Background: Type 1 interferons (IFN1) mediate defense against viruses but their role in regulating retrotransposon activities is unknown.
Results: LINE-1 retrotransposon induces IFN1, which in turn inhibits LINE-1 retrotransposition.
Conclusion: IFN1 regulate activities and propagation of LINE-1.
Significance: Given that retrotransposons alter the genome, IFN1 play a role in maintenance of genomic integrity.
Keywords: DNA damage, genomic instability, interferon, receptor, signaling, LINE-1, radiation, retrotransposon
Abstract
Type I interferons (IFN) including IFNα and IFNβ are critical for the cellular defense against viruses. Here we report that increased levels of IFNβ were found in testes from mice deficient in MOV10L1, a germ cell-specific RNA helicase that plays a key role in limiting the propagation of retrotransposons including Long Interspersed Element-1 (LINE-1). Additional experiments revealed that activation of LINE-1 retrotransposons increases the expression of IFNβ and of IFN-stimulated genes. Conversely, pretreatment of cells with IFN suppressed the replication of LINE-1. Furthermore, the efficacy of LINE-1 replication was increased in isogenic cell lines harboring inactivating mutations in diverse elements of the IFN signaling pathway. Knockdown of the IFN receptor chain IFNAR1 also stimulated LINE-1 propagation in vitro. Finally, a greater accumulation of LINE-1 was found in mice that lack IFNAR1 compared with wild type mice. We propose that LINE-1-induced IFN plays an important role in restricting LINE-1 propagation and discuss the putative role of IFN in preserving the genome stability.
Introduction
Type 1 interferons ((IFN)2 including IFNα and IFNβ) play a major role in anti-viral defenses (1). These cytokines act by interacting with the Type 1 IFN receptor that consists of IFNAR1 and IFNAR2 chains and mediates all cellular effects of IFN (2–4). Association of IFN with a heteromeric receptor leads to activation of Janus kinases TYK2 and JAK1 and phosphorylation-dependent activation of the Signal Transducers and Activators of Transcription (STAT1/2) proteins. These proteins interact with IRF9 to form a potent transcription factor that up-regulates the expression of several hundreds of IFN-stimulated genes, whose products both elicit anti-viral effects and contribute to the development of the inflammatory tissue injury (2, 3, 5–8). Since integration of viruses into host DNA induces genetic changes (9), it may be hypothesized that anti-viral cytokines including IFN can also play a role in maintaining the integrity of the host genome.
During evolution, eukaryotic genomes have been undergoing incessant modifications due to diverse events including the activities of mobile genetic elements (10). For example, the Long Interdispersed Element-1 (LINE-1) retrotransposons commonly found in many types of mammalian cells (11) have propagated to such extent that they constitute a substantial fraction of genome mass (12). Active propagation of LINE-1 and similar retrotransposons involves near random insertion of these elements into genomic DNA and, accordingly, may lead to gene disruption and an increase in genomic instability (13). Accordingly, germ cells guard their genome by developing a sophisticated and efficient system involving MOV10L1 RNA helicase and Piwi proteins that suppress propagation of LINE-1. Recent analysis of mice that lack Mov10l1 showed that spermatocytes from these animals exhibit an increase in LINE-1 activity, massive DNA damage, and post-meiotic proliferation arrest (14).
Here we report that germ cells from the Mov10l1 knock-out mice that express highly active LINE-1 also exhibit elevated expression of IFNβ. Using in vitro models of LINE-1 replication in cells we found that LINE-1 stimulates the expression and function of IFN and that the latter functions to suppress LINE-1 propagation. An increased rate of LINE-1 propagation was found in cells and mouse tissues deficient in IFN signaling. These results suggest that IFN produced in response to LINE-1 activities can restrict the very activities of these retrotransposons.
MATERIALS AND METHODS
Plasmids, siRNAs, and Other Reagents
The LINE-1-EGFP-puromycin reporter constructs (15, 16) pEF06R (which encodes the ORF2 protein with functional endonuclease) and pEF05J (encodes endonuclease-deficient ORF2) were kindly provided by Eline T. Luning Prak (University of Pennsylvania). Human IFNAR2 expression vector pMT2T-hIFNAR2-HA was a generous gift from John Krolewski (University of Rochester Medical Center). The sense strand sequences of siRNAs (Ambion) directed against target molecules were as follows: human RNaseL (5′-GGAAGUCUCUUGUCUGCAAtt-3′), human MOV10 (5′-GACCCUGACUGGAAAGUAUtt-3′), mouse IFNβ (5′-GAAUGAGACUAUUGUUGUAtt-3′), scrambled siRNA (siCon, Ambion Silencer® Negative Control No. 1). Human IFNβ (PBL Inc), and puromycin (Sigma) were purchased.
Cells, Cell Lines, Culture Conditions
Primary mouse embryonic fibroblasts (MEFs) were prepared from the embryos of wild-type C57Bl/6J mice as previously described (17). Briefly, embryos were collected from the pregnant mice on day 14–16 of gestation. Heads and internal organs were removed. Remaining tissue was minced and disassociated with 0.25% trypsin for 5 min. The cells were then plated in DMEM supplemented with 10% FBS (HyClone Laboratories), 100 units/ml penicillin, and 0.1 mg/ml streptomycin. Two hours later, the adherent MEFs (P0) were washed twice with phosphate-buffered saline (PBS) and cultured in the complete medium again. Cells were passaged every 2–3 days. Only P2 and P4 MEFs were used in this study. HeLa cells and mouse NIH3T3 cells were obtained from ATCC. Human fibrosarcoma 2fTGH cells and its derivatives (U1A, U3A, and U5A), kindly provided by George Stark, Cleveland Foundation, were maintained in DMEM supplemented with 10% (v/v) FBS, 100 units/ml penicillin and 0.1 mg/ml streptomycin. U1A-derived stable clones expressing either wild type (WT) or kinase-deficient (KR) Tyk2 (described in Refs. 18, 19, a gift from Sandra Pellegrini, Pasteur Institute, Paris, France) were grown in the same medium with addition of G418 (400 μg/ml).
Antibodies and Immunofluorescent Analysis
All the primary and secondary antibodies were diluted in blocking buffer (5% BSA and 0.1% Tween-20 in PBS). The following primary antibodies were used: anti- mouse IFNβ was purchased from Millipore. Anti-IRF7 was purchased from Abcam. Secondary antibody used in this study was Alexa Fluor 594 goat anti-rabbit IgG (H+L) (Invitrogen).
NIH3T3 cells were plated in 35 mm collagen-coated glass bottom dishes (MatTek Corporation). The next day, the cells were transfected with siRNA (control or against IFNβ) or treated with Abs (control or neutralizing antibody against IFNβ, from Leinco Technologies, both at 10 μg/ml). Twenty-four hours later, the cells were transfected with LINE-1-EGFP-puromycin reporter constructs. After 30 h, cells were fixed with 4% paraformaldehyde (Affymetrix) in PBS for 10 min at room temperature. The fixed cells were washed three times with PBS and permeabilized with 0.25% Triton X-100 in PBS for 10 min. After another PBS wash, cells were blocked by incubation with 5% BSA and 0.1% Tween-20 in PBS for 1 h at room temperature. The cells were then incubated with primary antibodies overnight at 4 °C. The cells were washed three times with PBS and incubated with secondary antibody for 1 h at 37 °C in the dark, washed twice more with PBS, and treated with DAPI (Sigma) (1 μg/ml) for 2 min. The cells were then washed twice and imaged using the Zeiss LSM710 confocal with a 40× objective lens. All images were processed and quantified using the Fuji software (20). A total of at least 25 fields of cells randomly selected from three independent experiments were scored per group for quantification of percentage of single or double positive cells for GFP, IFNβ, or IRF7 proteins in a double blind manner.
Antibodies and Immunoblots
Antibodies against IFNAR2 (Santa Cruz) and TYK2 (Cell Signaling) were purchased. Secondary antibodies conjugated to horseradish peroxidase were purchased from Millipore Bioscience Research Reagents. Immunoblotting procedures were described previously (17).
LINE-1 Activation Assays
LINE-1 activation assays were carried out by determining the percent of GFP-positive cells (by flow cytometry) indicative of LINE-1 expression and retrotransposition as previously described (15, 21) with minor modifications. To determine the role of IFNβ in LINE-1 retrotransposition, Hela cells were plated in a six-well tissue culture plate. The next day, Hela cells were transfected with LINE-1-EGFP-puromycin reporter construct using Lipofectamine 2000 (Invitrogen). Four hours later, the cells were washed and incubated with fresh medium with or without human IFNβ (500 units/ml for 20 h). After that, cells were washed twice, incubated with fresh medium without IFN for 24 h and then selected in this medium supplemented with puromycin (3 μg/ml) for 14 days.
To study the effect of endogenous MOV10 and RNaseL on LINE-1 retrotransposition, Hela cells were transfected with a control siRNA or siRNA against MOV10 or RNaseL using Lipofectamine RNAiMAX Transfection Reagent (Invitrogen). Twenty-four hours later, the cells were transfected with LINE-1-EGFP-puromycin reporter construct using Lipofectamine 2000 (Invitrogen) for four hours, washed, and incubated with fresh medium with or without human IFNβ (500 units/ml for 20 h). After that, cells were washed and incubated with fresh medium without IFNβ for 24 h. An aliquot of the cells without IFNβ treatment was used to analyze the mRNA levels of MOV10, RNaseL and IFNβ by QPCR. The remaining cells were re-plated and then subjected to puromycin selection (3 μg/ml for 72 h). To study the role of IFNAR1 in LINE-1 retrotransposition, Hela cells were transfected with indicated LINE-1-EGFP-puromycin reporter constructs and shRNA against IFNAR1 or against luciferase (control) using Lipofectamine 2000. After 48 h, cells were subjected to 14 days of puromycin selection prior to estimating percent of GFP-positive cells by flow cytometry.
In the experiments aimed to detect the effect of endogenous IFNβ signaling on LINE-1 retrotransposition, 2fTGH cells and 2fTGH-derivatives were co-transfected with LINE-1-EGFP-puromycin and mCherry plasmids (to normalize transfection efficiency) for 48 h and then analyzed by FACS to assess double-positive (GFP+/RFP+) cells. Cells were analyzed using LSRFortessa flow cytometer (BD Biosciences). Results were quantified using FlowJo 7.6 software.
Semi-quantitative RT-PCR and qPCR
In experiments to study the expression of LINE-1 or IFNβ in vivo, the testes collected from male mice (2-month-old Mov10l1−/− and Mov10l1−/− mice or 5-day-old Ifnar1+/+ and Ifnar1−/− mice) were flash-frozen and pulverized in liquid nitrogen, homogenized in Trizol reagent (Ambion, Life Technologies), and extracted with chloroform. In experiments to study the effect of LINE-1 retrotransposition on the expression of IFNβ and its targeted genes in vitro, primary MEFs were plated in 6-well tissue culture plate. The next day, the cells were transfected with LINE-1-EGFP-puromycin reporter constructs using Xfect™ Transfection Reagent (Clontech). After 30 h, total RNA of the cells were prepared with Trizol and chloroform.
Reverse transcription was carried out using Revertaid first strand cDNA synthesis kit (Thermo Scientific) and the cDNA was used for semi-quantitative RT-PCR and quantitative QPCR. Analyses of expression of genes were carried out using the following primers for targeted mouse molecules: Ifnb (FW, 5′-GTCAGAGTGGAAATCCTAAG-3′, REV, 5′-ACAGCATCTGCTGGTTGAAG-3′), Isg15 (FW, 5′-GGAACGAAAGGGGCCACAGCA-3′, REV, 5′- CCTCCATGGGCCTTCCCTCGA-3′), Irf7 (FW, 5′-CCACACCCCCATCTTCGA-3′, REV, 5′-CCTCCGAGCCCGAAACTC-3′), Mov10l1 (FW, 5′-TGTAGCAGTGCAGGACTGTTTTACC-3′, REV, 5′-CAACAATGGGTTATATGCACCGCAAG-3′), Mov10 (FW, 5′-GAGGTTCGAGAGTTTTCTGGC-3′, REV, 5′-GCGATCTTCATTCCATACAGCAT-3′), Apobec3 (FW, 5′-CAGAGCAGGTACTAAGGTTCCT-3′, REV, 5′-TTCTGGGTCCCGTATGTTGTA-3′), LINE-1 (FW, 5′-GAGAACATCGGCACAACAATC-3′, REV, 5′-TTTATTGGCGAGTTGAGACCA-3′), pri-let 7g (FW, 5′-GTACGGTGTGGACCTCATCA-3′, REV, 5′-TCTTGCTGTGTCCAGGAAAG-3′), RnaseL (FW, 5′-GTAAACGCCTGTGACAATATGGG-3′, REV, 5′-AGATGCGTAATAGCCTCCACAT-3′), β-actin (FW, 5′-AGAAGAGCTATGAGCTGCCT-3′, REV, 5′-TCATCGTACTCCTGCTTGCT-3′). For targeted human molecules: Ifnb (FW, 5′-AGCTCCAAGAAAGGACGAACAT-3′, REV, 5′-GCCCTGTAGGTGAGGTTGATCT-3′), β-actin (FW, 5′-AGAGCTACGAGCTGCCTGAC-3′, REV, 5′-CGTGGATGCCACAGGACT-3′). QPCRs were carried out by using Applied Biosystems 7500 Fast Real-Time PCR system.
Statistical Analyses
Every shown quantified result represents an average of at least three independent experiments carried out in either triplicate or quadruplicate and calculated as means ± S.E. The p values were calculated using the 2-tailed Student's t test.
RESULTS
LINE-1 Activities Stimulate IFN Expression and Signaling
We have previously reported a high level of LINE-1 mRNA expression in testes from mice whose spermatocytes lack MOV10L1 (14), RNA helicase, which is essential for silencing retrotransposons in the mouse male germline (14, 22, 23). Intriguingly, when compared with the testes from heterozygous animals, Mov10l1 knock-out tissues expressed noticeably increased mRNA levels of not only LINE-1 but also Ifnb (Fig. 1A). LINE-1 encodes ORF1 and ORF2 proteins; the latter harbors endonuclease activity and is capable of inducing double strand breaks (21, 24–26). Given the reports that DNA damage-inducing agents (such as ionizing radiation and anti-cancer drugs) can increase IFN expression (27, 28), we sought to determine whether such increase can be elicited in response to LINE-1 activation.
FIGURE 1.
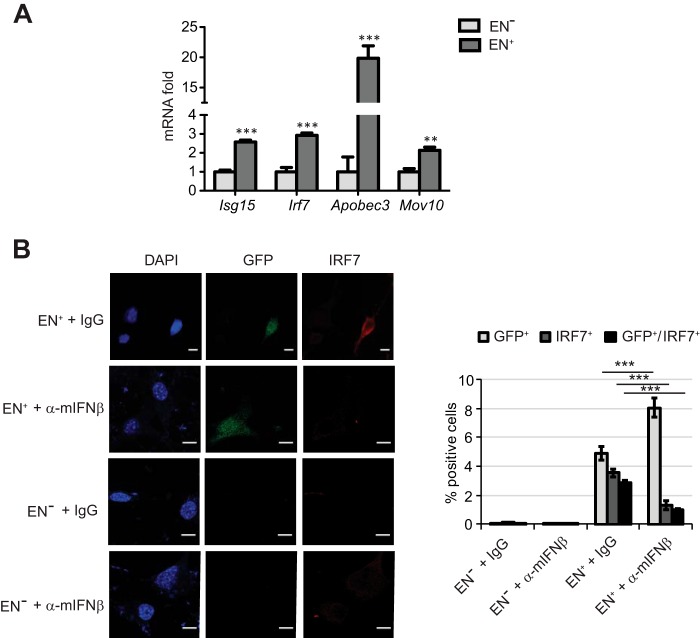
Retrotransposition of LINE-1 induces IFNβ expression. A, relative mRNA levels of indicated genes in the testes of 2-month-old Mov10l1 heterozygous or homozygous knock-out mice assessed by qPCR (levels in heterozygous mice taken as 1.0). Average from three independent experiments is shown as mean ± S.E. Here and thereafter: *, p < 0.05; **, p < 0.01; ***, p < 0.001. B, relative IFNβ mRNA levels (compared with Mock taken as 1.0) in mouse embryonic fibroblast cells transfected with LINE-1-expressing vectors that encode functional (EN+) or endonuclease-deficient (EN−) ORF2 protein. C, immunofluorescent analysis of IFNβ expression in NIH3T3 cells transfected with indicated RNAi oligos and indicated LINE-1 constructs (left panel; magnification bar: 10 μm). Right panel: quantification of percent of cells single or double positive for GFP and IFNβ proteins in total 25 fields randomly chosen from three independent experiments performed as described in left panel.
We transfected mouse embryonic fibroblast cells with LINE-1-expressing plasmids that enable detection of LINE-1 retrotransposition by expression of green fluorescent protein (GFP, (21)). Transfection of cells with LINE-1 whose ORF2 was competent in endonuclease activity (EN+) stimulated expression of Ifnb mRNA (Fig. 1B). Importantly, delivery of the EN− LINE-1 mutant whose ORF2 lacks the endonuclease function and, as a result, exhibits limited retrotransposition activity (29) led to a significantly lesser induction of Ifnb (Fig. 1B). These results suggest that endonuclease-dependent LINE-1 retrotransposition stimulates IFNβ expression.
To corroborate these results we used an immunocytofluorescence assay to assess the levels of IFNβ protein in NIH3T3 cells that received LINE-1 and where its retrotransposition could be monitored by GFP expression (21, 30). These studies showed that low yet detectable levels of IFNβ protein were observed predominantly in the GFP-positive cells that received endonuclease-competent LINE-1 (Fig. 1C). Targeting Ifnb mRNA with RNAi against this gene robustly decreased the number of IFNβ-positive cells indicating the specificity of IFNβ expression analysis. Together these results suggest that LINE-1 retrotransposons are capable of activating the production of IFNβ. Surprisingly, the overall number of cells that enabled LINE-1 retrotransposition (GFP-positive cells) was increased upon the knockdown of IFNβ (Fig. 1C) suggesting that, in turn, IFNβ may control LINE-1 activities.
We next sought to investigate whether LINE-1-induced IFNβ can function as an active cytokine. To this end, we measured mRNA levels of several known IFN-stimulated genes in mouse embryo fibroblasts transfected with LINE-1 plasmids. The expressions of Irf7, Isg15, Apobec3, and Mov10 were increased in cells that received endonuclease competent LINE-1 relative to the EN-deficient construct (Fig. 2A). Furthermore, the immunofluorescent analyses revealed an increase in IRF7 protein levels in the NIH3T3 cells that received the endonuclease-competent LINE-1 construct and became GFP-positive as a result of LINE-1 retrotransposition (Fig. 2B). Very few (if any) IRF7-positive cells were observed in cells receiving the endonuclease-deficient mutant of LINE-1. Importantly, the IRF7 levels in GFP-positive cells could be decreased upon treating cells with IFNβ-neutralizing antibodies (Fig. 2B). These results suggest that LINE-1 expression and retrotransposition can activate expression of IRF7 in a manner dependent upon functional IFNβ.
FIGURE 2.
LINE-1-induced IFNβ can up-regulate IFN-stimulated genes. A, relative mRNA levels of indicated IFN-stimulated genes in mouse embryonic fibroblasts transfected with indicated LINE-1 plasmids were assessed by qPCR (levels in EN−-transfected cells taken as 1.0). B, immunofluorescent analysis of IRF7 expression in NIH3T3 cells cultured in the presence of indicated antibodies (control IgG or neutralizing antibody against mouse IFNβ, 10 μg/ml for 30 h) and transfected with indicated LINE-1 constructs (left panel; magnification bar: 10 μm). Right panel: quantification of percent of cells single or double positive for GFP and IRF7 proteins in total 30 fields randomly chosen from three independent experiments performed as described in the left panel.
IFN Suppresses LINE-1 Activity
Products of IFN-stimulated genes induced by IFN, whose expression is triggered by viruses, will in turn limit the spread of these viruses (1). We next sought to determine whether the IFN produced in response to LINE-1 can affect the propagation of this retrotransposon. Quantification of immunofluorescence data showed that inactivation of IFNβ using either RNAi (Fig. 1C) or a neutralizing antibody (Fig. 2B) increases the number of GFP-positive cells that should have integrated LINE-1. Additional studies examining the effects of anti-IFNβ RNAi oligos and antibodies by immunofluorescence within the same experiment revealed an increase in GFP-positive cells upon IFNβ inactivation (Fig. 3A). These data suggest that endogenous IFNβ produced in response to LINE-1 activities may limit the efficacy of LINE-1 retrotransposition.
FIGURE 3.

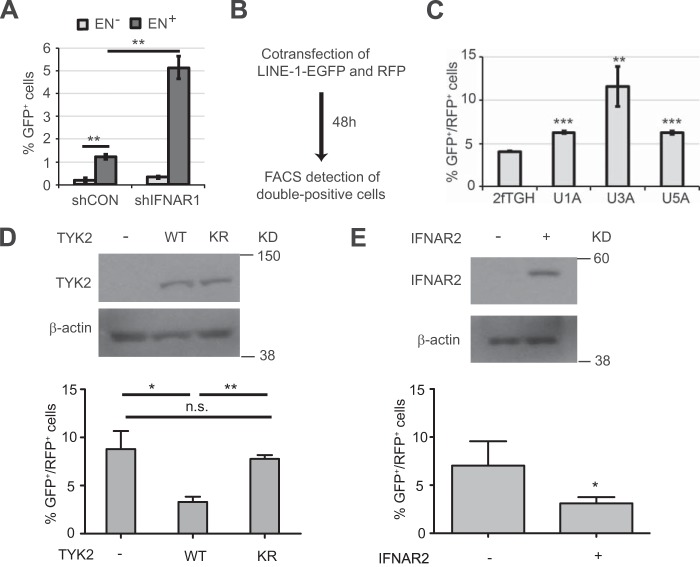
Exogenous IFNβ can decrease the efficacy of LINE-1 retrotransposition in a MOV10-dependent manner. A, appearance of GFP-positive cells NIH3T3 cells that received indicated RNAi (control or against mouse IFNβ) or antibodies (control IgG or neutralizing antibody against mouse IFNβ, 10 μg/ml for 30 h) prior to being transfected with EN+ LINE-1 construct. Magnification bar: 10 μm. B, schematic layout of the experiment described in panel C. C, HeLa cells were transfected with LINE-1-GFP plasmid, incubated for four hours after transfection and then treated with PBS or IFNβ (500 IU/ml for 20 h). After that, cells were washed and incubated with fresh medium without IFN for 24 h and then subjected to 14 days of puromycin selection (3 μg/ml). Percentage of GFP-positive cells was then determined by FACS analysis. D, representative FACS plot diagram (forward scattering versus GFP) of HeLa cells that received indicated RNAi oligos (siCon, siMOV10, or siRNaseL) prior to transfection with LINE-1-GFP plasmid, IFNβ treatment (as indicated), puromycin selection, and FACS analysis. E, quantification of data from three independent experiments (described in panel D). F, relative mRNA levels of indicated genes in HeLa cells from experiments described in panels C–D were assessed by qPCR. Levels in siCON-transfected cells were taken as 1.0.
To further determine the putative role of IFN in LINE-1 control, we used a standard LINE-1 retrotransposition assay in human HeLa cells (Fig. 3B and Refs. 16, 21). This assay allows to assess LINE-1 retrotransposition by using fluorescence-activated cytometry for measuring the efficacy of the recombination of the LINE-1-EGFP reporter (16, 21). We observed that efficacy of LINE-1 retrotransposition in HeLa cells was noticeably decreased after treatment of these cells with recombinant human IFNβ (Fig. 3C).
We next sought to investigate the mechanism by which exogenously added IFNβ may inhibit LINE-1 replication. A ubiquitously expressed paralogue of MOV10L1, MOV10 was found among IFN-inducible genes (31) and shown to suppress activities of LINE-1 and other retrotransposons (32). Importantly Mov10 mRNA levels were increased in mouse embryo fibroblasts that received endonuclease-competent LINE-1 (Fig. 2A).
The knockdown of MOV10 increased basal levels of LINE-1 retrotransposition and almost completely protected it from suppression by IFNβ (Fig. 3, D and E). Although knockdown of RNaseL (which was recently implicated in controlling the LINE-1 propagation (33)) increased the basal level of LINE-1 activities, the inhibitory effects of exogenous IFNβ were still observed in these cells (Fig. 3, D and E). The efficacy of knockdown of MOV10 and RNaseL were verified by qPCR (Fig. 3E). Intriguingly, we found that elevated LINE-1 activities in cells receiving RNAi against MOV10 were found despite the fact that these cells expressed high endogenous levels of Ifnb mRNA (Fig. 3E). These results indicate that MOV10 likely acts downstream of IFN signaling to restrict LINE-1 retrotransposition. Overall, these data support the critical role of MOV10 in the control of LINE-1 activities by IFNβ.
We next investigated the role of proximal mediators of IFN signaling in controlling LINE-1 propagation. The efficacy of retrotransposition of the endonuclease-competent LINE-1 in HeLa cells was robustly increased by knockdown of the IFNAR1 chain of IFN receptor (Fig. 4A). While this experiment suggests that IFN signaling may restrict LINE-1 activities, its interpretation is somewhat confounded due to a technical caveat that the assay involves puromycin selection (Fig. 3B). Given that both LINE-1 and shRNA (control or against IFNAR1) harbor the puromycin resistance marker, the actual effect of IFNAR1 knockdown on increasing the number of GFP-positive cells can be underestimated. To corroborate these data using an alternative assay that does not involve puromycin selection, we have used co-transfection of LINE-1 plasmid with a plasmid for expression of red fluorescent protein (mCherry, RFP) followed by FACS-based detection of double positive GFP+/RFP+ cells. Furthermore, given that shRNA reagents are capable of inducing IFN response (34), we aimed to avoid possible RNAi-elicited artifacts. To this end, we analyzed retrotransposition of LINE-1 in human isogenic cell lines that differ in status of various elements of the IFN pathway (35). Activation of LINE-1-EGFP was significantly higher in these cell lines that were lacking TYK2 (U1A), STAT1 (U3A) or IFNAR2 (U5A) compared with parental fibrosarcoma 2fTGH cells (Fig. 4, B and C). Furthermore, U1A cells that were reconstituted with wild type TYK2 (WT) noticeably decreased LINE-1 activities. This effect was not seen in U1A cells expressing the comparable levels of catalytically inactive TYK2 (KR) mutant (Fig. 4D). Furthermore, suppression of LINE-1 retrotransposition was also observed in U5A cells upon their reconstitution with IFNAR2 (Fig. 4E). These data collectively suggest that endogenous IFN signaling limits propagation of LINE-1 retrotransposon in vitro.
FIGURE 4.
Endogenous IFNβ signaling suppresses the replication of human LINE-1 retrotransposons in vitro. A, effect of IFNAR1 knockdown in HeLa cells on the retrotransposition of endonuclease-positive (EN+) or negative (EN−) LINE-1 assessed as in A. B, layout of the experiment described in panel C. C, parental 2fTGH human fibrosarcoma cells and derivative cell lines lacking various components of IFN signaling including TYK2 (U1A), STAT1 (U3A), and IFNAR2 (U5A) were co-transfected with LINE-1-GFP and RFP-expressing vectors. Retrotransposition of LINE-1 was analyzed as % of double positive cells. D, U1A cells and its derivative clones harboring wild type TYK2 (WT) or its catalytically inactive mutant (KR) were co-transfected with LINE-1-GFP and RFP-expressing vectors. Retrotransposition of LINE-1 was analyzed as % of double positive cells. Leves of TYK2 and β-actin (used as a loading control) were analyzed by immunoblotting. E, U5A cells were co-transfected with IFNAR2 expression plasmid or vector, LINE-1-GFP and RFP-expressing vectors. Retrotransposition of LINE-1 was analyzed as % of double positive cells. Levels of IFNAR2 and β-actin (used as a loading control) were analyzed by immunoblotting.
Next we sought to determine whether IFN plays an in vivo role in regulating LINE-1 activities. To this end, we analyzed LINE-1 mRNA levels in testes from wild type mice or animals lacking the IFNAR1 chain of IFN receptor using either semi-quantitative RT-PCR (Fig. 5A) or quantitative Q-PCR (Fig. 5B). As seen from experiments using both approaches, a noticeably greater amount of LINE-1 mRNA was recovered from tissues from Ifnar1−/− mice compared with that from wild type animals. These results collectively suggest that IFN plays a protective role against LINE-1 retrotransoposon activation and propagation.
FIGURE 5.
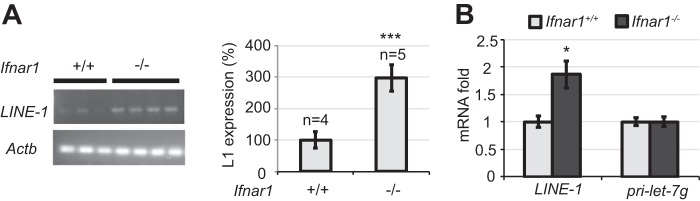
IFNβ suppresses the replication of mouse LINE-1 retrotransposons in vivo. A, expression of endogenous murine LINE-1 mRNA in the testes from indicated 5-day-old mice was assessed by RT-PCR. Each lane represents an independent individual mouse. Right panel: quantification of data combined from two independent experiments and presented as mean ± S.E. B, expression of endogenous murine LINE-1 mRNA in the testes from indicated mice assessed by qPCR. MicroRNA precursor pri-let-7g mRNA levels were used as a control. Data from three independent experiments have been combined and presented as mean ± S.E.
DISCUSSION
The results presented here link IFN production and activities with the regulation of LINE-1, a major class of retrotransposons, which is common in many types of mammalian cells and which contributes to genome evolution and instability (11). We propose that activation of LINE-1 triggers expression of low levels of IFN, which, in turn, restricts subsequent LINE-1 retrotransposition. This hypothesis is supported by a correlation between LINE-1 and IFNβ mRNA expression and the ability of exogenous LINE-1 to induce IFNβ and several IFN-stimulated genes in vitro (Figs. 1 and 2). Importantly, whereas added IFN suppresses LINE-1 retrotransposition (Fig. 3), endogenous IFN signaling appears to restrict the propagation of LINE-1 and the activity of this retrotransposon in cultured cells in a manner that involves engagement of IFNAR1/IFNAR2 and stimulation of the JAK-STAT signaling cascade (Fig. 4). Finally, the activities of LINE-1 in vivo are noticeably increased in IFNAR1-deficient mice (Fig. 5).
The counteracting relationship between LINE-1 and IFN activities are reminiscent of how virus-induced IFN plays a key role in mounting the anti-viral defenses. It is worth noting that the current paradigm highlights numerous mechanisms by which viruses limit the stimulation of IFN production or inhibit IFN signaling thus allowing propagation of these viruses (1, 36). While the nature of similar LINE-1-mediated mechanisms that allow this retrotransposon to ignore IFN signaling is yet to be determined, the putative existence of such mechanisms is supported by the very fact that LINE-1 and other retrotransposons have been propagating for hundreds of millions of years (37).
It remains to be seen whether IFN can also control other diverse retrotransposons whose activation may also lead to IFN induction. A very robust production of endogenous IFNβ and the ensuing induction of IFN-stimulated genes was observed concurrently with activation of Short Interspersed Nuclear Element (SINE) B1 and B2 in mouse fibroblasts treated with DNA demethylating agent 5-aza-2′-deoxycytidine (38). Intriguingly, these events along with cell death triggered by produced IFNβ were seen only in p53-deficient cells. Given the ability of recombinant IFNβ to induce p53 (39, 40) and the role of p53 in IFN-mediated anti-viral effects (41), it is also possible that the modes of negative regulation elicited by IFN may differ between retrotransposons and active viruses.
The mechanisms underlying the inhibitory effects of IFN on the replication of LINE-1 largely remain to be understood. Whereas IFN has been shown to induce the APOBEC3 members of anti-retroviral cytidine deaminases (42, 43), and this gene was robustly activated in response to LINE-1 (Fig. 2A), the role of these enzymes in suppressing LINE-1 replication remains to be established unequivocally (44–47). However, ubiquitously expressed MOV10 (an APOBEC-interacting paralogue of MOV10L1) was found among IFN-inducible genes (31) and has been shown to suppress activities of LINE-1 and other retrotransposons (32). Our current data demonstrating that MOV10 knockdown relieves suppression of LINE-1 retrotransposition by IFN (Fig. 3) indicate that MOV10 plays a critical role in the IFN-mediated control of LINE-1 activities.
It was hypothesized that an increase in activity of LINE-1 and other retrotransposons is associated with development of autoimmune diseases including systemic lupus erythematosus and Sjogren syndrome (48, 49). Interestingly, these diseases have also been characterized by increased production and activity of IFN (50, 51). While these published reports are consistent with our data linking LINE-1 activities and IFN production, the casual relationship between the activity of LINE-1 and IFN signaling in health and diverse diseases and the contribution of these mechanisms to normal and pathogenic processes needs to be further investigated.
Acknowledgments
We thank George Stark, John Krolewski, Sandra Pellegrini, and Eline T. Luning Prak for reagents, Ze'ev Ronai, Eric Lau, and the members of Diehl, Koumenis, Greenberg, Witze and Minn laboratories (at the University of Pennsylvania) for critical suggestions.
This work was supported by National Institutes of Health/National Cancer Institute Grants CA092900 and CA142425 (to S. Y. F.). Funding for open access charges is by the National Institutes of Health.
- IFN
- type I interferons
- LINE-1
- Long Interspersed Element-1
- MEF
- mouse embryonic fibroblast.
REFERENCES
- 1. Katze M. G., He Y., Gale M., Jr. (2002) Viruses and interferon: a fight for supremacy. Nature Reviews. Immunology 2, 675–687 [DOI] [PubMed] [Google Scholar]
- 2. Fuchs S. Y. (2013) Hope and fear for interferon: the receptor-centric outlook on the future of interferon therapy. J. Interferon and Cytokine 33, 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fuchs S. Y. (2012) Ubiquitination-mediated regulation of interferon responses. Growth Factors 30, 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piehler J., Thomas C., Garcia K. C., Schreiber G. (2012) Structural and dynamic determinants of type I interferon receptor assembly and their functional interpretation. Immunol. Rev. 250, 317–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Trinchieri G. (2010) Type I interferon: friend or foe? J. Exp. Med. 207, 2053–2063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Platanias L. C. (2005) Mechanisms of type-I- and type-II-interferon-mediated signalling. Nature Reviews. Immunology 5, 375–386 [DOI] [PubMed] [Google Scholar]
- 7. Bhattacharya S., Katlinski K. V., Reichert M., Takano S., Brice A., Zhao B., Yu Q., Zheng H., Carbone C. J., Katlinskaya Y. V., Leu N. A., McCorkell K. A., Srinivasan S., Girondo M., Rui H., May M. J., Avadhani N. G., Rustgi A. K., Fuchs S. Y. (2014) Triggering ubiquitination of IFNAR1 protects tissues from inflammatory injury. EMBO Mol. Med. 6, 384–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zheng H., Gupta V., Patterson-Fortin J., Bhattacharya S., Katlinski K., Wu J., Varghese B., Carbone C. J., Aressy B., Fuchs S. Y., Greenberg R. A. (2013) A BRISC-SHMT complex deubiquitinates IFNAR1 and regulates interferon responses. Cell Reports 5, 180–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Weitzman M. D., Lilley C. E., Chaurushiya M. S. (2010) Genomes in conflict: maintaining genome integrity during virus infection. Annu. Rev. Microbiol. 64, 61–81 [DOI] [PubMed] [Google Scholar]
- 10. Kumar A., Bennetzen J. L. (1999) Plant retrotransposons. Annu. Rev. Genet 33, 479–532 [DOI] [PubMed] [Google Scholar]
- 11. Kazazian H. H., Jr. (2004) Mobile elements: drivers of genome evolution. Science 303, 1626–1632 [DOI] [PubMed] [Google Scholar]
- 12. Lander E. S., Linton L. M., Birren B., Nusbaum C., Zody M. C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., Funke R., Gage D., Harris K., Heaford A., Howland J., Kann L., Lehoczky J., LeVine R., McEwan P., McKernan K., Meldrim J., Mesirov J. P., Miranda C., Morris W., Naylor J., Raymond C., Rosetti M., Santos R., Sheridan A., Sougnez C., Stange-Thomann N., Stojanovic N., Subramanian A., Wyman D., Rogers J., Sulston J., Ainscough R., Beck S., Bentley D., Burton J., Clee C., Carter N., Coulson A., Deadman R., Deloukas P., Dunham A., Dunham I., Durbin R., French L., Grafham D., Gregory S., Hubbard T., Humphray S., Hunt A., Jones M., Lloyd C., McMurray A., Matthews L., Mercer S., Milne S., Mullikin J. C., Mungall A., Plumb R., Ross M., Shownkeen R., Sims S., Waterston R. H., Wilson R. K., Hillier L. W., McPherson J. D., Marra M. A., Mardis E. R., Fulton L. A., Chinwalla A. T., Pepin K. H., Gish W. R., Chissoe S. L., Wendl M. C., Delehaunty K. D., Miner T. L., Delehaunty A., Kramer J. B., Cook L. L., Fulton R. S., Johnson D. L., Minx P. J., Clifton S. W., Hawkins T., Branscomb E., Predki P., Richardson P., Wenning S., Slezak T., Doggett N., Cheng J. F., Olsen A., Lucas S., Elkin C., Uberbacher E., Frazier M., Gibbs R. A., Muzny D. M., Scherer S. E., Bouck J. B., Sodergren E. J., Worley K. C., Rives C. M., Gorrell J. H., Metzker M. L., Naylor S. L., Kucherlapati R. S., Nelson D. L., Weinstock G. M., Sakaki Y., Fujiyama A., Hattori M., Yada T., Toyoda A., Itoh T., Kawagoe C., Watanabe H., Totoki Y., Taylor T., Weissenbach J., Heilig R., Saurin W., Artiguenave F., Brottier P., Bruls T., Pelletier E., Robert C., Wincker P., Smith D. R., Doucette-Stamm L., Rubenfield M., Weinstock K., Lee H. M., Dubois J., Rosenthal A., Platzer M., Nyakatura G., Taudien S., Rump A., Yang H., Yu J., Wang J., Huang G., Gu J., Hood L., Rowen L., Madan A., Qin S., Davis R. W., Federspiel N. A., Abola A. P., Proctor M. J., Myers R. M., Schmutz J., Dickson M., Grimwood J., Cox D. R., Olson M. V., Kaul R., Shimizu N., Kawasaki K., Minoshima S., Evans G. A., Athanasiou M., Schultz R., Roe B. A., Chen F., Pan H., Ramser J., Lehrach H., Reinhardt R., McCombie W. R., de la Bastide M., Dedhia N., Blocker H., Hornischer K., Nordsiek G., Agarwala R., Aravind L., Bailey J. A., Bateman A., Batzoglou S., Birney E., Bork P., Brown D. G., Burge C. B., Cerutti L., Chen H. C., Church D., Clamp M., Copley R. R., Doerks T., Eddy S. R., Eichler E. E., Furey T. S., Galagan J., Gilbert J. G., Harmon C., Hayashizaki Y., Haussler D., Hermjakob H., Hokamp K., Jang W., Johnson L. S., Jones T. A., Kasif S., Kaspryzk A., Kennedy S., Kent W. J., Kitts P., Koonin E. V., Korf I., Kulp D., Lancet D., Lowe T. M., McLysaght A., Mikkelsen T., Moran J. V., Mulder N., Pollara V. J., Ponting C. P., Schuler G., Schultz J., Slater G., Smit A. F., Stupka E., Szustakowski J., Thierry-Mieg D., Thierry-Mieg J., Wagner L., Wallis J., Wheeler R., Williams A., Wolf Y. I., Wolfe K. H., Yang S. P., Yeh R. F., Collins F., Guyer M. S., Peterson J., Felsenfeld A., Wetterstrand K. A., Patrinos A., Morgan M. J., de Jong P., Catanese J. J., Osoegawa K., Shizuya H., Choi S., Chen Y. J. (2001) Initial sequencing and analysis of the human genome. Nature 409, 860–921 [DOI] [PubMed] [Google Scholar]
- 13. Symer D. E., Connelly C., Szak S. T., Caputo E. M., Cost G. J., Parmigiani G., Boeke J. D. (2002) Human l1 retrotransposition is associated with genetic instability in vivo. Cell 110, 327–338 [DOI] [PubMed] [Google Scholar]
- 14. Zheng K., Wang P. J. (2012) Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet 8, e1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prak E. T., Dodson A. W., Farkash E. A., Kazazian H. H., Jr. (2003) Tracking an embryonic L1 retrotransposition event. Proc. Natl. Acad. Sci. U. S.A. 100, 1832–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rangwala S. H., Kazazian H. H., Jr. (2009) The L1 retrotransposition assay: a retrospective and toolkit. Methods 49, 219–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu J., HuangFu W. C., Kumar K. G., Qian J., Casey J. P., Hamanaka R. B., Grigoriadou C., Aldabe R., Diehl J. A., Fuchs S. Y. (2009) Virus-induced unfolded protein response attenuates antiviral defenses via phosphorylation-dependent degradation of the type I interferon receptor. Cell Host Microbe 5, 72–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gauzzi M. C., Velazquez L., McKendry R., Mogensen K. E., Fellous M., Pellegrini S. (1996) Interferon-alpha-dependent activation of Tyk2 requires phosphorylation of positive regulatory tyrosines by another kinase. J. Biol. Chem. 271, 20494–20500 [DOI] [PubMed] [Google Scholar]
- 19. Marijanovic Z., Ragimbeau J., Kumar K. G., Fuchs S. Y., Pellegrini S. (2006) TYK2 activity promotes ligand-induced IFNAR1 proteolysis. Biochem. J. 397, 31–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., Preibisch S., Rueden C., Saalfeld S., Schmid B., Tinevez J. Y., White D. J., Hartenstein V., Eliceiri K., Tomancak P., Cardona A. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Belgnaoui S. M., Gosden R. G., Semmes O. J., Haoudi A. (2006) Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int. 6, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Frost R. J., Hamra F. K., Richardson J. A., Qi X., Bassel-Duby R., Olson E. N. (2010) MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc. Natl. Acad. Sci. U. S.A. 107, 11847–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zheng K., Xiol J., Reuter M., Eckardt S., Leu N. A., McLaughlin K. J., Stark A., Sachidanandam R., Pillai R. S., Wang P. J. (2010) Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc. Natl. Acad. Sci. U. S.A. 107, 11841–11846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wallace N. A., Belancio V. P., Deininger P. L. (2008) L1 mobile element expression causes multiple types of toxicity. Gene 419, 75–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gilbert N., Lutz S., Morrish T. A., Moran J. V. (2005) Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol. Cell. Biol. 25, 7780–7795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gasior S. L., Wakeman T. P., Xu B., Deininger P. L. (2006) The human LINE-1 retrotransposon creates DNA double-strand breaks. J. Mol. Biol. 357, 1383–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim T., Kim T. Y., Song Y. H., Min I. M., Yim J., Kim T. K. (1999) Activation of interferon regulatory factor 3 in response to DNA-damaging agents. J. Biol. Chem. 274, 30686–30689 [DOI] [PubMed] [Google Scholar]
- 28. Brzostek-Racine S., Gordon C., Van Scoy S., Reich N. C. (2011) The DNA damage response induces IFN. J. Immunol. 187, 5336–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morrish T. A., Garcia-Perez J. L., Stamato T. D., Taccioli G. E., Sekiguchi J., Moran J. V. (2007) Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature 446, 208–212 [DOI] [PubMed] [Google Scholar]
- 30. Farkash E. A., Kao G. D., Horman S. R., Prak E. T. (2006) Gamma radiation increases endonuclease-dependent L1 retrotransposition in a cultured cell assay. Nucleic Acids Res. 34, 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Schoggins J. W., Wilson S. J., Panis M., Murphy M. Y., Jones C. T., Bieniasz P., Rice C. M. (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goodier J. L., Cheung L. E., Kazazian H. H., Jr. (2012) MOV10 RNA helicase is a potent inhibitor of retrotransposition in cells. PLoS Genet 8, e1002941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang A., Dong B., Doucet A. J., Moldovan J. B., Moran J. V., Silverman R. H. (2014) RNase L restricts the mobility of engineered retrotransposons in cultured human cells. Nucleic Acids Res. 42, 3803–3820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bridge A. J., Pebernard S., Ducraux A., Nicoulaz A. L., Iggo R. (2003) Induction of an interferon response by RNAi vectors in mammalian cells. Nature Genetics 34, 263–264 [DOI] [PubMed] [Google Scholar]
- 35. McKendry R., John J., Flavell D., Müller M., Kerr I. M., Stark G. R. (1991) High-frequency mutagenesis of human cells and characterization of a mutant unresponsive to both α and γ interferons. Proc. Natl. Acad. Sci. U. S.A. 88, 11455–11459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Galligan C. L., Murooka T. T., Rahbar R., Baig E., Majchrzak-Kita B., Fish E. N. (2006) Interferons and viruses: signaling for supremacy. Immunol. Res. 35, 27–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Furano A. V. (2000) The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons. Progr. Nucleic Acid Res. Mol. Biol. 64, 255–294 [DOI] [PubMed] [Google Scholar]
- 38. Leonova K. I., Brodsky L., Lipchick B., Pal M., Novototskaya L., Chenchik A. A., Sen G. C., Komarova E. A., Gudkov A. V. (2013) p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proc. Natl. Acad. Sci. U. S.A. 110, E89–E98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Moiseeva O., Mallette F. A., Mukhopadhyay U. K., Moores A., Ferbeyre G. (2006) DNA damage signaling and p53-dependent senescence after prolonged β-interferon stimulation. Mol. Biol. Cell 17, 1583–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Takaoka A., Hayakawa S., Yanai H., Stoiber D., Negishi H., Kikuchi H., Sasaki S., Imai K., Shibue T., Honda K., Taniguchi T. (2003) Integration of interferon-α/β signalling to p53 responses in tumour suppression and antiviral defence. Nature 424, 516–523 [DOI] [PubMed] [Google Scholar]
- 41. Muñoz-Fontela C., Macip S., Martínez-Sobrido L., Brown L., Ashour J., García-Sastre A., Lee S. W., Aaronson S. A. (2008) Transcriptional role of p53 in interferon-mediated antiviral immunity. J. Exp. Med. 205, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bonvin M., Achermann F., Greeve I., Stroka D., Keogh A., Inderbitzin D., Candinas D., Sommer P., Wain-Hobson S., Vartanian J. P., Greeve J. (2006) Interferon-inducible expression of APOBEC3 editing enzymes in human hepatocytes and inhibition of hepatitis B virus replication. Hepatology 43, 1364–1374 [DOI] [PubMed] [Google Scholar]
- 43. Tanaka Y., Marusawa H., Seno H., Matsumoto Y., Ueda Y., Kodama Y., Endo Y., Yamauchi J., Matsumoto T., Takaori-Kondo A., Ikai I., Chiba T. (2006) Anti-viral protein APOBEC3G is induced by interferon-α stimulation in human hepatocytes. Biochem. Biophys. Res. Commun. 341, 314–319 [DOI] [PubMed] [Google Scholar]
- 44. Lovsin N., Peterlin B. M. (2009) APOBEC3 proteins inhibit LINE-1 retrotransposition in the absence of ORF1p binding. Ann. N.Y. Acad. Sci. 1178, 268–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Niewiadomska A. M., Tian C., Tan L., Wang T., Sarkis P. T., Yu X. F. (2007) Differential inhibition of long interspersed element 1 by APOBEC3 does not correlate with high-molecular-mass-complex formation or P-body association. J. Virol. 81, 9577–9583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Esnault C., Heidmann O., Delebecque F., Dewannieux M., Ribet D., Hance A. J., Heidmann T., Schwartz O. (2005) APOBEC3G cytidine deaminase inhibits retrotransposition of endogenous retroviruses. Nature 433, 430–433 [DOI] [PubMed] [Google Scholar]
- 47. Peng G., Lei K. J., Jin W., Greenwell-Wild T., Wahl S. M. (2006) Induction of APOBEC3 family proteins, a defensive maneuver underlying interferon-induced anti-HIV-1 activity. J. Exp. Med. 203, 41–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nakkuntod J., Avihingsanon Y., Mutirangura A., Hirankarn N. (2011) Hypomethylation of LINE-1 but not Alu in lymphocyte subsets of systemic lupus erythematosus patients. Clin. Chim. Acta 412, 1457–1461 [DOI] [PubMed] [Google Scholar]
- 49. Crow M. K. (2010) Long interspersed nuclear elements (LINE-1): potential triggers of systemic autoimmune disease. Autoimmunity 43, 7–16 [DOI] [PubMed] [Google Scholar]
- 50. Bronson P. G., Chaivorapol C., Ortmann W., Behrens T. W., Graham R. R. (2012) The genetics of type I interferon in systemic lupus erythematosus. Curr. Opin. Immunol. 24, 530–537 [DOI] [PubMed] [Google Scholar]
- 51. Elkon K. B., Wiedeman A. (2012) Type I IFN system in the development and manifestations of SLE. Curr. Opin. Rheumatol. 24, 499–505 [DOI] [PubMed] [Google Scholar]