Abstract
We previously reported that the SbROMT3syn recombinant protein catalyzes the production of the methylated resveratrol derivatives pinostilbene and pterostilbene by methylating substrate resveratrol in recombinant E. coli. To further study the production of stilbene compounds in E. coli by the expression of enzymes involved in stilbene biosynthesis, we isolated three stilbene synthase (STS) genes from rhubarb, peanut, and grape as well as two resveratrol O-methyltransferase (ROMT) genes from grape and sorghum. The ability of RpSTS to produce resveratrol in recombinant E. coli was compared with other AhSTS and VrSTS genes. Out of three STS, only AhSTS was able to produce resveratrol from p-coumaric acid. Thus, to improve the solubility of RpSTS, VrROMT, and SbROMT3 in E. coli, we synthesized the RpSTS, VrROMT and SbROMT3 genes following codon-optimization and expressed one or both genes together with the cinnamate/4-coumarate:coenzyme A ligase (CCL) gene from Streptomyces coelicolor. Our HPLC and LC-MS analyses showed that recombinant E. coli expressing both ScCCL and RpSTSsyn led to the production of resveratrol when p-coumaric acid was used as the precursor. In addition, incorporation of SbROMT3syn in recombinant E. coli cells produced resveratrol and its mono-methylated derivative, pinostilbene, as the major products from p-coumaric acid. However, very small amounts of pterostilbene were only detectable in the recombinant E. coli cells expressing the ScCCL, RpSTSsyn and SbROMT3syn genes. These results suggest that RpSTSsyn exhibits an enhanced enzyme activity to produce resveratrol and SbROMT3syn catalyzes the methylation of resveratrol to produce pinostilbene in E. coli cells.
Keywords: metabolic engineering, pinostilbene, resveratrol, resveratrol O-methyltransferase, stilbene synthase
INTRODUCTION
Resveratrol (3,5,4′-trihydroxy-trans-stilbene) is a polyphenolic phytoalexin produced by a few plant species, such as grapes, peanuts, and berries, under biotic or abiotic stress conditions such as pathogen infection and UV irradiation. Resveratrol has diverse properties beneficial to health, including anti-inflammatory effects, anti-tumor activity, and anti-aging effects (Baur et al., 2006; Frémont, 2000; Ulrich et al., 2005). However, the beneficial effects of resveratrol are limited due to its instability when exposed to light and oxygen or in environments with harsh pH conditions. These stimuli may cause trans-to-cis isomerization or oxidation that leads to a reduction in the bioavailability and bioactivity of the compound (Walle et al., 2004). For this reason, it is important to develop resveratrol derivatives with enhanced stability. Furthermore, structure activity studies revealed that the substitution of hydroxyl groups of resveratrol with methoxy groups substantially potentiate its cytotoxic activity (Lee et al., 2003). The substitution of hydroxy with methoxy groups may give methylated resveratrol derivatives an increased lipophilicity compared to resveratrol, resulting in better bioavailability (Remsberg et al., 2008). For these reasons, the metabolic engineering of resveratrol and its methylated derivatives in plants and microbes is of great interest for the development of more stable and potent chemoprotective agents. There are various naturally occurring methylated derivatives, including pinostilbene (3,4′-dihydroxy-5-methoxy-trans-stilbene), pterostilbene (3,5-dimethoxy-4′-hydroxy-trans-stilbene), 3,4′,5-trimethoxystilbene, and desoxyrhapotigenin (3,5-dihydroxy-4′-methoxy-trans-stilbene) (Wang et al., 2010), but their biological activities remain largely unknown. One effective approach for stabilization of resveratrol is to produce methylated resveratrol derivatives via enzymatic O-methyltransaferases (OMTs). Plant OMTs constitute a large family of enzymes that methylate the hydroxyl groups of a variety of secondary metabolites including phenylpropanoids, flavonoids, and alkaloids. However, a few plant OMT genes have been isolated and characterized as possible resveratrol OMTs (ROMT) (Rimando et al., 2012; Schmidlin et al, 2008). We previously reported that SbROMT3syn catalyzed the methylation of resveratrol, yielding pinostilbene as the major product alongside pterostilbene as a minor product (Jeong et al., 2014). More studies are required to determine the enzymatic functions of various plant OMTs using biochemical approaches. Rhubarb (Rheum) species have been used as medicinal plants in Asian traditional medicine to treat constipation, inflammation and cancer (Foust, 1992; Lee et al., 2012). They are rich in polyketides, including stilbenes, phenylbutanoids, anthraquinones, and naphthalenes (Kashiwada et al., 1988). Six stilbene derivatives such as desoxyrhapontigenin, rhapontigenin, trans-resveratrol, piceatannol, piceatannol-3′-O-β-d-glucopyrano-side, and isorhapontin were isolated from rhubarb rhizomes. These stilbene derivatives showed a good anti-inflammatory activity (Lee et al., 2012). However, little is known about stilbene biosynthesis in rhubarb plants. Furthermore, there are still no reports on metabolic engineering of stilbene biosynthesis in microbes using stilbene biosynthetic genes from rhubarb plants. In plants, resveratrol is synthesized by STS action as a branch from the phenylpropanoid pathway (Sparvoli et al., 1994). As an intermediate, p-coumaric acid is converted to p-coumaroyl-CoA by 4-coumrate:coenzyme A ligase (4CL). STS catalyzes the condensation of one molecule of p-coumaroyl-CoA and three molecules of malonyl-CoA as substrates. Methylated resveratrol derivatives of pinostilbene and pterostilbene are produced by ROMT from resveratrol (Fig. 1). Extensive metabolic engineering studies have been carried out to increase the production of resveratrol in plants and microbes (Becker et al., 2003; Beekwilder et al., 2006; Delaunois et al., 2009; Horinouchi, 2009; Katsuyama et al., 2007; Melchior and Kindl, 1990; Ververidis et al., 2007; Watts et al., 2006; Zhang et al., 2006). However, little is known about the production of methylated resveratrol derivatives in recombinant microorganisms.
Fig. 1.
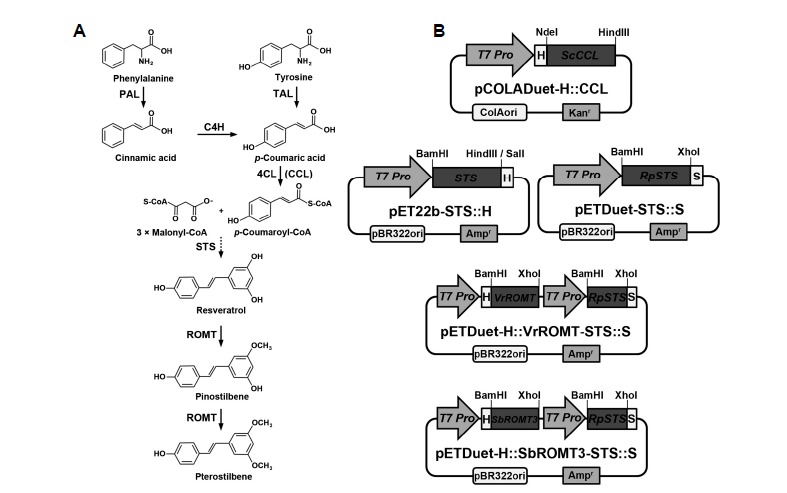
(A) Biosynthetic pathway of stilbene compound production from phenylalanine and (B) construction of recombinant plasmids carrying the genes (ScCCL, STS, the synthetic RpSTSsyn, VrROMTsyn, and SbROMT3syn) involved in stilbene biosynthesis. The sequential actions of PAL or TAL, C4H, 4CL (CCL), STS, and ROMT result in the conversion of phenylalanine to stilbenes, resveratrol, and its methylated derivatives. PAL, phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase; TAL, tyrosine ammonia-lyase; 4CL, 4-coumarate:coenzyme A ligase; CCL, cinnamate/4-coumarate:coenzyme A ligase; STS, stilbene synthase; ROMT, resveratrol O-methyltransferase; T7 Pro, T7 RNA polymerase promoter; H, His-tag; S, S-tag.
In the present study, we describe the production of resveratrol and its mono-methylated derivative pinostilbene as the major product from p-coumaric acid in E. coli through co-expression of multiple enzymes (CCL, STS, ROMT) responsible for stilbene biosynthesis. When p-coumaric acid was fed as the precursor, maximum levels of resveratrol (∼1.9 mg/L) and pinostilbene (∼2.4 mg/L) were produced by recombinant E. coli cells co-expressing ScCCL and RpSTSsyn, and ScCCL, RpSTSsyn and SbROMT3syn, respectively. These results suggest that RpSTSsyn is functional as a stilbene synthase and SbROMT3syn can be used as an enzyme to produce pinostilbene by methylating resveratrol in microorganisms.
MATERIALS AND METHODS
Materials
The E. coli strains DH5alpha and BL21-CodonPlus (DE3)-RIPL were used for cloning and expression, respectively. The Duet vectors of pETDuet-1 and pCOLADuet-1, pET-22b(+), and BL21-CodonPlus (DE3)-RIPL were purchased from Novagen. E. coli strains were cultured in Luria-Bertani (LB) medium and on agar supplemented with 50 μg/ml ampicillin or kanamycin. Pinostilbene and pterostilbene were purchased from Chromadex and resveratrol was purchased from Sigma-Aldrich. trans-p-Coumaric acid was purchased from Tokyo Chemical Industry. Methanol, acetonitrile, ethylacetate, and water were of HPLC grade and purchased from Burdick & Jackson.
DNA synthesis and manipulation
DNA manipulation and E. coli transformation protocols used during the preparation of plasmid vectors for recombinant CCL, STS, and ROMT experiments were carried out according to standard procedures (Sambrook et al., 1989). Full-length cDNAs for RpSTS, AhSTS, VrSTS, VrROMT and SbROMT3 were amplified from rhubarb (Rheum palmatum), peanut (Arachis hypogaea), grape (Vitis riparia), and sorghum (Sorghum bicolor) by homology-based PCR using PCR primers based on the RtSTS1 (accession number: AF508150), AhSTS (accession number: EU384706), VvSTS (accession number: EU156062), VvROMT (accession number: FM178870) and SbOMT3 (accession number: EF189708) genes, as previously described by Jeong et al. (2014). The primers were as follows: RpSTS-F (5′-TTAACATATCTGCTAGAGATGGCA-3′), RpSTS-R (5′-CAAGTTATTCAATGGTTTTCAGGT-3′); AhSTS-F (5′-ATGGTGTCTGTGAGTGGAATTCGC-3′), AhSTS-R (5′-TTATATGGCCACACTGCGGAGAAC-3′; VrSTS-F (5′-ATGGCTTCAGTTGAGGAAATCAGA-3′; VrSTS-R (5′-TTAATTTGTCACCATAGGAATGCTA-3′). The ScCCL gene was amplified from the actinomyces Streptomyces coelicolor as described by Choi et al. (2011). The obtained RpSTS, AhSTS, VrSTS, VrROMT, SbROMT3 and ScCCL genes were confirmed by nucleotide sequencing. For the expression of recombinant STS proteins in E. coli, the original STS genes were subcloned into the BamHI and HindIII or SalI sites of the pET-22b(+) bacterial expression vector to obtain pET22b-STS::H constructs (Fig. 1B). Furthermore, to enhance protein expression and solubility in E. coli, RpSTSsyn, VrROMTsyn and SbROMT3syn were synthesized following codon-optimization by replacing the plant codons with bacterial-preferred codons according to the manufacturer’s indications (Genscript). To facilitate the process of insert cloning into the pETDuet-1 bacterial expression vector, BamHI and XhoI restriction enzyme sites were introduced into the RpSTSsyn, VrROMTsyn and SbROMT3syn sequences. The codon-optimized and synthetic RpSTSsyn gene was cloned between the BglII and XhoI sites, and the VrROMTsyn and SbROMT3syn genes were introduced into the BamHI and SalI sites of the dual expression vector pETDuet-1. The ScCCL gene was cloned into the NdeI and HindIII sites of the dual expression vector pCOLADuet-1 (Fig. 1B). As a result, a His-tag was fused to the N-terminus of ScCCL, VrROMTsyn and SbROMT3syn, and an S-tag was fused to the C-terminus of RpSTSsyn. For the expression of His-tagged RpSTSsyn recombinant protein, the synthetic RpSTSsyn gene (BamHI-XhoI) was cloned into the BamHI and SalI sites of the pETDuet-1 vector.
Expression of recombinant CCL, STS and ROMT proteins in E. coli
The pET22b-STS::H, pCOLADuet-H::CCL, pETDuet-STS::S, pETDuet-H::STS and pETDuet-H::ROMT-STS::S constructs were transformed into E. coli strain BL21-CodonPlus (DE3)-RIPL for expression of the CCL, STS and ROMT proteins. Three transformants of each were cultured in 50 ml LB medium supplemented with ampicillin (50 μg/ml) at 37°C until the absorbance (600 nm) reached 0.6–0.7. Isopropyl-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.5 mM to stimulate induction of expression, and the bacteria were incubated for 4 h at 25°C. Induced culture cells were harvested and prepared by using BugBuster protein extraction reagent containing 0.2% (v/v) Lysonase Bioprocessing Reagent, according to the manufacturer’s indications (Novagen). Expression of the recombinant proteins was analyzed by 12% SDS-PAGE of the supernatant and pellet fractions. The promising recombinant E. coli transformant was selected for further experiments and stored in glycerol at −80°C for later use. Recombinant His-tagged proteins (CCL and STS) were affinity-purified using Ni-nitriloacetic acid (NTA) agarose (Qiagen) according to the manufacturer’s instructions. The molecular weights of the recombinant CCL and STS proteins were estimated by SDS-PAGE followed by Western blot analysis. The membranes were probed with mouse anti-6xHis monoclonal antibody horseradish peroxidase (HRP) conjugate (Clontech; 1:5,000 dilution) or HRP-conjugated rabbit anti-S-tag antibody (Bethyl; 1:10,000 dilution).
In vitro enzyme assays
The partially purified enzymes were assayed for CCL and STS activity with various phenolic substrates such as p-coumaric acid, caffeic acid, ferulic acid, and cinnamic acid. The reaction mixtures, in a total volumes of 0.5 ml, contained 100 mM Tris-HCl (pH 7.4), 5 mM MgCl2, 5 mM ATP, 0.3 mM malonyl-CoA, 0.3 mM CoA, 0.2 mM phenolic substrates, and the partially purified proteins (each 1.5 μM: His-ScCCL, 43 μg; His-RpSTSsyn, 33 μg). After incubation at 30°C for 3 h, the reaction mixtures were extracted with 0.5 ml of ethyl acetate. The solvent was removed by evaporation, the residual was dissolved in 40 μl methanol, and 20 μl of sample was subjected to HPLC.
Production of resveratrol and its methylated derivatives in E. coli
Recombinant E. coli BL21-CodonPlus (DE3)-RIPL cells harboring pCOLADuet-H::CCL/pET22b-STS::H, pCOLADuet-H::CCL/pETDuet-STS::S, pCOLADuet-H::CCL/pETDuet-H::VrROMT-STS::S, or pCOLADuet-H::CCL/pETDuet-H::SbROMT3-STS::S were induced by the addition of IPTG in 200 ml of modified M9 medium (M9 broth, 0.5% glycerol, 1 mM MgSO4, 0.125% yeast extract), as described previously (Jeong et al., 2014). To the cell suspension in modified M9 medium, 0.5 mM IPTG was added alongside 1 mM p-coumaric acid as a substrate, and the culture was further incubated at 28°C for 60 h. After incubation, 20 ml samples of the culture were taken at the indicated time points (0, 6, 12, 24, 36, 48, and 60 h) and adjusted to pH 9.0 using 5 N NaOH. The reaction mixtures were extracted with equal volumes of ethyl acetate, and subjected to HPLC and LC-MS.
RESULTS
Isolation of stilbene synthase genes from rhubarb
To get insights into stilbene biosynthesis in rhubarb plants, we attempted to isolate an STS gene (RpSTS) from rhubarb rhizomes (Rheum palmatum L. cv. Jangyeop) through a homology-based PCR approach. Nucleotide sequence analysis showed that the full-length RpSTS cDNA (accession no. JX673939) was 1,176 bp and encoded a protein of 391 amino acids with a predicted molecular weight of 42.8 kDa. The RpSTS cDNA sequence shared 94.7% and 93.4% identity to RtSTS at the nucleotide and amino acid levels, respectively. RpSTS had 66% and 65.7% similarity to AhSTS (accession no. AB027606) and VvSTS (accession no. EF192465) at the amino acid level, respectively. To compare the activity of RpSTS enzyme with other plant STS proteins in E. coli, we also amplified the AhSTS (accession no. JX673938) and VrSTS (accession no. JX673940) from peanut (Arachis hypogaea cv. Champyeong) and grape (Vitis riparia), respectively. Amino acid sequences of the RpSTS were aligned with the isolated AhSTS and VrSTS (Supplementary Fig. S1).
Heterologous expression of plant STS proteins and production of resveratrol in E. coli
ScCCL gene product was shown to have distinct plant 4CL activity as a functional homologue (Choi et al., 2011; Kaneko et al., 2003). Thus, we used the ScCCL gene for the production of p-coumaroyl-CoA from p-coumaric acid. To examine whether plant STS proteins are capable of producing resveratrol from p-coumaric acid, the pET22b-STS::H constructs were co-expressed with the pCOLADuet-H::CCL plasmid in E. coli (Fig. 1B). The expression of His-tagged STS and CCL proteins was confirmed by Western blot analysis with anti-His-tag antibody (Fig. 2A). The recombinant His-ScCCL protein corresponding to its theoretical molecular weight (MW) of 57.5 kDa was expressed well at 2 and 4 h after IPTG induction in the soluble and insoluble fractions. However, the most portions of the expressed STS recombinant proteins with the theoretical MW of 45.3 kDa were observed in the insoluble fraction. Only AhSTS recombinant protein was detectable at low level in the soluble fraction but not in the RpSTS and VrSTS. We further tested whether co-expression of STS and CCL recombinant proteins in E. coli cells can produce resveratrol from p-coumaric acids. E. coli cells containing pET22b-STS and pCOLADuet-CCL were cultured in modified M9 medium with 1 mM p-coumaric acid for 60 h after IPTG induction. The culture broth and bacterial cells were collected at the indicated time points and the extracted samples were subjected to HPLC analysis (Figs. 2B and 2C). HPLC analysis indicates that a major peak corresponding to resveratrol was detected at the same retention time (9.904 min) as that of standard resveratrol (9.887 min) only in the pET22b-AhSTS and pCOLADuet-CCL culture sample. Out of three STS constructs, only AhSTS was able to produce significant amounts of resveratrol from p-coumaric acids when added to the E. coli cells. The resveratrol level began to increase at 6 h followed by dramatic increase at 12 h, continuing to accumulate up to 60 h after the addition of p-coumaric acid. The highest level (3.3 mg/L) of resveratrol was detected at 48 h in the pET22b-AhSTS culture cells. By contrast, the other pET22b-RpSTS and pET22b-VrSTS failed to convert effectively p-coumaric acid to resveratrol. This differences among plant STS is likely due to the solubility of the STS recombinant proteins in E. coli. As shown in Fig. 2A, the AhSTS recombinant protein showed better expression than RpSTS and VrSTS in the soluble fraction. This suggests that the solubility of recombinant proteins is one of important factors for the better production of resveratrol in E. coli. Thus, to improve the solubility of recombinant RpSTS protein in E. coli, the original RpSTS gene was synthesized following codon-optimization using bacterial-preferred codon usage. In this paper, the codon-optimized synthetic RpSTSsyn was used for further experiments.
Fig. 2.
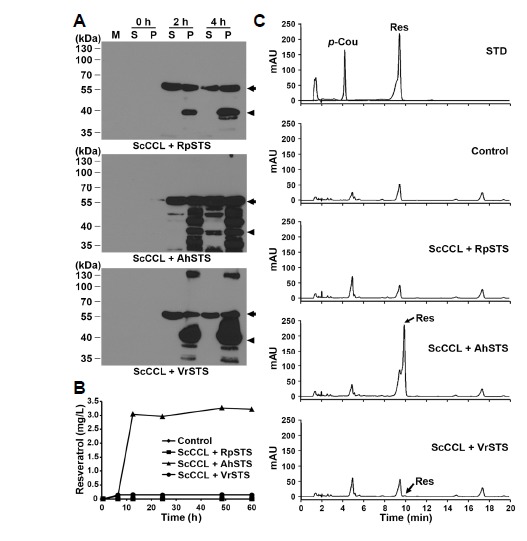
Expression of His-tagged plant STS and ScCCL recombinant proteins and time-course production of resveratrol in E. coli cells. (A) E. coli cells harboring both pCOLADuet-H::CCL and pET22b-STS::H (CCL+STS) constructs were grown in modified M9 media and induced by the addition of 0.5 mM IPTG at 25°C for 2 h and 4 h. Expression of His-tag fusion proteins was confirmed by Western blot analysis with anti-His-tag antibody. M, protein molecular marker in kDa; S, soluble fraction (20 μg); P, insoluble pellet fraction (1 μg). Arrows and arrowheads indicate the expressed ScCCL and plant STS recombinant proteins, respectively. (B) E. coli cells harboring CCL+STS constructs were cultured and harvested at the indicated time points after the addition of 1 mM p-coumaric acid to the pre-induced cells. E. coli cells carrying both pCOLADuet-1 and pET-22b were used as a control. Samples were extracted with ethyl acetate and subjected to HPLC analysis. E. coli cells were pre-induced to express recombinant proteins by the addition of 0.1 mM IPTG. (C) HPLC analysis was performed using a C18 reverse-phase column. Chromatogram STD represents the authentic standards of resveratrol (Res) and p-coumaric acid (p-Cou) with retention times of 9.887 and 4.907 min, respectively.
Expression of codon-optimized synthetic STS and ROMT proteins in E. coli
To improve the translational efficiency of the plant genes in a bacterial system, the RpSTS, VrROMT and SbROMT3 genes were synthesized following plant codon-optimization (Supplementary Fig. S3). The synthetic RpSTSsyn, VrROMTsyn and SbROMT3syn DNAs were introduced into the dual expression vector pETDuet-1 for the expression of the corresponding N-terminal His-tag or C-terminal S-tag fusion proteins (Fig. 1B). To examine the function of RpSTSsyn, VrROMTsyn and SbROMT3syn in E. coli, either pETDuet-STS or pETDuet-ROMT-STS was co-transformed into E. coli cells alongside pCOLADuet-CCL, and the bacteria were selected on LB media containing both kanamycin and ampicilin antibiotics. The most promising recombinant E. coli transformant was selected by SDS-PAGE and Western blot analyses, and used for further experiments (data not shown). Expression of recombinant CCL, STS and ROMT proteins was determined by SDS-PAGE and Western blot analysis with anti-His-tag or anti-S-tag antibody. As shown in Fig. 3, all the His-ScCCL, RpSTSsyn-S, His-VrROMTsyn and His-SbROMT3syn recombinant proteins corresponded to their theoretical molecular weights of 57.5, 45.3, 41.7 and 42.5 kDa, respectively, and were expressed at 2 and 4 h after IPTG induction in the soluble and insoluble fractions. Specific protein bands detected by His- or S-tag antibodies were detected, even in the presences of some degradation. All the recombinant CCL, STS and ROMT proteins appeared to be expressed in the both soluble and insoluble fractions of the E. coli cell lysate, but the major portions of the expressed proteins were observed in the insoluble fraction. The SbROMT3syn protein showed better expression than VrROMTsyn in the both soluble and insoluble fractions. Multiple protein bands hybridizing with His- or S-tag antibody were detected, which are likely to be the result of protein degradation. Producing soluble proteins at high levels in E. coli is still a major bottleneck for metabolic engineering. We observed that the codon-optimized RpSTSsyn improved the solubility in E. coli compared to the original RpSTS. However, we failed to obtain high-level expression of heterologous STS and ROMT recombinant proteins in the soluble fraction of E. coli, even with codon usage optimization and alterations affecting culture medium and temperature, IPTG induction, and the host strain.
Fig. 3.
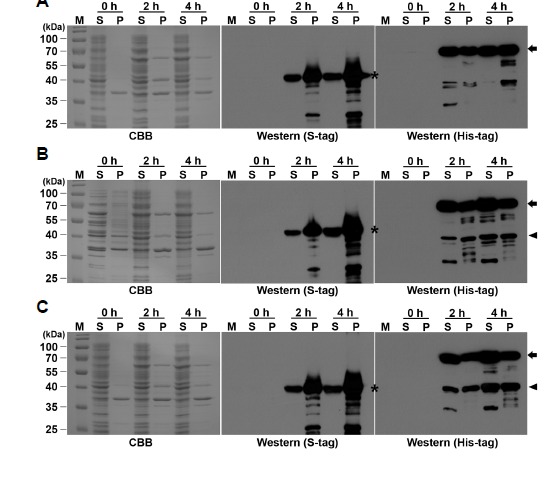
Expression of His-tagged CCL, VrROMTsyn and SbROMT3syn, and S-tagged STS recombinant proteins in E. coli cells. E. coli cells containing (A) CCL+STS, (B) CCL+STS-VrROMT, or (C) CCL+STS-SbROMT3 construct were grown and induced by the addition of 0.5 mM IPTG for 2 h and 4 h. Proteins were separated by 13% SDS-PAGE and stained with Coomassie brilliant blue (CBB). Expression of Histag or S-tag fusion proteins was confirmed by Western blot analysis with anti-His-tag or anti-S-tag antibodies. M, protein molecular marker in kDa; S, soluble fraction (15 μg); P, insoluble pellet fraction (1 μg). S-tagged STS, His-tagged CCL, and His-tagged ROMT recombinant proteins recognized by anti-S-tag and anti-His-tag antibody are shown with asterisks, arrows and arrowheads, respectively.
In vitro assays for the production of resveratrol from p-coumaric acid
To examine whether CCL and STS enzymes are capable of converting various cinnamate derivatives to resveratrol and its derivatives, the His-tagged CCL and STS recombinant proteins were purified using Ni-NTA agarose (Fig. 4A). Using the purified proteins, we conducted in vitro enzyme assays with p-coumaric acid, caffeic acid, ferulic acid, and cinnamic acid as phenolic substrates. As shown in Fig. 4B, the CCL and STS enzymes efficiently converted p-coumaric acid as a substrate to trans-resveratrol, when compared to the control without the enzymes. However, no reaction products corresponding to resveratrol and its derivatives were detected by HPLC when phenolic acids such as caffeic acid, ferulic acid, and cinnamic acid were used (data not shown). Here, we observed that the ScCCL enzyme catalyzed the conversion of cinnamic acid to the intermediate cinnamoyl-CoA, which was not further converted to resveratrol by the STS enzyme (data not shown). These results indicate that p-coumaric acid is a good substrate for CCL and STS in producing resveratrol derivatives in vitro, whereas the other compounds are very poor substrates for CCL and STS. Consistently, our results suggest that ScCCL efficiently converts p-coumaric acid to its coumaroyl-CoA ester, which in turn is synthesized to resveratrol by the action of RpSTSsyn. Kaneko et al. (2003) previously reported that the His-tagged ScCCL recombinant protein had distinct 4CL activity and possessed the ability to use p-coumarate as a substrate. ScCCL was also reported to have low activity toward caffeate and no activity toward ferulate. These results suggest that bacterial ScCCL in combination with RpSTS can be used to produce resveratrol and its derivatives from p-coumaric acid in E. coli.
Fig. 4.
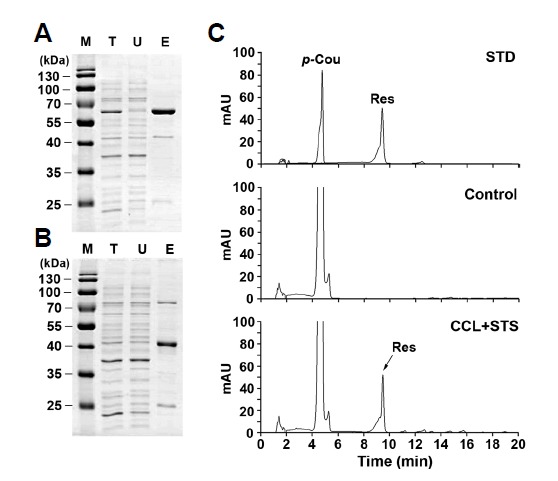
In vitro assays for production of resveratrol from p-coumaric acid by purified recombinant CCL and STS. His-tagged CCL (A) and STS (B) recombinant proteins were expressed in E. coli and affinity-purified using Ni-NTA agarose. Protein samples (2 μg) of total lysates (T) after induction, unbound fraction (U) after Ni-NTA bead incubation, and purified His-CCL or His-STS eluted from agarose beads (E) were separated by 13% SDS-PAGE and stained with CBB. The partially purified enzymes (each 1.5 μM) were assayed for CCL and STS activity with 0.2 mM p-coumaric acid as a precursor in the presence of 0.3 mM malonyl-CoA and 0.3 mM CoA. The reaction mixtures were incubated at 30°C for 3 h prior to extraction. Concentrated 20 μl samples were analyzed by HPLC. Chromatogram STD represents the authentic standards of p-coumaric acid (p-Cou) and resveratrol (Res) with retention times of 4.75 and 9.51 min, respectively. Chromatograms Control and CCL+ STS represent the samples produced from p-coumaric acid by in vitro assays without and with recombinant proteins, respectively.
Production of resveratrol and its methylated derivatives in recombinant E. coli
To further investigate the production of resveratrol and its methylated derivatives in E. coli by expression of major enzymes involved in stilbene biosynthesis, we reconstructed recombinant E. coli producing CCL, STS and ROMT (Fig. 1B). For production of resveratrol and pterostilbene, E. coli cells containing either pCOLADuet-CCL and pETDuet-STS, or pCOLADuet-CCL and pETDuet-ROMT-STS, were cultured in modified M9 medium with 1 mM p-coumaric acid for 60 hrs after IPTG induction. As a negative control, E. coli cells containing empty vectors of pETDuet-1 and pCOLADuet-1 were cultivated with p-coumaric acid. Increasing concentrations of p-coumaric acid up to 1 mM enhanced the resveratrol production, but not at higher concentrations than 1 mM (data not shown). Based on this result, 1 mM concentration of p-coumaric acid was used as a substrate for all subsequent experiments. The concentrations of resveratrol and pinostilbene produced in recombinant E. coli were determined by comparing with standard curves generated from known concentrations of resveratrol and pinostilbene. Figure 5 shows the time-course of resveratrol and pinostilbene production in recombinant E. coli cells. When p-coumaric acid was used as the precursor, resveratrol biosynthesis in recombinant E. coli cells carrying the constructs (CCL+STS, CCL+STS-VrROMT, or CCL+STS-SbROMT3) was first detected 6 h after addition of p-coumaric acid, and gradually increased up to 36 h. The highest levels of resveratrol produced in recombinant E. coli were 1.8 mg/L at 36 h in CCL+STS, 1.9 mg/L at 60 h in CCL+STS-VrROMT, and 1.2 mg/L at 24 h in CCL+STS-SbROMT3 samples. Resveratrol production in CCL+STS-SbROMT3 did not increase further after 24 h, but rather decreased. This is likely due to the consumption of resveratrol by conversion to pinostilbene by the action of SbROMT3syn. As shown in Fig. 5B, pinostilbene production in CCL+STS-SbROMT3 greatly increased from 24 h and the maximal level (2.4 mg/L) was reached at 48 h, after which production decreased. In addition, a very small amount of pterostilbene was detected at 48 h, only in CCL+STS-SbROMT3 (Fig. 6), indicating a low enzyme activity of recombinant SbROMT3syn in producing pterostilbene from resveratrol. As a result, the culture cells containing the pCOLADuet-CCL and pETDuet-SbROMT3-STS plasmids (CCL+STS-SbROMT3) were capable of producing both resveratrol and pinostilbene as the major products from p-coumaric acid along with the minor product of pterostilbene. This is consistent with our previous results showing that recombinant SbROMT3syn catalyzed the production of pinostilbene as the main compound from resveratrol with in vitro and in vivo methylation assays (Jeong et al., 2014). However, no pinostilbene and pterostilbene were produced in control, CCL+STS and CCL-STS-VrROMT. This indicates that VrROMTsyn has no or very poor enzyme activity toward resveratrol as a substrate.
Fig. 5.
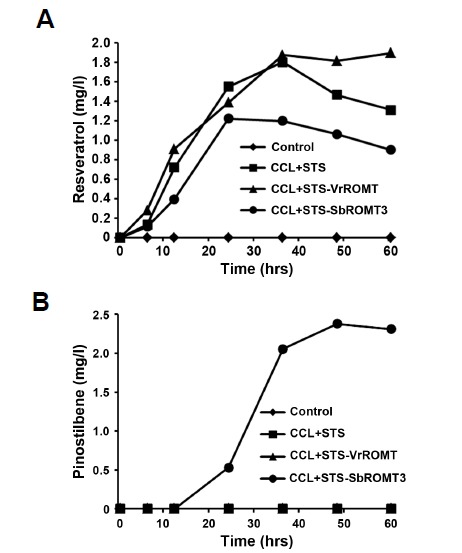
Time-course production of resveratrol and pinostilbene in recombinant E. coli. E. coli cells harboring CCL+STS, CCL+STS-VrROMT, or CCL+STS-SbROMT3 construct were cultured and harvested at the indicated time points after the addition of 1 mM p-coumaric acid to the pre-induced cells. E. coli cells carrying both pCOLADuet-1 and pETDuet-1 were used as a control. Samples were subjected to HPLC analysis. The amounts (mg/L) of resveratrol (A) and pinostilbene (B) at the indicated time points were measured by comparison with standard curves generated from known concentrations of resveratrol and pinostilbene.
Fig. 6.
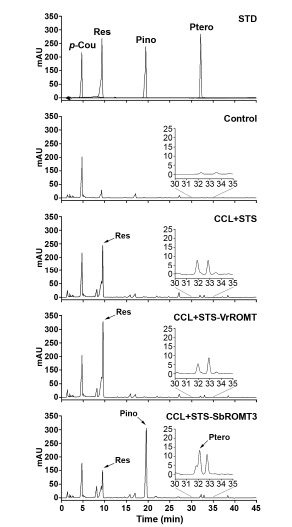
HPLC analysis of resveratrol and pinostilbene produced by recombinant E. coli. E. coli cells harboring CCL+STS, CCL+STS-VrROMT, or CCL+STS-SbROMT3 constructs were cultured for 48 h prior to extraction. Chromatogram STD represents the authentic standards of resveratrol (Res), pinostilbene (Pino), and pterostilbene (Ptero) with retention times of 9.423, 19.327, and 32.009 min, respectively. Chromatograms Control, CCL+STS, CCL+STS-VrROMT, and CCL+STS-SbROMT3 represent the samples produced from p-coumaric acid by recombinant E. coli. Insets indicate the chromatograms magnified to show small amounts of pterostilbene production.
Confirmation of resveratrol derivatives produced in recombinant E. coli
We further analyzed the 48 h culture cells by HPLC because of the high-level production of resveratrol, pinostilbene and pterostilbene at that time. When analyzing the culture cells, as shown in Fig. 6, a major peak corresponding to resveratrol was detected at the same retention times (9.694, 9.688 and 9.692 min in CCL+STS, CCL+STS-VrROMT and CCL+STS-SbROMT3 samples, respectively) as that of authentic resveratrol (9.423 min). Furthermore, another major peak corresponding to pinostilbene was observed at 19.825 min retention time, but only in CCL+ STS-SbROMT3. High-level production of resveratrol was detected in CCL+STS and CCL+STS-VrROMT, but the resveratrol level decreased after 24 h and the pinostilbene production instead increased in CCL+STS-SbROMT3 (Figs. 5 and 6). A very weak peak corresponding to pterostilbene, as shown in Fig. 6 (inset chromatograms), was also observed in CCL+STS-SbROMT3 only (retention time, 32.454 min). To confirm production of resveratrol, pinostilbene and pterostilbene in recombinant E. coli, subsequent analysis was carried out with LC-MS using 48 h cells. As shown in Fig. 7, the compounds in the peaks of the CCL+STS-SbROMT3 sample shown in Fig. 6 exhibited parent ion peaks [M+H]+ at m/z 229.2, 243.1 and 257.1. The corresponding mass spectra for these peaks revealed the compounds to be resveratrol, pinostilbene and pterostilbene, respectively. Collectively, the HPLC and LC-MS analyses indicate that recombinant E. coli cells co-expressing CCL, STS and ROMT proteins and fed with p-coumaric acid as the precursor are capable of producing stilbene compounds, such as resveratrol and pinostilbene as the major product, along with pterostilbene as a minor product.
Fig. 7.
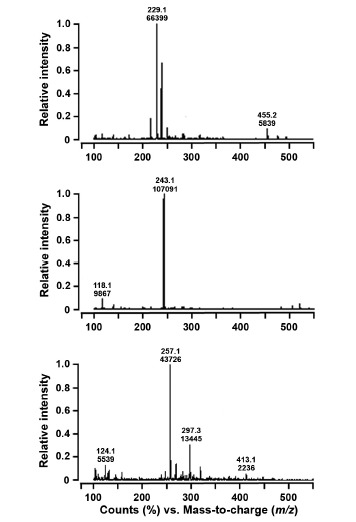
LC-MS analysis of resveratrol, pinostilbene and pterostilbene produced by recombinant E. coli cells. The molecular masses of resveratrol, pinostilbene and pterostilbene were analyzed with a linear ion trap quadrupole LC-MS at a positive mode of atmospheric pressure chemical ionization. Samples were taken from cell culture at 48 h after the addition of p-coumaric acid, and the extracted compounds were analyzed by LC-MS. The injection volume was 10 μl. Chromatograms (A), (B) and (C) represent the LC-MS spectra showing the peaks at m/z 229.2, 243.1 and 257.1 for reveratrol, pinostilbene and pterostilbene, respectively.
DISCUSSION
In the present study, we attempted to identify the RpSTS gene to study stilbene biosynthesis in rhubarb plants and examined its activity in heterologous system. We compared the ability of resveratrol production in E. coli by expressing the three original STS genes. Among them, we observed that co-expression of AhSTS together with ScCCL was able to produce resveratrol in E. coli. However, no or very low levels of resveratrol was produced in the RpSTS or VrSTS constructs. We realized that the most portions of the expressed STS recombinant proteins were observed in the insoluble fraction and AhSTS showed better expression than RpSTS and VrSTS in the soluble fraction. By contrast, RpSTS and VrSTS were not detectable in the soluble fraction. This solubility for AhSTS might increase the ability of resveratrol production in the recombinant E. coli cells. Thus, to improve the solubility of recombinant RpSTS protein, the originnal RpSTS gene was synthesized following codon-optimization.
We engineered E. coli to produce resveratrol by the expression of the codon-optimized RpSTS gene along with CCL. Furthermore, by the incorporation of SbROMT3 gene, we also developed the recombinant E. coli to produce both resveratrol and pinostilbene from p-coumaric acid. Our studies showed that RpSTS gene encodes a functional enzyme to catalyze the resveratrol production via condensation of p-coumaroyl-CoA and three malonyl-CoA molecules. Moreover, we observed that the SbROMT3 catalyzes the mono-methylation at the 3- or 5-position in resveratrol to yield pinostilbene. However, our data suggest that the amount of resveratrol and its methylated derivatives produced in recombinant E. coli is still low and further metabolic engineering is required to increase the amount to commercial levels. Several methylated resveratrol derivatives including pinostilbene and pterostilbene are found in some plants. It has been reported that the health-promoting activities of certain methylated resveratrol derivatives are more effective than those of unmodified resveratrol (Remsberg et al., 2008; Rimando et al., 2012; Schmidlin et al., 2008). Thus, metabolic engineering to increase their production has become an active filed of research. However, there are still a few reports about the production of methylated resveratrol derivatives in microbes (Jeong et al., 2014; Katsuyama et al., 2007). Only a few ROMT genes have been isolated and characterized to date. Thus, many more plant OMT genes involved in the formation of methylated resveratrol derivatives remain to be identified. Schmidlin et al. (2008) reported that VvROMT from grapevine (V. vinifera) was responsible for catalyzing the methylation of resveratrol to produce pterostilbene both in vitro and in planta. Rimando et al. (2012) demonstrated that SbOMT3 from sorghum was capable of utilizing resveratrol as a substrate, yielding pterostilbene as a major product via A-ring-specific 3,5-bis-O-methylation in vitro. In addition, the constitutive co-expression of SbOMT3 with the AhSTS3 from peanut was shown to induce the accumulation of pterostilbene in Arabidopsis and tobacco plants. By contrast, our data suggest that SbROMT3syn possessed high catalytic activity for transferring one methyl group to the hydroxyl group at the 3- or 5-position in resveratrol, thereby producing pinostilbene, even though it also has weak activity for methylation of both hydroxyl groups to generate pterostilbene. These methylated resveratrol derivatives are likely produced from resveratrol by the sequential 3,5-bis-O-methylation to form pinostilbene and pterostilbene. Indeed, by in vitro and in vivo methylation assays, we realized that higher levels of pterostilbene were generated from resveratrol when protein amounts and enzyme incubation times were increased (data not shown). Our data suggest that SbROMT3 in combination with RpSTSsyn are responsible for the production of pinostilbene from resveratrol.
Resveratrol possesses three hydroxyl groups at positions 3, 5, and 4′ that can possibly be methylated by OMT (Jeong et al., 2014; Katsuyama et al., 2007). In the previous and current reports, we found that SbROMT3syn has the enzyme activity to methylate the hydroxyl group at position 3 or 5 of resveratrol, thus yielding pinostilbene as the major product. However, no desoxyrhapontigenin (3,5-dihydroxy-4′-methoxy-trans-stilbene; 4′-O-methylresvratrol) or resveratrol trimethylether (3,4′,5-trimethoxy-trans-stilbene) derivatives were detected. Similarly, OsPMT also exhibited a preference for 3 or 5 methylation of resveratrol to produce pinostilbene (Katsuyama et al., 2007). This in vitro metabolic engineering of pinostilbene or pterostilbene biosynthesis by methylation of resveratrol in recombinant E. coli is superior in methylation selectivity than random methylation of hydroxyl groups by chemical synthesis. SbROMT3syn and OsPMT have high mono-methyl transferase activity for producing pinostilbene from resvertarol. However, little is known to date about the OMT enzymes that exhibit high di-methyl transferase activity to yield pterostilbene from resveratrol. Thus, for the high-level production of pterostilbene by metabolic engineering in E. coli, it is necessary to identify genes encoding the enzymes that catalyze the di-methylation of resvetarol to produce pterostilbene.
Supplementary materials
Acknowledgments
This work was supported by grants from the KRIBB Initiative Program and the Next-Generation BioGreen 21 Program (SSAC, grant #: PJ01107004), Rural Development Administration, Republic of Korea.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
REFERENCES
- Baur J.A., Pearson K.J., Price N.L., Jamieson H.A., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., et al. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J.V., Armstrong G.O., van der Merwe M.J., Lambrechts M.G., Vivier M.A., Pretorius I.S. Metabolic engineering of Saccharomyces cerevisiae for the synthesis of the wine-related antioxidant resveratrol. FEMS Yeast Res. 2003;4:79–85. doi: 10.1016/S1567-1356(03)00157-0. [DOI] [PubMed] [Google Scholar]
- Beekwilder J., Wolswinkel R., Jonker H., Hall R., de Vos C.H., Bovy A. Production of resveratrol in recombinant microorganisms. Appl. Environ. Microbiol. 2006;72:5670–5672. doi: 10.1128/AEM.00609-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi O., Wu C.Z., Kang S.Y., Ahn J.S., Uhm T.B., Hong Y.S. Biosynthesis of plant-specific phenylpropanoids by construction of an artificial biosynthetic pathway in Escherichia coli. J. Ind. Microbiol. Biotechnol. 2011;38:1657–1665. doi: 10.1007/s10295-011-0954-3. [DOI] [PubMed] [Google Scholar]
- Delaunois B., Cordelier S., Conreux A., Clément C., Jeandet P. Molecular engineering of resveratrol in plants. Plant Biotechnol. J. 2009;7:2–12. doi: 10.1111/j.1467-7652.2008.00377.x. [DOI] [PubMed] [Google Scholar]
- Foust C.M. Rhubarb: The wondrous drug. Princeton, NJ, USA: Princeton University Press; 1992. [Google Scholar]
- Frémont L. Biological effects of resveratrol. Life Sci. 2000;66:663–673. doi: 10.1016/s0024-3205(99)00410-5. [DOI] [PubMed] [Google Scholar]
- Horinouchi S. Combinatorial biosynthesis of plant medicinal polyketides by microorganisms. Curr. Opin. Chem. Biol. 2009;13:197–204. doi: 10.1016/j.cbpa.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Jeong Y.J., An C.H., Woo S.G., Jeong H.J., Kim Y.-M., Park S.-J., Yoon B.D., Kim C.Y. Production of pinostilbene compounds by the expression of resveratrol O-methyltransferase genes in Escherichia coli. Enzyme Microb. Tech. 2014;54:8–14. doi: 10.1016/j.enzmictec.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Kaneko M., Ohnishi Y., Horinouchi S. Cinnamate: coenzyme A ligase from the filamentous bacterium Streptomyces coelicolor A3(2) J. Bacteriol. 2003;185:20–27. doi: 10.1128/JB.185.1.20-27.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwada Y., Nonaka G., Nishioka I., Nishizawa M., Yamagishi T. Studies on rhubarb (Rhein Rhizoma). XIV. Isolation and characterization of stilbene glucosides from Chinese rhubarb. Chem. Pharm. Bull. 1988;36:1545–1549. [Google Scholar]
- Katsuyama Y., Funa N., Miyahisa I., Horinouchi S. Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem. Biol. 2007;14:613–621. doi: 10.1016/j.chembiol.2007.05.004. [DOI] [PubMed] [Google Scholar]
- Lee S.K., Nam K.A., Hoe Y.H., Min H.Y., Kim E.Y., Ko H., Song S., Lee T., Kim S. Synthesis and evaluation of cytotoxicity of stilbene analogues. Arch. Pharm. Res. 2003;26:253–257. doi: 10.1007/BF02976951. [DOI] [PubMed] [Google Scholar]
- Lee S.W., Hwang B.S., Kim M.H., Park C.S., Lee W.S., Oh H.M., Rho M.C. Inhibition of LFA-1/ICAM-1- mediated cell adhesion by stilbene derivatives from Rheum undulatum. Arch. Pharm. Res. 2012;35:1763–1770. doi: 10.1007/s12272-012-1008-8. [DOI] [PubMed] [Google Scholar]
- Melchior F., Kindl H. Grapevine stilbene synthase cDNA only slightly differing from chalcone synthase cDNA is expressed in Escherichia coli into a catalytically active enzyme. FEBS Lett. 1990;268:17–20. doi: 10.1016/0014-5793(90)80961-h. [DOI] [PubMed] [Google Scholar]
- Remsberg C.M., Yáñez J.A., Ohgami Y., Vega-Villa K.R., Rimando A.M., Davies N.M. Pharmacometrics of pterostilbene: preclinical pharmacokinetics and metabolism, anticancer, antiinflammatory, antioxidant and analgesic activity. Phytother. Res. 2008;22:169–179. doi: 10.1002/ptr.2277. [DOI] [PubMed] [Google Scholar]
- Rimando A.M., Pan Z., Polashock J.J., Dayan F.E., Mizuno C.S., Snook M.E., Liu C.J., Baerson S.R. In planta production of the highly potent resveratrol analogue pterostilbene via stilbene synthase and O-methyltransferase co-expression. Plant Biotechnol. J. 2012;10:269–283. doi: 10.1111/j.1467-7652.2011.00657.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E.F., Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd ed. 1,2, 3. NY, USA: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schmidlin L., Poutaraud A., Claudel P., Mestre P., Prado E., Santos-Rosa M., Wiedemann-Merdinoglu S., Karst F., Merdinoglu D., Hugueney P. A stress-inducible resveratrol O-methyltransferase involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol. 2008;148:1630–1639. doi: 10.1104/pp.108.126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparvoli F., Martin C., Scienza A., Gavazzi G., Tonelli C. Cloning and molecular analysis of structural genes involved in flavonoid and stilbene biosynthesis in grape (Vitis vinifera L.) Plant Mol. Biol. 1994;24:743–755. doi: 10.1007/BF00029856. [DOI] [PubMed] [Google Scholar]
- Ulrich S., Wolter F., Stein J.M. Molecular mechanisms of the chemopreventive effects of resveratrol and its analogs in carcinogenesis. Mol. Nutr. Food. Res. 2005;49:452–461. doi: 10.1002/mnfr.200400081. [DOI] [PubMed] [Google Scholar]
- Ververidis F., Trantas E., Douglas C., Vollmer G., Kretzschmar G., Panopoulos N. Biotechnology of flavonoids and other phenylpropanoid-derived natural products. Part II: Reconstruction of multienzyme pathways in plants and microbes. Biotechnol. J. 2007;2:1235–1249. doi: 10.1002/biot.200700184. [DOI] [PubMed] [Google Scholar]
- Walle T., Hsieh F., DeLegge M.H., Oatis J.E., Jr, Walle U.K. High absorption but very low bioavailability of oral resveratrol in humans. Drug Metab. Dispos. 2004;32:1377–1382. doi: 10.1124/dmd.104.000885. [DOI] [PubMed] [Google Scholar]
- Wang T.T., Schoene N.W., Kim Y.S., Mizuno C.S., Rimando A.M. Differential effects of resveratrol and its naturally occurring methylether analogs on cell cycle and apoptosis in human androgen-responsive LNCaP cancer cells. Mol. Nutr. Food Res. 2010;54:335–344. doi: 10.1002/mnfr.200900143. [DOI] [PubMed] [Google Scholar]
- Watts K.T., Lee P.C., Schmidt-Dannert C. Biosynthesis of plant-specific stilbene polyketides in metabolically engineered Escherichia coli. BMC Biotechnol. 2006;6:22. doi: 10.1186/1472-6750-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Li S.Z., Li J., Pan X., Cahoon R.E., Jaworski J.G., Wang X., Jez J.M., Chen F., Yu O. Using unnatural protein fusions to engineer resveratrol biosynthesis in yeast and Mammalian cells. J. Am. Chem. Soc. 2006;128:13030–13031. doi: 10.1021/ja0622094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


