Background: Mechanisms by which lysosomal biogenesis is regulated are poorly understood.
Results: Activation of PPARα stimulates lysosomal biogenesis via transcriptional up-regulation of TFEB.
Conclusion: These results delineate a novel role of PPARα in controlling lysosomal biogenesis.
Significance: Activation of PPARα may be of therapeutic benefit in lysosomal storage disorders.
Keywords: Astrocyte, Lysosomal Storage Disease, Neurodegeneration, Peroxisome proliferator-activated Receptor (PPAR), Retinoic Acid
Abstract
Lysosomes are ubiquitous membrane-enclosed organelles filled with an acidic interior and are central to the autophagic, endocytic, or phagocytic pathway. In contrast to its classical function as the waste management machinery, lysosomes are now considered to be an integral part of various cellular signaling processes. The diverse functionality of this single organelle requires a very complex and coordinated regulation of its activity with transcription factor EB (TFEB), a master regulator of lysosomal biogenesis, at its core. However, mechanisms by which TFEB is regulated are poorly understood. This study demonstrates that gemfibrozil, an agonist of peroxisome proliferator-activated receptor (PPAR) α, alone and in conjunction with all-trans-retinoic acid is capable of enhancing TFEB in brain cells. We also observed that PPARα, but not PPARβ and PPARγ, is involved in gemfibrozil-mediated up-regulation of TFEB. Reporter assay and chromatin immunoprecipitation studies confirmed the recruitment of retinoid X receptor α, PPARα, and PGC1α on the PPAR-binding site on the Tfeb promoter as well. Subsequently, the drug-mediated induction of TFEB caused an increase in lysosomal protein and the lysosomal abundance in cell. Collectively, this study reinforces the link between lysosomal biogenesis and lipid metabolism with TFEB at the crossroads. Furthermore, gemfibrozil may be of therapeutic value in the treatment of lysosomal storage disorders in which autophagy-lysosome pathway plays an important role.
Introduction
Lysosomes are membrane-bound organelles containing a host of hydrolytic enzymes that are highly active in acidic milieu (1–3). Classically identified as the waste management organelle, lysosomes have been shown to be involved in major cellular processes, including degradation, developmental programmed cell death, and nutritional responses (2, 4–8). The diverse roles and responses of the lysosome to different stimuli suggest a coordinated regulation of expression of lysosomal genes (9, 10). Recent studies provide modest information about the regulation of lysosomal genes (11, 12), but pattern discovery analysis for the lysosomal genes reveals the presence of a coordinated lysosomal expression and regulation element, which is a potential binding site for TFEB,2 a member of the microphthalmia-transcription factor E (MiT/TFE) subfamily of basic helix-loop-helix factors. These studies report a potential link between TFEB and lysosomal biogenesis (9, 10, 12). The regulation of Tfeb appears to be complex and dependent on cell type and stimuli. In differentiated osteoclasts, a receptor activator of NF-κB ligand-dependent signaling pathway induces TFEB activation and lysosomal biogenesis (13). Starvation or stress conditions may also activate TFEB, which otherwise is maintained in an inactivated state by mTORC1 (14, 15). One study also delineates that starvation-induced TFEB activity can play a vital role in lipid metabolism and that activated TFEB can also autoregulate its own gene expression (16). Therefore, we investigated the effect of lipid-lowering drugs on the status of TFEB and lysosomal biogenesis.
Gemfibrozil, an FDA-approved lipid-lowering drug, is used to decrease the risk of hyperlipidemia (17–19). However, gemfibrozil also regulates pathways responsible for inflammation, T-helper cell switching, cell migration, and oxidative stress (20–23). Here, we describe that gemfibrozil, alone and in conjunction with ATRA, is capable of enhancing TFEB via PPARα, but not PPARβ and PPARγ. We further demonstrate that the combination of gemfibrozil and ATRA enhances lysosomal biogenesis in brain cells as well as in cells from patients suffering from late infantile neuronal ceroid lipofuscinosis (LINCL), where the Cln2 gene is mutated.
MATERIALS AND METHODS
Reagents
DMEM/F-12 50:50, 1×, Hanks' balanced salt solution, and 0.05% trypsin were purchased from Mediatech (Washington, DC). Fetal bovine serum (FBS) was obtained from Atlas Biologicals (Fort Collins, CO). Antibiotic-antimycotic, gemfibrozil, ATRA, and GSK0660 were obtained from Sigma. GW7647, GW9662, and GW6471 were purchased from Tocris (Bristol, UK). Agarose, neurobasal media, B27 supplement, B27 supplement minus antioxidants, and Moloney murine leukemia virus-reverse transcriptase were obtained from Life Technologies, Grand Island, NY.
Isolation of Primary Mouse Astroglia
Astroglia were isolated from mixed glial cultures as described (24, 25) according to the procedure of Giulian and Baker (26). Briefly, on day 9, the mixed glial cultures were washed three times with Dulbecco's modified Eagle's medium/F-12 and subjected to shaking at 240 rpm for 2 h at 37 °C on a rotary shaker to remove microglia. After 2 days, the shaking was repeated for 24 h for the removal of oligodendroglia and to ensure the complete removal of all nonastroglial cells. The attached cells were seeded onto new plates for further studies.
Isolation of Primary Mouse Neurons
Fetal (E18 to E16) mouse neurons were prepared as described previously (27) with modifications. Whole brains were removed, and the cells were washed by centrifugation three times at 1000 rpm for 10 min, the pellet dissociated, and the cells plated at 10% confluence in 8-well chamber slides pretreated for >2 h with poly-d-lysine (Sigma). After 4 min, the nonadherent cell suspension was aspirated, and 500 ml of complete Neurobasal media supplemented with 2% B27 was added to each well. The cells were incubated for 4 days prior to experimentation. Double label immunofluorescence with β-tubulin and either GFAP or CD11b revealed that neurons were more than 98% pure (data not shown). The cells were stimulated with gemfibrozil in Neurobasal media supplemented with 2% B27 minus antioxidants for 24 h prior to methanol fixation and immunostaining.
Semi-quantitative Reverse Transcriptase-coupled Polymerase Chain Reaction (RT-PCR)
Total RNA was isolated from mouse primary astrocytes and human primary astrocytes using the RNeasy Qiagen (Valencia, CA) kit following the manufacturer's protocol. Semi-quantitative RT-PCR was carried out as described earlier (28) using oligo(dT)12–18 as primer and Moloney murine leukemia virus-RT in a 20-μl reaction mixture. The resulting cDNA was appropriately amplified using Promega Master Mix (Madison, WI) and the following primers (Life Technologies) for murine genes: mouse Tfeb, sense, 5′-AAC AAA GGC ACC ATC CTC AA-3′, and antisense, 5′-CAG CTC GGC CAT ATT CAC AC-3′; mouse Lamp2, sense, 5′-GGT GCT GGT CTT TCA GGC TTG ATT-3′, and antisense, 5′-ACC ACC CAA TCT AAG AGC AGG ACT-3′; mouse Limp2, sense, 5′-TGT TGA AAC GGG AGA CAT CA-3′, and antisense, 5′-TGG TGA CAA CCA AAG TCG TG-3′; mouse Npc1, sense, 5′-GGG ATG CCC GTG CCT GCA AT-3′, and antisense, 5′-CTG GCA GCT ACA TGG CCC CG-3′; mouse Gapdh, sense, 5′-GCA CAG TCA AGG CCG AGA AT-3′, and antisense, 5′-GCC TTC TCC ATG GTG GTG AA-3′.
Amplified products were electrophoresed on 2% agarose gels and visualized by ethidium bromide staining. Glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA was used as a loading control to ascertain that an equivalent amount of cDNA was synthesized from each sample.
Quantitative Real Time PCR
The mRNA quantification was performed using the ABI-Prism7700 sequence detection system (Applied Biosystems, Foster City, CA) using SYBR Select master mix (Applied Biosystems). The mRNA expression of the targeted genes was normalized to the level of Gapdh mRNA, and data were processed by the ABI Sequence Detection System 1.6 software.
Immunostaining of Cells
Immunocytochemistry was performed as described earlier (29). Briefly, mouse primary astrocytes or neurons cultured to 70–80% confluence in 8-well chamber slides were fixed with chilled methanol (Fisher Scientific, Waltham, MA) overnight, followed by two brief rinses with filtered PBS. Samples were blocked with 2% BSA (Fisher Scientific) in PBS containing Tween 20 (Sigma) and Triton X-100 (Sigma) for 30 min and incubated at room temperature under shaking conditions for 2 h in PBS containing the following anti-mouse primary antibodies: TFEB (1:1000; Abcam), GFAP, (1:1000; DAKO), LAMP2 (1:500, Abcam), NeuN (1:500, Millipore), and MAP2 (1:200, Millipore). After four 15-min washes in filtered PBS, the slides were further incubated with Cy2- or Cy5-labeled secondary antibodies (all 1:200; Jackson ImmunoResearch, West Grove, PA) for 1 h under similar shaking conditions. Following four 15-min washes with filtered PBS, cells were incubated for 4–5 min with 4′,6-diamidino-2-phenylindole (DAPI, 1:10,000; Sigma). The samples were run in an EtOH and xylene (Fisher) gradient, mounted, and observed under Olympus BX41 fluorescence microscope.
Immunostaining of Tissue Sections
After 60 days of treatment, mice were sacrificed, and their brains were fixed, embedded, and processed. Sections were made from different brain regions, and for immunofluorescence staining on fresh frozen sections, anti-mouse TFEB (1:500), anti-mouse LAMP2 (1:200), and anti-mouse NeuN (1:500) were used. The samples were mounted and observed under Olympus BX41 fluorescence microscope (30).
LysoTracker Staining
Fibroblasts cultured to 70–80% confluence were subjected to different stimuli under reduced serum (2%) DMEM followed by incubation with 75 nm LysoTracker Red DND99 (Life Technologies) for 45 min. Cells were then washed thoroughly with filtered PBS and mounted on glass slides and viewed under BX41 fluorescence microscope.
Immunoblotting
Western blotting was conducted as described earlier (23, 32) with modifications. Briefly, cells were scraped in 1× RIPA buffer; protein was measured using Bradford reagent, and SDS buffer was added and electrophoresed on NuPAGE® Novex® 4–12% BisTris gels (Life Technologies, Inc.); and proteins were transferred onto a nitrocellulose membrane (Bio-Rad) using the Thermo-Pierce Fast Semi-Dry Blotter. The membrane was then washed for 15 min in TBS plus Tween 20 (TBST) and blocked for 1 h in TBST containing BSA. Next, the membranes were incubated overnight at 4 °C under shaking conditions with the following 1° antibodies: TFEB (1:1000, Abcam), LAMP2 (1:500, Abcam), and β-actin (1:800; Abcam, Cambridge, MA). The next day, membranes were washed in TBST for 1 h, incubated in 2° antibodies against 1° antibody hosts (all 1:10,000; Jackson ImmunoResearch) for 1 h at room temperature, washed for 1 h more, and visualized under the Odyssey® infrared imaging system (Li-COR, Lincoln, NE).
Construction of Mouse Tfeb Promoter-driven Reporter Construct
Mouse genomic DNA isolated from primary mouse astrocytes was used as the template during PCR. The 5′-flanking sequence of the mouse TFEB (−916/+61) gene was isolated by PCR. Primers were designed from GenBankTM sequences as follows: Tfeb: sense, 5′-acgcgt CCA GGA GCC AGG GAC GGG GTA CAT CTC-3′, and antisense, 5′-agatct AAG GAG AAA CTG AGT CCG GGC AGA AGG-3′. The sense primer was tagged with an MluI restriction enzyme site, and the antisense primer was tagged with BglII. The PCR was performed using an Advantage-2 PCR kit (Clontech) according to the manufacturer's instruction. The resulting fragments were gel-purified and ligated into the PGEM-TEasy vector (Promega). These fragments were further subcloned into the PGL-3 Enhancer vector after digestion with the corresponding restriction enzymes and verification by sequencing ACGT Inc. DNA Sequencing Services.
Cloning of Tfeb Promoter and Site-directed Mutagenesis
Site-directed mutagenesis was done by using the site-directed mutagenesis kit (Stratagene). Two primers in opposite orientation were used to amplify the mutated plasmid in a single PCR. The primer sequence for mutated promoter site was as follows: mutated: sense, 5′-GCA ACA GCA AGT GCG GAT TTG AGG GGG GGG GAC GGT GGG-3′, and antisense, 5′-CCC ACC GTC CCC CCC CCT CAA ATC CGC ACT TGC TGT TGC-3′. The PCR product was precipitated with ethanol and then phosphorylated by T4 kinase. The phosphorylated fragment was self-ligated by T4 DNA ligase and digested with restriction enzyme DpnI to eliminate the nonmutated template. The mutated plasmid was cloned and amplified in Escherichia coli (DH5-α strain)-competent cells.
Assay of Tfeb Promoter-driven Reporter Activity
Cells plated at 50–60% confluence in 12-well plates were cotransfected with 0.25 μg of pTFEB(WT)-Luc, pTFEB(Mu)-Luc and using Lipofectamine Plus (Life Technologies, Inc.). After 24 h of transfection, cells were stimulated with different agents under serum-free conditions for 6 h. Firefly luciferase activities were analyzed in cell extracts using the luciferase assay system kit (Promega) in a TD-20/20 luminometer (Turner Designs) as described earlier (33, 34).
Chromatin Immunoprecipitation Assay
ChIP assays were performed using the method described by Nelson et al. (35), with certain modifications. Briefly, mouse primary astrocytes were stimulated by 10 μm gemfibrozil and 0.5 μm RA together for 6 h followed by fixing with formaldehyde (1.42% final volume) and quenching with 125 mm glycine. The cells were pelleted and lysed in IP buffer containing 150 mm NaCl, 50 mm Tris-HCl (pH 7.5), 5 mm EDTA, Nonidet P-40 (0.5% v/v), Triton X-100 (1.0% v/v). For 500 ml, 4.383 g of NaCl, 25 ml of 100 mm EDTA (pH 8.0), 25 ml of 1 m Tris-HCl (pH 7.5), 25 ml of 10% (v/v) Nonidet P-40 and 50 ml of 10% (v/v) Triton X-100 were added containing the following inhibitors: 10 μg/ml leupeptin, 0.5 mm phenylmethylsulfonyl fluoride (PMSF), 30 mm p-nitrophenyl phosphate, 10 mm NaF, 0.1 mm Na3VO4, 0.1 mm Na2MoO4, and 10 mm β-glycerophosphate. After one wash with 1.0 ml of IP buffer, the pellet was resuspended in 1 ml of IP buffer (containing all inhibitors), and sonicated and sheared chromatin was split into two fractions (one to be used as input). The remaining fraction was incubated overnight under rotation at 4 °C with 5–7 μg of anti-PPARα or anti-RXRα antibodies or anti-PGC1α or RNA polymerase or normal IgG (Santa Cruz Biotechnology) followed by incubation with protein G-agarose (Santa Cruz Biotechnology) for 2 h at 4 °C under rotation. Beads were then washed five times with cold IP buffer, and a total of 100 μl of 10% Chelex (10 g/100 ml of H2O) was added directly to the washed protein G beads and vortexed. After 10 min of boiling, the Chelex/protein G bead suspension was allowed to cool to room temperature. Proteinase K (100 μg/ml) was then added, and the beads were incubated for 30 min at 55 °C while shaking, followed by another round of boiling for 10 min. The suspension was centrifuged and supernatant collected. The Chelex/protein G beads fraction was vortexed with another 100 μl of water and centrifuged again, and the first and the second supernatants were combined. Eluate was used directly as a template in PCR. The following primers were used to amplify fragments flanking the RXR-binding element in the mouse Tfeb promoter: Set1, sense, 5′-GAA CAT TCC AGG TGG AGG CA-3′, and antisense, 5′-CCC CCA ACA CAT GCT TCT CT-3′; Set2, sense, 5′-GAG TCT CTC GGA GGA GGT GA-3′,m and antisense, 5′- ACT CCA GGC ATG CTT TCT CC-3′. The PCRs were repeated by using varying cycle numbers and different amounts of templates to ensure that results were in the linear range of PCR. The qRT-PCR was performed using the same primers and SYBR select MasterMix. Data were normalized to input and nonspecific IgG, and fold increase versus control was calculated.
Densitometric Analysis
Protein blots were analyzed using ImageJ (National Institutes of Health, Bethesda), and bands were normalized to their respective β-actin loading controls. Immunofluorescence quantification data are representative of the average fold change with respect to control for at least 25 different images per condition from three independent set of experiments.
Statistics
Values are expressed as means ± S.E. for at least three independent experiments. Statistical analyses for differences were performed via Student's t test. This criterion for statistical significance was p < 0.05.
RESULTS
Activation of PPARα and RXRα Induces Expression of TFEB in Mouse Primary Brain Cells
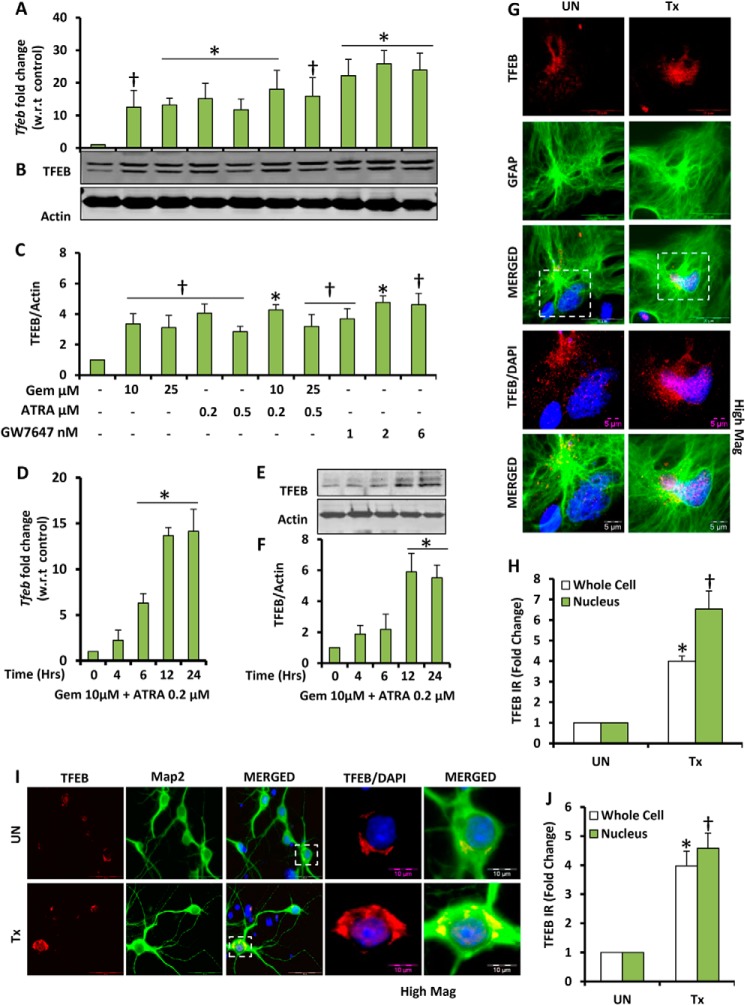
We examined whether gemfibrozil, an activator of PPARα and an FDA-approved drug for hyperlipidemia, could up-regulate the expression of TFEB in mouse brain cells. Because it has been known that PPARα and RXRα form a transcriptionally active complex (21, 36, 37), we used both gemfibrozil and ATRA to check whether there is any additive effect due to dual treatment. Mouse primary astrocytes were treated in serum-free media with single doses of gemfibrozil and ATRA and also in combination. Quantitative real time PCR data showed increased expression of Tfeb in all treatment groups. We observed 10–14-fold increase in the levels of Tfeb with single individual treatment of gemfibrozil and/or ATRA and ∼15–18-fold increase in combinatorial treatment, which was not found to be statistically significant with respect to individual treatment (Fig. 1A). Interestingly when a combination of both gemfibrozil and ATRA was used, we could achieve a similar level of expression of Tfeb at much lower doses of both compounds (10 and 0.2 μm, respectively) compared with 25 μm gemfibrozil and 0.5 μm ATRA. Treatment with GW7647, a high affinity and a high selectivity ligand of PPARα, also showed a significant increase in the levels of Tfeb mRNA (Fig. 1A), indicating that PPARα specifically could be responsible for stimulating the expression of Tfeb. The time point analysis with the combinatorial treatment showed that the Tfeb expression could be induced as early as 6 h up to 24 h (Fig. 1D). The qRT-PCR data for both dose and time were validated by Western blots, which showed a similar pattern of increase in TFEB levels (Fig. 1, B, C, E, and F). Furthermore, we used mouse primary astrocytes and neurons for treating with the combination of gemfibrozil and ATRA in serum-free media for 24 h and performed immunocytochemistry. The data showed a distinct increase in the levels of TFEB in both astrocytes and neurons as well as localization of TFEB in and around the nucleus (Fig. 1, G and I). The TFEB immunoreactivity was quantified using the ImageJ software, and we observed an ∼4-fold increase in the overall levels of TFEB and an ∼5–6-fold increase of TFEB localization of TFEB in the nucleus upon treatment (Fig. 1, H and J). It has been previously shown that starvation and nutrient deficiency lead to activation of TFEB. Therefore, in this study, all the untreated cells were maintained in serum-free conditions for the whole duration of the treatment as well, so that the baseline change in the levels of TFEB would remain the same between the groups.
FIGURE 1.
Gemfibrozil and retinoic acid up-regulate TFEB mRNA and protein levels in brain cells. A and B, mouse primary astrocytes were treated with different concentrations of gemfibrozil (Gem), ATRA, and GW7647 in serum-free DMEM/F-12 medium for 12 h followed by monitoring mRNA levels of Tfeb by qRT-PCR (A) and TFEB protein levels by immunoblot (B). C, densitometric analysis of the immunoblot for TFEB (relative to β-actin). D and E, mouse primary astrocytes were treated with a combination of gemfibrozil (10 μm) and ATRA (0.2 μm) for 4, 6, 12, and 24 h under similar culture conditions followed by monitoring of mRNA levels of TFEB by qRT-PCR (D) and protein levels by immunoblot (E). F, densitometric analysis for the immunoblot for TFEB. All results are representative of or the mean ± S.E. of at least three independent sets of experiments. †, <0.05 versus untreated control; *, p < 0.01 versus untreated control. G and I, mouse primary astrocytes (G) and mouse primary neurons (I) were treated with a combination of gemfibrozil and retinoic acid under serum-free conditions for 24 h and were double-labeled for TFEB (red)-GFAP (green) and TFEB (red)-Map2 (green), respectively. DAPI was used to stain nuclei. Scale bar, 20 μm (for G); scale bar, 5 μm for high magnification images (for G); scale bar, 50 μm (for I); scale bar, 10 μm for high magnification images (for I). H and J, quantification of TFEB immunoreactivity (TFEB IR) in whole cell and nucleus for mouse primary astrocytes (H) and mouse primary neurons (J) calculated as fold over control. At least 25 separate images per condition from three independent set of experiments are quantified using ImageJ. *, p < 0.05 versus untreated control (whole cell); †, p < 0.05 versus untreated control (nucleus). UN, DMSO-treated cells (used as control); Tx, treated with the combination of gemfibrozil (10 μm) and all-trans-retinoic acid (0.2 μm).
PPARα and RXRα Are Involved in the Drug-mediated Up-regulation of TFEB
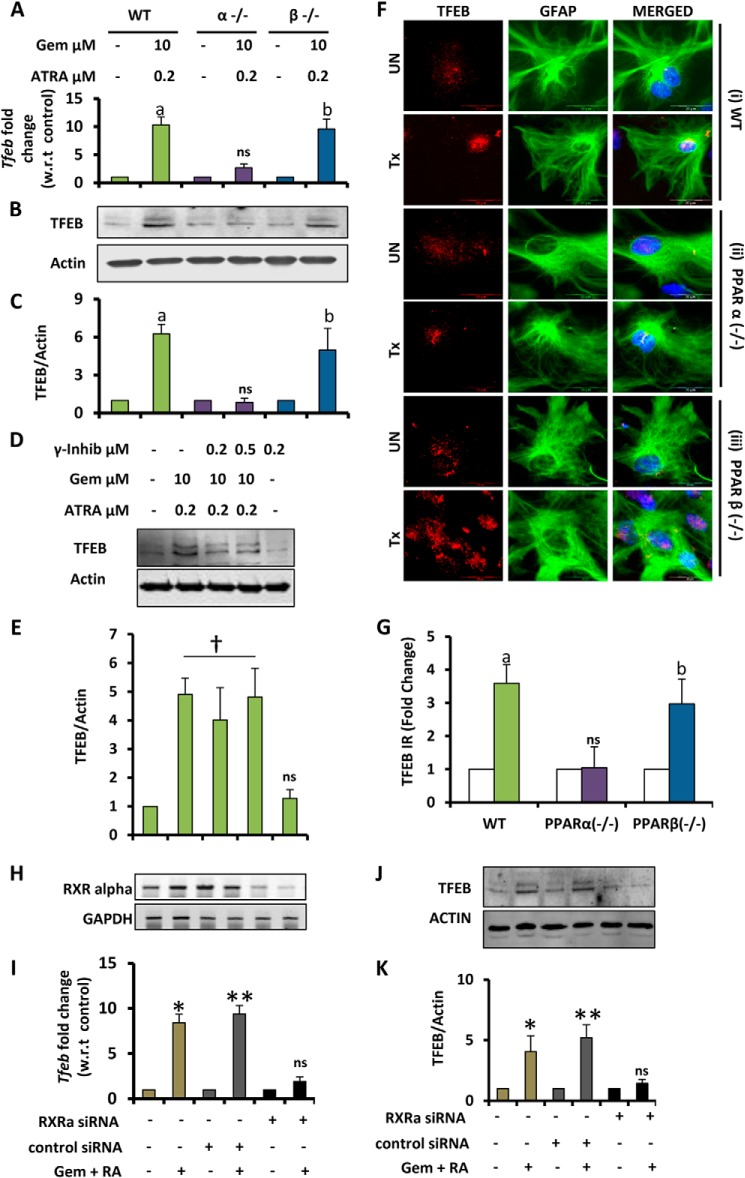
We tested our primary hypothesis that PPARα in conjunction with RXRα could be involved in the drug-mediated up-regulation of TFEB, by using primary astrocytes from PPARα−/− animals and knocking down RXRα in WT mouse primary astrocytes. Primary astrocytes obtained from WT, PPARα−/−, and PPARβ−/− mice were treated under similar conditions as above and checked for the mRNA and protein expression of TFEB. Both real time PCR and Western blots for TFEB showed that TFEB could be up-regulated in astrocytes from WT and PPARβ−/−, but not PPARα−/−, mice (Fig. 2, A–C). The findings were further confirmed by immunocytochemistry where we observed almost 3–4-fold increase in TFEB levels in astrocytes isolated from WT and PPARβ−/−, but not PPARα−/−, mice (Fig. 2, F and G). We also tested whether PPARγ was involved in this particular drug-mediated expression of TFEB. Western blotting for TFEB using pretreatment with PPARγ-specific inhibitors prior to treatment with the drugs indicated that gemfibrozil and ATRA may not be using the PPARγ-mediated pathway for the up-regulation of TFEB (Fig. 2, D and E). PPARα and RXRα have been known to form a transcriptional complex, and our data showed a marginal increase of TFEB in the presence of ATRA. Next, we examined whether ATRA exerted its effects via RXRα. WT mouse primary astrocytes were treated with RXRα-specific siRNA followed by treatment with the combination of gemfibrozil and ATRA. The data showed a successful knockdown of the RXRα gene (Fig. 2H), and consequently, the effect of drugs was partially abrogated in the absence of RXRα, which was evident from the levels of Tfeb mRNA after RXRα silencing (Fig. 2I). Western blotting also showed similar results with the TEFB levels being significantly less in RXRα-silenced cells compared with scrambled siRNA (Fig. 2, J and K). Taken together these data indicate that PPARα and RXRα could be involved in the up-regulation of TFEB by gemfibrozil and ATRA.
FIGURE 2.
Involvement of PPARα and RXRα in fibrate drug-mediated up-regulation of TFEB mRNA and protein. A and B, mouse primary astrocytes isolated from PPARα−/− and PPARβ−/− and wild type mouse were treated with a combination of gemfibrozil (Gem) (10 μm) and retinoic acid (0. 2 μm) in serum-free DMEM/F-12 for 24 h followed by monitoring the mRNA expression of Tfeb by real time PCR (A) and protein level of TFEB by immunoblot (B). C, densitometric analysis of TFEB levels (relative to β-actin) in PPARα−/− and PPARβ−/− and wild type astrocytes. a, p < 0.05 versus WT control; b, p < 0.05 versus PPARβ−/− control; ns, not significant with respect to PPARα−/− control. D, mouse primary astrocytes isolated from WT mice were pretreated with GW9662 for 30 min followed by treatment with gemfibrozil and retinoic acid under similar culture conditions followed by monitoring the levels of TFEB protein expression by immunoblot. E, densitometric analysis of immunoblot for TFEB (relative to β-actin) †, p <0.05 versus control; ns, not significant with respect to control. F, mouse primary astrocytes isolated from PPARα−/− and PPARβ−/− and WT mice were treated with 10 μm gemfibrozil and 0.2 μm retinoic acid in serum-free DMEM/F-12 for 24 h and double-labeled for TFEB (red) and GFAP (green). DAPI was used to stain nuclei. UN, no treatment. Scale bar, 20 μm. G, quantification of TFEB immunoreactivity (TFEB IR) for mouse primary astrocytes calculated as fold over control. At least 25 separate images per condition from three independent set of experiments are quantified using ImageJ. a, p < 0.05 versus WT control; b, p < 0.05 versus PPARβ−/− control; ns, not significant with respect to PPARα−/− control. H–J, mouse primary astrocytes were untransfected and transfected with scrambled siRNA (1.0 μg) or RXRα siRNA (1.0 μg) for 36 h followed by treatment with RA (0.2 μm) and gemfibrozil (10 μm) in combination for 24 h in serum-free DMEM/F-12 medium followed by RT-PCR for RXRα to check the level of gene silencing (H) and quantitative real time PCR for TFEB (J) and immunoblot for TFEB (J). K, densitometric analysis of immunoblot for TFEB (relative to β-actin). *, p < 0.05 versus untransfected control; **, p < 0.05 versus scrambled siRNA-transfected control; ns, not significant with respect to RXRα siRNA transfected control. All results are representative of or mean ± S.E. of at least three independent set of experiments. Tx, treated with the combination of gemfibrozil (10 μm) and all-trans-retinoic acid (0.2 μm).
PPARα/RXRα Heterodimer Transcriptionally Regulates TFEB Expression under Treatment Conditions
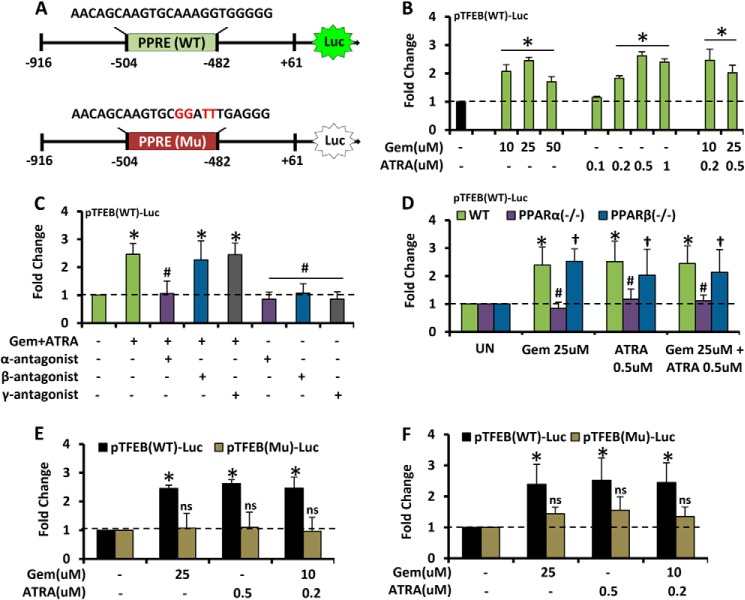
Next, we tested whether PPARα and RXRα transcriptionally regulated Tfeb expression. After analysis of the promoter site of Tfeb, we found the presence of a peroxisomal proliferator-response element (PPRE) about 480 bp upstream to the transcription start site of Tfeb. The Tfeb promoter (pTFEB(WT)) containing the PPRE was cloned into the pGL3 Enhancer vector. We also mutated the core sequence of the PPRE, and the mutated promoter construct (pTFEB(Mu)) was also cloned into the PGL3 vector. Gemfibrozil and ATRA, alone or in combination, markedly induced WT-TFEB-driven luciferase activity in BV-2 glial cells (Fig. 3B). Suppression of WT-TFEB-driven luciferase activity in (gemfibrozil + ATRA)-treated BV-2 glial cells (Fig. 3C) by GW6471 (an antagonist of PPARα), but neither GSK0660 (an antagonist of PPARβ) nor GW9662 (an antagonist of PPARγ), suggests the involvement of PPARα, but neither PPARβ nor PPARγ, in (gemfibrozil + ATRA)-induced activation of TFEB promoter. In primary astrocytes isolated from WT, PPARα−/−, and PPARβ−/− mice, we observed an increased luciferase activity in WT and PPARβ−/− but not in PPARα−/− astrocytes (Fig. 3D). Furthermore, when the construct with the mutated PPRE site (pTFEB(Mu)-Luc) was transfected into BV-2 and mouse primary astrocytes, we found a dramatic decrease in the luciferase activity in cells containing the mutant construct (Fig. 3, E and F). Taken together, these data indicate that the activation of PPARα plays an important role in the induction of Tfeb upon treatment with gemfibrozil and retinoic acid.
FIGURE 3.
PPARα transcriptionally regulates TFEB expression under treatment condition. A, map of wild type and mutated PPRE site of TFEB-luciferase promoter constructs. B, BV2 cells were transfected with pTFEB(WT)-Luc for 24 h followed by treatment with different concentrations of gemfibrozil (Gem) and retinoic acid alone and in combination and subjected to luciferase assay. *, p < 0.05 versus untreated control. C, BV2 cells were transfected with pTFEB(WT)-Luc for 24 h followed by pretreatment with PPARα, PPARβ, PPARγ antagonists (viz. GW6471, 250 nm; GSK0660, 200 nm, and GW9662, 4 nm) followed by treatment with gemfibrozil and retinoic acid and subjected to luciferase assay. *, p < 0.05 versus untreated control. #, p < 0.05 versus treatment. D, mouse primary astrocytes isolated from PPARα−/− and PPARβ−/− and wild type mice were transfected with pTFEB(WT)-Luc for 24 h followed by treatment with gemfibrozil and retinoic acid and subjected to luciferase assay. *, p < 0.05 versus untreated WT control; #, p not significant versus untreated PPARα−/− control; †, p < 0.05 versus untreated PPARβ−/− control. E and F, BV2 cells (E) and mouse primary astrocytes (F) were transfected with pTFEB(WT)-Luc and pTFEB(Mu)-Luc for 24 h followed by treatment with gemfibrozil and retinoic acid and subjected to luciferase assay. *, p < 0.05 versus untreated pTFEB(WT)-Luc transfected control. ns, not significant with respect to untreated pTFEB(Mu)-Luc transfected control. All results are mean ± S.E. of at least six sets of independent experiments.
Finally, we decided to investigate the actual DNA binding role of PPARα on the Tfeb promoter in this context. Because upon activation, PPARα, RXRα, and PGC1α form a complex, which initiates transcriptional activation of many genes (38–42), we investigated whether gemfibrozil and ATRA induced the recruitment of these molecules to the Tfeb promoter. Mouse primary astrocytes treated with gemfibrozil and retinoic acid for different time points from 30 to 240 min were subjected to ChIP analysis by immunoprecipitation of the chromatin fragments with anti-PPARα, -RXRα, -PGC1α, and -RNA polymerase antibodies. Both the semi-quantitative PCR and quantitative RT-PCR showed an increased enrichment of the amplicon over time with the pulldown by the specific antibodies (Fig. 4, B and C). Immunoprecipitation followed by PCR with normal IgG showed almost undetectable bands in RT-PCR, and PCR with total fragmented DNA showed uniform signal in RT-PCR, showing the uniformity and specificity of the results. In real time PCR, the Ct values were normalized to % input and further normalized with the IgG signal to get a signal over noise value, to verify the specificity of the results. The experiments were repeated at least three times under the same condition and cycles, and dilution of PCR products was adjusted to ensure that the data were in the linear range of the PCR. These results indicate that gemfibrozil and ATRA induce the recruitment of PPARα, RXRα, PGC1α, and RNA polymerase to the Tfeb promoter (Fig. 4D).
FIGURE 4.
Transcriptional activation of TFEB by PPARα-RXRα-PGC1α complex. A, map of PPRE on TFEB promoter with core sequence and amplicon length for ChIP. B and C, mouse astrocytes were treated with the combination of gemfibrozil (10 μm) and RA (0.2 μm) for 30, 60, 120, and 240 mins, and recruitment of PPARα, RXRα, PGC1α, and RNA polymerase (Pol) on the PPRE-binding site of Tfeb promoter was monitored by ChIP followed by RT-PCR (B) and qRT-PCR (C). IP, immunoprecipitation. Normal IgG was used as control. *, p < 0.05 versus untreated control. All results are representative of or mean ± S.E. at least there independent sets of experiments. D, schematic representation of the induction of lysosomal biogenesis by the activation of PPARα/RXRα.
Up-regulation of TFEB Leads to Increased Lysosomal Biogenesis
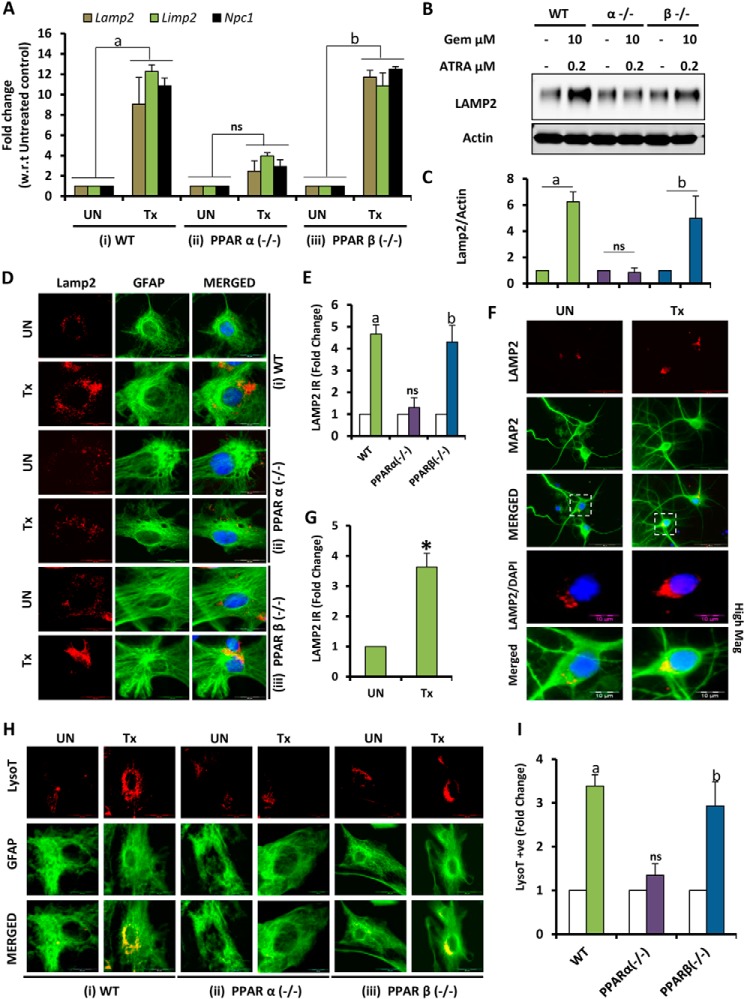
Because TFEB is the master regulator of lysosomal gene expression and biogenesis (9, 12, 16), we examined the effect of gemfibrozil and ATRA on lysosomal biogenesis. Primary astrocytes treated under the same conditions were subjected to mRNA analysis for some lysosomal markers like Lamp2, Limp2, and Npc1. Data showed elevated levels of these genes under treatment conditions in astrocytes isolated from WT and PPARβ−/−, but not in PPARα−/−, mice (Fig. 5A). Western blot analysis for LAMP2 in WT and KO cells showed a similar protein expression pattern (Fig. 5, B and C), which was further confirmed by immunocytochemistry for LAMP2 in WT and KO cells (Fig. 5, D and E). The increase in levels of LAMP2 was also observed in mouse primary neurons (Fig. 5, F and G). Furthermore, when cells were stained with LysoTracker Red, we observed increased lysosome content per cell in the case of drug-treated astrocytes isolated from WT and PPARβ−/−, but not PPARα−/−, mice (Fig. 5, H and I). These data suggest that gemfibrozil and ATRA can induce TFEB expression via PPARα:RXRα pathway, eventually leading to increased lysosomal biogenesis (Fig. 4D).
FIGURE 5.
PPARα-dependent up-regulation of TFEB induces lysosomal biogenesis. A and B, mouse primary astrocytes isolated from PPARα−/− and PPARβ−/− and wild type mouse were treated with combination of gemfibrozil (Gem) (10 μm) and retinoic acid (0. 2 μm) in serum-free DMEM/F-12 for 24 h followed by monitoring the mRNA expression of lysosomal genes (Lamp2, Limp2, and Npc1) by real time PCR (A) and protein level of LAMP2 by immunoblot (B). C, densitometric analysis of LAMP2 levels (relative to β-actin) in PPARα−/− and PPARβ−/− and wild type astrocytes. a, p < 0.05 versus WT control; b, p < 0.05 versus PPARβ−/− control; ns, not significant with respect to PPARα−/− control. All results are representative of or the mean ± S.E. of at least there independent sets of experiments. D, mouse primary astrocytes isolated from PPARα−/− and PPARβ−/− and WT mice were treated with 10 μm gemfibrozil and 0.2 μm retinoic acid in serum-free DMEM/F-12 for 24 h and double-labeled for LAMP2 (red) and GFAP (green). DAPI was used to stain nuclei. E, quantification of LAMP2 immunoreactivity (Lamp2 IR) for mouse primary astrocytes calculated as fold over control. a, p < 0.05 versus WT control; b, p < 0.05 versus PPARβ−/− control; ns, not significant with respect to PPARα−/− control. F, mouse primary neurons isolated from WT mice were treated with 10 μm gemfibrozil and 0.2 μm retinoic acid in serum-free DMEM/F-12 for 24 h and double-labeled for LAMP2 (red) and Map2 (green). DAPI was used to stain nuclei. G, quantification of LAMP2 immunoreactivity (Lamp2 IR) for mouse primary neurons calculated as fold over control. *, p < 0.05 versus untreated control. H, mouse primary astrocytes isolated from PPARα−/− and PPARβ−/− and WT mice were treated with 10 μm gemfibrozil and 0.2 μm retinoic acid in serum-free DMEM/F-12 for 24 h and double-labeled for LysoTracker Red (red) and GFAP (green). I, quantification of LysoTracker-positive signal (LysoT+ve) for mouse primary astrocytes calculated as fold over control. a, p < 0.05 versus WT control; b, p < 0.05 versus PPARβ−/− control; ns, not significant with respect to PPARα−/− control. UN, no treatment. Scale bar, 20 μm (for D–F), scale bar, 10 μm for high magnification images (for E). AT least 25 images per condition from three different sets of experiments were analyzed for all image quantification data using ImageJ. Tx, treated with the combination of gemfibrozil (10 μm) and all-trans-retinoic acid (0.2 μm).
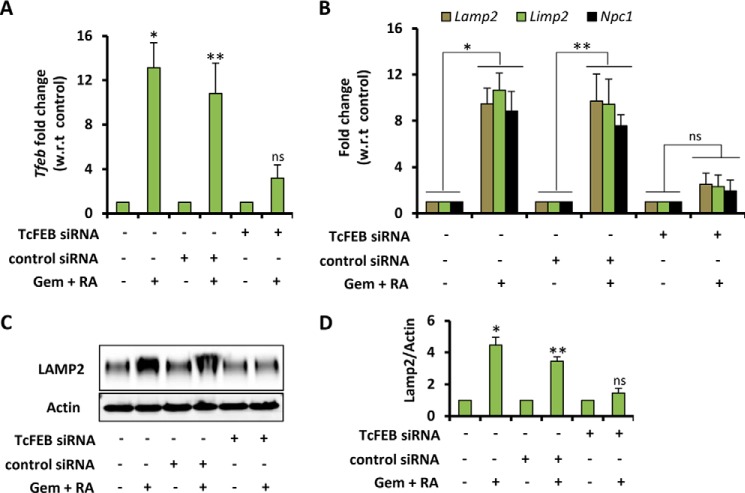
Knockdown of Tfeb Abrogates the Effect of Gemfibrozil and ATRA on Lysosomal Biogenesis
To establish the link between (gemfibrozil + ATRA)-mediated Tfeb induction and lysosomal biogenesis, a knockdown experiment was performed. WT mouse primary astrocytes were treated with Tfeb-specific siRNA followed by treatment with the combination of gemfibrozil and ATRA. The data showed a successful knockdown of Tfeb gene (Fig. 6A), and consequently the effects of drugs were significantly abrogated in the absence of Tfeb, which was evident from decreased levels of lysosomal gene (Lamp2, Limp2, and Npc1) mRNA after Tfeb silencing (Fig. 6B). The Western blot also showed similar results with the LAMP2 levels being significantly less in Tfeb-silenced cells as compared with scrambled siRNA (Fig. 6, C and D).
FIGURE 6.
Knockdown of TFEB abrogates the effect of gemfibrozil and ATRA on lysosomal biogenesis. A and B, mouse primary astrocytes were transfected with either scrambled siRNA (1. 0 μg) or Tfeb siRNA (1.0 μg) for 36 h followed by treatment with RA (0.2 μm) and gemfibrozil (Gem) (10 μm) in combination for 24 h in serum-free DMEM/F-12 medium followed by real time PCR for Tfeb to check the level of gene silencing (A) and real time PCR for lysosomal genes (Lamp2, Limp2, and Npc1) (B) and immunoblot for LAMP2 (C). D, densitometric analysis of immunoblot for LAMP2 (relative to β-actin). *, p < 0.05 versus untransfected control; **, p < 0.05 versus scrambled siRNA transfected control; ns, not significant with respect to Tfeb siRNA transfected control. All results are representative of or the mean ± S.E. of at least three independent set of experiments.
Agonists of PPARα and RXRα Induce Lysosomal Biogenesis in Vivo in the CNS of WT and PPARβ−/−, but Not in PPARα−/−, Mice
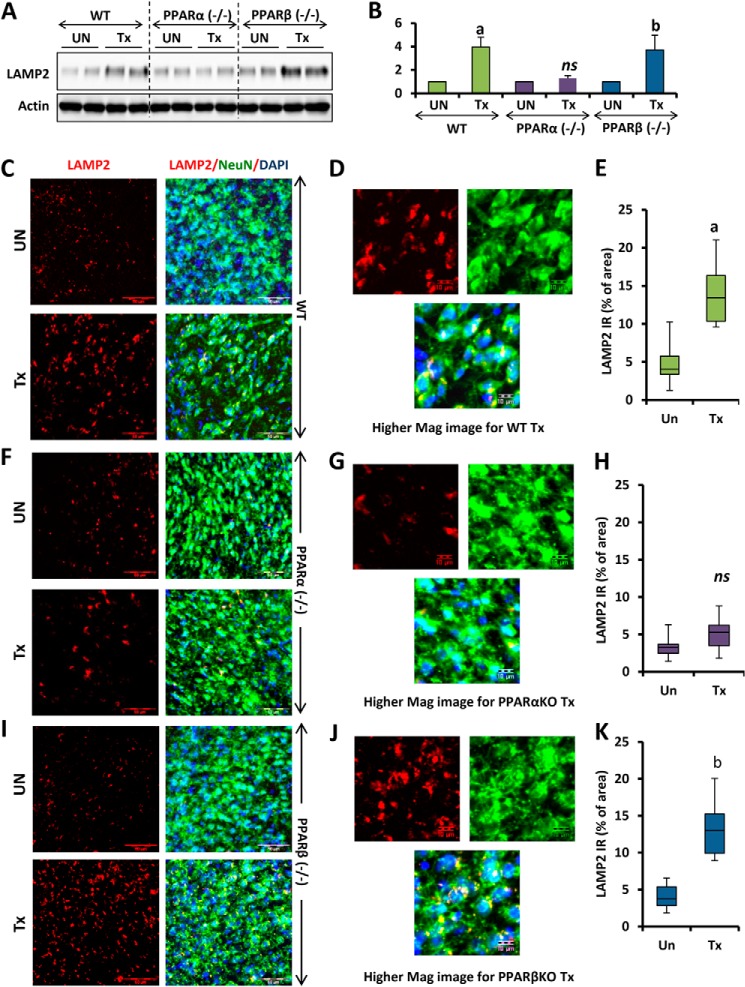
Once we confirmed the involvement of PPARα in the fibrate-mediated up-regulation of TFEB, we further checked whether the same results could be replicated in in vivo settings. WT, PPARα−/−, and PPARβ−/− mice from the same background were treated orally for 60 days with gemfibrozil (7.5 mg/kg body weight/day) and ATRA (0.1 mg/kg body weight/day) dissolved in 0.1% methylcellulose, which was also used as vehicle. At the end of the treatment, mice were sacrificed. Cortex was isolated from the brain of some animals and harvested and processed for immunoblot, and also the cerebral cortex from the rest of the animals was sectioned, and immunofluorescence was performed for the presence of TFEB. This in vivo data validated our cell culture findings as we observed remarkable elevation in the levels of TFEB in the cortex of WT, PPARβ−/−, and PPARα−/− mice after drug treatment (Fig. 7, A and B). The immunohistochemistry data also confirmed our biochemical findings as the TFEB-positive signal was more in WT and PPARβ−/− animals than in PPARα−/−-treated animals (Fig. 7, C, F, and I). We further quantified the TFEB-positive signals in at least 12 sections per group, and the values were represented as percentage of the total area. The quantitative analysis confirmed a significant increase in TFEB-positive signals in WT and PPARβ−/− animals, but not PPARα−/− animals (Fig. 7, E, H, and K). Other sections of the cortex from the same animals were subjected to immunohistochemistry for the presence of LAMP2. The results indicate increased LAMP2 immunoreactivity in WT and PPARβ−/−, but not PPARα−/−, animals (Fig. 8, C, F, and I). The quantitative data also suggested a significant increase in LAMP2-positive signals in WT and PPARβ−/−, but not PPARα−/−, animals (Fig. 8, E, H, and K). In this case also, the immunofluorescence data were confirmed by biochemical findings by performing immunoblot to check the levels of LAMP2 in the cortex of drug-treated animals (Fig. 8, A and B).
FIGURE 7.
Oral administration of gemfibrozil up-regulates TFEB in vivo in the cortex of WT and PPARβ−/−, but not PPARα−/−, mice. A, WT, PPARα−/−, and PPARβ−/− mice (n = 6 in each group) were treated with 7. 5 mg/kg body weight/day gemfibrozil and 0.1 mg/kg body weight of all-trans-retinoic acid (dissolved in 0.1% methylcellulose) or vehicle (0.1% methylcellulose) via gavage. After 60 days of treatment, mice were killed, and cortices from the animal brain were processed for immunoblot for TFEB (A). B, densitometric analysis of the immunoblot for TFEB (relative to β-actin). C, F, and I, cortical sections were double-labeled for TFEB (red) and NeuN (green). DAPI was used to visualize nucleus. D, G, and J, higher magnification images showing localization of TFEB and NeuN in the cortical neuron of mice from the treatment group (WT, PPARα−/−, and PPARβ−/−). E, H, and K, quantification of TFEB immunoreactivity (TFEB IR) in untreated and treated samples from each group (WT, PPARα−/−, and PPARβ−/−) expressed as percentage of area. a, p <0.05 versus WT control; b, p <0.05 versus PPARβ−/− control; ns, not significant with respect to PPARα−/− control. At least 12 sections from each group (two sections per animal) were quantified using ImageJ. Scale bar, 50 and 10 μm (for higher magnification images). Tx, animals fed orally with 7.5 mg/kg body weight/day gemfibrozil and 0.1 mg/kg body weight day of all-trans-retinoic acid (dissolved in 0.1% methylcellulose); UN, animals fed orally with 0.1% methylcellulose (used as vehicle) (Un can be replaced with Veh).
FIGURE 8.
Oral administration of gemfibrozil up-regulates LAMP2 in vivo in the cortex of WT and PPARβ−/−, but not PPARα−/−, mice. A, D, and G, WT, PPARα−/−, and PPARβ−/− mice (n = 6 in each group) were treated with 7. 5 mg/kg body weight/day gemfibrozil and 0.1 mg/kg body weight of all-trans retinoic acid (dissolved in 0.1% methylcellulose) or vehicle (0.1% methylcellulose) via gavage. After 60 days of treatment, mice were killed, and cortices from the animal brain were processed for immunoblot for LAMP2 (A). B, densitometric analysis of the immunoblot for LAMP2 (relative to β-actin). C, F, and I, cortical sections were double-labeled for LAMP2 (red) and NeuN (green). DAPI was used to visualize the nucleus. D, G, and J, higher magnification images showing localization of LAMP2 and NeuN in the cortical neuron of mice from the treatment group (WT, PPARα−/−, and PPARβ−/−). E, H, and K, quantification of LAMP2 immunoreactivity (LAMP2 IR) in untreated and treated samples from each group (WT, PPARα−/−, and PPARβ−/−) expressed as percentage of area. a, p < 0.05 versus WT control; b, p < 0.05 versus PPARβ−/− control; ns, not significant with respect to PPARα−/− control. At least 12 sections from each group (two sections per animal) were quantified using ImageJ. Scale bar, 50 and 10 μm (for higher magnification images). Tx, animals fed orally with 7.5 mg/kg body weight/day gemfibrozil and 0.1 mg/kg body weight day of all-trans-retinoic acid (dissolved in 0.1% methylcellulose); UN, animals fed orally with 0.1% methylcellulose (used as vehicle) (Un can be replaced with Veh).
Gemfibrozil and ATRA Induced Lysosomal Biogenesis in Fibroblasts of LINCL Patients
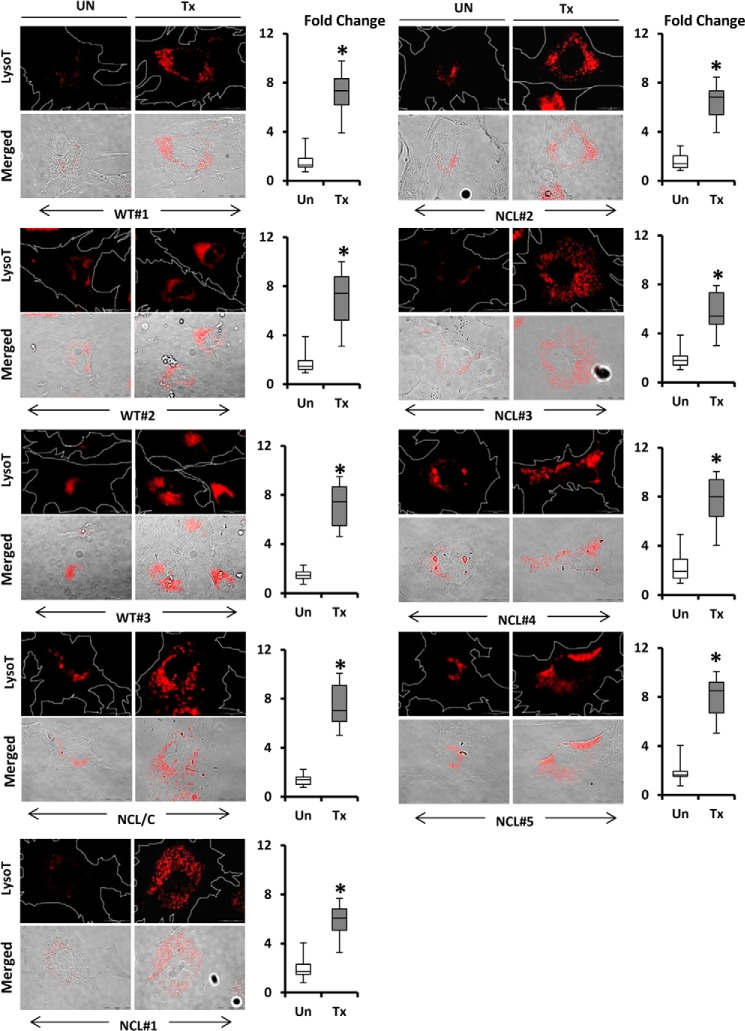
To test whether similar results could be replicated in patient cells, we obtained skin fibroblasts from normal and LINCL-affected patients and treated the cells with similar concentrations of gemfibrozil and ATRA in reduced (2%) serum-containing media. To account for any change resulting due to serum starvation, the untreated controls were kept in similar serum conditions for the length of the treatment (24 h). Fibroblasts were stained with LysoTracker Red, and we observed a similar pattern of increased lysosome accumulation in the cells across the board. Normal fibroblasts (WT#1 through WT#3) and fibroblasts from LINCL patients carrying Cln2 mutations (NCL#1 through NCL#5) as well as LINCL carrier (NCL/C) fibroblasts showed a similar increase in lysosomes per cell (Fig. 9). To normalize for the number and size of cells in the images, we calculated the LysoTracker +ve signals per unit area per cell and then performed a fold over control analysis. At least 25 fields per group were analyzed for LysoTracker-positive signals, and the results showed a significant increase in all fibroblasts irrespective of the disease status, although the basal level of lysosomes in the cell and the level of increase varied from cell to cell. These data suggest that the effect of the treatment is independent of the disease condition for LINCL patients.
FIGURE 9.
Up-regulation of TFEB induces lysosomal biogenesis in both normal and LINCL patient fibroblasts. Fibroblasts from healthy individuals (WT#1–3) and LINCL patients (NCL#1–5) and carrier of LINCL (NCL/C) were treated with gemfibrozil (10 μm) and retinoic acid (0. 2 μm) in reduced (2%) serum-containing DMEM for 24 h followed by staining with LysoTracker Red (red). Bright field microscopy was used for detecting cell morphology. Scale bar, 20 μm. Corresponding box plots represent fold change in the LysoTracker-positive signals in treated group versus control in each cell type. *, p < 0.05 versus untreated control. Regions of interest, white dotted lines, represent area of the cell. Fold change calculated as LysoTracker-positive signal per unit area per cell in treatment versus control. At least 25 individual images per condition per cell type were quantified using ImageJ. UN, DMSO-treated cells (used as control); Tx, treated with the combination of gemfibrozil (10 μm) and all-trans-retinoic acid.
DISCUSSION
Lysosomes are one of the major organelles in cells that not only act as the waste management machinery of the cell but also play significant roles in other biological processes like antigen presentation, regulation of certain hormones, bone remodeling, necrotic cell death, cell surface repair, and developmental and other signaling pathways (2, 43–47). To carry out these diverse functions, the biogenesis and activity of lysosomes need to be tightly regulated. According to recent findings, TFEB is a master regulator of lysosomal biogenesis (9, 12, 15). Over the years, different groups have underscored the role of lysosome in different disease scenarios (48–51). Lysosome-related genes are reported to be closely regulated in the orbital fat of patients suffering from Graves' ophthalmopathy, whereas down-regulation of lysosomal processing improved pegylated lipopolyplex-mediated gene transfection (51, 52). The increase in lysosomal biogenesis may not necessarily prove to be beneficial in all disease and cell types, but in some cases induction of the autophagy-lysosomal pathway could be helpful for cellular clearance of toxic wastes (53, 54). Over the past few years, TFEB has emerged as a potential therapeutic target for some lysosome-related diseases. According to Tsunemi et al. (55), activation of TFEB via PGC1α may result in increased htt turnover and the elimination of protein aggregates. There are reports suggesting a link between α-synuclein toxicity and impaired function of TFEB and identified TFEB as a target for neuroprotective therapy in Parkinson disease (56). TFEB activation has also been shown to enhance the folding, trafficking, and activity of a destabilized glucocerebrosidase variant in Gaucher disease. In case of Tay-Sachs disease, another LSD, TFEB, has been shown to rescue the activity of a β-hexosaminidase mutant. These findings describe TFEB as specific regulator of lysosomal proteostasis and a therapeutic target to rescue enzyme homeostasis in LSDs (57, 58). Furthermore, it is reported that induction of lysosomal exocytosis by TFEB overexpression can rescue pathological storage and restore normal cellular morphology in LSDs (59). Apart from LSDs, TFEB has been shown to induce lipid catabolism and clearance and may rescue obesity and metabolic syndrome in mice (15, 16). Together, TFEB is becoming a potentially important transcription factor for its role in not only lysosomal biogenesis but also due to its implications as a therapeutic target in various disease conditions. However, not many therapeutically viable compounds targeting TFEB activity have been identified. Recently, it has been shown that 2-hydroxypropyl-β-cyclodextrin, an FDA-approved excipient, promotes TFEB-mediated clearance of proteolipid aggregates in cells from patients suffering from LINCL (54). Another study reveals induction of TFEB levels and activity as well as lysosomal biogenesis by genistein (5,7-dihydroxy-3-(4-hydroxyphenyl)-4H-1-benzopyran-4-one), a potential drug for the use in substrate reduction therapy for mucopolysaccharidoses (60). Recent studies have linked TFEB, lysosomal biogenesis, and autophagy with lipid metabolism (14–16, 53, 61, 62). The potential interplay between TFEB and lipid metabolism led us to investigate the role of gemfibrozil and ATRA, which are potential activators of PPARα and RXRα, respectively.
Gemfibrozil, marketed as “Lopid,” is an FDA-approved drug prescribed for hyperlipidemia (17, 19), but it has been shown to have multiple beneficial effects (22). The ability of gemfibrozil to cross the blood-brain barrier has already been documented (20). We have previously reported that gemfibrozil in conjunction with ATRA could induce the levels of Cln2 gene in brain cells (63). We further investigated to see whether TFEB, the master regulator of lysosomal biogenesis, could be affected by the drugs. Our data indicate that gemfibrozil, alone or in conjunction with ATRA, could effectively induce the expression of TFEB in brain cells. Further investigation suggested the possible role of PPARα in the process. PPARα has been shown to play a significant role in different regulatory and modulatory pathways (64–68). Certain polyunsaturated fatty acids, oxidized derivatives, and lipid-modifying drugs of the fibrate family, including fenofibrate and gemfibrozil, are known to activate PPARα. Fibrate drugs replace the HSP90 repressor complex, which sequesters PPARα in the cytosol and helps to rescue the transcriptional activity of PPARα (21). Therefore, we assessed the role of the PPAR group of receptors for this phenomenon. Our data clearly indicate the involvement of PPARα, but not PPARβ and PPARγ, in the induction of TFEB by gemfibrozil. The in vitro studies were further validated by in vivo studies, where we used PPARα−/− and PPARβ−/− mice.
To delineate the mechanism of up-regulation of TFEB, we analyzed the promoter region of the Tfeb gene. Interestingly, the mouse Tfeb promoter was found to harbor a PPRE- and an RXR-binding site. According to previous studies, the PPAR/RXR heterodimer has been well known to show DNA binding activity (67). Together, the PPAR/RXR heterodimer regulates the transcription of genes for which products are involved in lipid homeostasis, cell growth, and differentiation (66, 69). We observed that the pathway of Tfeb up-regulation requires a cooperative effect of both PPARα and RXRα. Furthermore, the effects of both gemfibrozil and ATRA were abrogated in the absence of either RXRα or PPARα. The reporter assay results using both WT and mutated Tfeb promoter showed that the drugs induced luciferase activity driven by WT, but not PPRE-mutated, Tfeb promoter. Finally, the ChIP data indicated the recruitment of the PPARα and RXRα along with PGC1α and RNA Pol on the PPRE of the Tfeb promoter. The experimental data were critically analyzed along with incorporation of proper controls to ensure the specificity of the findings. Collectively, these data outline a unique mechanism where gemfibrozil, a known activator of PPARα, and ATRA, an agonist of RXRα, together can up-regulate TFEB in brain cells via the PPARα/RXRα heterodimer. Although one study reported that PPARγ-null trophoblast stem cells have lower levels of TFEB on day 4 of differentiation, our study using GW9662, a potent and known PPARγ antagonist, did not reveal any substantial involvement of PPARγ (31). This could possibly be due to variation in cell types, i.e. differentiating trophoblast stem cells versus matured primary brain astrocytes/neurons or differential levels of PPARα activation. In one comprehensive study by Settembre et al. (15), the authors reported that PPARα and PGC1α are targets of TFEB under starvation-induced stress and that TFEB is autoregulated in the case of starvation stress, but another study by Tsunemi et al. (55) places PGC1α upstream to TFEB in the Huntington disease scenario. It is quite possible that TFEB regulates lipid metabolism via PPARα and PGC1α, both of which have very significant roles in regulating lipid metabolism. In contrast, our data indicate that an activation of PPARα can induce the recruitment of the PPARα-RXRα-PGC1α complex on the TFEB promoter, thereby influencing lysosomal biogenesis as outlined in Fig. 4D. Although stress response directly regulates TFEB function, our finding suggests that activation of PPARα as well as RXRα by external stimuli can also regulate TFEB, which may in turn control the expression of PPARα or other genes responsible for lipid metabolism. However, more detailed studies are necessary to decipher the presence of any such feed-forward regulatory mechanism and the apparent bi-directional interplay between lipid metabolism and lysosomal biogenesis.
In summary, this study reports a novel finding that gemfibrozil, a lipid-lowering drug, can up-regulate lysosomal biogenesis in brain cells via PPARα-mediated activation of TFEB. Considering the importance of TFEB in certain disease scenarios, there is a growing interest in identifying TFEB as a therapeutic target. Therefore, the outcome of this investigation highlights undiscovered properties of PPARα, describes a new treatment option for LSDs, and reveals a more dynamic regulation of TFEB and fuel interest in understanding the link between the lipid metabolism pathway and lysosomal biogenesis.
Acknowledgments
We thank Dr. Avik Roy, Dr. Suresh Babu Rangaswamy, and Grant T. Corbett for their help in data analysis and suggestions in the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants AT6681 and NS083054. This work was also supported by Merit Award I01BX003033-01 from Veteran Affairs and funds from Noah's Hope, Hope for Bridgett, Cures Within Reach, and Two Hearts Rock.
- TFEB
- transcription factor EB
- PPAR
- peroxisome proliferator-activated receptor
- ATRA
- all-trans-retinoic acid
- LSD
- lysosomal storage disorder
- PPRE
- peroxisomal proliferator-response element
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- GFAP
- glial fibrillary acidic protein
- RXR
- retinoid X receptor
- FDA
- Food and Drug Administration
- RA
- retinoic acid
- LINCL
- late infantile neuronal ceroid lipofuscinosis
- qRT
- quantitative RT.
REFERENCES
- 1. De Duve C. (1963) The lysosome. Sci. Am. 208, 64–72 [DOI] [PubMed] [Google Scholar]
- 2. De Duve C., Wattiaux R. (1966) in Functions of Lysosomes (Mehta A., Beck M., Sunder-Plassmann G., eds) Annu. Rev. Physiol. 28, 435–492 [DOI] [PubMed] [Google Scholar]
- 3. Saftig P. (2006) in Fabry Disease: Perspectives from 5 Years of FOS (Mehta A., Beck M., Sunder-Plassmann G., eds) Chapter 3, Oxford PharmaGenesis, Oxford, UK: [PubMed] [Google Scholar]
- 4. Pérez-Sala D., Boya P., Ramos I., Herrera M., Stamatakis K. (2009) The C-terminal sequence of RhoB directs protein degradation through an endo-lysosomal pathway. PLoS One 4, e8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fuster J. J., González J. M., Edo M. D., Viana R., Boya P., Cervera J., Verges M., Rivera J., Andrés V. (2010) Tumor suppressor p27(Kip1) undergoes endolysosomal degradation through its interaction with sorting nexin 6. FASEB J. 24, 2998–3009 [DOI] [PubMed] [Google Scholar]
- 6. Korolchuk V. I., Saiki S., Lichtenberg M., Siddiqi F. H., Roberts E. A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F. M., O'Kane C. J., Deretic V., Rubinsztein D. C. (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boya P., Kroemer G. (2008) Lysosomal membrane permeabilization in cell death. Oncogene 27, 6434–6451 [DOI] [PubMed] [Google Scholar]
- 8. Martina J. A., Diab H. I., Lishu L., Jeong-A L., Patange S., Raben N., Puertollano R. (2014) The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci. Signal. 7, ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Palmieri M., Impey S., Kang H., di Ronza A., Pelz C., Sardiello M., Ballabio A. (2011) Characterization of the CLEAR network reveals an integrated control of cellular clearance pathways. Hum. Mol. Genet. 20, 3852–3866 [DOI] [PubMed] [Google Scholar]
- 10. Sardiello M., Palmieri M., di Ronza A., Medina D. L., Valenza M., Gennarino V. A., Di Malta C., Donaudy F., Embrione V., Polishchuk R. S., Banfi S., Parenti G., Cattaneo E., Ballabio A. (2009) A gene network regulating lysosomal biogenesis and function. Science 325, 473–477 [DOI] [PubMed] [Google Scholar]
- 11. Marschner K., Kollmann K., Schweizer M., Braulke T., Pohl S. (2011) A key enzyme in the biogenesis of lysosomes is a protease that regulates cholesterol metabolism. Science 333, 87–90 [DOI] [PubMed] [Google Scholar]
- 12. Settembre C., Di Malta C., Polito V. A., Garcia Arencibia M., Vetrini F., Erdin S., Erdin S. U., Huynh T., Medina D., Colella P., Sardiello M., Rubinsztein D. C., Ballabio A. (2011) TFEB links autophagy to lysosomal biogenesis. Science 332, 1429–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ferron M., Settembre C., Shimazu J., Lacombe J., Kato S., Rawlings D. J., Ballabio A., Karsenty G. (2013) A RANKL-PKCβ-TFEB signaling cascade is necessary for lysosomal biogenesis in osteoclasts. Genes Dev. 27, 955–969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Settembre C., Zoncu R., Medina D. L., Vetrini F., Erdin S., Erdin S., Huynh T., Ferron M., Karsenty G., Vellard M. C., Facchinetti V., Sabatini D. M., Ballabio A. (2012) A lysosome-to-nucleus signalling mechanism senses and regulates the lysosome via mTOR and TFEB. EMBO J. 31, 1095–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Settembre C., Fraldi A., Medina D. L., Ballabio A. (2013) Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat. Rev. Mol. Cell Biol. 14, 283–296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Settembre C., De Cegli R., Mansueto G., Saha P. K., Vetrini F., Visvikis O., Huynh T., Carissimo A., Palmer D., Klisch T. J., Wollenberg A. C., Di Bernardo D., Chan L., Irazoqui J. E., Ballabio A. (2013) TFEB controls cellular lipid metabolism through a starvation-induced autoregulatory loop. Nat. Cell Biol. 15, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Robins S. J., Collins D., Wittes J. T., Papademetriou V., Deedwania P. C., Schaefer E. J., McNamara J. R., Kashyap M. L., Hershman J. M., Wexler L. F., Rubins H. B. (2001) Relation of gemfibrozil treatment and lipid levels with major coronary events: VA-HIT: a randomized controlled trial. JAMA 285, 1585–1591 [DOI] [PubMed] [Google Scholar]
- 18. Rubins H. B., Robins S. J. (1992) Effect of reduction of plasma triglycerides with gemfibrozil on high-density-lipoprotein-cholesterol concentrations. J. Intern Med. 231, 421–426 [DOI] [PubMed] [Google Scholar]
- 19. Rubins H. B., Robins S. J., Collins D., Fye C. L., Anderson J. W., Elam M. B., Faas F. H., Linares E., Schaefer E. J., Schectman G., Wilt T. J., Wittes J. (1999) Gemfibrozil for the secondary prevention of coronary heart disease in men with low levels of high-density lipoprotein cholesterol. Veterans Affairs High-Density Lipoprotein Cholesterol Intervention Trial Study Group. N. Engl. J. Med. 341, 410–418 [DOI] [PubMed] [Google Scholar]
- 20. Dasgupta S., Roy A., Jana M., Hartley D. M., Pahan K. (2007) Gemfibrozil ameliorates relapsing-remitting experimental autoimmune encephalomyelitis independent of peroxisome proliferator-activated receptor-α. Mol. Pharmacol. 72, 934–946 [DOI] [PubMed] [Google Scholar]
- 21. Pahan K., Jana M., Liu X., Taylor B. S., Wood C., Fischer S. M. (2002) Gemfibrozil, a lipid-lowering drug, inhibits the induction of nitric-oxide synthase in human astrocytes. J. Biol. Chem. 277, 45984–45991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Roy A., Pahan K. (2009) Gemfibrozil, stretching arms beyond lipid lowering. Immunopharmacol. Immunotoxicol. 31, 339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corbett G. T., Roy A., Pahan K. (2012) Gemfibrozil, a lipid-lowering drug, upregulates IL-1 receptor antagonist in mouse cortical neurons: implications for neuronal self-defense. J. Immunol. 189, 1002–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brahmachari S., Pahan K. (2007) Sodium benzoate, a food additive and a metabolite of cinnamon, modifies T cells at multiple steps and inhibits adoptive transfer of experimental allergic encephalomyelitis. J. Immunol. 179, 275–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Saha R. N., Pahan K. (2007) Differential regulation of Mn-superoxide dismutase in neurons and astroglia by HIV-1 gp120: Implications for HIV-associated dementia. Free Radic. Biol. Med. 42, 1866–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Giulian D., Baker T. J. (1986) Characterization of ameboid microglia isolated from developing mammalian brain. J. Neurosci. 6, 2163–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jana M., Pahan K. (2005) Redox regulation of cytokine-mediated inhibition of myelin gene expression in human primary oligodendrocytes. Free Radic. Biol. Med. 39, 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khasnavis S., Jana A., Roy A., Mazumder M., Bhushan B., Wood T., Ghosh S., Watson R., Pahan K. (2012) Suppression of nuclear factor-κB activation and inflammation in microglia by a physically modified saline. J. Biol. Chem. 287, 29529–29542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khasnavis S., Pahan K. (2012) Sodium benzoate, a metabolite of cinnamon and a food additive, upregulates neuroprotective Parkinson disease protein DJ-1 in astrocytes and neurons. J. Neuroimmune Pharmacol. 7, 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dasgupta S., Jana M., Zhou Y., Fung Y. K., Ghosh S., Pahan K. (2004) Antineuroinflammatory effect of NF-κB essential modifier-binding domain peptides in the adoptive transfer model of experimental allergic encephalomyelitis. J. Immunol. 173, 1344–1354 [DOI] [PubMed] [Google Scholar]
- 31. Parast M. M., Yu H., Ciric A., Salata M. W., Davis V., Milstone D. S. (2009) PPARγ regulates trophoblast proliferation and promotes labyrinthine trilineage differentiation. PLoS One 4, e8055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saha R. N., Liu X., Pahan K. (2006) Up-regulation of BDNF in astrocytes by TNF-α: a case for the neuroprotective role of cytokine. J. Neuroimmune Pharmacol. 1, 212–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jana M., Jana A., Liu X., Ghosh S., Pahan K. (2007) Involvement of phosphatidylinositol 3-kinase-mediated up-regulation of IκBα in anti-inflammatory effect of gemfibrozil in microglia. J. Immunol. 179, 4142–4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jana M., Pahan K. (2012) Gemfibrozil, a lipid lowering drug, inhibits the activation of primary human microglia via peroxisome proliferator-activated receptor β. Neurochem. Res. 37, 1718–1729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nelson J. D., Denisenko O., Bomsztyk K. (2006) Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat. Protoc. 1, 179–185 [DOI] [PubMed] [Google Scholar]
- 36. Cullingford T. E., Bhakoo K., Peuchen S., Dolphin C. T., Patel R., Clark J. B. (1998) Distribution of mRNAs encoding the peroxisome proliferator-activated receptor α, β, and γ and the retinoid X receptor α, β, and γ in rat central nervous system. J. Neurochem. 70, 1366–1375 [DOI] [PubMed] [Google Scholar]
- 37. Nishizawa H., Manabe N., Morita M., Sugimoto M., Imanishi S., Miyamoto H. (2003) Effects of in utero exposure to bisphenol A on expression of RARα and RXRα mRNAs in murine embryos. J. Reprod. Dev. 49, 539–545 [DOI] [PubMed] [Google Scholar]
- 38. Chinetti G., Griglio S., Antonucci M., Torra I. P., Delerive P., Majd Z., Fruchart J. C., Chapman J., Najib J., Staels B. (1998) Activation of proliferator-activated receptors α and γ induces apoptosis of human monocyte-derived macrophages. J. Biol. Chem. 273, 25573–25580 [DOI] [PubMed] [Google Scholar]
- 39. Brun S., Carmona M. C., Mampel T., Viñas O., Giralt M., Iglesias R., Villarroya F. (1999) Activators of peroxisome proliferator-activated receptor-α induce the expression of the uncoupling protein-3 gene in skeletal muscle: a potential mechanism for the lipid intake-dependent activation of uncoupling protein-3 gene expression at birth. Diabetes 48, 1217–1222 [DOI] [PubMed] [Google Scholar]
- 40. Chinetti G., Lestavel S., Bocher V., Remaley A. T., Neve B., Torra I. P., Teissier E., Minnich A., Jaye M., Duverger N., Brewer H. B., Fruchart J. C., Clavey V., Staels B. (2001) PPAR-α and PPAR-γ activators induce cholesterol removal from human macrophage foam cells through stimulation of the ABCA1 pathway. Nat. Med. 7, 53–58 [DOI] [PubMed] [Google Scholar]
- 41. Kelly D. P. (2001) The pleiotropic nature of the vascular PPAR gene regulatory pathway. Circ. Res. 89, 935–937 [PubMed] [Google Scholar]
- 42. Boitier E., Gautier J. C., Roberts R. (2003) Advances in understanding the regulation of apoptosis and mitosis by peroxisome-proliferator activated receptors in pre-clinical models: relevance for human health and disease. Comp. Hepatol. 2, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pshezhetsky A. V., Ashmarina M. (2001) Lysosomal multienzyme complex: biochemistry, genetics, and molecular pathophysiology. Prog. Nucleic Acid Res. Mol. Biol. 69, 81–114 [DOI] [PubMed] [Google Scholar]
- 44. Karageorgos L. E., Isaac E. L., Brooks D. A., Ravenscroft E. M., Davey R., Hopwood J. J., Meikle P. J. (1997) Lysosomal biogenesis in lysosomal storage disorders. Exp. Cell Res. 234, 85–97 [DOI] [PubMed] [Google Scholar]
- 45. Weissmann G. (1967) The role of lysosomes in inflammation and disease. Annu. Rev. Med. 18, 97–112 [DOI] [PubMed] [Google Scholar]
- 46. Eskelinen E. L., Tanaka Y., Saftig P. (2003) At the acidic edge: emerging functions for lysosomal membrane proteins. Trends Cell Biol. 13, 137–145 [DOI] [PubMed] [Google Scholar]
- 47. Brignull L. M., Czimmerer Z., Saidi H., Daniel B., Villela I., Bartlett N. W., Johnston S. L., Meira L. B., Nagy L., Nohturfft A. (2013) Reprogramming of lysosomal gene expression by interleukin-4 and Stat6. BMC Genomics 14, 853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Neufeld E. F. (1991) Lysosomal storage diseases. Annu. Rev. Biochem. 60, 257–280 [DOI] [PubMed] [Google Scholar]
- 49. Gieselmann V. (1995) Lysosomal storage diseases. Biochim. Biophys. Acta 1270, 103–136 [DOI] [PubMed] [Google Scholar]
- 50. Appelqvist H., Wäster P., Kågedal K., Öllinger K. (2013) The lysosome: from waste bag to potential therapeutic target. J. Mol. Cell. Biol. 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 51. Bai J., Liu Y., Sun W., Chen J., Miller A. D., Xu Y. (2013) Down-regulated lysosomal processing improved pegylated lipopolyplex-mediated gene transfection. J. Gene Med. 15, 182–192 [DOI] [PubMed] [Google Scholar]
- 52. Chen M. H., Liao S. L., Chen M. H., Tsou P. L., Shih M. J., Chang T. C., Chuang L. M. (2008) Lysosome-related genes are regulated in the orbital fat of patients with Graves' ophthalmopathy. Invest. Ophthalmol. Vis. Sci. 49, 4760–4764 [DOI] [PubMed] [Google Scholar]
- 53. Sarkar S., Carroll B., Buganim Y., Maetzel D., Ng A. H., Cassady J. P., Cohen M. A., Chakraborty S., Wang H., Spooner E., Ploegh H., Gsponer J., Korolchuk V. I., Jaenisch R. (2013) Impaired autophagy in the lipid-storage disorder Niemann-Pick type C1 disease. Cell Rep. 5, 1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Song W., Wang F., Lotfi P., Sardiello M., Segatori L. (2014) 2-Hydroxypropyl-β-cyclodextrin promotes transcription factor EB-mediated activation of autophagy: implications for therapy. J. Biol. Chem. 289, 10211–10222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tsunemi T., Ashe T. D., Morrison B. E., Soriano K. R., Au J., Roque R. A., Lazarowski E. R., Damian V. A., Masliah E., La Spada A. R. (2012) PGC-1α rescues Huntington's disease proteotoxicity by preventing oxidative stress and promoting TFEB function. Sci. Transl. Med. 4, 142ra197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Decressac M., Mattsson B., Weikop P., Lundblad M., Jakobsson J., Björklund A. (2013) TFEB-mediated autophagy rescues midbrain dopamine neurons from α-synuclein toxicity. Proc. Natl. Acad. Sci. U.S.A. 110, E1817–E1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wang F., Segatori L. (2013) Remodeling the proteostasis network to rescue glucocerebrosidase variants by inhibiting ER-associated degradation and enhancing ER folding. PLoS One 8, e61418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Song W., Wang F., Savini M., Ake A., di Ronza A., Sardiello M., Segatori L. (2013) TFEB regulates lysosomal proteostasis. Hum. Mol. Genet. 22, 1994–2009 [DOI] [PubMed] [Google Scholar]
- 59. Medina D. L., Fraldi A., Bouche V., Annunziata F., Mansueto G., Spampanato C., Puri C., Pignata A., Martina J. A., Sardiello M., Palmieri M., Polishchuk R., Puertollano R., Ballabio A. (2011) Transcriptional activation of lysosomal exocytosis promotes cellular clearance. Dev. Cell 21, 421–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Moskot M., Montefusco S., Jakóbkiewicz-Banecka J., Mozolewski P., Węgrzyn A., Di Bernardo D., Węgrzyn G., Medina D. L., Ballabio A., Gabig-Cimińska M. (2014) The phytoestrogen genistein modulates lysosomal metabolism and Transcription Factor EB (TFEB) activation. J. Biol. Chem. 289, 17054-17069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu X., Grijalva A., Skowronski A., van Eijk M., Serlie M. J., Ferrante A. W., Jr. (2013) Obesity activates a program of lysosomal-dependent lipid metabolism in adipose tissue macrophages independently of classic activation. Cell Metab. 18, 816–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Singh R., Cuervo A. M. (2012) Lipophagy: connecting autophagy and lipid metabolism. Int. J. Cell Biol. 2012, 282041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ghosh A., Corbett G. T., Gonzalez F. J., Pahan K. (2012) Gemfibrozil and fenofibrate, Food and Drug Administration-approved lipid-lowering drugs, up-regulate tripeptidyl-peptidase 1 in brain cells via peroxisome proliferator-activated receptor α: implications for late infantile Batten disease therapy. J. Biol. Chem. 287, 38922–38935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Xu J., Racke M. K., Drew P. D. (2007) Peroxisome proliferator-activated receptor-α agonist fenofibrate regulates IL-12 family cytokine expression in the CNS: relevance to multiple sclerosis. J. Neurochem. 103, 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Xu J., Chavis J. A., Racke M. K., Drew P. D. (2006) Peroxisome proliferator-activated receptor-α and retinoid X receptor agonists inhibit inflammatory responses of astrocytes. J. Neuroimmunol. 176, 95–105 [DOI] [PubMed] [Google Scholar]
- 66. Krey G., Mahfoudi A., Wahli W. (1995) Functional interactions of peroxisome proliferator-activated receptor, retinoid-X receptor, and Sp1 in the transcriptional regulation of the acyl-coenzyme-A oxidase promoter. Mol. Endocrinol. 9, 219–231 [DOI] [PubMed] [Google Scholar]
- 67. Juge-Aubry C. E., Gorla-Bajszczak A., Pernin A., Lemberger T., Wahli W., Burger A. G., Meier C. A. (1995) Peroxisome proliferator-activated receptor mediates cross-talk with thyroid hormone receptor by competition for retinoid X receptor. Possible role of a leucine zipper-like heptad repeat. J. Biol. Chem. 270, 18117–18122 [DOI] [PubMed] [Google Scholar]
- 68. Roy A., Jana M., Corbett G. T., Ramaswamy S., Kordower J. H., Gonzalez F. J., Pahan K. (2013) Regulation of cyclic AMP response element binding and hippocampal plasticity-related genes by peroxisome proliferator-activated receptor α. Cell Rep. 4, 724–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Marcus S. L., Miyata K. S., Rachubinski R. A., Capone J. P. (1995) Transactivation by PPAR/RXR heterodimers in yeast is potentiated by exogenous fatty acid via a pathway requiring intact peroxisomes. Gene Expr. 4, 227–239 [PMC free article] [PubMed] [Google Scholar]