Background: CCAAT/Enhancer-binding Protein β (C/EBPβ) inhibits differentiation of muscle satellite cells and is rapidly down-regulated in early myogenesis.
Results: The E3 ubiquitin ligase Mouse double minute 2 homolog (Mdm2) targets C/EBPβ for degradation thereby promoting entry into myogenesis.
Conclusion: Mdm2 expression is necessary for entry into myogenesis.
Significance: Establishes a new role for Mdm2 in cellular differentiation.
Keywords: CCAAT-enhancer-binding protein (C/EBP), mouse double minute 2 homolog (MDM2), muscle regeneration, myogenesis, ubiquitylation (ubiquitination)
Abstract
Myogenesis is a tightly regulated differentiation process during which precursor cells express in a coordinated fashion the myogenic regulatory factors, while down-regulating the satellite cell marker Pax7. CCAAT/Enhancer-binding protein β (C/EBPβ) is also expressed in satellite cells and acts to maintain the undifferentiated state by stimulating Pax7 expression and by triggering a decrease in MyoD protein expression. Herein, we show that C/EBPβ protein is rapidly down-regulated upon induction of myogenesis and this is not due to changes in Cebpb mRNA expression. Rather, loss of C/EBPβ protein is accompanied by an increase in Mdm2 expression, an E3 ubiquitin ligase. We demonstrate that Mdm2 interacts with, ubiquitinates and targets C/EBPβ for degradation by the 26 S proteasome, leading to increased MyoD expression. Knockdown of Mdm2 expression in myoblasts using a shRNA resulted in high C/EBPβ levels and a blockade of myogenesis, indicating that Mdm2 is necessary for myogenic differentiation. Primary myoblasts expressing the shMdm2 construct were unable to contribute to muscle regeneration when grafted into cardiotoxin-injured muscle. The differentiation defect imposed by loss of Mdm2 could be partially rescued by loss of C/EBPβ, suggesting that the regulation of C/EBPβ turnover is a major role for Mdm2 in myoblasts. Taken together, we provide evidence that Mdm2 regulates entry into myogenesis by targeting C/EBPβ for degradation by the 26 S proteasome.
Introduction
The development of skeletal muscle is regulated by a tightly coordinated transcriptional cascade during which myogenic regulatory factors are expressed. This process is recapitulated in post-natal muscle growth and regeneration, where muscle stem cells, called satellite cells, are induced to differentiate. Satellite cells are defined by their anatomical location between the muscle fiber sarcolemma and the basement membrane and their expression of the paired box protein Pax7 (1, 2). A heterogeneous population, the majority of satellite cells also express the myogenic regulatory factor Myf5 (3). Although not the only source of myogenic precursors in skeletal muscle, satellite cells are recognized as the most important source of regenerative potential in this tissue (4). As satellite cells become activated, they progressively lose the expression of Pax7, and begin to express the myogenic regulatory factor MyoD (5). As the differentiating cells exit the cell cycle and terminally differentiate, the expression of myogenin is activated (a MyoD target gene), driving the expression of myosin heavy chain and other myocyte markers (5). The differentiation process culminates with the fusion of myocytes to form large multinucleated cells called myotubes in culture and myofibers in vivo.
In addition to the myogenic regulatory factors, the transcription factor CCAAT/Enhancer-binding protein β (C/EBPβ)2 is implicated in the regulation of adult myogenesis (6). C/EBPβ is localized to Pax7+ satellite cells in healthy muscle, and its expression is rapidly down-regulated in parallel with Pax7 as differentiation progresses (6). Persistent C/EBPβ expression blocks myogenesis, through suppression of MyoD protein expression and stimulation of Pax7 expression, and its loss promotes precocious differentiation even under growth conditions, as well as larger myofibers and enhanced repair after a single injury in vivo (6).
C/EBPβ is a member of the larger family of bzip transcription factors. Initially discovered as a regulator of IL-6 expression, C/EBPβ has been implicated in numerous differentiation processes including adipogenesis, osteoblastogenesis, mammary gland development, and female fertility (7–11). Cebpb is an intronless gene that produces a single mRNA from a single promoter (12). Differential initiation of translation results in 3 C/EBPβ proteins with identical carboxyl termini and variable amino termini. The full-length isoform (Liver Activating Protein, LAP*) and the second isoform (LAP), which lacks the first 21 amino acids, contain all 3 activation domains (13, 14). The shortest isoform (Liver Inhibitory Protein, LIP) lacks activation domains and acts as a dominant negative (13, 14). In normal skeletal muscle and SCs from young mice, only the LAP*/LAP isoforms are detected in Pax7+ cells, and are decreased with differentiation (6).
Protein expression can be regulated at the level of transcription, translation, and more rapidly via targeted degradation by the ubiquitin-proteasome system. The ubiquitin-proteasome system targets specific proteins for degradation by marking them with ubiquitin chains conjugated to lysine residues within the target protein sequence. The prey is recognized by an E3 ubiquitin ligase, an enzyme of one of four different classes (HECT, RING-finger, U-box, or PHD-finger), which transfers a ubiquitin moiety from an activated E2 enzyme to the target protein. Elongation of this chain to 4 ubiquitin subunits with specific lysine 48 linkages allows for recognition by the 26 S proteasome, recycling of the ubiquitin moieties and degradation of the targeted protein (15, 16).
Mouse double minute 2 homolog (Mdm2) is a RING finger family E3 ubiquitin ligase that is known for interacting with and targeting for degradation the oncogene p53 (17, 18). Blockade of p53 activities triggers progression through the cell cycle whereas high levels of p53 induce growth arrest and apoptosis (19). In addition to regulating p53 activity, Mdm2 can also interact with pRb, causing inhibition of its function, and the activation domain of E2F1, stimulating E2F1/DP1 transcriptional activity (20, 21). As such, high levels of Mdm2 trigger proliferation by inhibiting the activities of pRb and p53 and directly stimulating the activity of E2F1/DP1.
The Mdm2 knock-out is embryonic lethal, but can be rescued by concomitant loss of p53 expression. Indeed, the E3 ubiquitin ligase activity of Mdm2 is required to ensure normal development in mice, suggesting that the regulation of p53 levels is a major role for Mdm2 in vivo. However, Mdm2 has been implicated in activities independent of p53, such as the regulation of cellular differentiation, suggesting that Mdm2 has numerous substrates and/or activities other than that of an ubiquitin ligase. Herein, we identify Mdm2 as an E3 ubiquitin ligase that interacts with and regulates the expression of C/EBPβ in myoblasts. Specifically, we demonstrate that C/EBPβ can be ubiquitinated by Mdm2, and loss of Mdm2 in myoblasts results in higher than normal C/EBPβ protein levels, driving a blockade of myogenic differentiation.
EXPERIMENTAL PROCEDURES
Constructs and Reagents
GST-tagged full-length C/EBPβ and fragments corresponding to amino acids 1–296, 1–151, and 152–296 were cloned by PCR into the EcoR I/BamH I sites of pGEX-2T. Mdm2 was amplified by PCR from pCI-neo-Mdm2 (gift from Dr. Robert Haché) and inserted into Pet47B vector (Novagen).
Cell Culture
C2C12 mouse myoblasts were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) (Growth medium, GM) for maintenance. To induce differentiation, GM was replaced with DMEM containing 2% horse serum (differentiation medium, DM) when cells reached 80–90% confluence. Primary myoblasts were isolated from C57BL/6 mice or C/EBPβ conditional knock-out mice as described (6). Primary myoblasts were plated on matrigel-coated dishes in DMEM with 20% FBS and 10% HS with 10 ng/ml basic FGF and 2 ng/ml HGF (growth medium, GM). Differentiation was induced in DMEM containing 2% FBS and 2% HS (Differentiation medium, DM) at a confluency of 70–80%. Lentivirus containing a shRNA against mouse Mdm2 (shMdm2, sc-37263-V) or a scrambled shRNA (shScr, sc-108080) (Santa Cruz Biotechnology) were added to C2C12 cells or primary myoblasts at 30–40% confluency in GM according to manufacturer's instructions. Cultures were selected in 2 μm puromycin to create pooled stable cell lines beginning 48 h after transduction.
Western and Immunocytochemistry
Western blots were performed as described (22). Proteins were detected using anti-C/EBPβ E299 (Abcam), anti-Pax7 (DSHB), anti-α-actin (Sigma), anti-MDM2 C18, anti-MyoD 5.8A, anti-Myf5, anti-Myogenin, anti-p53, and anti-α-Tubulin (all from Santa Cruz Biotechnology). Immunocytochemistry was performed as described (6) using anti-MF20 (DSHB) and anti-Mouse-Cy3 (Invitrogen) antibodies to detect myosin heavy chain expression.
Protein-Protein Interaction Studies
GST-C/EBPβ protein and its fragments were expressed and isolated as described (23) and incubated with in vitro translated Mdm2 produced using the TNT T7 Quick-Coupled Transcription/Translation kit (Promega). Bound proteins were isolated by eluting with 2× SDS buffer, and eluates were separated by 8% SDS-PAGE gel. Mdm2 was detected by Western blotting. Immunoprecipitation of proteins from whole cell extracts from C2C12 cells was performed using anti-C/EBPβ antibody E299 (Abcam) or anti-Mdm2 antibody C18 (Santa Cruz Biotechnology) and co-precipitated C/EBPβ or Mdm2 was detected by Western blotting using the same antibodies.
In Vitro Ubiquitination Assay
The in vitro ubiquitination assay was performed as described (24). In each reaction, 10 μm of biotinylated ubiquitin U570 (Boston Biochem) and 20–50 μg of purified GST-C/EBPβ protein were added to cell extracts from shScr or shMdm2-expressing C2C12 cells. Reactions were incubated for 60 min at 37 °C. In the control experiments, biotinylated ubiquitin was not added to the reaction mixture. Ubiquitinated GST-C/EBPβ fusion proteins were purified from reaction mixtures by GST pull-down and detected by Western blotting using HRP-conjugated streptavidin (GE Healthcare).
Myoblast Transplantation
Muscle regeneration in 4-month-old C57BL/6 mice was induced by injecting 30 μl of 10 μm cardiotoxin (CTX) into the mid-belly of the right and left tibialis anterior (TA) muscles. The next day, 105 GFP+ myoblasts isolated from C57BL/6-Tg(UBC-GFP) mice (Jackson Labs) transduced to express shMdm2 or shScr were injected into the left and right TA of the CTX-injured TA muscle, respectively. Mice were sacrificed 2 weeks after transplantation, and the TAs were collected, flash frozen, and sectioned (8 μm thick) for analysis of GFP fluorescence.
Cell Cycle Analysis
106 C2C12 cells lentivirally transduced to express shMdm2 or scrambled control (shScr) cultured in growth medium were collected in PBS and fixed in 75% ethanol for 5 min on ice, washed, and rehydrated in PBS for 15 min at room temperature. Cells were then resuspended in 1 ml of propidium iodide staining solution (100 mm Tris, pH 7.4, 150 mm NaCl, 1 mm CaCl2, 0.5 mm MgCl2, and 0.1% Nonidet P-40) containing 2 μg/ml propidium iodide. DNA content was assessed by flow cytometry and compared with C2C12 in forced G1 following culture in medium lacking l-methionine.
RESULTS
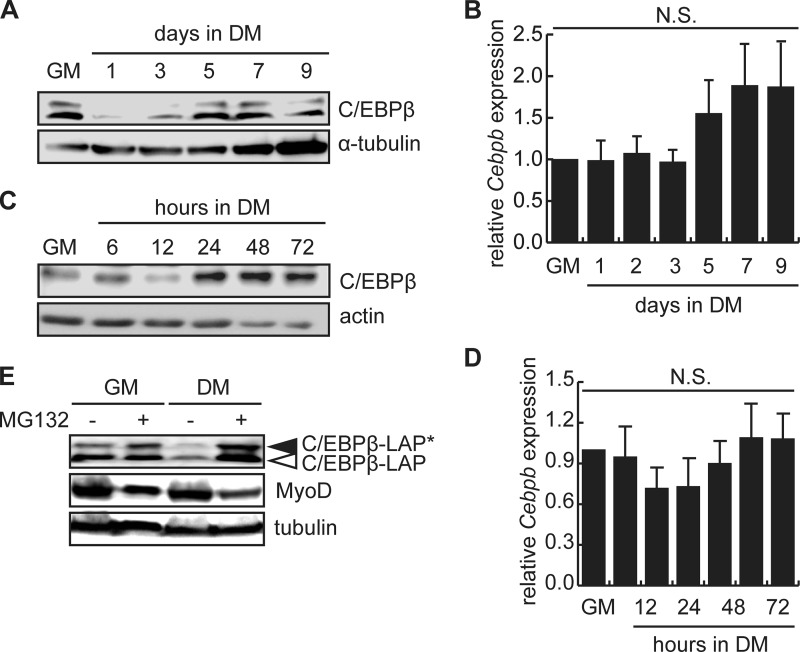
To examine changes in C/EBPβ expression in myoblasts during differentiation, we performed Western blotting and RT-qPCR in both the mouse myoblast cell line C2C12 and in primary myoblasts isolated from C57BL/6 mice (Fig. 1). In C2C12 cells, C/EBPβ expression was high in growth medium (GM) and was rapidly down-regulated after 1 day in differentiation medium (Fig. 1A). C/EBPβ expression remained low until day 5, when C/EBPβ expression increased again (Fig. 1A). Changes in C/EBPβ protein expression did not correlate with changes in Cebpb mRNA expression (Fig. 1B). In primary myoblasts, the differentiation timeline is much shorter, with large myotubes seen after only 48 h in low serum conditions. In this model, we observed a down-regulation of C/EBPβ protein expression after 12 h in differentiation medium, and similar to C2C12 cells, a second wave of C/EBPβ expression beginning at 24 h in differentiation medium and persisting to 72 h (Fig. 1C). Again, C/EBPβ expression varied without underlying changes in mRNA expression (Fig. 1D), suggesting that the regulation of C/EBPβ expression occurs at the level of mRNA translation or alternatively, at the level of protein turnover.
FIGURE 1.
C/EBPβ protein levels are regulated during myogenesis by the 26S proteasome. A, C/EBPβ LAP* and LAP protein expression in C2C12 myoblasts in growth medium (GM) or after induction to differentiate in low serum differentiation medium (DM) for the indicated for 1–9 days. α-Tubulin is a loading control. B, RT-qPCR analysis of Cebpb expression in GM and during differentiation. Error bars are the S.E., n = 3. N.S., not significant. C, C/EBPβ LAP expression in primary myoblasts isolated from C57BL/6 mice and cultured in GM or DM for the indicated time points in hours. Actin is a loading control. D, RT-qPCR analysis of Cebpb expression in primary myoblasts in GM and during differentiation. Error bars are the S.E., n = 3. N.S., not significant. E, C/EBPβ and MyoD protein expression in C2C12 myoblasts were cultured in GM or DM for 24 h in the presence of MG132 for 6 h in DM or 24 h in GM.
To explore the hypothesis that C/EBPβ levels are regulated at the level of protein turnover in myoblasts, C2C12 cells cultured in growth medium or induced to differentiate under low serum conditions for 24 h were treated with the 26 S proteasome inhibitor MG132 for either 6 or 24 h (Fig. 1E). Under growth conditions, where C/EBPβ expression is high, treatment with MG132 for 24 h had little impact on the expression of the most highly expressed activating isoform of C/EBPβ (LAP) but modestly increased C/EBPβ-LAP* expression (Fig. 1E). Expression of the inhibitory LIP isoform was not detected in primary myoblasts as previously observed (6). Under differentiation conditions, where vehicle-treated cells had low levels of C/EBPβ expression, a 6 h MG132 treatment prevented the rapid loss of C/EBPβ protein and increased the expression of both C/EBPβ-LAP and C/EBPβ-LAP* to levels superior to those observed in growth medium (Fig. 1E). Consistent with previous observations, in MG132-treated cells where C/EBPβ levels are high, MyoD expression was low. Interestingly, MG132 could not protect MyoD levels, suggesting that the regulation of MyoD expression by C/EBPβ is unlikely to be due to proteasomal degradation of MyoD (Fig. 1E). We have shown previously that the regulation is not due to changes in Myod1 mRNA expression (6).
To identify proteins involved in the accelerated turnover of C/EBPβ protein during myogenesis, we expressed a GFP-tagged C/EBPβ in C2C12 cells. Following 24 h in differentiation medium, GFP-C/EBPβ was precipitated and coprecipitated proteins were identified using mass spectroscopy. This screen identified the RING finger family E3 ubiquitin ligase Mdm2 as a C/EBPβ-interacting protein.
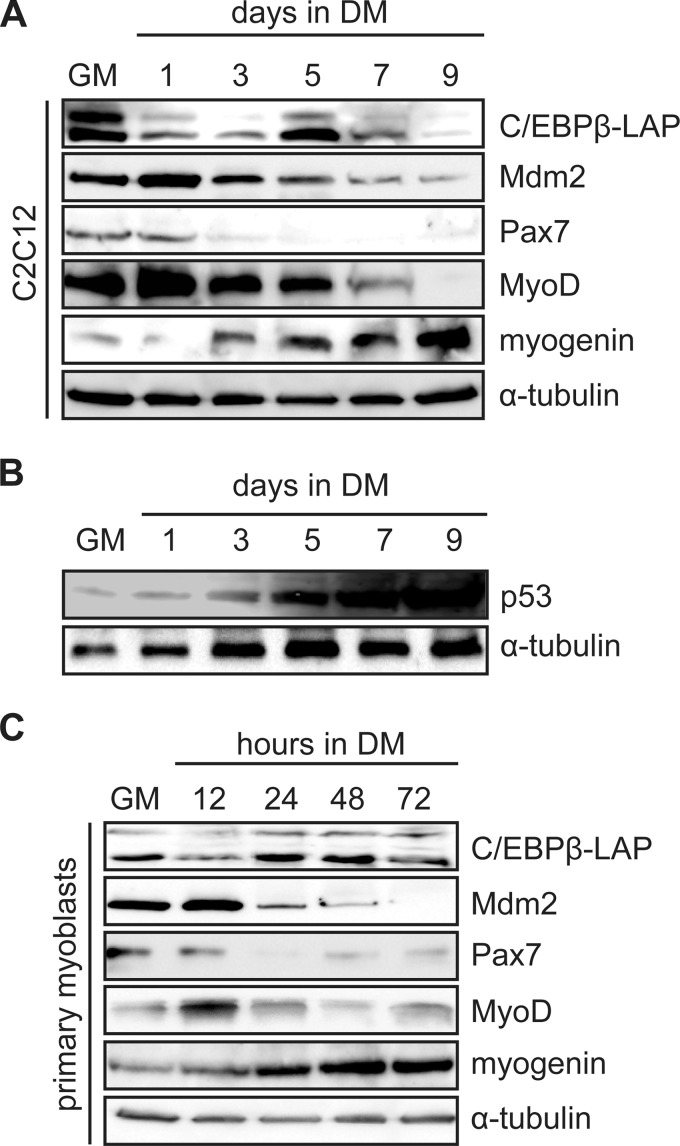
To characterize the Mdm2 expression pattern during myogenesis, C2C12 myoblasts were induced to differentiate in low serum conditions for 9 days (Fig. 2A) and extracts were analyzed by Western blotting. Mdm2 was readily detected in growth medium, and was up-regulated after 1 day in differentiation medium. Mdm2 expression was then slowly down-regulated as differentiation progressed, to its lowest levels at day 7 and 9 of differentiation (Fig. 2A). Correspondingly, p53 levels were lowest in GM and early differentiation and rose as differentiation progressed (Fig. 2B). Pax7 expression was also down-regulated on day 3 while MyoD expression was detected in GM and was up-regulated on day 1 of differentiation (Fig. 2A). MyoD levels then steadily decreased until day 9. Myogenin was up-regulated on day 3, and increased throughout the differentiation process.
FIGURE 2.
Mdm2 expression is regulated during myogenesis. A, C/EBPβ, Mdm2, and myogenic marker protein expression in C2C12 myoblasts cultured in GM or induced in DM for 1–9 days. α-Tubulin is a loading control. B, Western analysis of p53 expression in C2C12 myoblasts in growth medium (GM) or in differentiation medium (DM) for the time points indicated. α-Tubulin is used as a loading control. C, C/EBPβ, Mdm2, and myogenic marker protein expression in primary myoblasts cultured in GM or DM for the indicated time points in hours.
In primary myoblasts, Mdm2 levels were highest after 12 h in differentiation medium, and were rapidly down-regulated by 24 h of differentiation, a time point corresponding to the up-regulation of C/EBPβ expression (Fig. 2C). As in C2C12 cells, Pax7 levels were down-regulated early after induction to differentiate and remained low, though detectable at later time points. MyoD levels were induced at 12 h in DM, when C/EBPβ levels were low, and again decreased with the re-expression of C/EBPβ at 24 h of treatment (Fig. 2C). Myogenin expression was up-regulated at 12 h after induction to differentiate and continued to rise until 48 h in differentiation medium, remaining high at the 72 h time point (Fig. 2C). Taken together, this data demonstrates an inverse correlation between C/EBPβ and Mdm2 expression in early differentiation and reaffirms the negative influence of C/EBPβ on MyoD protein expression in early myogenesis.
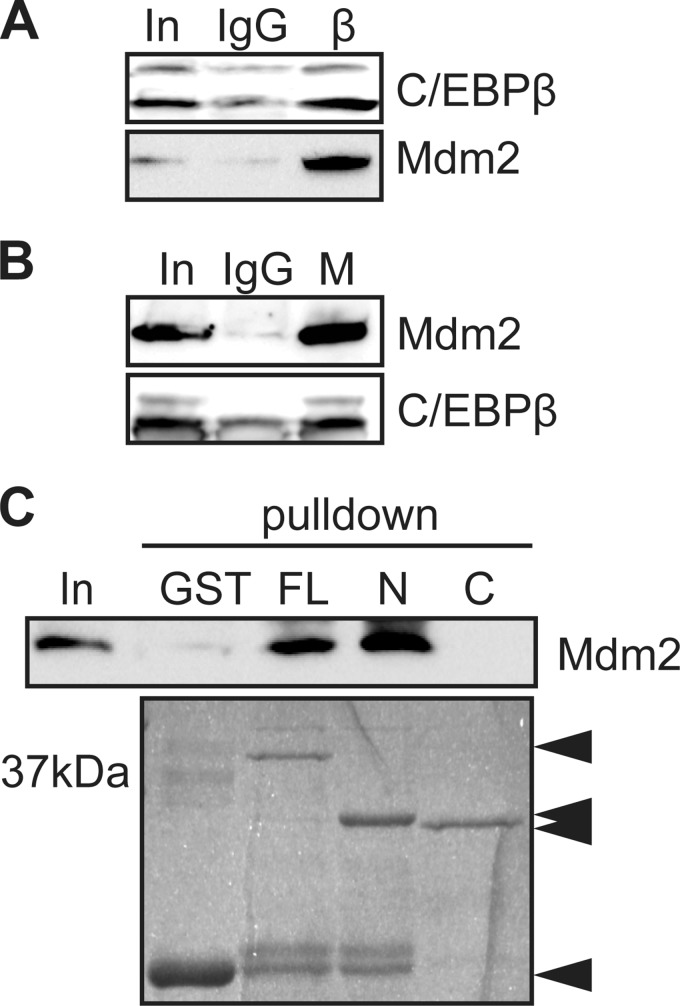
To validate the interaction between C/EBPβ and Mdm2 in vivo, C/EBPβ was immunoprecipitated from C2C12 myoblasts after 24 h in differentiation medium, and coprecipitated proteins were resolved by SDS-PAGE. While nonspecific type matched antibodies (IgG) did not co-precipitate Mdm2, Mdm2 was efficiently precipitated along with C/EBPβ (Fig. 3A). Immunoprecipitation of Mdm2 also enriched the coprecipitation of C/EBPβ under these same conditions (Fig. 3B). To determine which region of C/EBPβ interacts with Mdm2, GST-C/EBPβ (full-length, FL), GST-C/EBPβ1–151 (fragment N), and GST-C/EBPβ152–296 (fragment C) were incubated with in vitro translated Mdm2. As predicted, while GST alone did not precipitate Mdm2, full-length C/EBPβ efficiently precipitated the E3 ubiquitin ligase as did fragment N (amino acids 1–151), while the C-terminal half of C/EBPβ (fragment C, amino acids 152–296) did not precipitate Mdm2, suggesting that the interaction site between Mdm2 and C/EBPβ lies in the region spanning amino acid 1–151 of C/EBPβ (Fig. 3C).
FIGURE 3.
Mdm2 interacts with C/EBPβ. A, coimmunoprecipitation assay using anti-C/EBPβ (IP:β) antibody, or nonspecific type matched IgG to pulldown Mdm2. Input (In) is 10% of the material used for immunoprecipitation. B, coimmunoprecipitation assay using anti-Mdm2 (IP:M) antibody or nonspecific type matched IgG to pulldown C/EBPβ. C, GST pulldown assay using full-length GST-C/EBPβ (amino acids 1–296; FL), GST-C/EBPβ1–151 (C) GST-C/EBPβ152–296 (N), or GST alone as bait to precipitate in vitro translated Mdm2 detected by Western blotting. Migration of the GST constructs is indicated by Coomassie Blue staining (bottom).
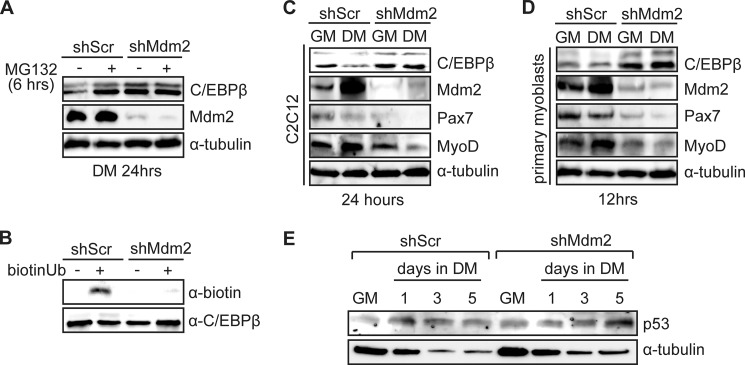
To determine if the physical interaction leads to the ubiquitination and degradation of C/EBPβ, we lentivirally transduced C2C12 myoblasts to express a shRNA against Mdm2 (shMdm2) or with a scrambled shRNA (shScr) and assessed C/EBPβ protein expression by Western blotting after 24 h in differentiation medium in the presence or absence of MG132 (Fig. 4A). In control cultures, a 6 h treatment with MG132 increased C/EBPβ expression when compared with vehicle-treated cells. In the shMdm2 cells, where Mdm2 expression was knocked down, C/EBPβ levels were increased in vehicle-treated cells to a level similar to MG132-treated control cultures, and this was not further enhanced by treatment with the proteasome inhibitor (Fig. 4A). These results suggested that Mdm2 targets C/EBPβ for degradation.
FIGURE 4.
Mdm2 ubiquitinates and targets C/EBPβ for degradation. A, C/EBPβ and Mdm2 protein expression in C2C12 cells lentivirally transduced to express shRNA against Mdm2 (shMdm2) or a scrambled shRNA (shScr) and induced to differentiate for 24 h during which the last 6 h included MG132 or vehicle as indicated. α-Tubulin is a loading control. B, in vitro ubiquitination assay in which biotinylated-ubiquitin and GST-C/EBPβ (target) was added to C2C12 cell extract from cells expressing shMdm2 or shScr. GST-C/EBPβ precipitates were analyzed for the addition of biotin-ubiquitin using strepavidin-HRP. C, C/EBPβ, Mdm2, Pax7, and MyoD protein expression in C2C12 myoblasts expressing shMdm2 or shScr in GM or DM for 24 h. D, C/EBPβ, Mdm2, Pax7, and MyoD protein expression in primary myoblasts expressing shMdm2 or shScr in GM or DM for 12 h. E, Western analysis of p53 expression in C2C12 myoblasts lentivirally transduced to express a shRNA against Mdm2 (shMdm2) or a scrambled shRNA control (shScr) cultured in GM or induced to differentiate in DM for the time points indicated.
To confirm ubiquitination of C/EBPβ by Mdm2, GST-C/EBPβ was incubated with biotinylated ubiquitin in whole cell extract from differentiating C2C12 cells lentivirally transduced to express the shMdm2 or with the scrambled control shRNA. Following pulldown of GST-C/EBPβ, ubiquitination was detected using HRP-conjugated streptavidin. Only in the presence of biotin-conjugated ubiquitin was a band corresponding to GST-C/EBPβ detected in control cultures (Fig. 4B). When GST-C/EBPβ was incubated in C2C12 extract in which Mdm2 was knocked down (shMdm2), we detected only a weak band corresponding to ubiquitin conjugation of C/EBPβ, suggesting that Mdm2 is the primary ubiquitin ligase targeting C/EBPβ in differentiating myoblasts (Fig. 4B). We then examined the expression of key myogenic markers in the C2C12 cell lines (shMdm2 and shScr) under growth conditions and after 24 h in differentiation medium. In cells in which Mdm2 was knocked down, C/EBPβ expression was increased in DM to a level comparable to shMdm2 cells and shScr cells in growth medium (Fig. 4C). As before, we also observed an increase in Mdm2 expression in DM in control cells, but also in shMdm2 cells, though the levels in this cell line were very low and did not induce further changes in C/EBPβ expression (Fig. 4C). Further, as expected, the switch to differentiation medium lead to a decrease in Pax7 expression and an increase in MyoD expression in controls, however in shMdm2 cells, Pax7 levels were lower than expected in growth medium, and this level was further reduced in DM despite high C/EBPβ levels, suggesting that loss of Mdm2 interferes with normal Pax7 expression independent of C/EBPβ (Fig. 4C). MyoD levels in the shMdm2 cells were comparable to the control cells in GM, but down-regulated in DM, corresponding to the high levels of C/EBPβ (Fig. 4C).
In primary myoblasts, loss of Mdm2 expression also resulted in higher levels of C/EBPβ which was more prominent than in C2C12 cells (Fig. 4D). We observed an increase in Mdm2 expression in control cells after 12 h in differentiation medium as compared with growth medium. Again, this corresponded to a decrease in C/EBPβ expression and Pax7 expression and an increase in MyoD expression (Fig. 4D). In shMdm2 primary myoblasts, we again observed an overall reduction in Pax7 expression as compared with shScr cells, along with a further decrease in differentiation conditions. MyoD levels were generally lower in the shMdm2 cells than in the controls, more so in DM conditions, consistent with the high C/EBPβ levels in these cells (Fig. 4D). Interestingly, loss of Mdm2 expression did not result in changes in p53 expression in growth medium or in early differentiation (Fig. 4E).
To assess Mdm2's role during myogenesis, we differentiated the C2C12-shMdm2 stable cells and controls (Fig. 5) for 7 days after which cells were immunostained for myosin heavy chain (MyHC) expression. While large myosin heavy chain positive myotubes could be observed in the control cultures, very few MyHC+ cells were observed in the shMdm2 cultures, and these largely appeared unfused (Fig. 5A). Indeed, the differentiation index (#nuclei in MyHC+ cells/total nuclei) was reduced ∼90% in shMdm2 cultures as compared with scrambled controls (Fig. 5B). This defect in differentiation was not due to reduced cell numbers in the shMdm2 cultures, as equal numbers of cells were induced for differentiation. Indeed, a significant increase in the number of cells in S phase was noted in these cultures without changes in the proportion of cells in G1 or G2/M phases of the cell cycle (Fig. 5C). Further, the average size of myotubes containing at least 2 nuclei (# nuclei in myotubes/# myotubes) was significantly reduced by ∼30% in the Mdm2 cells (Fig. 5D). Analysis of myogenic protein expression revealed that while control cultures expressed MyoD and myogenin after differentiation, the shMdm2 cultures had drastically reduced expression of these markers, along with high C/EBPβ expression (Fig. 5E). Taken together, these results suggest that Mdm2 expression is required for efficient myogenesis.
FIGURE 5.
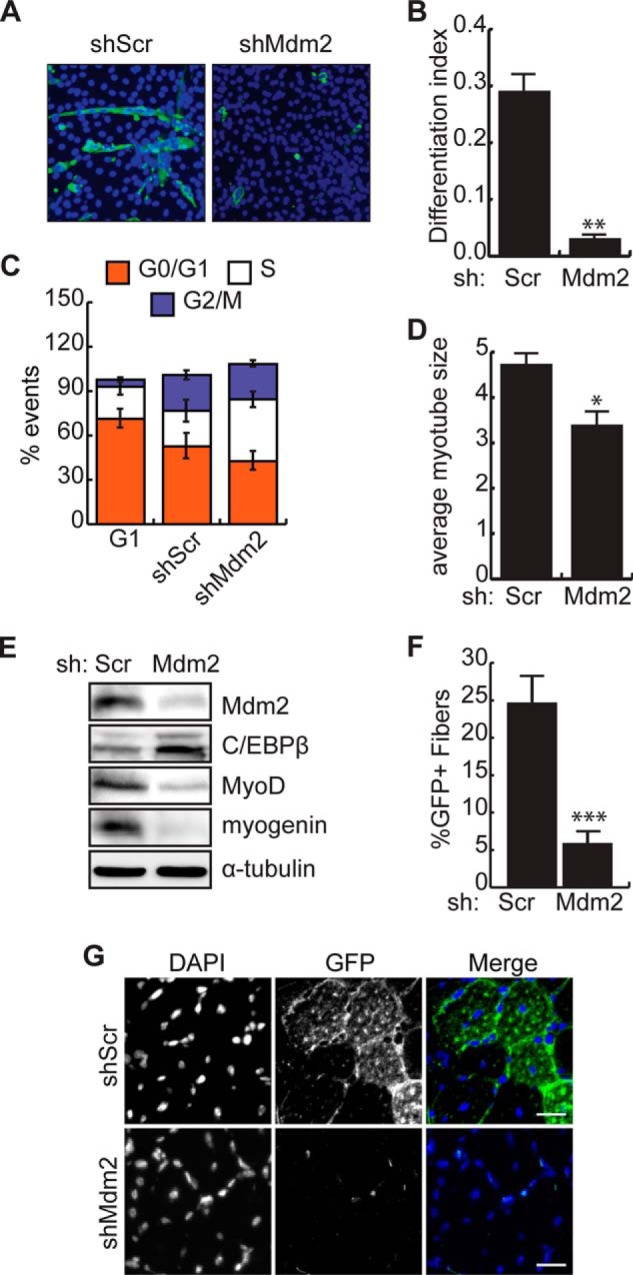
Loss of Mdm2 blocks myogenic differentiation and regeneration. A, immunocytochemistry for myosin heavy chain (MyHC) expression in C2C12 cells transduced to express shMdm2 or shScr and induced to differentiate for 7 days. DAPI stain reveals nuclei. B, differentiation index (no. nuclei in MyHC+ cells/total nuclei) for cells transduced and differentiated as in A. **, p < 0.01, n = 6. C, cell cycle analysis by flow cytometry in C2C12 cells forced into G1 arrest, or transduced as in A. Graph indicates the mean percentage of cells in each phase of the cell cycle ± S.D. The % cells in S-phase in shMdm2 cultures are significantly different (p = 0.044) from shScr controls. All other comparisons are not significantly different. D, average size of myotubes (no. nuclei in MyHC+ cells with nuclei>2/no. myotubes) for cells transduced and differentiated as in A. *, p < 0.05, n = 6. E, Mdm2, C/EBPβ, and myogenic marker protein expression in cells transduced and induced to differentiate as in A. F, percent of GFP+ fibers in regenerating muscle 2 weeks after injury with cardiotoxin and transplantation of primary myoblasts isolated from a GFP transgenic mouse transduced to express shMdm2 or shScr. ***, p < 0.001, n = 6. G, representative images of TA muscle from F. DAPI stains the nuclei. Scale bar, 25 μm.
To assess myogenesis in the absence of Mdm2 in vivo, we generated pooled stable cell lines expressing the shMdm2 or scrambled controls in primary myoblasts isolated from the GFP mouse. These cells were injected into host C57BL/6 muscle 24 h after injury with cardiotoxin to assess the contribution to regeneration (Fig. 5, F and G). Two weeks after grafting, the number of GFP+ fibers was scored and revealed that the scrambled control cells contributed to muscle repair, with on average 25% of the muscle fibers expressing GFP (Fig. 5, F and G). By contrast, only ∼5% of the muscle fibers in the muscle that received shMdm2-expressing myoblasts were GFP-positive, indicating that the loss of Mdm2 prevents myoblasts from participating in myogenesis in vivo.
To determine if loss of C/EBPβ could rescue differentiation in the shMdm2 cells, primary myoblasts were isolated from conditional null mice in which C/EBPβ was excised in Pax7+ cells (C/EBPβ−/−) and littermate controls, and were lentivirally transduced to express the shMdm2 construct or scrambled shRNA. Loss of Mdm2 expression in the WT background increased C/EBPβ expression 3 days after induction to differentiate in low serum conditions (Fig. 6A). Correspondingly, MyoD levels were lower in these cultures. Further, myogenin expression was inhibited, consistent with a decrease in myogenic differentiation. Loss of C/EBPβ expression resulted in an increase in both MyoD and myogenin expression in both control and shMdm2 cells, but for myogenin, not to the levels of controls (Fig. 6A).
FIGURE 6.
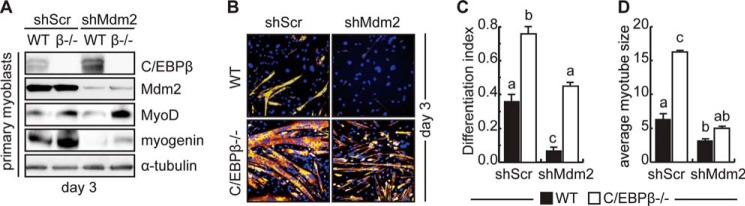
Loss of C/EBPβ rescues differentiation in cells lacking Mdm2. A, Mdm2, C/EBPβ, and myogenic marker protein expression in primary myoblasts from C/EBPβ conditional null mice (β−/−) or control littermates (WT) transduced to express shMdm2 or shScr and induced to differentiate for 3 days. B, immunocytochemistry for MyHC expression in differentiated cultures as in A. C, differentiation index for cells transduced and differentiated as in A. Means indicated with different letters are significantly different from one another, meeting a minimum cut-off of p < 0.05, n = 3. D, average myotube size for cells transduced and differentiated as in A.
Immunostaining for MyHC expression revealed that while loss of Mdm2 expression inhibited myogenesis in primary myoblasts, loss of C/EBPβ in the presence of Mdm2 enhanced myogenesis and resulted in larger myotubes as previously reported (Fig. 6, B–D) (6). Knock down of both C/EBPβ and Mdm2 restored the differentiation index to control levels, but not to the level observed in C/EBPβ−/− cells in the presence of Mdm2, consistent with our Western analysis (Fig. 6C). Further, while loss of C/EBPβ in the shMdm2 cells trended toward larger myotubes, the increase was not statistically significant. Taken together, these results suggest that the blockade of myogenesis imposed by loss of Mdm2 is mediated at least in part by C/EBPβ.
DISCUSSION
We and others have demonstrated that C/EBPβ is regulated by ubiquitination (25, 26) though the E3 ubiquitin ligase was unknown. We provide evidence that Mdm2 modifies and targets C/EBPβ for degradation by the 26 S proteasome and that this activity is required for myogenic differentiation. In addition to C/EBPβ, Pax7, Pax3, and MyoD are known to be regulated by the ubiquitin-proteasome system (27–29). However, while treatment with proteasome inhibitor protected C/EBPβ protein levels in early differentiation, MyoD levels were not protected. Rather, high C/EBPβ levels provoked a decline in MyoD expression. Since the regulation of MyoD expression is not due to changes in Myod1 mRNA expression (6) and is not mediated by the 26 S proteasome, a possible mechanism for regulation would be control of Myod1 translation, potentially through the C/EBPβ-dependent production of a miRNA. Indeed, Myod1 translation has been shown to be inhibited by miR-221/222 (30) though regulation of this miR by C/EBPβ has yet to be investigated. Further, while loss of Mdm2 protected C/EBPβ from degradation, Pax7 levels were abnormally low in both C2C12 and primary myoblasts, despite being a transcriptional target of C/EBPβ. These results suggest that Mdm2 plays a role in the maintenance of Pax7 expression in early myogenesis that is independent of C/EBPβ. Mdm2 has been shown to act as a regulator of gene transcription, for example by suppressing the activity of Smad transcription factors through sequestration of Smad4, by inhibiting MyoD-dependent transcription and by stimulating CREB-dependent transcription (31–34). Mdm2 can also inhibit mRNA translation and stability (35, 36).
High levels of Mdm2 have also been shown to block myogenesis in rhabdomyosarcoma cell lines and C2C12 myoblasts (37). Given the important roles attributed to Mdm2, it is perhaps not surprising that the expression of Mdm2 has to be tightly controlled to allow for normal cell growth and differentiation, with both over and underexpression negatively impacting differentiation. In contrast to the rhabdomyosarcoma model, in our experiments we studied Mdm2 expression at endogenous levels and in a loss of function model. When Mdm2 levels are below their normal expression early in differentiation, myogenesis is blocked and this is partially restored by loss of C/EBPβ, indicating that Mdm2 expression is required for myogenesis.
Mdm2 acts early in the myogenic differentiation program, with only low levels of Mdm2 expression seen once myogenin expression is up-regulated. Later in differentiation, corresponding to the low Mdm2 levels, C/EBPβ levels are again increased. In fibroblasts, p19(ARF) is involved in sustaining C/EBPβ levels in a p53-independent fashion, suggesting that a blockade of Mdm2 activity is protective for C/EBPβ, consistent with our results (38). While it is clear that failure to down-regulate C/EBPβ expression early in differentiation blocks myogenesis, and this is Mdm2-dependent, the function of C/EBPβ expression in terminal differentiation remains unclear. It is tempting to speculate that C/EBPβ may be playing a role in limiting cell fusion, such that loss of C/EBPβ results in large myotubes and larger fibers in vivo (6).
We have identified the amino acids 1–151 of mouse C/EBPβ to be required for the interaction with Mdm2. We have not yet identified the modified residue or residues, but computational prediction suggests that lysine 133 in the mouse C/EBPβ is the most likely target. Since the C/EBPβ-LIP isoform is not predicted to interact with Mdm2, Mdm2 may play a role in regulating the LAP/LIP ratios in the cell. A predominance of the LIP isoform has been shown to influence progression through the cell cycle (39), to block adipogenesis (40) and to promote oncogenesis (41). LIP is not expressed in primary myoblasts, and we did not observe an increase in LIP expression in cells lacking Mdm2, however, given its role in oncogenesis, it is of broader interest to understand the impact on cell growth of Mdm2-induced changes in cellular LAP/LIP ratios.
Footnotes
- C/EBP
- CCAAT-enhancer-binding protein
- MDM2
- mouse double minute 2 homolog
- CTX
- cardiotoxin
- TA
- tibialis anterior
- MyHC
- myosin heavy chain.
REFERENCES
- 1. Seale P., Sabourin L. A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M. A. (2000) Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786 [DOI] [PubMed] [Google Scholar]
- 2. Mauro A. (1961) Satellite cell of skeletal muscle fibers. J Biophys Biochem. Cytol 9, 493–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beauchamp J. R., Heslop L., Yu D. S., Tajbakhsh S., Kelly R. G., Wernig A., Buckingham M. E., Partridge T. A., Zammit P. S. (2000) Expression of CD34 and Myf5 defines the majority of quiescent adult skeletal muscle satellite cells. J. Cell Biol. 151, 1221–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sambasivan R., Yao R., Kissenpfennig A., Van Wittenberghe L., Paldi A., Gayraud-Morel B., Guenou H., Malissen B., Tajbakhsh S., Galy A. (2011) Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656 [DOI] [PubMed] [Google Scholar]
- 5. Wang Y. X., Rudnicki M. A. (2012) Satellite cells, the engines of muscle repair. Nat. Rev. Mol. Cell Biol. 13, 127–133 [DOI] [PubMed] [Google Scholar]
- 6. Marchildon F., Lala N., Li G., St-Louis C., Lamothe D., Keller C., Wiper-Bergeron N. (2012) CCAAT/enhancer binding protein β is expressed in satellite cells and controls myogenesis. Stem Cells 30, 2619–2630 [DOI] [PubMed] [Google Scholar]
- 7. Sterneck E., Tessarollo L., Johnson P. F. (1997) An essential role for C/EBPbeta in female reproduction. Genes Dev. 11, 2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cao Z., Umek R. M., McKnight S. L. (1991) Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 5, 1538–1552 [DOI] [PubMed] [Google Scholar]
- 9. Wiper-Bergeron N., St-Louis C., Lee J. M. (2007) CCAAT/Enhancer binding protein beta abrogates retinoic acid-induced osteoblast differentiation via repression of Runx2 transcription. Mol. Endocrinol. 21, 2124–2135 [DOI] [PubMed] [Google Scholar]
- 10. Tanaka T., Yoshida N., Kishimoto T., Akira S. (1997) Defective adipocyte differentiation in mice lacking the C/EBPβ and/or C/EBPδ gene. EMBO J. 16, 7432–7443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robinson G. W., Johnson P. F., Hennighausen L., Sterneck E. (1998) The C/EBPbeta transcription factor regulates epithelial cell proliferation and differentiation in the mammary gland. Genes Dev. 12, 1907–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ramji D. P., Foka P. (2002) CCAAT/enhancer-binding proteins: structure, function and regulation. Biochem. J. 365, 561–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Descombes P., Chojkier M., Lichtsteiner S., Falvey E., Schibler U. (1990) LAP, a novel member of the C/EBP gene family, encodes a liver-enriched transcriptional activator protein. Genes Dev. 4, 1541–1551 [DOI] [PubMed] [Google Scholar]
- 14. Ossipow V., Descombes P., Schibler U. (1993) CCAAT/enhancer-binding protein mRNA is translated into multiple proteins with different transcription activation potentials. Proc. Natl. Acad. Sci. U.S.A. 90, 8219–8223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chau V., Tobias J. W., Bachmair A., Marriott D., Ecker D. J., Gonda D. K., Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583 [DOI] [PubMed] [Google Scholar]
- 16. Hershko A., Ciechanover A. (1992) The ubiquitin system for protein degradation. Annu. Rev. Biochem. 61, 761–807 [DOI] [PubMed] [Google Scholar]
- 17. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 18. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 19. Vousden K. H., Lu X. (2002) Live or let die: the cell's response to p53. Nat Rev Cancer 2, 594–604 [DOI] [PubMed] [Google Scholar]
- 20. Xiao Z. X., Chen J., Levine A. J., Modjtahedi N., Xing J., Sellers W. R., Livingston D. M. (1995) Interaction between the retinoblastoma protein and the oncoprotein MDM2. Nature 375, 694–698 [DOI] [PubMed] [Google Scholar]
- 21. Martin K., Trouche D., Hagemeier C., Sørensen T. S., La Thangue N. B., Kouzarides T. (1995) Stimulation of E2F1/DP1 transcriptional activity by MDM2 oncoprotein. Nature 375, 691–694 [DOI] [PubMed] [Google Scholar]
- 22. Kim C. H., Kim K. H., Yoo Y. M. (2012) Melatonin-induced autophagy is associated with degradation of MyoD protein in C2C12 myoblast cells. J. Pineal Res. 53, 289–297 [DOI] [PubMed] [Google Scholar]
- 23. Birsoy K., Chen Z., Friedman J. (2008) Transcriptional Regulation of Adipogenesis by KLF4. Cell Metabolism 7, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alpi A. F., Pace P. E., Babu M. M., Patel K. J. (2008) Mechanistic insight into site-restricted monoubiquitination of FANCD2 by Ube2t, FANCL, and FANCI. Mol. Cell 32, 767–777 [DOI] [PubMed] [Google Scholar]
- 25. Liu Y., Zhang Y. D., Guo L., Huang H. Y., Zhu H., Huang J. X., Liu Y., Zhou S. R., Dang Y. J., Li X., Tang Q. Q. (2013) Protein inhibitor of activated STAT 1 (PIAS1) is identified as the SUMO E3 ligase of CCAAT/enhancer-binding protein beta (C/EBPβ) during adipogenesis. Mol. Cell. Biol. 33, 4606–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lechner S., Mitterberger M. C., Mattesich M., Zwerschke W. (2013) Role of C/EBPβ-LAP and C/EBPβ-LIP in early adipogenic differentiation of human white adipose-derived progenitors and at later stages in immature adipocytes. Differentiation 85, 20–31 [DOI] [PubMed] [Google Scholar]
- 27. Olguin H. C., Yang Z., Tapscott S. J., Olwin B. B. (2007) Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J. Cell Biol. 177, 769–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Joung H., Eom G. H., Choe N., Lee H. M., Ko J. H., Kwon D. H., Nam Y. S., Min H., Shin S., Kook J., Cho Y. K., Kim J. C., Seo S. B., Baik Y. H., Nam K. I., Kook H. (2014) Ret finger protein mediates Pax7-induced ubiquitination of MyoD in skeletal muscle atrophy. Cell. Signal. 26, 2240–2248 [DOI] [PubMed] [Google Scholar]
- 29. Boutet S. C., Disatnik M. H., Chan L. S., Iori K., Rando T. A. (2007) Regulation of Pax3 by proteasomal degradation of monoubiquitinated protein in skeletal muscle progenitors. Cell 130, 349–362 [DOI] [PubMed] [Google Scholar]
- 30. Tan S. B., Li J., Chen X., Zhang W., Zhang D., Zhang C., Li D., Zhang Y. (2014) Small molecule inhibitor of myogenic microRNAs leads to a discovery of miR-221/222-myoD-myomiRs regulatory pathway. Chem. Biol. 21, 1265–1270 [DOI] [PubMed] [Google Scholar]
- 31. Guo C. S., Degnin C., Fiddler T. A., Stauffer D., Thayer M. J. (2003) Regulation of MyoD activity and muscle cell differentiation by MDM2, pRb, and Sp1. J. Biol. Chem. 278, 22615–22622 [DOI] [PubMed] [Google Scholar]
- 32. Hallenborg P., Feddersen S., Francoz S., Murano I., Sundekilde U., Petersen R. K., Akimov V., Olson M. V., Lozano G., Cinti S., Gjertsen B. T., Madsen L., Marine J. C., Blagoev B., Kristiansen K. (2012) Mdm2 controls CREB-dependent transactivation and initiation of adipocyte differentiation. Cell Death Differ. 19, 1381–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sun P., Dong P., Dai K., Hannon G. J., Beach D. (1998) p53-independent role of MDM2 in TGF-β1 resistance. Science 282, 2270–2272 [DOI] [PubMed] [Google Scholar]
- 34. Yam C. H., Siu W. Y., Arooz T., Chiu C. H., Lau A., Wang X. Q., Poon R. Y. (1999) MDM2 and MDMX inhibit the transcriptional activity of ectopically expressed SMAD proteins. Cancer Res. 59, 5075–5078 [PubMed] [Google Scholar]
- 35. Gu L., Zhu N., Zhang H., Durden D. L., Feng Y., Zhou M. (2009) Regulation of XIAP translation and induction by MDM2 following irradiation. Cancer Cell 15, 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gu L., Zhang H., He J., Li J., Huang M., Zhou M. (2012) MDM2 regulates MYCN mRNA stabilization and translation in human neuroblastoma cells. Oncogene 31, 1342–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fiddler T. A., Smith L., Tapscott S. J., Thayer M. J. (1996) Amplification of MDM2 inhibits MyoD-mediated myogenesis. Mol. Cell. Biol. 16, 5048–5057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sebastian T., Johnson P. F. (2009) RasV12-mediated down-regulation of CCAAT/enhancer binding protein beta in immortalized fibroblasts requires loss of p19Arf and facilitates bypass of oncogene-induced senescence. Cancer Res. 69, 2588–2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Luedde T., Duderstadt M., Streetz K. L., Tacke F., Kubicka S., Manns M. P., Trautwein C. (2004) C/EBP beta isoforms LIP and LAP modulate progression of the cell cycle in the regenerating mouse liver. Hepatology 40, 356–365 [DOI] [PubMed] [Google Scholar]
- 40. Karagiannides I., Thomou T., Tchkonia T., Pirtskhalava T., Kypreos K. E., Cartwright A., Dalagiorgou G., Lash T. L., Farmer S. R., Timchenko N. A., Kirkland J. L. (2006) Increased CUG triplet repeat-binding protein-1 predisposes to impaired adipogenesis with aging. J. Biol. Chem. 281, 23025–23033 [DOI] [PubMed] [Google Scholar]
- 41. Luft F. C. (2015) C/EBPβ LIP induces a tumor menagerie making it an oncogene. J. Mol. Med. 93, 1–3 [DOI] [PubMed] [Google Scholar]