Background: In vitro the mARC-system catalyzes reductions of N-hydroxylated compounds as for instance N-hydroxylated base analogs.
Results: mARC2 knockdown increases N6-hydroxylaminopurine sensitivity of HeLa cells but N6-hydroxyadenosine detoxication is more effectively catalyzed by adenosine deaminase.
Conclusion: mARC2 plays a critical role in detoxication of N6-hydroxylaminopurine in HeLa cells.
Significance: Detoxication of mutagenic N-hydroxylated base analogs may constitute a physiological function of the mARC-system.
Keywords: apoptosis, cell metabolism, flow cytometry, nucleoside/nucleotide analogue, RNA interference (RNAi), MOSC, N-reduction, N6-hydroxyadenosine, N6-hydroxylaminopurine, mARC
Abstract
N-Hydroxylated nucleobases and nucleosides as N-hydroxylaminopurine (HAP) or N-hydroxyadenosine (HAPR) may be generated endogenously in the course of cell metabolism by cytochrome P450, by oxidative stress or by a deviating nucleotide biosynthesis. These compounds have shown to be toxic and mutagenic for procaryotic and eucaryotic cells. For DNA replication fidelity it is therefore of great importance that organisms exhibit effective mechanisms to remove such non-canonical base analogs from DNA precursor pools. In vitro, the molybdoenzymes mitochondrial amidoxime reducing component 1 and 2 (mARC1 and mARC2) have shown to be capable of reducing N-hydroxylated base analogs and nucleoside analogs to the corresponding canonical nucleobases and nucleosides upon reconstitution with the electron transport proteins cytochrome b5 and NADH-cytochrome b5 reductase. By RNAi-mediated down-regulation of mARC in human cell lines the mARC-dependent N-reductive detoxication of HAP in cell metabolism could be demonstrated. For HAPR, on the other hand, the reduction to adenosine seems to be of less significance in the detoxication pathway of human cells as HAPR is primarily metabolized to inosine by direct dehydroxylamination catalyzed by adenosine deaminase. Furthermore, the effect of mARC knockdown on sensitivity of human cells to HAP was examined by flow cytometric quantification of apoptotic cell death and detection of poly (ADP-ribose) polymerase (PARP) cleavage. mARC2 was shown to protect HeLa cells against the apoptotic effects of the base analog, whereas the involvement of mARC1 in reductive detoxication of HAP does not seem to be pivotal.
Introduction
N-Hydroxylated bases and nucleosides such as N6-hydroxylaminopurine (HAP),2 N6-hydroxyadenosine (HAPR), 2-amino-HAP, or N4-hydroxycytidine have been shown to possess toxic and mutagenic properties in procaryotic and eucaryotic cells (1, 2). As base analogs, their mutagenicity is attributed to their ability to act alongside the natural substrates in cellular nucleic acid metabolism and their eventual incorporation into DNA (3). The mechanism of activation is best explored for the N-hydroxylated adenine analogs. Just as adenine, HAP is postulated to be activated through the purine salvage pathway via initial conversion by adenosine phosphoribosyltransferase to the corresponding monophosphate (4, 5). HAPR, on the other hand, is believed to be converted to HAPMP by adenosine kinase (6). HAPMP may then be activated by kinases and ribonucleotide reductase resulting in deoxynucleoside triphosphate (dHAPTP), which can be a substrate for DNA polymerases (7, 8). Because of its ambivalent base pairing properties, incorporation can provoke mutations in subsequent replication cycles (7, 9). In vivo, N-hydroxylated base analogs can be generated from canonical nucleobases in the course of cellular metabolism via different pathways. HAP can be formed by cytochrome P450 catalyzed monooxygenation of adenine (10) or from oxidative stress (11). It has also been shown that HAPMP can be generated from IMP through de novo purine biosynthesis wherein hydroxylamine is provided instead of aspartate (12). Although it is conceivable that N-hydroxylated bases analogs could enter nucleotide pools in vivo by natural means, contamination by direct measurement could not be proven, yet (13). However, to ensure accuracy of DNA replication, it is of great importance for organisms to possess precise detoxication and repair mechanisms for protection from exogenous and endogenous base analogs. In Escherichia coli, mechanisms at several levels of the nucleic acid metabolism are described which prevent incorporation of HAP into DNA. Ycbx, a molybdoenzyme from the MOSC family, was found to be involved in detoxifying HAP by reduction to adenine (14). MOSC domain proteins form a small protein family in eu- and procaryotes with its members being characterized by the requirement of a molybdenum cofactor and a strictly conserved cysteine residue (15). If HAP enters nucleotide pools through the salvage pathway anyway, inosine triphosphate pyrophosphatase (ITPase) is postulated to prevent incorporation of HAPTP by sanitizing nucleotide pools and cleaving HAPTP or dHAPTP to its respective monophosphate derivate (8). Should dHAPTP be incorporated into DNA, endonuclease V recognizes and nicks HAP residues and initiates repair of these lesions (8). Repair systems of the adenine analogs in humans have been studied less extensively. However, it was shown that ITPase plays a critical role in the detoxication pathway in human cells. Furthermore, involvement of endonucleases is suggested, as HAP is able to induce DNA strand breaks in human cell lines (16, 17).
The mitochondrial amidoxime reducing component mARC is a recently discovered molybdenum enzyme in mammals which in concert with the electron transport proteins cytochrome b5 and NADH-cytochrome b5 reductase catalyzes the reduction of various N-hydroxylated compounds (18–20). Besides the eponymous mitochondrial localization, proteomic studies suggest a peroxisomal co-localization for mARC as well (21, 22). Alongside sulfite oxidase, aldehyde oxidase and xanthine oxidoreductase, mARC is one of four molybdenum-containing enzymes in humans and belongs to the MOSC family (23). The human genome codes for two mARC proteins (mARC1 and mARC2), which share a high degree of sequence similarity to each other. It is well accepted that the mARC-containing N-reductive enzyme system plays a major role in the activation of N-hydroxylated prodrugs as amidoximes, N-hydroxyguanidines or sulfhydroxamic acids (18, 24, 25). Though its physiological substrates, and therefore its physiological functions, are mostly unknown yet, mARC proteins are assumed to be involved in detoxication of mutagenic and toxic aromatic hydroxylamines like N-hydroxylated DNA-base analogs. This hypothesis is supported by a recent study that describes significant down-regulated expression of human mARC2 in colon tumors of human tissue samples (26). Indeed, we have shown previously that in vitro upon reconstitution with their electron transport proteins, both recombinant mARC proteins have the capability to reduce all hitherto examined N-hydroxylated nucleobases and nucleosides to their corresponding canonical compounds (27). Besides its involvement in metabolic detoxication other functions of mARC have been proposed. mARC has been suggested to be involved in NO metabolism with regulatory functions in the l-arginine-dependent biosynthesis of NO (28) or reduction of nitrite to NO under anaerobic conditions (29). Furthermore, mARC is brought into context with energy and lipid metabolism (30–32) as well as with metabolic disorders as diabetes mellitus (33).
Although both mARC proteins exhibit N-reductive activity in vitro in the reconstituted system as well as in human cells (34, 35), an evolutionary need for the function of each protein is likely. Accordingly, it has to be elucidated if functions of the molybdoenzymes are either overlapping or distinct.
In this study we determined whether this in vitro observed mARC-mediated N-reduction also acts as a physiological relevant detoxication pathway in human cell systems. By performing siRNA-mediated mARC knockdowns, the role of mARC in the cellular metabolism of exogenous HAP, as a model for N-hydroxylated nucleobase analogs, and HAPR, as a model for N-hydroxylated nucleoside analogs, was examined. Furthermore, flow cytometric analysis of Annexin V-PE and 7-AAD-stained cells as well as detection of caspase-mediated Poly (ADP-ribose) polymerase (PARP) cleavage was used to determine the influence of mARC knockdown on HAP-induced apoptotic effects on cells.
EXPERIMENTAL PROCEDURES
Cell Lines and Reagents
HEK-293 human embryonic kidney cells were purchased from Cell Lines Service (Eppelheim, Germany). HeLa cells were kindly provided by Prof. Dr. Holger Kalthoff (Institute for Experimental Cancer Research, Division of Molecular Oncology, UKSH Kiel, Germany). Minimum Essential Medium (MEM), Opti-MEM, sodium pyruvate solution, sodium bicarbonate solution, MEM non-essential amino acids, FBS, l-glutamine, HEPES buffer, Lipofectamine® RNAiMAX, Stealth Select RNAi® siRNA targeting human MOSC1 and Stealth RNAi® siRNA negative controls were obtained from Life Technologies (Carlsbad, CA). ON-Targetplus SMARTpool siRNA targeting human MOSC2 was purchased from Thermo Scientific (Waltham, MA). Trypsin/EDTA and Accutase were purchased from GE Healthcare Life Sciences (Chalfont St Giles, UK). Complete protease inhibitor mixture was acquired from Roche Applied Science (Mannheim, Germany). Human recombinant adenosine deaminase from E. coli was obtained from Sigma-Aldrich. HAP was obtained from Apollo Scientific Ltd. (Cheshire, UK). All other chemicals were purchased from Sigma-Aldrich, Merck KGaA (Darmstadt, Germany), or Roth (Karlsruhe, Germany). Methanol HPLC grade was purchased from J.T. Baker (Deventer, Netherlands). Anti-MOSC2 and anti-GAPDH antibodies were obtained from Sigma Life Science. Anti-MOSC1 antibody was purchased from Abgent (San Diego, CA), anti-PARP from Cell Signaling Technology (Danvers, MA) and anti-calnexin from Acris Antibodies GmbH (Herford, Germany).
Cell Culture
HEK-293 cells and HeLa cells were maintained in MEM supplemented with 10% (v/v) FBS, 2 mm l-glutamine, 0.1 mm non-essential amino acids, 1 mm sodium pyruvate, 18 mm sodium bicarbonate, and 20 mm HEPES. Cell lines were incubated at 37 °C in 5% (v/v) CO2.
siRNA Transfection
As described earlier optimal siRNA concentrations were evaluated and knockdown protocols validated (35). HEK-293 cells or HeLa cells were reverse transfected according to the manufacturer's transfection protocol from Invitrogen. Briefly, siRNA oligonucleotides were diluted in Opti-MEM in 24-well plates and incubated with transfection reagent to form liposome-siRNA complexes. The cells were trypsinized, counted, and suspended in culture medium. The cell suspension was then added to the liposome-siRNA complexes. Negative controls included a non-targeting siRNA control (scrambled siRNA) and transfection reagents without siRNA. Down-regulation of protein expression was attained 3 days after transfection and verified by Western blot analysis.
Cultivation in the Presence of HAP
To maintain the ability for active cell division, HeLa cells were trypsinized on day 3 after siRNA transfection and 75,000 cells/well (24 h assay) or 40,000 cells/well (48 h assay) were plated in 24-well plates. Cells were allowed to adhere for 4 h before HAP was added. Stock solutions of HAP were prepared in DMSO and heated slightly to facilitate dissolution. 10-μl aliquots of stock solutions were added to each well, which resulted in various concentrations of HAP and 1% (v/v) DMSO. Cells were cultivated and then assayed after 24 h or 48 h. As positive control for induced apoptosis cells were treated with 100 μm etoposide under the same conditions.
N-reductive Metabolism of HAP in HEK-293
For N-reduction studies in HEK-293 the culture medium was removed, and cells were carefully washed and pre-incubated with substrate-free incubation buffer (Hanks' balanced salt solution containing 10 mm HEPES, pH 7.4) at 37 °C for 10 min. After removing the substrate free incubation buffer, the vital cells were then incubated with HAP-containing incubation buffer (3 mm HAP, 0.5% (v/v) DMSO) at 37 °C for 180 min. After the designated time, the culture supernatant was carefully removed, centrifuged to eliminate cellular debris and analyzed by HPLC. Samples were separated isocratically on a Lichrospher® 60 RP-select B (125 × 4 mm) 5 μm C8 column with a RP-select B 4 mm × 4 mm guard column (Merck KGaA, Darmstadt, Germany). The mobile phase was composed of 10 mm octylsulfonate, 20 mm KH2PO4, pH 3.0 and 10% (v/v) acetonitrile. Flow-rate was kept at 1 ml/min, and detection was carried out at 262 nm. The retention times were 11.6 ± 0.5 min (HAP) and 15.1 ± 0.5 min (adenine).
Metabolism of HAPR in HEK-293
For metabolism studies in HEK-293, culture medium was removed, and cells were carefully washed and pre-incubated with substrate-free incubation buffer (Hanks' balanced salt solution containing 10 mm HEPES, pH 7.4) at 37 °C for 10 min. After removing the substrate free incubation buffer, the vital cells were then incubated with HAPR-containing incubation buffer at 37 °C for 120 min. For concurrent inhibition of adenosine deaminase, 10 μl of dipyridamole in ethanol/H2O (2:1) per well was added to the incubation medium. After the designated time, the culture supernatant was carefully removed, centrifuged to eliminate cellular debris and analyzed by HPLC. Samples were separated isocratically on a Lichrospher®60 RP-select B (125 × 4 mm) 5 μm C8 column with a RP-select B 4 mm × 4 mm guard column (Merck kGaA, Darmstadt, Germany). The mobile phase was composed of 10 mm octylsulfonate, 20 mm KH2PO4, pH 3.5, and 5% (v/v) acetonitrile. Flow-rate was kept at 1 ml/min, and detection was carried out at 260 nm. The retention times were 4.1 ± 0.1 (inosine), 9.5 ± 0.5 min (HAPR), and 17.2 ± 0.5 min (adenosine).
Total Cellular Protein Extraction
To harvest cellular protein, cells were detached by scraping and collected by centrifugation. Pellets were then washed with PBS and centrifuged again. PBS was removed, cells were resuspended in ice-cold lysis buffer (1% (v/v) Nonidet P-40, 150 mm NaCl, 50 mm Tris, pH 8.0, 1 mm EDTA, 2 mm DTT, protease inhibitor mixture) and were shaken 20 min on ice. After 20 s of sonification, cell lysates were cleared by centrifugation. The supernatant contained the total cellular protein extract.
Determination of Protein Concentrations
Protein concentrations were measured using the bicinchoninic acid (BCA) Protein Assay Kit from Pierce according to the manufacturer's instructions.
Western Blot Analysis
Samples were mixed with Laemmli sample buffer (36) and incubated for 6 min at 95 °C. Same amounts of protein for each sample of cell lysate were loaded and separated by SDS-PAGE. After electrophoresis, the proteins were transferred onto Hybond-P PVDF membranes (GE Healthcare, Chalfont St Giles, UK). Membranes were then blocked in TBS containing Tween-20 (TBST) and 5% (w/v) milk powder and incubated with primary antibodies at 4 °C overnight and then washed with TBST. Further they were incubated with HRP-labeled goat anti-rabbit IgG (Jackson ImmunoResearch Laboratories, Suffolk, UK). After washing with TBST, membranes were developed with the ECL Plus Western blotting Detection System (GE Healthcare). The protein levels of calnexin or GAPDH were used as loading controls.
Annexin V-PE/7-AAD Binding and Flow Cytometric Analysis
Apoptotic cell death was quantified using the PE Annexin V Apoptosis Detection Kit I from BD Pharmingen (Franklin Lakes, NJ). Culture medium was collected. For detachment, cells were incubated for 3 min with Accutase, carefully singularized by pipetting and added to the collected culture medium. Cell suspensions were centrifuged for 10 min, 600 × g and supernatants discarded. Cell pellets were resuspended in 100 μl of binding medium (culture medium supplemented with 0.5 mm CaCl2), transferred to flow cytometer tubes, mixed with 2 μl of Annexin V-PE and incubated for 10 min at room temperature in the dark. Subsequently, 3 μl of 7-AAD were added following another 5 min of incubation at room temperature in the dark. Cell suspensions were then analyzed by flow cytometry within 1 h using a Gallios 3L flow cytometer (Beckman Coulter, Brea, CA). Cytometer settings and gates were set on the basis of measurements with untreated and treated cells stained with only one dye. Analyses were carried out using Kaluza 1.2. Cells exhibiting high Annexin V-PE staining were regarded as apoptotic.
Hoechst Staining
Hoechst staining was used to detect changes in chromatin morphology as a typical characteristic of apoptosis (37, 38). A stock solution of Hoechst 33342 was prepared in aqua bidestillata. Aliquots were directly added to the culture medium (0.001% (w/v) Hoechst). After incubating for 10 min at 37 °C, cells were carefully washed twice and left in warm culture medium (without phenol red). Nuclear chromatin was afterward evaluated by fluorescence microscopy (Perkin Elmer precisely LS 55, Waltham, MA).
In Vitro Activity Assay with Human Recombinant Adenosine Deaminase
The adenosine deaminase catalyzed conversions of HAPR and adenosine were characterized by aerobic incubations at 37 °C in a shaking water bath. Incubation mixtures contained 0.05 mg human recombinant adenosine deaminase and various concentrations of HAPR or adenosine in a total volume of 150 μl of MES buffer (20 mm, pH 6.0). After pre-incubation for 3 min at 37 °C, the reaction was initiated by addition of enzyme and terminated after 15 min by addition of 150 μl of methanol. Precipitated proteins were eliminated by centrifugation. Samples were analyzed by HPLC as described above.
Data Analysis
Statistical analyses were carried out using the SigmaPlot 11 software (Systat Software Inc.). The significance of observed differences was evaluated by Student's unpaired t test or for multiple comparison by Bonferroni test. A probability less than 5% was considered to be significant. All experimental values are given as means ± S.D.
RESULTS
N-Reductive Detoxication Pathway of HAP in HEK-293
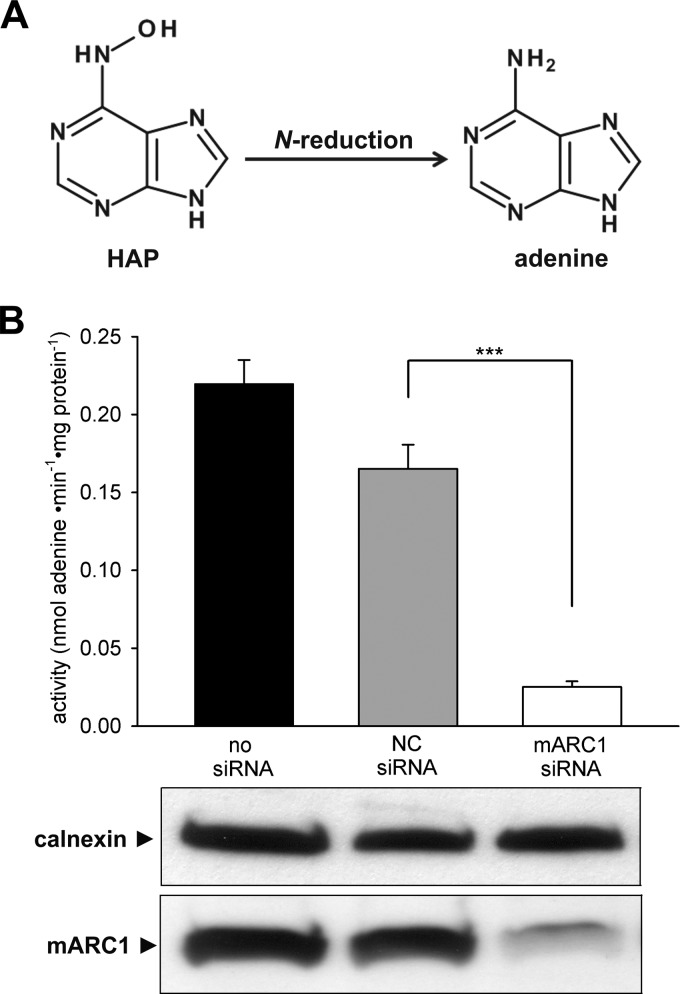
In previous studies, it was shown that the in vitro reconstituted mARC enzyme system is capable of reducing N-hydroxylated nucleobases to the corresponding canonical nucleobases and nucleosides (27). To find out if the mARC-mediated N-reduction depicts a physiological relevant detoxication pathway in living cell systems, metabolism studies with exogenous added HAP were carried out. In HEK-293 cells, reductive conversion of HAP to adenine occurs in a time-dependent and substrate-dependent manner (data not shown). Adenine formation increases linearly with time after a lag of 90 min. HAP reduction followed Michaelis-Menten kinetics (Vmax = 0.38 ± 0.01 nmol·min−1·mg protein−1). By siRNA-mediated down-regulation of mARC1, adenine formation decreased strongly to approx. 15% compared with the negative control (Fig. 1B). Knockdown of mARC2 was not tested due to the fact that mARC2 was shown to have no significant influence on N-reductive metabolism of N-hydroxylated substrates, attributed to its low level of protein expression in HEK-293 (35).
FIGURE 1.
N-Reductive metabolism of HAP in HEK-293. A, N-reduction of HAP to adenine. B, effect of mARC knockdown on HAP reduction. HEK-293 cells were transfected with 10 nm mARC1 or non-targeting (NC) siRNA. The siRNA-mediated down-regulations of the proteins of interest were verified by Western blot using anti-mARC1, or anti-calnexin antibody. Calnexin levels were used as loading control. N-Reductive activities were determined on day 3 after transfection as described under “Experimental Procedures.” Results are presented as means ± S.D. (n = 3). ***, p < 0.001.
Detoxication Pathway of HAPR in HEK-293
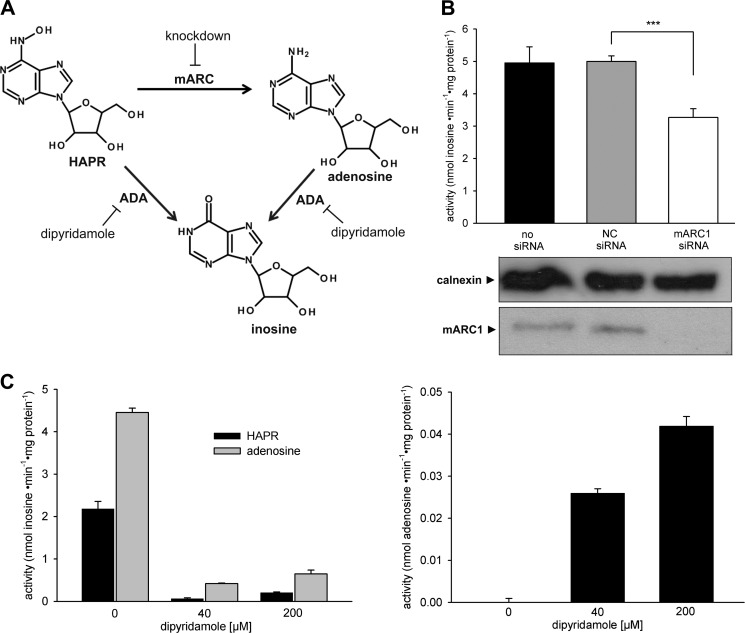
The in vitro reconstituted mARC-containing enzyme system is not only able to reduce N-hydroxylated base analogs like HAP but also N-hydroxylated nucleoside analogs as HAPR (27). But unlike for HAP, no reductive detoxication of HAPR to the corresponding canonical nucleoside adenosine could be detected in the cellular system. Instead, very efficient formation of inosine was observed (Vmax = 10.9 ± 0.5 nmol·min−1·mg protein−1, data not shown). It is known that adenosine has a short half-life in vivo as it is easily deaminated to inosine by adenosine deaminase (39, 40). Therefore, adenosine as an intermediate might not be detectable in the detoxication pathway of HAPR. On the other hand, direct dehydroxylamination of HAPR by ox adenosine deaminase has also been described (41). To find out which pathway (cf. Fig. 2A) takes place in human cell metabolism, mARC knockdown experiments and adenosine deaminase inhibition studies with dipyridamole (42–44) in HEK-293 were carried out. Knockdown of mARC1 in HEK-293 resulted in a decrease of inosine formation by 35% compared with the negative control (Fig. 2B). In Fig. 2C formation of inosine from HAPR or adenosine by HEK-293 metabolism with simultaneous inhibition of adenosine deaminase is shown. Without inhibition conversion rates with adenosine as substrate were 4.5 ± 0.1 nmol·min−1·mg protein−1 and with HAPR 2.2 ± 0.2 nmol·min−1·mg protein−1. By adding dipyridamole, conversion rates of both substrates were strongly reduced by 90–95% (Fig. 2C). Concurrent with adenosine deaminase inhibition, metabolism to adenosine could be observed (Fig. 2C). The effect of dipyridamole on N-reductive metabolism was checked on the basis of reduction of the model substrate benzamidoxime to benzamidine. N-reductive activity in HEK-293 was not affected by dipyridamole (data not shown).
FIGURE 2.
Metabolism of HAPR in HEK-293. A, possible inosine formation pathways from HAPR in human cells. B, effect of mARC knockdown on inosine formation from HAPR. HEK-293 cells were transfected with 10 nm mARC1 or non-targeting (NC) siRNA. The siRNA-mediated down-regulations of the proteins of interest were verified by Western blot using anti-mARC1, or anti-calnexin antibody. Calnexin levels were used as loading control. N-reductive activities were determined with 4 mm HAPR on day 3 after transfection as described under “Experimental Procedures.” Results are presented as means ± S.D. (n = 3). ***, p < 0.001. C, metabolism of HAPR or adenosine with concurrent inhibition of adenosine deaminase (ADA) in HEK-293. Formation of inosine from adenosine (2 mm) or from HAPR (2 mm) and formation of adenosine from HAPR (2 mm) with simultaneous incubation with dipyridamole as inhibitor were determined as described under “Experimental Procedures.”
Activity Assay with Recombinant Adenosine Deaminase
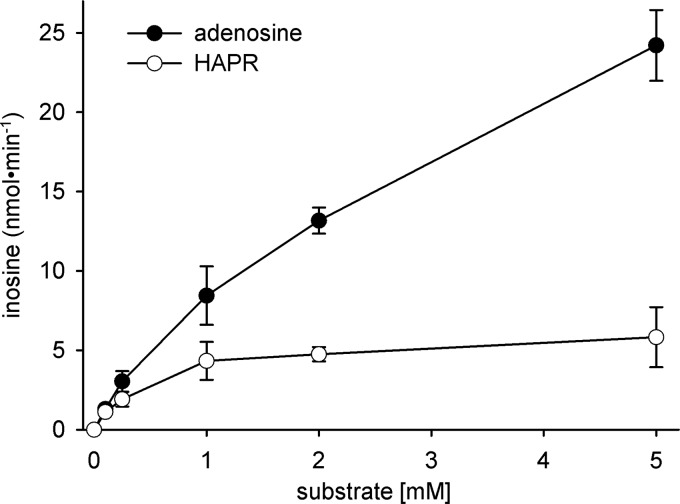
To further characterize the involvement of adenosine deaminase in detoxication of HAPR, incubations with recombinant adenosine deaminase were performed. In vitro, human recombinant adenosine deaminase is not only able to catalyze deamination of adenosine but also dehydroxylamination of HAPR (Fig. 3). Similar to metabolism in HEK-293 cells, conversion of HAPR is considerably lower than conversion of adenosine. Furthermore, we tested if addition of the mARC-containing enzyme system to the biotransformation assay could promote inosine formation by prior reduction to adenosine. However, a significant raise in inosine formation could not be measured under the prevailing conditions (data not shown).
FIGURE 3.
HAPR or adenosine as substrates of recombinant human adenosine deaminase. The dehydroxylamination of HAPR and the deamination of adenosine catalyzed by recombinant adenosine deaminase were characterized as described under “Experimental Procedures.”
HAP-induced Apoptosis in HeLa Cells with mARC Knockdown
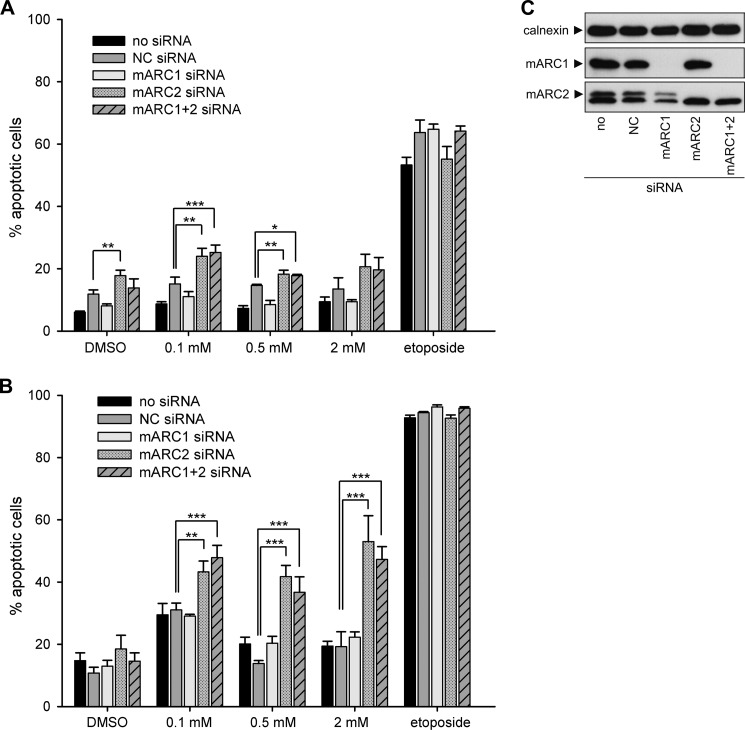
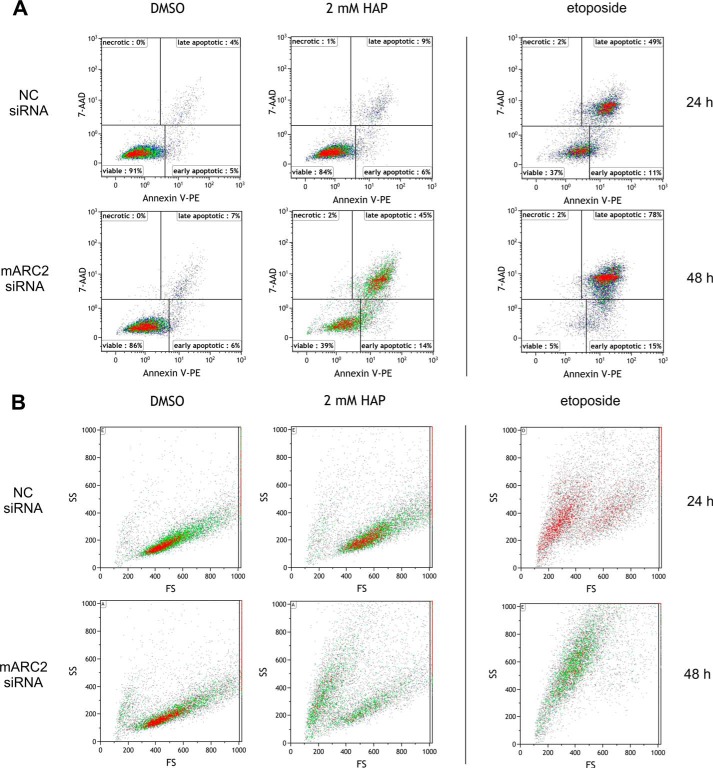
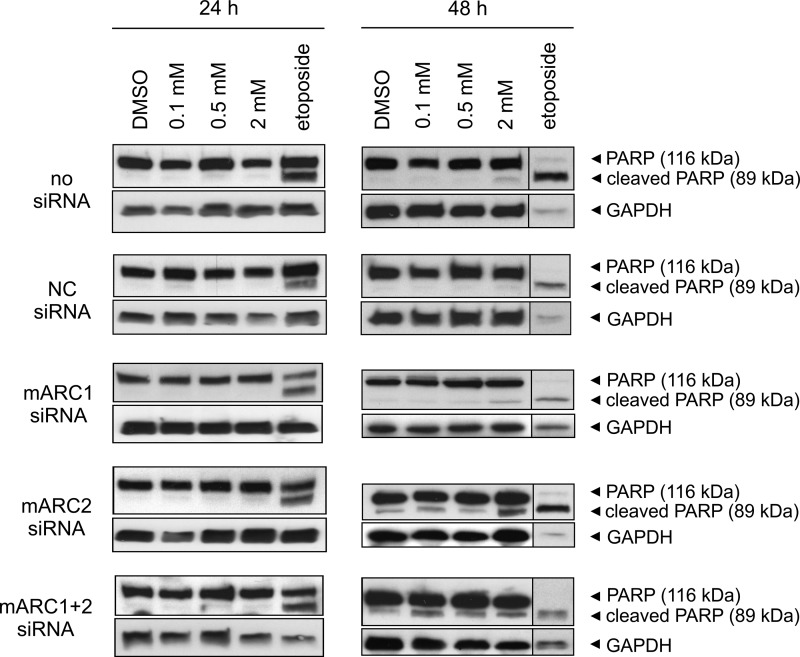
To examine the effect of mARC knockdown on sensitivity of HeLa cells toward HAP, cells with or without mARC knockdown were grown in the presence of increasing doses of HAP. mARC2 expression in HeLa is substantially higher than in HEK-293 cells. Therefore, both, mARC1 and mARC2 knockdown were evaluated. Down-regulation of mARC proteins were verified by Western blot (Fig. 5C). After 24 h and 48 h of cultivation, cells were compared regarding their apoptotic characteristics. For detection of apoptosis Annexin V-PE/7-AAD binding was quantified through flow cytometric analysis (Figs. 4 and 5). Additionally, caspase-mediated PARP cleavage as a hallmark of apoptosis was investigated by detection of 89-kDa PARP fragments in Western blot analysis (Fig. 6). Hoechst staining was used to visualize chromatin condensation during apoptosis (Fig. 7). No significant elevated levels of apoptosis could be observed after 24 h cultivation in the presence of HAP (Fig. 5A). Upon treatment with HAP for 48 h, 43–53% of cells with mARC2 or simultaneous mARC1 and mARC2 knockdown underwent apoptosis (versus 15–19% apoptotic cells in DMSO negative control). In control HeLa cells and cells with mARC1 knockdown HAP triggered apoptotic effects were less decisively. With 2 mm HAP 19–22% underwent apoptotic cell death (versus 11–15% apoptotic cells in DMSO negative control). Thus, the amount of apoptotic cells in HeLa with mARC2 or simultaneous mARC1 and 2 knockdown after 48 h cultivation in 2 mm HAP was increased by two times (Fig. 5B). Occurring apoptosis also becomes apparent when analyzing light scatter measurements (Fig. 4B). In the light scatter dotplots of cells with mARC2 or simultaneous mARC1 and 2 knockdown which were treated with HAP a second cell population with smaller forward scatter and higher side scatter can be observed. The same characteristics were seen in etoposide-treated cells. These patterns are typically seen with apoptotic cells due to associated decrease in cell size and formation of apoptotic vesicles (45). In Addition, using Hoechst staining, nuclei with morphological alterations due to chromatin condensation could be detected in cells with mARC2 and simultaneous mARC1 and 2 knockdown grown for 48 h in 2 mm HAP (Fig. 7). In accordance to these results, high levels of PARP fragments could be detected in etoposide-treated HeLa cells while no significant elevated PARP cleavage was observed in HAP-treated cells after 24 h of cultivation. After 48 h, however, in all HeLa cells grown in 2 mm HAP PARP fragments could be detected. Notably increased level of PARP cleavage products with even lower HAP dosages could be seen in cells with mARC2 or simultaneous mARC1 and 2 knockdown but not with control cells or cells with mARC1 knockdown (Fig. 6).
FIGURE 5.
Effect of mARC knockdown on HAP-induced apoptosis in HeLa. HeLa cells were grown in the presence of various concentrations of HAP or 1% (v/v) DMSO. Etoposide treatment (100 μm) was used as a positive control. Apoptotic cell death was quantified by flow cytometric analysis of Annexin V-PE/7-AAD-stained cells as described under “Experimental Procedures.” Results are presented as means ± S.D. (n = 3). *, p < 0.05; **, p < 0.01; ***, p < 0.001. A, evaluation after 24 h cultivation. B, evaluation after 48 h of cultivation. C, verification of mARC knockdown in HeLa. Down-regulation of the proteins of interest were verified by Western blot when cells were plated for HAP treatment using anti-mARC1, anti-mARC2, or anti-calnexin antibody. Calnexin levels were used as loading control.
FIGURE 4.
Flow cytometric analysis of Annexin V-PE/7-AAD stained HeLa cells. Representative data showing HeLa cells with or without mARC2 knockdown that were grown in the presence of 2 mm HAP or 1% (v/v) DMSO for 48 h. Etoposide treatment (100 μm, 24 h or 48 h) was used as a positive control (A) fluorescence dotplots (B) light scatter dotplots.
FIGURE 6.
Effect of mARC knockdown on HAP-induced PARP cleavage in HeLa. HeLa cells were grown in the presence of various concentrations of HAP or 1% (v/v) DMSO. Etoposide treatment (100 μm) was used as a positive control. Caspase-dependent apoptotic cell death was evaluated by immunoblot analysis of cleaved PARP using anti-PARP, or anti-GAPDH antibody. GAPDH levels were used as loading control. Analyzed lysates were pools of triplicates.
FIGURE 7.
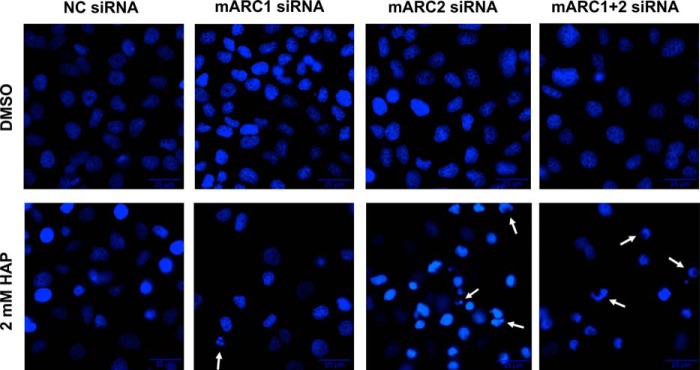
Nuclear morphology in HeLa after HAP treatment. HeLa cells with or without mARC knockdown were grown in the presence of 2 mm HAP or 1% (v/v) DMSO. Changes in chromatin morphology were evaluated by Hoechst staining as described under “Experimental Procedures.” Arrows indicate examples for nuclei with prominent chromatin condensation.
DISCUSSION
The mARC-containing three component enzyme system is responsible for the reduction of various N-hydroxylated compounds. Therefore, it is well accepted that it plays a major role in N-reductive metabolism (46). However, its endogenous substrates are as yet unknown. In previous studies it was demonstrated that in vitro the reconstituted recombinant mARC enzyme system has the ability to reduce all until now tested N-hydroxylated nucleobases and nucleosides (27). Accordingly, mARC proteins are assumed to be involved in metabolism to the corresponding canonical nucleobases or nucleosides. Since those analogs have been shown to be toxic and mutagenic in bacteria, yeast, and mammalian cells (4, 6, 16, 47), the question of physiological relevance of the mARC-mediated N-reduction as detoxication pathway is of particular interest. Therefore, detoxication studies with HAP as an adenine analog and with HAPR as an adenosine analog in human cells were performed. Initially, their metabolisms in HEK-293 cells were characterized and the involvement of mARC was examined via siRNA-mediated knockdown. For HAP, mARC-dependent N-reductive metabolism to adenine could be demonstrated. The N-hydroxylated nucleoside HAPR however, was very efficiently metabolized to inosine and not as expected to the corresponding nucleoside adenosine. In accordance to our findings, it is described that adenosine but also HAPR is quickly broken down to inosine in organisms (39, 40). It is known that this deamination of adenosine is catalyzed by adenosine deaminase (41). Therefore, it is conceivable that in cell metabolism HAPR is initially reduced to adenosine by the mARC system and subsequently deaminated to inosine through adenosine deaminase, or that HAPR acts as a direct substrate on adenosine deaminase as described by Rockwell and Maguire (41) (cf. Fig. 2A). The latter assumption is supported by the hemolytic effects caused by HAPR in humans (48). Similar effects are obtained with hydroxylamine, which would be released upon direct dehydroxylamination of HAPR. Our investigations in human cells demonstrate that metabolism of HAPR to inosine proceeds mARC-mediated to some degree, as mARC knockdown decreases inosine formation by 35% as compared with negative control (Fig. 2B). The involvement of adenosine deaminase was proven by the use of the adenosine deaminase inhibitor dipyridamole (42–44), leading to 90–95% inhibition of metabolism to inosine (Fig. 2C). Since dipyridamole did not have any impact on N-reductive activity of cells, interference with the mARC-system can be excluded. Concurrent to inhibition of adenosine deaminase, metabolism to adenosine could be detected. Supporting the results found with mARC knockdown this observation indicates the involvement of the mARC-containing N-reductive enzyme system in metabolism of HAPR. Beyond that, however, human recombinant adenosine deaminase is also able to catalyze direct dehydroxylamination of HAPR to inosine even though adenosine acts as a far better substrate (Fig. 3). Therefore, it can be assumed that in human cell systems metabolism to inosine is only to a small extent mARC-mediated via N-reduction to adenosine as an intermediate. In fact, direct dehydroxylamination of HAPR catalyzed by adenosine deaminase seems to be the primary and a much more efficient detoxication pathway.
In the following studies the effect of mARC knockdown on HAP sensitivity of human cells was examined. Preliminary investigation revealed that HEK-293 was not sensitive to HAP under any tested conditions. It has been reported previously that the response to HAP treatment can be cell line specific (16). HAP is assumed to exhibit its toxic and mutagenic effects under certain cellular conditions. For instance, variations in nucleotide pool size or differences in rate of cell division are supposably factors for variations in response to base analogs (49). Therefore, we chose to perform HAP toxicity studies with HeLa cells. For HeLa, a HAP-induced mutagenesis has already been described (17). Using flow cytometric analysis of Annexin V-PE and 7-AAD stained cells and by detecting caspase-mediated PARP cleavage, the impact of mARC knockdown on sensitivity to HAP was examined after 24 h and 48 h of cultivation in the presence of 0.1–2 mm HAP (Figs. 4–6). We observed no prominent induced effects after 24 h. After 48 h of treatment, however, HAP triggered apoptosis became evident. In conformity with the mechanism of mutagenesis via acting as a base analog described in literature (7, 9), cell division and therefore DNA replication seemed to be essential for HAP to exert its cytotoxic effects. Nevertheless, the increase of apoptotic cells within control cells without mARC knockdown and cells with mARC1 knockdown was rather modest. In the presence of HAP 10% more cells underwent apoptotic cell death. PARP cleavage could be detected with 2 mm HAP (Fig. 6). In contrast to that, sensitivity of cells with mARC2 or simultaneous mARC1 and 2 knockdown was increased strongly. Within those cells, 30% more cells revealed apoptotic characteristics compared with cultivations in media without HAP. Characteristic PARP fragments could already be detected under the influence of 0.1 mm HAP. We thus demonstrated that mARC2 plays a critical role in detoxication of HAP in human cell metabolism. Presumably mARC2-dependent reduction prevents contamination of nucleoside pools and subsequent incorporation into the DNA. Unlike ITPase, which prevents incorporation of HAPTP by cleaving it into the corresponding monophosphate, mARC-mediated reduction eliminates the base analog completely from the nucleotide pool and instead provides the corresponding canonical nucleobase to nucleic acid metabolism. Since mARC1 expression in HeLa cells is rather high and previous knockdown studies in human cells demonstrated that both mARC proteins are capable of reducing N-hydroxylated substrates, this finding is unexpected. The discrepancy between the involvement of mARC1 in N-reduction found in human cells and missing influence on sensitivity toward HAP in HeLa cells should be subject of further investigations. The fact that all up to now analyzed mammalian genomes harbor two mARC genes suggest the evolutionary need of each protein. However up to now, no major differences between mARC1 and mARC2 were found. Therefore, the investigation of this found difference is of great interest and of high relevance for the understanding of detoxication pathways and physiological functions of mARC within or even outside the N-reductive system.
In conclusion, mARC2 was shown to protect HeLa cells against the apoptotic effects of the base analog, whereas the involvement of mARC1 in reductive detoxication of HAP seems to be less pivotal. To explain the different roles of mARC1 and mARC2 in the detoxication pathway of HAP, regulatory mechanisms of the proteins need to be elucidated and alternative subcellular localizations of mARC as for instance the nucleus should be examined.
Acknowledgments
We thank Petra Köster, and Sven Wichmann for technical assistance. We thank Prof. Dr. Holger Kalthoff for providing HeLa cells.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG, Grant Number CL56/9-1, ME 1266/24-1).
- HAP
- N6-hydroxylaminopurine
- ADA
- adenosine deaminase
- HAPR
- N6-hydroxyadenosine
- ITPase
- inosine triphosphate pyrophosphatase
- mARC
- mitochondrial amidoxime reducing component
- MEM
- Minimum Essential Medium
- PARP
- poly (ADP-ribose) polymerase
- TBST
- TBS containing Tween-20.
REFERENCES
- 1. Khromov-Borisov N. N. (1997) Naming the mutagenic nucleic acid base analogs: the Galatea syndrome. Mutat. Res. 379, 95–103 [DOI] [PubMed] [Google Scholar]
- 2. Negishi K., Bessho T., Hayatsu H. (1994) Nucleoside and nucleobase analog mutagens. Mutat. Res 318, 227–238 [DOI] [PubMed] [Google Scholar]
- 3. Freese E. (1959) The specific mutagenic effect of base analogues on Phage T4. J. Mol. Biol. 1, 87–105 [Google Scholar]
- 4. Kozmin S. G., Schaaper R. M., Shcherbakova P. V., Kulikov V. N., Noskov V. N., Guetsova M. L., Alenin V. V., Rogozin I. B., Makarova K. S., Pavlov Y. I. (1998) Multiple antimutagenesis mechanisms affect mutagenic activity and specificity of the base analog 6-N-hydroxylaminopurine in bacteria and yeast. Mutat. Res. 402, 41–50 [DOI] [PubMed] [Google Scholar]
- 5. Krenitsky T. A., Neil S. M., Elion G. B., Hitchings G. H. (1969) Adenine phosphoribosyltransferase from monkey liver. Specificity and properties. J. Biol. Chem. 244, 4779–4784 [PubMed] [Google Scholar]
- 6. Burchenal J. H., Dollinger M., Butterbaugh J., Stoll D., Giner-Sorolla A. (1967) Studies of 6-N-hydroxylamino-9-beta-D-ribofuranosylpurine in mouse leukemia. Biochem. Pharmacol 16, 423–428 [DOI] [PubMed] [Google Scholar]
- 7. Abdul-Masih M. T., Bessman M. J. (1986) Biochemical studies on the mutagen, 6-N-hydroxylaminopurine. Synthesis of the deoxynucleoside triphosphate and its incorporation into DNA in vitro. J. Biol. Chem. 261, 2020–2026 [PubMed] [Google Scholar]
- 8. Burgis N. E., Brucker J. J., Cunningham R. P. (2003) Repair system for noncanonical purines in Escherichia coli. J. Bacteriol. 185, 3101–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pavlov Y. I., Suslov V. V., Shcherbakova P. V., Kunkel T. A., Ono A., Matsuda A., Schaaper R. M. (1996) Base analog N6-hydroxylaminopurine mutagenesis in Escherichia coli: genetic control and molecular specificity. Mutat. Res 357, 1–15 [DOI] [PubMed] [Google Scholar]
- 10. Clement B., Kunze T. (1990) Hepatic microsomal N-hydroxylation of adenine to 6-N-hydroxylaminopurine. Biochem. Pharmacol 39, 925–933 [DOI] [PubMed] [Google Scholar]
- 11. Simandan T., Sun J., Dix T. A. (1998) Oxidation of DNA bases, deoxyribonucleosides and homopolymers by peroxyl radicals. Biochem. J. 335 (Pt 2), 233–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lieberman I. (1956) Enzymatic synthesis of adenosine-5′-phosphate from inosine-5′-phosphate. J. Biol. Chem. 223, 327–339 [PubMed] [Google Scholar]
- 13. White R. H. (2010) The twists and turns of enzyme function. J. Bacteriol. 192, 2023–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kozmin S. G., Leroy P., Pavlov Y. I., Schaaper R. M. (2008) YcbX and yiiM, two novel determinants for resistance of Escherichia coli to N-hydroxylated base analogues. Mol. Microbiol. 68, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Anantharaman V., Aravind L. (2002) MOSC domains: ancient, predicted sulfur-carrier domains, present in diverse metal-sulfur cluster biosynthesis proteins including Molybdenum cofactor sulfurases. FEMS Microbiol. Lett. 207, 55–61 [DOI] [PubMed] [Google Scholar]
- 16. Waisertreiger I. S.-R., Menezes M. R., Randazzo J., Pavlov Y. I. (2010) Elevated levels of DNA strand breaks induced by a base analog in the human cell line with the P32T ITPA variant. J. Nucleic Acids 2010, 872180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Menezes M. R., Waisertreiger I. S.-R., Lopez-Bertoni H., Luo X., Pavlov Y. I. (2012) Pivotal role of inosine triphosphate pyrophosphatase in maintaining genome stability and the prevention of apoptosis in human cells. PLoS ONE 7, e32313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gruenewald S., Wahl B., Bittner F., Hungeling H., Kanzow S., Kotthaus J., Schwering U., Mendel R. R., Clement B. (2008) The fourth molybdenum containing enzyme mARC: cloning and involvement in the activation of N-hydroxylated prodrugs. J. Med. Chem. 51, 8173–8177 [DOI] [PubMed] [Google Scholar]
- 19. Havemeyer A., Grünewald S., Wahl B., Bittner F., Mendel R. R., Erdélyi P., Fischer J., Clement B. (2010) Reduction of N-hydroxy-sulfonamides, including N-hydroxy-valdecoxib, by the molybdenum-containing enzyme mARC. Drug Metab. Dispos 38, 1917–1921 [DOI] [PubMed] [Google Scholar]
- 20. Jakobs H. H., Froriep D., Havemeyer A., Mendel R. R., Bittner F., Clement B. (2014) The Mitochondrial Amidoxime Reducing Component (mARC): Involvement in Metabolic Reduction of N-Oxides, Oximes and N-hydroxy-amidinohdrazones. Chem. Med. Chem. 9, 2381–2387 [DOI] [PubMed] [Google Scholar]
- 21. Islinger M., Lüers G. H., Li K. W., Loos M., Völkl A. (2007) Rat liver peroxisomes after fibrate treatment. A survey using quantitative mass spectrometry. J. Biol. Chem. 282, 23055–23069 [DOI] [PubMed] [Google Scholar]
- 22. Wiese S., Gronemeyer T., Ofman R., Kunze M., Grou C. P., Almeida J. A., Eisenacher M., Stephan C., Hayen H., Schollenberger L., Korosec T., Waterham H. R., Schliebs W., Erdmann R., Berger J., Meyer H. E., Just W., Azevedo J. E., Wanders R. J. A., Warscheid B. (2007) Proteomics characterization of mouse kidney peroxisomes by tandem mass spectrometry and protein correlation profiling. Mol. Cell Proteomics 6, 2045–2057 [DOI] [PubMed] [Google Scholar]
- 23. Mendel R. R., Kruse T. (2012) Cell biology of molybdenum in plants and humans. Biochim. Biophys. Acta 1823, 1568–1579 [DOI] [PubMed] [Google Scholar]
- 24. Froriep D., Clement B., Bittner F., Mendel R. R., Reichmann D., Schmalix W., Havemeyer A. (2013) Activation of the anti-cancer agent upamostat by the mARC enzyme system. Xenobiotica 43, 780–784 [DOI] [PubMed] [Google Scholar]
- 25. Havemeyer A., Bittner F., Wollers S., Mendel R. R., Kunze T., Clement B. (2006) Identification of the missing component in the mitochondrial benzamidoxime prodrug-converting system as a novel molybdenum enzyme. J. Biol. Chem. 281, 34796–34802 [DOI] [PubMed] [Google Scholar]
- 26. Mikula M., Rubel T., Karczmarski J., Goryca K., Dadlez M., Ostrowski J. (2011) Integrating proteomic and transcriptomic high-throughput surveys for search of new biomarkers of colon tumors. Funct. Integr. Genomics 11, 215–224 [DOI] [PubMed] [Google Scholar]
- 27. Krompholz N., Krischkowski C., Reichmann D., Garbe-Schönberg D., Mendel R. R., Bittner F., Clement B., Havemeyer A. (2012) The Mitochondrial Amidoxime Reducing Component (mARC) Is Involved in Detoxification of N-Hydroxylated Base Analogues. Chem. Res. Toxicol. 25, 2443–2450 [DOI] [PubMed] [Google Scholar]
- 28. Kotthaus J., Wahl B., Havemeyer A., Kotthaus J., Schade D., Garbe-Schönberg D., Mendel R. R., Bittner F., Clement B. (2011) Reduction of N(ω)-hydroxy-L-arginine by the mitochondrial amidoxime reducing component (mARC). Biochem. J 433, 383–391 [DOI] [PubMed] [Google Scholar]
- 29. Sparacino-Watkins C. E., Tejero J., Sun B., Gauthier M. C., Thomas J., Ragireddy V., Merchant B. A., Wang J., Azarov I., Basu P., Gladwin M. T. (2014) Nitrite reductase and nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J. Biol. Chem. 289, 10345–10358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Neve E. P. A., Nordling A., Andersson T. B., Hellman U., Diczfalusy U., Johansson I., Ingelman-Sundberg M. (2012) Amidoxime reductase system containing cytochrome b5 type B (CYB5B) and MOSC2 is of importance for lipid synthesis in adipocyte mitochondria. J. Biol. Chem. 287, 6307–6317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jakobs H. H., Mikula M., Havemeyer A., Strzalkowska A., Borowa-Chmielak M., Dzwonek A., Gajewska M., Hennig E. E., Ostrowski J., Clement B. (2014) The N-Reductive System Composed of Mitochondrial Amidoxime Reducing Component (mARC), Cytochrome b5 (CYB5B) and Cytochrome b5 Reductase (CYB5R) Is Regulated by Fasting and High Fat Diet in Mice. PLoS ONE 9, e105371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Newton B. W., Cologna S. M., Moya C., Russell D. H., Russell W. K., Jayaraman A. (2011) Proteomic analysis of 3T3-L1 adipocyte mitochondria during differentiation and enlargement. J. Proteome Res. 10, 4692–4702 [DOI] [PubMed] [Google Scholar]
- 33. Malik A. N., Rossios C., Al-Kafaji G., Shah A., Page R. A. (2007) Glucose regulation of CDK7, a putative thiol related gene, in experimental diabetic nephropathy. Biochem. Biophys. Res. Commun. 357, 237–244 [DOI] [PubMed] [Google Scholar]
- 34. Wahl B., Reichmann D., Niks D., Krompholz N., Havemeyer A., Clement B., Messerschmidt T., Rothkegel M., Biester H., Hille R., Mendel R. R., Bittner F. (2010) Biochemical and spectroscopic characterization of the human mitochondrial amidoxime reducing components hmARC-1 and hmARC-2 suggests the existence of a new molybdenum enzyme family in eukaryotes. J. Biol. Chem. 285, 37847–37859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Plitzko B., Ott G., Reichmann D., Henderson C. J., Wolf C. R., Mendel R. R., Bittner F., Clement B., Havemeyer A. (2013) The involvement of mitochondrial amidoxime reducing components 1 and 2 and mitochondrial cytochrome b5 in N-reductive metabolism in human cells. J. Biol. Chem. 288, 20228–20237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 37. Toné S., Sugimoto K., Tanda K., Suda T., Uehira K., Kanouchi H., Samejima K., Minatogawa Y., Earnshaw W. C. (2007) Three distinct stages of apoptotic nuclear condensation revealed by time-lapse imaging, biochemical and electron microscopy analysis of cell-free apoptosis. Exp. Cell Res. 313, 3635–3644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ormerod M. G., Sun X. M., Brown D., Snowden R. T., Cohen G. M. (1993) Quantification of apoptosis and necrosis by flow cytometry. Acta Oncol 32, 417–424 [DOI] [PubMed] [Google Scholar]
- 39. Feng J. D., Yeung P. K. (2000) A simple high-performance liquid chromatography assay for simultaneous measurement of adenosine, guanosine, and the oxypurine metabolites in plasma. Ther. Drug Monit. 22, 177–183 [DOI] [PubMed] [Google Scholar]
- 40. Cristalli G., Vittori S., Eleuteri A., Grifantini M., Volpini R., Lupidi G., Capolongo L., Pesenti E. (1991) Purine and 1-deazapurine ribonucleosides and deoxyribonucleosides: synthesis and biological activity. J. Med. Chem 34, 2226–2230 [DOI] [PubMed] [Google Scholar]
- 41. Rockwell M., Maguire M. H. (1966) Studies on adenosine deaminase. I. Purification and properties of ox heart adenosine deaminase. Mol. Pharmacol 2, 574–584 [PubMed] [Google Scholar]
- 42. Pfleger K., Niederau D., Volkmer I. (1969) Ein Beitrag zum Wirkungsmechanismus von Dipyridamol: Hemmung der Adenosinaufnahme in Erythrocyten durch Dipyridamol. Naunyn Schmiedebergs Arch Pharmakol 265, 118–130 [PubMed] [Google Scholar]
- 43. Bunag R. D., Douglas C. R., Imai S., Berne R. M. (1964) Influence of a Pyrimidopyrimidine Derivative on Deamination of Adenosine by Blood. Circ. Res. 15, 83–88 [DOI] [PubMed] [Google Scholar]
- 44. Stafford A. (1966) Potentiation of adenosine and the adenine nucleotides by dipyridamole. Br. J. Pharmacol. Chemother. 28, 218–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dive C., Gregory C. D., Phipps D. J., Evans D. L., Milner A. E., Wyllie A. H. (1992) Analysis and discrimination of necrosis and apoptosis (programmed cell death) by multiparameter flow cytometry. Biochim. Biophys. Acta 1133, 275–285 [DOI] [PubMed] [Google Scholar]
- 46. Havemeyer A., Lang J., Clement B. (2011) The fourth mammalian molybdenum enzyme mARC: current state of research. Drug Metab. Rev 43, 524–539 [DOI] [PubMed] [Google Scholar]
- 47. Barrett J. C. (1981) Induction of gene mutation in and cell transformation of mammalian cells by modified purines: 2-aminopurine and 6-N-hydroxylaminopurine. Proc. Natl. Acad. Sci. U.S.A. 78, 5685–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Dollinger M. R., Krakoff I. H. (1975) Hemolysis induced by 6-N-hydroxylaminopurine riboside, an adenosine analogue. Clin. Pharmacol. Ther 17, 57–65 [DOI] [PubMed] [Google Scholar]
- 49. Haghdoost S., Sjölander L., Czene S., Harms-Ringdahl M. (2006) The nucleotide pool is a significant target for oxidative stress. Free Radic. Biol. Med. 41, 620–626 [DOI] [PubMed] [Google Scholar]