available with a non-covalent ligand bound to these pockets. It is therefore reasonable to suspect that limited sampling may explain why pMD identified a sub-pocket and not a druggable site at p4 (Fig 3).
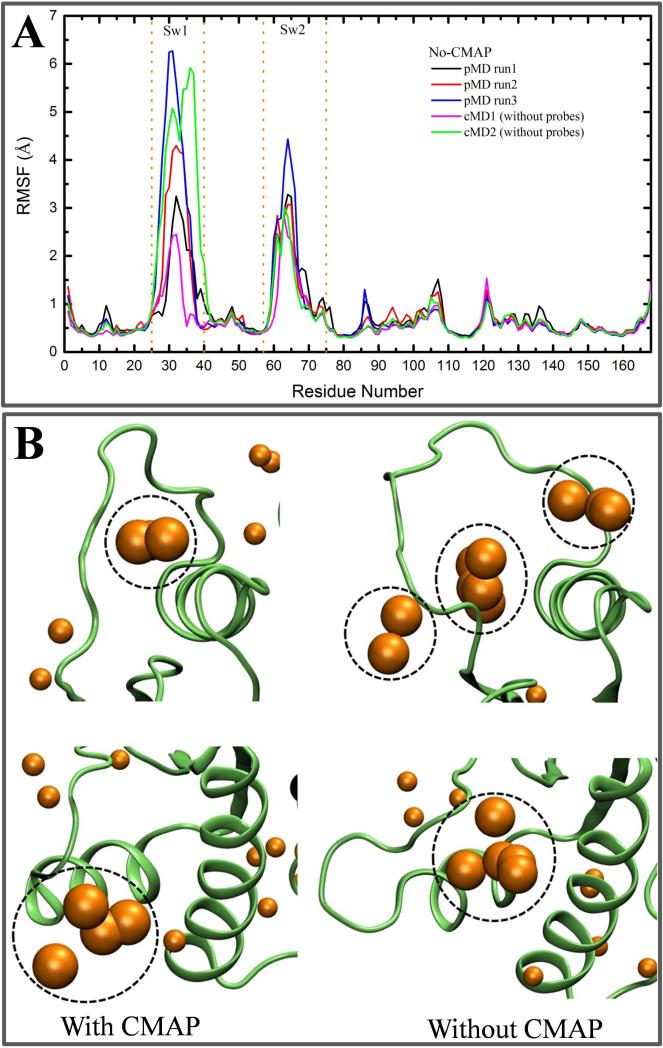
To test if our prediction of p4 would improve upon enhanced sampling, we took advantage of the fact that simulations with the CHARMM force field in the absence of CMAP often result in larger flexibility in loop regions54,63. To this end, we ran three probe-based and two normal MD simulations without CMAP correction. (We note that no-CMAP runs could yield unphysical structures; they are used here only for test of concept.) The simulations were stable and equilibrated within 10-40 ns with a mean backbone RMSD of 0.75-1.2 Å. However, comparison of the residue RMSFs from these simulations with those from the CMAP runs (Figs 2B vs. 6A) clearly shows enhanced fluctuations at either one or both of the switches. To quantify the degree to which the switch loops have opened, we monitored distances of Y32, T35 and G60 from the bound Mg2+ and GTP. This was because 31P-NMR and other biophysical studies have shown that GTP-Ras exists in at least two conformational sub-states that primarily differ in the way in which Y32, T35 and G60 are organized relative to the nucleotide16,64,65. Table 1 shows that simulations without CMAP indeed lead to a significantly more open switch conformation (state 1-like).
Figure 6. Enhanced flexibility facilitates opening of some pockets.
(A) Cα atom RMSF from two cMD and three pMD simulations without the CMAP dihedral correction. (B) Conformation and interaction spots around binding sites p4 (top) and p2 (bottom) derived from pMD simulations with (left) and without CMAP (right). Dotted black circles are the same as in Figure 3. The calculated grid free energies range from −1.57 to −0.50 kcal/mol for the simulations without CMAP and −2.03 to −0.50 kcal/mol for pMD with CMAP. The most flexible pMD simulation (blue in A) was used to calculate the grid binding free energies for the simulation without CMAP.
Since run 3 of the no-CMAP pMDs yielded the largest flexibility (Fig 6A), we analyzed interaction spots from this trajectory to examine if enhanced dynamics could lead to improved “druggability” prediction for some sites. We found that overall the CMAP and no-CMAP pMD yielded a similar distribution of interaction spots, especially in lobe 2 (not shown). However, there are some crucial differences around the switch loops. In particular, the trajectory without CMAP showed additional interaction spots (sub-pockets) near sw1 (Fig 6B). We believe these additional sub-pockets and sub-site2 are part of a true ligand-binding site (i.e., pocket p4). Fig 6B also shows some difference between CMAP and no-CMAP in that the location of interaction spots at p2 is deeper in the latter, suggesting that p2 has become slightly more pocket-like when the switches are more open. Taken together, these results imply that better sampling could facilitate binding site identification by pMD when pocket opening requires significant structural rearrangements.
Prediction of “other” hotspot surface patches
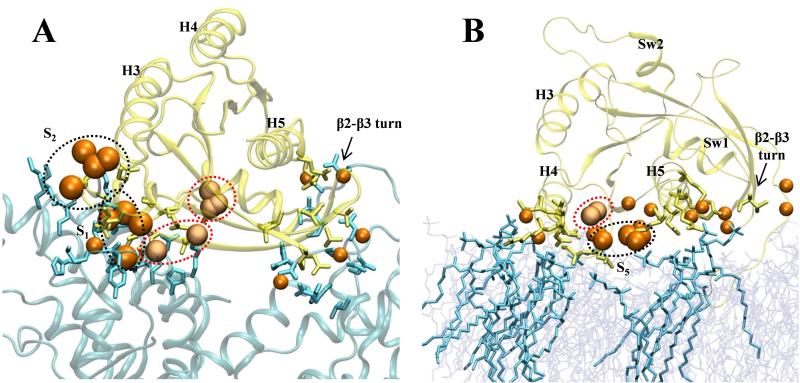
We now turn to the “other” pMD-predicted sites, which include S4 located between H3 and H4 and S5 at the N-terminus of H5 (Fig 3). The predicted maximum binding affinities for these sites are comparable to those of S1-S3 (Fig 3). However, no drug-like molecule that targets S4 has been reported, although small organic compounds such as dimethylformamide and glycerol were found to bind at this location25. Similarly, S5 has not previously been characterized as a ligand-binding site, apart from its ability to stabilize internal water molecules62. Moreover, MDpocket analysis (Figs 4A) suggested that these surfaces are likely unsuitable for ligand binding.
S4, S5 and sub-site3 are at the backside of the protein where there are many other interaction spots that did not satisfy our criteria for druggable site or sub-site. Another curious surface patch is the one near the β2-β3 turn where there is a cluster of interaction spots (Fig 3, bottom left of left panel). This turn is part of the so-called switch 3 and home to D47 and E49 whose mutation to alanine has been shown to result in a hyperactive H-Ras variant66. We thus hypothesized that S4 and S5 along with other proximal interaction spots, including those near the β2-β3 turn, may represent surfaces important for either dimerization or/and interaction with other biomolecules.
As an initial test of this hypothesis, we overlaid pMD-derived structures harboring these interaction spots with the crystal structure of the H-Ras:Sos complex (pdb id: 1NVV67). This revealed some overlap between the interaction spots at β2-β3 and part of the Sos surface that interacts with Ras (Fig 7A). However, since Ras interacts with Sos primarily via the switch loops, interaction spots at p1 and p2 dominate. To further test our hypothesis that the “other” hotspots are important for interaction with biomolecules, we overlaid the pMD-derived structure with a membrane-bound GTP-H-Ras (taken from ref19). There is a significant overlap in the interaction spots within S5 and sub-site3 as well as those at β2-β3 turn and H4 (Fig 7B). Finally, initial tests suggest that S4 might play an important role in Ras oligomerization (not shown). Although by no means conclusive, these observations suggest that pMD may be used to gain an initial qualitative picture of protein-protein and protein-membrane interfaces. It may also be possible to develop empirical relationships between probe occupancy and surface (often polar) residue's contribution to biomolecular binding in much the same way as Kd could be derived from ΔGgrid for small molecule binding (e.g.,42,45). If successful, this could expand the scope of pMD from predicting ligand-binding sites to predicting protein-protein/protein-membrane interfaces.
Conclusion
Ras has long been thought undruggable in part because there were no readily observable allosteric ligand-binding sites or interfaces on the structure17,68,69. A major goal of the current work was to identify ligand-binding sites and other cavities on G12D K-Ras to assist ongoing drug discovery efforts. Our additional goal was to obtain clues about previously uncharacterized surface patches that Ras may use to interact with other biomolecules.
To achieve these goals, we used the relatively fast and tractable probe-based MD. pMD is a better alternative to other approaches because it directly accounts for conformational flexibility, which is particularly important for identifying ligand-binding sites on flexible targets such as Ras proteins. Since the method has not been tested extensively, we carefully compared the predicted druggable sites and their affinities with literature data as well as recently reported structures of ligand/Ras complexes. In addition, we compared the pMD-predicted pockets with those from structure-based pocket identification schemes. We found that the method is robust and efficient, and yields information that could not be obtained by structure analysis alone. We also found that in some cases enhanced sampling and the probe molecules themselves facilitate pocket opening.
Regarding K-Ras, the consensus outcome of the current and previous studies is that it has four allosteric ligand-binding sites suitable for structure-based drug design. The probe-based simulations also suggest additional reactive surfaces that might be involved in protein-protein or protein-membrane interactions. These results highlight the potential of probe-based MD simulation not only as a realistic means of identifying ligand binding sites but also-- upon further development--as a more general method for searching the surface of biomolecules for any reactive region.
Acknowledgment
PP is supported by a postdoctoral training fellowship from the Keck center Computational Cancer Biology Training Program of the Gulf Coast Consortia (CPRIT Grant No. RP140113). We gratefully acknowledge the Texas Advanced Computing Center (TACC) for computational resources. This work was supported in part by grant from the National Institutes of Health General Medical Sciences (Grant No. R01GM100078).
References
- 1.Barbacid M. Ras genes. Annu Rev Biochem. 1987;56:779–827. doi: 10.1146/annurev.bi.56.070187.004023. [DOI] [PubMed] [Google Scholar]
- 2.Karnoub AE, Weinberg RA. Ras oncogenes: split personalities. Nat Rev Mol Cell Biol. 2008;9:517–531. doi: 10.1038/nrm2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox AD, Der CJ. Ras history: the saga continues. Small GTPases. 2010;1:2–27. doi: 10.4161/sgtp.1.1.12178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hancock JF, Peterson H, Marshall CJ. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell. 1990;63:133–139. doi: 10.1016/0092-8674(90)90294-o. [DOI] [PubMed] [Google Scholar]
- 5.Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49:4682–4689. [PubMed] [Google Scholar]
- 6.Pylayeva-Gupta Y, Grabocka E, Bar-Sagi D. Ras oncogenes: weaving a tumorigenic web. Nat Rev. 2011;11:761–774. doi: 10.1038/nrc3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prior IA, Lewis PD, Mattos C. A comprehensive survey of Ras mutations in cancer. Can Res. 2012;72:2457–2467. doi: 10.1158/0008-5472.CAN-11-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Forrester K, Almoguera C, Han K, Grizzle WE, Perucho M. Detection of high incidence of K-ras oncogenes during human colon tumorigenesis. Nature. 1987;327:298–303. doi: 10.1038/327298a0. [DOI] [PubMed] [Google Scholar]
- 9.Almoguera C, Shibata D, Forrester K, Martin J, Arnheim N, Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 10.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, Hong SM, Fu B, Lin MT, Calhoun ES, Kamiyama M, Walter K, Nikolskaya T, Nikolsky Y, Hartigan J, Smith DR, Hidalgo M, Leach SD, Klein AP, Jaffee EM, Goggins M, Maitra A, Iacobuzio-Donahue C, Eshleman JR, Kern SE, Hruban RH, Karchin R, Papadopoulos N, Parmigiani G, Vogelstein B, Velculescu VE, Kinzler KW. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 12.Gorfe AA. Mechanisms of allostery and membrane attachment in Ras GTPases: implications for anti-cancer drug discovery. Curr Med Chem. 2010;17:1–9. doi: 10.2174/092986710789957832. [DOI] [PubMed] [Google Scholar]
- 13.Baines AT, Xu D, Der CJ. Inhibition of Ras for cancer treatment: the search continues. Future Med Chem. 2011;3:1787–1808. doi: 10.4155/fmc.11.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- 15.Vo U, Embrey KJ, Breeze AL, Golovanov AV. 1H, 13C, and 15N resonance assignment for the human K-Ras at physiological pH. Biomol NMR Assign. 2013;7:215–219. doi: 10.1007/s12104-012-9413-y. [DOI] [PubMed] [Google Scholar]
- 16.Araki M, Shima F, Yoshikawa Y, Muraoka S, Ijiri Y, Nagahara Y, Shirono T, Kataoka T, Tamura A. Solution stucture of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers. J Biol Chem. 2011;286:39644–39653. doi: 10.1074/jbc.M111.227074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prakash P, Gorfe AA. Lessons from computer simulations of Ras proteins in solution and in membrane. Biochim Biophys Acta. 2013;1830:5211–5218. doi: 10.1016/j.bbagen.2013.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prakash P, Gorfe AA. Overview of simualtion studies on the enzymatic activity and conformational dynamics of the GTPase Ras. Mol Sim. 2014;40:839–847. doi: 10.1080/08927022.2014.895000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorfe AA, Hanzal-Bayer M, Abankwa D, Hancock JF, McCammon JA. Structure and dynamics of the full-length lipid-modified H-Ras protein in a 1,2-dimyristoylglycero-3- phosphocholine bilayer. J Med Chem. 2007;50:674–684. doi: 10.1021/jm061053f. [DOI] [PubMed] [Google Scholar]
- 20.Gorfe AA, Grant BJ, McCammon JA. Mapping the nucleotide and isoform-dependent structural and dynamical features of Ras proteins. Structure. 2008;16:885–896. doi: 10.1016/j.str.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grant BJ, Gorfe AA, McCammon JA. Ras conformational switching: simulating nucleotide-dependent conformational transitions with accelerated molecular dynamics. PLoS Comput Biol. 2009;5:e1000325. doi: 10.1371/journal.pcbi.1000325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buhrman G, Holzapfel G, Fetics S, Mattos C. Allosteric modulation of Ras positions Q61 for a direct role in catalysis. Proc Natl Acad Sci U S A. 2010;107:4931–4936. doi: 10.1073/pnas.0912226107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grant BJ, McCammon JA, Gorfe AA. Conformational selection in G proteins: Lessons from Ras and Rho. Biophys J. 2010;99:L87–L89. doi: 10.1016/j.bpj.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant BJ, Lukman S, Hocker HJ, Sayyah J, Brown JH, McCammon JA, Gorfe AA. Novel allosteric sites on Ras for lead generation. PLoS One. 2011;6:e25711. doi: 10.1371/journal.pone.0025711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buhrman G, O'Connor C, Zerbe B, Kearney BM, Napoleon R, Kovrigina EA, Vajda S, Kozakov D, Kovrigin EL, Mattos C. Analysis of Binding Site Hot Spots on the Surface of Ras GTPase. J Mol Biol. 2011;413:773–789. doi: 10.1016/j.jmb.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hocker HJ, Cho KJ, Chen CY, Rambahal N, Sagineedu SR, Sharri K, Stanslas J, Hancock JF, Gorfe AA. Andrographolide derivatives inhibit guanine nucleotide exchange and abrogate oncogenic Ras function. Proc Natl Acad Sci U S A. 2013;110:10201–10206. doi: 10.1073/pnas.1300016110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hocker HJ, Rambahal N, Gorfe AA. LIBSA--a method for the determination of ligand- binding preference to allosteric sites on receptor ensembles. J Chem Inf Model. 2014;54:530–538. doi: 10.1021/ci400474u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosnizeck IC, Graf T, Spoerner M, Tränkle J, Filchtinski D, Herrmann C, Gremer L, Vetter IR, Wittinghofer A, König B, Kalbitzer HR. Stabilizing a Weak Binding State for Effectors in the Human Ras Protein by Cyclen Complexes. Angew Chem Int Ed Engl. 2010;49:3830–3833. doi: 10.1002/anie.200907002. [DOI] [PubMed] [Google Scholar]
- 29.Maurer T, Garrenton LS, Oh A, Pitts K, Anderson DJ, Skelton NJ, Fauber BP, Pan B, Malek S, Stokoe D, Ludlam MJ, Bowman KK, Wu J, Giannetti AM, Starovasnik MA, Mellman I, Jackson PK, Rudolph J, Wang W, Fang G. Small-molecule ligands bind to a distinct pocket in Ras and inhibit SOS-mediated nucleotide exchange activity. Proc Natl Acad Sci U S A. 2012;109:5299–5304. doi: 10.1073/pnas.1116510109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun Q, Burke JP, Phan J, Burns MC, Olejniczak ET, Waterson AG, Lee T, Rossanese OW, Fesik SW. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosnizeck IC, Spoerner M, Harsch T, Kreitner S, Filchtinski D, Herrmann C, Engel D, König B, Kalbitzer HR. Metal–Bis(2-picolyl)amine Complexes as State 1(T) Inhibitors of Activated Ras Protein. Angew Chem Int Ed Engl. 2012;51:10647–10651. doi: 10.1002/anie.201204148. [DOI] [PubMed] [Google Scholar]
- 32.Shima F, Yoshikawa Y, Ye M, Araki M, Matsumoto S, Liao J, Hu L, Sugimoto T, Ijiri Y, Takeda A, Nishiyama Y, Sato C, Muraoka S, Tamura A, Osoda T, Tsuda K-I, Miyakawa T, Fukunishi H, Shimada J, Kumasaka T, Yamamoto M, Kataoka T. In silico discovery of small-molecule Ras inhibitors that display antitumor activity by blocking the Ras– effector interaction. Proc Natl Acad Sci U S A. 2013;110:8182–8187. doi: 10.1073/pnas.1217730110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ostrem JM, Peters U, Sos ML, Wells JA, Shokat KM. K-Ras(G12C) inhibitors allosterically control GTP affinity and effector interactions. Nature. 2013;503:548–551. doi: 10.1038/nature12796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Q, Phan J, Friberg AR, Camper DV, Olejniczak ET, Fesik SW. A method for the second-site screening of K-Ras in the presence of a covalently attached first-site ligand. J Biomol NMR. 2014;60:11–14. doi: 10.1007/s10858-014-9849-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inouye K, Mizutani S, Koide H, Kaziro Y. Formation of the Ras Dimer Is Essential for Raf-1 Activation. J Biol Chem. 2000;275:3737–3740. doi: 10.1074/jbc.275.6.3737. [DOI] [PubMed] [Google Scholar]
- 36.Tian T, Harding A, Inder K, Plowman S, Parton RG, Hancock JF. Plasma membrane nanoswitches generate high-fidelity Ras signal transduction. Nat Cell Biol. 2007;9:905–914. doi: 10.1038/ncb1615. [DOI] [PubMed] [Google Scholar]
- 37.Abankwa D, Gorfe AA, Hancock JF. Ras nanoclusters: molecular structure and assembly. Semin Cell Dev Biol. 2007;18:599–607. doi: 10.1016/j.semcdb.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Güldenhaupt J, Rudack T, Bachler P, Mann D, Triola G, Waldmann H, Kötting C, Gerwert K. N-Ras Forms Dimers at POPC Membranes. Biophys J. 2012;103:1585–1593. doi: 10.1016/j.bpj.2012.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cho KJ, Hancock JF. Ras nanoclusters: a new drug target? Small GTPases. 2013;4:57–60. doi: 10.4161/sgtp.23145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin WC, Iversen L, Tu HL, Rhodes C, Christensen SM, Iwig JS, Hansen SD, Huang WY, Groves JT. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc Natl Acad Sci U S A. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Hancock JF. Ras nanoclusters: Versatile lipid-based signaling platforms. Biochim Biophys Acta. 2014;S0167-4889:00333–4. doi: 10.1016/j.bbamcr.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 42.Seco J, Luque FJ, Barril X. Binding site detection and druggability index from first principles. J Med Chem. 2009;52:2363–71. doi: 10.1021/jm801385d. [DOI] [PubMed] [Google Scholar]
- 43.Guvench O, MacKerell AD. Computational Fragment-Based Binding Site Identification by Ligand Competitive Saturation. PLoS Comput Biol. 2009;5:e1000435. doi: 10.1371/journal.pcbi.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raman EP, Yu W, Guvench O, MacKerell AD. Reproducing Crystal Binding Modes of Ligand Functional Groups Using Site-Identification by Ligand Competitive Saturation (SILCS) Simulations. J Chem Inf Model. 2011;51:877–896. doi: 10.1021/ci100462t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bakan A, Nevins N, Lakdawala AS, Bahar I. Druggability Assessment of Allosteric Proteins by Dynamics Simulations in the Presence of Probe Molecules. J Chem Theory Comput. 2012;8:2435–2447. doi: 10.1021/ct300117j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Allen KN, Bellamacina CR, Ding X, Jeffery CJ, Mattos C, Petsko GA, Ringe D. An Experimental Approach to Mapping the Binding Surfaces of Crystalline Proteins. J Phys Chem. 1996;100:2605–2611. [Google Scholar]
- 47.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-based screening data. J Med Chem. 2005;48:2518–25. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 48.Brenke R, Kozakov D, Chuang GY, Beglov D, Hall D, Landon MR, Mattos C, Vajda S. Fragment-based identification of druggable ‘hot spots’ of proteins using Fourier domain correlation techniques. Bioinformatics. 2009;25:621–627. doi: 10.1093/bioinformatics/btp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interaction. J. Mol. Graph. 1995;13:323–330. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 50.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmidtke P, Bidon-Chanal A, Luque FJ, Barril X. MDpocket: open-source cavity detection and characterization on molecular dynamics trajectories. Bioinformatics. 2011;27:3276–3285. doi: 10.1093/bioinformatics/btr550. [DOI] [PubMed] [Google Scholar]
- 52.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FT, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. All- Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins. J Phys Chem B. 1998;102:586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 54.Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 55.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu W, He X, Vanommeslaeghe K, MacKerell AD. Extension of the CHARMM general force field to sulfonyl-containing compounds and its utility in biomolecular simulations. J Comput Chem. 2012;33:2451–2468. doi: 10.1002/jcc.23067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darden T, York D, Pedersen L. Particle mesh ewald: An Nlog(N) method for Ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 58.Tan YS, Śledź P, Lang S, Stubbs CJ, Spring DR, Abell C, Best RB. Using Ligand- Mapping Simulations to Design a Ligand Selectively Targeting a Cryptic Surface Pocket of Polo-Like Kinase 1. Angew Chem Int Ed Engl. 2012;51:10078–10081. doi: 10.1002/anie.201205676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 60.Upadhyaya P, Qian Z, Habir NA, Pei D. Direct Ras inhibitors identified from a structurally rigidified bicyclic peptide library. Tetrahedron. 2014;70:7714–7720. doi: 10.1016/j.tet.2014.05.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lee GM, Craik CS. Trapping moving targets with small molecules. Science. 2009;324:213–215. doi: 10.1126/science.1169378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Prakash P, Sayyed-Ahmad A, Gorfe AA. The role of conserved waters in conformational transitions of Q61H K-ras. PLoS Comput Biol. 2012;8:e1002394. doi: 10.1371/journal.pcbi.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buck M, Bouguet-Bonnet S, Pastor RW, MacKerell AD. Importance of the CMAP correction to the CHARMM22 protein force field: dynamics of hen lysozyme. Biophys J. 2006;90:L36–L38. doi: 10.1529/biophysj.105.078154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shima F, Ijiri Y, Muraoka S, Liao J, Ye M, Araki M, Matsumoto K, Yamamoto N, Sugimoto T, Yoshikawa Y, Kumasaka T, Yamamoto M, Tamura A, Kataoka T. Structural basis for conformational dynamics of GTP-bound Ras protein. J Biol Chem. 2010;285:22696–22705. doi: 10.1074/jbc.M110.125161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Spoerner M, Hozsa C, Poetzl JA, Reiss K, Ganser P, Geyer M, Kalbitzer HR. Conformational States of Human Rat Sarcoma (Ras) Protein Complexed with Its Natural Ligand GTP and Their Role for Effector Interaction and GTP Hydrolysis. J Biol Chem. 2010;285:39768–39778. doi: 10.1074/jbc.M110.145235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Abankwa D, Hanzal-Bayer M, Ariotti N, Plowman SJ, Gorfe AA, Parton RG, McCammon JA, Hancock JF. A novel switch region regulates H-ras membrane orientation and signal output. EMBO J. 2008;27:727–735. doi: 10.1038/emboj.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Margarit SM, Sondermann H, Hall BE, Nagar B, Hoelz A, Pirruccello M, Bar-Sagi D, Kuriyan J. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 68.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 69.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: mission possible?. Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]